Abstract
Recently, cell-mediated immune response in malignant neoplasms has become the focus in immunotherapy against cancer. However, in leukemia, most studies on the cytotoxic potential of T cells have concentrated only on T cells that recognize peptide antigens (Ag) presented by polymorphic molecules of the major histocompatibility complex (MHC). This ignores the great potential of unconventional T cell populations, which include gamma-delta T cells (γδ), natural killer T cells (NKT), and mucosal-associated invariant T cells (MAIT). Collectively, these T cell populations can recognize lipid antigens, specially modified peptides and small molecule metabolites, in addition to having several other advantages, which can provide more effective applications in cancer immunotherapy. In recent years, these cell populations have been associated with a repertoire of anti- or protumor responses and play important roles in the dynamics of solid tumors and hematological malignancies, thus, encouraging the development of new investigations in the area. This review focuses on the current knowledge regarding the role of unconventional T cell populations in the antitumor immune response in leukemia and discusses why further studies on the immunotherapeutic potential of these cells are needed.
1. Introduction
Leukemia comprises a heterogeneous group of hematological neoplasms, which can be classified into lymphoblastic or myeloid leukemias and divided into acute and chronic types, depending on the affected cell type, maturation stage, and blast count, respectively [1]. While acute leukemias are characterized by a deep block in hematopoietic differentiation and result in an overproduction of immature blasts, chronic leukemias are characterized by the excessive production of partially mature differentiated cells, for example, lymphocytes in chronic lymphocytic leukemia (CLL) and granulocytes in chronic myeloid leukemia (CML) [2, 3]. The hallmark of these neoplasms is the increase in leukemic cells (LCs) in the bone marrow (BM) and their release in the peripheral blood (PB) and in extramedullary sites [1].
The immunological mechanisms in patients with leukemia are not very well known. However, with the increasing advances in the field of immunotherapy, there has been great progress in research regarding the tumor microenvironment in leukemia. Studies have shown that LCs secrete factors that disrupt healthy BM niches, reprogramming and transforming them into “leukemic niches,” as well as inducing a disruption in balanced cytokine production, and favoring leukemic persistence and metastatic potential [4, 5]. However, despite the protumor microenvironment created by LCs, studies have reported that a specific immune response can be triggered and, therefore, contribute to the defense against the tumor, although not sufficient enough to control the neoplasia [6].
Several studies have described the immunotherapeutic potential of CD8 and CD4 T cells that recognize peptide antigens (Ag) presented by polymorphic major histocompatibility complex (MHC) class I and MHC class II molecules, respectively [7, 8]. Leaving aside, the populations of T cells are considered “unconventional,” which are also implicated in tumor immunity, although their role in them is not well understood. Collectively, these T cell populations differ from their conventional counterparts mainly in the way they recognize and respond to foreign molecules. Unlike MHC-reactive T cells, unconventional T cells generally show simplified patterns of the T cell antigen receptor (TCR) expression and usually target monomorphic Ag-presenting molecules and other ligands, where after their activation, they promote rapid and strong effector responses [9, 10]. These T cell populations include γδ T cells, NKT cells, and MAIT cells.
In this review, we describe the main characteristics of these T cell populations and explore their activities during the neoplastic process, as well as their relationship with the establishment of an antitumor immune response or tumor-favorable response, as described briefly in Table 1. A better understanding of the participation of these underexplored cells in tumor dynamics may provide a basis for the development of potential immunotherapeutic strategies in the field of leukemias.
Table 1.
The table indicates some functions triggered by unconventional T cells in the immune response against the tumor.
| Subsets | Role played in the immune response against cancer cells | Reference |
|---|---|---|
| γδ T cells | Mediate tumor regression by recognizing MIC-A/MIC-B and ULBPs through TCR and NKG2D | [34, 35, 51, 52, 90] |
| Antitumor potential increased by expression of NCRs | [36] | |
| Positive regulation of pAgs in LCs mediates the immune response through γδ TCR | [40, 41, 72] | |
| BTN3A/BTN2A1 increases the antitumor functions of γδ T cells in blood | [72–74] | |
| Mediate tumor regression by recognizing PVR and nectin-2 through TCR and DNAM-1 | [41] | |
| BTN3A-expressing LCs are recognized and destroyed by γδ T cells in blood through TCR | [69] | |
| Induce the maturation of DCs, which consequently enhance their activity against LCs | [29, 30] | |
| NKT cells | Mediates tumor regression by recognizing CD1d+ LCs through TCR | [133] |
| Induces direct destruction of tumor cells through granulysin | [124, 125] | |
| Induces direct tumor lysis through FasL | [111] | |
| They act in the immunovigilance during the initial phase of the neoplastic process | [131–134] | |
| MAIT cells | Induces direct cytotoxicity mediated by granzyme B and perforin in cancer cells that express MR1 | [162, 175] |
2. Subsets of Unconventional T Cells in Antileukemic Response
2.1. Gamma-Delta (γδ) T Cells
γδ T cells are developed in the thymus during lymphopoiesis and, by the expression of the TCR (T-cell receptor), composed of gamma (γ) and delta (δ) chains [11]. These cells are capable of providing a potent and lasting immune response through innate and adaptive mechanisms and stand out for their recognition and destruction of several tumors, regardless of major histocompatibility complex (MHC) expression [12]. In addition, these cells constitute up to 10% of circulating CD3+ cells and are classified according to the genetic rearrangements of the δ chain of the TCR into three main subsets: Vδ1, Vδ2, and Vδ3 (in humans) [13]. These lymphocytes are present in blood circulation, tissues, and mucosal membranes, which are strategic places to exercise their high cytotoxic power against infections and tumors [14].
γδ T cell lineage is evolutionarily conserved and has several pleiotropic functions. While these cells are naturally specialized in the secretion of proinflammatory mediators (Figure 1) [15], these lymphocytes can also adopt Th2-, Th9-, and Th17-like response phenotypes [16–18]. In the tumor microenvironment (TME), these lymphocytes express a diverse repertoire of recognition receptors that have received favorable prognostic value in several malignancies, including leukemia [19]. Based on several reports, it has been confirmed that γδ T cells are capable of triggering an immune response by direct and indirect mechanisms, for example, through the cytolytic synapse with the target cell, and the recruitment and stimulation of other immune cells necessary for the establishment of an antitumor response [12, 20].
Figure 1.
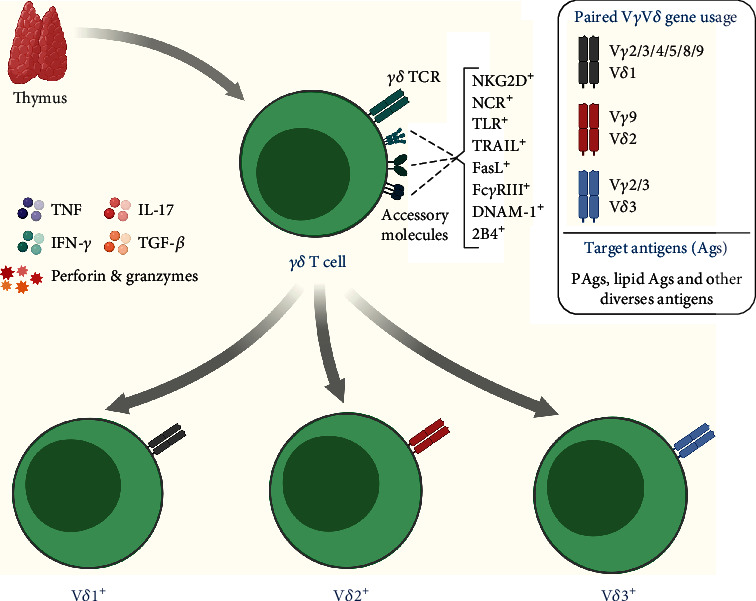
Overview of subpopulations, important receptors, and cytokines produced by γδ T cells. γδ T cells express some receptors that are essential for the tumor recognition and destruction, which gives a certain advantage when compared to other conventional T lymphocyte populations, either due to MHC independence or due to the high expression of the receptors mentioned in the image. The antileukemic recognition repertoire includes several molecules, such as NKG2D, NCRs, FasL, CD16, DNAM-1, and the TCR γδ itself.
These lymphocytes can induce neoplastic regression through cell-cell interaction or by secreting several soluble molecules (such as IFN-γ and TNF) that inhibit tumor expansion. These effector molecules induce an increase in the antitumor activity of other cytotoxic cells or positively regulate the expression of MHC-I by cancer cells [21]. In addition, γδ T cells stimulate somatic hypermutation and isotype switching in B cells [22–24] and possibly induce antibody-mediated immunity. Their effector functions also include the activation of macrophages and the recruitment and activation of CD8+ cytotoxic T cells and NK cells [25–28]. The immune role of these lymphocytes also includes stimulating the maturation of dendritic cells (DCs), and, in turn, DCs are able to potentiate their cytotoxic activity [29, 30]. Notably, cancer cells tend to express several stress-induced molecules or metabolic antigens that are recognized through γδ TCRs and accessory receptors, thereby, mediating a potent response against the tumor [31–34].
Without restrictions on MHC expression, γδ T cells recognize several antigens that are expressed in LCs, and generally include metabolic molecules and stress-induced molecules [32, 35–38]. Ligands, such as MIC-A/MIC-B and ULBPs, can be identified through the NKG2D receptor, which is expressed mainly in γδ T cells [34, 39]. Furthermore, some metabolites of the mevalonate pathway, known as phosphoantigens (pAgs), can be recognized directly through the TCRs and are highly regulated in LCs [40, 41]. In addition, other molecules assist in tumor recognition and possibly support the antileukemic response of γδ T cells, such as TLRs (toll-like receptors), DNAM-1 (DNAX Accessory Molecule-1), FasL (Fas ligand), FcγRIII, TRAIL (TNF-related apoptosis-inducing ligand), NCRs (Natural Cytotoxicity Receptors) such as NKp30, NKp44, and NKp46, and the 2B4 receptor [12, 36, 41–44].
Vδ1+ cells, which express TCR chains Vγ1 to Vγ11, respond preferentially in skin tissues, intestinal epithelium, lung, spleen, and liver, where they play crucial roles in maintaining epithelial tissue [14, 45]. It is known that these lymphocytes patrol the several tissues in search of stressed cells, derived from infections and tumorigenesis, thus, maintaining tissue homeostasis [46]. For this, the secretion of Th1 and Th17 cytokines is essential for immune surveillance [12, 47–49].
These cells, although uncommon in peripheral blood (~ 10% of blood γδ T cells), have a high diversity of tumor recognition and have demonstrated great potential against LCs [36, 50, 51]. Correia et al. demonstrated that Vδ1+ cells that expressed NCRs managed to destroy lymphoid and myeloid cancer cells through NKp30 and NKp44, which seemed to recognize antigens that are distinct from their classic ligands, such as the molecule B7-H6, which binds to NKp30 [36]. In the same study, the stable expression of these NCRs was associated with elevated levels of granzyme B and seemed to synergize greater cytotoxic activity against LCs [36]. Taking into account that in some leukemias, a high expression of members of the ULBP family is observed, as NKG2D ligands [52], Lança et al. demonstrated that the expression of ULBP1 in LCs is important for recognition by Vδ1 cells [51]. ULBP3 was confirmed by Poggi et al. who presented similar findings [52]. Therefore, the data suggest possible immunological participation of these cells and indicate a significant contribution to the antileukemic immune response.
These cells can also recognize lipid or metabolic antigens that are presented through MHC class I-like molecules, such as CD1 and MR1 [46, 53]. These and other characteristics of Vδ1 cells make them potential candidates for new immunotherapeutic approaches in several human tumors, and these have recently been explored in several experimental trials, including some against leukemia [54–56]. Although their roles are still poorly known, we know that Vδ1 cells demonstrate important functions in antitumor activity, and these deserve to be highlighted since they present characteristics that are different to Vγ9Vδ2 cells, either due to their high expression of NCRs or the nonsusceptibility to activation-induced cell death (AICD) [57–59]. The roles of this subset of γδ T cells in the environment of bone marrow and peripheral blood in vivo, in the context of leukemia, still need further investigation regarding the ligands and receptors of recognition that are engaged during the immune response.
The Vδ2+ subset, which pairs exclusively with the Vγ9 TCR chain, responds mainly in the blood, where it recognizes pAgs derived from bacteria and cancer cells [60]. Once activated, these lymphocytes secrete effector molecules such as IFN-γ, TNF, perforins, and granzymes and exert important cytotoxic activities in peripheral blood against pathogens and tumors [12, 61]. These cells make up as much as 95% of blood γδ T cells [50, 62–64] and generally respond to a wide variety of pAgs, such as IPP (isopentenyl pyrophosphate) and HMB-PP (4-hydroxy-3-methyl-but-2-enylpyrophosphate), which are intermediates of the mevalonate pathway in eukaryotes and prokaryotes [65–67]. The recognition of these pAgs occurs in the context of the butyrophilin (BTN) family of molecules, such as BTN3A1 and BTN2A1 [68–71], which can be detected in LCs, and mediates a potent immune response that can be used therapeutically [72]. These molecules can be recognized directly through the γδ TCR and are capable of triggering a Th1-like response against the target cells [73, 74].
The mechanism for recognizing pAgs is not yet clear, although several studies have recently expanded the information about how γδ T cells identify these molecules. Recent reports have pointed out that pAgs recognition is mediated by BTN-like molecules, which are expressed in cancer cells [72, 75] and are able to modulate the responses of conventional αβ T cells [76–79], and most notably, of γδ T cells [68–71, 80, 81]. The dependent detection of pAgs by Vγ9Vδ2 cells involves the entire structure of the TCR, which interacts with BTN molecules through the Vγ9 and Vδ2 TCR domains. Among the various molecules that make up the BTN family, the proteins BTN3A1 and BTN2A1 synergize the presentation of pAgs to γδ T cells, binding directly to the TCR Vγ9Vδ2 [68, 70, 82, 83].
Previously, it was thought that the unit expression of BTN3A1 performed the activation of these lymphocytes alone [84], but it is now clear that the BTN2A1 protein acts as a critical factor in the activation of Vδ2 cells [70]. For this to occur, it is necessary that pAgs bind to the intracellular domain (B30.2) of these proteins [85, 86]. After binding of pAgs, the intra- and extracellular domains of BTN3A1 and BTN2A1 undergo a conformational change [85–87] that allows the contact of the TCR Vγ9 chain with the BTN2A1 molecule, sending activation signals to γδ T cells [68]. In addition, the involvement of other molecules during the pAgs detection mechanism cannot be ignored, as recent reports suggest the molecular collaboration of CDR3, periplakin, and GTPase RhoB in this process [70, 88, 89].
In addition to direct and TCR-dependent recognition, other accessory molecules possibly support the antileukemic activity of Vδ2 cells against LCs, such as the DNAM-1 receptor that recognizes the PVR (poliovirus receptor) and Nectin-2 ligands, both expressed in LCs [41], and ULBP4 that can be recognized through NKG2D [90]. γδ T cells also express the 2B4 receptor (which recognizes the CD48 ligand), an accessory molecule that strengthens target effector interactions and is possibly related to increased cytotoxic activity against cancer cells [44]. One disadvantage of these Vδ2+ cells is their strong propensity for AICD upon prolonged exposure to antigens and the polarization of these lymphocytes towards a tumor-promoting phenotype, which limits the persistence and efficiency of the immune response [57, 59].
Little data is available regarding the receptors and ligands involved in the events of innate and adaptive immunity mediated by Vδ3+ cells, which express the TCR chains Vγ2, Vγ3, or Vγ8 and are a specific subset that responds mainly in the liver [91, 92]. The frequency of this cell population is low in peripheral blood (~0.2% of total circulating T cells) though it is high in the liver and intestinal region [91, 92]. Studies have reported that Vδ3+ cells are related to antiviral immunity, responding efficiently against cytomegalovirus [93], Epstein-Barr virus [94], hepatitis [95], and HIV infection [64]. So far, their roles in antitumor immunity are not clear, although some reports have identified an expansion of these lymphocytes in the peripheral blood of patients with leukemia [96]. Also, Vδ3 cells secrete IFN-γ, express NKG2D, FcγRIII, and CD161, and appear to respond in a restricted way to CD1d, functionally resembling iNKT cells, which recognize and destroy CD1d+ target cells [24, 91]. Subsequently, it was identified that these cells also respond to ANX-2 (annexin-2) [97], but their role in leukemia is still unexplored, although it has been identified that these cells expand during tumor progression [96].
γδ T cells have an enormous potential to regulate local immunity and remodel the tumor niche [45, 98, 99]. The evidence discussed so far suggests that these cells realize various antitumor activities through the direct identification of LCs through the γδ TCR or accessory receptors and by secreting soluble effector molecules against the tumor. Despite this, emerging knowledge about the molecular and cellular interaction between these lymphocytes and cancer cells suggests that, like conventional αβ T cells, γδ T cells may possibly not be exempt from the immunosuppression established by the TME [100–103]. However, the lack of knowledge about these cells in the leukemic microenvironment makes it difficult to elaborate larger and more comprehensive discussions about the possible crosstalk between this population of lymphocytes and the LCs in the bone marrow compartment and in extramedullary sites.
In addition, other subsets of γδ T cells that express other variable TCR chains (TCR domains Vδ5, Vδ6, and Vδ8) have been identified in other hematological malignancies, where, for example, these cells seemed to expand and respond to cancer cells in the blood of patients with lymphoma [104]. However, it is still unclear whether these cells respond against LCs and not have information available on the possible roles played by these γδ T cell subsets in leukemia antitumor immunity. In view of its enormous therapeutic applicability, further research is needed on the interaction of γδ T cells and their subsets in the leukemic microenvironment and on how this can impact the prognosis of patients.
2.2. Natural Killer T (NKT) Cells
Natural killer T cells (NKT) correspond to a population of innate-like T cells characterized by the expression of a TCR composed of alpha (α) and beta (β) chains similar to those of conventional T cells, in addition to specific surface markers of natural killer cells (NK), such as CD16+, CD56+, CD69+, CD161+, NKG2D+, and Ly49A+ [105, 106]. Another striking feature of these cells is their restriction to the CD1d molecule, which is an MHC class I-like molecule that presents lipid and glycolipid antigens [107–109].
NKT cells constitute approximately 0.001-1% of circulating lymphocytes (in humans) and are also present in the thymus, liver, intestine, and spleen [110, 111]. These cells are divided into two main subsets, the invariant natural killer T (iNKT) and natural killer T type II (NKTII) cells [112]. Both express the following transcription factors: T-bet [113, 114], PLZF [115], RORγt, GATA-3 [114], and NF-kB [116], which together grant high cellular plasticity and allow polarization for phenotypes of profile Th1, Th2, and Th17 [116–118]. In addition, iNKTs express the LEF-1 factor [119, 120], which is correlated with the regulation of the expression of the gene that encodes the CD1d molecule in antigen presenting cells (APCs). The LEF-1 factor also plays a crucial role as a regulator of the Wnt pathway, and it is possible that it influences the growth, development, differentiation, and functions of NKT cells [121].
In regards to their antitumor activity, NKT cells can act directly through cell-cell interaction, through the Fas receptor and its ligand (FasL) that trigger the activation of caspase enzymes and cause apoptosis of the target cells, in addition to the interaction of other receptors such as NKG2D, TRAIL, natural cytotoxicity receptors (NCRs), and their ligands (Figure 2) [111, 113, 122]. There is also the substantial release of perforins and granzymes A and B, in addition to the production of Granulysin that act in a similar way to perforins, forming pores in the plasma membrane and altering their permeability, which results in cell lysis [123, 124].
Figure 2.
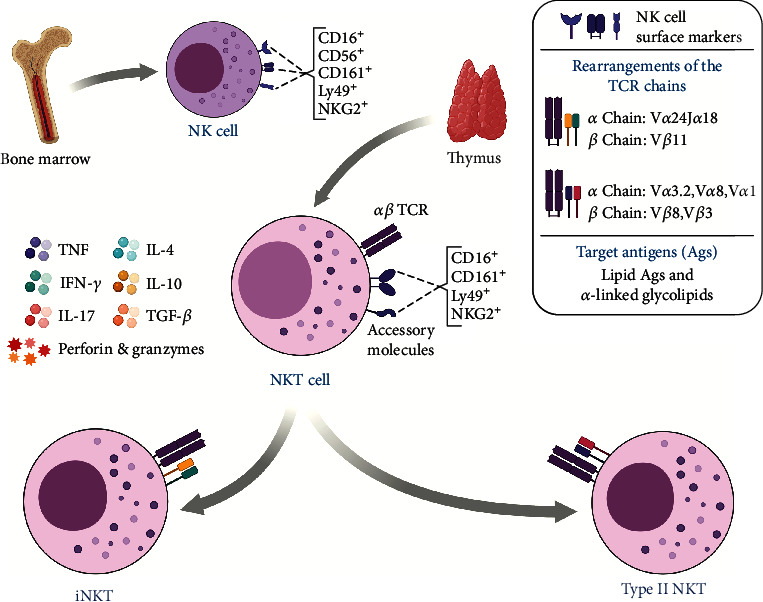
Overview of subpopulations, important receptors, and cytokines produced by NKT cells. Unlike conventional NK cells that mature in the bone marrow, NKT cells develop in the thymus and acquire an invariant or variant TCR, through which their subgroups are stratified. Recent studies show that these lymphocytes participate in the antitumor immune response by recognizing CD1d+ tumors and secreting Th1, Th2, and Th17 profile cytokines.
NKT cells also can act through indirect mechanisms by releasing a range of mediators, especially Th1 or Th2 profile cytokines, which can vary depending on the NKT subtype, a fact that will be discussed later [125]. The release of the aforementioned mediators can results in the immunoregulation of other cells of the immune system, for example, activating or inducing the maturation of DCs through interaction with CD1d or CD40/CD40L and IFN, respectively. After activation of DCs, they will be regulated positively, expressing costimulatory molecules such as CD86 and CD80, in addition to release cytokines such as IL-12, a pleiotropic cytokine that plays an essential role in Th1-type immune response against cancer. iNKT cells also activate CD8 cytotoxic T cells through the IFN, as well as conventional NK cells and macrophages that can act against cancer cells [125–128]. In addition, NKT cells can also stimulate B lymphocytes and induce increased secretion of IgG class antibodies, which can result in antibody-dependent cell cytotoxicity (ADCC) in cancer cells that will subsequently undergo cytotoxic cell-mediated cell lysis [111, 129].
Due to their secretory repertoire and the ability to activate and target other cells of the immune system, NKT cells contribute strongly to tumor immunovigilance. It is important to highlight that one of the main forms of recognition of cancer cells by NKT cells occurs through the interaction of the TCR with the antigen presented by the CD1d molecule, expressed by B cells, macrophages, DCs, and several types of cancer cells, in solid and hematological neoplasms [116, 117]. These cells have been shown to contribute to tumor surveillance and suppression, controlling the initial stage of the neoplastic process [130–133].
iNKT cells, also known as natural killer type I cells, have characteristics that differ from other subsets, such as reactivity to ∝-galactosylceramide (α-GalCer) and the more pronounced expression of the Th1 response profile [134]. Another characteristic of iNKT cells is related to its invariant TCR, represented in humans by a TCR-α chain (Vα24Jα18) and a TCR-β chain (Vβ11). iNKT cells recognize lipid antigens and glycolipids such as α-GalCer analogs, phospholipids, diacylglycerols, gangliosides (GD3), and glycosphingolipids [107, 112].
Studies show that iNKT cells are strongly reactive to α-GalCer, a synthetic glycolipid derived from the marine sponge Agelas mauritianus, which is capable of inducing an important immunomodulatory effect in iNKT cells and which stimulates the antitumor response mediated by cytotoxic cells (NK cells and TCD8+ lymphocytes) that are in a state of exhaustion or anergy resulting from the TME. Stimulation mediated by iNKT cells has been shown to promote the invigoration of cytotoxic cells and reverse the dysfunction presented by these cells [107, 135].
The α-GalCer antigen presented through the CD1d molecule is recognized by the TCR expressed in iNKT cells and induces the production of cytokines, such as IL-2, IL-12, and IL-21, which act on conventional NK cells and exhausted or anergic CD8 cytotoxic T cells, thus, reversing the dysfunction and the hyporesponsive character [125, 136]. In addition, α-GalCer-stimulated iNKT cells have been shown to incite the activation of APCs via CD40-CD40L signaling and also induce the production of IL-12 [125, 126, 129, 137].
A study by Fais et al., in patients with acute leukemia, evaluated the expression of the CD1d molecule in LCs, as well as the activity of iNKT cells in their recognition. Where, through incubating LCs with 100 ng/ml of α-GalCer for 4 hours, it was observed that the CD1d+ LCs associated with the α-GalCer antigen underwent apoptosis induced by iNKT cells, in addition to stimulating the production of cytokines INF-γ, TNF-α, and IL-5 [138]. Similar results were observed by Bojarska-Junak et al. in a study with BP samples from patients with chronic leukemia, in which iNKT cells stimulated in vitro for 24 hours with 100 ng/ml of α-GalCer showed greater intracellular expression of the INF-γ and IL-4 cytokines when compared with healthy volunteers. The study also pointed out that, among the two cytokines analyzed, the expression of IL-4 was greater than that of INF-γ, indicating that the iNKT cell may acquire a polarization for the Th2 response profile [139].
Another study showed that the frequency of iNKT cells was lower in patients with chronic leukemia compared to the healthy control group and that therapy for 14 days with IFN-γ and 1 μM IM (imatinib mesylate) in combination with 100 ng/ml of α-GalCer in these patients resulted in a significant increase in iNKT cells [140]. Similarly, Weinkove et al. demonstrated that LCs purified and stimulated in vitro with 200 ng/ml of α-GalCer for 5 days incited the proliferation of autologous and allogeneic iNKT cells, but not in a significant amount it was also demonstrated that there was a stimulation of IFN-γ production by iNKT cells in leukemic patients. However, it was observed that the prolonged culture of these cells resulted in the polarization of iNKT cells to a Th2 profile and resulted in high levels of cytokines associated with tumor tolerance [141]. Additional investigations should be carried out to identify how stimulation with α-GalCer can provide better results, associating the increased frequency of iNKT cells with polarization for a response profile against LCs.
Finally, in a longitudinal analysis carried out over 18 months with 22 patients that had been diagnosed with leukemia or myelodysplasia and who underwent HLA haploidentical stem cell transplantation for iNKT cell reconstitution, the existence of a correlation between the frequency of iNKT cells and disease remission was reported. The observed data showed that 14 patients whose iNKT cells were completely reconstituted showed remission, and 8 patients whose iNKT cells were not reconstituted showed recurrence, thus, associating the frequency of these cells with a better prognosis [142, 143]. Furthermore, the analysis of the frequency of iNKT cells in the circulation and bone marrow compartment of newly diagnosed patients with acute leukemia demonstrated that a low frequency of these cells is associated with a worse prognosis [144].
Unlike iNKT cells, NKT II cells have an αβ TCR variant and form diversified rearrangements. They are not reactive to α-GalCer [112, 125, 145]; however, they are restricted to CD1d [146] and recognize antigens such as β-glucosylceramide (β-GlcCer) and sulfatide, the latter being found in the plasma membrane of myelin, in the central nervous system, in the liver, in the pancreas, and in the kidneys [147–149]. Another characteristic of the NKT II cell is related to its response profile, which is more polarized for the Th2 profile [134, 150].
In contrast to studies on iNKT cells, the number of studies on the role of NKT II cells in leukemia is limited. However, some studies report that NKT II can act in a negative way by contributing to the suppression of surveillance and antitumor activity and, in some situations, contributing to neoplastic progression [111, 112, 151]. Such immunosuppression is mediated through the secretion of IL-13, which activates myeloid-derived suppressor cells (MDSCs), and these, consequently, suppress the activity of tumor-infiltrating cytotoxic cells [150]. It is also believed that this negative regulation of NKT II in the tumor occurs due to a cross-regulation between iNKT and NKT II, where the effect of Th2 cytokines produced by NKT II cells overlap the effect by the Th1 cytokines produced by iNKT, resulting in an immunosuppressed microenvironment. In addition, it has been observed that tumors grow faster when the frequency of NKT II cells is higher than that of iNKT cells, although this suppression mechanism still needs further investigation [111, 134].
It is important to note that, in addition to NKT cells, there are other populations of nonconventional T cells restricted to molecules of the CD1 family, more specifically group 1, composed of CD1a, CD1b, and CD1c, which have lipid antigens or glycolipids, of microbial origin or from the organism itself [152]. Similar to NKT cells, these CD1-restricted cell populations are considered an attractive target for studies in the field of cancer immunotherapy, especially in the context of hematological neoplasms, due to the diversified distribution of CD1 isotypes, where they have been shown to be expressed (CD1a, CD1b, and CD1c) in 75% of acute leukemia blasts [153].
2.3. Mucosal-Associated Invariant T (MAIT) Cells
MAIT cells correspond to a population of innate-like T cells and are characterized by the expression of a restricted TCR-α with a unique gene rearrangement pattern, namely, TRAV1-2-TRAJ33 / 12/20 (Vα7.2-Jα33 / 12/20 in humans), which pair with a limited repertoire of the TCR-β chain, predominantly from the TRBV6 and TRBV20 gene families [154–156], and form a semi-invariant TCR restricted to nonpeptide antigens presented by MHC-related protein 1 (MR1). MR1 is a monomorphic molecule that is highly conserved throughout the evolution of mammals [157, 158] and is capable of presenting metabolites derived from vitamin B2 that is synthesized by a variety of microorganisms for MAIT cells [159, 160].
Like most T cells, MAIT cells develop in the thymus [161] and are positively selected by the cortical thymocytes CD4+ CD8+ MR1+ [161, 162]. After the selection process, they undergo extrathymic maturation and integrate with different tissues [163]. In humans, MAIT cells represent up to 10% of peripheral blood T cells and are found in abundance in mucous tissues, mesenteric lymph nodes, and liver, where they can represent up to 45% of all T cells [164, 165].
Human MAIT cells can be immunophenotyped as CD3+ Vα7.2+ CD161HI [166] and can be categorized based on the expression of CD4 and CD8 coreceptors in five subsets: CD4+ CD8−, CD4+ CD8+, CD4− CD8−, CD4− CD8αα+ e CD4− CD8αβ+ (Figure 3); the last two (CD4− CD8αα+ and CD4− CD8αβ+) being the most abundant and collectively correspond to approximately 80% of MAIT cells [164]. Of note, the development of MR1 tetramers loaded with 5-OP-RU [5-(2-oxopropylideneamino)-6-d-ribitylaminouracil] marked a breakthrough in MAIT cell research and have allowed for the reliable identification of distinct phenotypical and functional MAIT cell subsets [167].
Figure 3.
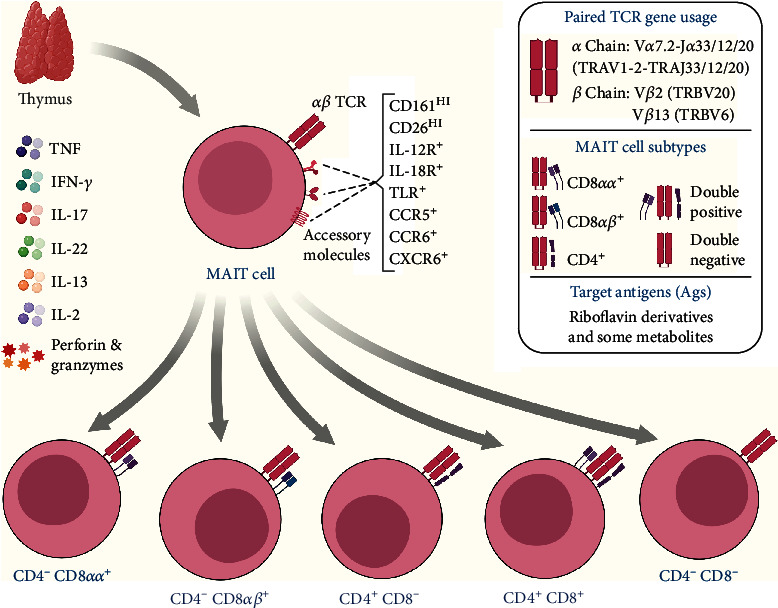
Overview of subpopulations, important receptors, and cytokines produced by MAIT cells. MAIT cells develop in the thymus, where they acquire a semi-invariant TCR, restricted to MR1. In humans, they can be categorized into five subsets based on the expression of the CD4 and CD8 coreceptors, with CD4− CD8αα+ or CD4− CD8αβ+ being the most abundant, collectively corresponding to approximately 80% of MAIT cells. They produce a repertoire of Th1 and Th17 cytokines (IFN-γ, TNF, IL-2, IL-17A, and IL-22), in addition to perforins and granzymes B, and express various cytokine, chemokine, and homing molecules.
In addition to the surface molecules mentioned above, MAIT cells also express CD25, CD26, CD44, and CD69, as well as IL-7R, IL-12R, IL-15R, and IL-18R cytokine receptors and PLZF, T transcription factors T-bet and RORγt, providing high plasticity and the ability to secrete mediators of Th1 profile and/or Th17 profile [168]. It is important to note that most MAIT cells have a memory-like phenotype effector [169] and can be quickly activated by mechanisms that do not depend on TCR stimulation [170]. This is due to the high expression of various cytokine and chemokine receptors, a property they share with other innate-like T cells (γδ and iNKT cells) [171].
After activation, these cells are able to release substantial quantities of perforin and granzymes, in addition to producing the proinflammatory cytokines TNF, IFNγ, and IL-17A, as well as CCL3 and CCL4, which are chemokines that are crucial for immune responses to infection and inflammation [172, 173]. The release of these mediators results in the destruction of infected cells and the activation of other immune cells, such as DCs, which consequently leads to the mobilization of conventional T cells and reflects in a cascade of immunological events [174, 175].
Due to some characteristics of MAIT cells, such as their high frequency in humans and their ability to rapidly secrete a repertoire of mediators that induces activation and regulation of other cells of the immune system, in addition to the ability to recognize antigens restricted to MR1, research on MAIT cells, as well as other populations of T cells restricted to MR1, has aroused great interest in the field of oncology. Currently, the functional role triggered by these innate-like T cells in cancer is unclear and has been the subject of several studies.
Investigations performed on patients with mucosal neoplasms have reported a reduction in the frequency of circulating MAIT cells and show a significant accumulation of these cells in the tumor tissue [176–178]. However, it is important to note that tumor-infiltrating MAIT cells were less able to produce IFN-γ in response to factors secreted by the TME [178]. In addition, a recent in vivo study by Yan et al. showed that MAIT cells exhibited a tumor-promoting function and promoted cancer metastasis through the suppression of cytotoxic cells (which was partly IL-17 dependent) after interaction with MR1 molecules expressed on cancer cells [179].
In contrast, in the study by Won et al., tumor-infiltrating MAIT cells were not compromised in the production of cytokines IFN-y, TNF-α, and IL-17. In addition, in the in vitro assay, it was observed that MAIT cells, isolated from PB from healthy individuals, not only had lymphokine-activated killer activity but also exhibited direct cytotoxicity in K562 cell lines through the degranulation of granzyme B and perforin [177].
To date, the involvement of MAIT cells in leukemia remains largely unexplored, and, in the context of other hematological neoplasms, the functional role of these cells is also limited. An in vitro study by Gherardin et al. demonstrated that multiple myeloma (MM) cell lines express the MR1 protein and are capable of presenting vitamin B metabolites to MAIT cells isolated from healthy donors, which, in response, induce the lysis of these myeloma cells with efficiency and kinetics similar to NK cells [164].
Another study observed the functional capacity of these lymphocytes by analyzing the cell phenotype in samples of PB and BM [180]. Favreau et al. reported that patients recently diagnosed with MM had a significant decrease in the frequency of MAIT cells when compared to healthy controls. They also highlighted a reduction in the MAIT CD8+ and double-negative (CD8− CD4−) phenotype, in addition to the functional impairment of the Th1 response profile, with fewer IFN-γ and TNFα producing MAIT cells [180]. Favreau et al. also demonstrated that circulating MAIT cells exhibited high levels of PD-1 and that their respective in vitro blockade resulted in the restoration of MAIT cell function and activation [180]. As a whole, these results highlight the important antitumor activity of MAIT cells and identify them as a potential immunotherapeutic target in MM.
Likewise, MR1 also represents an attractive target in immunotherapy against cancer, due to characteristics such as its monomorphic nature and functional expression found in several types of cancer cells [181]. In this regard, a recent study by Crowther et al. demonstrated that a human T cell clone potentially recognizes a specific cancer or associated metabolite, restricted to MR1, and mediates the lysis of different types of cancer cells, including LCs lineages, as such, it mediated in vivo leukemia regression and conferred longer survival in mice [182]. Moreover, other atypical MR1-restricted T cells, which respond to autoantigens, have been described [181, 183, 184]. This new group of T cells has been named MR1T and has been shown to recognize and eliminate a wide range of cancer cells that express MR1 [181]. However, more research is needed to elucidate the precise nature and function of these cells, as well as their activity in the context of TME.
3. Concluding Remarks and Perspectives
Unconventional T cells can promote tumor rejection and offer advantages that indicate them as being potential targets for T-cell-based immunotherapy. Although effective, it is important to note that unconventional T cells are not exempt from the influence of checkpoint receptors, since these cells positively regulate the inhibitory PD1 receptor on their cell surface after activation [185–187]. However, checkpoint blockade therapy using drugs based on anti-PD1 and anti-CTLA-4 is proving to be a powerful approach for preventing effector cells from entering into a state of anergy caused by cancer cells, thereby, providing a persistent immune response [188].
In the context of hematologic neoplasms, among the promising alternatives to conventional chemotherapy are the upcoming immunotherapies, in particular, the transfer of chimeric antigen receptor (CAR) T cells [189]. These autologous T cells, which are designed to express a CAR receptor against the CD19 antigen, are at the forefront of contemporary oncohematological therapies and lead to high rates of remission in B cell malignancies [190, 191]. It is important to highlight the obstacles, such as the complex and expensive individualized manufacturing process and the loss or modulation of the target CD19 antigen, that lead to resistance and relapse after therapy with CAR T cells.
In this scenario, unconventional T cells present themselves as possible solutions to overcome these obstacles, since they have a series of specific biological characteristics that can significantly expand and diversify the repertoire of CAR-based therapies, although they also have their limitations [192]. Few preclinical studies have investigated unconventional T cells as alternative platforms for CAR engineering, and, given this limitation, it is not surprising that there is a very low number of clinical trials that evaluate the viability of CAR γδ T therapies (B cell lymphoma, NCT02656147; and acute myeloid leukemia, NCT03885076) and CAR NKT (refractory B cell neoplasm, NCT03774654) in hematologic neoplasms that are in progress.
The fact is that the role of unconventional T cells during the neoplastic process is usually related to better immune surveillance or antitumor response in patients with leukemia, and that, to date, unconventional T cells are still largely underexplored. Factors, such as the absence of barriers related to histocompatibility, since the molecules that present Ag for unconventional T cells are monomorphic; activation by TCR dependent and independent mechanisms; ability to gather quick and powerful responses; in addition to the high frequency in specific tissues in humans, all demonstrate the need for further studies on the immunotherapeutic potential of these cells and, mainly, the translation of these studies into clinical trials.
Acknowledgments
This study was funded by Fundação de Amparo à Pesquisa do Estado do Amazonas (FAPEAM) (Pró-Estado Program-#002/2008, PAMEQ Program-#004/2019, and PAPAC Program-#005/2019), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). NDA, FMG, FSA, LAX, MSB, TLPR, and ISC received fellowships from CAPES and FAPEAM (PhD, Masters, and SI student). AM is a level 2 research fellow from CNPq.
Abbreviations
- AICD:
Activation-induced cell death
- ALL:
Acute lymphoblastic leukemia
- AML:
Acute myeloid leukemia
- ANX-2:
Annexin-2
- APCs:
Antigen-presenting cells
- BM:
Bone marrow
- BTN:
Butyrophilin
- BTN2A1:
Butyrophilin subfamily 2 member A1
- BTN3A:
Butyrophilin 3A
- CAR:
Chimeric antigen receptor
- CD:
Cluster of differentiation
- CDR3:
Complementarity-determining region–3
- CTLA-4:
Cytotoxic T-lymphocyte–associated antigen 4
- CLL:
Chronic lymphocytic leukemia
- CML:
Chronic myeloid leukemia
- DCs:
Dendritic cells
- DNAM-1:
DNAX accessory molecule-1
- Fas:
Fas Cell Surface Death Receptor
- FasL:
Fas ligand
- FcγRIII:
Fc-gamma receptor type III
- GM-CSF:
Granulocyte-macrophage colony-stimulating factor
- HIV:
Human immunodeficiency virus
- HMB-PP:
(E)-4-Hydroxy-3-methyl-but-2-enyl pyrophosphate
- IFN:
Interferon
- IL:
Interleukin
- iNKT:
Invariant NKT
- IPP:
Isopentenyl pyrophosphate
- LCs:
Leukemic cells
- MAIT:
Mucosal-associated invariant T
- MHC:
Major histocompatibility complex
- MIC-A:
MHC class I polypeptide-related sequence A
- MIC-B:
MHC class I polypeptide-related sequence B
- MM:
Multiple myeloma
- MR1:
MHC class I-related protein
- NCRs:
Natural cytotoxicity receptors
- NK:
Natural killer
- NKG2D:
Natural-killer group 2 member D
- NKT:
Natural killer T
- PB:
Peripheral blood
- PD1:
Programmed cell death protein 1
- pAgs:
Phosphoantigens
- PVR:
Poliovirus receptor
- TCR:
T-cell receptor
- TGF:
Transforming growth factor
- TLRs:
Toll-like receptors
- TME:
Tumor microenvironment
- TNF:
Tumor necrosis factor
- TRAIL:
TNF-related apoptosis-inducing ligand
- ULBP:
UL16-binding protein
- α-GalCer:
α-Galactosylceramide
- γδ T:
Gamma-delta T.
Additional Points
Cell-mediated immune response in malignant neoplasms is becoming the focus in immunotherapy against cancer. Unconventional T cells utilize a range of receptors and proinflammatory or regulatory profiles that directly contribute to the immune response in the leukemic microenvironment. The role played by unconventional T cells found in the leukemic medullary microenvironment may be related to the patient's prognosis. Cytokines and chemokines secreted by unconventional T cells include IL-1β, IL-4, IL-6, IL-10, IL-13, IL-17, IL-22, IL-23, IFN-γ, TNF-α, and TGF-β.
Disclosure
The funders had no role in study design, decision to publish, or preparation of the manuscript.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
References
- 1.Ehsanpour A., Saki N., Bagheri M., Behzad M. M., Abroun S. The expression of microvesicles in leukemia: prognostic approaches. Cell Journal. 2018;21:115–123. doi: 10.22074/cellj.2019.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Etten R. A. Mechanisms of transformation by the _BCR-ABL_ oncogene: new perspectives in the post-imatinib era. Leukemia Research. 2004;28:21–28. doi: 10.1016/j.leukres.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Van Etten R. A. Aberrant cytokine signaling in leukemia. Oncogene. 2007;26(47):6738–6749. doi: 10.1038/sj.onc.1210758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiarini F., Lonetti A., Evangelisti C., et al. Advances in understanding the acute lymphoblastic leukemia bone marrow microenvironment: from biology to therapeutic targeting. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2016;1863(3):449–463. doi: 10.1016/j.bbamcr.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 5.Huan J., Hornick N. I., Shurtleff M. J., et al. RNA trafficking by acute myelogenous leukemia exosomes. Cancer Research. 2013;73(2):918–929. doi: 10.1158/0008-5472.CAN-12-2184. [DOI] [PubMed] [Google Scholar]
- 6.Yotnda P., Mintz P., Grigoriadou K., Lemonnier F., Vilmer E., Langlade-Demoyen P. Analysis of T-cell defects in the specific immune response against acute lymphoblastic leukemia cells. Experimental Hematology. 1999;27:1375–1383. doi: 10.1016/S0301-472X(99)00083-1. [DOI] [PubMed] [Google Scholar]
- 7.Ahrends T., Borst J. The opposing roles of CD4+ T cells in anti-tumour immunity. Immunology. 2018;154:582–592. doi: 10.1111/imm.12941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maimela N. R., Liu S., Zhang Y. Fates of CD8+ T cells in tumor microenvironment. Computational and Structural Biotechnology Journal. 2019;17:1–13. doi: 10.1016/j.csbj.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Godfrey D. I., Uldrich A. P., McCluskey J., Rossjohn J., Moody D. B. The burgeoning family of unconventional T cells. Nature Immunology. 2015;16:1114–1123. doi: 10.1038/ni.3298. [DOI] [PubMed] [Google Scholar]
- 10.Godfrey D. I., Le Nours J., Andrews D. M., Uldrich A. P., Rossjohn J. Cell targets for cancer immunotherapy. Immunity. 2018;48:453–473. doi: 10.1016/j.immuni.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 11.Pellicci D. G., Koay H.-F., Berzins S. P. Thymic development of unconventional T cells: how NKT cells, MAIT cells and γδ T cells emerge. Nature Reviews Immunology. 2020;20(12):756–770. doi: 10.1038/s41577-020-0345-y. [DOI] [PubMed] [Google Scholar]
- 12.Zhao Y., Niu C., Cui J. Gamma-delta (γδ) T cells: friend or foe in cancer development. Journal of Translational Medicine. 2018;16(1):p. 3. doi: 10.1186/s12967-017-1378-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parker C. M., Groh V., Band H., et al. Evidence for extrathymic changes in the T cell receptor γ/δ repertoire. Journal of Experimental Medicine. 1990;171:1597–1612. doi: 10.1084/jem.171.5.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vantourout P., Hayday A. Six-of-the-best: unique contributions of γδ T cells to immunology. Nature Reviews Immunology. 2013;13:88–100. doi: 10.1038/nri3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wesch D., Glatzel A., Kabelitz D. Differentiation of resting human peripheral blood γδ T cells toward Th1- or Th2-phenotype. Cellular Immunology. 2001;212:110–117. doi: 10.1006/cimm.2001.1850. [DOI] [PubMed] [Google Scholar]
- 16.Ness-Schwickerath K. J., Jin C., Morita C. T. Cytokine requirements for the differentiation and expansion of IL-17A– and IL-22–producing human Vγ2Vδ2 T cells. The Journal of Immunology. 2010;184:7268–7280. doi: 10.4049/jimmunol.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caccamo N., La Mendola C., Orlando V., et al. Differentiation, phenotype, and function of interleukin-17-producing human vγ9vδ2 T cells. Blood. 2011;118:129–138. doi: 10.1182/blood-2011-01-331298. [DOI] [PubMed] [Google Scholar]
- 18.Peters C., Häsler R., Wesch D., Kabelitz D. Human Vδ2 T cells are a major source of interleukin-9. Proceedings of the National Academy of Sciences. 2016;113:12520–12525. doi: 10.1073/pnas.1607136113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gentles A. J., Newman A. M., Liu C. L., et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nature Medicine. 2015;21:938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo P. E., Pizzolato G., Corsale A. M., et al. γδ T cells and tumor microenvironment: from immunosurveillance to tumor evasion. Frontiers in Immunology. 2018;9 doi: 10.3389/fimmu.2018.01395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao Y., Yang W., Pan M., et al. γδ T cells provide an early source of interferon γ in tumor immunity. Journal of Experimental Medicine. 2003;198:433–442. doi: 10.1084/jem.20030584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horner A. A., Jabara H., Ramesh N., Geha R. S. γ/δ T lymphocytes express CD40 ligand and induce isotype switching in B lymphocytes. Journal of Experimental Medicine. 1995;181:1239–1244. doi: 10.1084/jem.181.3.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caccamo N., Battistini L., Bonneville M., et al. CXCR5 identifies a subset of Vγ9Vδ2 T cells which secrete IL-4 and IL-10 and help B cells for antibody production. The Journal of Immunology. 2006;177:5290–5295. doi: 10.4049/jimmunol.177.8.5290. [DOI] [PubMed] [Google Scholar]
- 24.Petrasca A., Melo A. M., Breen E. P., Doherty D. G. Human Vδ3 + γδ T cells induce maturation and IgM secretion by B cells. Immunology Letters. 2018;196:126–134. doi: 10.1016/j.imlet.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 25.Brandes M., Willimann K., Moser B. Professional antigen-presentation function by human gd T cells. Science. 2005;309(5732):264–268. doi: 10.1126/science.1110267. [DOI] [PubMed] [Google Scholar]
- 26.Muto M., Baghdadi M., Maekawa R., Wada H., Seino K. I. Myeloid molecular characteristics of human γδ T cells support their acquisition of tumor antigen-presenting capacity. Cancer Immunology, Immunotherapy. 2015;64:941–949. doi: 10.1007/s00262-015-1700-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mao C., Mou X., Zhou Y., et al. Tumor-activated T cells from gastric cancer patients induce the antitumor immune response of T cells via their antigen-presenting cell-like effects. Journal of Immunology Research. 2014;2014:10. doi: 10.1155/2014/593562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maniar A., Zhang X., Lin W., et al. Human γδ T lymphocytes induce robust NK cell-mediated antitumor cytotoxicity through CD137 engagement. Blood. 2010;116:1726–1733. doi: 10.1182/blood-2009-07-234211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Acker H. H., Anguille S., Van Tendeloo V. F., Lion E. Empowering gamma delta T cells with antitumor immunity by dendritic cell-based immunotherapy. Oncoimmunology. 2015;4(8, article e1021538) doi: 10.1080/2162402X.2015.1021538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martino A., Casetti R., Sacchi A., Poccia F. Central memory Vγ9Vδ2 T lymphocytes primed and expanded by Bacillus Calmette-Guérin-infected dendritic cells kill mycobacterial-infected monocytes. The Journal of Immunology. 2007;179:3057–3064. doi: 10.4049/jimmunol.179.5.3057. [DOI] [PubMed] [Google Scholar]
- 31.Simões A. E., Di Lorenzo B., Silva-Santos B. Molecular determinants of target cell recognition by human γδ T cells. Frontiers in Immunology. 2018;9:1–7. doi: 10.3389/fimmu.2018.00929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gomes A. Q., Correia D. V., Grosso A. R., et al. Identification of a panel of ten cell surface protein antigens associated with immunotargeting of leukemias and lymphomas by peripheral blood γδ t cells. Haematologica. 2010;95:1397–1404. doi: 10.3324/haematol.2009.020602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Weerdt I., Terpstra S., Hofland T., et al. Chronic lymphocytic leukemia (CLL) cells are susceptible to γδ-T cell mediated killing, provided CLL-derived γδ-T cell dysfunction can be reversed. Blood. 2015;126:2914–2914. doi: 10.1182/blood.v126.23.2914.2914. [DOI] [Google Scholar]
- 34.Gundermann S., Klinker E., Kimmel B., et al. A comprehensive analysis of primary acute myeloid leukemia identifies biomarkers predicting susceptibility to human allogeneic Vγ9Vδ2 T cells. Journal of Immunotherapy. 2014;37:321–330. doi: 10.1097/CJI.0000000000000043. [DOI] [PubMed] [Google Scholar]
- 35.Siegers G. M., Dhamko H., Wang X.-H., et al. Human Vδ1 γδ T cells expanded from peripheral blood exhibit specific cytotoxicity against B-cell chronic lymphocytic leukemia-derived cells. Cytotherapy. 2011;13:753–764. doi: 10.3109/14653249.2011.553595. [DOI] [PubMed] [Google Scholar]
- 36.Correia D. V., Fogli M., Hudspeth K., Gomes Da Silva M., Mavilio D., Silva-Santos B. Differentiation of human peripheral blood Vδ1+ T cells expressing the natural cytotoxicity receptor NKp30 for recognition of lymphoid leukemia cells. Blood. 2011;118:992–1001. doi: 10.1182/blood-2011-02-339135. [DOI] [PubMed] [Google Scholar]
- 37.Siegers G. M., Felizardo T. C., Mark Mathieson A., et al. Anti-leukemia activity of in vitro-expanded human gamma delta T cells in a xenogeneic Ph+leukemia model. PLoS One. 2011;6(2, article e16700) doi: 10.1371/journal.pone.0016700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siegers G. M. Anti-leukemia activity of human gamma delta T cells. Oncoimmunology. 2012;1:237–239. doi: 10.4161/onci.1.2.18231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silva-Santos B., Strid J. Working in “NK mode”: natural killer group 2 member D and natural cytotoxicity receptors in stress-surveillance by γδ T cells. Frontiers in Immunology. 2018;9:p. 24. doi: 10.3389/fimmu.2018.00851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pandyra A., Mullen P. J., Kalkat M., et al. Immediate utility of two approved agents to target both the metabolic mevalonate pathway and its restorative feedback loop. Cancer Research. 2014;74:4772–4782. doi: 10.1158/0008-5472.CAN-14-0130. [DOI] [PubMed] [Google Scholar]
- 41.Gertner-Dardenne J., Castellano R., Mamessier E., et al. Human Vγ9Vδ2 T cells specifically recognize and kill acute myeloid leukemic blasts. The Journal of Immunology. 2012;188:4701–4708. doi: 10.4049/jimmunol.1103710. [DOI] [PubMed] [Google Scholar]
- 42.Kalyan S., Wesch D., Kabelitz D. Aminobisphosphonates and toll-like receptor ligands: recruiting Vγ9Vδ2 T cells for the treatment of hematologic malignancy. Current Medicinal Chemistry. 2011;18(34):5206–5216. doi: 10.2174/092986711798184280. [DOI] [PubMed] [Google Scholar]
- 43.Mami-Chouaib F., Del Porto P., Delorme D., Hercend T. Further evidence for a gamma/delta T cell receptor-mediated TCT.1/CD48 recognition. Journal of Immunological. 1991;147(9):2864–2867. [PubMed] [Google Scholar]
- 44.Flament C., Bellagha K., Rosenthal-Allieri A., Chouaib S., Mami-Chouaib F. CD48 may serve as an accessory molecule for the activation of a subset of human γ/δ T cells. Human Immunology. 1996;46:82–92. doi: 10.1016/0198-8859(96)00010-9. [DOI] [PubMed] [Google Scholar]
- 45.Wu D., Wu P., Qiu F., Wei Q., Huang J. Human γδT-cell subsets and their involvement in tumor immunity. Cellular & Molecular Immunology. 2017;14:245–253. doi: 10.1038/cmi.2016.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davey M. S., Willcox C. R., Baker A. T., Hunter S., Willcox B. E. Recasting human Vδ1 lymphocytes in an adaptive role. Trends in Immunology. 2018;39:446–459. doi: 10.1016/j.it.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silva-Santos B., Serre K., Norell H. γδT cells in cancer. Nature reviews immunology. 2015;15:683–691. doi: 10.1038/nri3904. [DOI] [PubMed] [Google Scholar]
- 48.Wu Y. L., Ding Y. P., Tanaka Y., et al. γδ T cells and their potential for immunotherapy. International Journal of Biological Sciences. 2014;10:119–135. doi: 10.7150/ijbs.7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramstead A. G., Jutila M. A. Complex role of γδ T-cell-derived cytokines and growth factors in cancer. Journal of Interferon & Cytokine Research. 2012;32:563–569. doi: 10.1089/jir.2012.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kozbor D., Trinchieri G., Monos D. S., et al. Human TCR-γ+/δ+, CD8+ T lymphocytes recognize tetanus toxoid in an MHC-restricted fashion. Journal of Experimental Medicine. 1989;169:1847–1851. doi: 10.1084/jem.169.5.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lança T., Correia D. V., Moita C. F., et al. The MHC class Ib protein ULBP1 is a nonredundant determinant of leukemia/lymphoma susceptibility to γδ T-cell cytotoxicity. Blood. 2010;115:2407–2411. doi: 10.1182/blood-2009-08-237123. [DOI] [PubMed] [Google Scholar]
- 52.Poggi A., Venturino C., Catellani S., et al. Vδ1 T lymphocytes from B-CLL patients recognize ULBP3 expressed on leukemic B cells and up-regulated by trans-retinoic acid. Cancer Research. 2004;64:9172–9179. doi: 10.1158/0008-5472.CAN-04-2417. [DOI] [PubMed] [Google Scholar]
- 53.Willcox B. E., Mohammed F., Willcox C. R. γδ TCR recognition of MR1: adapting to life on the flip side. Trends in Biochemical Sciences. 2020;45:551–553. doi: 10.1016/j.tibs.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Almeida A. R., Correia D. V., Fernandes-Platzgummer A., et al. Delta one T cells for immunotherapy of chronic lymphocytic leukemia: clinical-grade expansion/differentiation and preclinical proof of concept. Clinical Cancer Research. 2016;22:5795–5804. doi: 10.1158/1078-0432.CCR-16-0597. [DOI] [PubMed] [Google Scholar]
- 55.Di Lorenzo B., Simões A. E., Caiado F., et al. Broad cytotoxic targeting of acute myeloid leukemia by polyclonal delta one T cells. Cancer Immunology Research. 2019;7:552–558. doi: 10.1158/2326-6066.CIR-18-0647. [DOI] [PubMed] [Google Scholar]
- 56.Garber K. γδ T cells bring unconventional cancer-targeting to the clinic - again. Nature Biotechnology. 2020;38:389–391. doi: 10.1038/s41587-020-0487-2. [DOI] [PubMed] [Google Scholar]
- 57.Gan Y. H., Lui S. S. N., Malkovsky M. Differential susceptibility of naïve and activated human γδ T cells to activation-induced cell death by T-cell receptor cross-linking. Molecular Medicine. 2001;7:636–643. doi: 10.1007/bf03401870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kabelitz D. Effector functions and control of human γδ T-cell activation. Microbes and Infection. 1999;1:255–261. doi: 10.1016/S1286-4579(99)80042-2. [DOI] [PubMed] [Google Scholar]
- 59.Meeh P. F., King M., O’Brien R. L., et al. Characterization of the γδ T cell response to acute leukemia. Cancer Immunology, Immunotherapy. 2006;55:1072–1080. doi: 10.1007/s00262-005-0094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Willcox C. R., Davey M. S., Willcox B. E. Development and selection of the human Vγ9Vδ2+ T-cell repertoire. Frontiers in Immunology. 2018;9:p. 1501. doi: 10.3389/fimmu.2018.01501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vyborova A., Beringer D. X., Fasci D., et al. γ9δ2T cell diversity and the receptor interface with tumor cells. Journal of Clinical Investigation. 2020;130(9):4637–4651. doi: 10.1172/jci132489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fonseca S., Pereira V., Lau C., dos Anjos Teixeira M., Bini-Antunes M., Lima M. Human peripheral blood gamma delta T cells: report on a series of healthy Caucasian Portuguese adults and comprehensive review of the literature. Cells. 2020;9:p. 729. doi: 10.3390/cells9030729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hinz T., Wesch D., Halary F., et al. Identification of the complete expressed human TCR V(γ) repertoire by flow cytometry. International Immunology. 1997;9:1065–1072. doi: 10.1093/intimm/9.8.1065. [DOI] [PubMed] [Google Scholar]
- 64.Kabelitz D., Hinz T., Dobmeyer T., et al. Clonal expansion of Vγ3/Vδ3-expressing γδ T cells in a HIV-1/2-negative patient with CD4 T-cell deficiency. British Journal of Haematology. 1997;96:266–271. doi: 10.1046/j.1365-2141.1997.d01-2027.x. [DOI] [PubMed] [Google Scholar]
- 65.Gruenbacher G., Thurnher M. Mevalonate metabolism in immuno-oncology. Frontiers in Immunology. 2017;8:p. 1714. doi: 10.3389/fimmu.2017.01714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moulin M., Alguacil J., Gu S., et al. Vγ9Vδ2 T cell activation by strongly agonistic nucleotidic phosphoantigens. Cellular and Molecular Life Sciences. 2017;74:4353–4367. doi: 10.1007/s00018-017-2583-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Herrmann T., Fichtner A. S., Karunakaran M. M. An update on the molecular basis of phosphoantigen recognition by Vγ9Vδ2 T cells. Cells. 2020;9:p. 1433. doi: 10.3390/cells9061433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rigau M., Ostrouska S., Fulford T. S., et al. Butyrophilin 2A1 is essential for phosphoantigen reactivity by gd T cells. Science. 2020;367, article eaay5516 doi: 10.1126/science.aay5516. [DOI] [PubMed] [Google Scholar]
- 69.Blazquez J. L., Benyamine A., Pasero C., Olive D. New insights into the regulation of γδ T cells by BTN3A and other BTN/BTNL in tumor immunity. Frontiers in Immunology. 2018;9:1–11. doi: 10.3389/fimmu.2018.01601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Karunakaran M. M., Willcox C. R., Salim M., et al. Butyrophilin-2A1 directly binds germline-encoded regions of the Vγ9Vδ2 TCR and is essential for phosphoantigen sensing. Immunity. 2020;52:487–498.e6. doi: 10.1016/j.immuni.2020.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Willcox C. R., Vantourout P., Salim M., et al. Butyrophilin-like 3 directly binds a human Vγ4+ T cell receptor using a modality distinct from clonally-restricted antigen. Immunity. 2019;51:813–825.e4. doi: 10.1016/j.immuni.2019.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Benyamine A., le Roy A., Mamessier E., et al. BTN3A molecules considerably improve Vγ9Vδ2T cells-based immunotherapy in acute myeloid leukemia. Oncoimmunology. 2016;5(10, article e1146843) doi: 10.1080/2162402X.2016.1146843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lafont V., Liautard J., Liautard J. P., Favero J. Production of TNF-α by human Vγ9Vδ2 T cells via engagement of FcγRIIIA, the low affinity type 3 receptor for the Fc portion of IgG, expressed upon TCR activation by nonpeptidic antigen. Journal of Immunology. 2001;166:7190–7199. doi: 10.4049/jimmunol.166.12.7190. [DOI] [PubMed] [Google Scholar]
- 74.deBarros A., Chaves-Ferreira M., D’Orey F., Ribot J. C., Silva-Santos B. CD70-CD27 interactions provide survival and proliferative signals that regulate T cell receptor-driven activation of human γδ peripheral blood lymphocytes. Eur J Immunol. 2011;41:195–201. doi: 10.1002/eji.201040905. [DOI] [PubMed] [Google Scholar]
- 75.Compte E., Pontarotti P., Collette Y., Lopez M., Olive D. Frontline: characterization of BT3 molecules belonging to the B7 family expressed on immune cells. European Journal of Immunology. 2004;34:2089–2099. doi: 10.1002/eji.200425227. [DOI] [PubMed] [Google Scholar]
- 76.Cubillos-Ruiz J. R., Martinez D., Scarlett U. K., et al. CD277 is a negative co-stimulatory molecule universally expressed by ovarian cancer microenvironmental cells. Oncotarget. 2010;1:329–338. doi: 10.18632/oncotarget.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yamazaki T., Goya I., Graf D., Craig S., Martin-Orozco N., Dong C. A butyrophilin family member critically inhibits T cell activation. Journal of Immunology. 2010;185:5907–5914. doi: 10.4049/jimmunol.1000835. [DOI] [PubMed] [Google Scholar]
- 78.Sarter K., Leimgruber E., Gobet F., et al. Btn2a2, a T cell immunomodulatory molecule coregulated with MHC class II genes. Journal of Experimental Medicine. 2016;213:177–187. doi: 10.1084/jem.20150435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Messal N., Mamessier E., Sylvain A., et al. Differential role for CD277 as a co-regulator of the immune signal in T and NK cells. European Journal of Immunology. 2011;41:3443–3454. doi: 10.1002/eji.201141404. [DOI] [PubMed] [Google Scholar]
- 80.Payne K. K., Mine J. A., Biswas S., et al. BTN3A1 governs antitumor responses by coordinating ab and gd T cells. Science. 2020;369:942–949. doi: 10.1126/science.aay2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Barros R. D. M., Roberts N. A., Dart R. J., et al. Epithelia use butyrophilin-like molecules to shape organ-specific γδ T cell compartments. Cell. 2016;167:203–218.e17. doi: 10.1016/j.cell.2016.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vavassori S., Kumar A., Wan G. S., et al. Butyrophilin 3A1 binds phosphorylated antigens and stimulates human γδ T cells. Nature Immunology. 2013;14:908–916. doi: 10.1038/ni.2665. [DOI] [PubMed] [Google Scholar]
- 83.Sandstrom A., Peigné C. M., Léger A., et al. The intracellular B30.2 domain of butyrophilin 3A1 binds phosphoantigens to mediate activation of human Vγ9Vδ2T Cells. Immunity. 2014;40:490–500. doi: 10.1016/j.immuni.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Harly C., Guillaume Y., Nedellec S., et al. Key implication of CD277/butyrophilin-3 (BTN3A) in cellular stress sensing by a major human γδ T-cell subset. Blood. 2012;120:2269–2279. doi: 10.1182/blood-2012-05-430470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gu S., Sachleben J. R., Boughter C. T., et al. Phosphoantigen-induced conformational change of butyrophilin 3A1 (BTN3A1) and its implication on Vγ9Vδ2 T cell activation. Proceedings of the National Academy of Sciences. 2017;114:E7311–E7320. doi: 10.1073/pnas.1707547114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang Y., Li L., Yuan L., et al. A structural change in butyrophilin upon phosphoantigen binding underlies phosphoantigen-mediated Vγ9Vδ2 T cell activation. Immunity. 2019;50:1043–1053.e5. doi: 10.1016/j.immuni.2019.02.016. [DOI] [PubMed] [Google Scholar]
- 87.Salim M., Knowles T. J., Baker A. T., et al. BTN3A1 discriminates γδ T cell phosphoantigens from nonantigenic small molecules via a conformational sensor in its B30.2 domain. ACS Chemical Biology. 2017;12:2631–2643. doi: 10.1021/acschembio.7b00694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sebestyen Z., Scheper W., Vyborova A., et al. RhoB mediates phosphoantigen recognition by Vγ9Vδ2 T cell receptor. Cell Reports. 2016;15:1973–1985. doi: 10.1016/j.celrep.2016.04.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rhodes D. A., Chen H.-C., Price A. J., et al. Activation of human γδ T cells by cytosolic interactions of BTN3A1 with soluble phosphoantigens and the cytoskeletal adaptor periplakin. Journal of Immunology. 2015;194:2390–2398. doi: 10.4049/jimmunol.1401064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kong Y., Cao W., Xi X., Ma C., Cui L., He W. The NKG2D ligand ULBP4 binds to TCRγ9/δ2 and induces cytotoxicity to tumor cells through both TCRγδ and NKG2D. Blood. 2009;114:310–317. doi: 10.1182/blood-2008-12-196287. [DOI] [PubMed] [Google Scholar]
- 91.Mangan B. A., Dunne M. R., O’Reilly V. P., et al. Cutting edge: CD1d restriction and Th1/Th2/Th17 cytokine secretion by human Vδ3 T cells. Journal of Immunology. 2013;191:30–34. doi: 10.4049/jimmunol.1300121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kenna T., Golden-Mason L., Norris S., Hegarty J. E., O’Farrelly C., Doherty D. G. Distinct subpopulations of γδ T cells are present in normal and tumor-bearing human liver. Clinical Immunology. 2004;113:56–63. doi: 10.1016/j.clim.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 93.Knight A., Madrigal A. J., Grace S., et al. The role of Vδ2-negative γδ T cells during cytomegalovirus reactivation in recipients of allogeneic stem cell transplantation. Blood. 2010;116:2164–2172. doi: 10.1182/blood-2010-01-255166. [DOI] [PubMed] [Google Scholar]
- 94.Farnault L., Gertner-Dardenne J., Gondois-Rey F., et al. Clinical evidence implicating gamma-delta T cells in EBV control following cord blood transplantation. Bone Marrow Transplant. 2013;48:1478–1479. doi: 10.1038/bmt.2013.75. [DOI] [PubMed] [Google Scholar]
- 95.Rajoriya N., Fergusson J., Leithead J. A., Klenerman P. Gamma delta T-lymphocytes in hepatitis C and chronic liver disease. Frontiers in Immunology. 2014;5:1–9. doi: 10.3389/fimmu.2014.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bartkowiak J., Kulczyck-Wojdala D., Blonski J. Z., Robak T. Molecular diversity of gammadelta T cells in peripheral blood from patients with B-cell chronic lymphocytic leukaemia. Neoplasma. 2002;49:86–90. [PubMed] [Google Scholar]
- 97.Marlin R., Pappalardo A., Kaminski H., et al. Sensing of cell stress by human γδ TCR-dependent recognition of annexin A2. Proceedings of the National Academy of Sciences. 2017;114:3163–3168. doi: 10.1073/pnas.1621052114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rei M., Pennington D. J., Silva-Santos B. The emerging protumor role of γδ T lymphocytes: implications for cancer immunotherapy. Cancer Research. 2015;75:798–802. doi: 10.1158/0008-5472.CAN-14-3228. [DOI] [PubMed] [Google Scholar]
- 99.Lo Presti E., Di Mitri R., Pizzolato G., Mocciaro F., Dieli F., Meraviglia S. γδ cells and tumor microenvironment: a helpful or a dangerous liason? Journal of Leukocyte Biology. 2018;103:485–492. doi: 10.1002/JLB.5MR0717-275RR. [DOI] [PubMed] [Google Scholar]
- 100.Lo P. E., Pizzolato G., Gulotta E., et al. Current advances in γδ T cell-based tumor immunotherapy. Frontiers in Immunology. 2017;8:p. 1401. doi: 10.3389/fimmu.2017.01401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Höpken U. E., Rehm A. Targeting the tumor microenvironment of leukemia and lymphoma. Trends in Cancer. 2019;5:351–364. doi: 10.1016/j.trecan.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 102.Lamble A. J., Lind E. F. Targeting the immune microenvironment in acute myeloid leukemia: a focus on T cell immunity. Frontiers in Oncology. 2018;8:p. 213. doi: 10.3389/fonc.2018.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Baryawno N., Przybylski D., Kowalczyk M. S., et al. A cellular taxonomy of the bone marrow stroma in homeostasis and leukemia. Cell. 2019;177:1915–1932.e16. doi: 10.1016/j.cell.2019.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang L., Xu M., Wang C., et al. The feature of distribution and clonality of tcr γ / δ subfamilies T cells in patients with b-cell non-Hodgkin lymphoma. Journal of Immunology Research. 2014;2014:6. doi: 10.1155/2014/241246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Abel A. M., Yang C., Thakar M. S., Malarkannan S. Natural killer cells: development, maturation, and clinical utilization. Frontiers in Immunology. 2018;9 doi: 10.3389/fimmu.2018.01869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Benlagha K., Kyin T., Beavis A., Teyton L., Bendelac A. A thymic precursor to the NK T cell lineage. Science. 2002;296:553–555. doi: 10.1126/science.1069017. [DOI] [PubMed] [Google Scholar]
- 107.Liao C.-M., Zimmer M. I., Wang C.-R. The functions of type I and type II natural killer T cells in inflammatory bowel diseases. Inflamm Bowel Dis. 2013;19:1330–1338. doi: 10.1097/MIB.0b013e318280b1e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wu L., Gabriel C. L., Parekh V. V., Van Kaer L. Invariant natural killer T cells: innate-like T cells with potent immunomodulatory activities. Tissue Antigens. 2009;73:535–545. doi: 10.1111/j.1399-0039.2009.01256.x. [DOI] [PubMed] [Google Scholar]
- 109.Godfrey D. I., MacDonald H. R., Kronenberg M., Smyth M. J., Van Kaer L. NKT cells: what’s in a name? Nature Reviews Immunology. 2004;4:231–237. doi: 10.1038/nri1309. [DOI] [PubMed] [Google Scholar]
- 110.Berzins S. P., Cochrane A. D., Pellicci D. G., Smyth M. J., Godfrey D. I. Limited correlation between human thymus and blood NKT cell content revealed by an ontogeny study of paired tissue samples. European Journal of Immunology. 2005;35:1399–1407. doi: 10.1002/eji.200425958. [DOI] [PubMed] [Google Scholar]
- 111.Waldowska M., Bojarska-Junak A., Roliński J. A brief review of clinical trials involving manipulation of invariant NKT cells as a promising approach in future cancer therapies. Central European Journal of Immunology. 2017;42:181–195. doi: 10.5114/ceji.2017.69361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kumar V., Delovitch T. L. Different subsets of natural killer T cells may vary in their roles in health and disease. Immunology. 2014;142:321–336. doi: 10.1111/imm.12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Townsend M. J., Weinmann A. S., Matsuda J. L., et al. T-bet regulates the terminal maturation and homeostasis of NK and V α 14 i NKT cells. Immunity. 2004;20:477–494. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 114.Altman J. B., Benavides A. D., Das R., Bassiri H. Antitumor responses of invariant natural killer T cells. Journal of Immunology Research. 2015;2015:10. doi: 10.1155/2015/652875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kovalovsky D., Uche O. U., Eladad S., et al. The BTB–zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nature Immunology. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Van Kaer L., Parekh V. V., Wu L. Invariant natural killer T cells: bridging innate and adaptive immunity. Cell Tissue Res. 2011;343:43–55. doi: 10.1007/s00441-010-1023-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lee P. T., Benlagha K., Teyton L., Bendelac A. Distinct functional lineages of human V α 24 natural killer T cells. Journal of Experimental Medicine. 2002;195:637–641. doi: 10.1084/jem.20011908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Moreira-Teixeira L., Resende M., Coffre M., et al. Proinflammatory environment dictates the IL-17 − producing capacity of human invariant NKT cells. The Journal of Immunology. 2020;186:5758–5765. doi: 10.4049/jimmunol.1003043. [DOI] [PubMed] [Google Scholar]
- 119.Wang H., Hogquist K. A. How lipid-specific T cells become effectors: the differentiation of iNKT subsets. Frontiers in Immunology. 2018;9 doi: 10.3389/fimmu.2018.01450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chen Q.-Y., Zhang T., Pincus S. H., et al. Human CD1D gene expression is regulated by LEF-1 through distal promoter regulatory elements. The Journal of Immunology. 2010;184:5047–5054. doi: 10.4049/jimmunol.0901912. [DOI] [PubMed] [Google Scholar]
- 121.Haseeb M., Pirzada R. H., Ain Q. U., Choi S. Wnt signaling in the regulation of immune cell and cancer therapeutics. Cells. 2019;8(11):p. 1380. doi: 10.3390/cells8111380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Strati F. License to kill : when iNKT cells are granted the use of lethal cytotoxicity. International Journal of Molecular Sciences. 2020;21:1–17. doi: 10.3390/ijms21113909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wu C., Li J., Li L., et al. Extracellular vesicles derived from natural killer cells use multiple cytotoxic proteins and killing mechanisms to target cancer cells. Journal of Extracellular Vesicles. 2019;8(1) doi: 10.1080/20013078.2019.1588538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sparrow E. Granulysin: the attractive side of a natural born killer. Immunology Letters. 2020;217:126–132. doi: 10.1016/j.imlet.2019.11.005. [DOI] [PubMed] [Google Scholar]
- 125.Bae E. A., Seo H., Kim I. K., Jeon I., Kang C. Y. Roles of NKT cells in cancer immunotherapy. Archives of Pharmacal Research. 2019;42(7):543–548. doi: 10.1007/s12272-019-01139-8. [DOI] [PubMed] [Google Scholar]
- 126.Kitamura H., Iwakabe K., Yahata T., et al. The natural killer T (NKT) cell ligand α-galactosylceramide demonstrates its immunopotentiating effect by inducing interleukin (IL)-12 production by dendritic cells and IL-12 receptor expression on NKT cells. Journal of Experimental Medicine. 1999;189:1121–1128. doi: 10.1084/jem.189.7.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bassiri H., Das R., Guan P., et al. iNKT cell cytotoxic responses control T-lymphoma growth in vitro and in vivo. Cancer Immunology Research. 2014;2:59–69. doi: 10.1158/2326-6066.CIR-13-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.McEwen-Smith R. M., Salio M., Cerundolo V. The regulatory role of invariant NKT cells in tumor immunity. Cancer Immunology Research. 2015;3:425–435. doi: 10.1158/2326-6066.CIR-15-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tan J., Xiao W., Wang L., He Y. Type І natural killer T cells : naturally born for fighting. Acta Pharmacologica Sinica. 2010;31:1123–1132. doi: 10.1038/aps.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Vivier E., Ugolini S., Blaise D., Chabannon C., Brossay L. Targeting natural killer cells and natural killer T cells in cancer. Natural Killer Cells. 2012;12:239–252. doi: 10.1038/nri3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gorini F., Azzimonti L., Delfanti G., et al. Invariant NKT cells contribute to chronic lymphocytic leukemia surveillance and prognosis. Blood. 2017;129:3440–3451. doi: 10.1182/blood-2016-11-751065. [DOI] [PubMed] [Google Scholar]
- 132.Metelitsa L. S., Weinberg K. I., Emanuel P. D., Seeger R. C. Expression of CD1d by myelomonocytic leukemias provides a target for cytotoxic NKT cells. Leukemia. 2003;17:1068–1077. doi: 10.1038/sj.leu.2402943. [DOI] [PubMed] [Google Scholar]
- 133.Fiedler T., Walter W., Reichert T. E., Maeurer M. J. Regulation of CD1d expression by murine tumor cells: escape from immunosurveillance or alternate target molecules? International Journal of Cancer. 2002;98:389–397. doi: 10.1002/ijc.10141. [DOI] [PubMed] [Google Scholar]
- 134.Terabe M., Berzofsky J. A. NKT cells in immunoregulation of tumor immunity: a new immunoregulatory axis. Trends Immunol. 2007;28:491–496. doi: 10.1016/j.it.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 135.Brossay L., Chioda M., Burdin N., et al. CD1d-mediated recognition of an α-galactosylceramide by natural killer T cells is highly conserved through mammalian evolution. Journal of Experimental Medicine. 1998;188:1521–1528. doi: 10.1084/jem.188.8.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Seo H., Jeon I., Kim B.-S., et al. IL-21-mediated reversal of NK cell exhaustion facilitates anti-tumour immunity in MHC class I-deficient tumours. Nature Communications. 2017;8:1–14. doi: 10.1038/ncomms15776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang Y., Springfield R., Chen S., et al. α-GalCer and iNKT cell-based cancer immunotherapy: realizing the therapeutic potentials. Frontiers in Immunology. 2019;10 doi: 10.3389/fimmu.2019.01126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fais F., Tenca C., Cimino G., et al. CD1d expression on B-precursor acute lymphoblastic leukemia subsets with poor prognosis. Leukemia. 2005;19:551–556. doi: 10.1038/sj.leu.2403671. [DOI] [PubMed] [Google Scholar]
- 139.Bojarska-Junak A., Waldowska M., Woś J., et al. Intracellular IL-4 and IFN-γ expression in iNKT cells from patients with chronic lymphocytic leukemia. Oncology Letters. 2017;15:1580–1590. doi: 10.3892/ol.2017.7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rossignol A., Levescot A., Jacomet F., et al. Evidence for BCR-ABL-dependent dysfunctions of iNKT cells from chronic myeloid leukemia patients. European Journal of Immunology. 2012;42:1870–1875. doi: 10.1002/eji.201142043. [DOI] [PubMed] [Google Scholar]
- 141.Weinkove R., Brooks C. R., Carter J. M., Hermans I. F., Ronchese F. Functional invariant natural killer T-cell and CD1d axis in chronic lymphocytic leukemia: implications for immunotherapy. Haematologica. 2013;98(3):376–384. doi: 10.3324/haematol.2012.072835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Dellabona P., Casorati G., de Lalla C., Montagna D., Locatelli F. On the use of donor-derived iNKT cells for adoptive immunotherapy to prevent leukemia recurrence in pediatric recipients of HLA haploidentical HSCT for hematological malignancies. Clinical Immunology. 2011;140(2):152–159. doi: 10.1016/j.clim.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 143.Casorati G., de Lalla C., Dellabona P. Invariant natural killer T cells reconstitution and the control of leukemia relapse in pediatric haploidentical hematopoietic stem cell transplantation. OncoImmunology. 2014;1:355–357. doi: 10.4161/onci.18399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Najera Chuc A. E., Cervantes L. A. M., Retiguin F. P., Ojeda J. V., Maldonado E. R. Low number of invariant NKT cells is associated with poor survival in acute myeloid leukemia. Journal of Cancer Research and Clinical Oncology. 2012;138:1427–1432. doi: 10.1007/s00432-012-1251-x. [DOI] [PubMed] [Google Scholar]
- 145.Dhodapkar M. V., Kumar V. Type II NKT cells and their emerging role in health and disease. The Journal of Immunology. 2017;198:1015–1021. doi: 10.4049/jimmunol.1601399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Nours L., Andrews D. M., Uldrich A. P., Rossjohn J., Godfrey D. I. Unconventional T cell targets for cancer immunotherapy. Immunity. 2018;48(3):453–473. doi: 10.1016/j.immuni.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 147.Terabe M., Berzofsky J. A. Tissue-specific roles of NKT cells in tumor immunity. Frontiers in Immunology. 2018;9:1–11. doi: 10.3389/fimmu.2018.01838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Blomqvist M., Rhost S., Teneberg S., et al. Multiple tissue-specific isoforms of sulfatide activate CD1d-restricted type II NKT cells. European Journal of Immunology. 2014;39:1726–1735. doi: 10.1002/eji.200839001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Arrenberg P., Halder R., Dai Y., Maricic I., Kumar V. Oligoclonality and innate-like features in the TCR repertoire of type II NKT cells reactive to a-linked self-glycolipid. Proceedings of the National Academy of Sciences. 2010;107(24):10984–10989. doi: 10.1073/pnas.1000576107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Singh A. K., Tripathi P., Cardell S. L. Type II NKT cells: an elusive population with immunoregulatory properties. Frontiers in Immunology. 2018;9:p. 1969. doi: 10.3389/fimmu.2018.01969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Godfrey D. I., Kronenberg M. Going both ways: immune regulation via CD1d-dependent NKT cells. Journal of Clinical Investigation. 2004;114:1379–1388. doi: 10.1172/JCI200423594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Le Nours J., Shahine A., Gras S. Molecular features of lipid-based antigen presentation by group 1 CD1 molecules. Seminars in Cell & Developmental Biology. 2018;84:48–57. doi: 10.1016/j.semcdb.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 153.Consonni M., de Lalla C., Bigi A., Dellabona P., Casorati G. Harnessing the CD1 restricted T cell response for leukemia adoptive immunotherapy. Cytokine & Growth Factor Reviews. 2017;36:117–123. doi: 10.1016/j.cytogfr.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 154.Porcelli S., Yockey C. E., Brenner M. B., Balk S. P. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4-8- alpha/beta T cells demonstrates preferential use of several V beta genes and an invariant TCR alpha chain. Journal of Experimental Medicine. 1993;178:1–16. doi: 10.1084/jem.178.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lantz O., Bendelac A. An invariant T cell receptor alpha chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4-8- T cells in mice and humans. Journal of Experimental Medicine. 1994;180:1097–1106. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tilloy F., Treiner E., Park S.-H., et al. An invariant T cell receptor α chain defines a novel TAP-independent major histocompatibility complex class Ib–restricted α/β T cell subpopulation in mammals. Journal of Experimental Medicine. 1999;189:1907–1921. doi: 10.1084/jem.189.12.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Treiner E., Duban L., Bahram S., et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. 2003;422:164–169. doi: 10.1038/nature01433. [DOI] [PubMed] [Google Scholar]
- 158.Yamaguchi H., Hirai M., Kurosawa Y., Hashimoto K. A highly conserved major histocompatibility complex class I-related gene in mammals. Biochem Biophys Res Commun. 1997;238:697–702. doi: 10.1006/bbrc.1997.7379. [DOI] [PubMed] [Google Scholar]
- 159.Eckle S. B. G., Corbett A. J., Keller A. N., et al. Recognition of vitamin B precursors and byproducts by mucosal associated invariant T cells. Journal of Biological Chemistry. 2015;290:30204–30211. doi: 10.1074/jbc.R115.685990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Corbett A. J., Eckle S. B. G., Birkinshaw R. W., et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. 2014;509:361–365. doi: 10.1038/nature13160. [DOI] [PubMed] [Google Scholar]
- 161.Martin E., Treiner E., Duban L., et al. Stepwise development of MAIT cells in mouse and human. PLoS Biology. 2009;7, article e1000054:3. doi: 10.1371/journal.pbio.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Seach N., Guerri L., Le Bourhis L., et al. Double positive thymocytes select mucosal-associated invariant T cells. Journal of Immunology. 2013;191(12):6002–6009. doi: 10.4049/jimmunol.1301212. [DOI] [PubMed] [Google Scholar]
- 163.Koay H.-F., Gherardin N. A., Enders A., et al. A three-stage intrathymic development pathway for the mucosal-associated invariant T cell lineage. Nature Immunology. 2016;17:1300–1311. doi: 10.1038/ni.3565. [DOI] [PubMed] [Google Scholar]
- 164.Gherardin N. A., Loh L., Admojo L., et al. Enumeration, functional responses and cytotoxic capacity of MAIT cells in newly diagnosed and relapsed multiple myeloma. Scientific Reports. 2018;8:p. 4159. doi: 10.1038/s41598-018-22130-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Godfrey D. I., Koay H.-F., McCluskey J., Gherardin N. A. The biology and functional importance of MAIT cells. Nature Immunology. 2019;20(9):1110–1128. doi: 10.1038/s41590-019-0444-8. [DOI] [PubMed] [Google Scholar]
- 166.Rahimpour A., Koay H. F., Enders A., et al. Identification of phenotypically and functionally heterogeneous mouse mucosal-associated invariant T cells using MR1 tetramers. Journal of Experimental Medicine. 2015;212:1095–1108. doi: 10.1084/jem.20142110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Reantragoon R., Corbett A. J., Sakala I. G., et al. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. Journal of Experimental Medicine. 2013;210:2305–2320. doi: 10.1084/jem.20130958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Toubal A., Nel I., Lotersztajn S., Lehuen A. Mucosal-associated invariant T cells and disease. Nature Reviews Immunology. 2019;19:643–657. doi: 10.1038/s41577-019-0191-y. [DOI] [PubMed] [Google Scholar]
- 169.Dusseaux M., Martin E., Serriari N., et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17–secreting T cells. Blood. 2011;117:1250–1259. doi: 10.1182/blood-2010-08-303339. [DOI] [PubMed] [Google Scholar]
- 170.Ussher J. E., Bilton M., Attwod E., et al. CD161 ++ CD8 + T cells, including the MAIT cell subset, are specifically activated by IL-12+IL-18 in a TCR-independent manner. European Journal of Immunology. 2014;44:195–203. doi: 10.1002/eji.201343509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Haeryfar S. M. M., Shaler C. R., Rudak P. T. Mucosa-associated invariant T cells in malignancies: a faithful friend or formidable foe? Cancer Immunol Immunother. 2018;67:1885–1896. doi: 10.1007/s00262-018-2132-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Serriari N.-E., Eoche M., Lamotte L., et al. Innate mucosal-associated invariant T (MAIT) cells are activated in inflammatory bowel diseases. Clinical & Experimental Immunology. 2014;176:266–274. doi: 10.1111/cei.12277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Leng T., Akther H. D., Hackstein C.-P., et al. TCR and inflammatory signals tune human MAIT cells to exert specific tissue repair and effector functions. Cell Reports. 2019;28:3077–3091.e5. doi: 10.1016/j.celrep.2019.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Salio M., Gasser O., Gonzalez-Lopez C., et al. Activation of human mucosal-associated invariant T cells induces CD40L-dependent maturation of monocyte-derived and primary dendritic cells. The Journal of Immunology. 2017;199:2631–2638. doi: 10.4049/jimmunol.1700615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kurioka A., Ussher J. E., Cosgrove C., et al. MAIT cells are licensed through granzyme exchange to kill bacterially sensitized targets. Mucosal Immunology. 2015;8:429–440. doi: 10.1038/mi.2014.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ling L., Lin Y., Zheng W., et al. Circulating and tumor-infiltrating mucosal associated invariant T (MAIT) cells in colorectal cancer patients. Scientific Reports. 2016;6, article 20358 doi: 10.1038/srep20358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Won E. J., Ju J. K., Cho Y.-N., et al. Clinical relevance of circulating mucosal-associated invariant T cell levels and their anti-cancer activity in patients with mucosal-associated cancer. Oncotarget. 2016;7:76274–76290. doi: 10.18632/oncotarget.11187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sundström P., Ahlmanner F., Akéus P., et al. Human mucosa-associated invariant T cells accumulate in colon adenocarcinomas but produce reduced amounts of IFN-γ. Journal of Immunology. 2015;195:3472–3481. doi: 10.4049/jimmunol.1500258. [DOI] [PubMed] [Google Scholar]
- 179.Yan J., Allen S., McDonald E., et al. MAIT cells promote tumor initiation, growth, and metastases via tumor MR1. Cancer Discovery. 2020;10:124–141. doi: 10.1158/2159-8290.CD-19-0569. [DOI] [PubMed] [Google Scholar]
- 180.Favreau M., Venken K., Faict S., et al. Both mucosal-associated invariant and natural killer T-cell deficiency in multiple myeloma can be countered by PD-1 inhibition. Haematologica. 2017;102:e266–e270. doi: 10.3324/haematol.2017.163758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lepore M., Kalinichenko A., Calogero S., et al. Functionally diverse human T cells recognize non-microbial antigens presented by MR1. Elife. 2017;6 doi: 10.7554/eLife.24476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Crowther M. D., Dolton G., Legut M., et al. Genome-wide CRISPR–Cas9 screening reveals ubiquitous T cell cancer targeting via the monomorphic MHC class I-related protein MR1. Nature Immunology. 2020;21:178–185. doi: 10.1038/s41590-019-0578-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Meermeier E. W., Laugel B. F., Sewell A. K., et al. Human TRAV1-2-negative MR1-restricted T cells detect S. pyogenes and alternatives to MAIT riboflavin-based antigens. Nature Communications. 2016;7, article 12506 doi: 10.1038/ncomms12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Gherardin N. A., Keller A. N., Woolley R. E., et al. Diversity of T cells restricted by the MHC class I-related molecule MR1 facilitates differential antigen recognition. Immunity. 2016;44:32–45. doi: 10.1016/j.immuni.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 185.Wu K., Feng J., Xiu Y., et al. Vδ2 T cell subsets, defined by PD-1 and TIM-3 expression, present varied cytokine responses in acute myeloid leukemia patients. International Immunopharmacology. 2020;80, article 106122 doi: 10.1016/j.intimp.2019.106122. [DOI] [PubMed] [Google Scholar]
- 186.Hoeres T., Holzmann E., Smetak M., Birkmann J., Wilhelm M. PD-1 signaling modulates interferon-γ production by gamma delta (γδ) T-cells in response to leukemia. Oncoimmunology. 2018;8 doi: 10.1080/2162402X.2018.1550618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Durgan K., Ali M., Warner P., Latchman Y. E. Targeting NKT cells and PD-L1 pathway results in augmented anti-tumor responses in a melanoma model. Cancer Immunol Immunother. 2011;60:547–558. doi: 10.1007/s00262-010-0963-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Melero I., Berman D. M., Aznar M. A., Korman A. J., Gracia J. L. P., Haanen J. Evolving synergistic combinations of targeted immunotherapies to combat cancer. Nature Reviews Cancer. 2015;15:457–472. doi: 10.1038/nrc3973. [DOI] [PubMed] [Google Scholar]
- 189.Tasian S. K. Acute myeloid leukemia chimeric antigen receptor T-cell immunotherapy: how far up the road have we traveled? Therapeutic Advances in Hematology. 2018;9:135–148. doi: 10.1177/2040620718774268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Jacoby E., Shahani S. A., Shah N. N. Updates on CAR T-cell therapy in B-cell malignancies. Immunological Reviews. 2019;290:39–59. doi: 10.1111/imr.12774. [DOI] [PubMed] [Google Scholar]
- 191.Salter A. I., Pont M. J., Riddell S. R. Chimeric antigen receptor–modified T cells: CD19 and the road beyond. Blood. 2018;131:2621–2629. doi: 10.1182/blood-2018-01-785840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Cortés-Selva D., Dasgupta B., Singh S., Grewal I. S. Innate and innate-like cells: the future of chimeric antigen receptor (CAR) cell therapy. Trends in Pharmacological Sciences. 2020;42:45–59. doi: 10.1016/j.tips.2020.11.004. [DOI] [PubMed] [Google Scholar]


