Abstract
Background
SARS-CoV-2 has caused a global pandemic, infecting millions of people. A safe, effective vaccine is urgently needed and remains a global health priority. Subunit vaccines are used successfully against other viruses when administered in the presence of an effective adjuvant.
Methods
We evaluated three different clinically tested adjuvant systems in combination with the SARS-CoV-2 pre-fusion stabilized (S-2P) spike protein using a one-dose regimen in mice.
Findings
Whilst spike protein alone was only weakly immunogenic, the addition of either Aluminum hydroxide, a squalene based oil-in-water emulsion system (SE) or a cationic liposome-based adjuvant significantly enhanced antibody responses against the spike receptor binding domain (RBD). Kinetics of antibody responses differed, with SE providing the most rapid response. Neutralizing antibodies developed after a single immunization in all adjuvanted groups with ID50 titers ranging from 86–4063. Spike-specific CD4 T helper responses were also elicited, comprising mainly of IFN-γ and IL-17 producing cells in the cationic liposome adjuvanted group, and more IL-5- and IL-10-secreting cells in the AH group.
Interpretation
These results demonstrate that adjuvanted spike protein subunit vaccine is a viable strategy for rapidly eliciting SARS-CoV-2 neutralizing antibodies and CD4 T cell responses of various qualities depending on the adjuvant used, which can be explored in further vaccine development against COVID-19.
Funding
This work was supported by the European Union Horizon 2020 research and innovation program under grant agreement no. 101003653.
Keywords: SARS-CoV-2, Alum, Subunit vaccine, CAF®01, MF59TM, Squalene emulsion, Neutralizing antibodies
Research in context.
Evidence before this study
A safe and effective vaccine against the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) causing COVID-19 disease is urgently needed. A number of SARS-CoV-2 vaccine candidates are currently evaluated. Many of these rely on novel vaccine technologies, e.g. RNA-, DNA- or adenovirus-vectors. Subunit vaccines composed of purified recombinant proteins are used successfully in licensed vaccines against Hepatitis virus B (HBV) and human papilloma virus (HPV) and a SARS-CoV-2 subunit vaccine could therefore be a safe and effective alternative. However, as subunit vaccines are poorly immunogenic on their own, adjuvants are required to boost vaccine immune responses. Adjuvants may differentially affect antibody secreting B cell response magnitude and breadth as well as CD4 T cell profiles. The optimal adjuvant for a SARS-CoV-2 subunit vaccine is currently unclear.
Added value of this study
Our study demonstrates that adjuvanted SARS-CoV-2 spike protein subunit vaccine can elicit neutralizing antibody responses after a single immunization and that the elicited T cell response profile depends on the type of adjuvant.
Implications of all the available evidence
This study supports the use of subunit vaccines containing adjuvanted SARS-CoV-2 spike protein for rapidly inducing neutralizing antibodies and T cell responses against SARS-CoV-2 and warrants further studies for determining the optimal adjuvant in animal challenge models.
Alt-text: Unlabelled box
1. Introduction
A safe and effective vaccine against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is urgently needed. Antibody responses are the best correlate of protection for most vaccines [1] and inducing such responses is a central goal for a SARS-CoV-2 vaccine. The receptor binding domain (RBD) of the spike protein is an attractive target for neutralization of coronaviruses [2,3], and neutralizing monoclonal antibodies (mAbs) targeting the RBD and blocking receptor binding have been isolated from COVID-19 convalescent patients [4,5] and offer protection in animal studies [6], [7], [8]. Several of these are now under evaluation as prophylactic or therapeutic antibodies to protect against clinical COVID-19 disease. Acute SARS CoV-2 infection also induced CD4 T cell responses directed against the membrane (M), nucleocapsid (N) and/or spike proteins in 100% of COVID-19 convalescent cases [9,10]. CD4 T cells shape the overall immune response, including antibody profiles and sustained humoral immunity [11], but may also reduce viral load by direct killing of infected cells [12]. Although the optimal vaccine profile remains unclear, a vaccine inducing both potent neutralizing antibody and CD4 T cell responses is likely to be protective.
The most progressed SARS-CoV-2 vaccine candidates rely on relatively novel vaccine technologies, including RNA- [13,14], DNA- [15,16], or adenovirus-vectored trimer proteins [17]. These strategies appear promising and have generated neutralizing antibodies in clinical trials [13,17] and protection against SARS-CoV-2 in preclinical models [15,18]. Preliminary data also demonstrate high efficacy for the RNA- and adenovirus vectored trimer proteins in clinical trials. However, the concurrent development of vaccines based on established vaccine platforms is prudent and particularly vaccines that show protection after a single immunization are desired. While inactivated virus vaccines are a well-established technology, and are being explored for SARS-CoV-2 [19], particular care must be paid to the potential exacerbation of lung immunopathology following viral infection, possibly mediated by T helper type 2 (Th2) responses, as has been reported for other inactivated vaccines e.g. against RSV [20] and in mouse models for SARS-CoV [21]. Another concern with inactivated vaccines is elicitation of antibodies against non-neutralizing antigen targets, which may exacerbate viral infection through antibody-dependent enhancement (ADE) [22]. Studies in animal challenge models and clinical trials will reveal whether there are any concerns with using inactivated vaccines against SARS-CoV-2. Another approach is to use purified recombinant proteins (subunit vaccines), which can effectively elicit high titers of neutralizing antibodies against SARS-CoV-2[23,24] and have demonstrated protective immunity for the highly successful hepatitis virus B (HBV) and human papilloma virus (HPV) vaccines, when administered in the presence of adjuvant.
Adjuvants are used to augment and orchestrate immune responses and influence affinity, specificity, magnitude and functional profile of B and T cell responses [25,26]. Although mechanisms of adjuvants may to some extent be translated across antigens and disease targets, antigen-specific responses have also been demonstrated, e.g. depending on antigen physicochemical properties, such as size and charge [27], [28], [29]. Adjuvants may also influence stability and integrity of antigens [28,30]. Importantly, to speed up SARS-CoV-2 vaccine development, a desired adjuvant should not only be safe and effective but also approved for clinical use or far in clinical development. In this study we tested prefusion-stabilized (S-2P) spike trimer [31,32] formulated in three different clinically tested adjuvant systems with diverse properties; Aluminium hydroxide (AH), which is in several licensed vaccines and is known to promote an antigen depot at the site of injection [28], an oil-in-water squalene emulsion (SE) adjuvant resembling MF59TM, which is licensed in a seasonal influenza vaccine, and a cationic liposome-based adjuvant (CAF®01), which has been tested in four phase 1 clinical trials and has been shown to induce strong CD4 T cell responses when tested with protein-based antigens [33,34].
2. Methods
2.1. Antigens and adjuvants
Both the SARS-CoV-2 S-2P stabilized spike trimer [32] antigen, and the RBD domain (RVQ-VNF) were produced by transient expression in freestyle 293-F cells, and purified first by affinity and then by size exclusion chromatography, as reported previously [24]. CAF®01 (DDA/TDB) was produced in house (Statens Serum Institut, Copenhagen, Denmark) [35], the AddaVaxTM oil-in-water squalene emulsion (SE, vac-adx-10) was from Invivogen (Toulouse, France) and aluminium oxyhydroxide (AH) (2% Alhydrogel®) was from Croda (Frederikssund, Denmark).
2.2. Characterization of prefusion-stabilized SARS-CoV-2 (S-2P) trimer and adjuvant formulations
A compatibility study of the spike trimer and adjuvant combinations was performed at room temperature (RT). Formulations were first analyzed visually for potential flocculation. Spike protein-adjuvant formulations were characterized for particle size and polydispersity index (PDI) by dynamic light scattering, by using the photon correlation spectroscopy technique. The surface charge of the particles was analyzed by measuring the zeta potential (laser-Doppler electrophoresis). Adjuvants alone or mixed with equal volumes of spike protein solutions were mixed and left to equilibrate for at least 10 min prior to the size and zeta potential measurements. Measurements were repeated after 24 h. For the size measurements, the samples were diluted 10 times, whereas for the zeta potential measurements, the samples were diluted 100 times in milli-Q water. The measurements were performed at 25°C by using a Zetasizer Nano ZS (Malvern Instruments, Worcestershire, UK) equipped with a 633 nm laser and 173° detection optics. Malvern DTS v.6.20 software was used for data acquisition and analysis. Particle size distribution was reflected in the PDI, which ranges from 0 for a monodisperse to 1.0 for a heterodisperse formulation.
2.3. Cryo-TEM
Formulations were prepared with CAF01 (1250 μg/ml DDA and 250 μg/ml TDB), SE (undiluted) and AH (2500 μg aluminium content/ml). The samples were analyzed directly or mixed 1:1 with antigen (25 μg/ml spike protein). Formulations with AH were further diluted 1:100 before analysis. Samples were prepared under controlled temperature (4 °C) and humidity conditions (100%) within an environmental vitrification system using a Vitrobot Mark IV (ThermoFisher). A sample droplet of 3 uL of formulation was deposited on a glow-discharged 300 mesh holey carbon grid. Excess liquid was blotted away for 3 s. The sample was immediately submerged into liquid ethane, resulting in the formation of a thin (10–500 nm) vitrified film. Samples were stored in liquid nitrogen and transferred to a Gatan 626 cryo-holder for imaging by cryo-TEM, using a Tecnai G2 T20 TWIN transmission electron microscope (ThermoFisher, USA). The sample temperature was constantly kept below −170°C. All observations were made in the bright field mode at an acceleration voltage of 200 kV. Images were recorded using a 4 × 4 CCD Eagle camera (ThermoFisher).
2.4. Antigen adsorption
Spike trimer was mixed with AH, using similar antigen and adjuvant concentrations as used for immunizations. In addition, formulations were prepared containing two-fold dilutions of antigen, whilst keeping the adjuvant concentration constant. Samples were centrifuged using an OptimalTM MAX-XP ultra-centrifuge (Beckman Coulter, Ramcon, Copenhagen, Denmark) at 135,700 g for 30 min. After centrifugation, the supernatant was quantified for non-adsorbed antigen by Micro BCA (Thermo Scientific, Wohlen, Switzerland). The antigen adsorption rates were extrapolated from results obtained when measuring antigen alone (in the absence of adjuvant).
2.5. Mice
Female C57Bl/6 wild type mice, 6–8 weeks old, were ordered from Harlan Laboratories (The Netherlands) and housed in the animal facilities at Statens Serum Institut, Denmark. Mice were housed with up to eight mice/cage.
2.6. Ethics
Mouse studies were conducted in accordance with the regulations set forth by the Danish National Committee for the Protection of Animals used for Scientific Purposes and in accordance with European Community Directive 2010/63/EU. The experiments have been approved by, and conducted in compliance with, the governmental Animal Experiments Inspectorate under license 2017-15-0201-01363.
2.7. Immunizations
Mice were randomized to different groups and given a single immunization subcutaneously (s.c.) at the base of the tail with 5 μg recombinant SARS-CoV-2 trimer in a volume of 200 μl TRIS buffer (pH 7.4) per immunization. Adjuvant doses were according to manufacturer's instructions: CAF01 (dose 250 μg/50 μg (DDA/TDB)), SE (dose of 100 μl 4.3% w/v squalene, 0.5% w/v Tween 80, 0.5% w/v Span 85 mixed 1:1 with antigen/PBS) and AH (dose of 500 μg aluminium content). The immunization studies were nonblinded.
2.8. Organ preparation
Mice were euthanized by CO2 (80%)/O2 (20%), using a flow rate of 3 l/min, followed by cervical dislocation. Inguinal lymph nodes (LNs) and spleens were filtered through a 70 μm nylon mesh (BD Biosciences). Lungs were dissociated via AutoMACS (C-tubes, Miltenyi). The cells were washed and prepared as previously described [36] and re-suspended in cell culture medium (RPMI-1640 supplemented with 5 × 10- [5] M 2-mercaptoethanol, 1% pyruvate, 1% HEPES, 1% (v/v) premixed penicillin-streptomycin solution (Invitrogen Life Technologies), 1 mM glutamine, and 10% (v/v) fetal calve serum (FCS)). Cell numbers were 2 × 105 cells/well for MSD cytokine profiling or 1 × 106 cells/well for flow cytometry (added in 200 μl of cell culture medium)
2.9. Cytokine profiling
The Mouse U-plex (IFN-γ, IL-17, IL-5, IL-13 and IL-10) was performed according to the manufacturer's instructions (Meso Scale Discovery) to measure CD4 T cell profiles after ex vivo re-stimulation of splenocytes with SARS-CoV-2 trimer antigen (2 µg/ml cell culture medium incubated for 72 or 96 hours at 37 °C and 5% CO2) [37]. The plates were analyzed on a Sector Imager 2400 system (Meso Scale Discovery) and calculation of cytokine concentrations was performed by 4-parameter logistic non-linear regression analysis of the standard curve.
2.10. ELISA for antibody responses
Maxisorp Plates (Nunc) were coated overnight with 0.05 μg/well SARS-CoV-2 trimer antigen (4 °C). After blocking, serum was added in PBS with 2% BSA, starting with a 30-fold dilution for antigen-specific IgG or IgG subclasses. Polyclonal HRP-conjugated secondary antibody (rabbit anti-mouse IgG (Thermofisher, RRID:AB_138451), Goat anti-mouse IgG1 (Southern Biotech, 1070-05), IgG2c (Invitrogen, RRID:AB_10983148), IgG3 (Thermofisher, RRID:AB_2536652) or rabbit anti mouse IgG2b (Invitrogen, RRID:AB_2533920) was diluted in PBS with 1% BSA. After 1 h of incubation, antigen-specific antibodies were detected using TMB substrate as described by the manufacturer (Kem-En-Tec Diagnostics), and the reaction was stopped with H2SO4. Antibody titers were determined as the highest serum dilution corresponding to a cut-off of ≥0.2 OD450.
2.11. Neutralization assay
Neutralization of SARS-CoV-2 was evaluated using a pseudotyped lentivirus neutralization assay. The assay used the same format as an assay validated for the evaluation of HIV neutralization [38], but with the use of SARS-CoV-2 spike and HEK293T cells engineered to express human ACE2, as previously described [39]. ID50 values were estimated by fitting a logistic curve in Prism 5 (Graphpad Software), bounded between 0% and 100%, and interpolating the dilution at which luciferase expression was reduced by 50% relative to wells in the absence of serum. The investigator performing the neutralization assay was blinded to the experimental groups.
2.12. Flow cytometry
One million cells were stained in PBS+1% FBS and FcBlock (BD) was added to block unspecific binding. Cells were stained with a live/dead marker (Fixable viability dye EF780, eBioscience) and a cocktail of antibodies against the following surface proteins: B220 PerCP-Cy5.5 (RA3-6B2, RRID:AB_394457) GL7 BV421 (GL7,
562967), GL7 FITC (GL7, 553666), IgD BV786 (11-26c.2a, RRID:AB_2738322) (All BD), CD38 PE-Cy7 (90, RRID:AB_11051806), CxCR5 BV421 (2G8, RRID:AB_2562128), CD4 APC (RM4-5, RRID:AB_469323) (eBioscience), PD-1 BV605 (29F.1A12, RRID:AB_11125371) (Biolegend). Cells were analysed on a BD Fortessa flow cytometer.
2.13. Statistical analysis
Differences between groups were analyzed by Kruskal-Wallis test, using Dunn´s multiple comparisons test with the unadjuvanted SARS-CoV-2 spike protein group as reference. Prism 8 software (GraphPad v8.2.1) was used for all statistical analyses.
2.14. Role of the funding source
The funding source had no role in study design, collection, analysis or interpretation of data or in the writing of the publication.
3. Results
3.1. Physicochemical characterization of adjuvanted spike trimer formulations
The SARS-CoV-2 spike trimer (spike protein) is the main determinant for viral entry into host cells [40]. The trimer protein is intrinsically metastable, but adding two proline mutations in the C-terminal S2 fusion domain resulted in a prefusion-stabilized spike antigen [32], which is very similar in structure to functional SARS-CoV-2 spike proteins on intact virions [41]. Highly homogenous spike glycoprotein trimers were produced and verified to maintain a trimeric pre-fusion conformation [39]. In order to evaluate compatibility of the prefusion-stabilized SARS-CoV-2 spike trimer protein (spike protein) with different adjuvant systems, we initially performed physicochemical characterization of spike protein formulated with CAF01, SE and AH. The full spike is a 139.125 kDa protein predicted to have an isoelectric point of 6.24 [42]. Based on the net charge of the spike protein, it may adsorb to the CAF01 and AH adjuvants via electrostatic and hydrophobic interactions and, additionally to AH via ligand exchange. When mixing CAF01 with spike protein, nanoparticle size increased from approx. 210 nm to 500 nm, suggesting that CAF01 interacted with spike protein. The PDI did not change upon mixture and the particles remained highly cationic (zp = +40 mV) (Fig 1a). Mixing trimer with SE adjuvant did not influence particle size (150 nm) and emulsion droplets remained largely monodisperse with no change in PDI. The SE adjuvant also remained net negatively charged in presence of spike protein. Addition of spike protein to AH particles increased size from approx. 1200 µm to 1800 µm, suggesting interaction between spike protein and AH, whilst the charge of the AH particles was unaffected.
Fig 1.
Characterization of vaccine formulations. Different adjuvant systems, cationic liposomes (CAF01), squalene emulsion (SE) and aluminium hydroxide (AH), were tested for compatibility with pre-fusion stabilized (S-2P) spike trimer protein. a) The particle size (left panels), polydispersity index (PDI, middle panels) were analyzed by dynamic light scattering and zeta potentials (Zp, right panels) of the adjuvant formulations were tested by laser-Doppler electrophoresis without the addition of spike protein (-Spike) and in the presence of spike protein, directly after mixture (+ Spike 0 h) and after 24 h (+ Spike, 24 h). Size was measured as diameter in nanometer (d.nm) and three replicates are shown. B) Adsorption of spike protein to aluminium hydroxide (AH). b) Cryo-TEM micrograph of the different adjuvant formulations in the absence or presence of spike protein (diluted 1:1 with the adjuvant formulation). Scale bar = 1 µm or 200 nm. C) Adsorbed spike retention onto AH was determined by measuring protein content recovered in the supernatant after ultracentrifugation.
To investigate in more detail how spike protein affected the morphology of the different adjuvants, we performed cryo-TEM. CAF01 contained both multilamellar and unilamellar spherical liposomes but also more elongated liposomes (fig 1b). This was largely similar when spike protein was added, although we noticed more clustering of liposomes and the appearance of rod-like structures, which could be collapsed liposomes binding the protein. As expected from the dynamic light scattering studies, we did not see noticeable changes in the SE droplets in the presence of spike protein. In contrast, with AH we observed larger aggregates in the formulation when spike protein was added compared to for AH alone. Thus, CAF01 and AH interacted with spike and formed aggregates as expected from the presence of electrostatic attractive forces, whereas SE with neutral surface charge did not interact with the protein.
Traditionally, the WHO has recommended that at least 80% of antigen is associated with AH, e.g. for diphtheria and pertussis vaccines [43]. The degree of antigen adsorption may affect both stability and structural integrity of antigens [30] and influence immune responses. Modifying antigens to increase adsorption to AH has led to increased antibody titers [29,44,45], illustrating the importance of interactions between antigens and AH. However, a very tight antigen-AH binding may compromise antibody responses [27,46]. Thus a certain degree of adsorption to AH seems beneficial. We tested adsorption capability of AH by measuring spike protein in the supernatant following centrifugation of the antigen−adjuvant mixture. Spike protein levels were below detection limit (2 µg/ml) in the supernatant following centrifugation of the AH-spike protein formulation, demonstrating that at least at the concentrations used for immunization, most if not all spike protein was adsorbed to AH (Fig 1c). We thus continued to evaluate spike trimer in combination with the different adjuvants in vivo.
3.2. Adjuvanted spike trimer induces neutralizing antibody responses after a single immunization
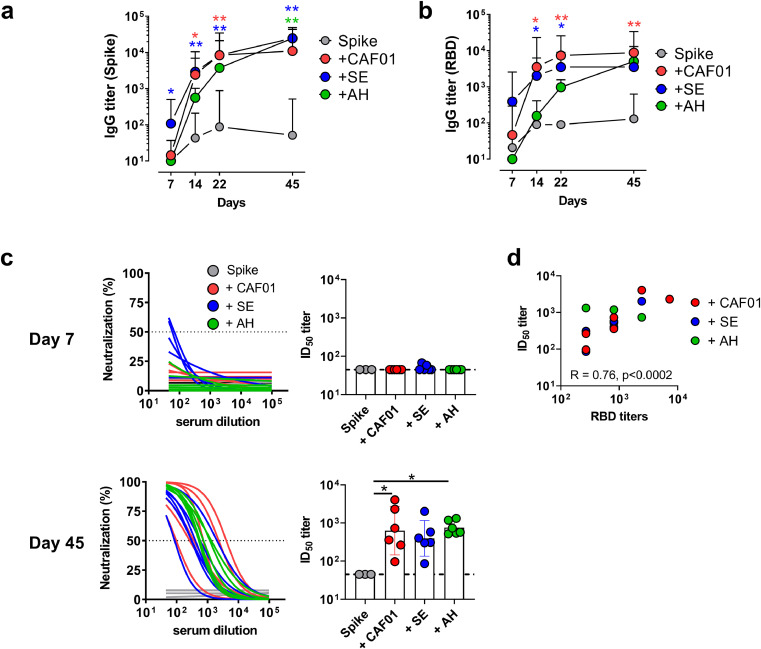
A pandemic vaccine against SARS-CoV-2 should preferably be effective after only a single immunization. To compare different adjuvant systems for stimulating immune responses to a single dose of SARS-CoV-2 spike trimer protein, we immunized mice subcutaneously with spike protein (5 µg) given alone or formulated in CAF01, SE or AH. To investigate if the formulations differed in their capacities to induce a rapid antibody response, we examined the kinetics of IgG antibody responses against the total spike protein and the RBD, which is the most important determinant for neutralization. Notably, spike-specific IgG antibody responses could be observed already at seven days post immunization in the SE adjuvanted group (significantly higher than in the spike alone group (p < 0.05, Kruskal-Wallis test)) (Fig 2a). All the adjuvants enhanced both total spike protein-specific and RBD-specific (Fig 2b) antibody responses, although AH induced a delayed antibody response compared to the CAF01 and SE adjuvants. At 45 days post immunization, when the experiment was terminated, the antibody titers were similar in all the adjuvanted groups (Fig. 2a and b). The anti-RBD IgG response consisted of IgG1 and IgG2b in all groups, whilst IgG2c was only detected in the CAF01 and SE adjuvanted groups and IgG3 was not detected in any of the groups (sFig1).
Fig 2.
Adjuvanted pre-fusion stabilized (S-2P) spike protein elicits neutralizing antibody responses after a single dose. Mice were immunized with a single dose of spike trimer protein alone or formulated in CAF01, aluminium hydroxide (AH) or squalene emulsion (SE). Kinetics of IgG antibody responses against spike protein (a) and the receptor binding domain (RBD) (b) were measured by ELISA. c) Neutralization of SARS-CoV-2 by pseudovirus neutralization assay measured at day 7 (upper panels) and 45 (lower panels). d) Correlation between IgG titers against RBD and neutralization titers (Spearman R and p-value indicated) (right panel). Bars show geometric mean ± 95% CI. Statistically significant differences are indicated by * and ** (Kruskal Wallis test, using the unadjuvanted spike trimer group as reference and significance levels of p < 0.05 and p < 0.01, respectively). Figures represent three to six mice per group from one experiment.
To investigate if the vaccine-elicited antibodies were capable of neutralizing SARS-CoV-2, we performed a homotypic SARS-CoV-2 pseudovirus neutralization assay. Little neutralizing activity was detected at seven days post immunization and only in the SE group. However, at day 45, neutralizing antibodies were found in all the adjuvanted groups, but not in the unadjuvanted spike protein group (Fig 2c). Neutralizing antibody responses in mice immunized with the CAF01 and AH adjuvanted formulations were significantly higher than in the spike protein alone group (p < 0.05, Kruskal-Wallis test). ID50 titers in the adjuvanted groups ranged from 86–4063 and there was a significant correlation between anti-RBD IgG antibody and neutralization titers (spearman R = 0.77, p = 0.0002) (Fig 2d). Thus, a single immunization with spike trimer in adjuvant was sufficient to induce neutralizing antibody responses.
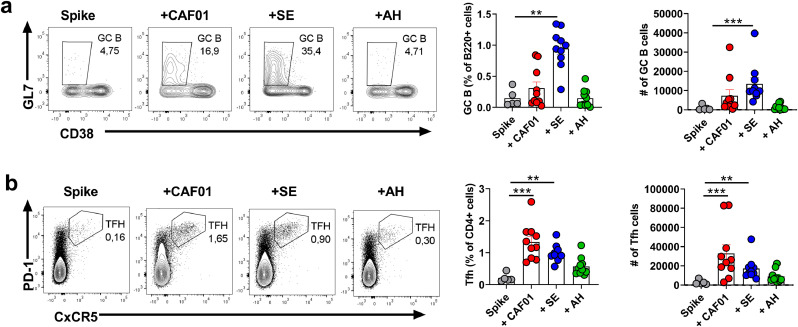
Antibody responses to T-dependent antigens, including antibody affinity maturation and class-switching, depend on germinal center (GC) formation. We evaluated GC responses by staining for GC B cells (B220+IgD-CD38-GL7+) and T follicular helper cells (Tfh) (B220-CD4+PD-1+CxCR5+) [47] at seven days post immunization with a single dose of spike trimer antigen alone or formulated in adjuvant. Only the SE adjuvant significantly boosted GC B cell responses compared to unadjuvanted trimer protein (p < 0.01, Kruskal-Wallis test) at seven days post immunization (Fig 3a). Tfh responses on the other hand were robustly induced in both the CAF01 and SE groups (significantly different from unadjuvanted trimer protein, p < 0.01, Kruskal-Wallis test) (Fig 3b). Notably, GL7 expression amongst Tfh cells was highest in the CAF01 group (sFig2). DC priming is sufficient to induce early Tfh differentiation and expression of CxCR5 and GL7 even without cognate B cell help [48], which may explain why GC B cell and Tfh responses did not correlate. Since adjuvants may differentially affect kinetics of GC responses [49], it is possible that GC initiation was delayed when spike protein was formulated in CAF01 and AH adjuvants compared to SE. To investigate GC persistence, we evaluated GC B cells at day 45, at which, there was still a tendency towards higher GC B cell responses in the adjuvanted groups, compared to trimer protein alone (only significant for the AH group, p < 0.05, Kruskal-Wallis test), indicating that the adjuvants promoted long-lived GCs (sFig3). Overall, the tested adjuvants differentially influenced timing of GC initiation in response to spike protein, however the antibody responses, including capacity to neutralize SARS-CoV-2, were largely similar at 45 days post immunization.
Fig 3.
Squalene emulsion elicits a rapid germinal center (GC) B cell response when formulated with pre-fusion stabilized (S-2P) spike protein. Mice were immunized with a single dose of unadjuvanted spike trimer protein or formulated in CAF01, aluminium hydroxide (AH) or squalene emulsion (SE) and GC B cell responses measured in the draining lymph node at seven days post immunization. a) GC B cell responses measured by flow cytometry (gated on Live B220+CD38−GL7+ cells). b) T follicular helper (Tfh) cells (gated on live B220-CD4+PD-1+CxCR5+ cells). Bars show mean ± SEM. Statistically significant differences are indicated by *, ** and *** (Kruskal Wallis test, using the spike trimer alone group as reference and significance levels of p < 0.05, p < 0.01 and p < 0.001 respectively). Figures represent five to ten mice per group from one experiment.
3.3. T cell profile in response to spike trimer protein is dependent on the adjuvant
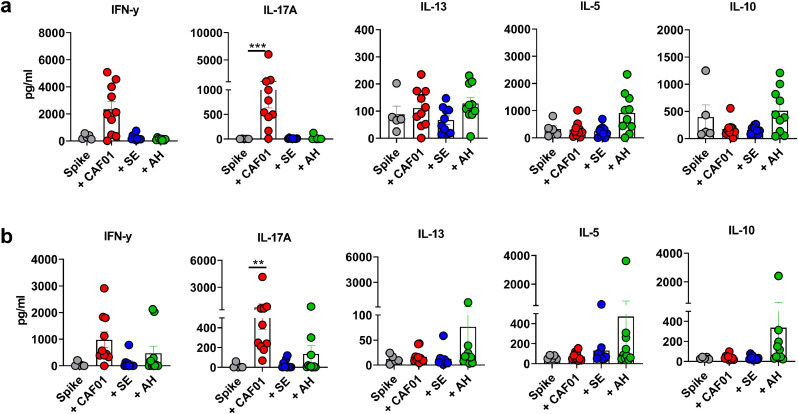
Vaccine-associated enhanced respiratory disease (VAERD) has been associated with CD4 T cell responses biased towards a Th2 profile in the context of RSV [50] and SARS-CoV [21,51]. To evaluate polarization of the vaccine-induced T cell responses, we re-stimulated splenocytes with either the RBD or total spike protein early (day 7) and late (day 45) after immunization. Compared to the group having received unadjuvanted spike protein, there was a tendency for increased IFN-γ production, suggesting Th1 responses, in the CAF01 group (not significant) at day 7 (Fig 4a). CAF01 also induced a Th17 response, which was significantly higher than in the unadjuvanted group (p < 0.01, Kruskal-Wallis test). Th1/Th17 cell responses were lower in the SE and AH groups and low IL-13, IL-5 and IL-10 responses were detected in all groups. Examining T cell responses in the lung (Fig 4b), CAF01 also induced a Th17 response, significantly higher than in the spike protein alone group (p < 0.01, Kruskal-Wallis test). We also detected IL-5 and IL-10 in the lungs of some of the mice in the AH group, although levels were not significantly different from in the unadjuvanted spike group. IFN-γ and IL-17 responses remained detectable in the CAF01 group at day 45 post immunization and were significantly higher compared to in the unadjuvanted spike protein group (p < 0.05, Kruskal-Wallis test) (sFig 4). In contrast to when re-stimulating with intact spike protein, we failed to detect any T cell responses when re-stimulating with the RBD (data not shown). To investigate if the observed T cell responses were mice strain-dependent, we repeated the studies using BALB/c mice, which are more Th2-prone. Similarly to in C57Bl/6 mice, spleen and lung Th17 responses were significantly higher in mice that had received spike protein formulated in CAF01 than when spike was given alone (sFig 5). In contrast, Th2 responses were generally low although there was a tendency for higher splenic Th2 responses in the AH group than in the other adjuvanted groups. Overall, the adjuvants differentially influenced the magnitude and profile of the elicited T helper response against the prefusion-stabilized SARS-CoV-2 spike protein.
Fig 4.
Adjuvants differentially influence CD4 T cell responses to pre-fusion stabilized (S-2P) spike protein. Mice were immunized with a single dose of unadjuvanted spike trimer protein or formulated in CAF01, aluminium hydroxide (AH) or squalene emulsion (SE). T cell responses were measured by stimulating a) splenocytes and b) lung cells with spike protein at seven days post immunization and measuring secreted cytokines in the supernatant. Bars show mean ± SEM. Statistically significant differences are indicated by ** and *** (Kruskal Wallis test, using the unadjuvanted spike trimer group as reference and significance levels of p < , respectively). The figure represents five to ten mice per group from one experiment.
4. Discussion
SARS-CoV-2 has caused more than 1 300 000 deaths worldwide to date, and triggered the deepest global recession in decades. A vaccine against SARS-CoV-2 is thus urgently needed. The immune profile required for vaccine-mediated protection against SARS-CoV-2 is currently unknown. Vaccine-induced antibody responses are the best correlate of protection for many infectious agents [1] and several anti-SARS-CoV-2 antibodies can neutralize the virus in vitro [4,52] and afford protection against SARS-CoV-2 when passively transferred to Syrian hamsters [53]. Vaccine-induced CD4 T cell responses may also play a role in protection against SARS-CoV-2, as CD4 T cells shape various immune effector functions, including orchestration of innate immunity, cytotoxic T cells and B cell functionality (reviewed in [54]). CD4 T cells can be segregated into functional subsets based on their cytokine production. Skewing of T cell responses towards a predominant type 2 helper T-cell (Th2) cell profile (producing IL-4, IL-5 or IL-13) has been associated with vaccine-enhanced respiratory disease, as seen in the context of RSV [20,50] and in a murine model of SARS-CoV [21,51,55]. A more balanced Th1/Th2 or a predominantly Th1-directed response on the other hand is generally considered favorable in anti-viral immunity [56].
Adjuvants can direct the T-helper response [36,57] and shape humoral immunity by influencing the magnitude, breadth and affinity of the antibody response [58] in addition to inducing specific antibody subclasses with different Fc-based effector functions [59]. Here we tested three different adjuvant systems together with the pre-fusion stabilized [60] SARS-CoV-2 (S-2P) spike trimer protein [18]. Encouragingly, all the adjuvants boosted neutralizing antibody responses after only a single immunization, supporting the possibility of a one-dose vaccine regimen. In contrast, spike protein alone did not induce detectable neutralization. Neutralization titers in the adjuvanted groups were of similar magnitude to those seen in mice given a single dose of the mRNA-1273 vaccine (encoding the stabilized (S-2P) trimer) [31], and in convalescent SARS-CoV-2 cases [61], although it may be difficult to compare across different assays. The adjuvants induced different T helper cell profiles, with CAF01 promoting a mixed IFN-γ (Th1)/IL-17 (Th17) response, whilst SE and AH displayed lower responses. Some of the mice that had received AH had lung T cells producing IL-5 and IL-10. Although Th2 responses in the SE and AH groups were low, possibly since the vaccine was given as a single dose, the adjuvant-directed T helper profile was similar to that described when tested with antigens from other disease targets [36]. However, the optimal T helper response in relation to COVID-19 is unclear. Convalescing COVID-19 cases had predominantly a Th1 response with little Th2 cytokines detected [9] and subjects with moderate COVID-19 disease had more IFN-γ+ Th1 cells than severe cases, which could imply a beneficial role of Th1 cells. The role for Th17 cells also remains elusive. High levels of peripheral blood CCR6+ CD4 T cells possibly representing Th17 cells were linked to severe disease in one case, although IL-17 expression was not directly examined [62]. IL-17 was identified amongst cytokines associated with SARS-CoV-2-induced pulmonary inflammation [63] and it has been proposed that Th17 cells may exacerbate lung immunopathology, possibly by facilitating eosinophil recruitment [64]. Th17 cells have also been described to regulate antibody glycosylation [65], which may influence Fc receptor binding and thus potentially antibody dependent enhancement (ADE). More studies focusing on the role of infection- and vaccine-induced T helper profiles in protection against SARS-CoV-2 are required to determine the optimal adjuvant-mediated T cell polarization.
DNA and RNA-based vaccine platforms are promising, due to their scalable and relatively rapid production, and preliminary data suggest that the SARS-CoV-2 mRNA based vaccines are highly effective for preventing COVID-19 infection. However, no nucleic-acid based vaccine has been approved for human use so far and these novel vaccine technology platforms require careful evaluation of efficacy and safety. Subunit vaccines based on proteins, are used successfully against other infections including HBV and HPV and should be considered as important components of the SARS-CoV-2 vaccine pipeline. Here we have demonstrated that three different clinically tested adjuvant systems could be formulated with the pre-fusion stabilized (S-2P) SARS-CoV-2 spike protein [31] to enhance neutralizing antibody titers after a single immunization in mice. Furthermore, the adjuvants differentially influenced T helper responses. Studies in animal challenge models are required to select the optimal adjuvanted SARS-CoV-2 subunit vaccine.
Declaration of Competing Interest
D.C. is co-inventor on patents on the cationic adjuvant formulations (CAF). All rights have been turned over to Statens Serum Institut, which is a non-profit government research facility. The rest of the authors declare that there are no competing interests.
Acknowledgments
Contributors
G.M., G.K.H., B.M., D.C. and G.K.P., designed research; D.S., L.H. J.Z and K.W. performed experiments; K.W., G.K.P., D.S, S.T.S, D.C. analyzed data; and G.M., G.K.H., B.M, D.C. and G.K.P. wrote the paper. All authors read and approved the final version of the manuscript, and have had access to the raw data. Katharina Wørzner, Signe Tandrup Schmidt, Dennis Christensen and Gabriel Kristian Pedersen can verify the accuracy of the raw data for the study.
Acknowledgements
We appreciate the expert technical assistance provided by the technicians at Center for Vaccine Research and the staff at the experimental animal facilities at Statens Serum Institut. We are also grateful for the expert assistance with CryoTEM microscope analysis at the Core Facility for Integrated Microscopy, Faculty of Health and Medical Sciences, University of Copenhagen. This work was supported by the European Union Horizon 2020 research and innovation program under grant agreement no. 101003653 (CoroNAb).
Data sharing statement
Study protocol and all data collected for the study, including raw data and data analysis will be made available to others upon request. All data will be available upon publication of the manuscript, by contacting the corresponding author. Data will be made available after approval of a proposal and with a signed data access agreement.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.103197.
Appendix. Supplementary materials
References
- 1.Plotkin SA. Correlates of protection induced by vaccination. Clin Vaccine Immunol. 2010;17(7):1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Du L, Zhao G, Kou Z. Identification of a receptor-binding domain in the S protein of the novel human coronavirus Middle East respiratory syndrome coronavirus as an essential target for vaccine development. J Virol. 2013;87(17):9939–9942. doi: 10.1128/JVI.01048-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong SK, Li W, Moore MJ, Choe H, Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J Biol Chem. 2004;279(5):3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ni L, Ye F, Cheng ML. Detection of SARS-CoV-2-Specific Humoral and Cellular Immunity in COVID-19 Convalescent Individuals. Immunity. 2020;52(6):971–977. doi: 10.1016/j.immuni.2020.04.023. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou D, Duyvesteyn HME, Chen CP. Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient. Nat Struct Mol Biol. 2020 doi: 10.1038/s41594-020-0480-y. [DOI] [PubMed] [Google Scholar]
- 6.Rogers TF, Zhao F, Huang D. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369(6506):956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao Y, Su B, Guo X. Potent Neutralizing Antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B Cells. Cell. 2020;182(1):73–84. doi: 10.1016/j.cell.2020.05.025. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi R, Shan C, Duan X. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584(7819):120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 9.Grifoni A, Weiskopf D, Ramirez SI. Targets of T Cell Responses to SARS-CoV-2 coronavirus in humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181(7):1489–1501. doi: 10.1016/j.cell.2020.05.015. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiskopf D, Schmitz KS, Raadsen MP. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol. 2020;5(48) doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kunzli M, Schreiner D, Pereboom TC. Long-lived T follicular helper cells retain plasticity and help sustain humoral immunity. Sci Immunol. 2020;5(45) doi: 10.1126/sciimmunol.aay5552. [DOI] [PubMed] [Google Scholar]
- 12.Sant AJ, McMichael A. Revealing the role of CD4(+) T cells in viral immunity. J Exp Med. 2012;209(8):1391–1395. doi: 10.1084/jem.20121517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson LA, Anderson EJ, Rouphael NG. An mRNA Vaccine against SARS-CoV-2 - preliminary report. N Engl J Med. 2020 doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erasmus JH, Khandhar AP, O'Connor MA. An Alphavirus-derived replicon RNA vaccine induces SARS-CoV-2 neutralizing antibody and T cell responses in mice and nonhuman primates. Sci Transl Med. 2020;12(555) doi: 10.1126/scitranslmed.abc9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu J, Tostanoski LH, Peter L. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020 doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith TRF, Patel A, Ramos S. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat Commun. 2020;11(1):2601. doi: 10.1038/s41467-020-16505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu FC, Guan XH, Li YH. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2020 doi: 10.1016/S0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corbett KS, Flynn B, Foulds KE. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N Engl J Med. 2020 doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao Q, Bao L, Mao H. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020;369(6499):77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castilow EM, Olson MR, Varga SM. Understanding respiratory syncytial virus (RSV) vaccine-enhanced disease. Immunol Res. 2007;39(1-3):225–239. doi: 10.1007/s12026-007-0071-6. [DOI] [PubMed] [Google Scholar]
- 21.Tseng CT, Sbrana E, Iwata-Yoshikawa N. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS One. 2012;7(4):e35421. doi: 10.1371/journal.pone.0035421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arvin AM, Fink K, Schmid MA. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature. 2020;584(7821):353–363. doi: 10.1038/s41586-020-2538-8. [DOI] [PubMed] [Google Scholar]
- 23.Keech C, Albert G, Cho I. Phase 1-2 Trial of a SARS-CoV-2 Recombinant Spike Protein Nanoparticle Vaccine. N Engl J Med. 2020 doi: 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mandolesi M, Sheward DJ, Hanke L. 2020. SARS-CoV-2 protein subunit vaccination elicits potent neutralizing antibody responses. 2020.07.31.228486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Del Giudice G, Rappuoli R, Didierlaurent AM. Correlates of adjuvanticity: a review on adjuvants in licensed vaccines. Semin Immunol. 2018 doi: 10.1016/j.smim.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Khurana S, Verma N, Yewdell JW. MF59 adjuvant enhances diversity and affinity of antibody-mediated immune response to pandemic influenza vaccines. Sci Transl Med. 2011;3(85) doi: 10.1126/scitranslmed.3002336. 85ra48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansen B, Sokolovska A, HogenEsch H, Hem SL. Relationship between the strength of antigen adsorption to an aluminum-containing adjuvant and the immune response. Vaccine. 2007;25(36):6618–6624. doi: 10.1016/j.vaccine.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 28.HogenEsch H, O'Hagan DT, Fox CB. Optimizing the utilization of aluminum adjuvants in vaccines: you might just get what you want. NPJ Vaccines. 2018;3:51. doi: 10.1038/s41541-018-0089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moyer TJ, Kato Y, Abraham W. Engineered immunogen binding to alum adjuvant enhances humoral immunity. Nat Med. 2020;26(3):430–440. doi: 10.1038/s41591-020-0753-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colaprico A, Senesi S, Ferlicca F. Adsorption onto aluminum hydroxide adjuvant protects antigens from degradation. Vaccine. 2020;38(19):3600–3609. doi: 10.1016/j.vaccine.2020.02.001. [DOI] [PubMed] [Google Scholar]
- 31.Corbett KS, Edwards DK, Leist SR. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020 doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wrapp D, Wang N, Corbett KS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abraham S, Juel HB, Bang P. Safety and immunogenicity of the chlamydia vaccine candidate CTH522 adjuvanted with CAF01 liposomes or aluminium hydroxide: a first-in-human, randomised, double-blind, placebo-controlled, phase 1 trial. Lancet Infect Dis. 2019;19(10):1091–1100. doi: 10.1016/S1473-3099(19)30279-8. [DOI] [PubMed] [Google Scholar]
- 34.van Dissel JT, Joosten SA, Hoff ST. A novel liposomal adjuvant system, CAF01, promotes long-lived Mycobacterium tuberculosis-specific T-cell responses in human. Vaccine. 2014;32(52):7098–7107. doi: 10.1016/j.vaccine.2014.10.036. [DOI] [PubMed] [Google Scholar]
- 35.Davidsen J, Rosenkrands I, Christensen D. Characterization of cationic liposomes based on dimethyldioctadecylammonium and synthetic cord factor from M. tuberculosis (trehalose 6,6 '-dibehenate) - a novel adjuvant inducing both strong CMI and antibody responses. Bba-Biomembranes. 2005;1718(1-2):22–31. doi: 10.1016/j.bbamem.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 36.Knudsen NPH, Olsen A, Buonsanti C. Different human vaccine adjuvants promote distinct antigenin-dependent immunological signatures tailored to different pathogens. Sci Rep-Uk. 2016;6 doi: 10.1038/srep19570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olsen AW, Follmann F, Erneholm K, Rosenkrands I, Andersen P. Protection Against Chlamydia trachomatis Infection and Upper Genital Tract Pathological Changes by Vaccine-Promoted Neutralizing Antibodies Directed to the VD4 of the Major Outer Membrane Protein. J Infect Dis. 2015;212(6):978–989. doi: 10.1093/infdis/jiv137. [DOI] [PubMed] [Google Scholar]
- 38.Sarzotti-Kelsoe M, Bailer RT, Turk E. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J Immunol Methods. 2014;409:131–146. doi: 10.1016/j.jim.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanke L, Vidakovics Perez L, Sheward DJ. An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction. Nat Commun. 2020;11(1):4420. doi: 10.1038/s41467-020-18174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang S, Hillyer C, Du L. Neutralizing Antibodies against SARS-CoV-2 and Other Human Coronaviruses. Trends Immunol. 2020;41(5):355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ke Z, Oton J, Qu K. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020 doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheller C, Krebs F, Minkner R, Astner I, Gil-Moles M, Watzig H. Physicochemical properties of SARS-CoV-2 for drug targeting, virus inactivation and attenuation, vaccine formulation and quality control. Electrophoresis. 2020;41(13-14):1137–1151. doi: 10.1002/elps.202000121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Organization World Health. 1977. Expanded Programme on I. Manual for the production and control of vaccines: pertussis vaccine. Geneva: World Health Organization. [Google Scholar]
- 44.Clapp T, Siebert P, Chen D, Jones Braun L. Vaccines with aluminum-containing adjuvants: optimizing vaccine efficacy and thermal stability. J Pharm Sci. 2011;100(2):388–401. doi: 10.1002/jps.22284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu F, Boutselis I, Borch RF, Hogenesch H. Control of antigen-binding to aluminum adjuvants and the immune response with a novel phosphonate linker. Vaccine. 2013;31(40):4362–4367. doi: 10.1016/j.vaccine.2013.07.019. [DOI] [PubMed] [Google Scholar]
- 46.Hansen B, Belfast M, Soung G. Effect of the strength of adsorption of hepatitis B surface antigen to aluminum hydroxide adjuvant on the immune response. Vaccine. 2009;27(6):888–892. doi: 10.1016/j.vaccine.2008.11.078. [DOI] [PubMed] [Google Scholar]
- 47.Tam HH, Melo MB, Kang M. Sustained antigen availability during germinal center initiation enhances antibody responses to vaccination. Proc Natl Acad Sci U S A. 2016;113(43) doi: 10.1073/pnas.1606050113. E6639-E48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kerfoot SM, Yaari G, Patel JR. Germinal center B cell and T follicular helper cell development initiates in the interfollicular zone. Immunity. 2011;34(6):947–960. doi: 10.1016/j.immuni.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pedersen GK, Worzner K, Andersen P, Christensen D. Vaccine adjuvants differentially affect kinetics of antibody and germinal center responses. Front Immunol. 2020;11(2302) doi: 10.3389/fimmu.2020.579761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim HW, Canchola JG, Brandt CD. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am J Epidemiol. 1969;89(4):422–434. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- 51.Honda-Okubo Y, Barnard D, Ong CH, Peng BH, Tseng CT, Petrovsky N. Severe acute respiratory syndrome-associated coronavirus vaccines formulated with delta inulin adjuvants provide enhanced protection while ameliorating lung eosinophilic immunopathology. J Virol. 2015;89(6):2995–3007. doi: 10.1128/JVI.02980-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klasse PJ, Moore JP. Antibodies to SARS-CoV-2 and their potential for therapeutic passive immunization. Elife. 2020:9. doi: 10.7554/eLife.57877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rogers TF, Zhao F, Huang D. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020 doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swain SL, McKinstry KK, Strutt TM. Expanding roles for CD4(+) T cells in immunity to viruses. Nat Rev Immunol. 2012;12(2):136–148. doi: 10.1038/nri3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bolles M, Deming D, Long K. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J Virol. 2011;85(23):12201–12215. doi: 10.1128/JVI.06048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Snell LM, Osokine I, Yamada DH, De la Fuente JR, Elsaesser HJ, Brooks DG. Overcoming CD4 Th1 Cell Fate Restrictions to Sustain Antiviral CD8 T cells and control persistent virus infection. Cell Rep. 2016;16(12):3286–3296. doi: 10.1016/j.celrep.2016.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Francica JR, Zak DE, Linde C. Innate transcriptional effects by adjuvants on the magnitude, quality, and durability of HIV envelope responses in NHPs. Blood Adv. 2017;1(25):2329–2342. doi: 10.1182/bloodadvances.2017011411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khurana S, Verma N, Yewdell JW. MF59 adjuvant enhances diversity and affinity of antibody-mediated immune response to pandemic influenza vaccines. Sci Transl Med. 2011;3(85) doi: 10.1126/scitranslmed.3002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boudreau CM, Yu WH, Suscovich TJ, Talbot HK, Edwards KM, Alter G. Selective induction of antibody effector functional responses using MF59-adjuvanted vaccination. J Clin Invest. 2020;130(2):662–672. doi: 10.1172/JCI129520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pallesen J, Wang N, Corbett KS. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc Natl Acad Sci U S A. 2017;114(35) doi: 10.1073/pnas.1707304114. E7348-E57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang X, Guo X, Xin Q. Neutralizing antibodies responses to SARS-CoV-2 in COVID-19 inpatients and convalescent patients. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu Z, Shi L, Wang Y. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang C, Wang Y, Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hotez PJ, Corry DB, Bottazzi ME. COVID-19 vaccine design: the Janus face of immune enhancement. Nat Rev Immunol. 2020;20(6):347–348. doi: 10.1038/s41577-020-0323-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pfeifle R, Rothe T, Ipseiz N. Regulation of autoantibody activity by the IL-23-TH17 axis determines the onset of autoimmune disease. Nat Immunol. 2017;18(1):104–113. doi: 10.1038/ni.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.