Abstract
SARS-CoV-2 is a highly contagious coronavirus causing the ongoing pandemic. Very recently its genomic RNA of ∼30 kb was decoded to be packaged with nucleocapsid (N) protein into phase separated condensates. Interestingly, viruses have no ability to generate ATP but host cells have very high ATP concentrations of 2–12 mM. A key question thus arises whether ATP modulates liquid-liquid phase separation (LLPS) of the N protein. Here we discovered that ATP not only biphasically modulates LLPS of the viral N protein as we previously found on human FUS and TDP-43, but also dissolves the droplets induced by oligonucleic acid. Residue-specific NMR characterization showed ATP specifically binds the RNA-binding domain (RBD) of the N protein with the average Kd of 3.3 ± 0.4 mM. The ATP-RBD complex structure was constructed by NMR-derived constraints, in which ATP occupies a pocket within the positive-charged surface utilized for binding nucleic acids. Our study suggests that ATP appears to be exploited by SARS-CoV-2 to promote its life cycle by facilitating the uncoating, localizing and packing of its genomic RNA. Therefore the interactions of ATP with the viral RNA and N protein might represent promising targets for design of drugs and vaccines to terminate the pandemic.
Keywords: Adenosine triphosphate (ATP), SARS-CoV-2, Nucleocapsid (N) protein, RNA-Binding domain (RBD), Liquid-liquid phase separation (LLPS), NMR spectroscopy
Graphical abstract
1. Introduction
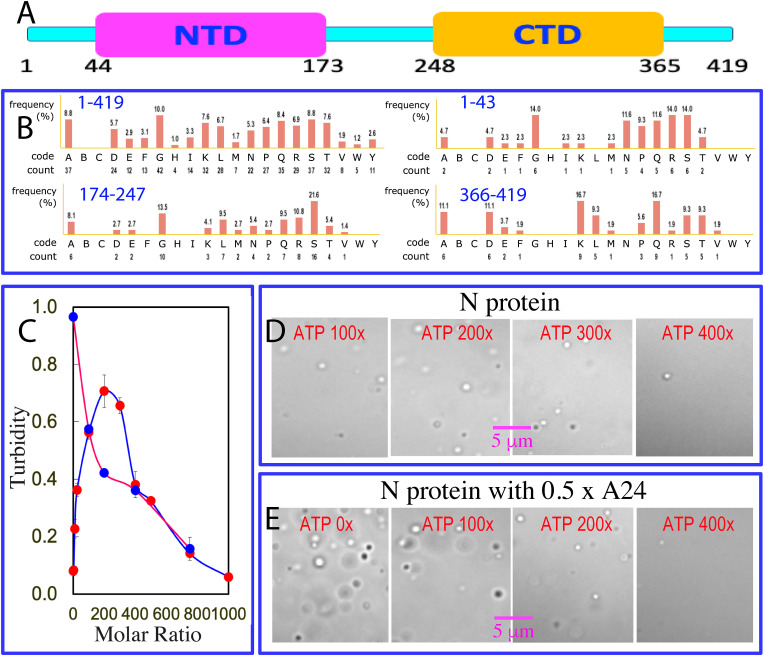
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is the etiologic agent of the ongoing pandemic [1], which is a highly contagious beta-coronavirus of a large family of positive-stranded RNA coronaviruses with ∼30 kb genomic RNA packaged into a membrane-enveloped virion. SARS-CoV-2 has four structural proteins: the spike (S) protein that recognizes cell receptors angiotensin converting enzyme-2 (ACE2), nucleocapsid (N) protein responsible for packing viral genomic RNA, membrane-associated envelope (E) and membrane (M) proteins [2]. Its N protein is a 419-residue RNA-binding protein composed of two well-folded domains, namely N-terminal domain (NTD) over residues 44–173 and C-terminal domain (CTD) over residues 248–365 (Fig. 1 A), as well as three intrinsically-disordered regions (IDRs) respectively over 1–43, 174–247 and 366–419 with low-complexity sequences (Fig. 1B). The N protein plays multifunctional roles in the coronavirus life cycle, which include to assemble genomic RNA into the viral RNA-protein complex, as well as to localize it to replicase-transcriptase complexes. Previous studies revealed that its NTD is an RNA-binding domain (RBD) while CTD functions to dimerize/oligomerize to form high-order structures [[3], [4], [5], [6], [7]]. Very recently, it was decoded that the N protein functions through liquid-liquid phase separation (LLPS), which could be significantly induced by dynamic and multivalent interactions with various nucleic acids [[5], [6], [7]]. In this context, the N-terminal RBD was proposed to bind the specific sites of the genomic RNA for initiating the assembly into the RNA-N-protein condensates. Strikingly, the NMR structure of the SARS-CoV-2 N protein has been now reported both in the free state and in complex with short RNA fragments [8].
Fig. 1.
ATP biphasically modulates LLPS of the SARS-CoV-2 N protein.
ATP (adenosine triphosphate) is best known as the universal energy currency for all living cells. Mysteriously the cellular concentrations of ATP are much higher than those required for its classic functions, ranging from 2 to 12 mM dependent of cell types [9,10]. Only recently it was decoded that at concentrations >5 mM, ATP functions as a biological hydrotrope to dissolve LLPS of RNA-binding proteins with the prion-like domains such as FUS [10]. Our NMR studies further identified that ATP can biphasically modulate LLPS of the intrinsically disordered domains (IDDs) of FUS and TDP-43 by specific binding to the Arg/Lys residues within IDDs: induction at low ATP concentrations but dissolution at high concentrations [11,12]. Moreover, we also found that ATP is capable of specifically binding the RNA-recognition motif (RRM) domains of FUS and TDP-43, as well as to the SYNCRIP acidic domain (AcD) with an all-helical fold which is a non-canonical RNA-binding domain [[13], [14], [15]].
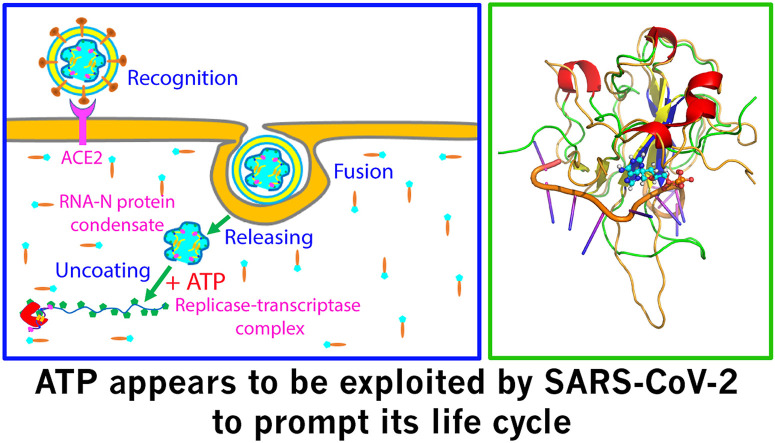
Intriguingly, viruses have no ability to generate ATP [16] and therefore upon release into the infected cell, the viral RNA-N-protein condensate of SARS-CoV-2 is anticipated to experience a sudden exposure to the environment with very high ATP concentrations. So two key questions of both fundamental and therapeutic significance arise: 1) does ATP have any effect on LLPS of the SARS-CoV-2 N protein? 2) can ATP specifically bind to its RBD domain whose structural fold is very different from those of RRM and AcD? In the present study, we first assessed the effect of ATP on LLPS of the SARS-CoV-2 N protein in the absence and in the pre-existence of 24-mer poly(A) (A24) as imaged by differential interference contrast (DIC) microscopy. Subsequently we characterized the binding of ATP to the RBD by NMR HSQC titrations, which led to determining the average dissociation constant (Kd) to be 3.3 ± 0.4 mM as well as to constructing the ATP-RBD complex structure. The results reveal for the first time: 1) ATP does biphasically modulate LLPS of the N protein. 2) ATP is capable of dissolving LLPS of the N protein induced by A24.3) ATP at biologically-relevant concentrations is able to specifically bind to a pocket within the RBD surface utilized to bind RNA. The results together imply that ATP appears to be exploited by SARS-CoV-2 to promote its life cycle ranging from initial uncoating of the genomic RNA from the released viral condensate to subsequent localizing of the genomic RNA to replicase-transcriptase complexes, and final packing of the genomic RNA with the N protein into the phase separated condensates for new virions.
2. Materials and methods
2.1. Preparation of recombinant SARS-CoV-2 nucleocapsid and its RBD proteins
The gene encoding 419-residue SARS-CoV-2 N protein was purchased from a local company (Bio Basic Asia Pacific Pte Ltd), which was cloned into an expression vector pET-28a with a TEV protease cleavage site between N protein and N-terminal 6xHis-SUMO tag used to enhance the solubility. Its RNA-binding domain (RBD) over residues 44–180 was also cloned into the same vector.
The recombinant N protein and its RBD domain were expression in E. coli cells BL21 with IPTG induction at 18 °C. Both proteins were found to be soluble in the supernatant. For NMR studies, the bacteria were grown in M9 medium with addition of (15NH4)2SO4 for 15N-labeling. The recombinant proteins were first purified by Ni2+-affinity column (Novagen) under native conditions and subsequently in-gel cleavage by TEV protease was conducted. The eluted fractions containing the recombinant proteins were further purified by FPLC chromatography system with a Superdex-75 column. The purity of the recombinant proteins was checked by SDS-PAGE gels and NMR assignment for RBD. The concentration of protein samples was determined by the UV spectroscopic method in the presence of 8 M urea [[11], [12], [13], [14], [15]]. ATP was purchased from Sigma-Aldrich as previously reported, and MgCl2 was added into ATP for stabilization by forming the ATP-Mg complex [[11], [12], [13], [14], [15]].
2.2. LLPS imaged by differential interference contrast (DIC) microscopy
The formation of liquid droplets was imaged on 50 μl of the N protein samples by DIC microscopy (OLYMPUS IX73 Inverted Microscope System with OLYMPUS DP74 Color Camera) as previously described [11,12]. The N protein samples were prepared at 20 μM in 25 mM HEPES buffer (pH 7.5) with 70 mM KCl with addition of ATP or A24 in the same buffer at different molar ratios. Subsequently, the N protein samples at 20 μM in the same buffer with the pre-existence of A24 at 1:0.5 were also imaged with further addition of ATP at different molar ratios. Turbidity, the absorption at 600 nm, were measured three times for each sample.
2.3. NMR characterizations of the binding of ATP to RBD
NMR experiments were conducted at 25 °C on an 800 MHz Bruker Avance spectrometer equipped with pulse field gradient units and a shielded cryoprobe as described previously [[11], [12], [13], [14], [15]]. For NMR HSQC titrations with ATP, two dimensional 1H–15N NMR HSQC spectra were collected on the 15N-labeled RBD samples at 50 μM in 10 mM phosphate buffer at pH 6.8 with 150 mM NaCl in the absence and in the presence of ATP in the same buffer at 0.5, 1, 2.5, 5, 7.5, 10, 15 and 20 mM respectively.
2.4. Calculation of CSD and data fitting
Sequential assignment was achieved based on the deposited chemical shifts (BMRB ID of 34511) [8]. To calculate chemical shift difference (CSD), HSQC spectra collected without and with ATP at different concentrations were superimposed. Subsequently, the shifted HSQC peaks were identified and further assigned to the corresponding RBD residues. The chemical shift difference (CSD) was calculated by an integrated index with the following formula [[11], [12], [13], [14], [15]]:
| CSD = ((Δ1H)2 + (Δ15N)2/4)1/2 |
In order to obtain residue-specific dissociation constant (Kd), we fitted the shift traces of the 11 residues with significant shifts (CSD > average + STD) by using the one binding site model with the following formula as we previously performed [[13], [14], [15]]:
| CSDobs = CSDmax {([P] + [L] + Kd) – [([P] + [L] + Kd)2–4[P][L]]1/2}/2[P] |
Here, [P] and [L] are molar concentrations of RBD and ligands (ATP) respectively.
2.5. Molecular docking
The three-dimensional model of the ATP-RBD complex was constructed with the NMR structure of the RBD domain of the N protein (PDB ID of 6YI3) [8] by use of HADDOCK software [17], which makes use of CSD data to derive the docking, allowing various degrees of flexibility. The ATP-RBD structure with the lowest energy score was selected for the detailed analysis and display by Pymol (The PyMOL Molecular Graphics System).
3. Results
3.1. ATP biphasically modulates LLPS of the N protein
The N protein of SARS-CoV-2 at 20 μM has a turbidity value of 0.08 (Fig. 1C) and showed no droplets as imaged by DIC. However, upon titration with ATP, the turbidity values of the N protein samples increased and reached the highest of 0.7 at 1:200 (N-protein:ATP). Further increase of ATP concentrations led to the decrease of the turbidity values and at 1:1000, the turbidity value became only 0.06. DIC characterization revealed that dynamic droplets could be observed at 1:25 and at 1:100, many droplets were formed with the diameters of some of ∼1 μm (Fig. 1D). At 1:200, more droplets were formed but further increase of ATP concentrations led to the reduction of droplet numbers. At 1:500, all droplets were dissolved. This set of results indicate that ATP does biphasically modulate LLPS of the SARS-CoV-2 N protein, as we previously observed on the Arg-containing IDRs of FUS and TDP-43 [11,12].
Very recently, it was shown that LLPS of the SARS-CoV-2 N protein could be induced by various nucleic acids regardless of their sequences [[5], [6], [7]]. Indeed, here we found that A24 also biphasically modulated LLPS of the N protein as ATP but the ratios required to induce and dissolve droplets were much lower than those of ATP. Addition of A24 even at 1:0.5 resulted a turbidity value of 0.97 (Fig. 1C) as well as the formation of a large number of dynamic droplets with the diameter of some even close to ∼2 μm (Fig. 1E). Interestingly, additional addition of ATP to this sample led to the monotonic reduction of the turbidity values, as well as disappearance of the droplets as imaged by DIC. At 1:750, the droplets were completely disappeared. The results indicate that; 1) ATP has the much lower capacity than A24 in inducing LLPS of the N protein; but 2) nevertheless, ATP is able to dissolve the droplets of the N protein induced by A24, implying that ATP and A24 most likely target the same sites of the N protein for biphasically modulating its LLPS, similar to what we previously observed on the biophasic effects of ATP and oligonucleic acids on LLPS of FUS and TDP-43 IDRs [11,12].
3.2. ATP specifically binds RBD at biologically-relevant concentrations
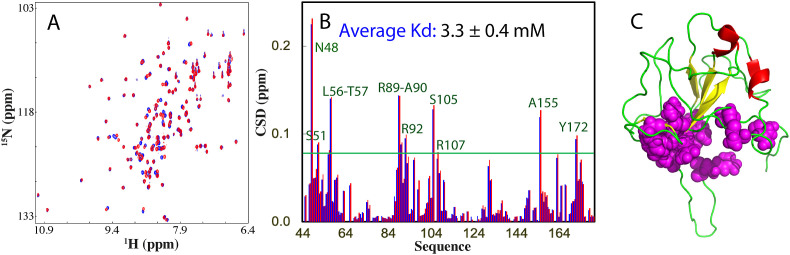
We then assessed whether ATP is able to bind the RBD of the SARS-CoV-2 N protein. As shown in Fig. 2 A, the 15N-labeled RBD (44–180) has a well-dispersed HSQC spectrum at 50 μM (Fig. 2A) in 10 mM sodium phosphate buffer containing 150 mM NaCl (pH 6.8), with most peaks superimposable to those previously reported in different buffer at pH 5.5 [8]. Subsequently, we collected HSQC spectra of RBD by titrating ATP at different concentrations up to 20 mM. Interestingly, at ATP concentrations <1 mM, only minor shifts of HSQC peaks were observed, indicating that the RBD has no significant binding with ATP at concentrations below mM.
Fig. 2.
ATP specifically binds RBD at biologically-relevant concentrations.
By superimposing HSQC spectra in the absence and in the presence of ATP at different concentrations, we found that only a small set of HSQC peaks showed large shifts upon adding ATP (Fig. 2A). We then calculated their chemical shift difference (CSD) induced by binding ATP at different concentrations and the results indicate that 11 residues have significant shifts which were largely saturated at 8 mM ATP (Fig. 2B). Strikingly, the 11 residues were distributed over the whole RND sequence which include Asn48, Ser51, Leu56, Thr57, Arg89, Ala90, Arg92, Ser105, Arg107, Ala155 and Tyr172. As such, by fitting their CSD traces to the one-site binding model as we performed on other proteins [[13], [14], [15]], we obtained the residue-specific dissociation constant (Kd) values of all 11 residues with the average value of 3.3 ± 0.4 mM (Fig. 2B). Remarkably, upon mapping the 11 residues back to the NMR structure of RBD, these residues are clustered together to form a pocket (Fig. 2C).
3.3. Visualization of the ATP-RBD complex
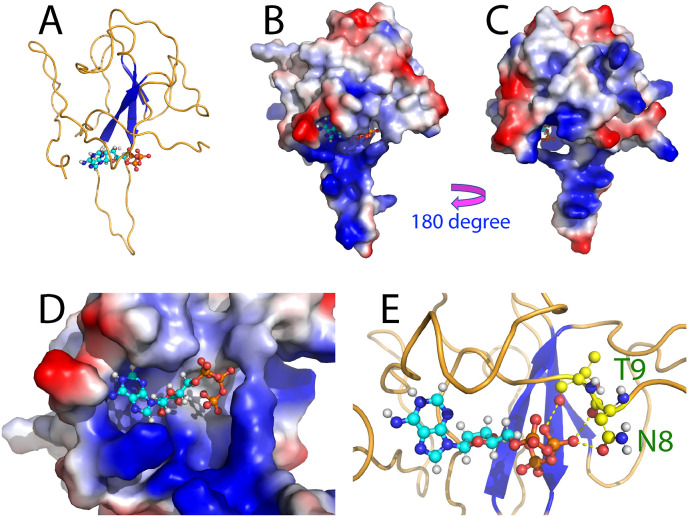
Due to the extremely low binding affinity with Kd at mM, it is impossible to determine the three-dimensional structure of the ATP-BRD complex by NMR spectroscopy or X-ray crystallography. Therefore, to visualize the complex structure, the constraints derived from NMR titrations were utilized to guide molecular docking with the well-established HADDOCK program [[13], [14], [15]]. Fig. 3 A presents the lowest-energy docking structure of the ATP-RBD complex, in which ATP occupies a pocket constituted by the residues with significant shifts of their HSQC peaks upon binding ATP (Fig. 2C). This pocket is within the large surface of the RBD structure which is highly positively-charged (Fig. 3B–D). In the complex, the purine ring of ATP appears to have π-cation interactions with several Arg residues. On the other hand, oxyanions of the β-phosphate of the triphosphate chain established three hydrogen bonds with RBD residues: two with Asn8 and one with Thr9 (Fig. 3E).
Fig. 3.
Three-dimensional model of the ATP-RBD complex.
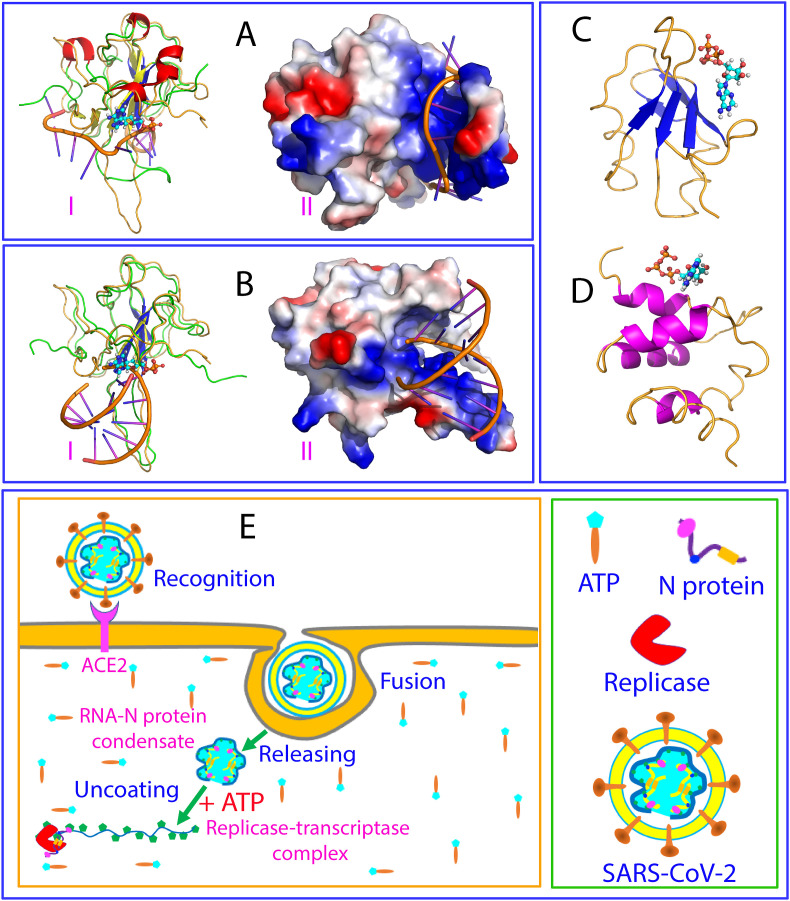
Very recently, the NMR structures of the RBD (44–180) of the SARS-CoV-2 N protein were reported also in complex with both single-stranded RNA (SsRNA) with a sequence of UCUCUAAACG (Fig. 4 A) and double-stranded RNA (DsRNA) with a sequence of CACUGAC (Fig. 4B). Noticeably, superimposition of the structures with that of the ATP-RBD complex revealed that ATP occupies a pocket within the large positively-charged surface which bind both SsRNA and DsRNA. Therefore, the viral RBD of the SARS-CoV-2 N protein represent the third fold which is capable of binding ATP with Kd of ∼mM, additional to the RRM (Fig. 4C) [13,14] and AcD folds (Fig. 4D) [15]. Noticeably, although they have very different structural folds, their ATP-binding pockets are all located within the large surfaces which are utilized by these folds to bind various nucleic acids (13–15).
Fig. 4.
ATP appears to be exploited by SARS-CoV-2 to prompt its life cycle.
4. Discussion
The catastrophic pandemic caused by SARS-CoV-2 has already resulted in infection of >81 millions and death of >1.7 millions. The SARS-CoV-2 N protein is not only the essential component of the packed viral genome, but also a key candidate for vaccine development due to its high expression in the infected cells [18,19]. Very recently, the N protein was identified to undergo LLPS as induced by various nucleic acids. Therefore, to understand the underlying mechanisms for its LLPS is not only essential for developing therapeutic agents, but also critical for design of effective vaccines as the immune response to the phase separated N protein could be very different from those to the soluble forms/fragments of N protein. However, due to the extreme challenge in biophysically characterizing LLPS of the SARS-CoV-2 N protein for which both folded domains and IDRs are involved in, currently the high-resolution mechanism still remains unknown for its LLPS, particularly induced by nucleic acids.
In the present study, for the first time, we found that like nucleic acids, ATP, which is not presented in the virions, does biphasically modulate LLPS of the N protein. On the other hand, as compared to A24, ATP has much weaker (∼400-time weaker) capacity in both inducing and dissolving LLPS of the N protein. Furthermore, although much higher concentrations are needed, ATP can dissolve LLPS induced by A24. This set of results thus not only indicates that ATP and nucleic acids modulate LLPS of the N protein by targeting the same sites as we previously observed on FUS and TDP-43 [11,12], but further implies that the interactions of ATP and A24 with the N proteins are specific binding, rather than non-specific electrostatic effects from the phosphate groups. As ATP has the triphosphate chain which is much more negatively-charged than the phosphate group in A24, ATP is expected to have higher capacity than A24 in biphasically modulating LLPS of the N protein if the modulating capacity is mainly resulting from the non-specific electrostatic effect. However, the observation is opposite: ATP has much weaker capacity than A24 in both inducing and dissolving LLPS of the N protein. Therefore, based on the residue-specific results we previously obtained that ATP biphasically modulates LLPS of the IDDs of FUS and TDP-43 by specifically binding Arg/Lys residues within IDRs [11,12], here we propose that ATP biphasically modulates LLPS of the SARS-CoV-2 N proteins mainly by its bivalent interaction with Arg/Lys residues within its three IDRs (Fig. 1B). Briefly ATP is capable of establishing the π-cation interaction between its puring ring and the sidechain cations of Arg/Lys as well as the electrostatic interaction between its triphosphate chain and the side chains of Arg/Lys located in its IDRs. This rationalize the result that A24 has much higher capacity than ATP, because A24 has the multivalent ability to bivalently interact with Arg/Lys residues of IDRs of the N protein, which thus leads to the significantly enhanced affinity [20]. On the other hand, due to the low numbers of Arg/Lys residues within three IDRs of the N protein (Fig. 1B), ATP modulates LLPS of the SARS-CoV-2 N protein unlikely by the direct mechanism as we found on the 156-residue RGG-rich IDD of FUS by which the dynamic and bivalent binding of ATP to 25 Arg and 4 Lys residues is sufficient to drive the formation of large dynamic complexes manifesting as liquid droplets. Instead, ATP modulates LLPS of the N protein most likely by the indirect mechanism as we found with the 150-residue TDP-43 prion-like domain with only 5 Arg and 1 Lys residue, by which the binding of ATP to Arg/Lys residues only acts to coordinate other forces, particularly the dimerization by a hydrophobic fragment, to drive LLPS. This may explain the previous reports that the dimerization domain of the N protein is essential for its RNA-induced LLPS [[5], [6], [7]].
Another novel and critical finding here is that although RBD of the SARS-CoV-2 N protein has a structural fold very different from those adopted by RRM as well as by AcD, it can also specifically bind ATP with similar Kd values of ∼mM at a pocket within the surface for binding nucleic acids [[13], [14], [15]]. The result not only establishes the SARS-CoV-2 RBD to be the first viral domain capable of binding ATP at biologically-relevant concentrations (∼mM), but also suggests that RBD may have an pivotal role in specifically regulating the uncoating, localizing and packing of the genomic RNA by the N protein. As illustrated in Fig. 4E, immediately after the infection, the SARS-CoV-2 will release its genomic RNA-N-protein condensate into the infected cell, which is tightly packed into the gel-like state [5]. With consideration that at this stage, one infected cell may only have one to several copies of the condensate, the ratios between ATP and N protein/genomic RNA are very high. Consequently ATP acts to facilitate the condensate to be uncoated, such as to transform the gel-like condensate into more dynamic liquid droplets or even homogenous solution. Furthermore, once new copies of viral RNA polymerase and N-protein are synthesized by the host cell machinery, the ratios will reduce, and ATP may enhance LLPS of the mixture of the viral genomic RNA and N proteins as well as the host cell replicases to form replicase-transcriptase complexes. Finally, after all components needed for the assembly of new virions are synthesized by the infected cell, the ratios between ATP and N protein/genomic RNA will be further reduced and therefore a large population of the ATP-RBD complex will become dissociated. As such, the ATP-unbound RBD of the SARS-CoV-2 N protein become available for binding the specific sites of the genomic RNA to initiate the packing process, which might be even enhanced by ATP at low molar ratios.
In summary, here for the first time we discovered that ATP not only biphasically modulates LLPS of the viral N protein of SARS-CoV-2, but also dissolves its LLPS induced by oligonucleic acid. Furthermore, ATP also specifically bind its RNA-binding domain with a fold very different from those of RRM and AcD of human proteins. SARS-CoV-2 appears to exploit the high ATP concentrations of host cells to promote its life cycle by facilitating its genomic RNA to be uncoated at the initial stage of infection, localized to replicase-transcriptase complexes for synthesis of other viral proteins, and packed with the N protein into virions at the last stage. Therefore, better understanding the mechanisms underly the interactions of ATP with the viral genomic RNA and N protein as well as with other components may help the design of drugs and effective vaccines to terminate the pandemic.
Acknowledgement
This study is supported by Ministry of Education of Singapore (MOE) Tier 1 Grants R-154-000-B45-114 and R-154-000-B92-114 to Jianxing Song.
Footnotes
Transparency document related to this article can be found online at https://doi.org/10.1016/j.bbrc.2021.01.018
(A) Domain organization of the SARS-CoV-2 nucleocapsid (N) protein composed of the N-terminal intrinsically-disordered region (IDR), RNA-binding domain (RBD) or N-terminal domain (NTD), middle IDR, dimerization domain or C-terminal domain (CTD) and C-terminal IDR. (B) Compositions of the N protein and its three IDRs. (C) Turbidity curves of the N protein at 20 μM without (blue) and with the pre-existence of A24 at 1:0.5 (red) upon additional addition of ATP at different ratios. DIC images of the N protein at 20 μM without (D) and with the pre-existence of A24 at 1:0.5 (E) upon additional addition of ATP at different ratios.
(A) 1H–15N NMR HSQC spectra of the 15N-labeled RBD (44–180) at 50 μM without (blue), and with ATP at 10 mM (red). (B) Residue-specific chemical shift difference (CSD) in the presence of ATP at 8 mM (blue) and 10 mM (red). 11 residues with significantly shifted HSQC peaks are labeled, which are defined as those with CSD values at 10 mM ATP >0.08 (average value + one standard deviation) (green line). The average dissociation constant (Kd) for 11 residues was indicated. (C) NMR structure of RBD (PDB ID of 6YI3) with 11 residues displayed in sphere.
(A) The lowest-energy docking model of the ATP-RBD complex with RBD in ribbon and ATP in ball-and-stick. (B)-(C) The ATP-RBD complex with RBD in the electrostatic potential surface and ATP in ball-and-stick. (D) Expanded view of the ATP-binding pocket of RBD. (E) The ATP-RBD complex showing hydrogen bonds of the β-phosphate of the triphosphate chain of ATP with sidechain/backbone atoms of Asn8 as well as with backbone atom of Thr9 of RBD (in yellow dotted lines).
(A) Superimposition of the ATP-RBD complex and NMR structure of the SsRNA-RBD complex (PDB ID of 7ACS) with RBD in ribbon (I) and in electrostatic potential surface (II). (B) Superimposition of the ATP-RBD complex and NMR structure of the dsRNA-RBD complex (PDB ID of 7ACT) with RBD in ribbon (I) and in electrostatic potential surface (II). (C) The complex structure of ATP with the TDP-43 RRM1 we previously determined. (D) The complex structure of ATP with the SYNCRIP AcD we previously determined. (E) A proposed scheme to illustrate that ATP appears to be exploited by SARS-CoV-2 to promote its life cycle including the initial uncoating of the RNA-N-protein condensate and subsequent localizing to forming replicase-transcriptase complex.
Transparency document
References
- 1.Zhou P., Yang X.L., Wang X.G., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Snijder E.J., Bredenbeek P.J., Dobbe J.C., et al. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McBride R., Van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verheije M.H., Hagemeijer M.C., Ulasli M., et al. The coronavirus nucleocapsid protein is dynamically associated with the replication-transcription complexes. J. Virol. 2010;84:11575–11579. doi: 10.1128/JVI.00569-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu S., Ye Q., Singh D., et al. The SARS-CoV-2 Nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. BioRxiv. 2020 doi: 10.1101/2020.07.30.228023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Savastano A., de Opakua A.I., Rankovic M., et al. Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. Nat. Commun. 2020;11:6041. doi: 10.1038/s41467-020-19843-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perdikari T.M., Murthy A.C., H Ryan V., et al. SARS-CoV-2 nucleocapsid protein phase-separates with RNA and with human hnRNPs. EMBO J. 2020 doi: 10.15252/embj.2020106478. Nov 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dinesh D.C., Chalupska D., Silhan J., et al. Structural basis of RNA recognition by the SARS-CoV-2 nucleocapsid phosphoprotein. BioRxiv. 2020 doi: 10.1101/2020.04.02.022194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lehninger’s Principles of Biochemistry. fifth ed. W.H. Freeman and Company; NewYork: 2005. pp. 502–503. [Google Scholar]
- 10.Patel A., Malinovska L., Saha S., et al. ATP as a biological hydrotrope. Science. 2017;356:753–756. doi: 10.1126/science.aaf6846. [DOI] [PubMed] [Google Scholar]
- 11.Kang J., Lim L., Lu Y., et al. A unified mechanism for LLPS of ALS/FTLD-causing FUS as well as its modulation by ATP and oligonucleic acids. PLoS Biol. 2019;17 doi: 10.1371/journal.pbio.3000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dang M., Lim L., Kang J., et al. ATP regulates TDP-43 pathogenesis by specifically binding to an inhibitory component of a delicate network controlling LLPS. BioRxiv. 2020 doi: 10.1101/2020.10.08.330829. [DOI] [Google Scholar]
- 13.Kang J., Lim L., Song J. ATP binds and inhibits the neurodegeneration-associated fibrillization of the FUS RRM domain. Commun Biol. 2019;2:223. doi: 10.1038/s42003-019-0463-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dang M., Kang J., Lim L., et al. ATP is a cryptic binder of TDP-43 RRM domains to enhance stability and inhibit ALS/AD-associated fibrillation. Biochem. Biophys. Res. Commun. 2020;522:247–253. doi: 10.1016/j.bbrc.2019.11.088. [DOI] [PubMed] [Google Scholar]
- 15.He Y., Kang J., Lim L., et al. ATP binds nucleic-acid-binding domains beyond RRM fold. Biochem. Biophys. Res. Commun. 2020;522:826–831. doi: 10.1016/j.bbrc.2019.11.180. [DOI] [PubMed] [Google Scholar]
- 16.Wessner D.R. The origins of viruses. Nature Education. 2010;3:37. [Google Scholar]
- 17.de Vries S.J., van Dijk A.D., Krzeminski M., et al. HADDOCK versus HADDOCK: new features and performance of HADDOCK2.0 on the CAPRI targets. Proteins. 2007;69:726–733. doi: 10.1002/prot.21723. [DOI] [PubMed] [Google Scholar]
- 18.Cascarina S.M., Ross E.D. A proposed role for the SARS-CoV-2 nucleocapsid protein in the formation and regulation of biomolecular condensates. Faseb. J. 2020;34:9832–9842. doi: 10.1096/fj.202001351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahlén G., Frelin L., Nikouyan N. The SARS-CoV-2 N protein is a good component in a vaccine. J. Virol. 2020;94 doi: 10.1128/JVI.01279-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song J., Ni F. NMR for the design of functional mimetics of protein-protein interactions: one key is in the building of bridges. Biochem. Cell. Biol. 1998;76:177–188. doi: 10.1139/bcb-76-2-3-177. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.