Abstract
Background
Asthma is a major chronic disease that poses a heavy burden on health care. To facilitate the allocation of care management resources aimed at improving outcomes for high-risk patients with asthma, we recently built a machine learning model to predict asthma hospital visits in the subsequent year in patients with asthma. Our model is more accurate than previous models. However, like most machine learning models, it offers no explanation of its prediction results. This creates a barrier for use in care management, where interpretability is desired.
Objective
This study aims to develop a method to automatically explain the prediction results of the model and recommend tailored interventions without lowering the performance measures of the model.
Methods
Our data were imbalanced, with only a small portion of data instances linking to future asthma hospital visits. To handle imbalanced data, we extended our previous method of automatically offering rule-formed explanations for the prediction results of any machine learning model on tabular data without lowering the model’s performance measures. In a secondary analysis of the 334,564 data instances from Intermountain Healthcare between 2005 and 2018 used to form our model, we employed the extended method to automatically explain the prediction results of our model and recommend tailored interventions. The patient cohort consisted of all patients with asthma who received care at Intermountain Healthcare between 2005 and 2018, and resided in Utah or Idaho as recorded at the visit.
Results
Our method explained the prediction results for 89.7% (391/436) of the patients with asthma who, per our model’s correct prediction, were likely to incur asthma hospital visits in the subsequent year.
Conclusions
This study is the first to demonstrate the feasibility of automatically offering rule-formed explanations for the prediction results of any machine learning model on imbalanced tabular data without lowering the performance measures of the model. After further improvement, our asthma outcome prediction model coupled with the automatic explanation function could be used by clinicians to guide the allocation of limited asthma care management resources and the identification of appropriate interventions.
Keywords: asthma, forecasting, machine learning, patient care management
Introduction
Background
About 8.4% of Americans have asthma [1]. Each year in the United States, asthma costs over US $50 billion and results in more than 2 million emergency department (ED) visits, about half a million inpatient stays, and more than 3000 deaths [1,2]. A major goal in managing patients with asthma is to reduce their hospital visits, including ED visits and inpatient stays. As employed by health plans in 9 of 12 metropolitan communities [3] and by health care systems such as Intermountain Healthcare, Kaiser Permanente Northern California [4], and the University of Washington Medicine, the state-of-the-art method for achieving this goal is to employ a predictive model to predict which patients with asthma are highly likely to have poor outcomes in the future. Once identified, such patients are enrolled in care management. Care managers then call these patients on the phone regularly and help them make appointments for health and related services. By offering such tailored preventive care properly, up to 40% of future hospital visits by patients with asthma can be avoided [5-8].
A care management program has limited enrollment capacity [9]. As a result, the effectiveness of the program depends critically on the accuracy of the predictive model. Not enrolling a patient who is likely to have future hospital visits in the program is a missed opportunity to improve the patient’s outcomes. Unnecessarily enrolling a patient who is likely to have no future hospital visit would increase health care costs and waste scarce care management resources with no potential benefit. The current models for predicting hospital visits in patients with asthma are inaccurate, with published sensitivity of ≤49% and an area under the receiver operating characteristic curve (AUC) ≤0.81 [4,10-22]. When employed for care management, these models miss more than half of the patients who will have future hospital visits and erroneously label many other patients as likely to have future hospital visits [23]. To address these issues, we recently built an extreme gradient boosting (XGBoost) [24] machine learning model to predict asthma hospital visits in the subsequent year in patients with asthma [23]. Compared with previous models, our model raised the AUC by at least 0.049. However, like most machine learning models, our model offers no explanation of its prediction results. This creates a barrier for use in care management, where care managers need to understand why a patient is at risk for poor outcomes to make care management enrollment decisions and identify suitable interventions for the patient.
Objectives
To overcome the abovementioned barrier, this study aims to develop a method to automatically explain the prediction results of our model and recommend tailored interventions without lowering any of the performance measures of our model, such as AUC, accuracy, sensitivity, specificity, positive predictive value, and negative predictive value.
In the following sections, we describe our methods and the evaluation results. A list of abbreviations adopted in this paper is provided at the end of the paper.
Methods
We used the same patient cohort, data set, prediction target, cutoff threshold for binary classification, method for data preprocessing, including data cleaning and data normalization, and method for partitioning the whole data set into the training and test sets that we described in our prior paper [23].
Ethics Approval and Study Design
This study consists of a secondary analysis of retrospective data and was evaluated and approved by the institutional review boards of the University of Washington Medicine, University of Utah, and Intermountain Healthcare.
Patient Population
Our patient cohort included all patients with asthma who received care at any Intermountain Healthcare facility between 2005 and 2018 and resided in Utah or Idaho as recorded at the visit. Intermountain Healthcare is the largest health care system in Utah and southeastern Idaho. It operates 185 clinics and 22 hospitals and provides care for approximately 60% of people living in that region. A patient was considered asthmatic in a specific year if in the encounter billing database, the patient had one or more asthma diagnosis codes during that year (International Classification of Diseases, ninth revision [ICD-9]: 493.0x, 493.1x, 493.8x, 493.9x; International Classification of Diseases, tenth revision [ICD-10]: J45.x) [12,25,26]. The only exclusion criterion from the analysis in any given year was patient death during that year.
Data Set
We used a structured clinical and administrative data set provided by the enterprise data warehouse of Intermountain Healthcare. The data set covered all visits by the patient cohort within Intermountain Healthcare between 2005 and 2018.
Prediction Target (the Dependent or Outcome Variable)
For each patient identified as asthmatic in a specific year, the outcome was whether any asthma hospital visit occurred in the subsequent year. In this paper, an asthma hospital visit refers to an ED visit or an inpatient stay at an Intermountain Healthcare facility with a principal diagnosis of asthma (ICD-9: 493.0x, 493.1x, 493.8x, 493.9x; ICD-10: J45.x). For training and testing the XGBoost model and automatic explanation method, data of every patient with asthma up to the end of every year were used to predict the patient’s outcome in the subsequent year.
Predictive Model and Features (Independent Variables)
Our recent XGBoost model [23] uses 142 features to predict asthma hospital visits in the subsequent year in patients with asthma. As listed in the multimedia appendix in our previous study [23], these features were computed from the structured attributes in our data set covering a wide range of categories, such as patient demographics, visits, medications, laboratory tests, vital signs, diagnoses, and procedures. Each input data instance for our model has these 142 features, targets a pair of a patient with asthma and a year, and is employed to predict the patient’s outcome in the subsequent year. We set the cutoff threshold for binary classification at the top 10% of patients with asthma having the largest predicted risk. These patients were predicted to incur asthma hospital visits in the subsequent year.
Automatic Explanation Method
Previously, we developed an automated method to offer rule-formed explanations for any machine learning model’s prediction results on tabular data and recommend tailored interventions without lowering the performance measures of the model [27,28]. Our method was initially demonstrated to predict the diagnosis of type 2 diabetes [27]. Later, other researchers successfully applied our method to predict death or lung transplantation in patients with cystic fibrosis [29], predict cardiac death in patients with cancer, and use predictions to manage preventive care, heart transplant waiting list, and posttransplant follow-ups in patients with cardiovascular diseases [30]. In our method, each rule used for providing explanations has a performance measure termed confidence that must be greater than or equal to a given minimum confidence threshold cmin. Our original automatic explanation method [27] was designed for reasonably balanced data, where distinct values of the outcome variable appear with relatively similar frequencies. Recently, we outlined an extension of this method [31,32] to handle imbalanced data, where one value of the outcome variable appears much less often than another. This data imbalance exists when predicting asthma hospital visits in patients with asthma, where only about 4% of the data instances are linked to future asthma hospital visits [23]. In our extended method, each rule used for providing explanations has a second performance measure termed commonality, which must be greater than or equal to a given minimum commonality threshold mmin. To date, no technique has been developed to efficiently mine the rules with commonality greater than or equal to mmin, compute their confidence, and eliminate those rules with confidence less than cmin in the extended method, despite such techniques being essential for handling large data sets. No guideline exists for setting the values of the parameters used in the extended method, although they greatly impact the performance of the extended method. The extended method has never been implemented in computer code. Moreover, the effectiveness of the extended method has not been evaluated or demonstrated.
In this study, we made the following innovative contributions:
We provide several techniques for efficiently mining the rules with commonality greater than or equal to mmin, computing their confidence, and eliminating those rules with confidence less than cmin in the extended automatic explanation method. This completes our extended method. Although our extended method was designed for imbalanced data, it can also be used on reasonably balanced data to improve the efficiency of mining the rules needed to provide automatic explanations. Among the existing automatic explanation methods for machine learning prediction results, our method is the only one that can automatically recommend tailored interventions [33,34]. This capability is desired for many medical applications.
We present a guideline to set the values of the parameters used in the extended method (see the Discussion section).
We completed the first computer coding implementation of the extended method and explained it in this paper.
We demonstrate the effectiveness of the extended method in predicting asthma hospital visits in patients with asthma.
Review of Our Original Automatic Explanation Method
Main Idea
Our automatic explanation method separates explanation and prediction by employing 2 models concurrently, each for a distinct purpose. The first model is used to make predictions and can be any model that takes continuous and categorical features as its inputs. Usually, we adopt the most accurate model as the first model to avoid lowering the performance measures of the model. The second model uses class-based association rules [35,36] mined from historical data to explain the prediction results of the first model rather than to make predictions. Before using a standard association rule mining method like Apriori to mine the rules [36], each continuous feature is first transformed into a categorical feature through automatic discretization [35,37]. Each rule shows a feature pattern associated with a value w of the outcome variable in the form of q1 AND q2 AND … AND qn→w. The values of n and w can change across rules. For binary classification distinguishing poor versus good outcomes, w is usually the poor outcome value. Every item qi (1≤i≤n) is a feature-value pair (f, u) showing feature f has value u or a value within u, depending on whether u is a value or a range. The rule points out that a patient’s outcome variable is inclined to have value w if the patient fulfills q1, q2, ..., and qn. An example rule is as follows:
The patient had ≥12 ED visits in the past year
AND the patient had ≥21 distinct medications in all asthma medication orders in the past year
→the patient will incur one or more asthma hospital visits in the subsequent year.
The Association Rule Mining and Pruning Processes
The association rule mining process is controlled by 2 parameters: the minimum support threshold smin and the minimum confidence threshold cmin [36]. For any rule l: q1 AND q2 AND … AND qn→w, the percentage of data instances satisfying q1, q2, ..., and qn and linking to w is termed l’s support showing l’s coverage. Among all data instances satisfying q1, q2, ..., and qn, the percentage of data instances linking to w is termed l’s confidence reflecting l’s precision. Our original automatic explanation method uses rules with support ≥smin and confidence ≥cmin. For binary classification distinguishing poor versus good outcomes, we usually focus on the rules that have right-hand sides containing the poor outcome value.
Usually, numerous association rules have support and confidence ≥smin and ≥cmin, respectively. To avoid overwhelming the users of the automatic explanation function with too many rules, we used 4 techniques to reduce the number of rules in the second model. First, only features adopted by the first model are used to form rules. Second, a clinician in the automatic explanation function’s design team checks all possible values and value ranges of these features and marks those that could possibly have a positive correlation with the values of the outcome variable reflecting poor outcomes. Only those marked values and value ranges of these features are allowed to show up in the rules. Third, the rules are limited to having no more than a given small number of items on their left-hand sides, as long rules are hard to understand. A typical value of this number is 4. Fourth, each more specific rule is dropped when there exists a more general rule with confidence that is not lower by more than a given threshold τ≥0. More specifically, consider 2 rules, l1 and l2, whose right-hand sides have the same value. The items on the left-hand side of l2 are a superset of those on the left-hand side of l1. We drop l2 if l1’s confidence is ≥l2’s confidence-τ.
For the association rules remaining after the rule-pruning process, a clinician in the automatic explanation function’s design team gathers zero or more interventions targeting the reason the rule presents. A rule is called actionable if one or more interventions are compiled for it. Usually, each intervention links to one of the feature-value pair items on the rule’s left-hand side. Such an item is called actionable. Thus, an actionable rule contains at least 1 actionable item. To expedite the intervention compilation process, the clinician can identify all of the actionable items and compile interventions for each of them. All of the interventions linking to the actionable items on a rule’s left-hand side are automatically connected to the rule.
Our automatic explanation method uses 2 types of knowledge manually compiled by a clinician: the values and value ranges of the features that could possibly have a positive correlation with the outcome variable’s values reflecting poor outcomes and the interventions for the actionable items. Our automatic explanation method is fully automatic, except for the knowledge compilation step.
The Explanation Method
For each patient for whom the first model predicts a poor outcome, we explain the prediction result by listing the association rules in the second model whose right-hand sides have the corresponding poor outcome value and whose left-hand sides are fulfilled by the patient, whereas ignoring the rules in the second model whose right-hand sides have a value that differs from the corresponding poor outcome value and whose left-hand sides are fulfilled by the patient. Every rule listed offers a reason why the patient is predicted to have a poor outcome. For each actionable rule listed, the linked interventions are displayed next to it. This helps the user of the automatic explanation function find tailored inventions suitable for the patient. Typically, the rules in the second model describe common reasons for poor outcomes. However, some patients will have poor outcomes for rare reasons not covered by these rules. Consequently, the second model can provide explanations for most, but not all, of the patients for whom the first model predicts poor outcomes.
The Previously Outlined Extension of the Original Automatic Explanation Method
Our original automatic explanation method was designed for reasonably balanced data and is unsuitable for imbalanced data, where one value of the outcome variable appears much less often than another. If the minimum support threshold smin is large on imbalanced data, we cannot obtain enough association rules for the outcome variable's rare values. Consequently, for a large portion of the first model's prediction results on these values, we cannot give any explanation. Conversely, if smin is too small, the rule mining process will generate too many rules as intermediate results, most of which will be filtered out in the end. This easily exhausts computer memory and makes the rule mining process extremely slow. In addition, many overfitted rules will be produced in the end, making it difficult for clinicians to examine the mined rules.
In our recently outlined extension of the original automatic explanation method [31,32] to handle imbalanced data, we replace support with value-specific support termed commonality [38]. For any rule l: q1 AND q2 AND ... AND qn→w, among all data instances linking to w, the percentage of data instances satisfying q1, q2, ..., and qn is termed l’s commonality showing l’s coverage within the context of w. Moreover, we replace the minimum support threshold smin with the minimum commonality threshold mmin. Instead of using rules whose support is ≥smin and whose confidence is ≥ the minimum confidence threshold cmin, we used rules whose commonality is ≥mmin and whose confidence is ≥cmin.
Each value of the outcome variable falls into one of 2 possible cases. In the first case, the value is interesting and represents an abnormal case. The prediction results of this value require attention and explanations. In the second case, the value is uninteresting and represents a normal case. The prediction results of this value require neither special attention nor explanation. Typically, each interesting value is a rare one reflecting poor outcomes. The second model contains only the association rules related to interesting values. To mine these rules, we proceeded in 2 steps:
Step 1: For each interesting value w, we applied a standard association rule mining method like Apriori [36] to the set Sw of data instances linking to w to mine the rules related to w and with support on Sw ≥ the minimum commonality threshold mmin. These rules have commonality ≥mmin on the set Sall of all data instances. As Sw is much smaller than Sall, mining these rules from Sw is much more efficient than first applying the association rule mining method to Sall to obtain the rules with support on Sall ≥mmin×|Sw|/|Sall|, and then filtering out those rules unrelated to w. Here, |S| denotes the cardinality of set S.
Step 2: For each rule mined from Sw, we compute its confidence on Sall. We keep it only if its confidence on Sall is ≥ the minimum confidence threshold cmin.
Techniques for Efficiently Mining the Association Rules Whose Commonality is ≥mmin, Computing Their Confidence, and Eliminating Those Rules Whose Confidence is <cmin in the Extended Automatic Explanation Method
When the set Sall of all data instances includes many data instances and features, we often find that the set Sw of data instances linking to an interesting value w contains many data instances, and the first model adopts many features. Without limiting the number of data instances in Sw and the number of features, numerous (eg, several billion) association rules would be mined from Sw in Step 1. This makes the computer easily run out of memory and the rule mining process extremely slow. In addition, many rules will be produced at the end, making it difficult for clinicians to examine them. To address this issue, we can use one or more of the following approaches:
We take a random sample of data instances Ssample from Sall and use Ssample rather than Sall to mine the rules [39].
Before the rule mining process starts, each data instance is transformed into a transaction. To reduce its size, we remove from the transaction those values and value ranges that the clinician in the automatic explanation function’s design team marks as not allowed to show up in any of the rules.
Instead of using all of the features adopted by the first model, we use only the top features to mine the rules. Usually, the top features contain most of the predictive power possessed by all features adopted by the first model [23]. If the machine learning algorithm used to build the first model is like XGBoost [24] or random forest, which automatically computes each feature’s importance value, the top features are those with the highest importance values. Otherwise, if the machine learning algorithm used to build the first model does not automatically compute each feature’s importance value, we can use an automatic feature selection method [40] such as the information gain method to choose the top features. Alternatively, we can use XGBoost or random forest to construct a model, automatically compute each feature’s importance value, and choose the top features with the highest importance values.
In the following, we focus on the case of using the set Sall of all data instances to mine the association rules. The case of using a random sample of data instances Ssample from Sall to mine the rules can be handled in a similar way. To compute the rules’ confidence values, we transformed Sall to the matrix format, with each row of the matrix linking to a distinct data instance and each column of the matrix linking to a distinct value or value range of a feature. For medical data, the matrix is often not very sparse. In this case, we can use a separate bitmap to represent each column of the matrix in a condensed manner. For each rule l: q1 AND q2 AND ... AND qn→w, we performed efficient bitmap operations to pinpoint the data instances satisfying q1, q2, ..., and qn and needed for computing l’s confidence.
Among all the mined association rules related to an interesting value w, we needed to identify those whose confidence on the set Sall of all data instances is ≥ the minimum confidence threshold cmin. To expedite the identification process, we proceeded as follows: for each rule l: q1 AND q2 AND ... AND qn→w, let lw denote the number of data instances satisfying q1, q2, ..., and qn and linking to w, and l¬w denote the number of data instances satisfying q1, q2, ..., and qn and not linking to w. Our key insight was that l’s confidence on Sall lw/(lw+l¬w) is <cmin if and only if l¬w is >Tl
lw/(lw+l¬w) is <cmin if and only if l¬w is >Tl lw×(1-cmin)/cmin. We partitioned Sall into 2 subsets: Sw containing all of the data instances linking to w and S¬w containing all of the data instances not linking to w. Using the bitmap method mentioned above, we went over all of the data instances in Sw to compute lw. Then, we went over the data instances in S¬w one by one to count the data instances satisfying q1, q2, ..., and qn and not linking to w. Once this count is >Tl, we know l’s confidence on Sall is <cmin, stop the counting process, and drop l. This saves the overhead of going through the remaining data instances in S¬w to compute l¬w. Otherwise, if this count is ≤Tl when we reach the last data instance in S¬w, we keep l, obtain l¬w, and compute l’s confidence on Sall, which must be ≥cmin.
lw×(1-cmin)/cmin. We partitioned Sall into 2 subsets: Sw containing all of the data instances linking to w and S¬w containing all of the data instances not linking to w. Using the bitmap method mentioned above, we went over all of the data instances in Sw to compute lw. Then, we went over the data instances in S¬w one by one to count the data instances satisfying q1, q2, ..., and qn and not linking to w. Once this count is >Tl, we know l’s confidence on Sall is <cmin, stop the counting process, and drop l. This saves the overhead of going through the remaining data instances in S¬w to compute l¬w. Otherwise, if this count is ≤Tl when we reach the last data instance in S¬w, we keep l, obtain l¬w, and compute l’s confidence on Sall, which must be ≥cmin.
Computer Coding Implementation
We implemented our extended automatic explanation method in computer code, using a hybrid of the C and R programming languages. As R is an interpreted language and inefficient at handling certain operations on large data sets, we wrote several parts of our code in C to improve our code’s execution speed. Considering that our asthma outcome variable is hard to predict, we limited the association rules to have at most 5 items on their left-hand sides (see the guideline in the Discussion section). We set the minimum confidence threshold cmin to 50% and the minimum commonality threshold mmin to 0.2%.
Data Analysis
The Training and Test Set Partitioning
As outcomes came from the subsequent year, our data set included 13 years of effective data (2005-2017) during the 14 years between 2005 and 2018. To mirror the practical use of our XGBoost model and our extended automatic explanation method, the 2005 to 2016 data were used as the training set to train our XGBoost model and mine the association rules used by our extended method. The 2017 data were used as the test set to evaluate the performance of our XGBoost model and extended method. We used the full set of 142 features to make predictions and the top 50 features that our XGBoost model [23] ranked with the highest importance values to mine the association rules. Our XGBoost model reached an AUC of 0.859 using the full set of 142 features [23] and an AUC of 0.857 using the top 50 features.
Presenting 5 Example Association Rules Used in the Second Model
To give the reader a concrete feeling of the association rules used in the second model, we randomly chose 5 example rules to present in this paper.
Performance Metrics
We evaluated the performance of our extended automatic explanation method in several ways. The main performance metric that we used to show our extended method’s explanation capability was the percentage of patients for whom our extended method could provide explanations among the patients with asthma whom our XGBoost model correctly predicted to incur asthma hospital visits in the subsequent year. We reported both the average number of rules and the average number of actionable rules fitting such a patient. A rule fits a patient if the patient fulfills all of the items on its left-hand side.
As shown in our previous study [27], multiple rules fitting a patient frequently differ from each other by a single feature-value pair item on their left-hand sides. When many rules fit a patient, the amount of nonredundant information embedded in them is often much less than the number of these rules. To give a full picture of the information richness of the automatic explanations provided for the patients, we present 3 distributions of the patients with asthma whom our XGBoost model correctly predicted to incur asthma hospital visits in the subsequent year: (1) by the number of rules fitting a patient, (2) by the number of actionable rules fitting a patient, and (3) by the number of distinct actionable items appearing in all of the rules fitting a patient.
Results
Our Patient Cohort’s Demographic and Clinical Characteristics
Every data instance targets a distinct pair of a patient with asthma and a year. Table 1 lists the demographic and clinical characteristics of our patient cohort between 2005 and 2016, which included 182,245 patients. Table 2 lists the demographic and clinical characteristics of our patient cohort in 2017, which included 19,256 patients. These 2 sets of characteristics are reasonably similar. Between 2005 and 2016, 3.59% (11,332/315,308) of data instances were related to asthma hospital visits in the subsequent year. In 2017, this percentage was 4.22% (812/19,256).
Table 1.
Demographic and clinical characteristics of the Intermountain Healthcare patients with asthma between 2005 and 2016.
| Characteristics | Data instances related to no asthma hospital visit in the subsequent year (n=303,976), n (%) | Data instances related to asthma hospital visits in the subsequent year (n=11,332), n (%) | Data instances (n=315,308), n (%) | ||||
| Gender | |||||||
|
|
Female | 181,928 (59.85) | 6163 (54.39) | 188,091 (59.65) | |||
|
|
Male | 122,048 (40.15) | 5169 (45.61) | 127,217 (40.35) | |||
| Age (years) | |||||||
|
|
≥65 | 46,260 (15.22) | 621 (5.48) | 46,881 (14.87) | |||
|
|
18 to 65 | 172,436 (56.73) | 5003 (44.15) | 177,439 (56.27) | |||
|
|
6 to <18 | 50,572 (16.64) | 2590 (22.86) | 53,162 (16.86) | |||
|
|
<6 | 34,708 (11.42) | 3118 (27.52) | 37,826 (12.00) | |||
| Ethnicity | |||||||
|
|
Non-Hispanic | 244,442 (80.41) | 8157 (71.98) | 252,599 (80.11) | |||
|
|
Hispanic | 27,014 (8.89) | 2279 (20.11) | 29,293 (9.29) | |||
|
|
Unknown or not reported | 32,520 (10.70) | 896 (7.91) | 33,416 (10.60) | |||
| Race | |||||||
|
|
White | 273,206 (89.88) | 9420 (83.13) | 282,626 (89.63) | |||
|
|
Native Hawaiian or other Pacific Islander | 3877 (1.28) | 411 (3.63) | 4288 (1.36) | |||
|
|
Black or African American | 5291 (1.74) | 460 (4.06) | 5751 (1.82) | |||
|
|
Asian | 2120 (0.70) | 77 (0.68) | 2197 (0.70) | |||
|
|
American Indian or Alaska Native | 2295 (0.76) | 214 (1.89) | 2509 (0.80) | |||
|
|
Unknown or not reported | 17,187 (5.65) | 750 (6.62) | 17,937 (5.69) | |||
| Duration of asthma (years) | |||||||
|
|
>3 | 76,810 (25.27) | 3666 (32.35) | 80,476 (25.52) | |||
|
|
≤3 | 227,166 (74.73) | 7666 (67.65) | 234,832 (74.48) | |||
| Insurance | |||||||
|
|
Self-paid or charity | 26,611 (8.75) | 1902 (16.78) | 28,513 (9.04) | |||
|
|
Public | 76,916 (25.30) | 3238 (28.57) | 80,154 (25.42) | |||
|
|
Private | 200,449 (65.94) | 6192 (54.64) | 206,641 (65.54) | |||
| Smoking status | |||||||
|
|
Never smoker or unknown | 251,501 (82.74) | 8952 (79.00) | 260,453 (82.60) | |||
|
|
Former smoker | 18,735 (6.16) | 569 (5.02) | 19,304 (6.12) | |||
|
|
Current smoker | 33,740 (11.10) | 1811 (15.98) | 35,551 (11.28) | |||
| Comorbidity | |||||||
|
|
Sleep apnea | 20,421 (6.72) | 471 (4.16) | 20,892 (6.63) | |||
|
|
Sinusitis | 14,164 (4.66) | 592 (5.22) | 14,756 (4.68) | |||
|
|
Premature birth | 5102 (1.68) | 440 (3.88) | 5542 (1.76) | |||
|
|
Obesity | 35,215 (11.58) | 1076 (9.50) | 36,291 (11.51) | |||
|
|
Gastroesophageal reflux | 54,887 (18.06) | 1309 (11.55) | 56,196 (17.82) | |||
|
|
Eczema | 4484 (1.48) | 443 (3.91) | 4927 (1.56) | |||
|
|
Cystic fibrosis | 447 (0.15) | 11 (0.10) | 458 (0.15) | |||
|
|
Chronic obstructive pulmonary disease | 12,496 (4.11) | 391 (3.45) | 12,887 (4.09) | |||
|
|
Bronchopulmonary dysplasia | 394 (0.13) | 35 (0.31) | 429 (0.14) | |||
|
|
Anxiety or depression | 55,245 (18.17) | 1716 (15.14) | 56,961 (18.07) | |||
|
|
Allergic rhinitis | 4534 (1.49) | 181 (1.60) | 4715 (1.50) | |||
| Asthma medication prescription | |||||||
|
|
Systemic corticosteroid | 129,318 (42.54) | 7324 (64.63) | 136,642 (43.34) | |||
|
|
Short-acting, inhaled beta-2 agonist | 121,983 (40.13) | 7545 (66.58) | 129,528 (41.08) | |||
|
|
Mast cell stabilizer | 114 (0.04) | 7 (0.06) | 121 (0.04) | |||
|
|
Long-acting beta-2 agonist | 1744 (0.57) | 69 (0.61) | 1813 (0.58) | |||
|
|
Leukotriene modifier | 33,187 (10.92) | 2320 (20.47) | 35,507 (11.26) | |||
|
|
Inhaled corticosteroid/long-acting beta-2 agonist combination | 42,796 (14.08) | 2196 (19.38) | 44,992 (14.27) | |||
|
|
Inhaled corticosteroid | 73,566 (24.20) | 4539 (40.05) | 78,105 (24.77) | |||
Table 2.
Demographic and clinical characteristics of the Intermountain Healthcare patients with asthma in 2017.
| Characteristics | Data instances related to no asthma hospital visit in the subsequent year (n=18,444), n (%) | Data instances related to asthma hospital visits in the subsequent year (n=812), n (%) | Data instances (n=19,256), n (%) | |
| Gender | ||||
|
|
Female | 11,001 (59.65) | 439 (54.06) | 11,440 (59.41) |
|
|
Male | 7443 (40.35) | 373 (45.94) | 7816 (40.59) |
| Age (years) | ||||
|
|
≥65 | 3833 (20.78) | 46 (5.67) | 3879 (20.14) |
|
|
18 to 65 | 9879 (53.56) | 386 (47.54) | 10,265 (53.31) |
|
|
6 to <18 | 3054 (16.56) | 181 (22.29) | 3235 (16.80) |
|
|
<6 | 1678 (9.10) | 199 (24.51) | 1877 (9.75) |
| Ethnicity | ||||
|
|
Non-Hispanic | 16,242 (88.06) | 618 (76.11) | 16,860 (87.56) |
|
|
Hispanic | 2020 (10.95) | 192 (23.65) | 2212 (11.49) |
|
|
Unknown or not reported | 182 (0.99) | 2 (0.25) | 184 (0.96) |
| Race | ||||
|
|
White | 17,025 (92.31) | 681 (83.87) | 17,706 (91.95) |
|
|
Native Hawaiian or other Pacific Islander | 299 (1.62) | 47 (5.79) | 346 (1.80) |
|
|
Black or African American | 361 (1.96) | 42 (5.17) | 403 (2.09) |
|
|
Asian | 195 (1.06) | 10 (1.23) | 205 (1.06) |
|
|
American Indian or Alaska Native | 146 (0.79) | 13 (1.60) | 159 (0.83) |
|
|
Unknown or not reported | 418 (2.27) | 19 (2.34) | 437 (2.27) |
| Duration of asthma (years) | ||||
|
|
>3 | 7734 (41.93) | 389 (47.91) | 8123 (42.18) |
|
|
≤3 | 10,710 (58.07) | 423 (52.09) | 11,133 (57.82) |
| Insurance | ||||
|
|
Self-paid or charity | 1136 (6.16) | 142 (17.49) | 1278 (6.64) |
|
|
Public | 4920 (26.68) | 208 (25.62) | 5128 (26.63) |
|
|
Private | 12,388 (67.17) | 462 (56.90) | 12,850 (66.73) |
| Smoking status | ||||
|
|
Never smoker or unknown | 13,956 (75.67) | 583 (71.80) | 14,539 (75.50) |
|
|
Former smoker | 2243 (12.16) | 83 (10.22) | 2326 (12.08) |
|
|
Current smoker | 2245 (12.17) | 146 (17.98) | 2391 (12.42) |
| Comorbidity | ||||
|
|
Sleep apnea | 2925 (15.86) | 78 (9.61) | 3003 (15.60) |
|
|
Sinusitis | 746 (4.04) | 34 (4.19) | 780 (4.05) |
|
|
Premature birth | 435 (2.36) | 41 (5.05) | 476 (2.47) |
|
|
Obesity | 3389 (18.37) | 116 (14.29) | 3505 (18.20) |
|
|
Gastroesophageal reflux | 3477 (18.85) | 71 (8.74) | 3548 (18.43) |
|
|
Eczema | 273 (1.48) | 34 (4.19) | 307 (1.59) |
|
|
Cystic fibrosis | 94 (0.51) | 1 (0.12) | 95 (0.49) |
|
|
Chronic obstructive pulmonary disease | 1033 (5.60) | 23 (2.83) | 1056 (5.48) |
|
|
Bronchopulmonary dysplasia | 12 (0.07) | 3 (0.37) | 15 (0.08) |
|
|
Anxiety or depression | 3815 (20.68) | 131 (16.13) | 3946 (20.49) |
|
|
Allergic rhinitis | 382 (2.07) | 10 (1.23) | 392 (2.04) |
| Asthma medication prescription | ||||
|
|
Systemic corticosteroid | 11,327 (61.41) | 693 (85.34) | 12,020 (62.42) |
|
|
Short-acting, inhaled beta-2 agonist | 13,046 (70.73) | 739 (91.01) | 13,785 (71.59) |
|
|
Mast cell stabilizer | 8 (0.04) | 0 (0.00) | 8 (0.04) |
|
|
Long-acting beta-2 agonist | 47 (0.25) | 5 (0.62) | 52 (0.27) |
|
|
Leukotriene modifier | 3364 (18.24) | 209 (25.74) | 3573 (18.56) |
|
|
Inhaled corticosteroid/long-acting beta-2 agonist combination | 4178 (22.65) | 222 (27.34) | 4400 (22.85) |
|
|
Inhaled corticosteroid | 6817 (36.96) | 424 (52.22) | 7241 (37.60) |
For each demographic or clinical characteristic, Table 3 presents the statistical test results on whether the data instances related to asthma hospital visits in the subsequent year and those related to no asthma hospital visit in the subsequent year had the same distribution. When the P value was ≥.05, the 2 sets of data instances had the same distribution. Otherwise, they had different distributions. All P values <.05 are shown in italics in Table 3.
Table 3.
For each demographic or clinical characteristic, the statistical test results on whether the data instances related to asthma hospital visits in the subsequent year and those related to no asthma hospital visit in the subsequent year had the same distribution.
| Characteristics | P value for the 2005-2016 data | P value for the 2017 data | |
| Gender | <.001 a, b | .002 a | |
| Age (years) | <.001 c | <.001 c | |
| Ethnicity | <.001 a | <.001 a | |
| Race | <.001 a | <.001 a | |
| Duration of asthma (years) | <.001 c | <.001 c | |
| Insurance category | <.001 a | <.001 a | |
| Smoking status | <.001 a | <.001 a | |
| Comorbidity | |||
|
|
Sleep apnea | <.001 a | <.001 a |
|
|
Sinusitis | .006 a | .91a |
|
|
Premature birth | <.001 a | <.001 a |
|
|
Obesity | <.001 a | .004 a |
|
|
Gastroesophageal reflux | <.001 a | <.001 a |
|
|
Eczema | <.001 a | <.001 a |
|
|
Cystic fibrosis | .21a | .20a |
|
|
Chronic obstructive pulmonary disease | <.001 a | <.001 a |
|
|
Bronchopulmonary dysplasia | <.001 a | .02 a |
|
|
Anxiety or depression | <.001 a | .002 a |
|
|
Allergic rhinitis | .38a | .13a |
| Asthma medication prescription | |||
|
|
Systemic corticosteroid | <.001 a | <.001 a |
|
|
Short-acting, inhaled beta-2 agonist | <.001 a | <.001 a |
|
|
Mast cell stabilizer | .29a | >.99a |
|
|
Long-acting beta-2 agonist | .67a | .11a |
|
|
Leukotriene modifier | <.001 a | <.001 a |
|
|
Inhaled corticosteroid/long-acting beta-2 agonist combination | <.001 a | .002 a |
|
|
Inhaled corticosteroid | <.001 a | <.001 a |
aP values obtained by performing the chi-square two-sample test.
bP values <.05 marked in italics.
cP values obtained by performing the Cochran-Armitage trend test [41].
The Number of Association Rules Left at Different Phases of Rule Mining and Pruning Processes
The association rules used in the second model were mined on the training set. Using the top 50 features that were ranked by our XGBoost model with the highest importance values, we obtained 559,834 association rules. Figure 1 presents the number of rules left versus the confidence difference threshold τ. Recall that each more specific rule is dropped when there exists a more general rule whose confidence is not lower by more than τ. Initially, when τ is small, the number of rules left decreases quickly as τ increases. Once τ becomes 0.15 or larger, the number of rules left approaches an asymptote. Accordingly, in our computer coding implementation, we set τ to 0.15, resulting in 132,816 remaining rules.
Figure 1.
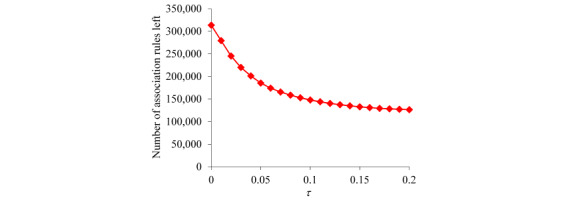
The number of association rules left versus τ.
A clinical expert on asthma (MJ) in our team marked the values and value ranges of the top 50 features that could possibly have a positive correlation with future asthma hospital visits. After dropping the rules including any other value or value range, 124,506 rules were left. Each rule explains why a patient is predicted to incur one or more asthma hospital visits in the subsequent year. Almost all (124,502/124,506, 100.00%) of these rules were actionable. The left-hand sides of these rules contain various combinations of 208 distinct items related to 50 features.
Example Association Rules in the Second Model
Table 4 presents 5 sample association rules randomly chosen from the 124,502 actionable rules used in the second model.
Table 4.
Five sample association rules.
| Item on the left-hand side of the rule | Implication of the item | Intervention compiled for the item | |
| Rule 1: The patient had ≥12 EDa visits in the past year AND the patient had ≥21 distinct medications in all of the asthma medication orders in the past year → the patient will incur one or more asthma hospital visits in the subsequent year. | |||
|
|
The patient had ≥12 ED visits in the past year | Having many ED visits reflects poor asthma control | Implement control strategies to avoid the need for emergency care |
|
|
The patient had ≥21 distinct medications in all of the asthma medication orders in the past year | Using many asthma medications reflects poor asthma control | Tailor prescribed asthma medications and help the patient maximize asthma control medication adherence |
| Rule 2: The patient had ≥9 distinct asthma medication prescribers in the past year AND the block group where the patient lives has a national health literacy score [42] ≤244 AND the patient had ≥21 distinct medications in all of the asthma medication orders in the past year → the patient will incur one or more asthma hospital visits in the subsequent year. | |||
|
|
The patient had ≥9 distinct asthma medication prescribers in the past year | Having many asthma medication prescribers reflects poor care continuity, which often leads to poor outcomes | Provide the patient with social resources to address social chaos that leads to ineffective access to health care |
|
|
The block group where the patient lives has a national health literacy score ≤244 | Having low health literacy is correlated with poor outcomes | Improve education access in the area where the patient lives to help increase health literacy |
| Rule 3: The patient had a total of ≥25 units of systemic corticosteroids ordered in the past year AND the patient had ≥12 ED visits in the past year AND the patient is Hispanic → the patient will incur one or more asthma hospital visits in the subsequent year. | |||
|
|
The patient had a total of ≥25 units of systemic corticosteroids ordered in the past year | Systemic corticosteroids are one type of asthma medication intended for short-term use to relieve acute asthma exacerbations. Using a lot of systemic corticosteroids reflects poor asthma control | Tailor prescribed asthma medications and help the patient maximize asthma control medication adherence |
|
|
The patient is Hispanic | In the US, Hispanic people have a disproportionately high rate of poor asthma outcomes | —b |
| Rule 4: The patient had ≥4 major visits for asthma in the past year AND the patient is between 11 and 35 years old AND the patient had no outpatient visit in the past year AND the average length of an inpatient stay of the patient in the past year is >1.75 and ≤2.95 days → the patient will incur one or more asthma hospital visits in the subsequent year. | |||
|
|
The patient had ≥4 major visits for asthma in the past year | As defined in our paper [23], a major visit for asthma is an inpatient stay or ED visit having an asthma diagnosis code, or an outpatient visit having a primary diagnosis of asthma. Intuitively, all else being equal, a patient having major visits for asthma has a higher likelihood of incurring future asthma hospital visits than a patient having only outpatient visits with asthma as a secondary diagnosis | Implement control strategies to avoid the need for emergency care |
|
|
The average length of an inpatient stay of the patient in the past year is >1.75 and ≤2.95 days | Having inpatient stays reflects poor asthma control | Implement control strategies to avoid the need for emergency care |
|
|
The patient had no outpatient visit in the past year | For good asthma management, a patient with asthma is supposed to see the primary care provider regularly. Having no outpatient visit often implies that the patient has no primary care provider | Help the patient obtain a primary care provider if the patient does not already have one |
| Rule 5: The patient had ≥4 major visits for asthma in the past year AND the patient's last ED visit is within the last 49 days AND the patient had between 6 and 8 distinct asthma medication prescribers in the past year AND the patient had a total of ≥36 units of asthma medications ordered in the past year AND >23.7% and ≤33.3% of families in the block group where the patient lives are below 150% of the federal poverty level → the patient will incur one or more asthma hospital visits in the subsequent year. | |||
|
|
The patient’s last ED visit is within the last 49 days | Having a recent ED visit reflects poor asthma control | Implement control strategies to avoid the need for emergency care |
|
|
The patient had a total of ≥36 units of asthma medications ordered in the past year | Taking many asthma medications reflects poor asthma control | Tailor prescribed asthma medications and help the patient maximize asthma control medication adherence |
|
|
>23.7% and ≤33.3% of families in the block group where the patient lives are below 150% of the federal poverty level | Poverty correlates with poor outcomes | Provide living wage programs in the area where the patient lives to increase resources for health care |
aED: emergency department.
bNot applicable.
Performance Measures Reached by the Extended Automatic Explanation Method
Our extended automatic explanation method was assessed on the test set. This method explained the prediction results for 92.4% (182/197) of the adults with asthma (age ≥18 years) and 87.5% (209/239) of the children with asthma (age <18 years) for whom our XGBoost model correctly predicted the occurrence of asthma hospital visits in the subsequent year. Combined, our extended method explained the prediction results for 89.7% (391/436) of the patients with asthma whom our XGBoost model correctly predicted to incur asthma hospital visits in the subsequent year. For each such patient, our extended method offered an average of 974.01 (SD 1600.48) explanations, 974.00 (SD 1600.47) of which were actionable. Each explanation came from 1 rule. When confined to using actionable rules, our extended method explained the prediction results for 89.7% (391/436) of the patients with asthma for whom our XGBoost model correctly predicted the occurrence of asthma hospital visits in the subsequent year.
For the patients for whom our extended automatic explanation method could offer explanations of our XGBoost model’s correct prediction results of incurring asthma hospital visits in the subsequent year, the average number of distinct actionable items appearing in all of the rules fitting a patient was 21.50 (SD 8.71). This number is much less than 974.01, the average number of actionable rules fitting such a patient.
For the patients with asthma whom our XGBoost model correctly predicted to incur asthma hospital visits in the subsequent year, Figure 2 shows the distribution of patients by the number of rules fitting a patient. This distribution has a long tail and is highly skewed toward the left. As the number of rules fitting a patient becomes larger, the number of patients to each of whom this number of rules apply is inclined to drop nonmonotonically. The largest number of rules fitting a patient is high, 9223, although only 1 patient fits such a high number of rules.
Figure 2.
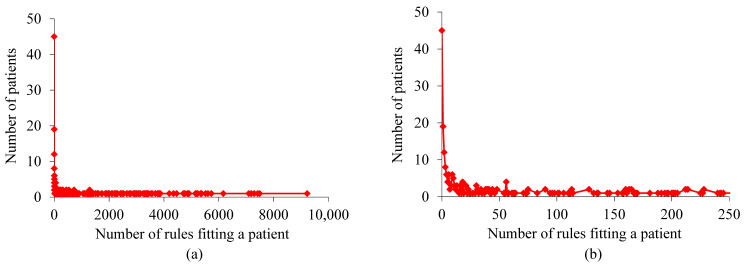
Distribution of patients by the number of rules fitting a patient for the patients with asthma whom our extreme gradient boosting model correctly predicted to incur asthma hospital visits in the subsequent year. (a) When no limit is placed on the number of rules fitting a patient. (b) When the number of rules fitting a patient is ≤250.
For the patients with asthma whom our XGBoost model correctly predicted to incur asthma hospital visits in the subsequent year, Figure 3 shows the distribution of patients by the number of actionable rules fitting a patient. This distribution is similar to that shown in Figure 2. The largest number of actionable rules fitting a patient is high, 9223, although only 1 patient fits such a high number of actionable rules.
Figure 3.
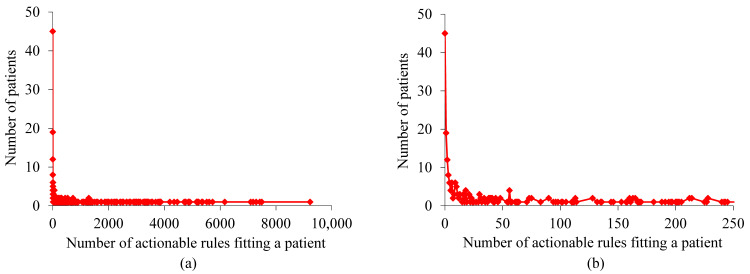
Distribution of patients by the number of actionable rules fitting a patient for the patients with asthma whom our extreme gradient boosting model correctly predicted to incur asthma hospital visits in the subsequent year. (a) When no limit is placed on the number of actionable rules fitting a patient. (b) When the number of actionable rules fitting a patient is ≤250.
For the patients with asthma whom our XGBoost model correctly predicted to incur asthma hospital visits in the subsequent year, Figure 4 exhibits the distribution of patients by the number of distinct actionable items appearing in all of the rules fitting a patient. The largest number of distinct actionable items appearing in all of the rules fitting a patient is 35, much smaller than the largest number of (actionable) rules fitting a patient. Frequently, 2 or more actionable items appearing in the rules fitting a patient link to the same set of interventions. For example, the intervention of tailoring prescribed asthma medications and helping the patient maximize asthma control medication adherence links to several value ranges of multiple medication-related features.
Figure 4.
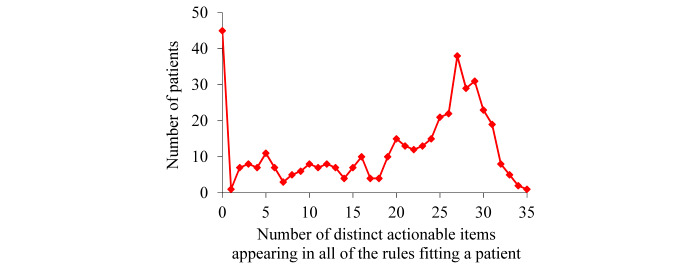
Distribution of patients by the number of distinct actionable items appearing in all of the rules fitting a patient for the patients with asthma whom our extreme gradient boosting model correctly predicted to incur asthma hospital visits in the subsequent year.
Our extended automatic explanation method could offer explanations for 69.2% (562/812) of patients with asthma who will incur asthma hospital visits in the subsequent year.
To evaluate the generalizability of our extended automatic explanation method for predicting asthma hospital visits, we tested our method on the University of Washington Medicine data and Kaiser Permanente Southern California data. The results we obtained are similar to the abovementioned results and are detailed in 2 separate papers [43,44].
Discussion
Principal Findings
We developed a method to automatically offer rule-formed explanations for any machine learning model’s prediction results on imbalanced tabular data without lowering the performance measures of the model. We showed that this method explained the prediction results for 89.7% (391/436) of the patients with asthma whom our XGBoost model correctly predicted to incur asthma hospital visits in the subsequent year. This percentage is high enough for routine clinical use of this method. After further improvement of its accuracy, our asthma outcome prediction model coupled with the automatic explanation function could be used for decision support to guide the allocation of limited asthma care management resources. This could help boost asthma outcomes and reduce resource use and costs.
Our extended automatic explanation method could offer explanations for 69.2% (562/812) of the patients with asthma who will incur asthma hospital visits in the subsequent year. This percentage is smaller than the success rate of 89.7% (391/436) for our extended automatic explanation method to explain the correct prediction results of our XGBoost model of incurring asthma hospital visits in the subsequent year. One possible reason is that the prediction results of the association rules are correlated with the prediction results of our XGBoost model. Among the patients with asthma who will incur asthma hospital visits in the subsequent year and on whom our XGBoost model gave incorrect predictions, many are difficult cases for any model to correctly predict or explain their outcomes. Among the patients with asthma whom our XGBoost model correctly predicted to incur asthma hospital visits in the subsequent year, many are easy cases for using association rules to explain the outcomes of these cases.
Asthma in adults differs from asthma in children. As shown in a previous study [23], the AUC of our XGBoost model for adults with asthma was 0.034 higher than that for children with asthma, that is, the outcome is easier to predict for adults with asthma than for children with asthma. Intuitively, the degree of difficulty in predicting the outcome is positively correlated with that of using association rules to explain the prediction results of the model, as each rule is a small predictive model. Hence, our extended automatic explanation method explained the prediction results for a larger portion of the adults with asthma than the children with asthma for whom our XGBoost model correctly predicted the occurrence of asthma hospital visits in the subsequent year.
A Guideline for Setting the Values of the Parameters Used in Our Extended Automatic Explanation Method
Our extended automatic explanation method has 4 parameters: the maximum number of items lmax allowed on the left-hand side of an association rule, the minimum commonality threshold mmin, the minimum confidence threshold cmin, and the confidence difference threshold τ. These parameters significantly affect the performance of the method. Our previous papers [31,32] outlined the method but gave no guideline for setting the values of these parameters. We offer such a guideline here.
The maximum number of items lmax allowed on the left-hand side of an association rule is usually small, as long rules are difficult to understand [35]. Our previous study [27] showed that for an outcome variable that is relatively easy to predict, an lmax of 4 works well for automatic explanation. When the outcome variable is hard to predict, we can increase lmax slightly to a number such as 5. Without making the rules too complex to understand, this helps ensure that the second model can provide explanations for a large portion of the data instances that the first model correctly predicts to take one of the interesting values of the outcome variable.
In the original paper [38] that proposed the concept of commonality for class-based association rules, mined rules were used to build a classifier. To maximize the accuracy of the classifier, the minimum commonality threshold mmin was set to 14%. However, this value is too high for automatic explanation. With such a high value, we cannot obtain enough rules for the outcome variable’s rare values. Consequently, for a large portion of the first model’s prediction results on these values, we cannot give any explanation. In addition, the mined rules tend to be too general and have low confidence, causing the users of the automatic explanation function to have little trust in the automatically generated explanations. To avoid these problems, for automatic explanation, we recommend setting mmin to a value much smaller than 14%. More specifically, our paper [27] showed that on reasonably balanced data, a minimum support threshold smin of 1% and a minimum confidence threshold cmin of 50% work well for automatic explanation. By definition, commonality is a value-specific support. Thus, we would expect mmin and smin to have relatively similar optimal values. Accordingly, we set mmin to a value close to 1% and cmin to a value close to 50%. Although a value close to 50% may not seem so high, it is already much larger than the percentage of data instances linking to an interesting value of the outcome variable. For instance, in our case of predicting asthma hospital visits in patients with asthma, this percentage is 4% [23]. Moreover, a value close to 50% is also much larger than our XGBoost model’s positive predictive value of 22.65%. The concrete values of mmin and cmin depend on the data set and are chosen to meet 2 goals simultaneously and as much as possible. First, the second model can provide explanations for a large portion of the data instances that the first model correctly predicts to take one of the interesting values of the outcome variable. Often, the harder the outcome variable is to predict, the smaller mmin and cmin need to be to meet this goal. Second, cmin is high enough for users of the automatic explanation function to trust the automatically generated explanations.
Recall that during the rule-pruning process, each more specific rule is dropped when there is a more general rule whose confidence is not lower by more than the confidence difference threshold τ. To determine the value of τ, we plot the number of rules left versus τ. As our previous paper [27] shows, initially when τ is small, the number of rules left decreases quickly as τ increases. Once τ becomes sufficiently large, the number of rules left approaches an asymptote. This is the place to set the value of τ to strike a balance between cutting the number of rules and retaining high-confidence rules.
Five Clarifications on Using the Automatic Explanation Function
In practice, our automatic explanation method could produce a paradox. Two patients both fulfilled the left-hand side of the same rule linking to a poor outcome. The first model correctly predicts one of them to have a poor outcome. The automatic explanation function displays the rule to explain this prediction result. Simultaneously, the first model correctly predicts a good outcome on the other patient, for whom the automatic explanation function shows nothing. In this case, one should not think that the automatic explanation function acts incorrectly because it behaves differently in these 2 patients; rather, this difference occurs because the second patient fulfills some items that are not in the rule. These items counter the risk induced by those on the rule’s left-hand side and reduce the second patient’s risk of having a poor outcome to a low level.
When using the automatic explanation function, one needs to remember that the function is intended to serve as a reminder system for decision support rather than a replacement for clinical judgment. The function is used to help the user quickly identify some reasons why a patient is predicted to have a poor outcome and some tailored interventions suitable for the patient. If successful, this helps the clinical user avoid substantial time laboriously reviewing the records of the patient to assess risk factors and devise interventions. This also helps reduce the number of interventions that are suitable for the patient, but the user forgets to consider. In the end, it is still the user who uses his or her own judgment to decide whether to use the prediction result of the first model and apply suggested interventions to the patient. If there is doubt about the appropriateness of the output of the function, the clinical user can always check the records of the patient to resolve the doubt before making the final decisions with the patient.
Different health care systems have different properties and practice patterns. Consequently, the association rules mined from the data of one health care system may or may not directly apply to or work well for another health care system. However, our automatic explanation method is general. It relies on no special property of a specific disease, patient cohort, prediction target, or health care system and can be applied to various predictive modeling problems and health care systems [27,29,30,43,44], regardless of whether the rules mined from the data of 1 health care system generalize to the data of another health care system. For any health care system, we would recommend mining rules from its own data whenever possible, rather than reusing the rules mined from the data of another health care system.
In our test case, the second model contained 124,506 association rules. The left-hand sides of these rules contain various combinations of 208 distinct items related to 50 features. Within 1 day, a clinician in our team (MJ) finished manually compiling the 2 types of knowledge needed by the automatic explanation function: the values and value ranges of the top 50 features that could possibly have a positive correlation with future asthma hospital visits and the interventions for the actionable items. The amount of time needed to perform this manual compilation is moderate and acceptable to the clinicians in our team.
Although many association rules could fit a patient, the total number of distinct items included on their left-hand sides is not large: at most 35. To avoid overwhelming the automatic explanation function’s user, we can use the rule diversification method in our paper [27] to rank these rules. The top few rules are likely to contain nonredundant information and are displayed by default.
Related Work
As described in a survey paper [33] and a book [34], other researchers previously proposed various methods for automatically explaining the prediction results of machine learning models. These methods often lower the performance measures of the model by replacing the original model with a less accurate model and usually give nonrule-formed explanations. Many of these methods work for only a specific machine learning algorithm rather than for all algorithms. Moreover, none of these methods can automatically recommend tailored interventions. In comparison, our extended automatic explanation method not only offers rule-formed explanations for the prediction results of any machine learning model on tabular data but also recommends tailored interventions without lowering the performance measures of the model [27]. Compared with nonrule-formed explanations, rule-formed explanations are easier to comprehend and can more directly recommend tailored interventions.
Hatwell et al [45] proposed a method to automatically provide rule-formed explanations for the prediction results of an AdaBoost model. This method does not work for non-AdaBoost machine learning algorithms. The rules are unknown before the prediction time and hence cannot be used to automatically recommend tailored interventions at prediction time. In comparison, the rules used in our extended automatic explanation method are precompiled beforehand and used to automatically recommend tailored interventions at prediction time.
Limitations
This study has 2 limitations that give interesting directions for future work:
Our data set contained no information on health care use of the patients outside of Intermountain Healthcare. Consequently, the features were computed using incomplete clinical and administrative data [46-49]. In addition, the prediction target was limited to asthma hospital visits at Intermountain Healthcare rather than asthma hospital visits anywhere. It would be interesting to see how the automatically generated explanations of the prediction results of the model would differ if we have access to more complete clinical and administrative data [50].
Our study used 1 predictive modeling problem, predicting asthma hospital visits as the test case. Although our original automatic explanation method [27] has been successfully applied to several predictive modeling problems [29,30], the generalizability of our extended automatic explanation method to other predictive modeling problems beyond predicting asthma hospital visits has not been evaluated. Conducting such evaluations would help inform the utility and refine the implementation of our extended method.
Conclusions
Using asthma outcome prediction as a demonstration case, this study shows for the first time the feasibility of automatically offering rule-formed explanations for the prediction results of any machine learning model on imbalanced tabular data without lowering the performance measures of the model. After further improvement, our asthma outcome prediction model coupled with the automatic explanation function could be used for decision support to guide the allocation of limited asthma care management resources. This could simultaneously help improve asthma outcomes and reduce resource use and cost.
Acknowledgments
The authors would like to thank Katherine A Sward for the useful discussions. GL, MJ, FN, SH, and BS were partially supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health under Award Number R01HL142503. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- AUC
area under the receiver operating characteristic curve
- ED
emergency department
- ICD-9
International Classification of Diseases, ninth revision
- ICD-10
International Classification of Diseases, tenth revision
- XGBoost
extreme gradient boosting
Footnotes
Authors' Contributions: GL was responsible for the paper. GL conceptualized and designed the study, performed the literature review and data analysis, and wrote the paper. MJ, FN, and BS provided feedback on various medical issues, contributed to conceptualizing the presentation, and revised the paper. SH took part in retrieving the Intermountain Healthcare data set and interpreting its detected peculiarities.
Conflicts of Interest: None declared.
References
- 1.Moorman JE, Akinbami LJ, Bailey CM, Zahran HS, King ME, Johnson CA, Liu X. National surveillance of asthma: United States, 2001-2010. Vital Health Stat 3. 2012 Nov;(35):1–58. [PubMed] [Google Scholar]
- 2.Nurmagambetov T, Kuwahara R, Garbe P. The economic burden of asthma in the United States, 2008-2013. Ann Am Thorac Soc. 2018 Mar;15(3):348–56. doi: 10.1513/AnnalsATS.201703-259OC. [DOI] [PubMed] [Google Scholar]
- 3.Mays GP, Claxton G, White J. Managed care rebound? Recent changes in health plans' cost containment strategies. Health Aff (Millwood) 2004;Suppl Web Exclusives:W4–427. doi: 10.1377/hlthaff.w4.427. [DOI] [PubMed] [Google Scholar]
- 4.Lieu TA, Quesenberry CP, Sorel ME, Mendoza GR, Leong AB. Computer-based models to identify high-risk children with asthma. Am J Respir Crit Care Med. 1998 Apr;157(4 Pt 1):1173–80. doi: 10.1164/ajrccm.157.4.9708124. [DOI] [PubMed] [Google Scholar]
- 5.Caloyeras JP, Liu H, Exum E, Broderick M, Mattke S. Managing manifest diseases, but not health risks, saved PepsiCo money over seven years. Health Aff (Millwood) 2014 Jan;33(1):124–31. doi: 10.1377/hlthaff.2013.0625. [DOI] [PubMed] [Google Scholar]
- 6.Greineder DK, Loane KC, Parks P. A randomized controlled trial of a pediatric asthma outreach program. J Allergy Clin Immunol. 1999 Mar;103(3 Pt 1):436–40. doi: 10.1016/s0091-6749(99)70468-9. [DOI] [PubMed] [Google Scholar]
- 7.Kelly CS, Morrow AL, Shults J, Nakas N, Strope GL, Adelman RD. Outcomes evaluation of a comprehensive intervention program for asthmatic children enrolled in medicaid. Pediatrics. 2000 May;105(5):1029–35. doi: 10.1542/peds.105.5.1029. [DOI] [PubMed] [Google Scholar]
- 8.Axelrod RC, Zimbro KS, Chetney RR, Sabol J, Ainsworth VJ. A disease management program utilizing life coaches for children with asthma. J Clin Outcomes Manag. 2001;8(6):38–42. [Google Scholar]
- 9.Axelrod RC, Vogel D. Predictive modeling in health plans. Dis Manag Health Out. 2003;11(12):779–87. doi: 10.2165/00115677-200311120-00003. [DOI] [Google Scholar]
- 10.Loymans RJ, Debray TP, Honkoop PJ, Termeer EH, Snoeck-Stroband JB, Schermer TR, Assendelft WJ, Timp M, Chung KF, Sousa AR, Sont JK, Sterk PJ, Reddel HK, Riet GT. Exacerbations in adults with asthma: a systematic review and external validation of prediction models. J Allergy Clin Immunol Pract. 2018;6(6):1942–1952.e15. doi: 10.1016/j.jaip.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 11.Loymans RJ, Honkoop PJ, Termeer EH, Snoeck-Stroband JB, Assendelft WJ, Schermer TR, Chung KF, Sousa AR, Sterk PJ, Reddel HK, Sont JK, Riet GT. Identifying patients at risk for severe exacerbations of asthma: development and external validation of a multivariable prediction model. Thorax. 2016 Sep;71(9):838–46. doi: 10.1136/thoraxjnl-2015-208138. [DOI] [PubMed] [Google Scholar]
- 12.Schatz M, Cook EF, Joshua A, Petitti D. Risk factors for asthma hospitalizations in a managed care organization: development of a clinical prediction rule. Am J Manag Care. 2003 Aug;9(8):538–47. https://www.ajmc.com/pubMed.php?pii=2500. [PubMed] [Google Scholar]
- 13.Eisner MD, Yegin A, Trzaskoma B. Severity of asthma score predicts clinical outcomes in patients with moderate to severe persistent asthma. Chest. 2012 Jan;141(1):58–65. doi: 10.1378/chest.11-0020. [DOI] [PubMed] [Google Scholar]
- 14.Sato R, Tomita K, Sano H, Ichihashi H, Yamagata S, Sano A, Yamagata T, Miyara T, Iwanaga T, Muraki M, Tohda Y. The strategy for predicting future exacerbation of asthma using a combination of the asthma control test and lung function test. J Asthma. 2009 Sep;46(7):677–82. doi: 10.1080/02770900902972160. [DOI] [PubMed] [Google Scholar]
- 15.Osborne ML, Pedula KL, O'Hollaren M, Ettinger KM, Stibolt T, Buist AS, Vollmer WM. Assessing future need for acute care in adult asthmatics: the Profile of Asthma Risk Study: a prospective health maintenance organization-based study. Chest. 2007 Oct;132(4):1151–61. doi: 10.1378/chest.05-3084. [DOI] [PubMed] [Google Scholar]
- 16.Miller MK, Lee JH, Blanc PD, Pasta DJ, Gujrathi S, Barron H, Wenzel SE, Weiss ST, TENOR Study Group TENOR risk score predicts healthcare in adults with severe or difficult-to-treat asthma. Eur Respir J. 2006 Dec;28(6):1145–55. doi: 10.1183/09031936.06.00145105. http://erj.ersjournals.com/cgi/pmidlookup?view=long&pmid=16870656. [DOI] [PubMed] [Google Scholar]
- 17.Peters D, Chen C, Markson LE, Allen-Ramey FC, Vollmer WM. Using an asthma control questionnaire and administrative data to predict health-care utilization. Chest. 2006 Apr;129(4):918–24. doi: 10.1378/chest.129.4.918. [DOI] [PubMed] [Google Scholar]
- 18.Yurk RA, Diette GB, Skinner EA, Dominici F, Clark RD, Steinwachs DM, Wu AW. Predicting patient-reported asthma outcomes for adults in managed care. Am J Manag Care. 2004 May;10(5):321–8. https://www.ajmc.com/pubMed.php?pii=2600. [PubMed] [Google Scholar]
- 19.Lieu TA, Capra AM, Quesenberry CP, Mendoza GR, Mazar M. Computer-based models to identify high-risk adults with asthma: is the glass half empty of half full? J Asthma. 1999 Jun;36(4):359–70. doi: 10.3109/02770909909068229. [DOI] [PubMed] [Google Scholar]
- 20.Schatz M, Nakahiro R, Jones CH, Roth RM, Joshua A, Petitti D. Asthma population management: development and validation of a practical 3-level risk stratification scheme. Am J Manag Care. 2004 Jan;10(1):25–32. https://www.ajmc.com/pubMed.php?pii=2474. [PubMed] [Google Scholar]
- 21.Grana J, Preston S, McDermott PD, Hanchak NA. The use of administrative data to risk-stratify asthmatic patients. Am J Med Qual. 1997;12(2):113–9. doi: 10.1177/0885713X9701200205. [DOI] [PubMed] [Google Scholar]
- 22.Forno E, Fuhlbrigge A, Soto-Quirós ME, Avila L, Raby BA, Brehm J, Sylvia JM, Weiss ST, Celedón JC. Risk factors and predictive clinical scores for asthma exacerbations in childhood. Chest. 2010 Nov;138(5):1156–65. doi: 10.1378/chest.09-2426. http://europepmc.org/abstract/MED/20472862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo G, He S, Stone BL, Nkoy FL, Johnson MD. Developing a model to predict hospital encounters for asthma in asthmatic patients: secondary analysis. JMIR Med Inform. 2020 Jan 21;8(1):e16080. doi: 10.2196/16080. https://medinform.jmir.org/2020/1/e16080/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen T, Guestrin C. XGBoost: A Scalable Tree Boosting System. Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; KDD'16; August 13-17, 2016; San Francisco, CA. 2016. pp. 785–94. [DOI] [Google Scholar]
- 25.Desai JR, Wu P, Nichols GA, Lieu TA, O'Connor PJ. Diabetes and asthma case identification, validation, and representativeness when using electronic health data to construct registries for comparative effectiveness and epidemiologic research. Med Care. 2012 Jul;50 Suppl:S30–S35. doi: 10.1097/MLR.0b013e318259c011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wakefield DB, Cloutier MM. Modifications to HEDIS and CSTE algorithms improve case recognition of pediatric asthma. Pediatr Pulmonol. 2006 Oct;41(10):962–71. doi: 10.1002/ppul.20476. [DOI] [PubMed] [Google Scholar]
- 27.Luo G. Automatically explaining machine learning prediction results: a demonstration on type 2 diabetes risk prediction. Health Inf Sci Syst. 2016;4:2. doi: 10.1186/s13755-016-0015-4. http://europepmc.org/abstract/MED/26958341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo G, Stone BL, Sakaguchi F, Sheng X, Murtaugh MA. Using computational approaches to improve risk-stratified patient management: rationale and methods. JMIR Res Protoc. 2015 Oct 26;4(4):e128. doi: 10.2196/resprot.5039. https://www.researchprotocols.org/2015/4/e128/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alaa AM, van der Schaar M. Prognostication and risk factors for cystic fibrosis via automated machine learning. Sci Rep. 2018 Jul 26;8(1):11242. doi: 10.1038/s41598-018-29523-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alaa AM, van der Schaar M. AutoPrognosis: Automated Clinical Prognostic Modeling via Bayesian Optimization With Structured Kernel Learning. Proceedings of 35th International Conference on Machine Learning; ICML'18; July 10-15, 2018; Stockholm, Sweden. 2018. pp. 139–48. [Google Scholar]
- 31.Luo G. A roadmap for semi-automatically extracting predictive and clinically meaningful temporal features from medical data for predictive modeling. Glob Transit. 2019;1:61–82. doi: 10.1016/j.glt.2018.11.001. http://europepmc.org/abstract/MED/31032483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo G, Stone BL, Koebnick C, He S, Au DH, Sheng X, Murtaugh MA, Sward KA, Schatz M, Zeiger RS, Davidson GH, Nkoy FL. Using temporal features to provide data-driven clinical early warnings for chronic obstructive pulmonary disease and asthma care management: protocol for a secondary analysis. JMIR Res Protoc. 2019 Jun 6;8(6):e13783. doi: 10.2196/13783. https://www.researchprotocols.org/2019/6/e13783/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guidotti R, Monreale A, Ruggieri S, Turini F, Giannotti F, Pedreschi D. A survey of methods for explaining black box models. ACM Comput Surv. 2019 Jan 23;51(5):93. doi: 10.1145/3236009. [DOI] [Google Scholar]
- 34.Molnar C. Interpretable Machine Learning. Morrisville, NC: lulu.com; 2020. [Google Scholar]
- 35.Liu B, Hsu W, Ma Y. Integrating classification and association rule mining. Proceedings of the 4th International Conference on Knowledge Discovery and Data Mining; KDD'98; August 27-31, 1998; New York City, NY. 1998. pp. 80–86. [Google Scholar]
- 36.Thabtah F. A review of associative classification mining. Knowl Eng Rev. 2007 Mar 1;22(1):37–65. doi: 10.1017/s0269888907001026. [DOI] [Google Scholar]
- 37.Fayyad UM, Irani KB. Multi-interval Discretization of Continuous-valued Attributes for Classification Learning. Proceedings of the 13th International Joint Conference on Artificial Intelligence; IJCAI'93; August 28-September 3, 1993; Chambéry, France. 1993. pp. 1022–9. [Google Scholar]
- 38.Paul R, Groza T, Hunter J, Zankl A. Inferring characteristic phenotypes via class association rule mining in the bone dysplasia domain. J Biomed Inform. 2014 Apr;48:73–83. doi: 10.1016/j.jbi.2013.12.001. https://linkinghub.elsevier.com/retrieve/pii/S1532-0464(13)00192-5. [DOI] [PubMed] [Google Scholar]
- 39.Han J, Kamber M, Pei J. Data Mining: Concepts and Techniques, 3rd ed. Waltham, MA: Morgan Kaufmann; 2011. [Google Scholar]
- 40.Witten IH, Frank E, Hall MA, Pal CJ. Data Mining: Practical Machine Learning Tools and Techniques, 4th ed. Burlington, MA: Morgan Kaufmann; 2016. [Google Scholar]
- 41.Agresti A. Categorical Data Analysis, 3rd ed. Hoboken, NJ: Wiley; 2012. [Google Scholar]
- 42.Health Literacy Data Map. US Health Literacy Scores. 2020. [2020-12-10]. http://healthliteracymap.unc.edu.
- 43.Tong Y, Messinger AI, Luo G. Testing the generalizability of an automated method for explaining machine learning predictions on asthma patients’ asthma hospital visits to an academic healthcare system. IEEE Access. 2020;8:195971–9. doi: 10.1109/access.2020.3032683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo G, Nau CL, Crawford WW, Schatz M, Zeiger RS, Koebnick C. Assessing the Generalizability of an Automatic Explanation Method for Machine Learning Prediction Results: A Secondary Analysis on Forecasting Asthma-related Hospital Visits in Patients With Asthma. UW Computer Sciences User Pages. [2020-12-10]. http://pages.cs.wisc.edu/~gangluo/explain_predict_hospital_use_for_asthma_KPSC.pdf.
- 45.Hatwell J, Gaber MM, Atif Azad RM. Ada-WHIPS: explaining AdaBoost classification with applications in the health sciences. BMC Med Inform Decis Mak. 2020 Oct 2;20(1):250. doi: 10.1186/s12911-020-01201-2. https://bmcmedinformdecismak.biomedcentral.com/articles/10.1186/s12911-020-01201-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bourgeois FC, Olson KL, Mandl KD. Patients treated at multiple acute health care facilities: quantifying information fragmentation. Arch Intern Med. 2010 Dec 13;170(22):1989–95. doi: 10.1001/archinternmed.2010.439. [DOI] [PubMed] [Google Scholar]
- 47.Finnell JT, Overhage JM, Grannis S. All health care is not local: an evaluation of the distribution of emergency department care delivered in Indiana. AMIA Annu Symp Proc. 2011;2011:409–16. http://europepmc.org/abstract/MED/22195094. [PMC free article] [PubMed] [Google Scholar]
- 48.Luo G, Tarczy-Hornoch P, Wilcox AB, Lee ES. Identifying patients who are likely to receive most of their care from a specific health care system: demonstration via secondary analysis. JMIR Med Inform. 2018 Nov 5;6(4):e12241. doi: 10.2196/12241. http://medinform.jmir.org/2018/4/e12241/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kern LM, Grinspan Z, Shapiro JS, Kaushal R. Patients' use of multiple hospitals in a major US city: implications for population management. Popul Health Manag. 2017 Apr;20(2):99–102. doi: 10.1089/pop.2016.0021. http://europepmc.org/abstract/MED/27268133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Samuels-Kalow ME, Faridi MK, Espinola JA, Klig JE, Camargo CA. Comparing statewide and single-center data to predict high-frequency emergency department utilization among patients with asthma exacerbation. Acad Emerg Med. 2018 Jun;25(6):657–67. doi: 10.1111/acem.13342. doi: 10.1111/acem.13342. [DOI] [PMC free article] [PubMed] [Google Scholar]


