Summary
To better understand cellular communication driving diverse behaviors, we need to uncover the molecular mechanisms of within-cell-type functional heterogeneity. While single-cell RNA sequencing (scRNAseq) has advanced our understanding of cell heterogeneity, linking individual cell phenotypes to transcriptomic data remains challenging. Here, we used a phenotypic cell sorting technique to ask whether phenotypically supervised scRNAseq analysis (pheno-scRNAseq) can provide more insight into heterogeneous cell behaviors than unsupervised scRNAseq. Using a simple 3D in vitro breast cancer (BRCA) model, we conducted pheno-scRNAseq on invasive and non-invasive cells and compared the results to phenotype-agnostic scRNAseq analysis. Pheno-scRNAseq identified unique and more selective differentially expressed genes than unsupervised scRNAseq analysis. Functional studies validated the utility of pheno-scRNAseq in understanding within-cell-type functional heterogeneity and revealed that migration phenotypes were coordinated with specific metabolic, proliferation, stress, and immune phenotypes. This approach lends new insight into the molecular systems underlying BRCA cell phenotypic heterogeneity.
Subject areas: Cell Biology, Complex System Biology, Transcriptomics
Graphical abstract

Highlights
-
•
Graph-based clusters do not identify collective migration phenotypes
-
•
Collective invasion is associated with stress response and proliferation
-
•
Non-invasive behavior is associated with plasticity and immunomodulation
-
•
Signatures corresponding to migration phenotypes are conserved across species
Cell Biology; Complex System Biology; Transcriptomics
Introduction
Cellular heterogeneity is indispensable for population-level survival strategies of multicellular organisms, such as bet-hedging in order to achieve a better chance of survival when faced with new stresses (Grimbergen et al., 2015). In cancer, the genetic, epigenetic, transcriptional, and proteomic differences among tumor cells can give rise to diverse phenotypes, some of which can persist in dysregulated environmental conditions, survive therapeutic attempts, and migrate away from the primary tumor to form metastases (Hinohara et al., 2018; Duan et al., 2018). Linking the heterogeneity observed in cellular genomics with phenotypic heterogeneity has significant potential to inform successful population-level treatments. However, the challenges associated with measuring phenotypic heterogeneity and isolating particular cells within the complex in vivo environment make it difficult to identify the most basic transcriptional modules regulating individual cell behaviors.
Fortunately, significant evidence suggests that physiologically relevant phenotypes of breast cancer (BRCA) cells can be studied in less complex in vitro systems by embedding the cells in 3D type I collagen (COL1) hydrogels. BRCA cell lines, organoids from mouse tumors, and organoids from human tumors embedded in this model system harbor the same pattern of differentiation markers as are observed in studies of mouse mammary tumor histology and human BRCA histology (Cheung et al., 2013). Studies have also shown that BRCA cells cultured in this model system upregulate a conserved transcriptional program of 70 genes that is predictive of poor prognosis in human BRCA and eight additional cancer types, with the highest predictive value in triple-negative breast cancer (hazard ratio = 3.85, Cox p value = 0.007) (Velez et al., 2017; Zhang et al., 2018). Thus, a growing body of evidence suggests that 3D culture of BRCA cells in COL1 is a relevant model system for studying physiologically relevant cancer phenotypes.
Importantly, BRCA cells embedded in a 3D COL1 matrix maintain heterogeneity. In particular, they can take on a range of migration phenotypes, from non-invasive to single-cell mesenchymal style migration to collective invasion (Velez et al., 2017), with the collective invasion phenotype being linked to the metastatic phenotype in vivo (Cheung et al., 2013, Cheung et al., 2016, Cheung and Ewald, 2016; Tabassum and Polyak, 2015; Aceto et al., 2014). To begin to define the molecular programs underlying BRCA cell migration heterogeneity, we sought a method capable of linking cell phenotype to gene expression programs. While advances in single-cell omics technologies have significantly improved our ability to characterize cell heterogeneity, these methods involve the sequencing of individual cells from a bulk sample and determining cell clusters solely based on differences in the molecular signature. However, the biological interpretation of these complex data is only at an early stage. Inferring cell state, function, and response to treatment from such data remains highly subjective and dependent on a priori knowledge (Choi and Kim, 2019). Cell subpopulations identified from analyzing sequencing data can only be validated with experiments after clusters have been defined, and this relies heavily on the assumption that transcriptomic data maps well to functional profiles. Partitions made from unsupervised clustering methods could potentially divide the sample into groups that may have no functional biological meaning, particularly for samples that are more similar as a whole, like cells of the same type. While standards and strategies are constantly evolving, there remains a lack of consensus on how to define cell types and subtypes based on sequencing data (Kiselev et al., 2019). The field of single-cell analysis is rapidly moving toward integrative, multi-scale measurements to improve the functional interpretability of single-cell data. Thus far, transcriptome measurements have been integrated with multiple omics (Chappell et al., 2018), genotype (Dixit et al., 2016; Jaitin et al., 2016), cell electrophysiology (Cadwell et al., 2016), lineage tracing (Kester and van Oudenaarden, 2018), and spatial information (Lein et al., 2017). To more concretely link phenotype to omics data, rare cell subpopulations may also be functionally sorted using innovative physical (Beri et al., 2020) or image-guided techniques (Konen et al., 2017).
Here, we explored whether traditional single-cell RNA sequencing (scRNAseq) followed by unsupervised clustering analysis would be capable of correctly inferring migration phenotype. This would inherently require that phenotypic regulators dominate the transcriptome of the cells to enable similarity-based clustering. However, we posited that other processes might dominate single-cell transcriptomes such that phenotypic regulators could represent a much smaller signal in the data. So, we used an image-guided phenotypic sorting technique to ask whether phenotypically supervised scRNAseq (hereafter referred to as pheno-scRNAseq for ease of reference) can provide more insight into the heterogeneous cell migration behaviors of MDA-MB-231 (MDA) BRCA cells than unsupervised scRNAseq. Photoconversion-based cell labeling followed by rapid dissociation into a single-cell suspension, fluorescence-activated cell sorting (FACS), and scRNAseq enabled direct comparison of phenotype labels with unsupervised transcriptional clustering. Unsupervised clustering was not able to correctly infer migration phenotype. Accordingly, pheno-scRNAseq revealed unique molecular programs associated with the migration state. Functional experiments targeting several identified genes validated that they play an active role in regulating migration behaviors. Specifically, perturbing HSP90AB1, DEK, and F3 regulated the collective invasion phenotype. Pheno-scRNAseq further revealed that collectively invasive cells exist in a “go and grow” state, where biosynthetic processes, proliferation, oxidative stress responses, and ER stress responses are upregulated. However, non-invasive cells limit proliferation and biosynthesis and are dominated by redox homeostasis and immunomodulatory gene expression programs. These relationships were recapitulated in mouse 4T1 BRCA cells to confirm that the results were not cell line specific. Our phenotypic sorting approach also enabled reseeding experiments that probed the stability of these cellular states, revealing that invasive cells remain in a stable migration state while non-invasive cells are plastic and capable of repopulating both phenotypes. Studies of other phenotypes and cell types of interest may benefit from the unique information provided by the pheno-scRNAseq approach.
Results
BRCA cells exhibit heterogeneous migration phenotypes
MDA cells embedded in 3D COL1 matrix take on at least two distinct collective phenotypes, which develop from single cells over the course of 7 days (Figure 1A). The majority of cells, approximately 81%, formed collectively invasive cell structures (Figure 1B), a morphology characterized by a low circularity index (Figure 1D). A smaller subset of tumor cells, approximately 19%, did not invade (Figure 1C), characterized by a high circularity index (Figure 1D). Based on their morphology, we termed the invasive structures as “networks” and the non-invasive structures as “spheroids.” Confocal microscopy revealed that collectively invading networks were tightly packed with cells (Figure 1B) while spherical structures were capable of forming hollow lumens reminiscent of normal breast epithelial acini (Figure 1C). Similar phenotypes were observed for mouse 4T1 mammary carcinoma cells embedded in 3D COL1 matrix (Figure 1E).
Figure 1.
BRCA cells exhibit heterogeneous migration phenotypes
(A) Representative brightfield image of MDA-MB-231 cells cultured in a 3D COL1 matrix after 7 days of culture. Scale bar, 200 μm.
(B and C) (B) Confocal z-slice of the network and (C) spheroid phenotypes. Scale bar, 100 μm.
(D) Quantification of the circularity of heterogeneous collective phenotypes (n = 25).
(E) Similar phenotypes are observed in 4T1 cells cultured in 3D type I collagen.
(F and G) Time-lapse microscopy depicting the different patterns of growth and morphogenesis of two structurally distinct multicellular phenotypes. (F) Single cells that eventually develop into networks display growth and migration that lead to eventual fusion into a multicellular network. (G) Single cells that eventually develop into spherical structures display localized growth and development with continual maintenance of the spherical shape. Scale bar, 50 μm.
(H) Quantification of the invasion of cells into the local extracellular matrix (ECM) depending on their collective phenotype (n = 13).
(I) Maximum invasion of each phenotype from the initial seeding point after 60 hr of culture (n = 13).
(J and K) Representative immunofluorescence z-slice images of networks (J) and spheroids (K) stained for COL4A1 and LAM5. Scale bar is 20 μm. Data are represented as mean ± standard error of the mean (SEM). Statistical significance was determined by the Student's t-test and is indicated as ∗, ∗∗, and ∗∗∗ for p ≤ 0.05, p ≤ 0.01, and p ≤ 0.001, respectively.
Time-lapse microscopy revealed that cells that developed into network structures (Video S1) began as single cells undergoing uniaxial elongation at early time points (Figure 1F). Proliferation appeared to support the eventual creation of a smooth, continuous, collectively invasive clonal network (Figure 1F). Cells that developed into spherical structures did not elongate at early time points (Figure 1G and Video S2). These cells maintained a predominantly rounded morphology. In this case, proliferation enabled the eventual creation of a smooth, rounded spheroid (Figure 1G).
The collective phenotypes also displayed differential aptitudes for invading into the local matrix. The network structures were significantly more invasive than the spheroids, displaying increased spreading away from the initial seeding point over the course of several days (Figure 1H). By 2.5 days, the cells that composed network structures invaded roughly three times farther into their surroundings compared to cells in spheroid structures (Figure 1I).
Since the formation of hollow acini by normal breast epithelial cells has previously been linked to basement membrane deposition (Wang et al., 2013), we assessed whether MDA cancer cells deposited matrix proteins into the COL1 microenvironment. Immunofluorescent staining revealed that both invasive and non-invasive subpopulations deposited their own cell-derived matrix consisting of basement membrane proteins laminin-5 (LAM5) and type IV collagen (COL4A1) (Figures 1J and 1K). COL4A1 appeared to be localized more toward the cell-extracellular matrix interface in spheroids compared to invasive networks, where localization was more heterogeneous and intracellular. For LAM5, expression in spheroids was low, while invasive networks displayed heterogeneous, intracellular expression.
Phenotypic cell sorting improves transcriptome-phenotype coupling
To begin to identify the mechanisms underlying collective migration heterogeneity in MDA cells, we next sought to separate the network cells from the spheroid cells and analyze their molecular differences. Morphologically, these invasive and non-invasive tumor cell subpopulations could be clearly differentiated based on their circularity (Figure 1D), so we subsequently used circularity as a metric by which we distinguished the two phenotypes. To sort the cells based on their phenotype for direct scRNAseq analysis, we devised a technique that enables fluorescent tagging of cells of interest (Figure 2A). Building on a method recently described by Konen et al., 2017, MDAs were first transduced with Dendra2-Lifeact (MDA-Dendra). Dendra is a photoconvertible protein that changes from green to red fluorescence upon stimulation with a 405-nm laser, facilitating targeted red fluorescent tagging of specific cells. MDA-Dendra cells were embedded sparsely and cultured in COL1 matrices for one week, allowing the development of clonal collective structures. Transduction with Dendra2-Lifeact did not significantly impact collective cell migration capabilities compared to wild-type MDA cells (Velez et al., 2017). To enable higher throughput photoconversion of each cell phenotype of interest while maintaining fine spatial resolution to target individual cell structures (see Transparent methods for details), we constructed a custom widefield microscope with a galvanometer scanner and laser power source. In one experiment, spheroid structures were selectively stimulated with the 405 nm laser, inducing red fluorescence, while unexposed cells continued to fluoresce green (Figure 2B). In a separate experiment and sample, network cell structures were similarly selectively photoconverted. Subsequent rapid digestion of the matrices and sorting of the cells by FACS enabled recovery of phenotypically pure populations for direct molecular analysis (Figures 2C and 2D).
Figure 2.
Phenotypic cell sorting improves transcriptome-phenotype coupling
(A) Schematic overview of our workflow for phenotypic cell sorting. MDA-Dendra cells are cultured in type I collagen, photoconverted, released from the matrix, and sorted based on red fluorescence for immediate scRNAseq or other downstream experiments.
(B) Images of multicellular MDA-Dendra structures before photoconversion (left) and after photoconversion (right). Scale bar, 100 μm.
(C) Fluorescent profile of a control gel, where no cells were photoconverted.
(D) Fluorescent profile of cells released from a gel after photoconversion. A fraction of cells exhibits greater red fluorescence compared to the control.
(E) A UMAP plot generated from the pooled transcriptomic signatures of the cells isolated by phenotypic cell sorting.
(F) Clusters identified based on unsupervised clustering methods.
(G) Clusters labeled by phenotype.
(H and I) Comparison of cells labeled by photoconversion (H) with non-photoconverted cells (I) that were scored by the metagene derived from the differentially expressed genes of the labeled cells. Data were corrected for batch effects and sequencing depth prior to UMAP projection.
Network and spheroid cells isolated by photoconversion were directly subjected to scRNAseq. A Uniform Manifold Approximation and Projection (UMAP) plot of the scRNAseq data from both cell phenotypes aggregated together was constructed (Figure 2E). Conventionally, unsupervised clustering would be used to identify two transcriptionally distinct subpopulations of cells (Figure 2F). However, with our phenotypic labels applied to the data (Figure 2G), it was evident that agnostic transcriptional clusters were not predictive of the functional cell migration phenotypes (compare Figures 2F, 2G, and S1). Even though there was a significant enrichment of network cells in the larger cluster (cluster 0) and of spheroid cells in the smaller cluster (cluster 1) (Fisher's exact test: odds ratio = 12.51, p value = 5.37 × 10−62), there was still a high number of spheroid cells that clustered together with network cells. Analysis on the basis of our phenotypic labels revealed a set of 178 genes that were differentially expressed (DE) between network and spheroid cells (Table S1), whereas analysis on unsupervised clusters highlighted a set of 528 differentially expressed genes (DEGs) (data not shown). Importantly, 70 of the genes identified using phenotypic labeling were not identified using unsupervised clustering (Table S1, highlighted genes).
To control for the effects of our labeling and sorting process, we also sequenced MDA-Dendra cells extracted from COL1 matrices that were not stimulated or sorted. Integrating the sorted and non-sorted cell data sets by normalizing for sequencing depth and correcting for batch effects demonstrated that both have a similar data structure (Figures 2H and 2I). We then constructed a metagene from the 178 DEGs between the network and spheroid cells and applied this metagene to score and label the non-sorted cells. The 178 gene signature placed phenotypic labels on the non-sorted cells in a similar state space compared to their sorted counterparts (Figures 2H and 2I), adding further evidence that stimulation and sorting did not significantly change the transcriptional profiles of the cells. We also investigated the DEGs between the sorted and non-sorted cells, which represent some combination of sequencing batch effects and photoconversion/sorting effects. This revealed 1127 DEGs, of which only 8 were associated with a response to UV. Further analysis showed that only 2 of these 8 UV response-associated genes were contained in our list of 178 DEGs that differentiated the network and spheroid cells. This suggested that the effects of the photoconversion and sorting process are negligible for our analysis.
Biological processes that differentiate collective cell phenotypes are conserved
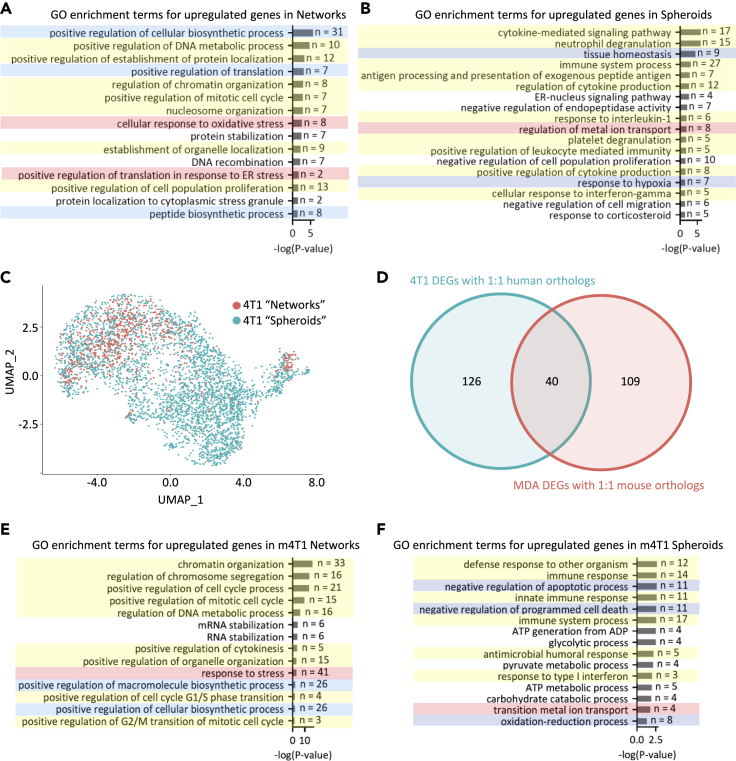
Gene expression analysis based on phenotypic labels revealed that 101 of the 178 DE genes were significantly upregulated in invasive network cells compared to non-invasive spheroid cells. Gene ontology (GO) analysis of these genes identified several enriched processes, including positive regulation of biosynthesis, translation in response to endoplasmic reticulum (ER) stress, response to oxidative stress, and positive regulation of proliferation (Figure 3A). Interestingly, the remaining 77 genes upregulated in the spheroid cells were enriched for several immunomodulatory processes, negative regulation of proliferation and migration, transcriptional regulation through ER-nucleus signaling pathways, and processes promoting homeostasis, including redox equilibrium (Figure 3B).
Figure 3.
Biological processes that differentiate collective cell phenotypes are conserved
(A) Highlighted significant GO enrichment terms based on the list of upregulated genes in the network cell population.
(B) Highlighted significant GO enrichment terms based on the list of upregulated genes in the spheroid cell population.
(C) UMAP of 4T1 mouse cells, labeled after scoring with the metagene derived from the DEGs between the MDA network and spheroid cells.
(D) Overlap between the 4T1 DEGs between labeled networks and spheroids with 1:1 human orthologs and the MDA DEGs between the MDA networks and spheroids with 1:1 mouse orthologs.
(E) Highlighted significant GO enrichment terms based on the list of upregulated genes in the labeled 4T1 network cell population. Similarly colored highlights between (A) and (E) denote similarly themed processes that were enriched.
(F) Highlighted significant GO enrichment terms based on the list of upregulated genes in the labeled 4T1 spheroid cell population. Similarly colored highlights between (B) and (F) denote similarly themed processes that were enriched.
To determine whether our findings extended to other BRCA cells, we performed scRNAseq on 4T1 murine mammary carcinoma cells. As shown in Figures 1E, 4T1s take on very similar migration phenotypes as MDAs, forming network and spheroid structures over seven days. UMAP projection and labeling by metagene scoring reveal a similar pattern to the MDA cells, where the spheroid phenotype is the predominant phenotype in the smaller cluster, while the network phenotype occupies the state space farthest from the lone cluster (Figure 3C). Differential expression analysis revealed 166 DEGs that describe the differences between scored 4T1 networks and spheroids with 1:1 human orthologs. When compared to the 149 MDA DEGs that had 1:1 mouse orthologs, 40 genes are shared between the mouse and human data sets (Figure 3D). This overlap was statistically significant (Fisher's exact test: odds ratio = 33.92, p value = 6.50 × 10−41). GO enrichment analyses of the murine cells also reveal similarly themed biological processes being enriched compared to their human counterparts, as shown in Figures 3E and 3F.
Pheno-scRNAseq provides unique and selective information
Since pheno-scRNAseq identified a smaller set of DEGs (178) than unsupervised scRNAseq (528), we hypothesized that phenotypic labeling may provide a more selective and more functionally relevant gene set than analyses on populations identified from unsupervised clustering. To further explore this idea, we asked whether certain biological processes were uniquely enriched by phenotype labeling or consistently enriched between supervised and unsupervised analyses. Such differences could provide important guidance for prioritizing gene modules to target in functional studies aimed at identifying meaningful associations with migration behaviors. In Figure S2, we show a Venn diagram comparing the detected DEGs using supervised pheno-scRNAseq analysis to those found using unsupervised scRNAseq analysis. We also display a Venn diagram comparing the significant GO enrichment terms detected using DEGs from supervised pheno-scRNAseq analysis to those found using DEGs from unsupervised scRNAseq analysis. A detailed list of the GO enrichment analysis is provided in Table S2. These plots reveal two distinct features of supervised analysis compared to unsupervised analysis. First, for our data set, supervised analysis narrows down the number of DEGs and GO enrichment terms that are found to be statistically significant (adjusted p value < 0.05, FDR <5%). The second feature can be seen visually in the left most partition in each Venn diagram. These are DEGs and GO terms that were uniquely detected using supervised analysis and not in unsupervised analysis. Together, these results suggest that supervised analysis is indeed more selective and provides unique information.
Invasive network cells are more proliferative and more sensitive to chemotherapy treatment
We set out to validate that gene modules identified by supervised analysis were functionally relevant. Of the GO terms that were uniquely identified by supervised analysis, we focused on “negative regulation of cell population proliferation” (GO:0008285, Figure 4A). Expression of this group of genes was higher in the spheroid population compared to the network population of cell. Furthermore, MKI67 (Ki-67), a known marker of proliferation, was uniquely identified as differentially regulated by supervised analysis (Table S1) and higher expressed in the network subpopulation. Immunostaining and quantification of Ki-67 showed that the percentage of actively proliferating cells was significantly higher in invasive networks (Figures 4B–4E). Cell cycle scoring on the transcriptional markers reveal that the spheroid subpopulation also occupies multiple cell cycle states and has not simply exited the cell cycle (Figure S3). Intriguingly, 34% of invasive networks contained leader cells that were Ki-67 positive (Figures 4C and 4F), suggesting that tip cells can be both invasive and proliferative (Figure 4C). The remaining 66% of networks had Ki-67-positive cells located randomly throughout the network (Figure 4F). Spherical structures were less proliferative, and many exhibited no staining for Ki-67 (Figures 4D and 4E). These results support our suggestion that supervised analysis provides distinct and important information that can effectively guide follow-up experiments compared to unsupervised analysis.
Figure 4.
Invasive network cells are more proliferative
(A) A heatmap of the list of genes detected by phenotypically guided DE analysis that are in the GO:0008285 term. Spheroids display upregulation of genes associated with “negative regulation of cell population proliferation”.
(B and C) Immunofluorescent staining of Ki-67 in the network cell population. Scale bar, 50 μm. (C) Some network structures display Ki-67 staining at the tips of the structures.
(D) Immunofluorescent staining of Ki-67 in the spheroid cell population. Many spheroids displayed no staining. Scale bar, 50 μm.
(E) Quantification of the percentage of cells in each collective phenotype that stained positively for Ki-67 (n = 36).
(F) Quantification of the percent of networks that had a tip cell which stained positively for Ki-67 (n = 36).
(G and H) Brightfield and fluorescence images after treatment with paclitaxel of spheroids (G) and networks (H). Scale bar is 20 μm.
(I) Quantification of cell death after treatment with paclitaxel (n = 13). Spheroids show a statistically significant decrease in sensitivity compared to networks.
Data are represented as mean ± standard error of the mean (SEM). For (E) and (F), statistical significance was determined by the Student's t-test and is indicated as ∗, ∗∗, and ∗∗∗ for p ≤ 0.05, p ≤ 0.01, and p ≤ 0.001, respectively. For (I), statistical significance was determined by analysis of variance (ANOVA) followed by post-hoc analyses (Tukey) and is indicated as ∗, ∗∗, and ∗∗∗ for p ≤ 0.05, p ≤ 0.01, and p ≤ 0.001, respectively.
To further validate the functional relevance of this difference in proliferative state, we performed a cytotoxicity assay with a widely used chemotherapy drug that acts on the cell division process: paclitaxel. Paclitaxel stabilizes tubulin polymerization, which results in cell cycle arrest (Weaver, 2014). We hypothesized that the spheroid cells, which express relatively lower Ki-67 and exhibit less Ki-67 staining, should be less sensitive to this drug. We cultured MDAs in 3D COL1 for one week, followed by three days of drug treatment with either 1 μM paclitaxel or vehicle control. Figures 4G and 4H show representative images of dead cell staining in treated networks versus spheroids. Representative images of vehicle control cells are shown in Figure S4. Cell death was significantly greater in drug-treated conditions compared to vehicle, and network cells died more than the spheroid cells (Figures 4I and S4). These data further confirm that the spheroid subpopulation is less proliferative and more resistant to chemotherapy (Figure 4). Taken together, these experiments serve as validation of the functional relevance of a biological process that was uniquely identified by supervised analysis, not by unsupervised analysis.
Functional validation of additional processes identified by pheno-scRNAseq
Of the 77 genes upregulated in the spheroid cells compared to network cells, GO analysis indicated significant enrichment of several immunomodulatory cellular processes (Figure 3B), including components of the innate, adaptive, and cytokine immune signaling machinery. A heatmap of the DEGs corresponding to the GO term “immune system process” (GO: 0002376) is shown Figure 5A. In particular, human leukocyte antigen class I (HLA-I) gene expression was significantly upregulated in spheroid cells and downregulated in network cells (Table S1). Unexpectedly, immunofluorescence staining of HLA-A revealed distinct patterns of localization in each cell phenotype (Figures 5B and 5C). Spheroid cells appeared to properly localize HLA-A to their plasma membrane (Figure 5B), whereas invasive cells seemed to localize HLA-A near the nucleus (Figure 5C). Quantification of this staining pattern demonstrated that spheroid cells had approximately three-fold less perinuclear co-localization of HLA-A compared to network cells (Figure 5D).
Figure 5.
Spheroid cells display proper antigen localization
(A) A heatmap of the list of genes detected by phenotypically guided DE analysis that are in the GO:0002376 term. Spheroids display upregulation of genes associated with “immune system process”.
(B) Immunofluorescent staining of HLA-A in the spheroid cell population. Spheroid cells display membrane localization of HLA-A. Scale bar, 50 μm.
(C) Immunofluorescent staining of HLA-A in the network cell population. Many network cells display perinuclear staining of HLA-A. Scale bar, 50 μm.
(D) Quantification of the perinuclear staining of HLA-A within each collective phenotype (n = 8). Data are represented as mean ± standard error of the mean (SEM). Statistical significance was determined by the Student's t-test and is indicated as ∗, ∗∗, and ∗∗∗ for p ≤ 0.05, p ≤ 0.01, and p ≤ 0.001, respectively.
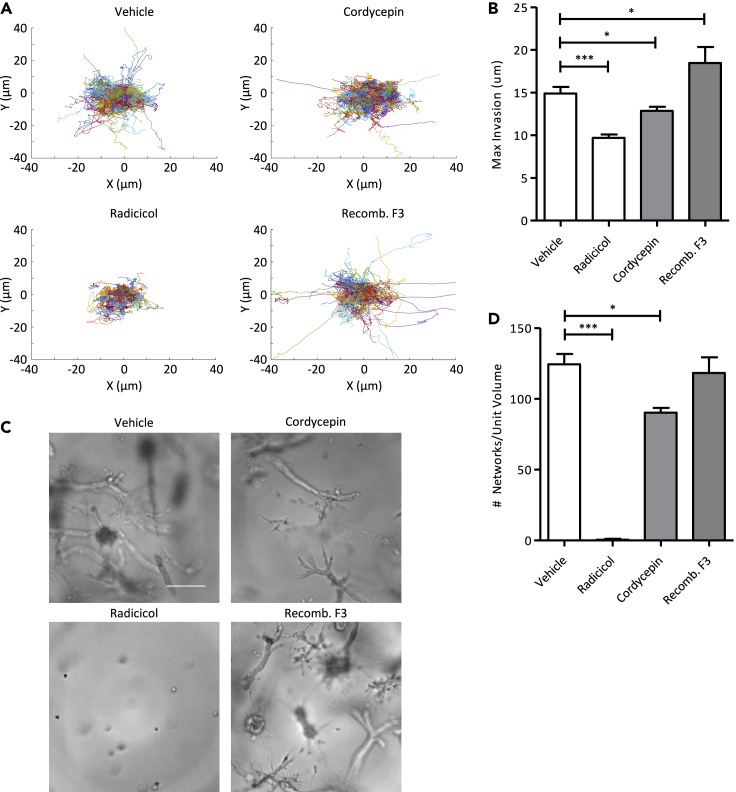
Of the 101 genes upregulated in the network cells compared to the spheroid cells, we selected three targets for functional studies: HSP90AB1, DEK, and F3. This was motivated by the fact that GO enrichment analysis based on supervised DEGs identified many stress response terms, many of which contained HSP90AB1 or F3 (Table S1). In addition, several GO enrichment terms identified were associated with the regulation of gene expression to which DEK is functionally associated. Based on their upregulation in networks, we hypothesized that their inhibition would negatively impact cell invasion and/or network formation, while their activation would positively promote cell invasion or network formation. Indeed, this is what we observed. Inhibition of HSP90AB1 by radicicol and DEK by cordycepin significantly reduced the invasive potential of MDA cells and also reduced the rate of formation of collectively invasive networks (Figure 6). Conversely, supplementing the culture media with recombinant F3, which was upregulated in the network cells, increased the invasiveness of MDA cells (Figures 6A and 6B). However, adding recombinant F3 did not increase the rate of network formation (Figures 6C and 6D). Thus, pheno-scRNAseq supervised gene expression analysis successfully predicted the roles that several DEGs played in regulating cell migration behavior.
Figure 6.
Inhibition of upregulated genes in the network phenotype reduces invasion
(A) Spider plots of cell trajectories during drug treatment.
(B) Quantification of the maximum invasion of cells within each drug condition (n = 59). Inhibition of the upregulated genes in the network phenotype reduced invasion. Adding recombinant F3, an upregulated network gene, increased invasion.
(C) Representative brightfield images of the cells after 7 days of drug treatment. Scale bar is 200 μm.
(D) Quantification of the number of network structures after 7 days of drug treatment (n = 18). Inhibition of the upregulated network genes reduced the formation of network structures. Data are represented as mean ± standard error of the mean (SEM). Statistical significance was determined by analysis of variance (ANOVA) followed by post-hoc analyses (Tukey) and is indicated as ∗, ∗∗, and ∗∗∗ for p ≤ 0.05, p ≤ 0.01, and p ≤ 0.001, respectively.
Phenotypic sorting enables analysis of phenotype stability
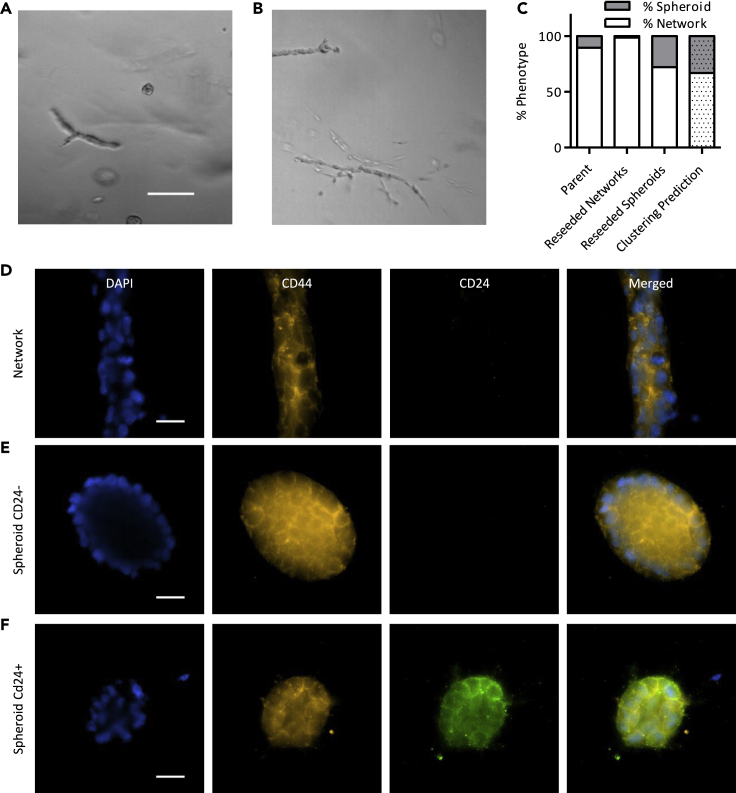
Since cells that were labeled as belonging to the spheroid phenotype fell into two distinct transcriptional clusters based on unsupervised analysis, we wondered if the spheroid cells could be phenotypically unstable. To determine the phenotypic stability of MDAs that formed spheroids, we labeled, sorted, and re-embedded spheroid cells as single cells (Figure 7A). This resulted in ~75% of the spheroid cells converting into network cells (Figure 7C). Conversely, sorting and re-embedding invasive network cells as single cells resulted in near 100% invasive network formation (Figures 7B and 7C). This suggests that spheroid cells retain the capacity to switch into a proliferative and invasive state, but network cells are less capable of reverting into a non-proliferative, non-invasive state. To assess whether one of these states represented a more cancer stem cell-like state, we immunostained for CD24 and CD44. Both phenotypes display a CD44 + profile (Figures 7C and 7D). However, CD24 staining was negative for the network cells (Figure 7C) and mixed for the spheroid cells (Figures 7D and 7E). A positive control was done for CD24 on Nalm6 cells to confirm the absence of CD24 staining (Figure S5). When comparing the spheroid population in the smaller lone cluster to the ones in the larger cluster, we found a list of surface markers that could potentially be used to sort the two subpopulations in future experiments (Table S3).
Figure 7.
Phenotypic sorting enables analysis of phenotype stability
(A) Representative brightfield image of reseeded spheroid cells after 7 days of culture in 3D type I collagen. Scale bar, 50 μm.
(B) Representative brightfield image of reseeded network cells after 7 days of culture in 3D type I collagen.
(C) Quantification of the phenotypes that arise after reseeding from sorted populations (n = 18). Invasive network cells largely reform network structures, while non-invasive spheroids may either form network or spheroid structures.
(D) CD44 immunostaining of network cells.
(E) CD44 staining of spheroid cells that are CD24 negative.
(F) CD44 staining of spheroid cells that are CD24 positive.
Discussion
Our study demonstrates that unsupervised transcriptional clustering analysis does not necessarily separate cells according to specific functions, and thus may represent lost opportunities to decipher functional intra-tumor heterogeneity. Precision sorting prior to scRNA-seq, as implemented here via Dendra2-based phenotype marking, can help overcome limitations of unsupervised clustering. Analysis based on unsupervised clustering resulted in the identification of 528 DEGs, whereas analysis based on phenotypic labeling of each cell subpopulation resulted in the identification of 178 DEGs, and only 108 genes overlapped. Thus, roughly 80% of the DEGs identified by unsupervised clustering were not specific to the observed cell phenotypes. Importantly, phenotypic labeling allowed for the identification of 70 unique DEGs that were not detected by unsupervised clustering. We validated the functional relevance of one of these genes, Ki-67. This suggests that phenotypic labeling allows for a more direct and relevant approach to investigating the molecular regulators of functional heterogeneity within a given cell type. However, the results of our sorting and reseeding experiments could support the view that traditional unsupervised clustering analysis is more likely to give information about how cells could behave, rather than how they are currently behaving (Tanay and Regev, 2017).
Phenotypically supervised scRNAseq based on the invasion phenotype of BRCA cells enabled us to gain a system-level view of distinct cancer cell states. Compared to confocal microscopy-based approaches, our setup significantly increased the number of cells that can be labeled and sorted within the limited time frame before photoconverted protein turnover. A previous study was only able to photoconvert and sort tens of cells (Konen et al., 2017), demanding growth-based amplification of cells prior to omics analyses. Given the plasticity and rapid adaptation of cancer cells in response to changes in their environment, which occurs on short timescales that cannot be explained by genetic evolution or clonal selection (Sharma et al., 2010; Shaffer et al., 2017), the requirement for cell amplification could skew omics data and complicate our ability to directly link omics data to phenotypes. Our custom setup enabled us to photoconvert thousands of cells, resulting in a ten-fold increase in throughput while maintaining high spatial precision through the optimization of the photoconversion parameters. This higher throughput approach powered our statistical analysis, enabling us to directly compare the differences between unsupervised and phenotypically supervised scRNAseq, and led us to discover distinct biological processes that are linked to specific migration states. Both our transcriptional and functional evidence suggests that collective invasion is associated with anabolic metabolism, proliferation, redox stress, and ER stress. Conversely, collective acini formation is associated with lower levels of proliferation, quality control mechanisms, homeostasis, and immunomodulatory functions. Such knowledge may provide opportunities to design multiplexed therapeutic cocktails that take advantage of the susceptibilities of each complex cancer phenotype.
In conclusion, the field of single-cell analysis is rapidly moving toward integrative, multi-scale measurements to improve the interpretability and actionable value of single-cell data. Our approach to enabling phenotypically supervised scRNAseq of BRCA cells reveals specific stress response and immunologic cellular processes that are coordinated with invasive and non-invasive phenotypes. Further, our 3D culture system and phenotypic cell sorting approach enable in vitro modeling of key aspects of phenotypic plasticity, which may provide a useful platform for the mechanistic dissection of these processes and identification of strategies that could effectively treat heterogeneous tumors. Understanding this plasticity may reveal ways to selectively target seemingly non-proliferative, non-invasive tumor cells while leaving quiescent noncancerous cells (such as normal stem cells) unharmed. For example, given the plasticity of the non-proliferative spheroid cells, it may be possible to identify mechanisms capable of sensitizing them to chemotherapies or preventing a phenotypic switch to a more invasive, more proliferative state. Our system also offers the opportunity to dissect the mechanisms underlying tumor cell plasticity in the expression and localization of MHC class I molecules, which could inform therapeutic strategies complementary to immunotherapies.
Limitations of the study
While our pheno-scRNAseq method improves on prior methods for assessing within-cell-type heterogeneity of BRCA cells by providing higher throughput and additional metadata for supervised analysis, many questions remain. Several of the biological processes that were associated with migration phenotypes were not explored functionally here, and the in vivo relevance of our findings remains to be determined. Future work will address these concerns by translating these experiments into mouse BRCA cell lines for syngeneic mouse studies.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Stephanie I. Fraley (sifraley@ucsd.edu).
Material availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and code availability
The accession number for the sequencing data reported in this paper is Gene Expression Omnibus: GSE158844.
Code generated during this study are available from the Lead Contact upon request.
Methods
All methods can be found in the accompanying Transparent methods supplemental file.
Acknowledgments
We would like to thank Khoi Le for experimental assistance in the development of pheno-scRNAseq. We would like to acknowledge the Human Embryonic Stem Cell Core Facility at UCSD for technical support with flow cytometry, the IGM Genomic Core for assistance with single-cell RNAseq experimentation, and the UCSD School of Medicine Microscopy Imaging Core at UCSD for microscopy support. This work was supported by a Burroughs Wellcome Fund Career Award at the Scientific Interface to S.I.F. (1012027), NSF CAREER Award to S.I.F. (1651855), ACS Institutional Research Grant (15-172-45-IRG) provided through the Moores Cancer Center, NCI R01 (NCI 5R01CA238042), and funding supporting the UCSD School of Medicine Microscopy Core (NS047101). This publication includes data generated at the UC San Diego IGM Genomics Center utilizing an Illumina NovaSeq 6000 that was purchased with funding from a National Institutes of Health SIG grant (#S10 OD026929). All relevant data are available within the article and supplementary files or from the corresponding author upon request.
Authors contribution
K.C. and S.I.F. designed the experiments. K.C. performed most experiments and data analysis. K.O. and H.C. assisted with scRNAseq data analysis. R.L.C. conducted matrix deposition staining. J.S., S.M., and W.C. assisted with cell tracking data. Data interpretation was conducted by K.C., K.O., H.C., and S.I.F. All authors contributed to writing or editing the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: January 22, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2020.101991.
Supplemental information
A positive log (fold change) indicates higher expression in the network subpopulation, while a negative log (fold change) indicates higher expression in the spheroid subpopulation. Highlighted genes were identified exclusively by using phenotypic labels.
The false discovery rate is reported based on the input of differentially expressed genes obtained from phenotypic clusters (Supervised Analysis) and graph-based clusters (Unsupervised Analysis).
References
- Aceto N., Bardia A., Miyamoto D.T., Donaldson M.C., Wittner B.S., Spencer J.A., Yu M., Pely A., Engstrom A., Zhu H. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158:1110–1122. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beri P., Popravko A., Yeoman B., Kumar A., Chen K., Hodzic E., Chiang A., Banisadr A., Placone J.K., Carter H. Cell adhesiveness serves as a biophysical marker for metastatic potential. Cancer Res. 2020;80:901–911. doi: 10.1158/0008-5472.CAN-19-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell C.R., Palasantza A., Jiang X., Berens P., Deng Q., Yilmaz M., Reimer J., Shen S., Bethge M., Tolias K. Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq. Nat. Biotechnol. 2016;34:199–203. doi: 10.1038/nbt.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell L., Russell A.J.C., Voet T. Single-cell (Multi)omics technologies. Annu. Rev. Genomics Hum. Genet. 2018;19:15–41. doi: 10.1146/annurev-genom-091416-035324. [DOI] [PubMed] [Google Scholar]
- Cheung K.J., Ewald A.J. A collective route to metastasis: seeding by tumor cell clusters. Science. 2016;352:167–169. doi: 10.1126/science.aaf6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K.J., Gabrielson E., Werb Z., Ewald A.J. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell. 2013;155:1639–1651. doi: 10.1016/j.cell.2013.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K.J., Padmanaban V., Silvestri V., Schipper K., Cohen J.D., Fairchild A.N., Gorin M.A., Verdone J.E., Pienta K.J., Bader J.S. Polyclonal breast cancer metastases arise from collective dissemination of keratin 14-expressing tumor cell clusters. Proc. Natl. Acad. Sci. U S A. 2016;113:E854–E863. doi: 10.1073/pnas.1508541113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.H., Kim J.K. Dissecting cellular heterogeneity using single-cell RNA sequencing. Mol. Cells. 2019;42:189–199. doi: 10.14348/molcells.2019.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit A., Parnas O., Li B., Chen J., Fulco C.P., Jerby-Arnon L., Marjanovic N.D., Dionne D., Burks T., Raychowdhury R. Perturb-seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell. 2016;167:1853–1866.e17. doi: 10.1016/j.cell.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan M., Hao J., Cui S., Worthley D.L., Zhang S., Wang Z., Shi J., Liu L., Wang X., Ke A. Diverse modes of clonal evolution in HBV-related hepatocellular carcinoma revealed by single-cell genome sequencing. Cell Res. 2018;28:359–373. doi: 10.1038/cr.2018.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimbergen A.J., Siebring J., Solopova A., Kuipers O.P. Microbial bet-hedging: the power of being different. Curr. Opin. Microbiol. 2015;25:67–72. doi: 10.1016/j.mib.2015.04.008. [DOI] [PubMed] [Google Scholar]
- Hinohara K., Wu H.J., Sebastien V., McDonald T.O., Igarashi K.J., Yamamoto K.N., Madsen T., Fassl A., Egri S.B., Papanastasiou M. KDM5 histone demethylase activity links cellular transcriptomic heterogeneity to therapeutic resistance. Cancer Cell. 2018;34:939–953.e9. doi: 10.1016/j.ccell.2018.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaitin D.A., Weiner A., Yofe I., Lara-Astiaso D., Keren-Shaul H., David E., et Salame T.M., Tanay A., van Oudenaarden A., Amit I. Dissecting immune circuits by linking CRISPR-pooled screens with single-cell RNA-seq. Cell. 2016;167:1883–1896 e15. doi: 10.1016/j.cell.2016.11.039. [DOI] [PubMed] [Google Scholar]
- Kester L., van Oudenaarden A. Single-cell transcriptomics meets lineage tracing. Cell Stem Cell. 2018;23:166–179. doi: 10.1016/j.stem.2018.04.014. [DOI] [PubMed] [Google Scholar]
- Kiselev V.Y., Andrews T.S., Hemberg M. Challenges in unsupervised clustering of single-cell RNA-seq data. Nat. Rev. Genet. 2019;20:273–282. doi: 10.1038/s41576-018-0088-9. [DOI] [PubMed] [Google Scholar]
- Konen J., Summerbell E., Dwivedi B., Galior K., Hou Y., Rusnak L., Chen A., Saltz W., Zhou L.H., Vertino P. Image-guided genomics of phenotypically heterogeneous populations reveals vascular signalling during symbiotic collective cancer invasion. Nat. Commun. 2017;8:15078. doi: 10.1038/ncomms15078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein E., Borm L.E., Linnarsson S. The promise of spatial transcriptomics for neuroscience in the era of molecular cell typing. Science. 2017;358:64–69. doi: 10.1126/science.aan6827. [DOI] [PubMed] [Google Scholar]
- Shaffer S.M., Dunagin M.C., Torborg S.R., Torre E.A., Emert B., Krepler C., Beqiri M., Sproesser K., Brafford P.A., Xiao M. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature. 2017;546:431–435. doi: 10.1038/nature22794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S.V., Lee D.Y., Li B., Quinlan M.P., Takahashi F., Maheswaran S., McDermott U., Azizian N., Zou L., Fischbach M.A. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabassum D.P., Polyak K. Tumorigenesis: it takes a village. Nat. Rev. Cancer. 2015;15:473–483. doi: 10.1038/nrc3971. [DOI] [PubMed] [Google Scholar]
- Tanay A., Regev A. Scaling single-cell genomics from phenomenology to mechanism. Nature. 2017;541:331–338. doi: 10.1038/nature21350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velez D.O., Tsui B., Goshia T., Chute C.L., Han A., Carter H., Fraley S.I. 3D collagen architecture induces a conserved migratory and transcriptional response linked to vasculogenic mimicry. Nat. Commun. 2017;8:1651. doi: 10.1038/s41467-017-01556-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Lacoche S., Huang L., Bin X., Muthuswamy S. Rotational motion during three-dimensional morphogenesis of mammary epithelial acini relates to laminin matrix assembly. Proc. Natl. Acad. Sci. U S A. 2013;110:163–168. doi: 10.1073/pnas.1201141110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver B.A. How Taxol/paclitaxel kills cancer cells. Mol. Biol. Cell. 2014;25:2677–2681. doi: 10.1091/mbc.E14-04-0916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Bojorquez-Gomez A., Velez D.O., Xu G., Sanchez K.S., Shen J.P., Chen K., Licon K., Melton C., Olson K.M. A global transcriptional network connecting noncoding mutations to changes in tumor gene expression. Nat. Genet. 2018;50:613–620. doi: 10.1038/s41588-018-0091-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A positive log (fold change) indicates higher expression in the network subpopulation, while a negative log (fold change) indicates higher expression in the spheroid subpopulation. Highlighted genes were identified exclusively by using phenotypic labels.
The false discovery rate is reported based on the input of differentially expressed genes obtained from phenotypic clusters (Supervised Analysis) and graph-based clusters (Unsupervised Analysis).
Data Availability Statement
The accession number for the sequencing data reported in this paper is Gene Expression Omnibus: GSE158844.
Code generated during this study are available from the Lead Contact upon request.