Abstract
Acute lung inflammatory diseases severely affect the patients' recovery and outcomes worldwide. Unregulated acute inflammatory response is fundamentally central to acute lung inflammation including acute lung injury (ALI) and acute respiratory distress syndrome (ARDS). To limit the potentially deleterious effects of acute lung inflammation, complex transcriptional and posttranscriptional regulatory networks have been explored, which often involves long noncoding RNAs (lncRNA). LncRNAs are RNAs that longer than 200 nucleotides, functioning as scaffolds or decoys in the cytoplasm or nucleus. By now, lncRNAs have been found to join in all major cellular processes including cell proliferation, metabolism, stress response or death. Extensive advance over the last decade furthermore indicated a fundamental role of lncRNAs in acute lung inflammation. This article reviews and summarizes the current knowledge on lncRNA in acute lung inflammatory response.
Keywords: LncRNAs, Acute lung inflammation, Acute respiratory distress syndrome, Pyroptosis
Graphical abstract
1. Introduction
Being one of main target organs of pro-inflammatory mediators released and secreted globally during trauma, sepsis, and major surgery, the lung was at high risk of acute lung inflammatory diseases. The unregulated acute inflammatory response would cause acute lung injury (ALI) and its severe form, known as acute respiratory distress syndrome (ARDS), which lead to high morbidity and mortality [1]. Acute lung inflammation was characterized fundamentally by dysfunction of the barrier properties of the pulmonary epithelium and endothelium due to direct pulmonary insults as well as indirect systemic inflammatory responses. Although a variety of anti-inflammation pharmacotherapy have been applied, the morbidity and outcome of ALI/ARDS patients were still poor. It is of particular importance to explore the new mechanism of acute lung inflammation for early diagnosis and treatment.
To limit the potentially deleterious effects of acute lung inflammation, complex transcriptional and posttranscriptional regulatory networks have been explored, which often involves long noncoding RNAs (lncRNA). LncRNAs, regarded as transcripts exceed 200 nucleotides with little low coding potential, were considered as the waste of biological metabolism in the past. But in recent decades, lncRNAs were found to involve in diverse biological processes, such as cell differentiation, proliferation, apoptosis, pyroptosis, and participate in various stress responses [2]. It was reported that lncRNAs might regulate gene expression by several different ways, including binding to microRNAs, mRNAs or proteins via the regulation of epigenetic modifications, transcriptional and posttranscriptional processing [3].
At present, lncRNAs have been found abnormally expressed in many inflammatory and infectious diseases. It was reported that lncRNAs might regulate the expression of multiple genes and activate the signaling pathways in the development of inflammatory diseases and pathogenesis. For instance, lncRNA XLOC_010280 can modulate the inflammatory reaction of eosinophilic granulocytes and the expression of chemokine ligand 18 (CCL18), to promote the development of inflammatory polyps [4].
More importantly, emerging evidence has revealed that lncRNAs play important roles in acute lung inflammation [5], including inflammation resolution of ALI/ARDS [6]. Since that many differentially expressed lncRNAs have been identified in ALI/ARDS recently, they may potentially serve as new therapeutic strategy for acute lung inflammation [7]. This article aims to review and summarize the specific function and mechanism of lncRNAs in acute lung inflammation and further explore their therapeutic potential in acute lung inflammation.
2. Overview of LncRNAs
Long non-coding RNA (lncRNAs) are RNAs that longer than 200 nucleotides with low coding potential. Mediated by RNA polymerase II [8], more than 90 thousand lncRNAs have been released in different databases [9,10]. Remarkably, the number of lncRNAs is increasing dramatically because of the continuous maturity of sequencing technology. However, it still lacks definitive standard for lncRNAs classification. The relatively common classification is based on the location of nearby encoded protein genes, according to which lncRNAs are divided into antisense lncRNAs, sense lncRNAs, intergenic lncRNAs, intron region lncRNAs, circRNAs and enhancer lncRNAs (eRNAs) [11]. Other reported classifications of lncRNAs include: 1) by subcellular localization or origin, e.g. mitochondrial, cytoplasmic, or nuclear; 2) by different regulatory mechanisms, e.g. scaffolding lncRNA (HOTAIR); 3) by association with specific biological processes; 4) encoded within specific DNA regulator elements, e.g. in rDNA loci (PAPAS), centromeres, telomeres (TERRA); and 5) length, e.g. very long lincRNA (vlincRNA) [2]. As the number of functional lncRNAs increased, the categorization standard of lncRNAs is being renewed continuously.
Besides the categorization standard, the function of lncRNAs is also being updated, which denies many traditional views. For instance, lncRNAs have no protein-coding potential due to their lack of gene-specific open reading frames (ORFs) in our traditional opinion. However, this should be updated since that recent researches showed that lncRNAs also own encoding ability. For example, lncRNA 00961 was found to encode SPAR and negatively regulate mTORC1 during muscle regeneration [12]. Ruiz-Orera also identified several lncRNAs which could translate RNA into proteins [3].
3. Regulation mechanism of lncRNAs
Researchers have conducted large numbers of extensive and in-depth explorations to clarify the mechanism of lncRNAs in different diseases comprehensively. LncRNAs were found to regulate genes at three levels including the epigenetic, transcriptional and posttranscriptional levels. 1) Epigenetic level: mainly including chromatin modification, genomic imprinting and dosage compensation [13,14]. 2) Transcriptional level: binding with target proteins in cis-acting elements or in a complexes form [15]. (3) Posttranscriptional level: regulating gene expression via mRNAs degradation, splicing and translation.
Recent studies also found lncRNAs to identify and bind to specific RNA or DNA sequences via base-pairing interactions and fold into secondary or higher order structures to modulate their interaction with proteins [16]. In addition, they could exert functions in both cytoplasm and nucleus, via different mechanisms including chromosome architecture modulation, genomic regions regulation, chromatin-modifying complexes interaction, nuclear domains conformation, transcriptional enhancers activation, the transcriptional machinery interference, and structural formation, nuclear bodies maintenance [17]. Of note, lncRNAs frequently acted as competitive endogenous RNAs (ceRNAs) to protect stable mRNA expression [18]. The special mechanism of ceRNAs indicated that non-coding and coding RNAs could interact via competing for miRNA binding, thus reciprocally affecting their respective expression levels. As time goes on, more advances in the regulating mechanisms of lncRNAs will be reported in the near future to help us understanding its entire regulation network.
4. Emerging research hotspots of acute lung inflammation
Acute lung inflammation is one of the main features in the pathogenesis of many acute lung diseases, such as ALI, ARDS and so on. Irrespective of the initial cause, ALI/ARDS is fundamentally characterized by severe inflammatory responses, intense epithelial/endothelial barrier damage, as well as alveolar edema [19]. Although there are some effective strategies to alleviate acute lung inflammation, the outcome of patients with acute lung inflammation has not yet improved and the underlying mechanism has not yet been clarified [16].
Multiple pulmonary cells are involved in acute lung inflammation, including epithelial cells, labrocyte, macrophages, neutrophils, mononuclear cells and T cells [20,21]. Since that alveolar macrophages (AMs) play central roles in both the initiation and resolution of acute inflammatory responses, we have focused on its underlying mechanisms in acute lung inflammation. Different from classic macrophages, AMs are derived from yoke-sac procurers in fetal monocytes and comprise the majority of inflammatory cells in the lung [22]. With inflammatory invasions, bone marrow derived monocytes are recruited to the bronchoalveolar space and differentiated into AMs [23] to form the first-line host defense against infection, acting as antigen-presenting cells and releasing pro-inflammatory cytokines to propagate the inflammatory responses in ALI/ARDS [24]. The functional phenotype of AMs is modulated by different microenvironment of the lung [19], which mainly including two distinct subsets: classically activated M1 macrophages (M1) and alternatively activated M2 macrophages (M2). Accumulating evidence showed that in the early stage of acute lung inflammation, large amounts of M1 secrete molecules IL-6, TNF-α, and IL-1 to propagate inflammatory responses and mediate host defense against various bacteria or viruses, while M2 secrete biological mediators TGF-β, IL-10, and chemokines engaged in infection, immune-regulation and tissue reconstruction [25]. Notably, M2 has four subtypes (M2a, M2b, M2c, and M2d) and the regulatory macrophages (M2b) produce a lot of anti-inflammatory cytokine, such as IL-10, to control the excessive inflammations and auto-immune reactions [26]. The modulation of M1/M2 macrophage polarization has been proven to be a potential therapeutic target for acute lung inflammatory disorders.
Besides AMs, increasing reports also highlight the complexity and important function of interstitial macrophages (IMs) in lung inflammation. Like AMs, IMs are phagocytic cells and thus can be considered as the second-line defense against invading microorganisms [27]. Up to now, three distinct subpopulations of IMs, including IM1, IM2, IM3, have been identified based on their phenotypical and functional properties [28]. Recent evidence supports that IMs play a vital in the second line of defense through their antigen presentation and phagocytosis abilities in bacterial or viral lung inflammation [29]. For instance, a CD169+ IMs subset substantially located to the bronchovascular tree was recently reported to regulate lung inflammation via regulating production of inflammatory cytokines and innate immune cell infiltration [30]. Moreover, inducing metabolic and epi-genetic reprogramming of lung IMs toward an anti-inflammatory phenotype could reduce inflammatory lung injury [31] and protect from airway allergic inflammation [32]. Although our understanding of the IMs' spatial and anatomical positioning and functions is still limited, emerging evidence suggests that they play vital roles in maintaining immune and tissue homeostasis in lung inflammation [33,34].
Multiple types of cell injury and programmed cell death were also found in acute lung inflammation, mainly includes cellular apoptosis, pyroptosis, autophagy, necrosis. Among which pyroptosis is a newly discovered type of programmed cell death. Including a typical pathway dependent on Caspase-1 activation, and an atypical pathway dependent on Caspase 4/5 (in human) and Caspase 11 (in mice) activation, pyroptosis played an important role in inflammatory resolution of acute lung inflammation [35]. Recent studies have shown that inflammasomes activation and inflammasome-dependent pyroptosis play a vital role in acute lung inflammation [36], including classical and nonclassical pathways (Fig. 1 ). For instance, recent studies revealed that Dihydromyricetin protected CLP-induced ALI via inhibition of Nod-like receptor protein 3 (NLRP3) inflammasome activation and pyroptosis [1]. Another study also showed that Ang1 exerted beneficial role of phosgene-induced ALI through inhibiting NLRP3 inflammasome activation and pyroptosis [37].
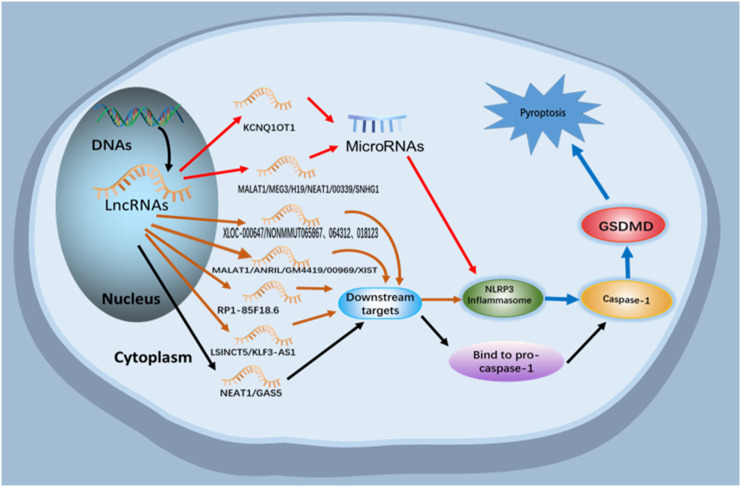
Fig. 1.
Relationship between lncRNAs and pyroptosis.
LncRNAs participate in the regulation of pyroptosis by indirectly acting on miRNAs and downstream targets or directly binding to pre-caspase-1, thus regulating the pathological process of pyroptosis related diseases.
5. Regulation of LncRNAs in acute lung inflammation
Conventional research on acute lung inflammation often involved investigations on gene expression, specific protein or protein related signaling pathways, etc. The discovery of lncRNA reveals a new insight on the pathogenesis of acute lung inflammation, which will open a new era for researchers to develop specific effective therapeutics. Several lncRNAs have been identified being involved in the procedure of acute lung inflammation, through modulating miRNAs or downstream targets or directly regulate pyroptosis [38,39]. We concluded the regulation of reported lncRNAs in acute lung inflammation as shown in Table 1 and Fig. 2 .
Table 1.
Role of lncRNAs in acute lung inflammation.
| LncRNA | Models | Cell types | Mechanisms | Ref# |
|---|---|---|---|---|
| MALAT1 | Sepsis | Alveolar macrophages | p38MAPK/p65NF-κB pathway | [6] |
| ALI | Alveolar macrophages | Sponging miR-146a | [7] | |
| ARDS | Aberrant fibroblast | miR-425/PETN axis | [77] | |
| HPME | HPMEC | miR-150-5p/ICAM-1 | [78] | |
| ALI | Alveolar type II epithelial A549 cells | Sponging miR175p/FOXA1 | [79] | |
| ALI | Alveolar macrophages | miR-149/MYD88/NF-kB signaling | [43] | |
| ALI | Lung fibroblasts WI-38 cells | TLR4/NF-kB/p38 MAPK pathway | [80] | |
| NEAT1 | Sepsis | Lung epithelial cell | Targeting miR-125a | [72] |
| ALI | Alveolar epithelial cells | HMGB1/RAGE/NF-kB signaling | [48] | |
| CASC2 | ALI | Lung epithelial cells | miR-144-3p/AQP1 axis | [49] |
| CASC9 | Sepsis | Alveolar macrophages | miR-195-5p/PDK4 axis | [50] |
| GAS5 | ALI | Alveolar epithelial cells | Targeting miR429/DUSP1axis | [51] |
| ARDS | Lung epithelial A549 cells | miR-200C-3p/ACE2 axis | [52] | |
| FOXD3-AS1 | HALI | Lung epithelial cells | Sponging miR150 | [53] |
| NANCI | HALI | Alveolar epithelial cells | NANCI/NKX2.1 signaling | [54] |
| GADD7 | HALI | Type II alveolar epithelial cells | miR-125a/MFN1axis | [55] |
| SNHG16 | ALI | Pulmonary epithelial A549 cells | miR-370-3p/IGF2 axis | [56] |
| ALI | Lung fibroblasts WI-38 cells | miR-146a-5p/CCL5 axis | [57] | |
| SNHG14 | ALI | Alveolar macrophages | miR-34c-3p/WISP1 axis | [58] |
| HAGLROS | ALI | Lung fibroblasts WI-38 cells | miR-100/NF-kB axis | [59] |
| XIST | ALI | Lung fibroblasts WI-38 cells | miR-370-3p/TLR4 axis | [60] |
| PGD | Bronchoalveolar lavage fluid cells | miR-21/NET/IL-12A axis | [61] | |
| MEG3 | ALI | Lung fibroblast WI-38 cells and human PMVECs cells | miR-4262/KLF4 axis | [62] |
| MEG3–4 | ALI | Alveolar macrophages and lung epithelial cells | miR-138/IL-1β axis | [63] |
| THRIL | Sepsis | Alveolar epithelial cells | As biomarkers | [64] |
| IL7R | ARDS | Alveolar epithelial cell | As biomarkers | [65] |
Fig. 2.
Regulation of lncRNAs in acute lung inflammation.
LncRNAs participate in the regulation of pyroptosis and apoptosis by indirectly acting on miRNAs and downstream targets in acute lung inflammation.
5.1. LncRNA MALAT1
Metastasis associated lung adenocarcinoma transcript 1 (MALAT1), a lncRNA located in more than 40 kb away on mice chromosome 19 from Neat1, is reported essential in the regulation of acute lung inflammation, especially in septic-lung injury [6]. For example, MALAT1 activates p38 MAPK/p65 NF-κB signaling pathway to evoke and intensify inflammation in septic-lung injury model [40]. Dai L et al. reported that MALAT1 was a potential biomarker to predict elevated ARDS risk and correlate with severe disease condition and raised mortality in sepsis patients [6].
MALAT1 is reported to participate in pulmonary pathogeneses via regulating aberrant macrophage activation. For example, MALAT1 upregulated in LPS-treated AMs and promoted pro-inflammatory M1 phenotype AMs activation and suppressed the alternative M2 phenotype AMs activation. Furthermore, MALAT1 knockdown attenuates pro-inflammatory activation of AMs [6], and MALAT1 overexpression aggravates LPS induced inflammatory response in murine AMs by sponging miR-146a [7].
Besides miR-146a, MALAT1 was also found to regulate acute lung inflammation via sponging or binding other miRNAs. A study showed that over expression of MALAT1 aggravating acute lung inflammation through sponging miR-425 to modulate PETN expression, and then enhance aberrant fibroblast proliferation [7] and apoptosis in LPS-induced ARDS [41]. In addition, MALAT1 binded to miR-150-5p and upregulated ICAM-1 expression to cause HPMEC apoptosis in both ARDS patients and LPS-treated HPME [41]. Similarly, MALAT1 knockdown was found to alleviate LPS induced ALI via targeting the miR 175p/FOXA1 axis [42].
Furthermore, MALAT1 can regulate acute lung inflammation through pyroptosis via different ways. A recent study showed that MALAT1 regarded as a pro-inflammatory factor in LPS-ALI model to regulate AM cell pyroptosis and inflammatory response via the miR-149/MyD88/NF-kB axis [43]. In addition, Li H et al. reported a lipoxin receptor agonist, BML-111, could significantly modulate the expression of MALAT1 and regulate the inflammatory resolution via TLR4/NF-kB/p38 MAPK signaling pathways [44], which also provided a potential target for acute lung inflammation.
Kölling M and his colleague also reported that MALAT1 knockdown could ameliorate histopathologic changes in lung injury in hypoxia-induced pulmonary hypertension [45]. All the increasing studies confirmed that MALAT1 knockdown might be potential therapeutic way to alleviate acute lung inflammation [7].
5.2. LncRNA NEAT1
Recently, lncRNA nuclear paraspeckle assembly transcript 1 (NEAT1) has grown up to be a topic of concern due to its pleiotropic function in inflammation-related diseases. It has been indicated there was positive correlation between high levels of NEAT1 and increased severity and unfavorable prognoses in patients with sepsis [46]. Groot M. also showed that high expression of NEAT1 and low expression miR-125a were associated with increased ARDS risk, higher 28-day mortality in sepsis patients and acute lung inflammation [47]. Nevertheless, the function and molecular mechanism of NEAT1 in acute lung inflammation remain elusive. In a recent study, Hongchao Zhou and his colleague confirmed that NEAT1 may modulate alveolar epithelial cells (AECs) pyroptosis and reduce inflammatory response to against LPS-induced ALI/ARDS by suppressing HMGB1/RAGE-NF-κB signaling [48]. Furthermore, NEAT1 inhibition alleviates cell apoptosis and promotes proliferation in sepsis-induced ALI. All the studies provide a theoretical basis for the therapy of NEAT1 in acute lung inflammation.
5.3. LncRNA CASC2, CASC9, GAS5
Increasing studies confirmed that lncRNAs can act as miRNA decoys to sequestrate the miRNAs and thus favors expression of their repressed target mRNAs in acute lung inflammation. For instance, cancer susceptibility candidate 2 (CASC2), which is a lncRNA downregulated in multiple cancer types, was recently found acting as a competitive endogenous RNA that regulates AQP1 expression by sponging to miR-144-3p to protect lung epithelial cell from apoptosis in acute lung injury [49]. Besides CASC2, down-regulation of cancer susceptibility candidate 9 (CASC9) was also reported to regulate miR-195-5p/PDK4 axis and exacerbate ALI in sepsis [50]. Likewise, growth arrest special 5 (GAS5) also acted as a competitive endogenous RNA to promote DUSP1 expression via sponging miR-429 to suppress inflammation and alveolar epithelial cell apoptosis [51]. Another study showed that GAS5 down regulation reduces ACE2 expression via increasing miR-200c-3p to promote A549 cells apoptosis in ARDS [52].
5.4. LncRNA FOXD3-AS1, NANCI, GADD7
Hyperoxia-induced acute lung injury (HALI) is a kind of iatrogenic pulmonary dysfunction as a result of prolonged exposure to hyperoxia-induced oxidative stress. Several lncRNAs have been found significantly altered in the presence of high concentration of oxygen. As Zhang et al. demonstrated, FOXD3 antisense RNA 1 (FOXD3-AS1) could down-regulate miRNA-150 to promote oxidative stress-induced lung epithelial cell apoptosis and cell death [53]. In addition, lncRNA Nkx2.1-associated noncoding intergenic RNA (NANCI) was also reported to decrease in HALI [54]. Guoyue Liu et al. found that agmatine protected hyperoxia-induced lung injury and inhibited cells apoptosis via decreasing lncRNA growth arrested DNA-damage inducible gene 7 (GADD7) to indirectly decrease MFN1 expression, due to the presence of the competitive binding of lncRNA GADD7 and MFN1 to miR-125a [55].
5.5. LncRNA SNHG16, SNHG14, HAGLROS
Increasing studies employed LPS-induced ALI model to confirm the role of lncRNAs in inflammatory resolution and provide potential targets for the treatment of acute lung inflammation. Take small nucleolar RNA host gene 16 (SNHG16) as an example, it modulates IGF2 expression to promote LPS-induced acute pneumonia in A549 cells by targeting miR-370-3p [56]. A recent study showed that SNHG16 regulates LPS-induced apoptosis of WI-38 cells and inflammatory response in acute lung inflammation via targeting miR-146a-5p/CCL5 expression [57]. Besides SNHG16, inhibiting small nucleolar RNA host gene 14 (SNHG14) was found to protect against LPS-induced ALI through miRNA-34c-3p-dependent inhibition of WISP1 [58]. Down-regulation of haglr opposite strand (HAGLROS) alleviated LPS-induced inflammation in WI-38 cells through modulating miR-100/NF-κB axis to increase cell viability, inhibit apoptosis and autophagy [59].
5.6. LncRNA XIST, MEG3, MEG3–4
Besides HAGLROS, lncRNA X inactive-specific transcript (XIST) has also emerged as a new modulator in LPS-induced ALI. For example, it was showed that XIST deficiency alleviated LPS-induced apoptosis of WI-38 cells and inflammatory response via regulating miR-370-3p/TLR4 in acute pneumonia [60]. Li J et al. also confirmed that XIST increased IL-12A via binding to miR-21 to induce NET formation and accelerated primary graft dysfunction (PGD) after lung transplantation, which suggested that XIST and NET inhibition might be beneficial for the therapy of PGD [61]. Besides that, the lncRNA maternally expressed gene 3 (MEG3), acting as a ceRNA and sponging miRNAs, was reported to affect various cell processes such as apoptosis, proliferation and angiogenesis. A report showed that knockdown of MEG3 intensified LPS induced lung injury via miR-4262 mediated down-regulation of KLF4 [62]. Moreover, among 10 transcriptional subtypes of MEG3, transcriptional 4 (MEG3–4) encodes the least abundant subtype in the lungs of mice. It was reported that MEG3–4 acted as miRNA decoy, increasing IL-1β abundance to modulate inflammatory responses in lung cells and tissues of ALI mice via competitively binding to miR-138 [63].
5.7. Other newly identified lncRNAs
Many new lncRNAs have been identified to be biomarkers for clinical outcomes of ARDS/ALI. Recent evidence showed that lncRNA THRIL was upregulated in patients with ARDS [47] and it was recommended as biomarker that positively correlated with mortality in sepsis-induced ARDS patients [64]. While plasma Lnc-IL7R was significantly downregulated in ARDS patients [65], and it has been verified to negatively regulate IL7R expression to inhibit LPS-induced acute lung inflammation [66]. LncRNA A_30_P01029194 and A_30_P01029806 might also serve as candidate biomarkers in ALI [67]. Another genome-wide transcriptome analysis revealed that lncRNA NONMMUT065867, NONMMUT064312 and NONMMUT018123 were closely related to the elevated NLRP3 expression and ARDS [68].
5.8. Promise and challenges of lncRNAs therapy in acute lung inflammation
LncRNAs have served as promising targets for acute lung inflammation by regulating miRNAs, targets or proteins, and many studies performed in LPS-induced ALI models have revealed the potential targeting lncRNAs to regulate acute lung inflammation, which provided a strong therapeutic basis for acute lung inflammation. Moreover, lncRNAs were also reported to play important roles in mesenchymal stem cells (MSCs) and MSC-derived exosomes treatment of acute lung inflammation [[69], [70], [71]]. However, researchers have not obtained sufficient evidence to confirm the therapeutic effects of these treatments on acute lung inflammation in clinical trials. For instance, plasma lnc-IL7R was preliminarily suggested as a biomarker in ARDS patients [72], but the sample size was small that still needs further confirmation. In addition, although numerous clinical trials of MSCs transplantation for ARDS including COVID-19 ARDS treatment have been registered and reported [[73], [74], [75]], whether lncRNAs are involved in the treatment still need further exploration. Moreover, before targeting the lncRNAs therapy for acute lung inflammation, several crucial challenges persist and need to be addressed. Firstly, since lncRNAs are poorly conserved among different species, it is difficult to confirm role of the selected lncRNAs in patients with acute lung inflammation in animal models, thus limits the exploration of the mechanism of lncRNAs in regulating acute lung inflammation [76]. Secondly, most current studies have focused on the role of lncRNAs as ceRNA. However, expression of most lncRNAs is generally not high in cells, in contrast to highly expressed miRNAs. It is difficult to coordinate the sponge function of ceRNAs because miRNAs offer only 1 binding site per lncRNA.
6. Conclusions and prospects
Acute lung inflammation including ALI/ARDS still lacks specific therapeutic targets. More and more lncRNAs have been found to play important roles in pathogenesis of ALI/ARDS and may serve as new diagnostic and treatment biomarkers for ALI/ARDS. Increasing evidence shows that lncRNAs can bind to microRNAs, mRNAs or proteins to regulate epigenetic modifications, transcriptional and posttranscriptional processing in epithelial cells, AMs, neutrophils and so on, which provides potential therapeutic targets for ALI/ARDS and basis for drug therapy.
However, the underlying mechanisms and therapeutic potential of lncRNAs in acute lung inflammation still need further investigation and confirmation in the future.
CRediT authorship contribution statement
CJC and YFH conceived the structure of this manuscript, drafted initial manuscript and also revised the manuscript; YWF and WLH collected all information and composed sections related to role of lncRNAs; ZQY and GJL provided the comments on this manuscript. All authors have read and approved the submission of this manuscript.
Declaration of competing interest
The authors declare that they have no competing interests.
Acknowledgments
Acknowledgments
Not applicable
Funding
The study was supported in part by the National Natural Science Foundation of China (Grant No. 81770649), the Science and Technology Program of Guangzhou (201704020049), the Postdoctoral Science Foundation of China (Grant No. 2019M663260), the Fundamental Research Funds for the Central Universities of China (Grant No. 20ykpy20) and Basic and Applied Basic Research Foundation of Guangdong Province (Grant No. 2019A1515110020).
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
References
- 1.Wang Y.C., Liu Q.X., Zheng Q., et al. Dihydromyricetin alleviates sepsis-induced acute lung injury through inhibiting NLRP3 inflammasome-dependent pyroptosis in mice model. Inflammation. 2019;42(4):1301–1310. doi: 10.1007/s10753-019-00990-7. [DOI] [PubMed] [Google Scholar]
- 2.Jarroux J., Morillon A., Pinskaya M. History, discovery, and classification of lncRNAs. Adv. Exp. Med. Biol. 2017;1008:1–46. doi: 10.1007/978-981-10-5203-3_1. [DOI] [PubMed] [Google Scholar]
- 3.Ruiz-Orera J., Messeguer X., Subirana J.A., et al. Long non-coding RNAs as a source of new peptides. Elife. 2014;3 doi: 10.7554/eLife.03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liao K., Xu J., Yang W., et al. The research progress of LncRNA involved in the regulation of inflammatory diseases. Mol. Immunol. 2018;101:182–188. doi: 10.1016/j.molimm.2018.05.030. [DOI] [PubMed] [Google Scholar]
- 5.Schulte L.N., Bertrams W., Stielow C., et al. ncRNAs in inflammatory and infectious diseases. Methods Mol. Biol. 2019;1912:3–32. doi: 10.1007/978-1-4939-8982-9_1. [DOI] [PubMed] [Google Scholar]
- 6.Cui H., Banerjee S., Guo S., et al. Long noncoding RNA Malat1 regulates differential activation of macrophages and response to lung injury. JCI Insight. 2019:4(4). doi: 10.1172/jci.insight.124522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai L., Zhang G., Cheng Z., et al. Knockdown of LncRNA MALAT1 contributes to the suppression of inflammatory responses by up-regulating miR-146a in LPS-induced acute lung injury. Connect. Tissue Res. 2018;59(6):581–592. doi: 10.1080/03008207.2018.1439480. [DOI] [PubMed] [Google Scholar]
- 8.Brannan C.I., Dees E.C., Ingram R.S., et al. The product of the H19 gene may function as an RNA. Mol. Cell. Biol. 1990;10(1):28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frankish A., Diekhans M., Ferreira A.M., et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47(D1):D766–D773. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang S., Zhang L., Guo J., et al. NONCODEV5: a comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2018;46(D1):D308–D314. doi: 10.1093/nar/gkx1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uchida S., Dimmeler S. Long noncoding RNAs in cardiovascular diseases. Circ. Res. 2015;116(4):737–750. doi: 10.1161/CIRCRESAHA.116.302521. [DOI] [PubMed] [Google Scholar]
- 12.Matsumoto A., Pasut A., Matsumoto M., et al. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature. 2017;541(7636):228–232. doi: 10.1038/nature21034. [DOI] [PubMed] [Google Scholar]
- 13.Gelbart M.E., Kuroda M.I. Drosophila dosage compensation: a complex voyage to the X chromosome. Development. 2009;136(9):1399–1410. doi: 10.1242/dev.029645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schoenherr C.J., Levorse J.M., Tilghman S.M. CTCF maintains differential methylation at the Igf2/H19 locus. Nat. Genet. 2003;33(1):66–69. doi: 10.1038/ng1057. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X.D., Huang G.W., Xie Y.H., et al. The interaction of lncRNA EZR-AS1 with SMYD3 maintains overexpression of EZR in ESCC cells. Nucleic Acids Res. 2018;46(4):1793–1809. doi: 10.1093/nar/gkx1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salmena L., Poliseno L., Tay Y., et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rusconi F., Battaglioli E., Venturin M. Psychiatric disorders and lncRNAs: a synaptic match. Int. J. Mol. Sci. 2020:21(9). doi: 10.3390/ijms21093030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim Y.K., Furic L., Desgroseillers L., et al. Mammalian Staufen1 recruits Upf1 to specific mRNA 3'UTRs so as to elicit mRNA decay. Cell. 2005;120(2):195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 19.Lee H., Abston E., Zhang D., et al. Extracellular vesicle: an emerging mediator of intercellular crosstalk in lung inflammation and injury. Front. Immunol. 2018;9:924. doi: 10.3389/fimmu.2018.00924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashbaugh D.G., Bigelow D.B., Petty T.L., et al. Acute respiratory distress in adults. Lancet. 1967;2(7511):319–323. doi: 10.1016/s0140-6736(67)90168-7. [DOI] [PubMed] [Google Scholar]
- 21.Máca J., Jor O., Holub M., et al. Past and present ARDS mortality rates: a systematic review. Respir. Care. 2017;62(1):113–122. doi: 10.4187/respcare.04716. [DOI] [PubMed] [Google Scholar]
- 22.Arora S., Dev K., Agarwal B., et al. Macrophages: their role, activation and polarization in pulmonary diseases. Immunobiology. 2018;223(4–5):383–396. doi: 10.1016/j.imbio.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan S.Y., Krasnow M.A. Developmental origin of lung macrophage diversity. Development. 2016;143(8):1318–1327. doi: 10.1242/dev.129122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joshi N., Walter J.M., Misharin A.V. Alveolar macrophages. Cell Immunol. 2018;330:86–90. doi: 10.1016/j.cellimm.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 25.Forbes A., Pickell M., Foroughian M., et al. Alveolar macrophage depletion is associated with increased surfactant pool sizes in adult rats. J Appl Physiol (1985) 2007;103(2):637–645. doi: 10.1152/japplphysiol.00995.2006. [DOI] [PubMed] [Google Scholar]
- 26.Gordon S., Plüddemann A., Martinez Estrada F. Macrophage heterogeneity in tissues: phenotypic diversity and functions. Immunol. Rev. 2014;262(1):36–55. doi: 10.1111/imr.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schyns J., Bureau F., Marichal T. Lung interstitial macrophages: past, present, and future. J Immunol Res. 2018;2018:5160794. doi: 10.1155/2018/5160794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibbings S.L., Thomas S.M., Atif S.M., et al. Three unique interstitial macrophages in the murine lung at steady state. Am. J. Respir. Cell Mol. Biol. 2017;57(1):66–76. doi: 10.1165/rcmb.2016-0361OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hussell T., Bell T.J. Alveolar macrophages: plasticity in a tissue-specific context. Nat Rev Immunol. 2014;14(2):81–93. doi: 10.1038/nri3600. [DOI] [PubMed] [Google Scholar]
- 30.Ural B.B., Yeung S.T., Damani-Yokota P., et al. Identification of a nerve-associated, lung-resident interstitial macrophage subset with distinct localization and immunoregulatory properties. Sci Immunol. 2020:5(45). doi: 10.1126/sciimmunol.aax8756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou B., Magana L., Hong Z., et al. The angiocrine Rspondin3 instructs interstitial macrophage transition via metabolic-epigenetic reprogramming and resolves inflammatory injury. Nat. Immunol. 2020;21(11):1430–1443. doi: 10.1038/s41590-020-0764-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sabatel C., Radermecker C., Fievez L., et al. Exposure to bacterial CpG DNA protects from airway allergic inflammation by expanding regulatory lung interstitial macrophages. Immunity. 2017;46(3):457–473. doi: 10.1016/j.immuni.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 33.Chakarov S., Lim H.Y., Tan L., et al. Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science. 2019:363(6432). doi: 10.1126/science.aau0964. [DOI] [PubMed] [Google Scholar]
- 34.Schyns J., Bai Q., Ruscitti C., et al. Non-classical tissue monocytes and two functionally distinct populations of interstitial macrophages populate the mouse lung. Nat. Commun. 2019;10(1):3964. doi: 10.1038/s41467-019-11843-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Linkermann A., Stockwell B.R., Krautwald S., et al. Regulated cell death and inflammation: an auto-amplification loop causes organ failure. Nat Rev Immunol. 2014;14(11):759–767. doi: 10.1038/nri3743. [DOI] [PubMed] [Google Scholar]
- 36.Guo H., Callaway J.B., Ting J.P. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat. Med. 2015;21(7):677–687. doi: 10.1038/nm.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He D.K., Chen J.F., Shao Y.R., et al. Adenovirus-delivered angiopoietin-1 ameliorates phosgene-induced acute lung injury via inhibition of NLRP3 inflammasome activation. Inhal. Toxicol. 2018;30(4–5):187–194. doi: 10.1080/08958378.2018.1492648. [DOI] [PubMed] [Google Scholar]
- 38.Li J., Tian H., Yang J., et al. Long noncoding RNAs regulate cell growth, proliferation, and apoptosis. DNA Cell Biol. 2016;35(9):459–470. doi: 10.1089/dna.2015.3187. [DOI] [PubMed] [Google Scholar]
- 39.Bär C., Chatterjee S., Thum T. Long noncoding RNAs in cardiovascular pathology, diagnosis, and therapy. Circulation. 2016;134(19):1484–1499. doi: 10.1161/CIRCULATIONAHA.116.023686. [DOI] [PubMed] [Google Scholar]
- 40.Ding Y., Guo F., Zhu T., et al. Mechanism of long non-coding RNA MALAT1 in lipopolysaccharide-induced acute kidney injury is mediated by the miR-146a/NF-κB signaling pathway. Int. J. Mol. Med. 2018;41(1):446–454. doi: 10.3892/ijmm.2017.3232. [DOI] [PubMed] [Google Scholar]
- 41.Al Zoubi S., Chen J., Murphy C., et al. Linagliptin attenuates the cardiac dysfunction associated with experimental sepsis in mice with pre-existing type 2 diabetes by inhibiting NF-κB. Front. Immunol. 2018;9:2996. doi: 10.3389/fimmu.2018.02996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tian H., Wu M., Zhou P., et al. The long non-coding RNA MALAT1 is increased in renal ischemia-reperfusion injury and inhibits hypoxia-induced inflammation. Ren. Fail. 2018;40(1):527–533. doi: 10.1080/0886022X.2018.1487863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang W.J., Zeng X.Y., Jiang S.L., et al. Long non-coding RNA MALAT1 sponges miR-149 to promote inflammatory responses of LPS-induced acute lung injury by targeting MyD88. Cell Biol. Int. 2019:1–10. doi: 10.1002/cbin.11235. [DOI] [PubMed] [Google Scholar]
- 44.Li H., Shi H., Ma N., et al. BML-111 alleviates acute lung injury through regulating the expression of lncRNA MALAT1. Arch. Biochem. Biophys. 2018;649:15–21. doi: 10.1016/j.abb.2018.04.016. [DOI] [PubMed] [Google Scholar]
- 45.Kölling M., Genschel C., Kaucsar T., et al. Hypoxia-induced long non-coding RNA Malat1 is dispensable for renal ischemia/reperfusion-injury. Sci. Rep. 2018;8(1):3438. doi: 10.1038/s41598-018-21720-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang Q., Huang C., Luo Y., et al. Circulating lncRNA NEAT1 correlates with increased risk, elevated severity and unfavorable prognosis in sepsis patients. Am. J. Emerg. Med. 2018;36(9):1659–1663. doi: 10.1016/j.ajem.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 47.Groot M., Zhang D., Jin Y. Long non-coding RNA review and implications in lung diseases. JSM Bioinform Genom Proteom. 2018:3(2). [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou H., Wang X., Zhang B. Depression of lncRNA NEAT1 antagonizes LPS-evoked acute injury and inflammatory response in alveolar epithelial cells via HMGB1-RAGE signaling. Mediat. Inflamm. 2020;2020:8019467. doi: 10.1155/2020/8019467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H., Shi H., Gao M., et al. Long non-coding RNA CASC2 improved acute lung injury by regulating miR-144-3p/AQP1 axis to reduce lung epithelial cell apoptosis. Cell Biosci. 2018;8:15. doi: 10.1186/s13578-018-0205-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang H.R., Guo X.Y., Liu X.Y., et al. Down-regulation of lncRNA CASC9 aggravates sepsis-induced acute lung injury by regulating miR-195-5p/PDK4 axis. Inflamm. Res. 2020;69(6):559–568. doi: 10.1007/s00011-020-01316-2. [DOI] [PubMed] [Google Scholar]
- 51.Li J., Liu S. LncRNA GAS5 suppresses inflammatory responses and apoptosis of alveolar epithelial cells by targeting miR-429/DUSP1. Exp. Mol. Pathol. 2020;113:104357. doi: 10.1016/j.yexmp.2019.104357. [DOI] [PubMed] [Google Scholar]
- 52.Li H.B., Zi P.P., Shi H.J., et al. Role of signaling pathway of long non-coding RNA growth arrest-specific transcript 5/microRNA-200c-3p/angiotensin converting enzyme 2 in the apoptosis of human lung epithelial cell A549 in acute respiratory distress syndrome. Zhonghua Yi Xue Za Zhi. 2018;98(41):3354–3359. doi: 10.3760/cma.j.issn.0376-2491.2018.41.013. [DOI] [PubMed] [Google Scholar]
- 53.Zhang D., Lee H., Haspel J.A., et al. Long noncoding RNA FOXD3-AS1 regulates oxidative stress-induced apoptosis via sponging microRNA-150. FASEB J. 2017;31(10):4472–4481. doi: 10.1096/fj.201700091R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y., Cheng H.P., Bao T.P., et al. Expression of long non-coding RNA NANCI in lung tissues of neonatal mice with hyperoxia-induced lung injury and its regulatory effect on NKX2.1. Zhongguo Dang Dai Er Ke Za Zhi. 2017;19(2):215–221. doi: 10.7499/j.issn.1008-8830.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu G., Mei H., Chen M., et al. Protective effect of agmatine against hyperoxia-induced acute lung injury via regulating lncRNA gadd7. Biochem. Biophys. Res. Commun. 2019;516(1):68–74. doi: 10.1016/j.bbrc.2019.04.164. [DOI] [PubMed] [Google Scholar]
- 56.Zhang J., Mao F., Zhao G., et al. Long non-coding RNA SNHG16 promotes lipopolysaccharides-induced acute pneumonia in A549 cells via targeting miR-370-3p/IGF2 axis. Int. Immunopharmacol. 2020;78:106065. doi: 10.1016/j.intimp.2019.106065. [DOI] [PubMed] [Google Scholar]
- 57.Zhou Z., Zhu Y., Gao G., et al. Long noncoding RNA SNHG16 targets miR-146a-5p/CCL5 to regulate LPS-induced WI-38 cell apoptosis and inflammation in acute pneumonia. Life Sci. 2019;228:189–197. doi: 10.1016/j.lfs.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 58.Zhu J., Bai J., Wang S., et al. Down-regulation of long non-coding RNA SNHG14 protects against acute lung injury induced by lipopolysaccharide through microRNA-34c-3p-dependent inhibition of WISP1. Respir. Res. 2019;20(1):233. doi: 10.1186/s12931-019-1207-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu M., Han T., Shi S., et al. Long noncoding RNA HAGLROS regulates cell apoptosis and autophagy in lipopolysaccharides-induced WI-38 cells via modulating miR-100/NF-κB axis. Biochem. Biophys. Res. Commun. 2018;500(3):589–596. doi: 10.1016/j.bbrc.2018.04.109. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y., Zhu Y., Gao G., et al. Knockdown XIST alleviates LPS-induced WI-38 cell apoptosis and inflammation injury via targeting miR-370-3p/TLR4 in acute pneumonia. Cell Biochem. Funct. 2019;37(5):348–358. doi: 10.1002/cbf.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li J., Wei L., Han Z., et al. Long non-coding RNA X-inactive specific transcript silencing ameliorates primary graft dysfunction following lung transplantation through microRNA-21-dependent mechanism. EBioMedicine. 2020;52:102600. doi: 10.1016/j.ebiom.2019.102600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li X., Zhang Q., Yang Z. Silence of MEG3 intensifies lipopolysaccharide-stimulated damage of human lung cells through modulating miR-4262. Artif Cells Nanomed Biotechnol. 2019;47(1):2369–2378. doi: 10.1080/21691401.2019.1623233. [DOI] [PubMed] [Google Scholar]
- 63.Li R., Fang L., Pu Q., et al. MEG3–4 is a miRNA decoy that regulates IL-1β abundance to initiate and then limit inflammation to prevent sepsis during lung infection. Sci. Signal. 2018:11(536). doi: 10.1126/scisignal.aao2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Y., Fu X., Yu B., et al. Long non-coding RNA THRIL predicts increased acute respiratory distress syndrome risk and positively correlates with disease severity, inflammation, and mortality in sepsis patients. J. Clin. Lab. Anal. 2019;33(6) doi: 10.1002/jcla.22882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wan B., Xu W.J., Xu W.N., et al. Plasma long noncoding RNA IL-7R as a prognostic biomarker for clinical outcomes in patients with acute respiratory distress syndrome. Clin. Respir. J. 2018;12(4):1607–1614. doi: 10.1111/crj.12717. [DOI] [PubMed] [Google Scholar]
- 66.Barata J.T., Durum S.K., Seddon B. Flip the coin: IL-7 and IL-7R in health and disease. Nat. Immunol. 2019;20(12):1584–1593. doi: 10.1038/s41590-019-0479-x. [DOI] [PubMed] [Google Scholar]
- 67.Huang Z., Liu H., Zhang X., et al. Transcriptomic analysis of lung tissues after hUC-MSCs and FTY720 treatment of lipopolysaccharide-induced acute lung injury in mouse models. Int. Immunopharmacol. 2018;63:26–34. doi: 10.1016/j.intimp.2018.06.036. [DOI] [PubMed] [Google Scholar]
- 68.Zhong Y., Wang Y., Zhang C., et al. Identification of long non-coding RNA and circular RNA in mice after intra-tracheal instillation with fine particulate matter. Chemosphere. 2019;235:519–526. doi: 10.1016/j.chemosphere.2019.06.122. [DOI] [PubMed] [Google Scholar]
- 69.Meng S.S., Xu X.P., Chang W., et al. LincRNA-p21 promotes mesenchymal stem cell migration capacity and survival through hypoxic preconditioning. Stem Cell Res Ther. 2018;9(1):280. doi: 10.1186/s13287-018-1031-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang X., Liu H., Wang S., et al. LncRNA analysis of lung tissues after hUC-MSCs and FTY720 treatment of lipopolysaccharide-induced acute lung injury in mouse models. Int. Immunopharmacol. 2019;71:68–75. doi: 10.1016/j.intimp.2019.03.017. [DOI] [PubMed] [Google Scholar]
- 71.Li Y., Yin Z., Fan J., et al. The roles of exosomal miRNAs and lncRNAs in lung diseases. Signal Transduct Target Ther. 2019;4:47. doi: 10.1038/s41392-019-0080-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang Y., Yang L., Liu Z., et al. Long noncoding RNA NEAT 1 and its target microRNA-125a in sepsis: correlation with acute respiratory distress syndrome risk, biochemical indexes, disease severity, and 28-day mortality. J. Clin. Lab. Anal. 2020;34(12) doi: 10.1002/jcla.23509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Walter J., Ware L.B., Matthay M.A. Mesenchymal stem cells: mechanisms of potential therapeutic benefit in ARDS and sepsis. Lancet Respir. Med. 2014;2(12):1016–1026. doi: 10.1016/S2213-2600(14)70217-6. [DOI] [PubMed] [Google Scholar]
- 74.Matthay M.A., Calfee C.S., Zhuo H., et al. Treatment with allogeneic mesenchymal stromal cells for moderate to severe acute respiratory distress syndrome (START study): a randomised phase 2a safety trial. Lancet Respir. Med. 2019;7(2):154–162. doi: 10.1016/S2213-2600(18)30418-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Askenase P.W. COVID-19 therapy with mesenchymal stromal cells (MSC) and convalescent plasma must consider exosome involvement: do the exosomes in convalescent plasma antagonize the weak immune antibodies? J Extracell Vesicles. 2020;10(1) doi: 10.1002/jev2.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.He D., Zheng J., Hu J., et al. Long non-coding RNAs and pyroptosis. Clin. Chim. Acta. 2020;504:201–208. doi: 10.1016/j.cca.2019.11.035. [DOI] [PubMed] [Google Scholar]
- 77.Wang L., Liu J., Xie W., et al. Overexpression of MALAT1 relates to lung injury through sponging miR-425 and promoting cell apoptosis during ARDS. Can. Respir. J. 2019;2019:1871394. doi: 10.1155/2019/1871394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yao M.Y., Zhang W.H., Ma W.T., et al. Long non-coding RNA MALAT1 exacerbates acute respiratory distress syndrome by upregulating ICAM-1 expression via microRNA-150-5p downregulation. Aging (Albany NY) 2020;12(8):6570–6585. doi: 10.18632/aging.102953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wei S., Wang K., Huang X., et al. Knockdown of the lncRNA MALAT1 alleviates lipopolysaccharide-induced A549 cell injury by targeting the miR-17-5p/FOXA1 axis. Mol. Med. Rep. 2019;20(2):2021–2029. doi: 10.3892/mmr.2019.10392. [DOI] [PubMed] [Google Scholar]
- 80.Zhu W., Men X. Negative feedback of NF-κB signaling by long noncoding RNA MALAT1 controls lipopolysaccharide-induced inflammation injury in human lung fibroblasts WI-38. J. Cell. Biochem. 2020;121(2):1945–1952. doi: 10.1002/jcb.29429. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.