Summary
Reactive oxygen species (ROS) are naturally produced by several redox reactions during plant regular metabolism such as photosynthesis and respiration. Due to their chemical properties and high reactivity, ROS were initially described as detrimental for cells during oxidative stress. However, they have been further recognized as key players in numerous developmental and physiological processes throughout the plant life cycle. Recent studies report the important role of ROS as growth regulators during plant root developmental processes such as in meristem maintenance, in root elongation, and in lateral root, root hair, endodermis, and vascular tissue differentiation. All involve multifaceted interplays between steady-state levels of ROS with transcriptional regulators, phytohormones, and nutrients. In this review, we attempt to summarize recent findings about how ROS are involved in multiple stages of plant root development during cell proliferation, elongation, and differentiation.
Subject areas: Biological Sciences, Plant Biology, Plant Development, Plant Physiology
Graphical abstract
Highlights
-
•
ROS are signaling molecules that choreograph root structures and anatomies
-
•
MYB36 acts in lateral root development, by manipulating the ROS balance
-
•
ROS do not act in isolation but rather as part of a large and complex redox signaling
-
•
ROS-producing or ROS-scavenging machineries are copresent simultaneously within cells
Biological Sciences; Plant Biology; Plant Development; Plant Physiology
Introduction
Reactive oxygen species
The emergence of complex multicellular life was interconnected with the buildup of oxygen on Earth. Oxygen accumulation resulted from the emergence of oxygenic photosynthesis and consequently enabled the evolution of aerobic life. This evolving era is characterized by high-energy yielding metabolic pathways (Ward et al., 2019) such as aerobic respiration and oxidative phosphorylation. These pathways, where oxygen serves as the final acceptor of electrons, are more thermodynamically efficient in the evolving complexity than other nonaerobic pathways with nonoxygenic oxidants. This denotes oxygen as the most energetic oxidant in biological life (Reinhard et al., 2016). The reduction of oxygen by the ATP-producing electron transport chains, in addition to other metabolic pathways in which molecular oxygen is the ultimate oxidizing agent, can generate various levels of reactive oxygen species (ROS) (see Box 1). Therefore, ROS are constitutive products of aerobic metabolism (Singh et al., 2016; Rabinovitch et al., 2017), and it is definite that the emergence and permanence of aerobic life was never “ROS-free” (Mittler, 2017).
Box 1. ROS homeostasis concepts.
ROS homeostasis box
-
•
What are ROS? ROS are highly reactive molecules that originate from oxygen (O2) where each has its distinct life space and chemical properties (Ozcan and Ogun, 2015).
-
•
ROS molecules: ROS are in various forms among which are the radicals having free electrons such as superoxide (O2.-), hydroxyl radical (OH•), peroxyl (RO2-), and alkoxyl (RO-); others are nonradicals as hypochlorous acid (HOCl), ozone (O3), singlet oxygen (1O2), and hydrogen peroxide (H2O2) (Mhamdi and Van Breusegem, 2018b). Each type of ROS molecule has distinct chemical properties: singlet oxygen can oxidize lipids, proteins, and guanidine residues of DNA; superoxide like singlet oxygen has a half-life time of 1-4 μs and reacts with Fe-S proteins; and hydroxyl radicals are extremely reactive and unstable with a half-life time of 1 ns (Mittler, 2017; Waszczak et al., 2018). In contrast, hydrogen peroxide is the most stable ROS (more than 1 ms) and, therefore, is the predominant ROS involved in cellular signaling.
-
•
ROS homeostasis is controlled by the delicate balance between ROS production and scavenging, maintaining a threshold boundary between redox potential and cytotoxicity (Saini et al., 2018).
-
•
ROS in the organelles. ROS are produced in the mitochondria by mitochondrial respiration, in the chloroplasts during photosynthesis, in the peroxisome-localized photorespiratory reactions, and in the apoplast by NADPH oxidases such as the respiratory burst oxidase homologs (RBOHs), peroxidases, and other oxidases.
-
•ROS in the apoplast. Two enzymatic families contribute to the apoplastic ROS production. In addition, other families of enzymes are also able to modify the ROS homeostasis in the apoplast such as oxalate oxidases, polyamine oxidases, quinone oxidases, lipoxygenases, etc.
-
1.The NADPH oxidases also called respiratory burst oxidative homologs (RBOHs) (Orman-Ligeza et al., 2016) consist of transmembrane proteins which produce superoxide O2.-. In Arabidopsis thaliana, ten genes have been asserted to encode RBOHs (RBOHA-RBOHJ) (Marino et al., 2012).
-
2.The class III peroxidase family (PRX) consists of apoplastic proteins that oxidize various substrates by using H2O2 as an electron acceptor but can also regulate the local concentration of H2O2 or produce ROS (Francoz et al., 2015). In Arabidopsis thaliana, 73 members of this family have been found (Tognolli et al., 2002), but only few of them have been characterized.
-
1.
-
•
Apoplastic ROS can be channeled into the cytoplasm by multiple aquaporin of the PIP-type (Tian et al., 2016).
-
•ROS scavenging is a two-armed mechanism:
-
1.The first arm consists of the antioxidative enzymes as the superoxide dismutase (SOD), catalase (CAT), ascorbate peroxidase (APx), monodehydroascorbate reductase (MDHAR), dehydroascorbate reductase (DHAR), glutathione reductase (GR), glutathione peroxidase (GPx), and class III peroxidase (PRX) (Das and Roychoudhury, 2014; Choudhary et al., 2020).
-
2.The second arm comprises nonenzymatic antioxidants such as ascorbic acid (ASC), reduced glutathione (GSH), carotenoids, flavonoids, and proline (Eltayeb et al., 2007).
-
1.
See also Table 1 for more details.
ROS homeostasis
Earlier on, our understanding to ROS was restricted to the lethal manifestations of the disparity between ROS production and scavenging (Sies, 1986, 1997). This disparity leads to oxidative damage to different molecules and cellular components as nucleic acids, proteins, and membranes and cell death (Das and Roychoudhury, 2014; Anjum et al., 2015). Therefore, as a part of their evolutionary adaptations, plants have developed important ROS-detoxifying machineries (see Box 1). In planta, a large battery of antioxidative enzymes was identified in addition to various antioxidants that form together a double-armed ROS-scavenging mechanism (Das and Roychoudhury, 2014). Together, they forage ROS and counterattack the elevations in their concentrations, preventing oxidative bursts and the consequent detrimental manifestations (Mittler et al., 2004) and maintaining a threshold boundary between redox potential and cytotoxicity, referred to as ROS homeostasis (Saini et al., 2018). In Arabidopsis thaliana, there exists a highly dynamic and complementary network of more than 150 genes that have been identified and correlated to ROS homeostasis (Miller et al., 2009). Few examples related to root development are listed in Table 1. On another side, ROS have been lately recognized for playing fundamental roles as signaling molecules engaged in plant development. They are currently considered as key actors that control cell proliferation, patterning, and differentiation during plant growth (Schippers et al., 2016; Tsukagoshi, 2016; Tognetti et al., 2017). They are also involved in the perception of environmental signals, their transduction, and their translation into physiological responses. In fact, changes in ROS homeostasis are signals that can trigger growth modifications (Miller et al., 2010; Das and Roychoudhury, 2014; Saini et al., 2018). These modifications derive from the association between ROS homeostasis and subsequent adjustments of gene expression. Hence, being fundamental in plant growth and development, ROS detection in time and space is required to better understand their roles and regulations. This triggered the development of several dyes, probes, and, recently, genetically encoded sensors to detect ROS, mostly as H2O2 (see Box 2).
Table 1.
Selected examples of components that regulate ROS homeostasis: ROS scavengers, ROS producers, and an ROS sensor involved in root development processes
| Gene name/protein | Subcellular localization | Functional evidence, mutant analysis, phenotypes | References |
|---|---|---|---|
| ROS scavengers | |||
| Superoxide dismutase (SOD) | Chloroplast, mitochondria, peroxisome, and cytosol | Defense against oxidative damage. It eradicates O2− by producing O2 and H2O2 | (Saini et al., 2018) |
| Catalase (CAT) CAT1, CAT2, CAT3 |
Peroxisome and mitochondria in pollen and seeds; photosynthetic tissues and roots | Removes H2O2 and transforms it into H2O and O2. | (Das and Roychoudhury, 2014) |
| Ascorbate peroxidase (APX) | Chloroplast and cytosol | Redundant to CAT. Transforms H2O2 into H2O and dehydroascorbate (DHA) | (Sharma and Dubey, 2004) |
| Monodehydroascorbate reductase (MDHAR) | Colocalized with APX in the peroxisome | Replenishes the ascorbic acid (AA) reservoir | (Das and Roychoudhury, 2014) |
| Dehydroascorbate reductase (DHAR) | Intracellular and in the apoplast | Reduces DHA to AA using glutathione (GSH) as a reductant | (Eltayeb et al., 2007) |
| Glutathione reductase (GR) | Chloroplast, mitochondria and cytosol | Maintain a high cellular GSH:GSSG ratio, reducing glutathione disulfide (GSSG) into glutathione (GSH). | (Das and Roychoudhury, 2014) |
| Guaiacol peroxidase (GP) | Active both intracellular and extracellular | Remove of H2O2 during either regular metabolism or oxidative burst. | (Saini et al., 2018) |
| RBOHs | |||
| RBOHF (AT1G64060) RBOHD (AT5G47910) |
Roots: endodermis, lateral roots and vascular tissues; stems, seedlings, inflorescences, leaves and guard cells. RBOHF is phosphorylated by CIPK26, binding Ca+2. Besides interacts with SRC2. It is upregulated by aba. RBOHD can interacts with BIK1 and PBL13 Atrbohf and Atrbohd control ROI spatial production |
Are required for CS formation, producing H2O2. The number and density of LR increase in the loss-of-function mutants of rbohd and rbohf, also show lower H2O2 and O2- concentration in the root cap, MZ and EZ as compared to the WT. But has and increase of O2- in the DZ increasing LR density in rbohd and rbohf. RBOHD and RBOHF negatively regulate LRP formation and LR emergence by downregulating the peroxidase activity | (Lee et al., 2013; Li et al., 2014; Franke, 2015; Pedreira et al., 2011) |
| RBOHC (AT5G51060) | In roots, is expressed in the epidermis, TZ, EZ, DZ and in elongating root hairs. RBOHC actively produces OH− in the apoplast at the tip of the root hair. | It is required for the production of reactive oxygen species that regulate cell expansion through the activation of Ca2+ channels. His expression is increased in potassium starvation. Participates in root hair elongation. rbohc mutants shows shorter root hairs | (Mhamdi and Van Breusegem, 2018a, 2018b; Foreman et al., 2003) |
| RBOHE (AT1G19230) | Expressed in roots: lateral root; inflorescences, leaves and stems. GUS reporter shows accumulation of RBOHE at the starting site of the lateral root | Loss-of-function mutant rbohe delays the development of the lateral root | (Chapman et al., 2019) |
| Apoplastic type-III peroxidases (PRXs) | |||
| PRX72 (AT5G66390) | Roots endodermis; and xylem | PRX72 expression is upregulated when the xylem vessel elements are actively forming, playing a role in xylem formation. In prx72 mutants, lignin levels were 40% less compared to the WT. It has been observed that PRX72 expression is increased in the lac4-2 lac11 lac17 triple mutants, implying a possible functional linkage between PRXs and LACs |
(Herrero et al., 2013; Zhao et al., 2013) |
| PRX17 (AT2G22420) | Root endodermis and xylem. Is regulated by the MADS-box transcription factor AGL15 and interacts with GRF3.35S:PRX17 over accumulates xylem compared to WT |
Participates in lignin polymerization of TEs or indirectly via ROS signaling. Loss-of-function mutant, prx17, has lower lignin content in most organs | (Cosio et al., 2017; Herrero et al., 2013; Hoffmann et al., 2020) |
| PRX64 (AT5G42180) | Root endodermis Upregulated by cold |
Uses the H2O2 for monolignols oxidation, catalyzing lignin formation | (Lee et al., 2013; Franke, 2015) |
| PRX37 (AT4G08770) | Root endodermis and vascular bundles. Interacts with RGS1 and NAC3 |
Participates in phenolic "cross-linking" activity during lignin deposition in the cell wall. Overexpression of prx37 shows a dwarf phenotype with smaller plants and delayed development, and a increase in the amount of esterified phenolic products | (Pedreira et al., 2011) |
| PRX01 (AT1G05240) PRX44 (AT4G26010) PRX73 (AT5G67400) |
Root hair. Coexpressed with other genes as extensins and glycoproteins in the cell wall | Participation in cell wall expansion in root hairs and ROS homeostasis. PRX01 is involved in the lignification of cells walls. Double mutants of t prx44 prx73, prx01 prx44, and prx01 prx73, as well as the triple null mutant, prx01 prx44 prx73 showed shorter root hairs. PRX1 and PRX73 are upregulated in the overexpressors of RSL4 and downregulated in rsl4 KO mutants | (Mangano et al., 2017; Marzol et al., 2020) |
| PRX07 (AT1G30870) PRX57 (AT5G17820) |
Lateral roots. Root hair PRX57: Positively light-regulated during the first stage of development, upregulated by cold treatment, and downregulated by salicylic acid |
PRX7 participates in root hair elongation, is target of RSL4. PRX7 and PRX57 were detected to be auxin-independent. KO mutants prx7 and prx57 have lower LR density and number due to changes of ROS balance in the initiation stage | (Manzano et al., 2014; Vijayakumar et al., 2016) |
| Glutathione peroxidase (GPX) family | Lateral root | gpx mutants develop larger lateral root primordia, indicating that the GPX family plays a role in regulating root architecture, especially GPX1 and GPX7, having a critical role in development and formation of the lateral root | (Choudhary et al., 2020) |
| ROS sensor | |||
| HYDROGEN PEROXIDE-INDUCED Ca2+ INCREASES 1 (HPCA1) (AT5G49760) | HPCA1 is localized to the plasma membrane and is activated by H2O2 via covalent modification of extracellular cysteine residues, which leads to autophosphorylation of HPCA1. | HPCA1 encodes an LRR-RK that contains sites for extracelular peroxide sensing. This leads to the activation of intracellular kinase activity leading to calcium influx and hence activation of downstream signaling pathways. This extracelular peroxide sensing mechanism by HPCA1 is unique to plants. | (Wu et al., 2020) |
| ROS regulators | |||
| Retarded root growth (RRG) (AT1G69380) | Root meristem, primary and lateral root tips. Also present in leaves and pollen. In root tips, is preferentially expressed in QC cells, cortex/endodermis stem cells and endodermal and stele cells of root meristems | Promote cellular division in the RAM. KO mutants, rrg-1 and rrg-2 revealed a short-root phenotype compared to the WT. Required for the maintenance of mitochondrial structure. Positive regulator of cell division and but negative regulator of cell expansion in the postembryonic root meristem | (Zhou et al., 2011) |
| Prohibitin3 (PHB3) (AT5G40770) | Expressed in proliferative tissues, vasculature, stems, leaves, and roots. Preserves redox homeostasis in the mitochondria |
Inhibits the cell cycle. Maintains the stem cell niche in QC and activate the cellular division in the proximal meristem. ROS production is induced and subsequently inhibits PHB3 expression. In phb3 mutants, genes implicated in ROS production showed higher expression (i.e. AOX1 and NA | (Kong et al., 2018) |
| Casparian strip membrane domain proteins (CASPs) CASP1 (AT2G36100) |
Root endodermis. Interacts with CASP2, CASP3, CASP4, and CASP5. Along with CASP2, it is required for the localization of ESB1 | CASPs coordinate the proper functions of the different components in the lignin-polymerizing machinery. Their interaction with PRX64 and RBOHF and determine their precise localization at the CSs development site. CASP1 guides this localization by interacting with SGN1 and SGN3 | (Alassimone et al., 2016; Li et al., 2018a) |
| MYB domain protein 36 (MYB36) (AT5G57620) | Leaves, roots endodermis, lateral root and seedlings. Directly upregulated by SCR | It promotes differentiation of the endodermis during root development. Regulates the spatial expression of genes involved in positioning and build of CSs in the root endodermis (i.e., CASPs, PRX64, and ESB1). Loss-of-function mutants, myb36, show deficit CS, with an abnormal distribution of lignin in endodermal cells. The mutant alleles myb36-5 and myb36-2 had less LRs verse the WT |
(Cui et al., 2014; Kamiya et al., 2015; Liberman et al., 2015; Fernández-Marcos et al., 2017) |
| Like sex four 2 (LSF2) (AT3G10940) | Widely expressed, especially in root hair | lsf2-1 KO mutants showed lower O2-generation rate but higher levels of H2O2 upon oxidative stress treatments. LSF2 interacts with MPK8, which regulates the cytosolic redox balance | (Zhao et al., 2016) |
| Feronia (FER) (AT3G51550) | Widely expressed, especially in root hairs. It is induced by brassinosteroids. | FER acts as an upstream regulator for the Rac/Rop-signaling pathway that controls ROS-mediated root hair development. fer and rhd2 mutants shows short root hairs, and a decreased in ROS accumulation. FER interacts with ROP2, promoting radical hair growth upstream of RBOHC, interacts with RopGEF1, RALF1, ROP2 and BRL3 | (Chapman et al., 2019) |
| Root hair defective six like4 (RSL4) (AT1G27740) | Root hair, leaves, and flowers. Promotes post mitotic cell growth in root hair cells. Induced by jasmonate (JA) and low-phosphate conditions |
RSL4 is involved in the regulation of root hair elongation. KO rsl4 plants have no root hair. RSL4 is controlled by ARFs, inducing localized ROS synthesis. Has targets genes that promotes root hair growth, like GBF2, EXO70A1, CPK11 and SAC1. RSL4 is a direct transcriptional target of RHD6. RSL4 can bind to the RHE in the regulatory region of PRX1, PRX44, PRX60, and PRX73 | (Mangano et al., 2017; Chapman et al., 2019) |
| Hypoxia responsive ERF genes(HRE1 and HRE2) Ethylene response factor genes (ERF and RAP2) |
Lateral roots | KO mutants rap2, hre1, hre2, erf have more LRs compared to the WT, these mutants affect the delayed emergence in LR development | (Shukla et al., 2019) |
| UPBEAT1 (UPB1) (AT2G47270) | Expressed in the root vascular tissue and in root hairs and lateral root caps. 35S:UPB1-3YFP, showed reduced root length and a decrease in cortex cell number in the MZ | UPB1 manipulates the ROS homeostasis, regulating cellular division and differentiation. upb1 KO mutants had more and longer emerging LRs compared to WT, another mutant upb1-1, shows significant increase in the number of cortical cells, indicating enlargement of the meristem. UPB1 controls the expression of several peroxidases (PRX39, 40, and 57) |
(Tsukagoshi et al., 2010; Manzano et al., 2014) |
Box 2. Tool examples for ROS detection in plant cells.
Tools for ROS detection
-
•
Diaminobenzidine (DAB) and nitro blue tetrazolium (NBT) staining. Both staining methods are used to visualize H2O2 and superoxide radicals. DAB and NBT are not specific or direct measurements of both ROS, and color formation does not always reflect the levels of the desired ROS. NBT staining can reflect the presence of ascorbate or the activity of dehydrogenases, while DAB brownish color accumulates in the presence of higher peroxidase activity. Dihydroethidium (DHE) is also used to detect O2.-.
-
•
Dichlorofluorescein (DCF)-derived fluorescent dyes. DCF is widely used for quantification of H2O2 although it quantifies total ROS. This method is not very reliable and is not specific. Dichlorofluorescin (DCFH) does not react with H2O2 or other ROS directly. ROS imaging can be further complicated by the presence of endogenous autofluorescent compounds. Other fluorescent dyes exist such as BES-H2O2-Ac to detect H2O2 (Tsukagoshi et al., 2010).
-
•
Amplex Red (AR) and Amplex Ultra Red (AUR) fluorescent dyes. AR is used for cytoplasmic H2O2, while AUR is employed for extracellular-apoplastic H2O2 since it cannot go through the plasma membrane. Both, AU/AUR, undergoes one electron oxidation and yields the fluorescent product resorufin, a reaction with 1:1 (AR/AUR: H2O2) stoichiometry, in which concentration of horseradish peroxidase and pH play important roles. These two dyes have been used to track H2O2 levels in several plant tissues (Rhee et al., 2010; Deng et al., 2011; Sang et al., 2012; Chakraborty et al., 2016; Tian et al., 2016).
-
•
Genetically encoded probes. Genetically encoded sensors have been engineered to measure H2O2 which have several advantages over the dyes because they are noninvasive, can be targeted in time and space (at tissues and cellular level), and are stable over time. To name some of the most widely used in plant cells are roGFP1 (Schwarzländer et al., 2008), roGFP-GSH (Bratt et al., 2016), GRX1-roGFP2 (Gutscher et al., 2008; Schwarzländer et al., 2008), roGFP-Orp1 (Dagda et al., 2017), and HyPer (Costa et al., 2010; Hernández-Barrera et al., 2015; Mullineaux et al., 2018).
Several tools have been developed for the detection and visualization of ROS in plant tissues. For more details on limitation and controls to be included for their correct use, please see Noctor and Foyer, 2016; Mhamdi and Van Breusegem, 2018a; Fichman et al., 2019.
ROS production and scavenging
In plants, ROS are constitutively produced during different biochemical reactions located in various cellular compartments, i.e., chloroplast, mitochondria, and the peroxisome in addition to the apoplast. In the chloroplasts, ROS production is associated with the light-dependent reactions and is due to the leakage of electrons (Ivanov et al., 2018). Nevertheless, in the mitochondria, ROS production is relatively lower and is related to the respiratory chain (Marchi et al., 2012; Kröller-Schön et al., 2014). In addition, peroxisomes are also sites for hydrogen peroxide production during different processes as the glycolate oxidase reaction, fatty acid β-oxidation by acyl Co-A oxidase, and an enzymatic reaction of flavin oxidases. Furthermore, with reduced CO2 availability during photosynthesis, RUBISCO binds O2 and oxygenates ribulose-1,5-bisphosphate into glycolate. The latter is subsequently oxidized to glyoxylate in the peroxisome with concomitant production of hydrogen peroxide. Likewise, superoxide radicals are produced by xanthine oxidase in the peroxisomal matrix and by NADPH:ferricyanide reductase in the peroxisomal membrane (Del Río and López-Huertas, 2016). Moreover, two enzymatic families contribute to the apoplastic ROS production in a controlled manner: the respiratory burst oxidative homologs (RBOHs) (Orman-Ligeza et al., 2016) and the class III peroxidases (PRXs) (Francoz et al., 2015).
ROS released in the chloroplast during photosynthesis represent the biggest pool of ROS among the different compartments. These ROS are implicated in signaling processes such as the regulation programmed cell death (PCD) (Shapiguzov et al., 2012). In addition, ROS produced in mitochondria during oxidative phosphorylation are associated to roles in redox signaling, retrograde signaling, plant hormone action, and PCD during pathogen attacks (Huang et al., 2016). First, these ROS may possibly adjust protein functions in the mitochondrial matrix by oxidizing specific protein thiols as for alternative oxidases (AOXs) (Yoshida et al., 2013) and for several tricarboxylic acid cycle (TCA) enzymes (Daloso et al., 2015). Second, they are probably involved in the mitochondrial retrograde regulation (MRR) where the mitochondria communicate back their functional status to the nucleus. The observation of MRR responses shows remarkable overlaps with transcriptional responses to different ROS triggers and abiotic stress, and the induction of MRR with complex III inhibitor antimycin A increased ROS production (Van Aken et al., 2016). Moreover, the interaction between hormonal signaling that regulates all layers of plant development and cellular functions and mitochondrial signaling, including mitochondrial ROS signaling, has been recently reviewed (Berkowitz et al., 2016). Lastly, these ROS trigger the execution of PCD during pathogen attacks, where various mutants in mitochondrial functions have been reported to have diverse PCD or pathogen defense responses (Huang et al., 2016). To exemplify, Arabidopsis mosaic death 1, MOD1, codes for an enoyl-acyl carrier protein (ACP) reductase essential for fatty acid biosynthesis in the chloroplast and negatively regulates PCD in Arabidopsis. Recently, a study demonstrated that mitochondrial complex I is responsible for the ROS production that leads to PCD in the mod1 mutant (Wu et al., 2015). Furthermore, a comprehensive chloroplast-mitochondria communication pathway that regulates the generation of mitochondrial ROS and triggers PCD has been elucidated (Zhao et al., 2018). In turn, apoplastic ROS are engaged in plant development and responses to extrinsic signals. These ROS regulate plant cell division and cell elongation rate, and hence the whole plant growth (Podgórska et al., 2017); they are also implicated in regulating stomatal closure, cell wall lignification, and callose deposition in response to external signals (Podgórska et al., 2017; Qi et al., 2017; Janku et al., 2019; Farvardin et al., 2020). Obviously, ROS signaling encompasses several pathways within and between various organelles and cellular compartments. An elaborated understanding of all these pathways and their interconnections is still lacking, and their investigation remains a big challenge for plant biology research.
To sum up, in plants, ROS production is an inevitable part of the basal metabolism and occurs in various cellular compartments. Remarkably, ROS production may be suddenly enhanced in response to environmental cues; this is defined as the “oxidative burst” (Choudhary et al., 2020). The oxidative burst is one of the earliest and most important physiological responses to stresses (Saini et al., 2018); it fires various signaling pathways leading to transcriptional changes in the stress-related genes, resulting in a defensive response (Baxter et al., 2014; Sewelam et al., 2016; Zhang et al., 2016). ROS overproduction may lead to an irreversible oxidative damage as the degradation of biomolecules such as lipids, proteins, nucleic acids, and carbohydrates (Chapman et al., 2017). This may lead to physiological disruption or even cellular death (Anjum et al., 2015; Choudhary et al., 2020). Therefore, the normal cellular homeostasis requires maintaining the delicate balance between ROS production and scavenging (Mittler, 2017). ROS scavenging is a two-armed mechanism (Eltayeb et al., 2007) that shields the various cellular compartments against any provisional oxidative damage. Furthermore, many actors in ROS scavenging are also implicated in plant growth and development by taking part in processes such as mitosis, elongation, senescence, and PCD (Noctor et al., 2018).
ROS sensing
Understanding how plants sense ROS and translate their signals into physiological responses was thoroughly investigated. One mechanism to sense ROS is the oxidative post-translational modification (Ox-PTM) of proteins, which was recently reviewed (Waszczak et al., 2015). Numerous proteins with Cys residues have been identified and reviewed as targets of Ox-PTMs such as enzymes of the Calvin cycle or signal transduction enzymes such as transcription factors, kinases, phosphatases, and RNA-binding proteins (Dietz, 2014; Waszczak et al., 2015; Mittler, 2017; Sies et al., 2017). Apoplastic H2O2 is sufficiently stable to penetrate into the cytoplasm via aquaporin membrane proteins where it covalently modifies proteins. However, a recent breakthrough discovery characterized a cell surface ROS sensor, the HPCA1 (HYDROGEN PEROXIDE-INDUCED Ca2+ INCREASES 1) identified as the first known cell-surface-specific sensor for H2O2 in plants (Wu et al., 2020). HPCA1 encodes a leucine-rich-repeat receptor kinase (LRR-RK) that contains two special pairs of Cys residues in its extracellular domain. The thiol groups of Cys residues are targets for oxidation to sulfenic acids by H2O2, and these sites are the place for extracellular H2O2 sensing. More importantly, this covalent modification leads to the activation of HPCA1 intracellular kinase activity by its autophosphorylation, which subsequently triggers through unknown mechanisms Ca2+-channel gating and Ca2+ influx, followed by activation of downstream signaling pathways (Wu et al., 2020). Remarkably, the extracellular H2O2 sensing mechanism by HPCA1 does not resemble any H2O2 receptors or sensors previously reported for other eukaryotic organisms, highlighting that plants have evolved a unique manner to deal with ROS. Since H2O2 is also produced in several organelles, other H2O2 sensors may act together with HPCA1 to transmit organelle-specific redox messages to the nucleus, along with messages arising from the apoplastic face of the plasma membrane. On another hand, HPCA1 was recently identified to be involved in quinone sensing which triggers defense-related gene expression by elevating cytoplasmic Ca2+ and activating MAPKs (mitogen-activated protein kinase) signaling pathway named as CARD1 (CANNOT RESPOND TO DMBQ 1) (Laohavisit et al., 2020). The recognition of quinone molecules by CARD1 is also based on Cys residues in its apoplastic ectodomain. Yet, it is still unclear how extracellular quinone and H2O2 sensing are related to each other in the HPCA1/CARD1 proteins.
ROS recap
During the past years, several reviews recapped the recent findings about ROS as signaling molecules during natural metabolism or in response to exogenous factors. In 2004, the mechanisms of generation and removal of ROS during development and under biotic and abiotic stresses were reported, in addition to new insights into their complexity and roles from genetic analyses of ROS-detoxifying and ROS-signaling mutants (Apel and Hirt, 2004). Moreover, the important role of ROS waves as signaling molecules in and across different cells, the dynamics and specificity of these signals, and their cross talk with other networks were thoroughly interpreted (Mittler et al., 2011). Furthermore, upon zooming into the ROS gene network, the RBOHs were identified as a key protein at the center of this ROS signaling network. In fact, RBOHs demonstrated important functions in the cellular biological processes as integrating calcium signaling and protein phosphorylation necessary for controlled ROS production (Suzuki et al., 2011). In 2012, the influence of apoplastic and chloroplastic ROS on nuclear gene regulation and transcriptional reprogramming was pinpointed (Shapiguzov et al., 2012). Also, the oxidative burst after pathogen recognition and the consequent metabolic and proteomic fluxes during plant defense were described (O’Brien et al., 2012). Soon after came the findings that stress responses in plants are mediated by a spatiotemporal coordination between ROS and other signals to confer systemic acquired resistance or systemic acquired acclimation (Baxter et al., 2014). Additionally, numerous updates conferred the role of ROS during abiotic stresses, such as drought (Noctor et al., 2014), salinity (Bose et al., 2014), and anoxia (Gonzali et al., 2015). Further advancement in understanding the signaling aspect of ROS has been made in the past five years (Dietz et al., 2016; Sewelam et al., 2016; Mittler, 2017; Qi et al., 2017; Waszczak et al., 2018). Recently, the regulatory actions of ROS during plant development have also been extensively studied, yet many are still not well known. In fact, ROS have been shown to be involved in cell proliferation and differentiation, PCD, seed germination, organ growth, flower and pollen tube development, and leaf senescence (Singh et al., 2016; Mhamdi and Van Breusegem, 2018a).
ROS & roots
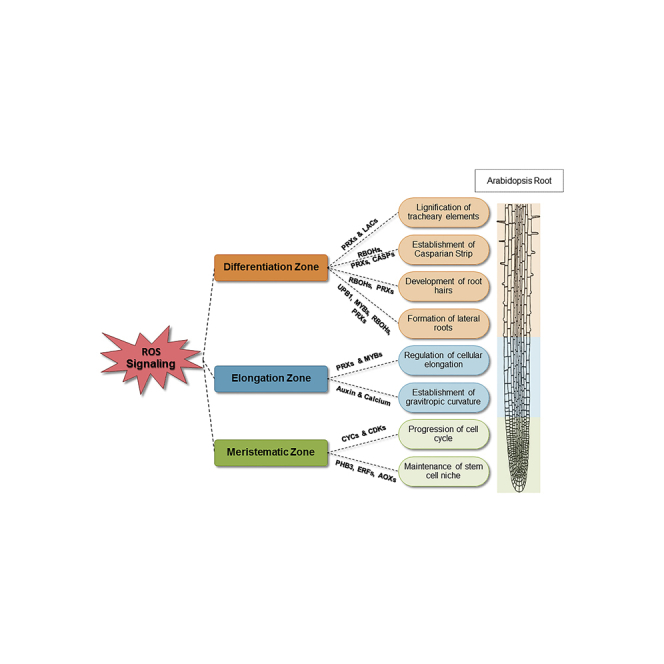
However, less is known about the involvement of ROS in roots growth and development despite the knowledge of their fundamentality. As roots grow into dark heterogeneous soils, their investigation by researchers is indeed challenging and limited, although these obstacles are being surpassed, thanks to the developing root imaging technologies (Mooney et al., 2012; Mairhofer et al., 2013; Rellán-Álvarez et al., 2015). In A. thaliana, the plant root architecture is established by the cross talk between two key players: the genetic background and the environmental signals (Wachsman et al., 2015). Similar to the aerial part of the plant, the primary development and formatting follow genetically programmed intrinsic patterns (Novoplansky, 2019), but the fate of root morphology is strongly adjusted by extrinsic environmental signals. These adjustments by exogenous cues reflect the phenotypic plasticity of plants, which allow them to cope with the fact that they are sessile organisms existing in a continuously changing environment (Simard, 2018). In A. thaliana, the root consists of three major zones: the meristematic zone (MZ), the elongation zone (EZ), and the differentiation zone (DZ). The root morphology is set up by highly orchestrated and dynamic balances between these zones. In fact, during this establishment of root architecture, ROS are involved as signaling molecules that choreograph root structures and anatomies (Tsukagoshi, 2016) through various developmental processes (Czarnocka and Karpiński, 2018). Several other molecules have been characterized for their role in establishing and maintaining such balances as phytohormones and nutrients (Majda and Robert, 2018; Mangano et al., 2018; Noctor et al., 2018; Choudhary et al., 2020). Additionally, roots represent a perfect model for studying plant growth since all the developmental stages that a cell goes through are present. In fact, the stem cell phase is present at the root tip where cell proliferation occurs. Stem cells and their derived cells divide and shift away from a mitotically inactive quiescent center (QC). Afterward, the proliferative phase ceases and cells start to elongate and differentiate until the fully differentiated phase in the DZ (Street et al., 2016). Therefore, this review aims to address the multiple roles of ROS during plant root development, elongation, and differentiation, focusing on the recently uncovered molecular mechanisms.
Role of ROS balances in the meristematic and elongation root zones during proliferation and elongation
ROS & progression of cell cycle
Root growth and development result from the constant generation of cells in its MZ (De Veylder, Beeckman and Inzé, 2007). Afterward, these dividing cells acquire specialized functions during differentiation where they undergo orchestrated modifications. A cell cycle controls the number of dividing cells and the duration of these divisions which determines the growth rate of the root. A proper cell cycle is hence highly important for a proper root growth because it maintains the MZ size and prevents the premature differentiation (González-García et al., 2011). In fact, the molecular mechanisms that coordinate the phase-to-phase transitions in the cell cycle have been thoroughly studied. The roles of cyclins (CYCs) and cyclin-dependent kinases (CDKs) as key actors in these transitions have been highlighted (Hamdoun et al., 2016; Tamirisa et al., 2017). In addition, ROS have been found to influence these actors and govern the interphasic transition in the cell cycle (Mhamdi and Van Breusegem, 2018b). First, the cell cycle stops owing to changes in the redox homeostasis. CYCs expression is controlled by the transcription factor TEOSINTE BRANCHED1-CYCLOIDEA-PROLIFERATING CELL FACTOR1 (TCP), which contains a redox-sensitive cysteine residue that determines its activity. When ROS levels increase, a disulfide bond is formed, preventing the binding of TCP on the CYC promoters (Viola et al., 2016). Moreover, CYCs and CDKs expression patterns have been correlated with the ascorbate:glutathione (ASC:GSH) cycle and their redox status (Figure 1). Several glutathione-deficit mutants revealed a cell cycle arrest as in root meristem less 1 (rml1) (Schnaubelt et al., 2015). The concentrations of ROS, glutathione, and ascorbate in various organelles were found synchronized with transition events in the cell cycle. Accordingly, ascorbate-deficit mutants had slower cell cycles than the wild-type (WT) ones (Tognetti et al., 2017; Ortiz-Espín et al., 2017). In addition, further studies reveal imperative roles of ROS in the cell plate synthesis during cytokinesis, the last stage of the cell cycle. During this process, the key to a proper patterning of the plant cell wall (PCW) is held by a dynamic network of filamentous structures, i.e., the cytoskeleton (Szymanski and Staiger, 2018). ROS have been shown as actors during this morphogenesis. Modification of ROS homeostasis disturbed the cytoskeleton structure and formed many atypical tubulin polymers (Livanos et al., 2012). This aberrant formation negatively affected the cytokinesis and the cell plate synthesis. Similarly, Arabidopsis mutated for RBOHs, ROS-producing enzymes, develop disorganized cytoskeletal tubulin and hence the cytokinesis is disturbed (Foreman et al., 2003; Livanos et al., 2017). Therefore, ROS are essential regulators of the cell cycle, hence contributing to the regulation of MZ size and progressive root growth.
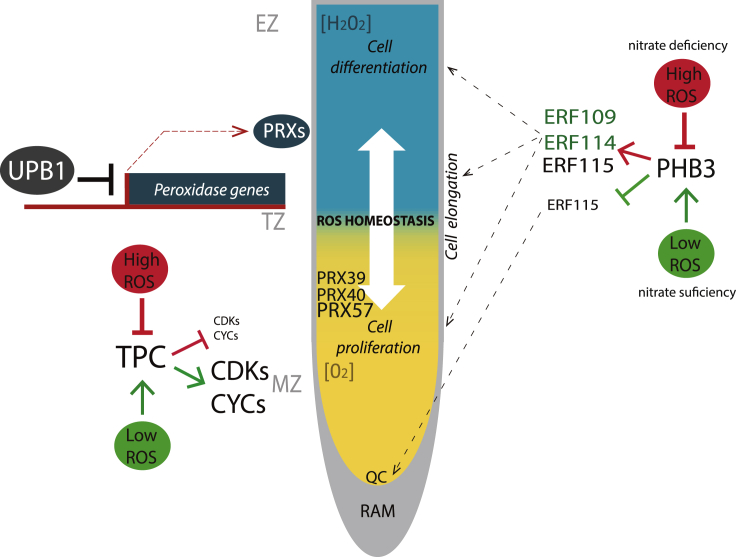
Figure 1.
ROS control the equilibrium between cell proliferation and differentiation in roots
The transcription factor UPBEAT1 (UPB1) modulates the balance between cell proliferation and differentiation by directly regulating the expression of a subset of peroxidases (PRXs; PRX39, PRX40, and PRX57). These PRXs are highly expressed at the boundary of the meristematic and elongation zone. In Col-0 root tips, O2- accumulates in the meristem (yellow area), whereas H2O2 accumulates in the elongation zone (blue area) and UPB1 represses expression of PRXs in the elongation zone. In mutants upb1-1, there is a major production of O2- owing to upregulated expression of the PRXs, so the transition zone shifts in favor of the proliferation zone, while in UPB1 overexpressors, the PRXs are downregulated, so with less PRXs, H2O2 cannot be catalyzed and the differentiation zone expands. PROHIBITIN3 (PHB3) maintains the root stem cell niche maintenance: under nitrate deficiency, ROS production is induced and subsequently inhibits PHB3 expression. This triggers ROS-responsive factors (ERF109, ERF114, and ERF115) leading to cellular proliferation and differentiation. On the contrary, nitrate sufficiency reduces ROS concentrations. Therefore, PHB3 becomes active and inhibits the underlying ETHYLENE RESPONSE FACTOR 115 (ERF115), thus maintaining the pluripotency of the QC. Simultaneously, ETHYLENE RESPONSE FACTOR 109 (ERF109) and ETHYLENE RESPONSE FACTOR 114 (ERF114) are induced to promote cellular elongation and differentiation in the elongation and differentiation zones, respectively. Cyclins (CYCs) and cyclin-dependent kinases (CDKs) and CYCs regulate the size of MZ, controlling the cell cycle. The cell cycle stops owing to changes in the redox homeostasis. CYCs are controlled by the transcription factor TEOSINTE BRANCHED1-CYCLOIDEA-PROLIFERATING CELL FACTOR1 (TCP). When ROS levels increase owing to an oxidative burst, a disulfide bond is formed preventing the binding of TCP on the CYC promoters. Moreover, CYC and CDK expression patterns have been correlated with the ASC:GSH cycle and their redox status. ASC: ascorbic acid; EZ: elongation zone; GSH: reduced glutathione; RAM: root apical meristem; QC: quiescent centre; TZ: transition zone
ROS & maintenance of stem cell niche
ROS are involved in maintaining the stem cell identity in the root apical meristem (RAM) and regulating the organogenesis (Wany et al., 2018). For this aim, they form extensive networks with other signaling molecules as nitric oxide (NO) in addition to several phytohormones (auxin, cytokinin, ethylene, etc.) and their associated signaling pathways (Wany et al., 2018). As well, ROS fine-tunes the stem cell fate in the RAM (Yang et al., 2018). PROHIBITIN3 (PHB3) maintains the root stem cell niche maintenance. In phb3 mutants, the transcript of the cell cycle inhibitor retinoblastoma RBR is higher than the WT. However, both KNOLLE, a member of SYP11 syntaxin gene family, and cyclin-dependent protein kinase (CYCB1) which are markers of the cell cycle are downregulated. These results confirm the role of PHB3 in the RAM size maintenance. Moreover, phb3 mutants show an increased division rate in the quiescent center (QC) (>80%) compared with the WT (∼5%) which indicates the role of PHB3 in the QC identity maintenance. In fact, this PHB3 regulation is in cross talk with the redox status since hydrogen peroxide and superoxide overaccumulate in the phb3 plants (Kong et al., 2018). In the RAM, ROS production is regulated according to the nutrient availability in the soil. Under nitrate deficiency, ROS production is induced and subsequently inhibits PHB3 expression. This triggers ROS-responsive factors (ERF109, ERF114, and ERF115) leading to cellular proliferation and differentiation (Figure 1). On the contrary, nitrate sufficiency promotes the production of glutathione antioxidant which reduces ROS concentrations. Therefore, PHB3 becomes active and inhibits the underlying ETHYLENE RESPONSE FACTOR 115 (ERF115), thus maintaining the pluripotency of the QC (Figure 1) (Kong et al., 2018). Simultaneously, ETHYLENE RESPONSE FACTOR 109 and 114 (ERF109 and ERF114) are induced to promote cellular elongation and differentiation in the elongation and differentiation zones, respectively (Wany et al., 2018). Moreover, PHB3 plays a role in preserving redox homeostasis in the mitochondria. In Arabidopsis phb3 mutants, genes implicated in ROS production and signaling such as alternative oxidases (AOXs), ALTERNATIVE NAD(P)H DEHYDROGENASE 1 (NDA1), and NAD(P)H DEHYDROGENASE B2 (NDB2) showed higher expression (Kong et al., 2018). These studies suggest a vital role for ROS in conserving the stem cell niche, which enables a continued growth of the root.
ROS & cell elongation
The dividing cells enter a new phase of cellular elongation in the EZ. This elongation is mostly dependent on the structure and plasticity of the primary cell walls (PCWs). The dynamics of the PCW are associated with the properties of its components, polysaccharides and proteins (Schmidt et al., 2016). In our scope, numerous studies reported the role of apoplastic compounds in controlling this cellular expansion in the EZ. Cellular expansion rates are tightly related to the balance between cell wall stiffening and loosening, where both are controlled by ROS homeostasis (Francoz et al., 2015). Primarily, during their peroxidative cycle, PRXs oxidize several PCW components and consequently enhance PCW stiffening by directly affecting its components' cross-links (Dunand et al., 2007; Marzol et al., 2020). PRXs indeed catalyze the formation of cross-links between phenolics and extensins. In this sense, the cell wall stiffens, and hence the cellular elongation capacity is reduced (Francoz et al., 2015). Thus, on the one hand, elevated levels of H2O2 in the apoplast were found to increase cell wall rigidity. However, in their hydroxylic cycle, PRXs produce ROS which enhance the nonenzymatic PCW loosening and thus promote cellular elongation. In this sense, auxin-induced RBOHs produce O2.- into the PCW. There, these short-living molecules are rapidly dismutated into H2O2. Then, H2O2 is utilized by PRXs to produce OH−- (Majda and Robert, 2018; Marzol et al., 2020). In fact, the produced OH− catalyzes the oxidative cleavage of xyloglucan and other pectins, which loosens the PCW and promotes cellular expansion (Schmidt et al., 2016). These contrasted findings suggest that ROS have a dual role in determining the cell wall status and reveal the effects of hydroxyl:peroxide (OH−:H2O2) balance in the apoplast (Schmidt et al., 2016). Yet, it is unclear how these opposite effects on cell wall polymers are coordinated during cell elongation (Lee et al., 2018; Francoz et al., 2019). In addition, a transcription factor of the MYB family, MYB30, which regulates the expression of several genes involved in the transport of very-long-chain fatty acids (VLCFAs), has been identified to regulate cellular expansion in the root in response to ROS (Mabuchi et al., 2018). The transfer DNA insertion line myb30-1, which has reduced levels of MYB30, showed a higher root elongation rate than the WT upon H2O2 treatment. By counting the number and measuring the length of cortical cells in the MZ and EZ, it was proved that this increase in elongation rate is due to longer cells in the EZ and not to an increase in cells number (Mabuchi et al., 2018). Also, the overexpression of MYB30 in myb30 mutants resulted in shorter roots under control conditions. These results suggested that MYB30 is sufficient to inhibit root elongation. The observation of MYB30 expression by transcriptional and translational fusions revealed that MYB30 is induced by H2O2 and expressed in the MZ and EZ (Mabuchi et al., 2018).
ROS at the interface between proliferation and differentiation
An additional complexity in the redox regulation of the developmental processes is presented at the transition zone (TZ) of the primary root at the interface between the MZ and the EZ (Singh et al., 2016). At the root TZ, UPBEAT1 (UPB1), a bHLH TF, regulates the ROS homeostasis between two ROS species, i.e., superoxide (O2.-) and peroxide (H2O2), to direct growth either toward proliferation in the MZ or toward differentiation in the upper zones (Figure 1). It was detected that a high (O2.-:H2O2) ratio stimulates cellular division that requires advanced O2 concentrations. However, a low (O2.-:H2O2) ratio gives preference to cellular differentiation apart from the MZ (Tsukagoshi et al., 2010). The upb1 mutant shows a significant increase in the number of cortical cells, indicating enlargement of the meristem. An overexpression of UPB1 showed reduced root length and a decrease in the cortex cell number in the MZ (Tsukagoshi et al., 2010). UPB1 acts as a regulator of root growth through modulation of the transition from cell proliferation to elongation as well as plays a role in controlling cell size. UPB1 has 166 putative direct target genes, among which are several TFs. It also inhibits expression of several peroxidase genes (PRX39, PRX40, and PRX57), where PRX57 is of higher importance in this context as it is colocalized at the TZ (Figure 1). This inhibition of PRX expression leads to an accumulation of H2O2, changing the redox homeostasis in favor of differentiation rather than proliferation. Hence, upb1 mutants develop longer roots than the WT (Tsukagoshi et al., 2010).
ROS & differential elongation
ROS are required to establish the gravitropic curvature response of plant roots. This response is an asymmetrical cellular elongation, where gravity stimulates the arrest of elongation at the lower flank of the root (Singh et al., 2016). In fact, gravitropism is a versatile and multistep process. First, gravity sensing begins in specialized columella cells containing statoliths, i.e., plastids rich in starch, in the root cap (Blancaflor et al., 1998; Leitz et al., 2009). Afterward, an auxin-dependent complex signaling pathway (Zhang and Friml, 2020), which also involves calcium (Ca2+), translocates this message into the EZ, where the curvature response occurs (Su et al., 2017). ROS are required to establish the root elongation in response to gravitropism (Swarup et al., 2013). In Zea mays roots, gravistimulation induces higher ROS production in the apex and the lower cortex than in the upper zones. A prolonged stimulation leads to consequent ROS accumulation in the upper zones, particularly in the EZ. This asymmetric distribution of ROS suggests that ROS are actors in developing the gravitropic response of roots. In fact, the gravity-dependent redistribution of auxin triggers the root bending responses by stimulating ROS generation (Joo et al., 2001). Auxin via phosphatidylinositol 3-kinase (PI3K)-dependent pathway activates transmembrane RBOHs and triggers subsequent ROS production at the bending region (Krieger et al., 2016). The produced ROS inhibit cellular elongation by modulating the cell wall stiffening.
Role of ROS during root differentiation
ROS & tracheary elements
ROS are implicated in the differentiation of the tracheary elements, particularly in their lignification process. In A. thaliana, vasculature is arranged into a central stele, where xylem and phloem are interspersed with undifferentiated procambial cells and surrounded by the pericycle. These longitudinal vessels transport water and nutrients through the roots to the upper plant parts. Accordingly, they develop reinforcing and lignified secondary wall thickenings to cope with the negative water potential and prevent cellular collapse (Seagull, 2016). Lignin formation has been extensively studied as part of xylem differentiation. Lignin is a polyphenolic polymer generated by the radical coupling of aromatic monolignol, which is oxidized by ROS-dependent PRXs (PRX72 and PRX17) and laccases (LAC4) (Tsukagoshi, 2016; Fujita et al., 2020; Hoffmann et al., 2020). In addition, a putative role of ROS as a lignification regulator is derived from the roles of these PRXs acting during xylem development. The cationic peroxidase from Zinnia elegans (ZePRX) catalyzes the last step of lignification in Zinnia elegans. The ortholog in A. thaliana, PRX72, is upregulated in the xylem vessel elements, suggesting a role in xylem formation. In prx72 mutants, lignin levels were 40% less than in the WT. It has been observed that PRX72 expression is increased in the lac4 lac11 lac17 triple mutants, implying a possible functional linkage between PRXs and LACs during monolignol coupling (Zhao et al., 2013). However, increased PRX72 expression is not sufficient to rescue the lignin defect in the triple mutant, indicating that LACs and PRXs can still have distinct or complementary roles during monolignol assembly (Rojas-Murcia et al., 2020). Moreover, PRX17 is also involved in lignified tissue formation (Figure 2) and, more precisely, in regulating the kinetics of lignified wall formation (Herrero et al., 2013; Hoffmann et al., 2020). Its corresponding loss-of-function mutant, prx17, has lower lignin content in most organs, whereas xylans were overaccumulated in the PRX17 overexpressor compared with the WT. Additionally, three MYBPLANT motifs have been localized in the PRX17 promoter; MYBPLANT-binding sites specify the expression of genes in lignifying cells (Cosio et al., 2017). However, it remains puzzling whether PRX17 acts directly in vessel lignification or indirectly via ROS signaling.
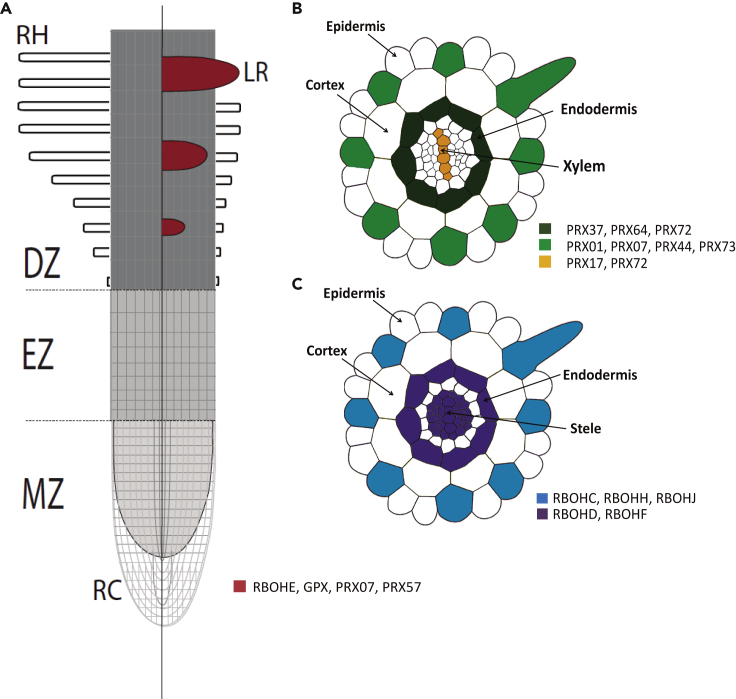
Figure 2.
RBOHs and PRXs are important molecular players in several ROS-mediated developmental processes in roots
(A) RBOHE accumulates at the initiation site of the LR, while LR development in the loss-of-function mutant rbohe is delayed. GPXs are also expressed in the LR. gpx mutants develop larger lateral root primordia, indicating that the GPX family plays a role in regulating root architecture, especially GPX1 and GPX7, having a critical role in the development and formation of the lateral root. PRX07 and PRX57 control LR growth by maintaining ROS balance during the initiation stage in an auxin-independent manner. prx07 and prx57 have lower LR density and number. DZ: differentiation zone; EZ: elongation zone, LR: lateral root; MZ: meristematic zone; RC: root cap; RH: root hair.
(B) Localization of expression of various peroxidases. PRX17 is expressed in the xylem and root endodermis and participates in the lignin polymerization. prx17 mutants have lower lignin content in most organs. PRX37 is expressed in the root endodermis, and it participates in the phenolic cross-linking activity during lignin deposition in the cell wall. The overexpression of PRX17 results in a dwarf phenotype with small plants and delayed development. PRX64 is expressed in the root endodermis; it utilizes peroxide for a monolignol oxidation during lignin formation. PRX72 is expressed in the root endodermis and the xylem. Its expression is upregulated when the xylem is actively forming. The lignin levels in prx72 mutants are 40% less compared with those in the WT.
(C) RBOHC and PRX01, PRX07, PRX44, and PRX73 are expressed in the trichoblast cells in the epidermis. They are key enzymes responsible for root hair elongation by controlling the ROS homeostasis at the tip of the root hair cell. RBOHD and RBOHF are active in the mature root region especially in the vascular cylinder and LRP. RBOHD is expressed in the endodermis and procambium; the number and density of LR is higher in rbohd mutants than in the WT. RBOHF is required during CS formation in the endodermis, where its localization is coordinated by several CASPs and participates in lignin polymerization.
ROS & root endodermis
Root endodermis is the innermost cortical layer that surrounds the vascular stele in the root that resembles a polarized epithelium and functions in nutrient uptake and stress resistance. The development and differentiation of this layer are triggered by both intrinsic and extrinsic cues such as abiotic stresses, nutritional signals, and abscisic acid (ABA)-dependent metabolism (Lee et al., 2013). Its primary function is to form a selective barrier that filters the passage of the soil-absorbed water, solutes, and nutrients into the plant's transporting vessels. This vital role is acknowledged to the hydrophobic coating of lignified Casparian strips (CSs) and its suberin lamellae (Cohen et al., 2020). CSs have belt-like structures middling the anticlinal cell walls of the endodermal cells. They seal the extracellular spaces by tying the cell walls between adjacent cells (Franke, 2015). CS formation is tightly regulated by ROS and their mediated enzymes RBOHs and PRXs (Figure 3). Nevertheless, the regulatory mechanisms underlying the endodermal selectivity are still under investigation.
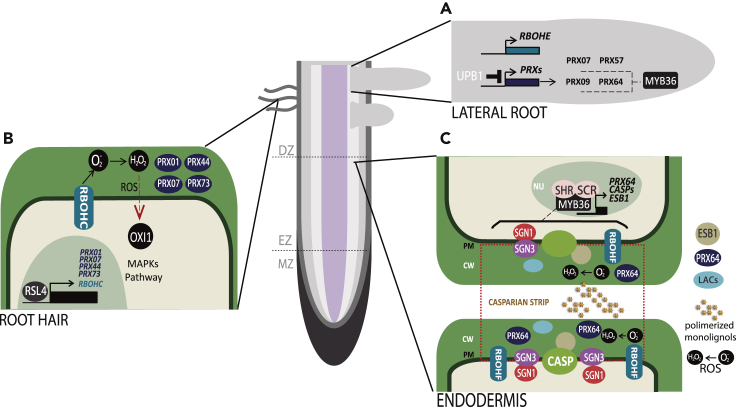
Figure 3.
Selected examples of ROS-mediated root developmental process
(A) In lateral roots (LRs), UPBEAT1, a basic helix-loop-helix TF expressed at the TZ, directly regulates peroxidases that alter the distribution of ROS. Cell proliferation requires high levels of O2, whereas high H2O2 induces differentiation. UPB1 is transcriptionally enriched during the early stages of lateral root primordia (LRP) when ROS significantly accumulate in the emerging LR. UPB1 overexpressors have more LRPs. upb1 has more emerging LRs than WT. UPB1 controls the expression of a subset of PRXs genes that manipulate the ROS balance. Besides, MYB36 acts in stages V and VI of LR development, by manipulating the ROS balance via the regulation of at least PRX09 and PRX64. (B) In growing root hairs, RBOHC is the key enzyme that produces ROS in the apoplast. RBOHC-mediated ROS leads to the activation of a MAPK cascade through OXI1 during cell elongation of root hairs. At the transcriptional level, RSL4, a bHLH transcription factor, regulates the expression of several class III PRXs (PRX01, PRX44, and PRX73), which participate in root hair growth and ROS homeostasis. In addition, PRX07 is also required for root hair elongation. (C) In inner root tissues, SHORTROOT (SHR) transcription factor (TF) moves from the stele to the endodermis where it interacts with another TF SCARECROW (SCR), and then the SHR:SCR complex specifies the endodermal cell identity and activates the transcription of MYB36. MYB36 regulates the biosynthetic apparatus for Casparian strips (CSs). It also plays role in the lateral root primordia differentiation and lateral root initiation. MYB36 regulates the expression of several underlying genes such as CASPS, PRX64, and ESB1 to trigger the proper endodermis development. CASP is responsible for the proper localization at the CS development site by the interaction with two receptor-like kinases SCHENGEN1 (SGN1) and SCHENGEN3 (SGN3). ESB1 stabilizes the CASP proteins scaffold. RBOHF generates superoxide that mutates into peroxide. PRX64 and LACs oxidize monolignols and catalyze lignin formation.
In Arabidopsis, CS formation sites are determined by the localization of the transmembrane protein family, CS membrane domain proteins (CASPs). CASPs are explicitly expressed in the endodermis (Li et al., 2018a, Li et al., 2018b) and localize specifically at the Casparian strip membrane domain (CSD) (Barbosa et al., 2019), where CASP1 guides CS-specific localization by interacting with two receptor-like kinases SCHENGEN1 (SGN1) and SCHENGEN3 (SGN3) (Alassimone et al., 2016). CASPs act as a transmembrane scaffold (Barbosa et al., 2019) and coordinate the proper localization of the different actors of the lignin-polymerizing machinery: RBOHF, PRX64, and ENHANCED SUBERIN 1 (ESB1) at the CSD (Li et al., 2018a). RBOHFs function downstream the formation of the localized CASP scaffold. It generates superoxide (O2.-) which readily mutates into H2O2. Subsequently, PRX64 utilizes the produced H2O2 for monolignol oxidation, hence catalyzing lignin formation. Additionally, ESB1 forms a part of this assembly; it stabilizes the CASP protein scaffold and, contrariwise, requires the CASP complex for its CS localization (Lee et al., 2013; Franke, 2015).
Moreover, MYB36 has been revealed to regulate the expression of CASPs, PRX64, and ESB1 (Kamiya et al., 2015). Loss-of-function mutants, myb36, show deficit CS and hence lose the apoplastic barrier in this region of the root. Beforehand, during the endodermal differentiation, MYB36 expression is driven by the interaction of two upstream TFs, SHORTROOT (SHR) and SCARECROW (SCR). SHR and SCR are responsible for the initial specification of endodermal cell identity (Liberman et al., 2015; Li et al., 2018b). Apart from PRXs, MYB36 may also target the endodermis-enriched LACs. LACs are glycosylated, multi-copper enzymes that catalyze the oxidation of various phenolic substrates using O2 as final electron acceptor, not requiring H2O2 (Rojas-Murcia et al., 2020). Previously, the role of LACs in lignification in the vascular tissue was reported as the overall lignin content was reduced in both lac and prx mutants (Pfister et al., 2014; von Wangenheim et al., 2017; Wunderling et al., 2018). However, the role of LACs in the endodermis and whether some cell types preferentially use LACs and others PRXs were addressed recently (Rojas-Murcia et al., 2020). In fact, the CS was devoid in the quintuple mutant prx03 prx09 prx39 prx72 prx64, while it showed no difference in the lac1 3 5 7 8 9 12 13 16 nonuple mutant compared with the WT. Together, these findings suggest that PRXs play very specific roles in various contexts, and LACs are redundant for lignification of primary cell walls but are required for lignification of secondary cell walls (Rojas-Murcia et al., 2020). Additionally, other RBOH and PRX enzymes are also known to be involved in ROS homeostasis and cell wall remodeling in the endodermis, including apoplastic RBOHD, PRX37, and PRX72. PRX37 participates in phenolic "cross-linking" activity during lignin deposition (Pedreira et al., 2011), and PRX72 is involved in lignin biosynthesis when plant growth has ceased (Figure 2) (Kim et al., 2010; Fernández-Pérez et al., 2015; Tognetti et al., 2017; Fujita et al., 2020).
ROS & root hair
Presence of root hairs marks the DZ of the primary root and is among its exhaustively studied features. In fact, root hairs are tube-like extensions projecting from single specialized epidermal cells. They massively increase the surface area of the root and hence improve its absorption potential of water and soil-borne nutrients. ROS play a critical role in root hair development (Sundaravelpandian et al., 2013; Zhao et al., 2016; Mhamdi and Van Breusegem, 2018b; Choudhary et al., 2020). In Arabidopsis, RBOHC is the key enzyme responsible for root hair elongation under ROS regulation although RBOHH and RBOHJ also contribute to a much lower extent (Mangano et al., 2017). RBOHC actively produces superoxide, and the hydroxyl radicals formed downstream of superoxide are the specific ROS that stimulates root hair elongation (Chapman et al., 2019), and expectedly, rbohc mutants has much shorter root hairs than the WT (Foreman et al., 2003). ROS stimulate ROS-dependent calcium gates, creating a Ca2+ gradient from the tip inward. This gradient drives the apical elongation of the root hair (Mhamdi and Van Breusegem, 2018b). RBOHC trafficking, transport to its correct subcellular compartment, and localization are complex processes regulated by the receptor-like kinase FERONIA (FER) and members of the Rho family of plant GTPases (ROP) (Chapman et al., 2019). The FER extracellular domain binds to the pectin components of the cell wall and activates Ca2+ channels that maintain the integrity of the cell wall during growth. fer and rbohc mutants possess similar phenotypes, with short root hairs and decreased ROS accumulation. Pull-down assays that map protein-protein interactions suggest that FER interacts with ROP2, promoting radical hair growth upstream of RBOHC (Chapman et al., 2019). Another actor on the ROS homeostasis during root hair development is the LIKE SEX FOUR 2 (LSF2) gene encoding a phosphatase. This gene was identified for its novel role in redox regulation (Zhao et al., 2016). lsf2 Arabidopsis mutants showed lower superoxide generation rate but higher levels of hydrogen peroxide upon oxidative stress treatments. LSF2 interacts with the mitogen-activated protein kinase 8 (MPK8) which regulates the cytosolic redox balance (Zhao et al., 2016). Besides RBOHC, another component of ROS signaling in growing root hair is the OXIDATIVE BURST INDUCIBLE1 (OXI1) kinase. OXI1 is a Ser-Thr kinase, which plays a role in elongation of root hairs. It was proposed that RBOHC-mediated ROS lead to the activation of a MAPK cascade through OXI1 during cell elongation of root hairs (Figure 3) (Rentel et al., 2004).
Moreover, auxin triggers a higher ROS level in the root hairs and enhances their growth by ROOT HAIR DEFECTIVE SIX LIKE4 (RSL4) activation (Mangano et al., 2017). RSL4 encoding a bHLH TF has been identified as a vital regulator of root hair development (Yi et al., 2010; Mangano et al., 2017; Pires et al., 2013). Furthermore, it is known that the expression of RSL4 is controlled by several auxin response factors (ARFs), including ARF5, ARF7, ARF8, and ARF19; this ARF-RSL network can induce localized ROS synthesis (Figure 3), promoting the development of root hairs (Mangano et al., 2017, Mangano et al., 2018; Chapman et al., 2019). Moreover, in addition to RBOHC, the key enzyme involved in root hair growth RBOHH and RBOHJ have been identified as important enzymes in this process. These RBOHs have root hair cis-elements (RHE) in their genes' promoters highly similar to the RSL4 response elements (RSL-RE). It was suggested that RSL4 binds at least one RHE region in the RBOHC and RBOHJ promoters and hence directly regulates their expression (Mangano et al., 2017). In parallel, among the thirty-four putative targets of RSL4, PRX07 has been identified which codes the PRXs required for root hair elongation (Vijayakumar et al., 2016). PRX regulatory regions include several putative RHE motifs. Chromatin immunoprecipitation (ChIP) experiment indicated that RSL4 can bind to the RHE in the regulatory region of PRX01, PRX44, PRX60, and PRX73. Consistently, PRX01 and PRX73 were upregulated in the overexpressors of RSL4 and downregulated in rsl4 mutants (Mangano et al., 2017). It is proposed that the phenotypes observed for peroxidase null mutants, especially prx01, prx44, and prx73, are due to their participation in root hair growth and ROS homeostasis (Figure 3) (Mangano et al., 2017). These three genes in turn are highly coexpressed with other root hair-specific genes such as extensins and glycoproteins, which jointly participate in the expansion of the cell wall (Mangano et al., 2018; Marzol et al., 2020). Double mutants prx44 prx73, prx01 prx44, and prx01 prx73, as well as the triple null mutant, prx01 prx44 prx73, showed shorter root hairs, possibly due to the high accumulation of H2O2 in the apoplast, affecting ROS homeostasis, the root hair growth, and the thickness of the cell wall (Marzol et al., 2020). ROOT HAIR DEFECTIVE SIX LIKE 2 (RSL2), a close relative of RSL4, is also involved in root hair growth in an ROS-dependent manner involving PRXs and RBOHC enzymes, suggesting that RSL2-RSL4 may act together to regulate ROS homeostasis (Mangano et al., 2017, Mangano et al., 2018). In addition, the subunit of the mediator complex PHYTOCHROME AND FLOWERING TIME1 (PFT1)/MED25 triggers H2O2 production by the upregulation of a subset of PRX expression and regulates the equilibrium between H2O2 and during root hair differentiation and cell elongation (Sundaravelpandian et al., 2013).
ROS & lateral roots
Plants have the capacity of the de novo development of new organs in which new cells are enrolled in the formation of primordia organs (Fernández-Marcos et al., 2017). Lateral roots (LRs) serve as an example of this feature and choreograph the dynamic architecture of the root system. LRs enhance the absorption of water and nutrients from the neighboring soil environment. They also contribute to the root acclimation to various biotic and abiotic constraints. LRs are intrinsically diverse as they have contrasted morphologies and anatomies, such as having different lengths and orientations (Muller et al., 2019). LRs are initiated when pericycle founder cells undergo several rounds of anticlinal divisions to create one-layered primordia of about ten equal cells; this is termed stage I. Next, the cells divide periclinally, forming an inner and an outer layer denoting stage II. Further anticlinal and periclinal divisions create dome-shaped primordia (stages III–VII) that eventually emerge at stage VIII from the parental root (Malamy and Benfey, 1997; Péret et al., 2009; Banda et al., 2019). In fact, the establishment of lateral root primordia (LRP) from the root pericycle is auxin dependent. These LRPs further develop into LRs by orchestrated divisions of the lateral root founder cells (Fernández-Marcos et al., 2017).
Redox signaling is notably involved in LR development by a complex genetic network that has been thoroughly studied (Manzano et al., 2014; Reyt et al., 2015; Orman-Ligeza et al., 2016). First, SKP2B (S-phase kinase-associated protein2), a marker for LR development, is coexpressed with many ROS-related genes. UPB1 is transcriptionally enriched during the early stages of LRPs, and ROS significantly accumulate in the emerging LR. In addition, plants overexpressing UPB1 have more LRPs in stages IV and V. However, in stage VIII, when LRPs emerge from the epidermis, their number was significantly reduced in the UPB1 overexpressors. Coherently, upb1 mutants had more emerging LRs than WT (Manzano et al., 2014). UPB1 controls the expression of a subset of PRX genes that manipulate the ROS balance (Figure 3). Altogether, this suggests that ROS play a role in the early development of LRPs. Moreover, a potassium cyanide treatment, which inhibits PRX activity, reduced LR emergence. Diaminobenzidine and nitro blue tetrazolium staining revealed, respectively, that H2O2 and superoxide are present in all the developmental stages of LRs (Manzano et al., 2014).
In addition, auxin plays an important role in LR formation and consequently in choreographing the root system architecture (Petricka et al., 2012). To exemplify, modifying auxin distribution by nutrient availability, as iron (Fe), influences the LR elongation (Giehl et al., 2012). Also, Fe and ROS homeostasis are intimately linked, with ferritins as major actors in this interaction, which suggests a cross talk between ROS homeostasis and auxin distribution during LR development (Reyt et al., 2015). Moreover, LR development is dependent on the interaction between auxin with other phytohormones, especially with ABA (Singh et al., 2016). Auxin and ABA functions antagonistically where the former triggers cellular expansion; however, the latter maintains a balance between proliferation and differentiation in the LRPs. Both hormones trigger ROS production by promoting cell expansion of growing roots. When indole acetic acid (IAA) is degraded by peroxidases, the formation of LRs is inhibited (Cosio et al., 2009; Choudhary et al., 2020). Furthermore, gpx mutants develop larger LRM, indicating that the GPx family plays a role in regulating root architecture, especially GPX01 and GPX07 that regulate LR development and formation (Figure 2) (Choudhary et al., 2020). Furthermore, two RBOHs from Arabidopsis, RBOHD and RBOHF, are suggested to negatively regulate LRP formation and LR emergence, which may be explained by the decrease in peroxidase activity (Li et al., 2014). Initially, these two RBOH genes are known to be expressed in the vascular cylinder and LRP in primary roots. LR number and density significantly increase in their corresponding loss-of-function double mutant rbohd/f (Li et al., 2014). In fact, this mutant has lower H2O2 concentration and generation rate in the root cap, MZ, EZ, and DZ than the WT. Similarly, it has lower O2.- concentration and generation rate in the root cap, MZ, and EZ, but not in the DZ. Surprisingly, it produces more superoxide than the WT in the DZ, which enhances the LR density. This increase in LR density in the mutant is credited to the enhanced peroxidase activity in the DZ of the rbohd/f mutant. In fact, the peroxidase activity in rbohd1/f1 and rbohd2/f2 were higher than in the WT (Li et al., 2014). In addition, it was observed that an rbohe loss-of-function mutant delays the development of the LR. Through the use of the β-glucuronidase (GUS) reporter gene staining, an accumulation of RBOHE is observed at the starting site of the LR, showing the possible participation of this enzyme in its development (Figure 2) (Chapman et al., 2019).
Additionally, MYB36, a previously identified advocate in CS formation during endodermal differentiation (Kamiya et al., 2015; Liberman et al., 2015), was also identified as an actor in LR development (Fernández-Marcos et al., 2017). Initially, MYB36 protein was revealed to accumulate in the root endodermis. Its overexpression leads to shorter primary roots, whereas three allelic myb36 mutants develop longer roots (Fernández-Marcos et al., 2017). This phenotype results from the increase in the number of cells in the meristem, where ROS balance is known to regulate the meristematic boundaries (Kamiya et al., 2015; Liberman et al., 2015). The new role of MYB36 in LR development was first discovered in 2017 (Fernández-Marcos et al., 2017). Two allelic mutants of myb36 had less LRs than the WT. A spatiotemporal tracking of the expression pattern of this gene showed that it was not expressed in the endodermal cells adjacent to the LRPs. However, it was cell-autonomously enriched in the lateral root primordia boundary (LRPB) at stage V of the LR development. LRPs in the two mutants were flat, and they failed to form into domes and hence failed to penetrate the overlying cellular layers. Further spatiotemporal analyses of the expression pattern of MYB36 and its corresponding PRX targets confirmed that MYB36 acts in stages V and VI of LR development, by manipulating the ROS balance via the regulation of at least PRX09 and PRX64 (Fernández-Marcos et al., 2017). Finally, supplementary forward genetics approaches identified another ROS-related genetic basis affecting the LR phenotype. PRX07 and PRX57 were explicitly identified as the actors in LR growth among other peroxidases (Figure 2) since prx07 and prx57 mutants have lower LR density owing to changes of ROS balance in the initiation stage. Interestingly, despite that LRP establishment depends on auxin, PRX07 and PRX57 were detected to be auxin-independent (Manzano et al., 2014). Together, all these studies highlight the importance of ROS homeostasis in shaping and growing LRs.
Decoding ROS signal
As demonstrated above, ROS production and scavenging naturally takes place in various cellular compartments and is surveilled by the profound ROS gene network to maintain their homeostasis (Das and Roychoudhury, 2014; Singh et al., 2016; Mittler, 2017). It is worth noting that ROS do not occur in isolation but rather as part of a large and complex redox signaling that involves many components. However, the different ROS levels in the different cellular compartments may be contemplated as ROS signatures in resemblance to the famous Ca2+ signature. In fact, Ca2+ signaling is implicated in countless cellular activities, where varying spatiotemporal distribution, vibration amplitude, and frequency of Ca2+ determine a distinct signature. This specific signature, triggered by a particular stimulus, results in a precise adaptive response (Yuan et al., 2017). As for ROS, different natures and intensities of stimuli lead to the formation of ROS signatures, which are decoded by ROS sensors to generate a stimulus-specific signal, and consequently to customize an acclimation response in the plant cell. This signature varies in compliance with the cell type, developmental stage, and environmental status (Choudhury et al., 2017), and its decoding can alter protein structures or functions via PTMs. For instance, these alterations can affect TFs leading to modifications in their transcription (Dietz et al., 2016).
This concept is exceptionally important, as it explains how plants are able to decrypt different ROS signals provoked by, chemically, the same molecules, and then elaborate a specific response to each particular signature (Choudhury et al., 2017). It is notable how these ROS-producing or ROS-scavenging machineries are copresent simultaneously within cells or even organelles in widely ranging proportions and activities. What turns them on or off remains an intriguing question. They are surely dependent on the ambient O2 concentration and on the exogenous environmental cues. Furthermore, they are also dependent on the cell type, localization, and status, which is subsequently depending on the gene expression pattern. Thus, an optimal pattern of oxidants and antioxidants exists for a given physiological condition (Sies et al., 2017).
Conclusion
Recent studies investigated the vital roles of ROS in plant root developmental processes. In summary, during cell proliferation, ROS concentrations regulate the cell cycle since oxidative bursts can block cyclin expression. In the MZ, ROS act to conserve the stem cell niche in the QC, hence allowing a sustained growth of the root. In the EZ, apoplastic ROS balances determine the elongation rates of cells in the root EZ. In fact, apoplastic ROS can promote either PCW stiffening or loosening, by catalyzing either the cross-links between PCW components or their oxidative cleavage, respectively. At the transition zone, ROS balances give preference to either proliferation or differentiation according to UPB1 regulation. These aforementioned mechanisms reveal a vital role of ROS during the plant root primary growth. Moreover, ROS are involved in modulating the differentiation stage of development at multiple levels. In the tracheary elements, ROS regulate xylem lignification by the functioning of the ROS-producing enzymes, PRX72 and PRX17. Similarly, PRX64 and RBOHF are essential for the proper formation of the CS in the root endodermis. In addition, in the epidermis, ROS trigger cell growth in root hairs. Last but not least, numerous ROS-related genes are implicated in LR formation. ROS production and scavenging are two co-occurring processes that are minutely adjusted to maintain the manifold “ROS homeostasis” (Mittler et al., 2004; Suzuki et al., 2012; Mittler, 2017; Mhamdi and Van Breusegem, 2018b). This homeostasis is essential for regulating the multiple stages of plant root development and hence choreographing the final root architecture. Furthermore, this redox balance is central to life since redox practices encompass all fundamental processes, from bioenergetics to metabolism and life functions (Sies et al., 2017). In our scope, ROS homeostasis has been shown as a requisite for proper root growth (Tsukagoshi, 2016; Choudhary et al., 2020), development (Mhamdi and Van Breusegem, 2018b), and response to stimuli (Miller et al., 2010; Liu and He, 2016; Zhang et al., 2016). In view of the above, ROS of underground plant cells are multitaskers in root development.
Future key issues
-
a.
Apoplastic ROS-sensing mechanism by cell surface receptors has been recently proposed to be mediated by HPCA1 (Laohavisit et al., 2020; Wu et al., 2020). The downstream post-translational modifications relative to the kinase domain of CARD1/HPCA1 were also described. However, the further detailed downstream signaling components still to be identified.
-
b.
ROS fluctuations are tightly connected with Ca2+ signals (e.g. polar growing cells like root hairs). However, it is not clear how these two signals are interconnected. In this context, it is known that Ca2+ activates RBOHs, but which Ca2+ channels are activated by ROS (e.g. H2O2) remain unknown. In addition, it is unknown if this process is direct and not mediated by other components (e.g. CARD1/HPCA1 or other kinases).
-
c.
How apoplastic ROS pool is coordinated with the “cytoplasmic” and “organellar” ROS pools in order to maintain a coherent ROS homeostasis in the plant cells along growth and development? The use of multiple genetically encoded ROS sensors targeted to different subcellular zones will allow us to get a high-resolution spatial map distribution of ROS species.
Acknowledgments
The authors are thankful to Université Paul Sabatier-Toulouse III (France) and CNRS for supporting their research work. This work was supported by the French Laboratory of Excellence project “TULIP” (ANR-10-LABX-41; ANR-11-IDEX-0002-02) and by grants from ANPCyT Argentina (PICT2016-0132 and PICT2017-0066) and Fondo Nacional de Desarrollo Científico y Tecnológico (1200010) and Instituto Milenio iBio – Iniciativa Científica Milenio, MINECON Chile to JME.
Author contributions
A.E. analyzed the bibliography, wrote the paper, and helped on the figures’ design. Y.d.C.R.G. analyzed the bibliography and helped on the figures’ design. A.E, J.M.E., and Ch.D. analyzed the bibliography, supervised the project, and wrote the paper. This manuscript has not been published and is not under consideration for publication elsewhere. All the authors have read the manuscript and have approved this submission.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Christophe Dunand, Email: dunand@lrsv.ups-tlse.fr.
José Manuel Estevez, Email: jestevez@leloir.org.ar.
References
- Alassimone J., Fujita S., Doblas V.G., van Dop M., Barberon M., Kalmbach L., Vermeer J.E., Rojas-Murcia N., Santuari L., Hardtke C.S. Polarly localized kinase SGN1 is required for Casparian strip integrity and positioning. Nat. Plants. 2016;2:1–10. doi: 10.1038/nplants.2016.113. [DOI] [PubMed] [Google Scholar]
- Anjum N.A., Sofo A., Scopa A., Roychoudhury A., Gill S.S., Iqbal M., Lukatkin A.S., Pereira E., Duarte A.C., Ahmad I. Lipids and proteins—major targets of oxidative modifications in abiotic stressed plants. Environ. Sci. Pollut. Res. 2015;22:4099–4121. doi: 10.1007/s11356-014-3917-1. [DOI] [PubMed] [Google Scholar]
- Apel K., Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- Banda J., Bellande K., von Wangenheim D., Goh T., Guyomarc’h S., Laplaze L., Bennett M.J. Lateral root formation in arabidopsis: a well-ordered LRexit. Trends Plant Sci. 2019;24:826–839. doi: 10.1016/j.tplants.2019.06.015. [DOI] [PubMed] [Google Scholar]
- Barbosa I.C.R., Rojas-Murcia N., Geldner N. The Casparian strip—one ring to bring cell biology to lignification? Curr. Opin. Biotechnol. 2019;56:121–129. doi: 10.1016/j.copbio.2018.10.004. [DOI] [PubMed] [Google Scholar]
- Baxter A., Mittler R., Suzuki N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014;65:1229–1240. doi: 10.1093/jxb/ert375. [DOI] [PubMed] [Google Scholar]
- Berkowitz O., De Clercq I., Van Breusegem F., Whelan J. Interaction between hormonal and mitochondrial signalling during growth, development and in plant defence responses. Plant Cell Environ. 2016;39:1127–1139. doi: 10.1111/pce.12712. [DOI] [PubMed] [Google Scholar]
- Blancaflor E.B., Fasano J.M., Gilroy S. Mapping the functional roles of cap cells in the response of arabidopsis primary roots to gravity. Plant Physiol. 1998;116:213–222. doi: 10.1104/pp.116.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose J., Rodrigo-Moreno A., Shabala S. ROS homeostasis in halophytes in the context of salinity stress tolerance. J. Exp. Bot. 2014;65:1241–1257. doi: 10.1093/jxb/ert430. [DOI] [PubMed] [Google Scholar]
- Bratt A., Rosenwasser S., Meyer A., Fluhr R. Organelle redox autonomy during environmental stress. Plant Cell Environ. 2016;39:1909–1919. doi: 10.1111/pce.12746. [DOI] [PubMed] [Google Scholar]
- Chakraborty K., Bishi S.K., Goswami N., Singh A.L., Zala P.V. Differential fine-regulation of enzyme driven ROS detoxification network imparts salt tolerance in contrasting peanut genotypes. Environ. Exp. Bot. 2016;128:79–90. [Google Scholar]
- Chapman J.M., Muhlemann J.K., Gayomba S.R., Muday G.K. RBOH-dependent ROS synthesis and ROS scavenging by plant specialized Metabolites to modulate plant development and stress responses. Physiol. Behav. 2017;176:139–148. doi: 10.1021/acs.chemrestox.9b00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J.M., Muhlemann J.K., Gayomba S.R., Muday G.K. RBOH-dependent ROS synthesis and ROS scavenging by plant specialized Metabolites to modulate plant development and stress responses. Chem. Res. Toxicol. 2019;32:370–396. doi: 10.1021/acs.chemrestox.9b00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary A., Kumar A., Kaur N. ROS and oxidative burst: roots in plant development. Plant Divers. 2020;42:33–43. doi: 10.1016/j.pld.2019.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury F.K., Rivero R.M., Blumwald E., Mittler R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2017;90:856–867. doi: 10.1111/tpj.13299. [DOI] [PubMed] [Google Scholar]
- Cohen H., Fedyuk V., Wang C., Wu S., Aharoni A. SUBERMAN regulates developmental suberization of the Arabidopsis root endodermis. Plant J. 2020;102:431–447. doi: 10.1111/tpj.14711. [DOI] [PubMed] [Google Scholar]
- Cosio C., Ranocha P., Francoz E., Burlat V., Zheng Y., Perry S.E., Ripoll J.J., Yanofsky M., Dunand C. The class III peroxidase PRX17 is a direct target of the MADS-box transcription factor AGAMOUS-LIKE15 (AGL15) and participates in lignified tissue formation. New Phytol. 2017;213:250–263. doi: 10.1111/nph.14127. [DOI] [PubMed] [Google Scholar]
- Cosio C., Vuillemin L., De Meyer M., Kevers C., Penel C., Dunand C. An anionic class III peroxidase from zucchini may regulate hypocotyl elongation through its auxin oxidase activity. Planta. 2009;229:823–836. doi: 10.1007/s00425-008-0876-0. [DOI] [PubMed] [Google Scholar]
- Costa A., Drago I., Behera S., Zottini M., Pizzo P., Schroeder J.I., Pozzan T., Lo Schiavo F. H2O2 in plant peroxisomes: an in vivo analysis uncovers a Ca2+-dependent scavenging system. Plant J. 2010;62:760–772. doi: 10.1111/j.1365-313X.2010.04190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H., Kong D., Wei P., Hao Y., Torii K.U., Lee J.S., Li J. SPINDLY, ERECTA, and its ligand STOMAGEN have a role in redox-mediated cortex proliferation in the arabidopsis root. Mol. Plant. 2014;7:1727–1739. doi: 10.1093/mp/ssu106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnocka W., Karpiński S. Friend or foe? Reactive oxygen species production, scavenging and signaling in plant response to environmental stresses. Free Radic. Biol. Med. 2018;122:4–20. doi: 10.1016/j.freeradbiomed.2018.01.011. [DOI] [PubMed] [Google Scholar]
- Dagda R.K., Rice M., Merrill R.A., Flippo K.H., Strack S. Techniques to investigate mitochondrial function in neurons. Neuromethods. 2017;123:1–27. [Google Scholar]
- Daloso D.M., Müller K., Obata T., Florian A., Tohge T., Bottcher A., Riondet C., Bariat L., Carrari F., Nunes-Nesi A. Thioredoxin, a master regulator of the tricarboxylic acid cycle in plant mitochondria. Proc. Natl. Acad. Sci. U S A. 2015;112:E1392–E1400. doi: 10.1073/pnas.1424840112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K., Roychoudhury A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2014;2:1–13. [Google Scholar]
- Deng B., Deng S., Sun F., Zhang S., Dong H. Down-regulation of free riboflavin content induces hydrogen peroxide and a pathogen defense in Arabidopsis. Plant Mol. Biol. 2011;77:185–201. doi: 10.1007/s11103-011-9802-0. [DOI] [PubMed] [Google Scholar]
- Dietz K.J. Redox regulation of transcription factors in plant stress acclimation and development. Antioxid. Redox Signal. 2014;21:1356–1372. doi: 10.1089/ars.2013.5672. [DOI] [PubMed] [Google Scholar]
- Dietz K.J., Mittler R., Noctor G. Recent progress in understanding the role of reactive oxygen species in plant cell signaling. Plant Physiol. 2016;171:1535–1539. doi: 10.1104/pp.16.00938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunand C., Crèvecoeur M., Penel C. Distribution of superoxide and hydrogen peroxide in Arabidopsis root and their influence on root development: possible interaction with peroxidases. New Phytol. 2007;174:332–341. doi: 10.1111/j.1469-8137.2007.01995.x. [DOI] [PubMed] [Google Scholar]
- Eltayeb A.E., Kawano N., Badawi G.H., Kaminaka H., Sanekata T., Shibahara T., Inanaga S., Tanaka K. Overexpression of monodehydroascorbate reductase in transgenic tobacco confers enhanced tolerance to ozone, salt and polyethylene glycol stresses. Planta. 2007;225:1255–1264. doi: 10.1007/s00425-006-0417-7. [DOI] [PubMed] [Google Scholar]
- Farvardin A., González-hernández A.I., Llorens E., García-agustín P., Scalschi L., Vicedo B. The apoplast: a key player in plant survival. Antioxidants. 2020;9:1–26. doi: 10.3390/antiox9070604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Marcos M., Desvoyes B., Manzano C., Liberman L.M., Benfey P.N., del Pozo J.C., Gutierrez C. Control of Arabidopsis lateral root primordium boundaries by MYB36. New Phytol. 2017;213:105–112. doi: 10.1111/nph.14304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Pérez F., Pomar F., Pedreño M.A., Novo-Uzal E. The suppression of AtPrx52 affects fibers but not xylem lignification in Arabidopsis by altering the proportion of syringyl units. Physiologia Plantarum. 2015;154:395–406. doi: 10.1111/ppl.12310. [DOI] [PubMed] [Google Scholar]
- Fichman Y., Miller G., Mittler R. Whole-plant live imaging of reactive oxygen species. Mol. Plant. 2019;12:1203–1210. doi: 10.1016/j.molp.2019.06.003. [DOI] [PubMed] [Google Scholar]
- Foreman J., Demidchik V., Bothwell J.H., Mylona P., Miedema H., Torres M.A., Linstead P., Costa S., Brownlee C., Jones J.D. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth , Nat. 2003;422:442–446. doi: 10.1038/nature01485. [DOI] [PubMed] [Google Scholar]
- Francoz E., Ranocha P., Le Ru A., Martinez Y., Fourquaux I., Jauneau A., Dunand C., Burlat V. Pectin demethylesterification generates platforms that anchor peroxidases to remodel plant cell wall domains. Dev. Cell. 2019;48:261–276.e8. doi: 10.1016/j.devcel.2018.11.016. [DOI] [PubMed] [Google Scholar]
- Francoz E., Ranocha P., Nguyen-Kim H., Jamet E., Burlat V., Dunand C. Roles of cell wall peroxidases in plant development. Phytochemistry. 2015;112:15–21. doi: 10.1016/j.phytochem.2014.07.020. [DOI] [PubMed] [Google Scholar]
- Franke R.B. ‘Caspary’s conductor’. Proc. Natl. Acad. Sci. U S A. 2015;112:10084–10085. doi: 10.1073/pnas.1512640112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita S., De Bellis D., Edel K.H., Köster P., Andersen T.G., Schmid-Siegert E., Dénervaud Tendon V., Pfister A., Marhavý P., Ursache R. SCHENGEN receptor module drives localized ROS production and lignification in plant roots. EMBO J. 2020;39:1–18. doi: 10.15252/embj.2019103894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giehl R.F.H., Lima J.E., von Wirén N. Localized iron supply triggers lateral root elongation in Arabidopsis by altering the AUX1-mediated auxin distribution. Plant Cell. 2012;24:33–49. doi: 10.1105/tpc.111.092973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-García M.P., Vilarrasa-Blasi J., Zhiponova M., Divol F., Mora-García S., Russinova E., Caño-Delgado A.I. Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development. 2011;138:849–859. doi: 10.1242/dev.057331. [DOI] [PubMed] [Google Scholar]
- Gonzali S., Loreti E., Cardarelli F., Novi G., Parlanti S., Pucciariello C., Bassolino L., Banti V., Licausi F., Perata P. Universal stress protein HRU1 mediates ROS homeostasis under anoxia. Nat. Plants. 2015;1:1–9. doi: 10.1038/nplants.2015.151. [DOI] [PubMed] [Google Scholar]
- Gutscher M., Pauleau A.L., Marty L., Brach T., Wabnitz G.H., Samstag Y., Meyer A.J., Dick T.P. Real-time imaging of the intracellular glutathione redox potential. Nat. Methods. 2008;5:553–559. doi: 10.1038/nmeth.1212. [DOI] [PubMed] [Google Scholar]
- Hamdoun S., Zhang Z., Gill M., Kumar N., Churchman M., Larkin J.C., Kwon A., Lu H. Differential roles of two homologous cyclin-dependent kinase inhibitor genes in regulating cell cycle and innate immunity in arabidopsis1[OPEN] Plant Physiol. 2016;170:515–527. doi: 10.1104/pp.15.01466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Barrera A., Velarde-Buendía A., Zepeda I., Sanchez F., Quinto C., Sánchez-Lopez R., Cheung A.Y., Wu H.M., Cardenas L. Hyper, a hydrogen peroxide sensor, indicates the sensitivity of the Arabidopsis root elongation zone to aluminum treatment. Sensors (Switzerland) 2015;15:855–867. doi: 10.3390/s150100855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero J., Fernández-Pérez F., Yebra T., Novo-Uzal E., Pomar F., Pedreño M.Á., Cuello J., Guéra A., Esteban-Carrasco A., Zapata J.M. Bioinformatic and functional characterization of the basic peroxidase 72 from Arabidopsis thaliana involved in lignin biosynthesis. Planta. 2013;237:1599–1612. doi: 10.1007/s00425-013-1865-5. [DOI] [PubMed] [Google Scholar]
- Hoffmann N., Benske A., Betz H., Schuetz M., Samuels A.L. Laccases and peroxidases co-localize in lignified secondary cell walls throughout stem development. Plant Physiol. 2020;184:806–822. doi: 10.1104/pp.20.00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Van Aken O., Schwarzländer M., Belt K., Millar A.H. The roles of mitochondrial reactive oxygen species in cellular signaling and stress response in plants. Plant Physiol. 2016;171:1551–1559. doi: 10.1104/pp.16.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov B.N., Borisova-Mubarakshina M.M., Kozuleva M.A. Formation mechanisms of superoxide radical and hydrogen peroxide in chloroplasts, and factors determining the signalling by hydrogen peroxide. Funct. Plant Biol. 2018;45:102–110. doi: 10.1071/FP16322. [DOI] [PubMed] [Google Scholar]
- Janku M., Luhová L., Petrivalský M. On the origin and fate of reactive oxygen species in plant cell compartments. Antioxidants. 2019;8:105. doi: 10.3390/antiox8040105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo J.H., Bae Y.S., Lee J.S. Role of auxin-induced reactive oxygen species in. Plant Physiol. 2001;126:1055–1060. doi: 10.1104/pp.126.3.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya T., Borghi M., Wang P., Danku J.M.C., Kalmbach L., Hosmani P.S., Naseer S., Fujiwara T., Geldner N., Salt D.E. The MYB36 transcription factor orchestrates Casparian strip formation. Proc. Natl. Acad. Sci. U S A. 2015;112:10533–10538. doi: 10.1073/pnas.1507691112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.J., Ciani S., Schachtman D.P. A peroxidase contributes to ros production during Arabidopsis root response to potassium deficiency. Mol. Plant. 2010;3:420–427. doi: 10.1093/mp/ssp121. [DOI] [PubMed] [Google Scholar]
- Kong X., Tian H., Yu Q., Zhang F., Wang R., Gao S., Xu W., Liu J., Shani E., Fu C. PHB3 maintains root stem cell niche identity through ROS-responsive AP2/ERF transcription factors in arabidopsis. Cell Rep. 2018;22:1350–1363. doi: 10.1016/j.celrep.2017.12.105. [DOI] [PubMed] [Google Scholar]
- Krieger G., Shkolnik D., Miller G., Fromm H. Reactive oxygen species tune root tropic responses. Plant Physiol. 2016;172:1209–1220. doi: 10.1104/pp.16.00660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröller-Schön S., Steven S., Kossmann S., Scholz A., Daub S., Oelze M., Xia N., Hausding M., Mikhed Y., Zinssius E. Molecular mechanisms of the crosstalk between mitochondria and NADPH oxidase through reactive oxygen species - studies in white blood cells and in animal models. Antioxid. Redox Signal. 2014;20:247–266. doi: 10.1089/ars.2012.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laohavisit A., Wakatake T., Ishihama N., Mulvey H., Takizawa K., Suzuki T., Shirasu K. Quinone perception in plants via leucine-rich-repeat receptor-like kinases. Nature. 2020;587:92–97. doi: 10.1038/s41586-020-2655-4. [DOI] [PubMed] [Google Scholar]
- Lee Y., Rubio M.C., Alassimone J., Geldner N. A mechanism for localized lignin deposition in the endodermis. Cell. 2013;153:402–412. doi: 10.1016/j.cell.2013.02.045. [DOI] [PubMed] [Google Scholar]
- Lee Y., Yoon T.H., Lee J., Jeon S.Y., Lee J.H., Lee M.K., Chen H., Yun J., Oh S.Y., Wen X. A lignin molecular brace controls precision processing of cell walls critical for surface integrity in arabidopsis. Cell. 2018;173:1468–1480.e9. doi: 10.1016/j.cell.2018.03.060. [DOI] [PubMed] [Google Scholar]
- Leitz G., Kang B.H., Schoenwaelder M.E.A. Statolith sedimentation kinetics and force transduction to the cortical endoplasmic reticulum in gravity-sensing Arabidopsis columella cells. Plant Cell. 2009;21:843–860. doi: 10.1105/tpc.108.065052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Sun L., Zhang L., Song Y., Hu P., Li C., Hao F.S. AtrbohD and AtrbohF negatively regulate lateral root development by changing the localized accumulation of superoxide in primary roots of Arabidopsis. Planta. 2014;241:591–602. doi: 10.1007/s00425-014-2204-1. [DOI] [PubMed] [Google Scholar]
- Li P., Yu Q., Gu X., Xu C., Qi S., Wang H., Zhong F., Baskin T.I., Rahman A., Wu S. Construction of a functional casparian strip in non-endodermal lineages is orchestrated by two parallel signaling systems in Arabidopsis thaliana. Curr. Biol. 2018;28:2777–2786.e2. doi: 10.1016/j.cub.2018.07.028. [DOI] [PubMed] [Google Scholar]
- Li P., Yang M., Chang J., Wu J., Zhong F., Rahman A., Qin H., Wu S. Spatial expression and functional analysis of casparian strip regulatory genes in endodermis reveals the conserved mechanism in tomato. Front. Plant Sci. 2018;9:832. doi: 10.3389/fpls.2018.00832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman L.M., Sparks E.E., Moreno-Risueno M.A., Petricka J.J., Benfey P.N. MYB36 regulates the transition from proliferation to differentiation in the Arabidopsis root. Proc. Natl. Acad. Sci. U S A. 2015;112:12099–12104. doi: 10.1073/pnas.1515576112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., He C. Regulation of plant reactive oxygen species (ROS) in stress responses: learning from AtRBOHD. Plant Cell Rep. 2016;35:995–1007. doi: 10.1007/s00299-016-1950-x. [DOI] [PubMed] [Google Scholar]
- Livanos P., Galatis B., Quader H., Apostolakos P. Disturbance of reactive oxygen species homeostasis induces atypical tubulin polymer formation and affects mitosis in root-tip cells of Triticum turgidum and Arabidopsis thaliana. Cytoskeleton. 2012;69:1–21. doi: 10.1002/cm.20538. [DOI] [PubMed] [Google Scholar]
- Livanos P., Galatis B., Quader H., Apostolakos P. ROS homeostasis as a prerequisite for the accomplishment of plant cytokinesis. Protoplasma. 2017;254:569–586. doi: 10.1007/s00709-016-0976-9. [DOI] [PubMed] [Google Scholar]
- Mabuchi K., Maki H., Itaya T., Suzuki T., Nomoto M., Sakaoka S., Morikami A., Higashiyama T., Tada Y., Busch W. MYB30 links ROS signaling, root cell elongation, and plant immune responses. Proc. Natl. Acad. Sci. U S A. 2018;115:E4710–E4719. doi: 10.1073/pnas.1804233115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mairhofer S., Zappala S., Tracy S., Sturrock C., Bennett M.J., Mooney S.J., Pridmore T.P. Recovering complete plant root system architectures from soil via X-ray μ-Computed Tomography. Plant Methods. 2013;9:1–7. doi: 10.1186/1746-4811-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majda M., Robert S. The role of auxin in cell wall expansion. Int. J. Mol. Sci. 2018;19:951. doi: 10.3390/ijms19040951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy J.E., Benfey P.N. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development. 1997;124:33–44. doi: 10.1242/dev.124.1.33. [DOI] [PubMed] [Google Scholar]
- Mangano S., Denita-Juarez S.P., Choi H.S., Marzol E., Hwang Y., Ranocha P., Velasquez S.M., Borassi C., Barberini M.L., Aptekmann A.A. Molecular link between auxin and ROS-mediated polar growth. Proc. Natl. Acad. Sci. U S A. 2017;114:5289–5294. doi: 10.1073/pnas.1701536114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangano S., Martínez Pacheco J., Marino-Buslje C., Estevez J.M. How does pH fit in with oscillating polar growth? Trends Plant Sci. 2018;23:479–489. doi: 10.1016/j.tplants.2018.02.008. [DOI] [PubMed] [Google Scholar]
- Manzano C., Pallero-Baena M., Casimiro I., De Rybel B., Orman-Ligeza B., Van Isterdael G., Beeckman T., Draye X., Casero P., del Pozo J.C. The emerging role of reactive oxygen species signaling during lateral root development. Plant Physiol. 2014;165:1105–1119. doi: 10.1104/pp.114.238873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchi S., Giorgi C., Suski J.M., Agnoletto C., Bononi A., Bonora M., De Marchi E., Missiroli S., Patergnani S., Poletti F. Mitochondria-Ros crosstalk in the control of cell death and aging. J. Signal Transduct. 2012;2012:1–17. doi: 10.1155/2012/329635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino D., Dunand C., Puppo A., Pauly N. A burst of plant NADPH oxidases. Trends Plant Sci. 2012;17:9–15. doi: 10.1016/j.tplants.2011.10.001. [DOI] [PubMed] [Google Scholar]
- Marzol E., Borassi C., Ranocha P., Aptekman A.A., Bringas M., Pennington J., Paez-Valencia J., Martínez Pacheco J., Rodríguez Garcia D.R., Rondón Guerrero Y.D.C. Class III peroxidases PRX01, PRX44, and PRX73 potentially target extensins during root hair growth in Arabidopsis thaliana. bioRxiv. 2020:1–13. doi: 10.3390/ijms23105375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhamdi A., Van Breusegem F. Reactive oxygen species in plant development. Development. 2018;145:dev164376. doi: 10.1242/dev.164376. [DOI] [PubMed] [Google Scholar]
- Mhamdi A., Van Breusegem F. Reactive oxygen species in plant development. Development. 2018;145:1–28. doi: 10.1242/dev.164376. [DOI] [PubMed] [Google Scholar]
- Mittler R. ROS are good. Trends Plant Sci. 2017;22:11–19. doi: 10.1016/j.tplants.2016.08.002. [DOI] [PubMed] [Google Scholar]
- Miller G., Schlauch K., Tam R., Cortes D., Torres M.A., Shulaev V., Dangl J.L., Mittler R. The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci. Signal. 2009;2:ra45. doi: 10.1126/scisignal.2000448. [DOI] [PubMed] [Google Scholar]
- Miller G., Suzuki N., Ciftci-Yilmaz S., Mittler R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010;33:453–467. doi: 10.1111/j.1365-3040.2009.02041.x. [DOI] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Gollery M., Van Breusegem F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Suzuki N., Miller G., Tognetti V.B., Vandepoele K., Gollery M., Shulaev V., Van Breusegem F. ROS signaling: the new wave? Trends Plant Sci. 2011;16:300–309. doi: 10.1016/j.tplants.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Mooney S.J., Pridmore T.P., Helliwell J., Bennett M.J. Developing X-ray computed tomography to non-invasively image 3-D root systems architecture in soil. Plant and Soil. 2012;352:1–22. [Google Scholar]
- Muller B., Guédon Y., Passot S., Lobet G., Nacry P., Pagès L., Wissuwa M., Draye X. Lateral roots: random diversity in adversity. Trends Plant Sci. 2019;24:810–825. doi: 10.1016/j.tplants.2019.05.011. [DOI] [PubMed] [Google Scholar]
- Mullineaux P.M., Exposito-Rodriguez M., Laissue P.P., Smirnoff N. ROS-dependent signalling pathways in plants and algae exposed to high light: comparisons with other eukaryotes. Free Radic. Biol. Med. 2018;122:52–64. doi: 10.1016/j.freeradbiomed.2018.01.033. [DOI] [PubMed] [Google Scholar]
- Noctor G., Foyer C.H. Intracellular redox compartmentation and ROS-related communication in regulation and signaling. Plant Physiol. 2016;171:1581–1592. doi: 10.1104/pp.16.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G., Mhamdi A., Foyer C.H. The roles of reactive oxygen metabolism in drought: not so cut and dried. Plant Physiol. 2014;164:1636–1648. doi: 10.1104/pp.113.233478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G., Reichheld J.P., Foyer C.H. ROS-related redox regulation and signaling in plants. Semin. Cell Dev. Biol. 2018;80:3–12. doi: 10.1016/j.semcdb.2017.07.013. [DOI] [PubMed] [Google Scholar]
- Novoplansky A. What plant roots know? Semin. Cell Dev. Biol. 2019;92:126–133. doi: 10.1016/j.semcdb.2019.03.009. [DOI] [PubMed] [Google Scholar]
- O’Brien J.A., Daudi A., Butt V.S., Bolwell G.P. Reactive oxygen species and their role in plant defence and cell wall metabolism. Planta. 2012;236:765–779. doi: 10.1007/s00425-012-1696-9. [DOI] [PubMed] [Google Scholar]
- Orman-Ligeza B., Parizot B., de Rycke R., Fernandez A., Himschoot E., van Breusegem F., Bennett M.J., Périlleux C., Beeckman T., Draye X. RBOH-mediated ROS production facilitates lateral root emergence in Arabidopsis. Development. 2016;143:3328–3339. doi: 10.1242/dev.136465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Espín A., Sánchez-Guerrero A., Sevilla F., Jiménez A. Ascorbic Acid in Plant Growth, Development Stress Tolerance. Springer, Cham; 2017. The role of ascorbate in plant growth and development; pp. 25–45. [Google Scholar]
- Ozcan A., Ogun M. Biochemistry of reactive oxygen and nitrogen species. Basic principles and clinical significance of oxidative stress. 2015;3:37–58. [Google Scholar]
- Pedreira J., Herrera M.T., Zarra I., Revilla G. The overexpression of AtPrx37, an apoplastic peroxidase, reduces growth in Arabidopsis. Physiologia Plantarum. 2011;141:177–187. doi: 10.1111/j.1399-3054.2010.01427.x. [DOI] [PubMed] [Google Scholar]
- Péret B., De Rybel B., Casimiro I., Benková E., Swarup R., Laplaze L., Beeckman T., Bennett M.J. Arabidopsis lateral root development: an emerging story. Trends Plant Sci. 2009;14:399–408. doi: 10.1016/j.tplants.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Petricka J.J., Winter C.M., Benfey P.N. Control of arabidopsis root development. Annu. Rev. Plant Biol. 2012;63:563–590. doi: 10.1146/annurev-arplant-042811-105501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister A., Barberon M., Alassimone J., Kalmbach L., Lee Y., Vermeer J.E., Yamazaki M., Li G., Maurel C., Takano J. A receptor-like kinase mutant with absent endodermal diffusion barrier displays selective nutrient homeostasis defects. Elife. 2014;3:e03115. doi: 10.7554/eLife.03115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires N.D., Yi K., Breuninger H., Catarino B., Menand B., Dolan L. Recruitment and remodeling of an ancient gene regulatory network during land plant evolution. Proc. Natl. Acad. Sci. U S A. 2013;110:9571–9576. doi: 10.1073/pnas.1305457110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podgórska A., Burian M., Szal B. Extra-cellular but extra-ordinarily important for cells: apoplastic reactive oxygen species metabolism. Front. Plant Sci. 2017;8:1–20. doi: 10.3389/fpls.2017.01353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J., Wang J., Gong Z., Zhou J.M. Apoplastic ROS signaling in plant immunity. Curr. Opin. Plant Biol. 2017;38:92–100. doi: 10.1016/j.pbi.2017.04.022. [DOI] [PubMed] [Google Scholar]
- Rabinovitch R.C., Samborska B., Faubert B., Ma E.H., Gravel S.P., Andrzejewski S., Raissi T.C., Pause A., St.-Pierre J., Jones R.G. AMPK maintains cellular metabolic homeostasis through regulation of mitochondrial reactive oxygen species. Cell Rep. 2017;21:1–9. doi: 10.1016/j.celrep.2017.09.026. [DOI] [PubMed] [Google Scholar]
- Reinhard C.T., Planavsky N.J., Olson S.L., Lyons T.W., Erwin D.H. ‘Earth’s oxygen cycle and the evolution of animal life’. Proc. Natl. Acad. Sci. U S A. 2016;113:8933–8938. doi: 10.1073/pnas.1521544113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rellán-Álvarez R., Lobet G., Lindner H., Pradier P.L., Sebastian J., Yee M.C., Geng Y., Trontin C., LaRue T., Schrager-Lavelle A. GLO-Roots: an imaging platform enabling multidimensional characterization of soil-grown root systems. Elife. 2015;4:e07597. doi: 10.7554/eLife.07597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentel M.C., Lecourieux D., Ouaked F., Usher S.L., Petersen L., Okamoto H., Knight H., Peck S.C., Grierson C.S., Hirt H. OXI1 kinase is necessary for oxidative burst-mediated signalling in Arabidopsis. Nature. 2004;427:858–861. doi: 10.1038/nature02353. [DOI] [PubMed] [Google Scholar]
- Reyt G., Boudouf S., Boucherez J., Gaymard F., Briat J.F. Iron- and ferritin-dependent reactive oxygen species distribution: impact on arabidopsis root system architecture. Mol. Plant. 2015;8:439–453. doi: 10.1016/j.molp.2014.11.014. [DOI] [PubMed] [Google Scholar]
- Del Río L.A., López-Huertas E. ROS generation in peroxisomes and its role in cell signaling. Plant Cell Physiol. 2016;57:1364–1376. doi: 10.1093/pcp/pcw076. [DOI] [PubMed] [Google Scholar]
- Rhee S.G., Chang T.S., Jeong W., Kang D. Methods for detection and measurement of hydrogen peroxide inside and outside of cells. Mol. Cells. 2010;29:539–549. doi: 10.1007/s10059-010-0082-3. [DOI] [PubMed] [Google Scholar]
- Rojas-Murcia N., Hématy K., Lee Y., Emonet A., Ursache R., Fujita S., De Bellis D., Geldner N., Rojas N. High-Order Mutants Reveal an Essential Requirement for Peroxidases but Not Laccases in Casparian Strip Lignification. bioRxiv. 2020 doi: 10.1073/pnas.2012728117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini P., Gani M., Kaur J.J., Godara L.C., Singh C., Chauhan S.S., Ghosh M.K. Abiotic Stress-Mediated Sensing and Signaling in Plants: An Omics Perspective. Springer, Singapore; 2018. Reactive oxygen species (ROS): a way to stress survival in plants; pp. 127–153. [Google Scholar]
- Sang S., Li X., Gao R., You Z., Lü B., Liu P., Ma Q., Dong H. Apoplastic and cytoplasmic location of harpin protein Hpa1 Xoo plays different roles in H2O2 generation and pathogen resistance in Arabidopsis. Plant Mol. Biol. 2012;79:375–391. doi: 10.1007/s11103-012-9918-x. [DOI] [PubMed] [Google Scholar]
- Schippers J.H.M., Foyer C.H., van Dongen J.T. Redox regulation in shoot growth, SAM maintenance and flowering. Curr. Opin. Plant Biol. 2016;29:121–128. doi: 10.1016/j.pbi.2015.11.009. [DOI] [PubMed] [Google Scholar]
- Schmidt R., Kunkowska A.B., Schippers J.H.M. Role of reactive oxygen species during cell expansion in leaves. Plant Physiol. 2016;172:2098–2106. doi: 10.1104/pp.16.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaubelt D., Queval G., Dong Y., Diaz-Vivancos P., Makgopa M.E., Howell G., De Simone A., Bai J., Hannah M.A., Foyer C.H. Low glutathione regulates gene expression and the redox potentials of the nucleus and cytosol in Arabidopsis thaliana. Plant Cell Environ. 2015;38:266–279. doi: 10.1111/pce.12252. [DOI] [PubMed] [Google Scholar]
- Schwarzländer M., Fricker M.D., Müller C., Marty L., Brach T., Novak J., Sweetlove L.J., Hell R., Meyer A.J. Confocal imaging of glutathione redox potential in living plant cells. J. Microsc. 2008;231:299–316. doi: 10.1111/j.1365-2818.2008.02030.x. [DOI] [PubMed] [Google Scholar]
- Seagull R.W. Elsevier; 2016. The Plant Cytoskeleton, Reference Module in Food Science. [Google Scholar]
- Sewelam N., Kazan K., Schenk P.M. Global plant stress signaling: reactive oxygen species at the cross-road. Front. Plant Sci. 2016;7:1–21. doi: 10.3389/fpls.2016.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiguzov A., Vainonen J.P., Wrzaczek M., Kangasjärvi J. ROS-talk - how the apoplast, the chloroplast, and the nucleus get the message through. Front. Plant Sci. 2012;3:1–9. doi: 10.3389/fpls.2012.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P., Dubey R.S. Ascorbate peroxidase from rice seedlings: properties of enzyme isoforms, effects of stresses and protective roles of osmolytes. Plant Sci. 2004;167:541–550. [Google Scholar]
- Shukla V., Lombardi L., Iacopino S., Pencik A., Novak O., Perata P., Giuntoli B., Licausi F. Endogenous hypoxia in lateral root primordia controls root architecture by antagonizing auxin signaling in arabidopsis. Mol. Plant. 2019;12:538–551. doi: 10.1016/j.molp.2019.01.007. [DOI] [PubMed] [Google Scholar]
- Sies B.H. Biochem. Oxidative Stress. 1986;25:1058–1071. [Google Scholar]
- Sies H. Oxidative stress: oxidants and antioxidants. Exp. Physiol. 1997;82:291–295. doi: 10.1113/expphysiol.1997.sp004024. [DOI] [PubMed] [Google Scholar]
- Sies H., Berndt C.P., Jones D., Jones D.P. Oxidative stress. Annu. Rev. Biochem. 2017;36:147–154. doi: 10.1146/annurev-biochem-061516-045037. [DOI] [PubMed] [Google Scholar]
- Simard S.W. Memory and Learning in Plants. Springer; Cham: 2018. Mycorrhizal networks facilitate tree communication, learning, and memory; pp. 191–213. [Google Scholar]
- Singh R., Singh S., Parihar P., Mishra R.K., Tripathi D.K., Singh V.P., Chauhan D.K., Prasad S.M. Reactive oxygen species (ROS): beneficial companions of plants’ developmental processes. Front. Plant Sci. 2016;7:1–19. doi: 10.3389/fpls.2016.01299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street I.H., Mathews D.E., Yamburkenko M.V., Sorooshzadeh A., John R.T., Swarup R., Bennett M.J., Kieber J.J., Schaller G.E. Cytokinin acts through the auxin influx carrier AUX1 to regulate cell elongation in the root. Development (Cambridge) 2016;143:3982–3993. doi: 10.1242/dev.132035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S.H., Gibbs N.M., Jancewicz A.L., Masson P.H. Molecular mechanisms of root gravitropism. Curr. Biol. 2017;27:R964–R972. doi: 10.1016/j.cub.2017.07.015. [DOI] [PubMed] [Google Scholar]
- Sundaravelpandian K., Chandrika N.N.P., Tsai Y.H., Schmidt W. PFT1-controlled ROS balance is critical for multiple stages of root hair development in Arabidopsis. Plant Signaling Behav. 2013;8:10–12. doi: 10.4161/psb.24066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Koussevitzky S., Mittler R., Miller G. ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 2012;35:259–270. doi: 10.1111/j.1365-3040.2011.02336.x. [DOI] [PubMed] [Google Scholar]
- Suzuki N., Miller G., Morales J., Shulaev V., Torres M.A., Mittler R. Respiratory burst oxidases: the engines of ROS signaling. Curr. Opin. Plant Biol. 2011;14:691–699. doi: 10.1016/j.pbi.2011.07.014. [DOI] [PubMed] [Google Scholar]
- Swarup R., Wells D.M., Bennett M.J. Plant Roots: The Hidden Half. Fourth Edition. Vol. 37. 2013. Root gravitropism; pp. 265–278. [Google Scholar]
- Szymanski D., Staiger C.J. The actin cytoskeleton: functional arrays for cytoplasmic organization and cell shape control. Plant Physiol. 2018;176:106–118. doi: 10.1104/pp.17.01519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamirisa S., Vudem D.R., Khareedu V.R. A cyclin dependent kinase regulatory subunit (CKS) gene of pigeonpea imparts abiotic stress tolerance and regulates plant growth and development in Arabidopsis. Front. Plant Sci. 2017;8:1–15. doi: 10.3389/fpls.2017.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S., Wang X., Li P., Wang H., Ji H., Xie J., Qiu Q., Shen D., Dong H. Plant aquaporin AtPIP1;4 links apoplastic H2O2 induction to disease immunity pathways. Plant Physiol. 2016;171:1635–1650. doi: 10.1104/pp.15.01237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognetti V.B., Bielach A., Hrtyan M. Redox regulation at the site of primary growth: auxin, cytokinin and ROS crosstalk. Plant Cell Environ. 2017;40:2586–2605. doi: 10.1111/pce.13021. [DOI] [PubMed] [Google Scholar]
- Tognolli M., Penel C., Greppin H., Simon P. Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene. 2002;288:129–138. doi: 10.1016/s0378-1119(02)00465-1. [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H. Control of root growth and development by reactive oxygen species. Curr. Opin. Plant Biol. 2016;29:57–63. doi: 10.1016/j.pbi.2015.10.012. [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H., Busch W., Benfey P.N. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell. 2010;143:606–616. doi: 10.1016/j.cell.2010.10.020. [DOI] [PubMed] [Google Scholar]
- De Veylder L., Beeckman T., Inzé D. The ins and outs of the plant cell cycle. Nat. Rev. Mol. Cell Biol. 2007;8:655–665. doi: 10.1038/nrm2227. [DOI] [PubMed] [Google Scholar]
- Van Aken O., de Clercq I., Ivanova A., Law S.R., van Breusegem F., Millar A.H., Whelan J. Mitochondrial and chloroplast stress responses are modulated in distinct touch and chemical inhibition phases. Plant Physiol. 2016;171:2150–2165. doi: 10.1104/pp.16.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayakumar P., Datta S., Dolan L. ROOT HAIR DEFECTIVE SIX-LIKE4 (RSL4) promotes root hair elongation by transcriptionally regulating the expression of genes required for cell growth. New Phytol. 2016;212:944–953. doi: 10.1111/nph.14095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola I.L., Camoirano A., Gonzalez D.H. Redox-dependent modulation of anthocyanin biosynthesis by the TCP transcription factor TCP15 during exposure to high light intensity conditions in arabidopsis1[OPEN] Plant Physiol. 2016;170:74–85. doi: 10.1104/pp.15.01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wangenheim D., Goh T., Dietrich D., Bennett M.J. Plant biology: building barriers in roots. Curr. Biol. 2017;27:R172–R174. doi: 10.1016/j.cub.2017.01.060. [DOI] [PubMed] [Google Scholar]
- Wachsman G., Sparks E.E., Benfey P.N. Genes and networks regulating root anatomy and architecture. New Phytol. 2015;208:26–38. doi: 10.1111/nph.13469. [DOI] [PubMed] [Google Scholar]
- Wany A., Foyer C.H., Gupta K.J. Nitrate, NO and ROS signaling in stem cell homeostasis. Trends Plant Sci. 2018;23:1041–1044. doi: 10.1016/j.tplants.2018.09.010. [DOI] [PubMed] [Google Scholar]
- Ward L.M., Stamenković V., Hand K., Fischer W.W. Follow the oxygen: comparative histories of planetary oxygenation and opportunities for aerobic life. Astrobiology. 2019;19:811–824. doi: 10.1089/ast.2017.1779. [DOI] [PubMed] [Google Scholar]
- Waszczak C., Akter S., Jacques S., Huang J., Messens J., Van Breusegem F. Oxidative post-translational modifications of cysteine residues in plant signal transduction. J. Exp. Bot. 2015;66:2923–2934. doi: 10.1093/jxb/erv084. [DOI] [PubMed] [Google Scholar]
- Waszczak C., Carmody M., Kangasjärvi J. Reactive oxygen species in plant signaling. Development. 2018;145:209–236. doi: 10.1146/annurev-arplant-042817-040322. [DOI] [PubMed] [Google Scholar]
- Wu J., Sun Y., Zhao Y., Zhang J., Luo L., Li M., Wang J., Yu H., Liu G., Yang L. Deficient plastidic fatty acid synthesis triggers cell death by modulating mitochondrial reactive oxygen species. Cell Res. 2015;25:621–633. doi: 10.1038/cr.2015.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Yuan C., Jiang Z., Xu Y., Xie L., Huang F., Wan D., Ni J., Yuan F., Wu X. Hydrogen peroxide sensor HPCA1 is an LRR receptor kinase in Arabidopsis. Nature. 2020;578:577–581. doi: 10.1038/s41586-020-2032-3. [DOI] [PubMed] [Google Scholar]
- Wunderling A., Ripper D., Barra-Jimenez A., Mahn S., Sajak K., Targem M.B., Ragni L. A molecular framework to study periderm formation in Arabidopsis. New Phytol. 2018;219:216–229. doi: 10.1111/nph.15128. [DOI] [PubMed] [Google Scholar]
- Yang S., Yu Q., Zhang Y., Jia Y., Wan S., Kong X., Ding Z. ROS: the fine-tuner of plant stem cell fate. Trends Plant Sci. 2018;23:850–853. doi: 10.1016/j.tplants.2018.07.010. [DOI] [PubMed] [Google Scholar]
- Yi K., Menand B., Bell E., Dolan L. A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nat. Genet. 2010;42:264–267. doi: 10.1038/ng.529. [DOI] [PubMed] [Google Scholar]
- Yoshida K., Noguchi K., Motohashi K., Hisabori T. Systematic exploration of thioredoxin target proteins in plant mitochondria. Plant Cell Physiol. 2013;54:875–892. doi: 10.1093/pcp/pct037. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Friml J. Auxin guides roots to avoid obstacles during gravitropic growth. New Phytol. 2020;225:1049–1052. doi: 10.1111/nph.16203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P., Jauregui E., Du L., Tanaka K., Poovaiah B.W. ‘Calcium signatures and signaling events orchestrate plant–microbe interactions’. Curr. Opin. Plant Biol. 2017;38:173–183. doi: 10.1016/j.pbi.2017.06.003. [DOI] [PubMed] [Google Scholar]
- Zhang M., Smith J.A.C., Harberd N.P., Jiang C. The regulatory roles of ethylene and reactive oxygen species (ROS) in plant salt stress responses. Plant Mol. Biol. 2016;91:651–659. doi: 10.1007/s11103-016-0488-1. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Luo L., Xu J., Xin P., Guo H., Wu J., Bai L., Wang G., Chu J., Zuo J. Malate transported from chloroplast to mitochondrion triggers production of ROS and PCD in Arabidopsis thaliana. Cell Res. 2018;28:448–461. doi: 10.1038/s41422-018-0024-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Nakashima J., Chen F., Yin Y., Fu C., Yun J., Shao H., Wang X., Wang Z.Y., Dixon R.A. LACCASE is necessary and nonredundant with PEROXIDASE for lignin polymerization during vascular development in Arabidopsis. Plant Cell. 2013;25:3976–3987. doi: 10.1105/tpc.113.117770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Sokolov L.N., Ye J., Tang C.Y., Shi J., Zhen Y., Lan W., Hong Z., Qi J., Lu G.H. The LIKE SEX FOUR2 regulates root development by modulating reactive oxygen species homeostasis in Arabidopsis. Scientific Rep. 2016;6:1–11. doi: 10.1038/srep28683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Li Q., Chen X., Liu J., Zhang Q., Liu Y., Liu K., Xu J. The arabidopsis RETaRDED ROOT GROWTH gene encodes a mitochondria-localized protein that is required for cell division in the root meristem. Plant Physiol. 2011;157:1793–1804. doi: 10.1104/pp.111.185827. [DOI] [PMC free article] [PubMed] [Google Scholar]