Summary
This review describes recent research that has advanced our understanding of the role of immune cells in the tumor microenvironment (TME) using advanced 3D in vitro models and engineering approaches. The TME can hinder effective eradication of tumor cells by the immune system, but immunotherapy has been able to reverse this effect in some cases. However, patient-to-patient variability in response suggests that we require deeper understanding of the mechanistic interactions between immune and tumor cells to improve response and develop novel therapeutics. Reconstruction of the TME using engineered 3D models allows high-resolution observation of cell interactions while allowing control of conditions such as hypoxia, matrix stiffness, and flow. Moreover, patient-derived organotypic models are an emerging tool for prediction of drug efficacy. This review highlights the importance of modeling and understanding the immune TME and describes new tools for identifying new biological targets, drug testing, and strategies for personalized medicine.
Subject areas: Components of the Immune System, Cancer, Bioengineering
Graphical abstract
Components of the Immune System; Cancer; Bioengineering
Introduction: the intersection of immune cells, cancer, and engineering
Cancer is still a leading cause of death, despite significant progress made in cancer therapy over the last decade and improvements in life expectancy of patients in several developed countries (Bray et al., 2018). Notably, immunotherapy has proved to be effective for eradicating primary and metastatic lesions for some tumor types, by promoting the immune system’s elimination of cancerous cells (Farkona et al., 2016). Cancer immunotherapy includes several methods to boost the immune system’s response to cancer such as cytokines, vaccines, oncolytic viruses, adoptive cell therapy, and immune checkpoints. The United States Food and Drug Administration has approved a number of immune checkpoint inhibitors in adjuvant or first-line setting for several solid tumors (Vaddepally et al., 2020). However, response to immunotherapy varies strongly from person to person due to several factors, including the diversity of the tumor microenvironment (TME) among patients (Galon and Bruni, 2019). So called “hot” tumors that respond well to immunotherapy typically have a high number of infiltrating lymphocytes and present neoantigens, such as tumors with high mutational burden (Goodman et al., 2017). In contrast, in a “cold” tumor, lymphocytes are excluded or absent from the TME, likely through immunosuppression, or the tumor has low mutational burden. Lack of tumor antigens is typically due to the process of immunoediting during cancer development, in which some of the most immunogenic tumor cells are killed by the immune system (Yarchoan et al., 2017).
The TME consists of many elements that can influence diffusion of oxygen and nutrients, as well hindering efficient drug delivery and immune surveillance. These features can include fibrosis or desmoplasia, interstitial fluid pressure, vascularization, or lack thereof including lymphatic vasculature, hypoxia, and altered pH/metabolic pathways. All these features involve interaction between the tumor and its surroundings in ways that influence prognosis and response to therapy. Few of these features can be mimicked with 2D cell culture, so advances in 3D culture are vital to unraveling the complex and interlaced role of the TME.
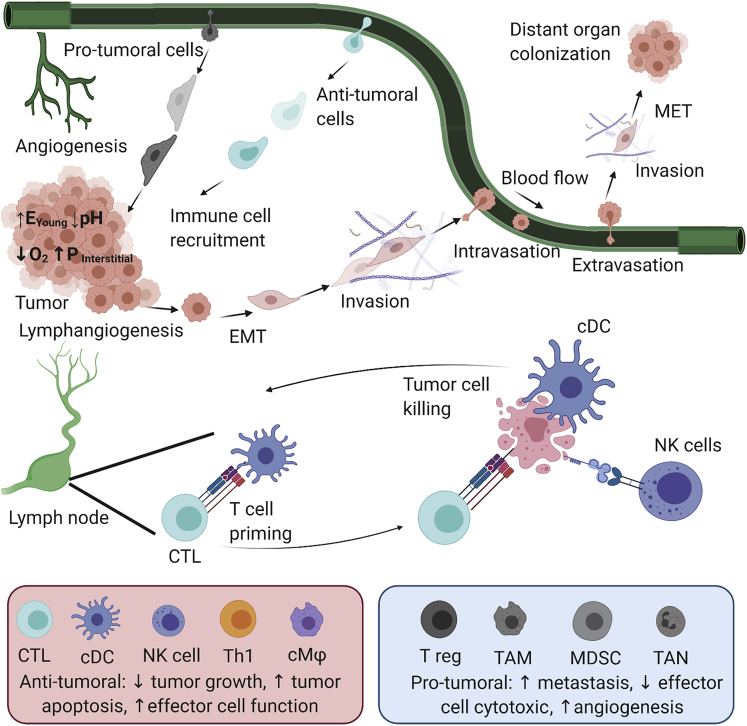
Through the course of tumorigenesis, cancer cells can acquire phenotypic traits that allow them to avoid immune recognition or destruction and recruit inflammatory immune cells that support their progression (Hanahan and Weinberg, 2011). The immune landscape surrounding the tumor changes gradually from anti-tumoral to pro-tumoral by transformative chemical and physical signaling from the growing tumor (Gonzalez et al., 2018). Clinical data showed that subsets of infiltrating lymphocytes that are associated with a better patient survival rate include cytotoxic T cells (CTL), conventional dendritic cells (cDC), natural killer (NK) cells, and Th1 helper T cells (Galon and Bruni, 2019). Other immune cell types are considered pro-tumoral, as they promote tumor cell metastasis and suppress anti-tumoral responses, including immunosuppressive tumor-associated macrophages (TAM), myeloid-derived suppressor cells (MDSC), and regulatory T cells (Treg) (Galon and Bruni, 2019). Figure 1 illustrates the anti-tumoral or pro-tumoral behaviors of some immune cell types and the microenvironmental conditions that affect tumor development and dissemination, including processes such as tumor cell killing, immune cell recruitment, angiogenesis, lymphangiogenesis, and tumor cell metastasis. Physical conditions in solid tumors such as increased extracellular matrix stiffness, vascular perfusion and permeability, and interstitial flow, as well as hypoxia and low pH - byproducts of tumor metabolism, also contribute to the transformation of the TME (Seager et al., 2017). Understanding the physical interaction between cancer and stromal cells is therefore essential to guide immunotherapy research to modify the TME to become more anti-tumoral.
Figure 1.
Infiltrating immune cells and physical conditions regulate several cellular events during tumor development and dissemination
Bottom left box: anti-tumoral leukocytes such as cytotoxic T lymphocytes (CTL), conventional dendritic cells (cDC), natural killer (NK) cells, type 1 helper T cell (Th1), and cytotoxic macrophage (cMΦ). Anti-tumoral immune cells infiltrate to the tumor and kill tumor cells by different mechanisms: anti-tumoral cytokine secretion, phagocytosis, or tumor-specific cytotoxicity. Bottom right box: pro-tumoral cells such as regulatory T cells (T reg), tumor-associated macrophages (TAMs), myeloid-derived suppressor cells (MDSCs), and tumor-associated neutrophils (TANs) secrete pro-tumoral cytokines that promote tumor cell survival and proliferation and induce angiogenesis or lymphangiogenesis. They also support tumor cell metastasis and suppress anti-tumoral immune cell recruitment and function in both primary and metastatic sites. Physical conditions within the tumor such as ECM stiffness, low pH, and additional factors such as hypoxia and high interstitial fluid pressure also tend to suppress the recruitment and function of anti-tumoral leukocytes.
In the age of immunotherapy when the array of therapeutics to target the TME is becoming increasingly diverse and sophisticated, the need for models that integrate human immune components is rapidly increasing. Despite recent advances in xenograft or humanized mice, the development of cancer immunotherapy drugs relies heavily on syngeneic mouse models that have an intact immune system (Olson et al., 2018). Engineered 3D in vitro systems have the advantage of using human cells and offer a complexity similar to in vivo while retaining tight control over physical parameters and cell composition. Therefore, 3D in vitro systems can be used to recapitulate the physiological state of the cells within a body, allowing the study of both chemical and physical signaling between cells in the TME (Huh et al., 2011; Sontheimer-Phelps et al., 2019). Finally, owing to the capability of multiplexing, microfluidic systems can be used as a tool for the evaluation of drug efficacy, either for drug development or in the clinic for patient diagnostics and stratification. Here we highlight several models that have been used to study the mechanistic interaction between tumor cells and immune cells and their implications in drug testing in some recent publications.
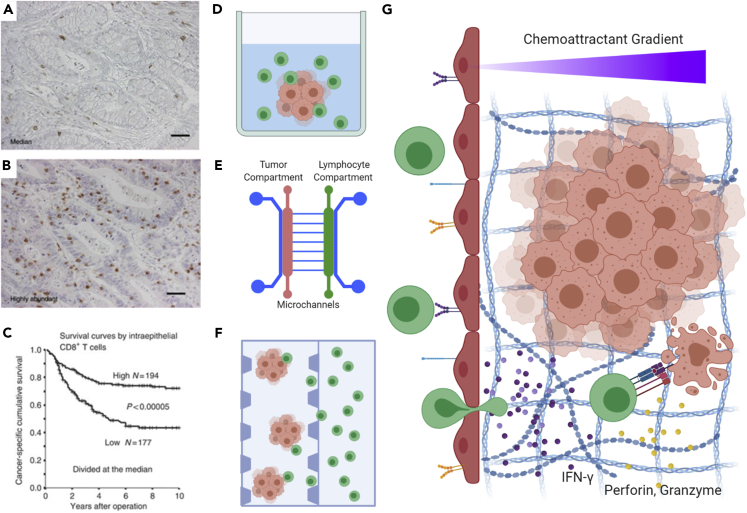
Effective response to immunotherapy relies on the response of the adaptive immune system to initiate a sequence of T cell priming via antigen presentation, recruitment of lymphocytes to a lesion (especially by CD8+, cytotoxic, effector T cells), followed by recognition and killing of cancer cells (Farhood et al., 2019). Indeed, the abundance of tumor infiltrating lymphocytes correlates with overall survival (Figures 2A–2C). However, the difficulty of predicting which subset of patients will benefit from immune checkpoint blockade has led to a need for new in vitro systems capable of modeling different aspects of the therapeutic response in the complex setting of the TME. Several of these models attempt to understand chemoattractant gradients and factors influencing the migration of lymphocytes in order to improve our understanding of how T cells are recruited to or excluded from solid tumors. Other models focus on the direct interactions between immune cells and cancer cells, dissecting the processes of T cell activation and cytotoxicity in simplified 3D, in vitro, engineered systems.
Figure 2.
Adaptive immunity in cancer focuses on the role of T lymphocytes in the TME
(A and B) Immunohistochemical staining of CD8+ lymphocytes in colorectal cancer illustrating examples of “immune excluded” (panel A) versus “immune infiltrated” (panel B) tumors.
(C) Kaplan-Meier curve revealing the association between survival and the number of infiltrating lymphocytes observed.
(D–F) Three-dimensional, in vitro approaches to studying tumor-immune interactions including organoid culture in multiwall plates with lymphocytes in suspension (D), microfluidic devices with tumor and lymphocyte compartments separated by microchannels (E), and microfluidic devices with tumor organoids embedded in hydrogel channels immediately adjacent to a lymphocyte compartment (F).
(G) Studies focusing on the trajectory and activity of T cells in the cancer microenvironment, including chemoattractant gradients to recruit T cells, T cell adhesion and extravasation through the vascular endothelium, migration through extracellular matrix, and interaction with cancer cells resulting in the secretion of proteins such as IFN-γ, perforin, and granzymes, culminating tumor cell killing.
Panels A–C adapted from (Chiba et al., 2004).
Innate immune cells play important roles in the constant fight against pathogens and in wound healing. However, in cancer, they are usually hijacked or functionally compromised by tumor cells (Gonzalez et al., 2018). Anti-tumoral, innate immune cells, such as dendritic cells, link the innate and adaptive immune response through antigen presentation, priming of T cells, and cytokine secretion. Furthermore, an effective adaptive immune response to immunotherapy treatments such as immune checkpoint blockade is often dictated by how tumor innate immunity shapes the immune microenvironment (Petitprez et al., 2020). Macrophages are particularly relevant due to their plasticity that results in polarization into phenotypes that tend to be pro-tumoral (M2) or anti-tumoral (M1), as well as their ability to guide other processes pertinent to the TME, such as angiogenesis, fibrosis, and inflammation (Long and Beatty, 2013). Microfluidic devices allow us to recapitulate selected aspects of the TME, while decoupling the effects of chemical and physical conditions to observe interactions and signaling between tumor and immune cells. In the following section, we discuss the use of 3D models to reveal the range of roles played by T-lymphocytes of the adaptive immune system, several types of innate immune cells (monocytes, macrophages, neutrophils, dendritic cells, and natural killer cells), and physical conditions throughout the processes of tumor development and dissemination.
Migration, extravasation, and angiogenesis
In the context of the immune environment in cancer, the process of migration is of particular importance in order to understand the cues to which immune cells respond and how immune cell infiltration of tumors occurs. Furthermore, several studies have also explored the role that immune cells can play in promoting or inhibiting the migration and metastasis of cancer cells, both in the TME as well as the vascular system. In vitro systems also possess the ability to control several microenvironmental factors such as chemokine gradients, hypoxia, and matrix composition. In comparison to conventional methods for quantifying cell migration such as transwell assays, microfluidic systems offer improved visualization and characterization of cell motion and interactions using microscopy.
Immune cell migration and infiltration
Immune cells in the peripheral blood are recruited into a tumor by both chemical and physical signaling (Seager et al., 2017). Using microfluidic 3D cell culture, several reports have demonstrated that the interplay between tumor cells and the TME can affect immune cell migration. One group used a microfluidic device to co-culture human pancreatic adenocarcinoma (Panc1) and blood monocyte-derived macrophages in separate gel chambers (Figure 3A), to show that tumor cells enhanced macrophage migration through the secretion chemokines IL-8 and CCL2 (Lee et al., 2020). The device used also had the capability of introducing interstitial flow by imposing a pressure gradient across the gel region. This was utilized to demonstrate that interstitial flow independently increased the directedness and speed of macrophage migration through activation of Rho-kinase, highlighting the effect of physical stimuli in immune cell migration and recruitment. An earlier, similar study had also demonstrated that interstitial flow polarized macrophages into a pro-tumoral phenotype through STAT3/6 activation and enhanced the capability of macrophages to migrate and further promote cancer cell migration (Li et al., 2018). Furthermore, a microfluidic model of bladder cancer revealed that lactate was responsible for increased macrophage chemotaxis through Matrigel as well as inhibiting anti-tumoral M1 polarization, instead promoting pro-tumoral polarization toward M2 (Zhao et al., 2015).
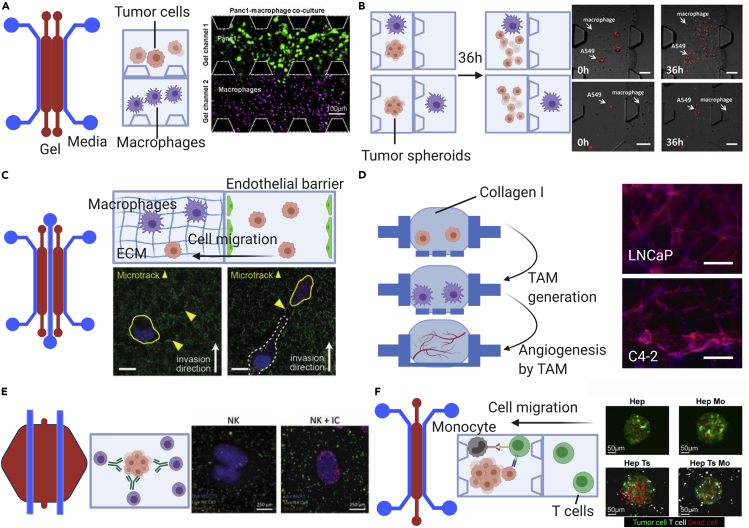
Figure 3.
Three-dimensional models employed to study different cellular events during tumor progression and dissemination
(A) Immune cell recruitment. Microfluidic device with two adjacent gel channels (red) flanked by media channels (blue) is used for the study of macrophage migration in the presence of tumor cells. Tumor cells and macrophages are suspended in collagen I in separate gel channels. Single-cell analysis of macrophage migration demonstrates that macrophages have higher speed and directedness when co-cultured with tumor cells compared with controls.
(B) Tumor cell dissemination. A device similar to that in (A) was used to study epithelial-mesenchymal transition (EMT). Tumor spheroids are suspended in gel, and macrophages are either suspended together in the same or an adjacent but different channel than the tumor spheroid. After 36 h, tumor spheroids cultured with M2a macrophages, but no other sub-type, dispersed more readily when in contact with the tumor as opposed to separated. Scale bars: 100 μm.
(C) Tumor cell invasion at the metastatic site. First, monocytes are introduced into the endothelialized center channel and allowed to extravasate. Two days later, tumor cells are introduced into the same channel. Monocyte migration created microtracks that facilitated tumor cell invasion into the ECM. Scale bars:10 µm.
(D) Angiogenesis. Open-top, stackable, microfluidic devices demonstrate different effects of tumor cell phenotypes on angiogenesis through induction of TAMs. This study showed that different tumor prostate cancer phenotypes resulted in different vessel morphologies through modulation of TAM phenotypes. Scale bars: 150 μm.
(E) Cytotoxicity. Effects of antibody drugs on tumor cell killing by NK cells is recapitulated by a microfluidic platform that has tumor cells and NK cells in gel suspension, and drugs diffuse from the media channel. This study reports the capability of NK cells to induce tumor cell apoptosis.
(F) Immunosuppression. Effector T cell suppression by monocytic cells are modeled by capturing tumor-killing efficacy within a microfluidic device. Tumor cells and monocytes are suspended in the gel channel, whereas effector T cells are introduced into the media channel. T cells migrate then to the gel channel, toward tumor cells and kill them. However, the presence of monocytic cells in the gel impeded the killing efficacy. Using check point blockade targeting PD-L1/PD-1 signaling, T cell cytotoxicity is restored.
Figures are adapted from the following publications: (A–F) panels are respectively from Lee et al., 2020; Bai et al., 2015; Kim et al., 2019a; Yu et al., 2019; Ayuso et al., 2019; Lee et al., 2018):
In a series of studies using the C57BL/6 mouse model and B16F10 melanoma, one research group examined and quantified the migration of immune cells harvested from the spleen, toward cancer cells in an adjacent compartment separated by microchannels 500 μm long, with cross sections of 10 × 12 μm (Businaro et al., 2013; Agliari et al., 2014; Mattei et al., 2014). Using time-lapse microscopy, the trajectories of splenocytes from wild-type mice and mice lacking a transcription factor related to immune cell differentiation were compared (Businaro et al., 2013). They determined that normal splenocytes were able to achieve coordinated migration toward the cancer cell compartment of the device but that cells from the knockout mice were not. The same device was also used to study the migration of peripheral blood mononuclear cells (PBMC) toward doxorubicin-treated MDA-MB-231 (a human breast cancer cell line), revealing weaker migration of PBMC from individuals with mutations in formyl peptide receptor 1 (FPR1), a protein-sensing annexin A1 from dying cells (Biselli et al., 2017). Dendritic cell migration and phagocytosis of tumor cells was also tracked and characterized using the same microfluidic system (Parlato et al., 2017). This study used the SW-620 colorectal cancer cell line embedded in collagen I gel in combination with dendritic cells derived from PBMC and stimulated with interferon-α (IFN-α) and added to a separate fluid compartment. Through tracing dendritic cell migration, this study demonstrated that, compared with untreated SW620 cells, IFN-stimulated DCs tended to migrate more toward SW620 cells treated by a combination of IFN-α and an epigenetic drug targeting histone deacetylase (romidepsin).
In addition to the ability to observe and track cell migration using in vitro models, especially microfluidic devices (see Figures 2D–2F), they can be configured in a variety of ways and customized to address different research questions that would not be possible to address through in vivo studies. Some designs contain multiple cell reservoirs and migratory channels or porous layers to establish a chemotactic gradient, whereas others offer simple controlled environments for observing lymphocytes under flow. One example is that of a microfluidic device coated with adhesion molecules, through which T cells (the Jurkat leukemia line) were introduced by flow, either with or without chemokine gradients (Kim et al., 2012, Kwasny et al., 2011). Another simple microfluidic device observed PBMC-derived T cell chemotaxis in 2D, in response to gradients of CCL19 or CXCL12 created with a simple Y-shaped geometry (Lin and Butcher, 2006). Increasing the complexity slightly, another group used a microfluidic model to test the ability of immunomodulatory nanoparticles embedded along with ovarian cancer to stimulate the infiltration of dendritic cells and T cells through an adjacent endothelial-coated channel (Wimalachandra et al., 2019).
Other groups have developed microfluidic devices with more complex arrangements in order to accommodate multiple cell types. A perfusable vascular microfluidic model was used recently to demonstrate Jurkat T cell adhesion in a study of endothelial activation of the cGAS/STING pathway via tumor-derived 2′3′-cGAMP (Campisi et al., 2020). In a study of human breast cancer cell lines MCF-7 and MDA-MB-231, researchers created cancer spheroids and surrounded them with human umbilical vein endothelial cells (HUVEC) to mimic the process of extravasation (Aung et al., 2020). T cells from the TALL-104 line were found to cross the HUVEC boundary more readily when THP-1 monocytes were included, highlighting the importance of crosstalk between different populations of immune cells. They attributed T cell migration to chemokines secreted by the spheroids and monocytes, including CCL4, CCL5, CCL11, and CXCL9. Using the same device, they were also able to examine the influence of gel stiffness, model diffusion around the spheroids, and visualize hypoxia with a fluorescent detection dye. In a study by Mitra et al., the authors used a microfluidic device to generate a chemokine gradient (CCL19) to direct dendritic cell migration toward a compartment of T cells (Mitra et al., 2013). They found that mature DCs from the MUTZ-3 cell line activated T cells from PBMC more strongly than immature DCs. Other studies that combined the assessment of T cell migration and activation used CAR-T cells model to evaluation infiltration and cytotoxicity under different oxygenation states and found lower CAR-T cell infiltration under hypoxic conditions (Ando et al., 2019; Pavesi et al., 2017). Collectively, these reports demonstrate how tumors can influence the mobility of PBMC and antigen-presenting cells such as macrophages and dendritic cells, facilitating their recruitment into the TME.
Tumor cell dissemination
Tumor cell migration in a matrix
Several studies have highlighted the role of innate immune cells, primarily TAMs, in dissemination of tumor cells through the extracellular matrix. Generally, TAMs promote tumor cell migration, and several studies have sought to demonstrate this phenomenon in 3D microfluidic devices and to identify the factors that directly promote dissemination of tumor cells. One study determined that collagen stiffness inhibited migration of breast cancer cell lines from spheroids but inclusion of TAMs increased cell mobility (Yuan et al., 2019). In inflammatory breast cancer, Allen et al. came to the conclusion that macrophages were able to “prime” cancer cells for migration by pre-treating cancer cells in microfluidic devices with macrophage-conditioned medium and then subjecting them to an independent chemotactic signal (serum) (Allen et al., 2016). Cancer cells exposed to media from macrophages were significantly more migratory, and the authors showed that this response required RhoC, as knocking out RhoC abrogated the response.
Another study used a microfluidic device to demonstrate that TAMs derived from human blood and polarized into different sub-types differentially promoted tumor epithelial-mesenchymal transition and migration of A579 tumor cell aggregates (Bai et al., 2015). The TAMs and tumor organoids were embedded in collagen I, within either the same channel or an adjacent gel channel of the microfluidic device (Figure 3B). Only when the M2a subtype macrophages were co-cultured with tumor cells within the same channel and allowed to contact with the tumor cells, tumor expression of E-cadherin decreased, and tumor aggregates dispersed more readily. In a similar study, macrophages from several sources (RAW 264.7 macrophages, primary mouse or human macrophages) and breast cancer cell lines (MDA-MB-231, MDA-MB-435) were co-cultured within two adjacent gel channels (Li et al., 2017). TAMs enhanced the persistence and speed of cancer cell migration through tumor necrosis factor alpha (TNF-α) and transforming growth factor β 1 (TGF-β1) signaling, suggesting that dual blockade of TNF-α and TGF-β1 could be a potential treatment to ameliorate the effect of macrophages on tumor cell migration. TNF-α was also identified as a key cytokine secreted by macrophages in a study examining lung cancer migration in response to conditioned media from myofibroblasts and macrophages (Hsu et al., 2012). However, in this case, TNF-α from macrophages inhibited the ability of myofibroblasts to promote cancer cell migration through the reduction of α-smooth muscle actin and TGF-β by myofibroblasts.
At metastatic sites, monocytic cells also favor tumor cell migration. In one study, the metastatic site was modeled in a microfluidic device with a human dermal microvascular endothelial cell monolayer-coated channel sandwiched between two collagen gel channels (Figure 3C). THP-1 (a human monocytic cell line) or primary monocytes were flowed into the endothelial channel prior to introduction of tumor cells MDA-MB-231 (Kim et al., 2019a). Monocytes broke endothelial tight junctions by secretion of matrix metalloproteinase 9 and created “microtracks” in the underlying matrix that enhanced cancer cell extravasation and invasion.
Cancer cell extravasation and metastasis
Microfluidic devices that include a vascular component are able to capture and observe certain rare events that occur during the metastatic cascade such as intravasation and extravasation of cancer cells. They also facilitate the study of the critical role played by immune cells in these potentially rate-limiting processes. Several strategies have been employed to create in vitro vascular structures, and these have been extensively reviewed elsewhere (Cochrane et al., 2019; Sontheimer-Phelps et al., 2019). The simplest geometry uses a monolayer of endothelial cells to enable the observation of cancer or immune cells crossing a vascular barrier. One such study examined the role of immune cells in metastasis to observe the process by which macrophages assist tumor cells intravasation (Zervantonakis et al., 2012). Tumor cells (HT1080 fibrosarcoma or MDA-MB-231) and Raw 264.7 macrophages were suspended in a collagen I gel channel coated with a monolayer of HUVEC or microvascular endothelial cells. The authors found that macrophages promoted tumor cell trans-endothelial migration roughly 5-fold compared with control conditions lacking macrophages and that secretion of TNF-α, rather than direct cell-cell contact, was a critical factor due to its ability to impair endothelial barrier function. A similar study of breast cancer spheroids and TAMs embedded in adjacent collagen channels with an endothelial monolayer obtained results that supported the earlier findings of macrophage promotion of migration by MDA-MB-231 breast cancer cells (Mi et al., 2019).
Recently, researchers have also studied extravasation of circulating tumor cells by perfusing tumor cells into 3D microvascular networks (Coughlin and Kamm, 2020). In particular, a microfluidic vasculature with physiological levels of flow was employed to demonstrate that primary neutrophils clustered in the vicinity of tumor cells (A375-MA2 melanoma) in the microvessels and enhanced their rate of extravasation, mediated by a combination of neutrophil-secreted IL-8 and tumor-cell-secreted CXCL-1 (Chen et al., 2018). Compared with standard 2D assays such as transwells, microfluidic systems recapitulate physiological conditions more faithfully and allow quantification of metastatic processes in 3D through confocal microscopy.
Angiogenesis in the cancer microenvironment
As a tumor grows, it induces angiogenesis to access more nutrients and oxygen (Hanahan and Weinberg, 2011). Macrophages are one cell type that has the capability to induce angiogenesis due to their role in wound healing (Gonzalez et al., 2018). Therefore, their presence in a tumor can promote neovascularization and help cancer cells to access the bloodstream (Riabov et al., 2014). To decouple the effect of TAMs and tumor cells on angiogenesis, researchers created multilayered, open-top, suspended, microfluidic layers of 2D and 3D cell cultures (Yu et al., 2019). This method allowed the assembly and separation of microfluidic chambers (Figure 3D). First, TAMs were generated by stacking one gel layer containing THP-1 monocytes on top of another gel layer containing LNCaP or C4-2 prostate cancer. LNCaP tumor cells polarized THP-1-derived macrophages to a pro-inflammatory phenotype, whereas C4-2 cell lines polarized them to an anti-inflammatory phenotype. When the macrophage layer was removed and joined with a layer containing HUVECs suspended in a hydrogel, it induced vessel formation. Macrophages exposed to LNCaP and C4-2 resulted in different vessel morphologies, highlighting the effect of different tumor phenotypes on the TME.
In models that incorporate an endothelial monolayer, sprouting angiogenesis can be quantified to determine the angiogenic influence of cancer cells and macrophages. An in vitro breast cancer model showed that activated macrophages and interstitial flow increased production of vascular endothelial growth factor (VEGF) and sprouting of the endothelial monolayer (Song et al., 2019). In another system, the angiogenic capability of macrophages was demonstrated by the induction of endothelial cell sprouting when macrophages were co-cultured with glioblastoma cells (Cui et al., 2018). Using a device similar to the one shown in Figure 3E, researchers coated one media channel with C166 mouse endothelial cells and loaded the other with Raw 264.7 macrophages and GL261 or CT-2A glioblastoma cells that were subsequently allowed to migrate into the gel. This enabled signaling between endothelial cells and macrophages via cytokines such as TGF-β and IL-1 that induced angiogenic sprouting toward the tumor cells and macrophages.
Immune recognition of cancer and elimination
Activation of T cells and cytotoxicity
Once lymphocytes infiltrate the TME, they must receive activation signals in order to carry out cell killing. Standard approaches for quantifying T cell activation include limiting dilution studies, enzyme-linked immune absorbent spot (ELISPOT), and fluorescent MHC/peptide tetramers in conjunction with flow cytometry (Bercovici et al., 2000). However, these approaches are not easily performed within 3D environments that permit real-time imaging or cellular interactions with tumor organoids. Therefore, many groups have developed alternative approaches for assessing T cell activation and cytotoxicity that can be tracked serially and often combined with the quantification of additional outcome variables. Three-dimensional systems for assessing T cell activation typically combine cancer cell spheroids and T cells, either in microwell plates or embedded in a hydrogel such as Matrigel or collagen. Methods for quantifying T cell activation and cytotoxicity range from very straightforward measurements of cytokine concentrations and size changes in the cancer spheroids (Hoffmann et al., 2015) to more sophisticated markers of activation, such calcium sensitive fluorescent dye (Mitra et al., 2013) or T cells engineered to express luciferase inducible by TCR, PD-1, and NFAT (Zboralski et al., 2017). One group used droplet microfluidics to monitor T cell activation of single cells in real time with T cells engineered to express GFP upon activation (Segaliny et al., 2018). Primary T cells can be isolated from PBMCs, including subpopulations of T cells such as regulatory T cells, as was done in a study from 2015 that described methods for 3D culture of breast cancer spheroids embedded in Matrigel with T-Regs and NK cells derived from PBMCs (Augustine et al., 2015). Other models combining T cells with cancer cell line spheroids have included colorectal carcinoma, prostate cancer, glioblastoma multiforme, non-small cell lung cancer, and pancreatic adenocarcinoma (Zboralski et al., 2017; Hoffmann et al., 2015; Sadovska et al., 2018; Florczyk et al., 2012).
Three-dimensional, co-culture approaches to studying T cell activation in the context of cancer are valuable because of differences observed between activation in 2D versus 3D culture environments, with 3D culture more faithfully recapitulating in vivo cellular responses. Dangles et al. used a 3D model of bladder cancer to demonstrate that tumor infiltrating lymphocytes experience reduced levels of T cell activation compared with the same cells cultured in 2D (Dangles et al., 2002). Feder-Mengus et al. also observed less T cell activation in 3D culture, which they were able to attribute to lower expression of HLA class 1 and increased production of lactic acid by spheroids compared with 2D culture (Feder-Mengus et al., 2007). In this model, which combined melanoma spheroids and primary T cells in microwell plates, the authors observed that most T cells remained around the periphery and did not invade the spheroids. These studies, in particular, highlight the need for in vitro experimental systems that are more representative of in vivo systems.
Due to the difficulty of developing autologous models of the adaptive immune response, several methods have been utilized to model T cell activation in the TME. One approach is to use syngeneic cells and tissue derived from mice such as the C57BL/6 or MMTVneu strains. Thus, experiments can be repeated and cells and tissue harvested from several animals without inducing allogeneic reactions. One group used this approach to study the inhibitory effect of cancer-associated fibroblasts on T cell activation in mammary carcinoma spheroids (Phan-Lai et al., 2013). Others used the B16F10 melanoma model due to its well-established role in cancer immunotherapy research (Businaro et al., 2013; Agliari et al., 2014; Mattei et al., 2014). A different approach is the combination of melanoma cell lines expressing melanoma antigen expressed by T cells (Melan-A/MART-1) and HLA -A∗0201-restriced, MART-1-specific T cells from peripheral blood lymphocytes. This approach has been used to compare the activation of T cells (via interferon-γ production) co-cultured with cell lines expressing or lacking the MART-1 antigen in 3D spheroid culture (Feder-Mengus et al., 2007). Yet another method is to use matched, patient-specific tumor tissue and immune cells, a method known as “patient-derived organotypic” models or PDOs, which will be discussed in more detail in a later section.
Another approach to T-cell-mediated eradication of cancer is the use of T cells engineered to express chimeric antigen receptors or CAR-T cells. Although CAR-T therapy has been successful for some hematological malignancies, the technology has shown limited efficacy in solid tumors (Hong et al., 2020). Thus, advances in 3D models allows researchers to understand some of the fundamental differences in using CAR-T cells in the complex TME. One study used lung or breast cancer spheroids to interrogate the infiltration of CAR-T cells and their ability to eliminate layers of cancer cells, while tracking the increase of IL-2 and IFN-γ in the devices (Wallstabe et al., 2019). Two other groups used microfluidic devices with a chamber for tumor cells and adjacent channels to introduce TCR-T cells or CAR-T cells (Ando et al., 2019; Pavesi et al., 2017). Both studies subjected the devices to reduced oxygen concentrations and observed similar results of reduced T cell infiltration and cytotoxicity under hypoxic conditions, pointing to a possible mechanism of failure in engineered T cell treatment of solid malignancies, which are prone to hypoxia.
T cells are known to be a particularly mechanosensitive cell type that requires mechanotransduction at the immunological synapse, the complex formed between the T cell receptor and the MHC molecule bearing the antigen peptide on the tumor cell (Li et al., 2020). Mechanotransduction and mechanosensitive activation of CAR-T cells has also been exploited using sophisticated in vitro methods, such as one system that combined genetically engineered genetic circuits with ultrasound stimulation to activate T cells (Pan et al., 2018). The authors used the Piezo1 gene to transduce transcriptional expression of the CAR upon demand using low-frequency ultrasound (2 MHz) and Arg-Gly-Asp (RGD)-coated microbubbles that modulated Piezo1 through integrin-mediate cell membrane tension. Piezo1 is a ion channel responsible that results in calcium influx in response to mechanical stimulation and is involved in enhancing activation of both T cells and macrophages (Liu et al., 2018; Solis et al., 2019). Furthermore, cytotoxicity of T cells is correlated to the level of force they exert at the immunological synapse by improving perforin-mediated pore formation in target cells (Basu et al., 2016). Basu and colleagues calculated forces at the immunological synapse by measuring the deflections of 6 × 0.7 μm PDMS micropillars coated with antigen and ICAM1 to immobilize the cells. T cells can also sense and respond to the mechanical properties of their environment, as CD4+ T cells in stiffer alginate hydrogels showed higher migration velocity, activation, and proliferation compared with 3D culture in gel 1 order of magnitude softer (Majedi et al., 2020). Better understanding of the mechanisms of T cell mechanosensitivity is also likely to promote the development of improved T cell expansion methodology by modulating the rigidity of vehicles for CD3 and CD28 antibody presentation (Dang et al., 2018; Lambert et al., 2017).
Innate immune cell cytotoxicity
The capability of NK cells to rapidly recognize and kill tumor cells that lack MHC-I makes them a promising immunotherapy target. This led Ayuso and colleagues to study NK cell chemotaxis and cytotoxicity using a microfluidic device (Ayuso et al., 2019). When MCF7 tumor spheroids and NK cells were suspended together in collagen I (Figure 3E) the NK cells migrated toward the tumor spheroid. Once inside, NK cells destroyed tumor cells in the periphery as well as in the core. Using EPCAM antibody–IL2 conjugates, cytotoxicity of NK cells was enhanced, mostly at the periphery. To increase the throughput, 96-well plates containing tumor spheroids were employed to investigate NK cell infiltration and cytotoxicity (Giannattasio et al., 2015, Sherman et al., 2018). Another study found that NK cell cytotoxic efficacy depended on the number of NK cells at the periphery of the tumor relative to the number of tumor cells (Christakou et al., 2015). Researchers introduced ultrasound standing waves inside a microfluidic device to generate HepG2 hepatocellular carcinoma spheroids and characterized tumor spheroid collapse under NK cell attacks. They found that a ratio of 1:10 NK cell to tumor cells was needed to eradicate tumor cells after 5 days of co-culture.
Therapeutic testing
Many of the studies that quantified lymphocyte migration, activation, and cytotoxicity described herein also incorporated therapeutics into their platforms. Some researchers used established drugs to induce predictable responses to chemotherapy (Biselli et al., 2017) or immune checkpoint blockade (Zboralski et al., 2017), whereas others leveraged their models to test novel therapeutics with simplified real-time monitoring of response compared with using animal models. One of these studies used spheroids created with the hanging drop method with mixed cancer cells and fibroblasts, followed by the addition of PBMC and treatment with a new immunocytokine (Herter et al., 2017). The authors observed increased lymphocyte infiltration and cytokine production in the treated spheroids. They also went on to test a T cell bispecific antibody with and without combination treatment with IL-2 (Herter et al., 2017). Figure 2G summarizes several of the mechanisms covered by these in vitro studies that allow us to combine the imaging and quantification of lymphocyte migration, activation, and cytotoxicity with therapeutics to gain a deeper understanding of how immune cells and cancer interact, ultimately leading to the design of more effective treatments.
Immunosuppression
The innate immune system can also interact with the adaptive immune system during tumor progression, and systemic immunosuppression can play a role in the development of cancer (Jiang et al., 2010). Therefore, modulating the innate immune system to augment the adaptive immune response represents a new immunotherapy strategy. DNA sensing is an innate immunity mechanism for eradication of intracellular pathogens, but it can also play a role in immune surveillance of cancer. This process is defective in KRAS-LKB1-mutated lung cancer through STING silencing, hindering T cell recruitment (Kitajima et al., 2019). Restoring STING can promote immune cell recruitment, as demonstrated using a microfluidic assay (Kitajima et al., 2019). In this report, LKB1-reconstituted, STING-low H1355 lung tumor spheroids were suspended in a collagen I gel region, flanked by two media channels. CXCR3+ Jurkat T cells were introduced into one media channel, and they migrated and invaded the gel channel in response to higher CXCL10 expression from LKB1-reconstituted H1355 spheroids. A similar microfluidic assay was employed to evaluate T cell recruitment following a treatment of PTEN-null triple-negative breast tumor cells with an STING antagonist (Ritter et al., 2020).
The interaction between innate and adaptive immune cells within the TME has been studied using a similar single-gel channel device containing tumor aggregates and monocytes in collagen I and T cells that were added to the adjacent media channel (Figure 3F) (Lee et al., 2018). T cells and monocytes were isolated from healthy donor's human PBMCs. T cells were then transfected to express a T cell receptor (TCR) recognizing hepatitis-B-virus-specific antigen expressed by a human HCC cell line HepG2. Results demonstrated that monocytes express PD-L1 when co-cultured with tumor cells and TCR T cells, thus decreasing T cells' cytotoxic activity toward tumor cells. Using checkpoint blockade targeting PD-L1/PD-1 signaling, HBV-specific TCR T cell cytotoxicity was restored.
Microfluidic devices are ideal for studies of biomechanic interplay in the TME, as physiological parameters such as matrix stiffness, shear stress, and flows can each be independently controlled. Although much progress has been made, this technology is still not widely used as a pre-clinical model during drug development due to several practical difficulties such as lack of standardization and difficulties including additional immune components such as lymph nodes or the spleen. In the future, incorporation of more immune cell types such as MDSCs, Th1 cells, B reg, mast cells, eosinophils, and basophils can be expected (Rigoni et al., 2018; Zamarron and Chen, 2011). Also, more studies are needed to study the roles of other mechanical cues such as vascular flow or extracellular matrix stiffness in the formation of a tumor inflammatory niche. Enhancing anti-tumor innate immune response represents a class of emerging immunotherapies that can potentially combine with immunotherapy targeting the adaptive arm of the immune system. Combination immunotherapies targeting myeloid cells and lymphoid cells may pave the way for more effective treatment for cancer (Moynihan and Irvine, 2017).
Patient-derived organotypic models
Establishment of patient-derived tumor organoids
Patient-derived organotypic models contain cellular material and tissues from a single individual biopsy or surgical resection specimen. Organoids from patient tissue are typically treated in one of two ways, either using the air-liquid interface method or fully embedded in hydrogel (Figure 4A). These organotypic models are distinct from cancer cell line organoids, not only because they are patient-derived, primary cancer cells, but also because they retain stromal cell populations such as cancer-associated fibroblasts and immune cells, including lymphoid and myeloid cells (Neal et al., 2018; Jenkins et al., 2018). Importantly, when tumor fragments are used, they also retain some of the patient's own ECM and the signaling that derives from it. Several studies using patient-derived organotypic spheroids have focused on immune cell behavior, including one study in colorectal cancer that isolated infiltrating immune cells at the endpoint and analyzed their gene expression (Vellinga et al., 2016). Similar methodologies have also been used in murine models (Nozaki et al., 2016; Moore et al., 2018). Another study using human colorectal carcinoma specimens reported a 60% success rate for organoid creation from resections or core-needle biopsies, including the ability to successfully passage and cryopreserve the organoids (Dijkstra et al., 2018) and another achieved successful culture and passage of patient-derived organoids in 73% of samples, with 80% successful cryopreservation (Neal et al., 2018) (Figures 4B and 4C).
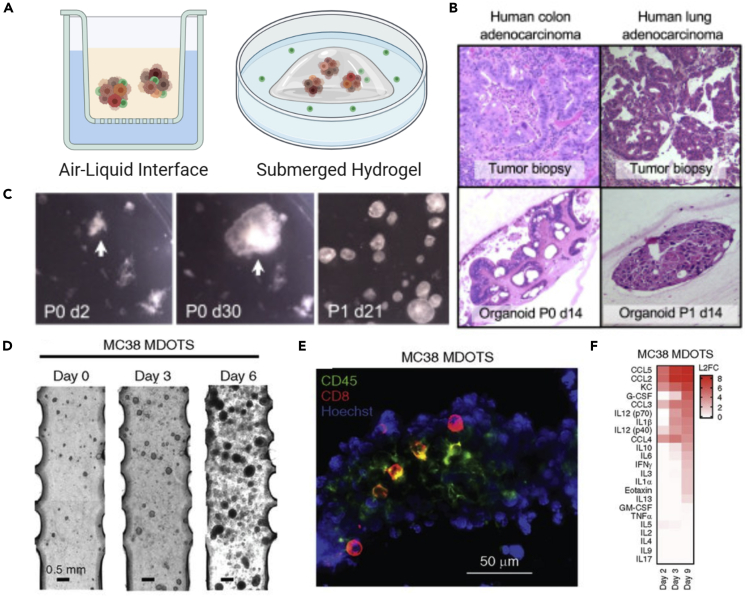
Figure 4.
Approaches and characterization of patient-derived organotypic models in culture
(A) Common methods for patient-derived organoid culture include the air-liquid interface in which organoids are cultured in collagen and medium diffuses through a permeable support or the approach where organoids are embedded in Matrigel and surrounded by cell culture medium.
(B) Histology sections comparing matched biopsies and patient-derived organoids from colon and lung adenocarcinomas.
(C) Stereomicroscopy images of patient-derived colon adenocarcinoma organoids at day 2 and 30 of culture and after passaging.
(D) Phase contrast images of murine-derived organotypic tumor spheroids (MDOTS) embedded in a collagen gel compartment of a microfluidic device. (E) Fluorescent staining of CD8 T cells and CD45 immune population within a murine organotypic model of colorectal cancer.
(F) In vitro culture permits repeated media sampling for cytokine profiling throughout several days of culture to identify changes through time or in response to treatment.
Panels (B and C) adapted from (Neal et al., 2018) and (D–F) from (Jenkins et al., 2018).
To study tumor-immune interactions in patient-derived organoid models, some groups have used cancer types more likely to contain immune cells, such as mismatch repair deficient and microsatellite-unstable colorectal carcinomas (Dijkstra et al., 2018; Vellinga et al., 2016). Others isolate T cells from peripheral blood lymphocytes before integrating them with the organoids (Dijkstra et al., 2018; Tsai et al., 2018). One benefit of using organotypic spheroids with intact immune populations is to more accurately recreate the immune-tumor microenvironment. Compared with circulating immune cells, patient-derived organotypic models reveal immunosuppression in the solid TME. A study of 100 human samples representing 19 cancer types and 28 distinct subtypes revealed that tumor-infiltrating immune populations had higher levels of MDSC and double-negative T cells (CD4−/CD8−) and lower levels of NK cells and monocytes (Finnberg et al., 2017). The same study reported that CD45+ cells remained viable after 8 days of 3D culture but that the proportion of T cells (CD3+) dropped in same time frame. However, another group found that adding IL-2 to the culture medium maintained the infiltrating T cells, including CD4+ and CD8+ subsets, for a month, but not longer than 60 days (Neal et al., 2018). Furthermore, patient-derived organotypic models preserve the original T cell receptor repertoire of the tissue sample, making them ideal for studies of antigen-specific tumor recognition (Neal et al., 2018).
Testing therapeutics in patient-derived organoids
Patient-derived organotypic models have been used to advance the goal of “personalized medicine”, the effort to customize diagnosis and treatment for a specific individual's tumor. Given the heterogeneous response to therapy, one major aim of patient-derived organotypic modeling is testing therapeutic efficacy. With the parallelization of testing combined with the ability to repeatedly sample and image these systems, several drugs can be compared simultaneously and results achieved within days to weeks rather than the weeks to months required to assess clinical response to therapy according to RECIST criteria (Litiere et al., 2017). Several groups have used patient-derived organotypic models to test radiation or therapeutics and predict efficacy, but not all of these studies have included or examined the immune cell populations (Tiriac et al., 2018; Mazzocchi et al., 2018; Kim et al., 2019b; Ooft et al., 2019; Kaaijk et al., 1997). For example, resistance to gemcitabine increased under co-culture with cancer-associated fibroblasts in pancreatic organotypic models, a notably fibrotic tumor type (Tsai et al., 2018). Another group tested response to paclitaxel in 163 samples from patients with breast cancer and discovered a pro-apoptotic role for activation of the cGAS/STING innate immunity pathway and type 1 interferons in response to antimitotic chemotherapy (Lohard et al., 2020).
In addition to looking at the direct response of epithelial cancer cells to chemotherapy, other researchers have taken a closer look at changes in immune cell populations and response to immune checkpoint blockade. These groups have treated organotypic spheroids, derived from both murine and human samples, with immune checkpoint blockade and quantified readouts such as the number and proportion of cytotoxic T cells (CD8+), T cell activation (through PCR or cytokine sampling of targets such as interferon-γ, perforin-1, granzyme-B), and immunofluorescent analysis of tumor cell killing (Jenkins et al., 2018; Aref et al., 2018; Neal et al., 2018). These groups found that immunotherapy tended to increase the number of T cells in the organoid, as well as the fraction of T cells that were CD8+. A different model was designed to culture organoids from non-small cell lung cancer specimens under constant flow rather than embedded in gel (Beckwith et al., 2019). After treatment with anti-PD-1 antibodies, fluorescent staining and confocal microscopy imaging revealed that regions of tumor cell death co-localized with tumor infiltrating lymphocytes.
Studies in ex vivo murine organotypic models (Figures 4D–4F) have demonstrated that they can recapitulate in vivo drug sensitivities, lending credibility for performing similar studies with human organotypic models (Jenkins et al., 2018). Cytokine profiling of conditioned media in murine organotypic models was also able to identify robust features of response to immune checkpoint blockade (PD-1), suggesting that a similar approach could be used to assess response to checkpoint inhibition in patient-derived organotypic models (Jenkins et al., 2018). Future approaches to increasing the versatility of organotypic models may combine patient-derived organoids with humanized mice to study therapeutic approaches such as vaccination, which cannot yet be recapitulated in vitro (Bartucci et al., 2016).
Conclusion and perspective
Three-dimensional, in vitro models of the tumor microenvironment must faithfully capture the reality of human, in situ cancers to be relevant tools in cancer research. These complex in vitro systems such as microfluidic models possess many advantages, including the ability to observe and quantify cell motion through time, non-destructive sampling (such as cytokine analysis), as well as control over matrix stiffness and composition, interstitial flow, chemical gradients, and hypoxia. However, they must also mimic the true tumor microenvironment to generate results that will translate to the clinic. Organoid culture of 3D cell assemblies has become an increasingly common methodology in bioengineering labs in order to incorporate cellular cues that are unique in 3D, including cell adhesion, gradients and matrix binding of solutes, stiffness, and direct interaction with matrix proteins through integrins (and the resultant signaling that occurs (Griffith and Swartz, 2006)). Culture in 3D is thought to recapitulate in vivo gene expression more accurately than 2D culture, including pathways important to immune response such as reduced tumor expression of HLA molecules and reduced T cell activation in 3D (Yamada and Cukierman, 2007; Dangles et al., 2002; Feder-Mengus et al., 2007).
Pre-clinical studies have often failed to predict results of clinical trials for new drugs, which has been attributed to the relative simplicity of animal models in comparison to heterogeneous human cancers (Tentler et al., 2012). Patient-derived xenografts (PDX) were developed to maintain the complexity of human tumors after excision from the body, and patient-derived organoid systems can be viewed as a similar approach with the benefits of access and visibility provided by in vitro culture, as well as being more efficient and lower in cost than the maintenance of large-scale animal colonies. Furthermore, over time, the stromal compartment of PDX models tends to drift toward that of the murine host, complicating immunotherapy studies (Tentler et al., 2012). Therefore, patient-derived organoids might be a better choice for studies focused on the immune and stromal compartments of the tumor microenvironment, but these models also incur limitations in immune cell viability over long-term culture. As a result, groups using patient-derived organotypic models must be aware of limitations to immune cell longevity in culture and identify strategies to optimally maintain the diverse immune populations observed in the tumor microenvironment (Finnberg et al., 2017; Neal et al., 2018).
The range of questions and applications that have been studied using engineering approaches to observe, measure, and quantify the interactions between cancer and immune cells is broad and widespread. The studies described in this review have incorporated many different immune cell types from varying sources including cell lines, mouse models, peripheral blood mononuclear cells, and patient-derived infiltrating lymphocytes. Similarly, many different cancer types and sources have also been reported. However, despite the breadth and diversity in these studies, we have only scratched the surface of possibilities, and many fruitful avenues of inquiry remain.
For drug discovery and pre-clinical testing, microfluidic tools offer valuable insight into the mechanism in which drugs can target stromal cells in the TME. More efforts in this field are needed to mimic the physiological conditions of the TME with increasing fidelity. Moreover, to be widely used in the clinic and biological laboratories, microfluidic models need to be designed to be compatible with high-throughput molecular analysis such as gene sequencing or mass spectrometry. High-throughput and single-cell gene expression profiling will help us understand better the evolution of tumor cell heterogeneity as well as the change of the immune landscape when patient-derived organotypic models are cultured within a microfluidic TME. We also foresee that more in vitro engineered models will be used during pre-clinical studies of new drugs and that new insights into correlation between in vitro models and in vivo response to immunotherapy will be gained. Hopefully, 3D engineered systems will help elucidate the relationships between tumor molecular profiles and immune cell landscapes to identify predictive markers, therefore guiding personalized immunotherapy, lowering its cost, and increasing its effectiveness.
Several unexplored features of the cancer immune microenvironment remain available for development. For example, researchers have only begun to utilize the sophisticated 3D models described throughout this review to model antigen presentation in cancer (Rosa et al., 2016; Shim et al., 2019). This represents an excellent opportunity of exploration for combining oncology studies, with recent results reported in the creation of lymphatic system and lymph node models (Sun et al., 2019; Osaki et al., 2018; Zhou et al., 2020). Another key component of the cancer microenvironment is the vasculature, through which circulating immune cells must travel to reach a lesion. A few of the studies in this review included immune cell transmigration across an endothelium (Aung et al., 2020; Zervantonakis et al., 2012) or immune-stimulated neoangiogenesis (Yu et al., 2019; Cui et al., 2018), but few groups have looked at the circulation of immune cells in such models (Chen et al., 2018; Boussommier-Calleja et al., 2019; Campisi et al., 2020). However, as techniques for developing vascular microphysiological models improve, so does the opportunity to explore this aspect of immune cell recruitment in cancer. Novel vascularized tumor models such as the one described by Haase et al. demonstrate how these systems can be used to reveal the influence of cancer cell lines on the microenvironment and lead to significant differences in vascular morphology and permeability, as well as response to therapy (Haase et al., 2020).
Although several of the studies described here incorporated immune checkpoint inhibitor drugs or other forms of immunotherapy, standardized protocols for such testing has yet to emerge. Instead, much of the focus has been on using these models to understand the impact of these treatments in multicellular, 3D models of the TME. Before microfluidic and organoid culture methods can be used as a surrogate for in vivo pre-clinical testing, more effort must be directed toward validation and improved consistency of results. It will be critical to repeat studies such as that by Jenkins et al., which evaluated the correlation between response to PD-1 blockade in murine-derived organotypic spheroids and in vivo response of the same cancer line and drug (Jenkins et al., 2018). Drug testing in microphysiological tumor-immune models will increase as organoid platforms become more widespread and validation data accumulate, both in order to test new therapeutics and also to better understand mechanisms of resistance to therapy. Patient-derived organotypic models are also likely to become more prevalent, particularly for identifying effective therapy regimes (Ivanova et al., 2020). The establishment of biobanks of patient-derived organoids will broaden the reach of this approach, especially if it can be combined with matched peripheral blood samples for the isolation of circulating immune cells (Palechor-Ceron et al., 2019; van de Wetering et al., 2015; Jacob et al., 2020). In addition, as more drugs are developed with the ability to modify the TME rather than the tumor itself, these engineered models hold significant value for interrogating the influence of the microenvironment and the ability to combine, include, or exclude particular cell types results with more straightforward hypothesis testing than using animal models (Murciano-Goroff et al., 2020).
Finally, as our capability to mimic the immune microenvironment of cancer improves through the development of increasingly realistic and sophisticated 3D models, more researchers will begin to combine these approaches with other techniques such as single-cell sequencing or advanced microscopy methods, lending even greater clarity to our understanding of immune-tumor cell interactions and immunotherapy and paving the way for translation of these methods into the clinic.
Acknowledgments
We thank Vivian Vu for her assistance in preparing Figure 1. Figures created using Biorender.com. This work was supported by the National Institutes of Health through grant U01CA214381. SES is supported by fellowship K00CA212227 from the National Cancer Institute. HTN is supported by a Swiss National Science Foundation postdoctoral fellowship (SNSF-P400PB_186779).
Author contributions
All authors contributed to writing and editing this manuscript.
Declaration of interests
RDK is a co-founder and has a significant financial interest in AIM Biotech, a company that manufactures microfluidic systems and has received research funding from Elstar, Amgen and Biogen. DAB is an inventor on patents related on manipulating, culturing, and evaluating tumor spheroids. DAB is a consultant for N of One and Tango Therapeutics, has received honoraria from Loxo Oncology and Madalon Consulting, research grants from BMS, Novartis, Lilly, and Gilead Sciences, and is a co-founder and on the scientific advisory board of Xsphera Biosciences Inc.
References
- Agliari E., Biselli E., De Ninno A., Schiavoni G., Gabriele L., Gerardino A., Mattei F., Barra A., Businaro L. Cancer-driven dynamics of immune cells in A microfluidic environment. Sci. Rep. 2014;4:6639. doi: 10.1038/srep06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen S.G., Chen Y.-C., Madden J.M., Fournier C.L., Altemus M.A., Hiziroglu A.B., Cheng Y.-H., Wu Z.F., Bao L., Yates J.A. Macrophages enhance migration in inflammatory breast cancer cells via rhoc gtpase signaling. Sci. Rep. 2016;6:39190. doi: 10.1038/srep39190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando Y., Siegler E.L., Ta H.P., Cinay G.E., Zhou H., Gorrell K.A., Au H., Jarvis B.M., Wang P., Shen K. Evaluating car-T cell therapy in A hypoxic 3d tumor model. Adv. Healthc. Mater. 2019;8:E1900001. doi: 10.1002/adhm.201900001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aref A.R., Campisi M., Ivanova E., Portell A., Larios D., Piel B.P., Mathur N., Zhou C., Coakley R.V., Bartels A. 3d microfluidic ex vivo culture of organotypic tumor spheroids to model immune checkpoint blockade. Lab Chip. 2018;18:3129–3143. doi: 10.1039/c8lc00322j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustine T.N., Dix-Peek T., Duarte R., Candy G.P. Establishment of A heterotypic 3d culture system to evaluate the interaction of treg lymphocytes and nk cells with breast cancer. J. Immunol. Methods. 2015;426:1–13. doi: 10.1016/j.jim.2015.07.003. [DOI] [PubMed] [Google Scholar]
- Aung A., Kumar V., Theprungsirikul J., Davey S.K., Varghese S. An engineered tumor-on-A-chip device with breast cancer-immune cell interactions for assessing T cell recruitment. Cancer Res. 2020;80:263–275. doi: 10.1158/0008-5472.CAN-19-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayuso J.M., Truttschel R., Gong M.M., Humayun M., Virumbrales-Munoz M., Vitek R., Felder M., Gillies S.D., Sondel P., Wisinski K.B. Evaluating natural killer cell cytotoxicity against solid tumors using A microfluidic model. Oncoimmunology. 2019;8:1553477. doi: 10.1080/2162402X.2018.1553477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J., Adriani G., Dang T.M., Tu T.Y., Penny H.X.L., Wong S.C., Kamm R.D.R.D., Thiery J.P. Contact-dependent carcinoma aggregate dispersion by M2a macrophages via icam-1 and ß2 integrin interactions. Oncotarget. 2015;6:25295–25307. doi: 10.18632/oncotarget.4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartucci M., Ferrari A.C., Kim I.Y., Ploss A., Yarmush M., Sabaawy H.E. Personalized medicine approaches in prostate cancer employing patient derived 3d organoids and humanized mice. Front. Cell Dev. Biol. 2016;4:64. doi: 10.3389/fcell.2016.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu R., Whitlock B.M., Husson J., Le Floc’h A., Jin W., Oyler-Yaniv A., Dotiwala F., Giannone G., Hivroz C., Biais N. Cytotoxic T cells use mechanical force to potentiate target cell killing. Cell. 2016;165:100–110. doi: 10.1016/j.cell.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckwith A.L., Velásquez-García L.F., Borenstein J.T. Microfluidic model for evaluation of immune checkpoint inhibitors in human tumors. Adv. Healthc. Mater. 2019;8:1900289. doi: 10.1002/adhm.201900289. [DOI] [PubMed] [Google Scholar]
- Bercovici N., Duffour M.T., Agrawal S., Salcedo M., Abastado J.P. New methods for assessing T cell responses. Clin. Diagn. Lab. Immunol. 2000;7:859–864. doi: 10.1128/cdli.7.6.859-864.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biselli E., Agliari E., Barra A., Bertani F.R., Gerardino A., De Ninno A., Mencattini A., Di Giuseppe D., Mattei F., Schiavoni G. Organs on chip approach: a tool to evaluate cancer -immune cells interactions. Sci. Rep. 2017;7:12737. doi: 10.1038/s41598-017-13070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussommier-Calleja A., Atiyas Y., Haase K., Headley M., Lewis C., Kamm R.D. The effects of monocytes on tumor cell extravasation in A 3d vascularized microfluidic model. Biomaterials. 2019;198:180–193. doi: 10.1016/j.biomaterials.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. A. Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- Businaro L., De Ninno A., Schiavoni G., Lucarini V., Ciasca G., Gerardino A., Belardelli F., Gabriele L., Mattei F. Cross talk between cancer and immune cells: exploring complex dynamics in A microfluidic environment. Lab Chip. 2013;13:229–239. doi: 10.1039/c2lc40887b. [DOI] [PubMed] [Google Scholar]
- Campisi M., Sundararaman S.K., Shelton S.E., Knelson E.H., Ivanova E., Cañadas I., Yoshida R., Osaki T., Lee S.W., Thai T. Tumor-derived cgamp regulates activation of the vasculature. Front. Immunol. 2020;11:2090. doi: 10.3389/fimmu.2020.02090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.B., Hajal C., Benjamin D.C., Yu C., Azizgolshani H., Hynes R.O., Kamm R.D. Inflamed neutrophils sequestered at entrapped tumor cells via chemotactic confinement promote tumor cell extravasation. Proc. Natl. Acad. Sci. U S A. 2018;115:7022–7027. doi: 10.1073/pnas.1715932115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba T., Ohtani H., Mizoi T., Naito Y., Sato E., Nagura H., Ohuchi A., Ohuchi K., Shiiba K., Kurokawa Y., Satomi S. Intraepithelial Cd8+ T cell-count becomes A prognostic factor After A longer follow-up period in human colorectal carcinoma: possible association with suppression of micrometastasis. Br. J. Cancer. 2004;91:1711–1717. doi: 10.1038/sj.bjc.6602201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christakou A.E., Ohlin M., Onfelt B., Wiklund M. Ultrasonic three-dimensional on-chip cell culture for dynamic studies of tumor immune surveillance by natural killer cells. Lab Chip. 2015;15:3222–3231. doi: 10.1039/c5lc00436e. [DOI] [PubMed] [Google Scholar]
- Cochrane A., Albers H.J., Passier R., Mummery C.L., van den Berg A., Orlova V.V., van der Meer A.D. Advanced in vitro models of vascular biology: human induced pluripotent stem cells and organ-on-chip technology. Adv. Drug Deliv. Rev. 2019;140:68–77. doi: 10.1016/j.addr.2018.06.007. [DOI] [PubMed] [Google Scholar]
- Coughlin M.F., Kamm R.D. The use of microfluidic platforms to probe the mechanism of cancer cell extravasation. Adv. Healthc. Mater. 2020;9:1901410. doi: 10.1002/adhm.201901410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X., Morales R.T., Qian W., Wang H., Gagner J.P., Dolgalev I., Placantonakis D., Zagzag D., Cimmino L., Snuderl M. Hacking macrophage-associated immunosuppression for regulating glioblastoma angiogenesis. Biomaterials. 2018;161:164–178. doi: 10.1016/j.biomaterials.2018.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang A.P., De Leo S., Bogdanowicz D.R., Yuan D.J., Fernandes S.M., Brown J.R., Lu H.H., Kam L.C. Enhanced activation and expansion of T cells using mechanically soft elastomer fibers. Adv. Biosyst. 2018;2:1700167. doi: 10.1002/adbi.201700167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangles V., Validire P., Wertheimer M., Richon S., Bovin C., Zeliszewski D., Vallancien G., Bellet D. Impact of human bladder cancer cell architecture on autologous T-lymphocyte activation. Int. J. Cancer. 2002;98:51–56. doi: 10.1002/ijc.10140. [DOI] [PubMed] [Google Scholar]
- Dijkstra K.K., Cattaneo C.M., Weeber F., Chalabi M., Van De Haar J., Fanchi L.F., Slagter M., Van Der Velden D.L., Kaing S., Kelderman S. Generation of tumor-reactive T cells by Co-culture of peripheral blood lymphocytes and tumor organoids. Cell. 2018;174:1586–1598 E12. doi: 10.1016/j.cell.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhood B., Najafi M., Mortezaee K. Cd8(+) cytotoxic T lymphocytes in cancer immunotherapy: a review. J. Cell Physiol. 2019;234:8509–8521. doi: 10.1002/jcp.27782. [DOI] [PubMed] [Google Scholar]
- Farkona S., Diamandis E.P., Blasutig I.M. Cancer immunotherapy: the beginning of the end of cancer? BMC Med. 2016;14:73. doi: 10.1186/s12916-016-0623-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder-Mengus C., Ghosh S., Weber W.P., Wyler S., Zajac P., Terracciano L., Oertli D., Heberer M., Martin I., Spagnoli G.C., Reschner A. Multiple mechanisms underlie defective recognition of melanoma cells cultured in three-dimensional architectures by antigen-specific cytotoxic T lymphocytes. Br. J. Cancer. 2007;96:1072–1082. doi: 10.1038/sj.bjc.6603664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnberg N.K., Gokare P., Lev A., Grivennikov S.I., Macfarlane A.W.T., Campbell K.S., Winters R.M., Kaputa K., Farma J.M., Abbas A.E. Application of 3d tumoroid systems to define immune and cytotoxic therapeutic responses based on tumoroid and tissue slice culture molecular signatures. Oncotarget. 2017;8:66747–66757. doi: 10.18632/oncotarget.19965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florczyk S.J., Liu G., Kievit F.M., Lewis A.M., Wu J.D., Zhang M. 3d porous chitosan-alginate scaffolds: a new matrix for studying prostate cancer cell-lymphocyte interactions in vitro. Adv. Healthc. Mater. 2012;1:590–599. doi: 10.1002/adhm.201100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galon J., Bruni D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 2019;18:197–218. doi: 10.1038/s41573-018-0007-y. [DOI] [PubMed] [Google Scholar]
- Giannattasio A., Weil S., Kloess S., Ansari N., Stelzer E.H., Cerwenka A., Steinle A., Koehl U., Koch J. Cytotoxicity and infiltration of human NK cells in in vivo-like tumor spheroids. BMC Cancer. 2015;15:351. doi: 10.1186/s12885-015-1321-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez H., Hagerling C., Werb Z. Roles of the immune system in cancer: from tumor initiation to metastatic progression. Genes Dev. 2018;32:1267–1284. doi: 10.1101/gad.314617.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman A.M., Kato S., Bazhenova L., Patel S.P., Frampton G.M., Miller V., Stephens P.J., Daniels G.A., Kurzrock R. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol. Cancer Ther. 2017;16:2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith L.G., Swartz M.A. Capturing complex 3d tissue physiology in vitro. Nat. Rev. Mol. Cell. Biol. 2006;7:211–224. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- Haase K., Offeddu G.S., Gillrie M.R., Kamm R.D. Endothelial regulation of drug transport in A 3d vascularized tumor model. Adv. Funct. Mater. 2020:2002444. doi: 10.1002/adfm.202002444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Herter S., Morra L., Schlenker R., Sulcova J., Fahrni L., Waldhauer I., Lehmann S., Reislander T., Agarkova I., Kelm J.M. A novel three-dimensional heterotypic spheroid model for the assessment of the activity of cancer immunotherapy agents. Cancer Immunol. Immunother. 2017;66:129–140. doi: 10.1007/s00262-016-1927-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann O.I., Ilmberger C., Magosch S., Joka M., Jauch K.W., Mayer B. Impact of the spheroid model complexity on drug response. J. Biotechnol. 2015;205:14–23. doi: 10.1016/j.jbiotec.2015.02.029. [DOI] [PubMed] [Google Scholar]
- Hong M., Clubb J.D., Chen Y.Y. Engineering car-T cells for next-generation cancer therapy. Cancer Cell. 2020;38:473–488. doi: 10.1016/j.ccell.2020.07.005. [DOI] [PubMed] [Google Scholar]
- Hsu T.-H., Kao Y.-L., Lin W.-L., Xiao J.-L., Kuo P.-L., Wu C.-W., Liao W.-Y., Lee C.-H. The migration speed of cancer cells influenced by macrophages and myofibroblasts Co-cultured in A microfluidic chip. Integr. Biol. 2012;4:177–182. doi: 10.1039/c2ib00112h. [DOI] [PubMed] [Google Scholar]
- Huh D., Hamilton G.A., Ingber D.E. From 3d cell culture to organs-on-chips. Trends Cell Biol. 2011;21:745–754. doi: 10.1016/j.tcb.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova E., Kuraguchi M., Xu M., Portell A.J., Taus L., Diala I., Lalani A.S., Choi J., Chambers E.S., Li S. Use of <Em>Ex</Em> <Em>Vivo</Em> patient-derived tumor organotypic spheroids to identify combination therapies for <Em>Her2</Em> mutant non–small cell lung cancer. Clin. Cancer Res. 2020;26:2393–2403. doi: 10.1158/1078-0432.CCR-19-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F., Salinas R.D., Zhang D.Y., Nguyen P.T., Schnoll J.G., Wong S.Z.H., Thokala R., Sheikh S., Saxena D., Prokop S. A patient-derived glioblastoma organoid model and biobank recapitulates inter-and intra-tumoral heterogeneity. Cell. 2020;180:188–204. E22. doi: 10.1016/j.cell.2019.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins R.W., Aref A.R., Lizotte P.H., Ivanova E., Stinson S., Zhou C.W., Bowden M., Deng J., Liu H., Miao D. Ex vivo profiling of Pd-1 blockade using organotypic tumor spheroids. Cancer Discov. 2018;8:196–215. doi: 10.1158/2159-8290.CD-17-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Villeneuve P., Wielgosz A., Schaubel D., Fenton S., Mao Y. The incidence of cancer in A population-based cohort of Canadian heart transplant recipients. Am. J. Transplant. 2010;10:637–645. doi: 10.1111/j.1600-6143.2009.02973.x. [DOI] [PubMed] [Google Scholar]
- Kaaijk P., Troost D., Sminia P., Hulshof M.C., Van Der Kracht A.H., Leenstra S., Bosch D.A. Hypofractionated radiation induces A decrease in cell proliferation but No histological damage to organotypic multicellular spheroids of human glioblastomas. Eur. J. Cancer. 1997;33:645–651. doi: 10.1016/s0959-8049(96)00503-5. [DOI] [PubMed] [Google Scholar]
- Kim H., Chung H., Kim J., Choi D.H., Shin Y., Kang Y.G., Kim B.M., Seo S.U., Chung S., Seok S.H. Macrophages-triggered sequential remodeling of endothelium-interstitial matrix to form pre-metastatic niche in microfluidic tumor microenvironment. Adv. Sci. 2019;6:1900195. doi: 10.1002/advs.201900195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.K., Moon W.K., Park J.Y., Jung H. Inflammatory mimetic microfluidic chip by immobilization of cell adhesion molecules for T cell adhesion. Analyst. 2012;137:4062–4068. doi: 10.1039/c2an35424a. [DOI] [PubMed] [Google Scholar]
- Kim M., Mun H., Sung C.O., Cho E.J., Jeon H.J., Chun S.M., Jung D.J., Shin T.H., Jeong G.S., Kim D.K. Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat. Commun. 2019;10:3991. doi: 10.1038/s41467-019-11867-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima S., Ivanova E., Guo S., Yoshida R., Campisi M., Sundararaman S.K., Tange S., Mitsuishi Y., Thai T.C., Masuda S. Suppression of sting associated with Lkb1 loss in kras-driven lung cancer. Cancer Discov. 2019;9:34–45. doi: 10.1158/2159-8290.CD-18-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwasny D., Kiilerich-Pedersen K., Moresco J., Dimaki M., Rozlosnik N., Svendsen W.E. Microfluidic device to study cell transmigration under physiological shear stress conditions. Biomed. Microdevices. 2011;13 doi: 10.1007/s10544-011-9559-x. [DOI] [PubMed] [Google Scholar]
- Lambert L.H., Goebrecht G.K., De Leo S.E., O’connor R.S., Nunez-Cruz S., Li T.-D., Yuan J., Milone M.C., Kam L.C. Improving T cell expansion with A soft touch. Nano Lett. 2017;17:821–826. doi: 10.1021/acs.nanolett.6b04071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.W.L., Adriani G., Ceccarello E., Pavesi A., Tan A.T., Bertoletti A., Kamm R.D., Wong S.C. Characterizing the role of monocytes in T cell cancer immunotherapy using A 3d microfluidic model. Front. Immunol. 2018;9:416. doi: 10.3389/fimmu.2018.00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.W.L., Seager R.J., Litvak F., Spill F., Sieow J.L., Leong P.H., Kumar D., Tan A.S.M., Wong S.C., Adriani G. Integrated in silico and 3d in vitro model of macrophage migration in response to physical and chemical factors in the tumor microenvironment. Integr. Biol. 2020;12:90–108. doi: 10.1093/intbio/zyaa007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Hebert J.D., Lee T.A., Xing H., Boussommier-Calleja A., Hynes R.O., Lauffenburger D.A., Kamm R.D. Macrophage-secreted TNFα and TGFβ1 influence migration speed and persistence of cancer cells in 3D tissue culture via independent pathways. Cancer Research. 2017;77(2):279–290. doi: 10.1158/0008-5472.CAN-16-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Ma C., Cai H., Chen W. The car T-cell mechanoimmunology at A glance. Adv. Sci. 2020;7:2002628. doi: 10.1002/advs.202002628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Serrano J.C., Xing H., Lee T.A., Azizgolshani H., Zaman M., Kamm R.D. Interstitial flow promotes macrophage polarization toward an M2 phenotype. Mol. Biol. Cell. 2018;29:1927–1940. doi: 10.1091/mbc.E18-03-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F., Butcher E.C. T cell chemotaxis in a simple microfluidic device. Lab Chip. 2006;6:1462–1469. doi: 10.1039/b607071j. [DOI] [PubMed] [Google Scholar]
- Litiere S., Collette S., De Vries E.G., Seymour L., Bogaerts J. Recist - learning from the past to build the future. Nat. Rev. Clin. Oncol. 2017;14:187–192. doi: 10.1038/nrclinonc.2016.195. [DOI] [PubMed] [Google Scholar]
- Liu C.S.C., Raychaudhuri D., Paul B., Chakrabarty Y., Ghosh A.R., Rahaman O., Talukdar A., Ganguly D. Cutting edge: Piezo1 mechanosensors optimize human T cell activation. J. Immunol. 2018;200:1255–1260. doi: 10.4049/jimmunol.1701118. [DOI] [PubMed] [Google Scholar]
- Lohard S., Bourgeois N., Maillet L., Gautier F., Fetiveau A., Lasla H., Nguyen F., Vuillier C., Dumont A., Moreau-Aubry A. Sting-dependent paracriny shapes apoptotic priming of breast tumors in response to anti-mitotic treatment. Nat. Commun. 2020;11:259. doi: 10.1038/s41467-019-13689-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long K.B., Beatty G.L. Harnessing the antitumor potential of macrophages for cancer immunotherapy. Oncoimmunology. 2013;2:E26860. doi: 10.4161/onci.26860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majedi F.S., Hasani-Sadrabadi M.M., Thauland T.J., Li S., Bouchard L.-S., Butte M.J. T Cell Activation Is Modulated by the 3d Mechanical Microenvironment. Biomaterials. 2020;252:120058. doi: 10.1016/j.biomaterials.2020.120058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattei F., Schiavoni G., De Ninno A., Lucarini V., Sestili P., Sistigu A., Fragale A., Sanchez M., Spada M., Gerardino A. A multidisciplinary study using in vivo tumor models and microfluidic cell-on-chip approach to explore the cross-talk between cancer and immune cells. J. Immunotoxicol. 2014;11:337–346. doi: 10.3109/1547691X.2014.891677. [DOI] [PubMed] [Google Scholar]
- Mazzocchi A.R., Rajan S.A.P., Votanopoulos K.I., Hall A.R., Skardal A. In vitro patient-derived 3d mesothelioma tumor organoids facilitate patient-centric therapeutic screening. Sci. Rep. 2018;8:2886. doi: 10.1038/s41598-018-21200-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S., Liu Z., Du Z., Yi X., Sun W. Three-dimensional microfluidic tumor–macrophage system for breast cancer cell invasion. Biotechnol. Bioeng. 2019;116:1731–1741. doi: 10.1002/bit.26961. [DOI] [PubMed] [Google Scholar]
- Mitra B., Jindal R., Lee S., Xu Dong D., Li L., Sharma N., Maguire T., Schloss R., Yarmush M.L. Microdevice integrating innate and adaptive immune responses associated with antigen presentation by dendritic cells. RSC Adv. 2013;3:16002–16010. doi: 10.1039/C3RA41308J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore N., Doty D., Zielstorff M., Kariv I., Moy L.Y., Gimbel A., Chevillet J.R., Lowry N., Santos J., Mott V. A multiplexed microfluidic system for evaluation of dynamics of immune-tumor interactions. Lab Chip. 2018;18:1844–1858. doi: 10.1039/c8lc00256h. [DOI] [PubMed] [Google Scholar]
- Moynihan K.D., Irvine D.J. Roles for innate immunity in combination immunotherapies. Cancer Res. 2017;77:5215–5221. doi: 10.1158/0008-5472.CAN-17-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murciano-Goroff Y.R., Warner A.B., Wolchok J.D. The future of cancer immunotherapy: microenvironment-targeting combinations. Cell Res. 2020;30:507–519. doi: 10.1038/s41422-020-0337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal J.T., Li X., Zhu J., Giangarra V., Grzeskowiak C.L., Ju J., Liu I.H., Chiou S.H., Salahudeen A.A., Smith A.R. Organoid modeling of the tumor immune microenvironment. Cell. 2018;175:1972–1988 E16. doi: 10.1016/j.cell.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki K., Mochizuki W., Matsumoto Y., Matsumoto T., Fukuda M., Mizutani T., Watanabe M., Nakamura T. Co-culture with intestinal epithelial organoids allows efficient expansion and motility analysis of intraepithelial lymphocytes. J. Gastroenterol. 2016;51:206–213. doi: 10.1007/s00535-016-1170-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson B., Li Y., Lin Y., Liu E.T., Patnaik A. Mouse models for cancer immunotherapy research. Cancer Discov. 2018;8:1358–1365. doi: 10.1158/2159-8290.CD-18-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooft S.N., Weeber F., Dijkstra K.K., Mclean C.M., Kaing S., Van Werkhoven E., Schipper L., Hoes L., Vis D.J., Van De Haar J. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci. Transl. Med. 2019;11:eaay2574. doi: 10.1126/scitranslmed.aay2574. [DOI] [PubMed] [Google Scholar]
- Osaki T., Serrano J.C., Kamm R.D. Cooperative effects of vascular angiogenesis and lymphangiogenesis. Regenerative Eng. Transl. Med. 2018;4:120–132. doi: 10.1007/s40883-018-0054-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palechor-Ceron N., Krawczyk E., Dakic A., Simic V., Yuan H., Blancato J., Wang W., Hubbard F., Zheng Y.-L., Dan H. Conditional reprogramming for patient-derived cancer models and next-generation living biobanks. Cells. 2019;8:1327. doi: 10.3390/cells8111327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Yoon S., Sun J., Huang Z., Lee C., Allen M., Wu Y., Chang Y.-J., Sadelain M., Shung K.K. Mechanogenetics for the remote and noninvasive control of cancer immunotherapy. Proc. Natl. Acad. Sci. U S A. 2018;115:992–997. doi: 10.1073/pnas.1714900115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlato S., de Ninno A., Molfetta R., Tosch E., Salerno D., Mencattini A., Romagnoli G., Fragale A., Roccazzello L., Buoncervello M. 3D Microfluidic model for evaluating immunotherapy efficacy by tracking dendritic cell behaviour toward tumor cells. Sci. Rep. 2017;7:1–16. doi: 10.1038/s41598-017-01013-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi A., Tan A.T., Koh S., Chia A., Colombo M., Antonecchia E., Miccolis C., Ceccarello E., Adriani G., Raimondi M.T. A 3d microfluidic model for preclinical evaluation of tcr-engineered T cells against solid tumors. JCI Insight. 2017;2:e89762. doi: 10.1172/jci.insight.89762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitprez F., Meylan M., De Reynies A., Sautes-Fridman C., Fridman W.H. The tumor microenvironment in the response to immune checkpoint blockade therapies. Front. Immunol. 2020;11:784. doi: 10.3389/fimmu.2020.00784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan-Lai V., Florczyk S.J., Kievit F.M., Wang K., Gad E., Disis M.L., Zhang M. Three-dimensional scaffolds to evaluate tumor associated fibroblast-mediated suppression of breast tumor specific T cells. Biomacromolecules. 2013;14:1330–1337. doi: 10.1021/bm301928u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riabov V., Gudima A., Wang N., Orekhov A., Mickley A., Kzhyshkowska J. Role of tumor associated macrophages in tumor angiogenesis and lymphangiogenesis. Front. Physiol. 2014;5:75. doi: 10.3389/fphys.2014.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigoni A., Colombo M.P., Pucillo C. Mast cells, basophils and eosinophils: from allergy to cancer. Semin. Immunol. 2018;35:29–34. doi: 10.1016/j.smim.2018.02.001. [DOI] [PubMed] [Google Scholar]
- Ritter J.L., Zhu Z., Thai T.C., Mahadevan N.R., Mertins P., Knelson E.H., Piel B.P., Han S., Jaffe J.D., Carr S.A., Barbie D.A., Barbie T.U. Phosphorylation of Rab7 by TBK1/IKKε regulates innate immune signaling in triple-negative breast cancer. Cancer Res. 2020;80(1):44–56. doi: 10.1158/0008-5472.CAN-19-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa P.M., Gopalakrishnan N., Ibrahim H., Haug M., Halaas Ø. The intercell dynamics of T cells and dendritic cells in A lymph node-on-A-chip flow device. Lab Chip. 2016;16:3728–3740. doi: 10.1039/c6lc00702c. [DOI] [PubMed] [Google Scholar]
- Sadovska L., Zandberga E., Sagini K., Jekabsons K., Riekstina U., Kalnina Z., Llorente A., Line A. A novel 3d heterotypic spheroid model for studying extracellular vesicle-mediated tumour and immune cell communication. Biochem. Biophys. Res. Commun. 2018;495:1930–1935. doi: 10.1016/j.bbrc.2017.12.072. [DOI] [PubMed] [Google Scholar]
- Seager R.J., Hajal C., Spill F., Kamm R.D., Zaman M.H. Dynamic interplay between tumour, stroma and immune system can drive or prevent tumour progression. Converg. Sci. Phys. Oncol. 2017;3:034002. doi: 10.1088/2057-1739/aa7e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segaliny A.I., Li G., Kong L., Ren C., Chen X., Wang J.K., Baltimore D., Wu G., Zhao W. Functional tcr T cell screening using single-cell droplet microfluidics. Lab Chip. 2018;18:3733–3749. doi: 10.1039/c8lc00818c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman H., Gitschier H.J., Rossi A.E. A novel three-dimensional immune oncology model for high-throughput testing of tumoricidal activity. Front. Immunol. 2018;9:857. doi: 10.3389/fimmu.2018.00857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S., Belanger M.C., Harris A.R., Munson J.M., Pompano R.R. Two-way communication between ex vivo tissues on A microfluidic chip: application to tumor–lymph node interaction. Lab Chip. 2019;19:1013–1026. doi: 10.1039/c8lc00957k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis A.G., Bielecki P., Steach H.R., Sharma L., Harman C.C., Yun S., De Zoete M.R., Warnock J.N., To S.F., York A.G. Mechanosensation of cyclical force by Piezo1 is essential for innate immunity. Nature. 2019;573:69–74. doi: 10.1038/s41586-019-1485-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C., Gao D., Yuan T., Chen Y., Liu L., Chen X., Jiang Y. Microfluidic three-dimensional biomimetic tumor model for studying breast cancer cell migration and invasion in the presence of interstitial flow. Chin. Chem. Lett. 2019;30:1038–1042. [Google Scholar]
- Sontheimer-Phelps A., Hassell B.A., Ingber D.E. Modelling cancer in microfluidic human organs-on-chips. Nat. Rev. Cancer. 2019;19:65–81. doi: 10.1038/s41568-018-0104-6. [DOI] [PubMed] [Google Scholar]
- Sun W., Luo Z., Lee J., Kim H.J., Lee K., Tebon P., Feng Y., Dokmeci M.R., Sengupta S., Khademhosseini A. Organ-on-A-chip for cancer and immune organs modeling. Adv. Healthc. Mater. 2019;8:1801363. doi: 10.1002/adhm.201900754. [DOI] [PubMed] [Google Scholar]
- Tentler J.J., Tan A.C., Weekes C.D., Jimeno A., Leong S., Pitts T.M., Arcaroli J.J., Messersmith W.A., Eckhardt S.G. Patient-derived tumour xenografts as models for oncology drug development. Nat. Rev. Clin. Oncol. 2012;9:338–350. doi: 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiriac H., Belleau P., Engle D.D., Plenker D., Deschenes A., Somerville T.D.D., Froeling F.E.M., Burkhart R.A., Denroche R.E., Jang G.H. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov. 2018;8:1112–1129. doi: 10.1158/2159-8290.CD-18-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S., Mcolash L., Palen K., Johnson B., Duris C., Yang Q., Dwinell M.B., Hunt B., Evans D.B., Gershan J., James M.A. Development of primary human pancreatic cancer organoids, matched stromal and immune cells and 3d tumor microenvironment models. BMC Cancer. 2018;18:335. doi: 10.1186/s12885-018-4238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaddepally R.K., Kharel P., Pandey R., Garje R., Chandra A.B. Review of indications of fda-approved immune checkpoint inhibitors per nccn guidelines with the level of evidence. Cancers (Basel) 2020;12:738. doi: 10.3390/cancers12030738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Wetering M., Francies H.E., Francis J.M., Bounova G., Iorio F., Pronk A., Van Houdt W., Van Gorp J., Taylor-Weiner A., Kester L. Prospective derivation of A living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellinga T.T., Den Uil S., Rinkes I.H., Marvin D., Ponsioen B., Alvarez-Varela A., Fatrai S., Scheele C., Zwijnenburg D.A., Snippert H. Collagen-rich stroma in aggressive colon tumors induces mesenchymal gene expression and tumor cell invasion. Oncogene. 2016;35:5263–5271. doi: 10.1038/onc.2016.60. [DOI] [PubMed] [Google Scholar]
- Wallstabe L., Gottlich C., Nelke L.C., Kuhnemundt J., Schwarz T., Nerreter T., Einsele H., Walles H., Dandekar G., Nietzer S.L., Hudecek M. Ror1-Car T cells are effective against lung and breast cancer in advanced microphysiologic 3d tumor models. JCI Insight. 2019;4:e126345. doi: 10.1172/jci.insight.126345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimalachandra D.C., Li Y., Liu J., Shikha S., Zhang J., Lim Y.-C., Zhang Y. Microfluidic-based immunomodulation of immune cells using upconversion nanoparticles in simulated blood vessel–tumor system. ACS Appl. Mater. Inter. 2019;11:37513–37523. doi: 10.1021/acsami.9b15178. [DOI] [PubMed] [Google Scholar]
- Yamada K.M., Cukierman E. Modeling tissue morphogenesis and cancer in 3d. Cell. 2007;130:601–610. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Yarchoan M., Johnson B.A., 3rd, Lutz E.R., Laheru D.A., Jaffee E.M. Targeting neoantigens to augment antitumour immunity. Nat. Rev. Cancer. 2017;17:569. doi: 10.1038/nrc.2017.74. [DOI] [PubMed] [Google Scholar]
- Yu J., Berthier E., Craig A., De Groot T.E., Sparks S., Ingram P.N., Jarrard D.F., Huang W., Beebe D.J., Theberge A.B. Reconfigurable open microfluidics for studying the spatiotemporal dynamics of paracrine signalling. Nat. Biomed. Eng. 2019;3:830–841. doi: 10.1038/s41551-019-0421-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan T., Gao D., Li S., Jiang Y. Co-culture of tumor spheroids and monocytes in A collagen matrix-embedded microfluidic device to study the migration of breast cancer cells. Chin. Chem. Lett. 2019;30:331–336. [Google Scholar]
- Zamarron B.F., Chen W. Dual roles of immune cells and their factors in cancer development and progression. Int. J. Biol. Sci. 2011;7:651–658. doi: 10.7150/ijbs.7.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zboralski D., Hoehlig K., Eulberg D., Fromming A., Vater A. Increasing tumor-infiltrating T cells through inhibition of Cxcl12 with nox-A12 synergizes with Pd-1 blockade. Cancer Immunol. Res. 2017;5:950–956. doi: 10.1158/2326-6066.CIR-16-0303. [DOI] [PubMed] [Google Scholar]
- Zervantonakis I.K., Hughes-Alford S.K., Charest J.L., Condeelis J.S., Gertler F.B., Kamm R.D. Three-dimensional microfluidic model for tumor cell intravasation and endothelial barrier function. Proc. Natl. Acad. Sci. U S A. 2012;109:13515–13520. doi: 10.1073/pnas.1210182109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Wang D., Xu T., Liu P., Cao Y., Wang Y., Yang X., Xu X., Wang X., Niu H. Bladder cancer cells Re-educate tams through lactate shuttling in the microfluidic cancer microenvironment. Oncotarget. 2015;6:39196. doi: 10.18632/oncotarget.5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R., Weathered R., Kurtanich T., Mezyk-Kopec R., Mansurov A., Hubbell J., Swartz M.A. Development and validation of a novel organotypic in vitro model of the tumor-lymph-immune interface for predicting immunotherapy resistance. Cancer Res. 2020;78:68. [Google Scholar]