Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is responsible for coronavirus disease 2019 (COVID-19). Since its emergence, the COVID-19 pandemic has not only distressed medical services but also caused economic upheavals, marking urgent the need for effective therapeutics. The experience of combating SARS-CoV and MERS-CoV has shown that inhibiting the 3-chymotrypsin-like protease (3CLpro) blocks the replication of the virus. Given the well-studied properties of FDA-approved drugs, identification of SARS-CoV-2 3CLpro inhibitors in an FDA-approved drug library would be of great therapeutic value. Here, we screened a library consisting of 774 FDA-approved drugs for potent SARS-CoV-2 3CLpro inhibitors, using an intramolecularly quenched fluorescence (IQF) peptide substrate. Ethacrynic acid, naproxen, allopurinol, butenafine hydrochloride, raloxifene hydrochloride, tranylcypromine hydrochloride, and saquinavir mesylate have been found to block the proteolytic activity of SARS-CoV-2 3CLpro. The inhibitory activity of these repurposing drugs against SARS-CoV-2 3CLpro highlights their therapeutic potential for treating COVID-19 and other Betacoronavirus infections.
Keywords: SARS-CoV-2 3CL protease, antiviral, repurposing drugs, FRET, 3CLpro inhibitors
Introduction
Coronavirus disease 2019 (COVID-19), resulting from severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, has distressed medical services and economies worldwide and has had profound psychological effects since its emergence1,2. Among COVID-19 patients, about 81% have no or mild symptoms, with severe symptoms in 14% and critical illness in 5%2. The clinical manifestations of SARS-CoV-2 infection often include, but are not limited to, fever, cough, fatigue, muscle soreness and abdominal pain, similar to severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV)2. Risk factors for becoming critically ill with COVID-19 include cardiovascular disease, diabetes and obesity; however, healthy people of any age can become critically ill with COVID-19, although the current data suggest that individuals over 65 years of age, particularly men, are more likely to have severe symptoms3. Because SARS-CoV-2 infection has become a global pandemic, causing severe damage to public health4, there is a desperate need for effective therapeutics.
SARS-CoV-2, an enveloped, positive-sense, single-stranded RNA (+ssRNA) Betacoronavirus (β CoVs), is quite similar to SARS-CoV2,5,6. The genome of SARS-CoV-2 is about 30 kb, in which open reading frames (ORF) 1a and 1 b encode two polyproteins (pps), pp1a and pp1ab2. To complete the lifecycle of SARS-CoV-2, successful proteolytic processing of pp1a and pp1ab is required to yield a total of 16 non-structural proteins (nsp1–16)2. The consensus functions of these virus-encoded proteolytic proteins are found in all β CoVs, specifically papain-like protease (PLpro) and chymotrypsin-like protease (3CLpro)2. In particular, the substrate binding site of SARS-CoV-2 3CLpro is highly conserved across the β CoVs suggesting the therapeutic potential of 3CLpro inhibitors for SARS-CoV-2 and other β CoVs2,7. In addition, alignment of the genomic sequences of SARS-CoV-2, SARS-CoV and MERS-CoV reveals a high-level conservation of the proteolytic sites and proteolytic enzymes2,8,9.
A member of the cysteine protease family, the active SARS-CoV-2 3CLpro comprises two identical monomers, each with three structural domains; the first two domains (domain I: 8–101 and II: 102–184) form a chymotrypsin fold, and the third (domain III: 201–303) forms a globular α-helical structure, with an identity of 96% to SARS-CoV 3CLpro7,10. In particular, the catalytic dyad of SARS-CoV-2 3CLpro includes H41 and C145 in domains I and II, respectively7; meanwhile, dimerisation and formation of the S1 subsite of the substrate binding site involve the interaction between the N-terminal residue (N-finger) of one polypeptide and the E166 residue of the other11. Consistently, the most variable regions of 3CLpro in known β CoVs were found to be situated in domain III and the surface loops, indicating that the proteolytic activity is mainly governed by domains I and II7.
Inhibition of the activity of 3CLpro in SARS-CoV-2 is regarded as a plausible approach to block its replication. Screening of FDA-approved drugs for SARS-CoV-2 3CLpro inhibitors has been conducted in silico and in vitro7, identifying two FDA-approved drugs (disulfiram and carmofur), and five preclinical or investigational compounds as promising antiviral agents against 3CLpro. In this study, we screened a library consisting of 774 FDA-approved drugs for potential SARS-CoV-2 3CLpro inhibitors. To evaluate the extent of inhibition of SARS-CoV-2 3CLpro, a fluorogenic peptide with intramolecularly quenched fluorescence (IQF) was used as the substrate for the protease. Subject to the inhibitory effect, the half maximal inhibitory concentrations of the repurposing existing drugs of interest were characterised, along with analysis of docking poses in the substrate binding site of SARS-CoV-2 3CLpro.
Materials and methods
Drug library
The SCREEN-WELL® FDA v. 2.0 Approved Drug Library (BML-2843–0100) was purchased from ENZO Life Sciences Inc., NY, USA, and comprises 774 clinical drugs with well-studied bioactivity, safety and bioavailability.
Construction of pET28b(+)-SARS-CoV-2-3CLpro
A published sequence of SARS-CoV-2 3CLpro11 was chemically synthesised and cloned into an yT&A plasmid by Genomics, Taiwan. The insert, encoding full length SARS-CoV-2 3CLpro, was amplified from the yT&A plasmid using ExcelTaq™ Taq DNA polymerase (Smobio, Taiwan), primer 5′-ATGGGTCGGGATCCCAGTGGTTTTAGAAA-3′ and primer 5′-GGTGCTCGAGTTCATCTAGTTATTGGAAAGTAACACCTGAG-3′ and cloned into a T7-based pET-28b(+) plasmid (Thermo Fisher Scientific, MA, USA) digested with BamHI and XhoI (New England Biolabs, MA, USA). Plasmid extraction from E. coli DH5α cells was carried out using Presto™ Mini Plasmid kits or Geneaid™ Midi Plasmid kits. The amplicon was purified using a PCR clean-up DNA/RNA extraction kit (Viogene, Taiwan). The insert sequence of the pET28b(+) DNA plasmid was verified by the National Yang-Ming University Genome Research Center, Taiwan.
Protein expression and purification
The SARS-CoV-2 3CLpro was purified using the His-tag at its N-terminal, using a nickel column from GE healthcare, IL, USA, following the procedure described previously12. The purified protein was resolved by SDS-PAGE and the image quantification with Multi Gauge densitometry (Fujifilm, Japan) characterised the protein purity to be over 95%. Biochemical protein quantification was performed using Bio-Rad protein assays (CA, USA), with the measurements at 595 nm in a SPARK® multimode microplate reader (TECAN, Switzerland).
Protease activity assays using IQF peptide substrates
An Edans-Dabcyl FRET platform was established, following a published protocol13. Briefly, a consensus cleavage sequence recognised by SARS-CoV-2 3CLpro was synthesised by Genomics, Taiwan, with Dabcyl at the N-terminus and Edans at the C-terminus, Dabcyl-TSAVLQ↓SGFRKME-Edans. In protease activity assays, 0.25 µM protease was incubated with 1.25 µM peptide substrate for three hours. Assays were conducted in triplicate in Eppendorf® black 96-well microplates (MA, USA) using an assay buffer containing 12 mM Tris-HCl (pH 7.5), 120 mM NaCl, 0.1 mM EDTA and 1 mM dithiothreitol (DTT), in a final volume of 100 µL. The fluorescence signal at 538 nm, at a bandwidth of 15 nm, emitted from the cleaved IQF peptide substrate after excitation at 355 nm, at a bandwidth of 10 nm, was recorded by a SPARK® multimode microplate reader (TECAN, Switzerland). The relative fluorescence units (RFU) at a gain of 131 were calculated using Spark® Control Magellan™ v2.2 software.
Dose-response curve analysis
SARS-CoV-2 3CLpro was incubated with drugs at 0–100 µM for an hour at 37 °C. Then, 1.25 µM IQF peptide substrate was added to the mixture to a final volume of 100 µL and incubated at 37 °C for another three hours, prior to detection. With the same parameters applied in protease activity assays, the RFU readouts obtained from the SPARK® multimode microplate reader (TECAN, Switzerland) were normalised to the negative control (vehicle only) in each assay plate. After drug treatment at a concentration between 0–100 µM, points of relative protease activity were fitted to a normalised dose-response model in GraphPad Prism 7.03 for IC50 characterisation, where .
Molecular docking
For molecular docking, the interaction profile of a compound in the substrate binding site of SARS-CoV-2 3CLpro was simulated in GEMDOCK: molecular docking tool14. Retrieving the crystal structure of SARS-CoV-2 main protease from the Protein Data Bank (PDB ID: 6LU77), the substrate binding site of SARS-CoV-2 3CLpro was defined by an 8 Å-radius sphere around the bound peptide-like inhibitor PRD_002214. The 3D drug structures (SDF files) from DrugBank15 were converted to MOL files by Open Babel16.
Statistical analysis
Data collected in the study were analysed and plotted with GraphPad Prism 7.03 (GraphPad software) when a minimum of N = 3 independent samples was obtained. Values were expressed as the mean ± standard mean error (SEM) if not otherwise specified.
Results
Screening a 774 FDA-approved drug library against 3CLpro activity
A compound library of FDA-approved drugs was screened for SARS-CoV-2 3CLpro inhibitory activity using an IQF peptide substrate. A flowchart of the screening procedure is shown in Figure 1. To identify compounds as potential SARS-CoV-2 3CLpro inhibitors, 774 FDA-approved drugs were screened at 20 µM in the high-throughput, initial screening. Among these 774 FDA-approved drugs, twenty potentially active compounds were found, including seven drugs with superior inhibitory activity against SARS-CoV-2 3CLpro. The twenty most active SARS-CoV-2 3CLpro inhibitors are listed in Table 1, with their IC50 values. Briefly, ethacrynic acid, naproxen, allopurinol, butenafine hydrochloride, raloxifene hydrochloride, tranylcypromine hydrochloride, and saquinavir mesylate led to 50% inhibition on SARS-CoV-2 3CLpro activity at concentrations below 10 µM. In addition, triptorelin acetate, goserelin acetate, rocuronium bromide, bisacodyl, armodafinil, and clobetasol propionate had an IC50 value of 10–20 µM, followed sequentially by seven moderate SARS-CoV-2 3CLpro inhibitors: sirolimus (rapamycin), colistin sulphate, cetirizine, bexarotene, cefpodoxime proxetil, clindamycin palmitate hydrochloride and oxaliplatin.
Figure 1.
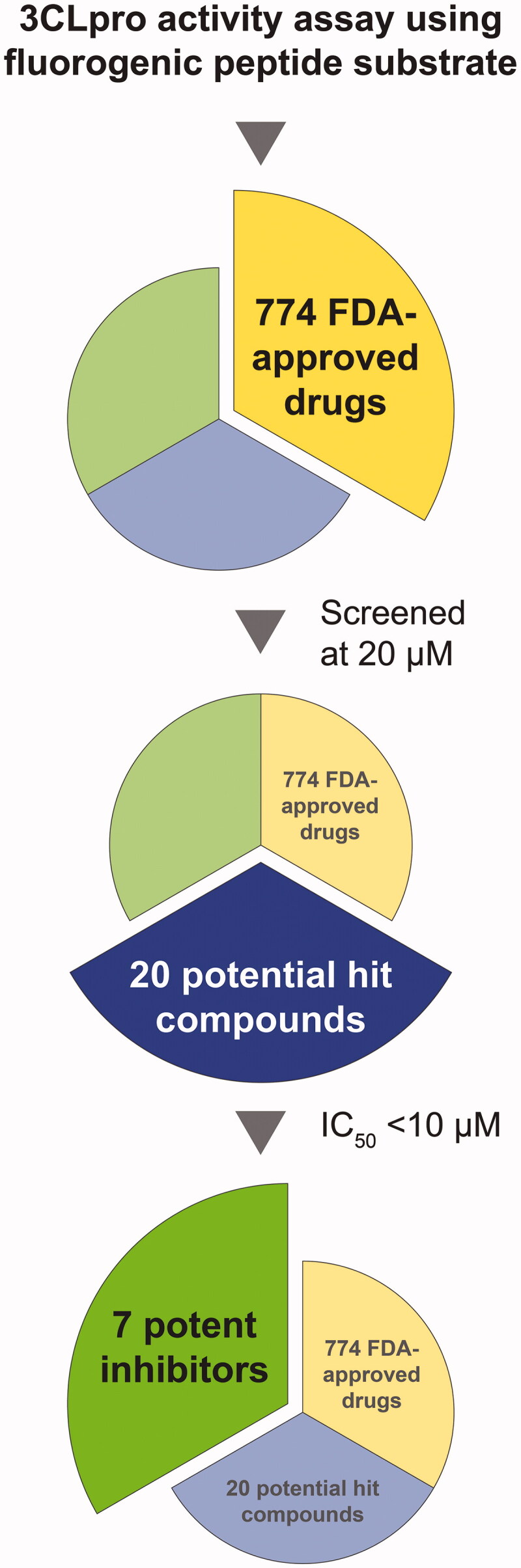
Flowchart of identification of SARS-CoV-2 3CLpro inhibitors in a library of 774 FDA-approved drugs. An initial screening was performed to evaluate the inhibition of SARS-CoV-2 3CLpro activity by FDA-approved drugs at 20 µM. Subsequently, IC50 characterisation was performed to pinpoint the more effective drugs. Twenty potential hit compounds were found, of which seven had a more pronounced effect in inhibiting SARS-CoV-2 3CLpro.
Table 1.
Inhibition of SARS-CoV-2 3CLpro activity by FDA-approved drugs.
| Compounds | IC50 (µM) <10 µM |
|---|---|
| Ethacrynic acid | 1.11 |
| Naproxen | 3.45 |
| Allopurinol | 3.77 |
| Butenafine hydrochloride | 5.40 |
| Raloxifene hydrochloride | 5.61 |
| Tranylcypromine hydrochloride | 8.64 |
| Saquinavir mesylate | 9.92 |
| Compounds | IC50 (µM) <50 µM |
|---|---|
| Triptorelin acetate | 10.12 |
| Goserelin acetate | 12.02 |
| Rocuronium bromide | 17.47 |
| Bisacodyl | 17.51 |
| Armodafinil | 17.87 |
| Clobetasol propionate | 18.09 |
| Sirolimus (Rapamycin) | 22.30 |
| Colistin sulphate | 23.20 |
| Cetirizine | 25.58 |
| Bexarotene | 26.49 |
| Cefpodoxime proxetil | 32.43 |
| Clindamycin palmitate hydrochloride | 33.21 |
| Oxaliplatin | 47.13 |
SARS-CoV-2 3CLpro inhibitors of therapeutic potentials
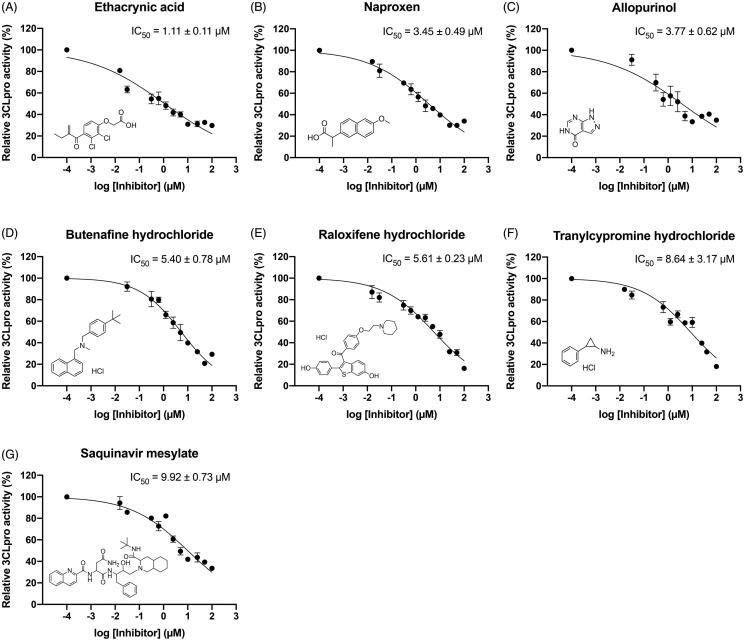
Regarding the therapeutic potential of the seven potent SARS-CoV-2 3CLpro inhibitors, dose–response curves of ethacrynic acid, naproxen, allopurinol, butenafine hydrochloride, raloxifene hydrochloride, tranylcypromine hydrochloride and saquinavir mesylate are shown in Figure 2, with their IC50 values and chemical structures. The measured IC50 values were 1.11 ± 0.11, 3.45 ± 0.49, 3.77 ± 0.62, 5.40 ± 0.78, 5.61 ± 0.23, 8.64 ± 3.17, and 9.92 ± 0.73 µM, respectively. Interestingly, despite the different protease family, saquinavir mesylate, an inhibitor of aspartate proteases17, was able to inhibit SARS-CoV-2 3CLpro, a cysteine protease.
Figure 2.
Dose-response curves of potent SARS-CoV-2 3CLpro inhibitors. The inhibitory activity of (A) Ethacrynic acid, (B) Naproxen, (C) Allopurinol, (D) Butenafine hydrochloride, (E) Raloxifene hydrochloride, (F) Tranylcypromine hydrochloride, and (G) Saquinavir mesylate against SARS-CoV-2 3CLpro are shown, along with a depiction of the chemical structure. Data (N = 3) are expressed as the mean ± SEM.
Molecular modelling of identified inhibitors in the substrate binding site of SARS-CoV-2 3CLpro
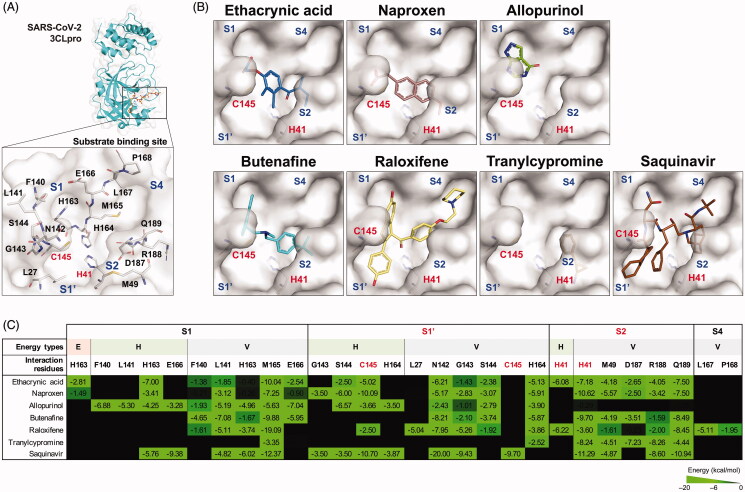
To elucidate the inhibitory mechanism of the identified SARS-CoV-2 3CLpro inhibitors, molecular docking was performed to simulate the binding model in the substrate binding site of SARS-CoV-2 3CLpro. As shown in Figure 3(A), the substrate binding site of SARS-CoV-2 3CLpro can be divided into four subsites7, where the S1 subsite comprises L27, N142, G143, S144, C145 and H164, the S1' subsite consists of H163, F140, L141, E166 and M165, the S2 subsite includes H41, M49, D187, R188 and Q189, and the S4 subsite is made up of L167 and P168. A way to disrupt the catalytic function of SARS-CoV-2 3CLpro is to occlude the access of the substrate to the Cys-His catalytic dyad (C145 and H41)18. The molecular docking results revealed that the identified inhibitors interacted with the Cys-His catalytic dyad, along with other residues, in the substrate binding site of SARS-CoV-2 3CLpro (Figure 3(B,C)). Specifically, ethacrynic acid and naproxen form a stable electrostatic force with H163 through the carboxyl group, and hydrogen bonding and van der Waals force with the catalytic dyad and other residues in the S1, S1' and S2 subsites. Butenafine interacted with the catalytic residue H41 and other residues in the S1, S1’ and S2 subsites through van der Waals force alone. Raloxifene and saquinavir filled all four subsites of SARS-CoV-2 3CLpro, binding to the catalytic residues C145 and H41, and the enclosed hydrophobic residues N142, G143, L141, M165, M49, L167, P168, R188 and Q189, resembling to the binding mode of Michael acceptor inhibitors7. As for compounds of a low heavy atom count, allopurinol and tranylcypromine occupied deeply in the S1 and the S2 subsite, respectively. Taken together, the identified inhibitors docked into up to four subsites of the substrate binding site of SARS-CoV-2 3CLpro, interacting with the catalytic dyad and other residues involving in substrate binding.
Figure 3.
Interaction forces between the identified inhibitors and the substrate binding residues of SARS-CoV-2 3CLpro. (A) The substrate binding site of SARS-CoV-2 3CLpro. S1, S1’, S2 and S4 subsites are labelled in blue. Catalytic residues (red) H41 and C145, and other substrate binding residues (black) are labelled. (B) Molecular docking of seven SARS-CoV-2 3CLpro inhibitors. Substrate binding subsites (blue) and catalytic residues (red) H41 and C145 are labelled. (C) Interaction profiles of seven SARS-CoV-2 3CLpro inhibitors. The interaction energy (kcal/mol) positively correlates with the brightness of the colour (bright green). Catalytic residues H41 and C145 are labelled in red. E: electrostatic force (red fill); H: hydrogen binding force (green fill); V: van der Waals force (gray fill).
Discussion
Coronaviruses, known for the crown-like appearance of the virions in electron microscopy, are enveloped + ssRNA viruses with the largest known genome size among RNA viruses. The genome encodes structural proteins (e.g. spike glycoproteins), non-structural proteins (e.g. papain-like protease (PLpro) and 3-chymotrypsin-like protease (3CLpro), helicase, RNA-dependent RNA polymerase), and accessory proteins2,5,6. SARS-CoV-2, a recently identified coronavirus, is responsible for the COVID-19 pandemic. In terms of societal demographics, the most vulnerable individuals are adults above 65 years of age, those with underlying conditions, and the economically disadvantaged3,19. Meanwhile, it has been determined that lymphopenia and elevated cytokine production resulting from SARS-CoV-2-induced immunopathology are responsible for disease progression and increased severity20. Based on the experience with SARS-CoV and MERS-CoV, active approaches to fight SARS-CoV-2 infection can be divided into three groups: (i) agents targeting the virus, (ii) agents targeting the host response, and (iii) spike-based vaccines2. Although the preliminary clinical data of vaccine development showed promise19,21, agents directly inhibiting viral replication remain of great interest. The current knowledge of β CoVs highlights the pivotal role of 3CLpro in viral replication and transcription and the value of developing broad-spectrum anti-β CoVs drugs in this regard2. Thus, 3CLpro inhibition has been regarded as a molecular approach in anti-SARS drug discovery and development7,13,22. Here, we screened a drug library consisting of 774 FDA-approved drugs for potential SARS-CoV-2 3CLpro inhibitors, using a protease-specific IQF peptide substrate.
Recently, treatment of severe COVID-19 patients with the HIV protease inhibitors lopinavir-ritonavir had no obvious efficacy beyond standard care6 but the final determination of their efficacy for COVID-19 patients requires further clinical study23. The use of hydroxychloroquine sulphate, an antimalarial agent, in severe or critically ill COVID-19 patients showed contradictory results in clinical trials24,25, and it is suggested to be more effective in early infection. Remdesivir, a nucleotide analogue prodrug in phase III clinical trials for Ebola virus infection, showed therapeutic promise for treating severe COVID-19 patients, with shortened recovery times26–28. Dexamethasone, a corticosteroid, was found to reduce the 28-day mortality of COVID-19 patients receiving either invasive mechanical ventilation or oxygen alone29. Based on the therapeutic experience against viruses, the most effective therapy for SARS-CoV-2 infection would most likely require a cocktail of agents targeting different stages of viral infection30. Indeed, combining lopinavir-ritonavir with two other agents helped alleviate symptoms, and a shortened viral shedding period was reported in mild-to-moderate COVID-19 patients10.
Utilisation of FDA-approved drug library is an effective and ideal tool for drug repurposing in antiviral research7,31, such as zika virus32, human rhinovirus33, and hepatitis B virus34. Regarding the possibility of using FDA-approved drugs for anti-SARS-CoV-2 therapy, we identified twenty potentially active drugs and these are listed in Table 1. Several of those drugs were previously reported to have antiviral activity. For example, ethacrynic acid derivatives have been shown to inhibit SARS-CoV 3CLpro activity by binding directly to the active site35. Naproxen was reported to be incorporated into the RNA-binding groove of the nucleoprotein of influenza A virus, suggesting its potential role in antiviral research36. The therapeutic potential of tranylcypromine for herpes simplex virus 1 (HSV-1) infection was evaluated because of its inhibitory activity against the histone-modifying enzyme, lysine-specific demethylase 137. Raloxifene, a selective oestrogen receptor modulator, was reported to inhibit Ebola virus infection38. Saquinavir, the first HIV protease inhibitor made available in the market, was shown to be ineffective for inhibiting SARS-CoV replication39,40. Sirolimus blocked stages after the reverse transcription event in activated human T cells infected by human immunodeficiency virus 1 (HIV-1)41. Cetirizine, an antihistamine reported to inhibit the replication of respiratory syncytial virus (RSV) and the expression of interleukin-8 (IL-8), has an unknown property in reducing of RSV infectivity42. Bexarotene was shown to inhibit the expression of the hepatitis C virus core protein43. As for those that have not been mentioned, they have not yet been evaluated in antiviral research.
Importantly, a systematic review of the current evidence for non-steroidal anti-inflammatory drugs (NSAIDs) in the management of COVID-19 suggests that naproxen may be worthy of further investigation in clinical trials, because of its positive effects in controlling the symptoms of coryza, rhinovirus infection and influenza-related pneumonia44. On the other hand, the inhibitory activity of saquinavir against SARS-CoV-2 3CLpro denoted in this study matched the result from in silico molecular docking models reported previously45. Furthermore, sirolimus, a moderate SARS-CoV-2 3CLpro inhibitor identified in this study, was suggested to help prevent progression to severe forms of COVID-19 by mitigating the SARS-CoV-2-induced cytokine storm30,46. Last, but not least, bexarotene, a moderate SARS-CoV-2 3CLpro inhibitor, was shown to have broad-spectrum anticoronavirial activity in a study published recently47.
Taken together, we found several potent SARS-CoV-2 3CLpro inhibitors in a library of 774 FDA-approved drugs, including ethacrynic acid, naproxen, allopurinol, butenafine hydrochloride, raloxifene hydrochloride, tranylcypromine hydrochloride, and saquinavir mesylate. These drugs exert SARS-CoV-2 3CLpro inhibition by obscuring the accessibility of the C145-H41 catalytic dyad via hydrogen bonding and van der Waals force. Including the forces mentioned, the carboxyl group of ethacrynic acid and naproxen form an additional electrostatic force to H163 in the substrate binding site of SARS-CoV-2 3CLpro. Although ethacrynic acid had the best inhibitory activity against SARS-CoV-2 3CLpro, repurposing naproxen and sirolimus for COVID-19 treatment shows promise in that they have anti-inflammatory and immunosuppressive activities, respectively, which may help address the immunopathology induced by SARS-CoV-2 infection. Our identification of potent SARS-CoV-2 3CLpro inhibitors among FDA-approved drugs highlights their potential for treating COVID-19 and other diseases caused by β CoVs.
Funding Statement
This work was supported by research grant [MOST 109–2327-B-010–006] – from the Ministry of Science and Technology, Taiwan.
Author contributions
W.C.C., M.S.H., and Y.T.C performed the experiments. Y.G.T. provided the compounds. J.M.Y. and C.H. designed the experiments. W.C.C., H.C.H. and C.H. were primarily responsible for writing the manuscript. All authors contributed to manuscript editing and approved the final version.
Disclosure statement
The authors declare no conflicts of interest.
References
- 1.King JS. Covid-19 and the need for health care reform. N Engl J Med 2020;382:e104. [DOI] [PubMed] [Google Scholar]
- 2.Chen B, Tian EK, He B, et al. Overview of lethal human coronaviruses. Signal Transduct Target Ther 2020;5:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berlin DA, Gulick RM, Martinez FJ.. Severe Covid-19. N Engl J Med 2020. [DOI] [PubMed] [Google Scholar]
- 4.Ienca M, Vayena E.. On the responsible use of digital data to tackle the COVID-19 pandemic. Nat Med 2020;26:463–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khailany RA, Safdar M, Ozaslan M.. Genomic characterization of a novel SARS-CoV-2. Gene Rep 2020;19:100682. ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao B, Wang Y, Wen D, et al. A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N Engl J Med 2020;382:1787–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin Z, Du X, Xu Y, et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors . Nature 2020;582:289–93. [DOI] [PubMed] [Google Scholar]
- 8.Hegyi A, Friebe A, Gorbalenya AE, Ziebuhr J.. Mutational analysis of the active centre of coronavirus 3C-like proteases. J Gen Virol 2002;83:581–93. [DOI] [PubMed] [Google Scholar]
- 9.Needle D, Lountos GT, Waugh DS.. Structures of the Middle East respiratory syndrome coronavirus 3C-like protease reveal insights into substrate specificity. Acta Crystallogr D Biol Crystallogr 2015;71:1102–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hung IF-N, Lung K-C, Tso EY-K, et al. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet 2020;395:1695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L, Lin D, Sun X, et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 2020;368:409–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuang WF, Chow LP, Wu MH, Hwang LH.. Mutational and inhibitive analysis of SARS coronavirus 3C-like protease by fluorescence resonance energy transfer-based assays. Biochem Biophys Res Commun 2005;331:1554–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jo S, Kim S, Shin DH, Kim MS.. Inhibition of SARS-CoV 3CL protease by flavonoids. J Enzyme Inhib Med Chem 2020;35:145–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang JM, Chen CC.. GEMDOCK: a generic evolutionary method for molecular docking. Proteins 2004;55:288–304. [DOI] [PubMed] [Google Scholar]
- 15.Wishart DS, Feunang YD, Guo AC, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 2018;46:D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Boyle NM, Banck M, James CA, et al. Open Babel: an open chemical toolbox. J Cheminform 2011;3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghosh AK, Osswald HL, Prato G.. Recent progress in the development of HIV-1 protease inhibitors for the treatment of HIV/AIDS. J Med Chem 2016;59:5172–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang H, Xie W, Xue X, et al. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol 2005;3:e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heaton PM. The Covid-19 vaccine-development multiverse. N Engl J Med 2020;383:1986–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang L, Liu S, Liu J, et al. COVID-19: immunopathogenesis and Immunotherapeutics. Signal Transduct Target Ther 2020;5:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson LA, Anderson EJ, Rouphael NG, et al. An mRNA vaccine against SARS-CoV-2 – preliminary report. N Engl J Med 2020;383:1920–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen C-N, Lin CPC, Huang K-K, et al. Inhibition of SARS-CoV 3C-like protease activity by theaflavin-3,3'-digallate (TF3). Evid Based Complement Alternat Med 2005;2:209–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao B, Zhang D, Wang C.. A trial of lopinavir-ritonavir in Covid-19. Reply. N Engl J Med 2020;382:e68. [DOI] [PubMed] [Google Scholar]
- 24.Taccone FS, Gorham J, Vincent JL.. Hydroxychloroquine in the management of critically ill patients with COVID-19: the need for an evidence base. Lancet Respir Med 2020;8:539–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molina JM, Delaugerre C, Le Goff J, et al. No evidence of rapid antiviral clearance or clinical benefit with the combination of hydroxychloroquine and azithromycin in patients with severe COVID-19 infection. Med Mal Infect 2020;50:384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grein J, Ohmagari N, Shin D, et al. Compassionate use of remdesivir for patients with severe Covid-19. N Engl J Med 2020;382:2327–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Zhang D, Du G, et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet 2020;395:1569–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beigel JH, Tomashek KM, Dodd LE, et al. Remdesivir for the treatment of Covid-19 – final report. N Engl J Med 2020;383:1813–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Group RC, et al. Dexamethasone in hospitalized patients with Covid-19 – preliminary report. N Engl J Med 2020. [Google Scholar]
- 30.Wu R, et al. An update on current therapeutic drugs treating COVID-19. Curr Pharmacol Rep 2020;1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang P, Liu Y, Zhang G, et al. Screening and identification of lassa virus entry inhibitors from an FDA-approved drug library. J Virol 2018;92:e00954-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrows NJ, Campos RK, Powell ST, et al. A screen of FDA-approved drugs for inhibitors of zika virus infection. Cell Host Microbe 2016;20:259–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shim A, Song J-H, Kwon B-E, et al. Therapeutic and prophylactic activity of itraconazole against human rhinovirus infection in a murine model. Sci Rep 2016;6:23110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sekiba K, Otsuka M, Ohno M, et al. Inhibition of HBV transcription from cccDNA with nitazoxanide by targeting the HBx-DDB1 interaction. Cell Mol Gastroenterol Hepatol 2019;7:297–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghosh AK, Xi K, Johnson ME, et al. Progress in anti-SARS coronavirus chemistry, biology and chemotherapy. Annu Rep Med Chem 2007;41:183–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lejal N, Tarus B, Bouguyon E, et al. Structure-based discovery of the novel antiviral properties of naproxen against the nucleoprotein of influenza A virus. Antimicrob Agents Chemother 2013;57:2231–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao H-W, Lin P-H, Shen F-H, et al. Tranylcypromine reduces herpes simplex virus 1 infection in mice. Antimicrob Agents Chemother 2014;58:2807–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johansen LM, et al. FDA-approved selective estrogen receptor modulators inhibit Ebola virus infection. Sci Transl Med 2013;5:190ra179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.la Porte CJ. Saquinavir, the pioneer antiretroviral protease inhibitor. Expert Opin Drug Metab Toxicol 2009;5:1313–22. [DOI] [PubMed] [Google Scholar]
- 40.Wu C-Y, Jan J-T, Ma S-H, et al. Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc Natl Acad Sci USA 2004;101:10012–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Donia M, McCubrey JA, Bendtzen K, Nicoletti F.. Potential use of rapamycin in HIV infection. Br J Clin Pharmacol 2010;70:784–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prince GA. An update on respiratory syncytial virus antiviral agents. Expert Opin Investig Drugs 2001;10:297–308. [DOI] [PubMed] [Google Scholar]
- 43.Murakami Y, Fukasawa M, Kaneko Y, et al. Retinoids and rexinoids inhibit hepatitis C virus independently of retinoid receptor signaling. Microbes Infect 2014;16:114–22. [DOI] [PubMed] [Google Scholar]
- 44.Yousefifard M, et al. Non-steroidal anti-inflammatory drugs in management of COVID-19; a systematic review on current evidence. Int J Clin Pract 2020;e13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hall DC, Jr., Ji HF.. A search for medications to treat COVID-19 via in silico molecular docking models of the SARS-CoV-2 spike glycoprotein and 3CL protease. Travel Med Infect Dis 2020;35:101646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Omarjee L, Janin A, Perrot F, et al. Targeting T-cell senescence and cytokine storm with rapamycin to prevent severe progression in COVID-19. Clin Immunol 2020;216:108464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan S, Chan JFW, Chik KKH, et al. Discovery of the FDA-approved drugs bexarotene, cetilistat, diiodohydroxyquinoline, and abiraterone as potential COVID-19 treatments with a robust two-tier screening system. Pharmacol Res 2020;159:104960. [DOI] [PMC free article] [PubMed] [Google Scholar]