Abstract
Background
Two hyphomycetous species were collected from leaves of Smilax china (Liliales, Smilacaceae) and Cremastra appendiculata (Asparagales, Orchidaceae). ITS barcoding indicated that they belong to the genus Zasmidium.
New information
Morphological data in combination with molecular phylogenetic analyses based on ITS, LSU and rpb2 confirmed that our Chinese strains represented a new species, Zasmidium liboense and a new record of Z. citri-griseum.
Keywords: one new species, asexual morph, Dothideomycetes , Mycosphaerellaceae , taxonomy
Introduction
The fungi of southern Asian are extremely diverse (Hyde et al. 2018, Cheek et al. 2020). During a survey of fungal diversity in ornamental plants in south-western China from 2017 to 2019, more than 2000 strains were obtained, which represented asexual morphs of both Ascomycota and Basidiomycota. Some new taxa were previously described by our research group as pathogens or endophytes (e.g. Liang et al. 2018, Long et al. 2019, Sun et al. 2020, Wijesinghe et al. 2020, Zhang et al. 2020).
The genus Zasmidium was established by Fries (1849) with Z. cellare (Pers.) Fr. as the type species. It is currently placed in the order Capnodiales within the Dothideomycetes (Hongsanan et al. 2020, Wijayawardene et al. 2020). Arzanlou et al. (2007) showed that Zasmidium was the oldest name for Stenella-like hyphomycetes within Mycosphaerellaceae, which are characterised by conidiogenous loci and conidia with truncate hila (Bensch et al. 2012). Hence, many former Stenella species were transferred to Zasmidium (Braun et al. 2010, Kamal 2010). Up to now, the number of accepted species in the genus is about 150 (Wijayawardene et al. 2020).
In this paper, we report on Zasmidium species found on medicinal plants in China. One new species (Zasmidium liboense) and one new Chinese record (Z. citri-griseum) are reported, based on evidence from morphology and molecular phylogeny.
Materials and methods
Samples collection and fungal strains isolation
The samples were collected in Xishuangbanna City, Yunnan Province, China. In order to obtain pure cultures, diseased leaf pieces of Smilax china (Liliales, Smilacaceae) and Cremastra appendiculata (Asparagales, Orchidaceae) were surface-disinfected following the method of Zhang et al. (2020). The strains were isolated using the single-spore method (Chomnunti et al. 2014). Colonies growing from single spores were transferred to potato-dextrose agar (PDA) and incubated at room temperature (28ºC). The holotype was deposited in the Herbarium of Department of Plant Pathology, Agricultural College, Guizhou University (HGUP). The ex-type cultures were deposited in the Culture Collection at the Department of Plant Pathology, Agriculture College, Guizhou University, P.R. China (GUCC) and the Mae Fah Luang University Culture Collection (MFLUCC) in Thailand.
Morphological description
Morphological culture characters were recorded after 2–3 weeks of growth on PDA. Microscopic slides were prepared in lactophenol. Light microscopy observations were made using a BX53 compound microscopy (Olympus, Tokyo, Japan) at 1000× magnification. The morphology was observed using a compound microscope (OLYMPUS BX53) showing all necessary details of morphology and ontogeny of reproductive propagules. Measurements were made of 30 structures for conidia, hila and conidiophores. The new species name was submitted to MycoBank (www.mycobank.org).
DNA extraction, PCR amplification and sequencing
Fungal cultures were grown on PDA at 28°C. When the whole Petri-dish (90 mm diam.) was nearly covered, fresh mycelia were scraped from the surface with sterilised scalpels. Genomic DNA was extracted using Fungus Genomic DNA Extraction Kit (Biomiga #GD2416, San Diego, California, USA) and following the manufacturer's instructions. PCR amplification of the internal transcribed spacer (ITS) region and the large subunit (LSU) of the ribosomal RNA gene was performed in a 25-μl reaction volume system as in Liang et al. (2018). Primers V9G and ITS4 (White et al. 1990, de Hoog and van den Ende 1998) were used to amplify the ITS and LSU1Fd and LR5 for the LSU (Vilgalys and Hester 1990, Crous et al. 2009). In addition, one protein-coding gene fragment, RNA polymerase II second largest subunit (rpb2), was amplified with the primers fRPB2-5F and fRPB2-7cR (Liu et al. 1999). Purification and sequencing of the PCR amplicons were undertaken by SinoGenoMax (Beijing, China). The resulting DNA sequences were submitted to NCBI GenBank (https://www.ncbi.nlm.nih.gov/genbank/) and their accession numbers are provided in Table 1.
Table 1.
Taxa used for molecular phylogenetic analyses and their GenBank accession numbers. (T) = ex-type strain.
| Species name | Strain number | GenBank Accession numbers | ||
| LSU | ITS | rpb2 | ||
| Zasmidium angulare | CBS 132094(T) = CPC 19042 = GA2 27B1a | JQ622096 | JQ622088 | MF951690 |
| Zasmidium anthuriicola | CBS 118742(T) | FJ839662 | FJ839626 | MF951691 |
| Zasmidium arcuatum | CBS 113477(T) | EU041836 | EU041779 | MF951692 |
| Zasmidium aucklandicum | CPC 13569 | MF951280 | MF951409 | MF951733 |
| Zasmidium biverticillatum | CBS 335.36 | EU041853 | EU041796 | – |
| Zasmidium cellare | CBS 146.36N(T) = ATCC 36951 = IFO4862 = IMI 044943 = LCP 52.402 = LSHBBB274 = MUCL 10089 | EU041878 | EU041821 | MF951693 |
| Zasmidium cerophillum | CBS 103.59(T) of Acrotheca cerophila = MUCL10034 | GU214485 | EU041798 | MF951694 |
| Zasmidium citri-griseum | CBS 122455 = CPC 15289 = X126 | KF902151 | KF901792 | MF951695 |
| GUCC 1507.3 | MT712179 | MT683372 | MT700485 | |
| Zasmidium commune | CBS 142530(T) | KY979820.1 | NR_156003.1 | – |
| Zasmidium corymbiae | CBS 145047(T) | NG_066279.1 | NR_161118.1 | MK047534.1 |
| Zasmidium daviesiae | CBS 116002 = VPRI 31767 | FJ839669 | FJ839633 | MF951698 |
| Zasmidium ducassei | BRIP 53367(T) | – | NR_164517.1 | – |
| Zasmidium elaeocarpi | CBS 142187(T) = CPC 16642 | MF951263 | MF951398 | MF951699 |
| Zasmidium eucalypticola | CBS 142186(T) = CPC 15149 | MF951265 | MF951400 | MF951701 |
| Zasmidium eucalyptorum | CBS 118500(T) = CPC 11174 | MF951266 | KF901652 | MF951702 |
| Zasmidium fructicola | CBS 139625(T) = CPC 24487 = ZJUM 80 | KP895922 | KP896052 | MF951703 |
| Zasmidium fructigenum | CBS 139626(T) = CPC 24471 = ZJUM 36 | KP895926 | KP896056 | MF951704 |
| Zasmidium grevilleae | CBS 124107(T) = CPC 14761 | FJ839670 | FJ839634 | MF951705 |
| Zasmidium gupoyu | CBS 122099 = RoKi 3022 | MF951267 | MF951401 | MF951706 |
| Zasmidium hakeae | CBS 142185(T) = CPC 15577 | MF951268 | MF951402 | MF951707 |
| Zasmidium hakeicola | CBS 144590(T) | NG_066335.1 | NR_163384.1 | MK442687.1 |
| Zasmidium indonesianum | CBS 139627(T) = CPC 15300 | KF902086 | KF901739 | MF951710 |
| Zasmidium iteae | CBS 113094(T) = RoKi 1279 | MF951271 | MF951405 | MF951711 |
| Zasmidium liboense sp. nov. | GUCC 1720.2 | MT712180 | MT683373 | MT700486 |
| Zasmidium lonicericola | CBS 125008(T) of Cladosporium lonicericola = CPC11671 | KF251787 | KF251283 | MF951712 |
| Zasmidium musae | CBS 121384 = CIRAD 41 = X877 | MF951272 | EU514292 | MF951713 |
| Zasmidium musae-banksii | CBS 121710(T) = X1100 | EU041852 | EU041795 | MF951716 |
| Zasmidium musicola | CBS 122479(T) = X1019 | MF951275 | EU514294 | MF951717 |
| Zasmidium musigenum | CBS 190.63 = MUCL 9557 | EU041857 | EU041800 | MF951718 |
| Zasmidium nocoxi | CBS 125009(T) = CPC 14044 | KF251788 | KF251284 | MF951719 |
| Zasmidium pitospori | CBS 122274 = ICMP 17098 | MF951276 | MF951406 | MF951720 |
| Zasmidium podocarpi | CBS 142529 | KY979821.1 | NR_156004.1 | – |
| Zasmidium proteacearum | CBS 116003 = VPRI 31812 | FJ839671 | FJ839635 | MF951721 |
| Zasmidium pseudoparkii | CBS 110999(T) = CPC 1087 | JF700965 | DQ303023 | MF951723 |
| Zasmidium pseudotsugae | rapssd | EF114704 | EF114687 | – |
| Zasmidium pseudovespa | CBS 121159(T) = AC0466 | KF901836 | MF951407 | MF951724 |
| Zasmidium queenslandicum | CBS 122475(T) = X1084 | MF951277 | EU514295 | MF951725 |
| Zasmidium scaevolicola | CBS 127009(T) = CPC 17344 | KF251789 | KF251285 | MF951726 |
| Zasmidium schini | CBS 142188(T) = CPC 19516 | MF951278 | MF951408 | MF951727 |
| Zasmidium sp. | CBS 118494 = CPC 11004 | MF951279 | DQ303039 | MF951728 |
| Zasmidium strelitziae | CBS 121711(T) = X1029 | EU041860 | EU041803 | MF951729 |
| Zasmidium suregadae | P36 | KC677939.1 | KC677914.1 | – |
| Zasmidium syzygii | CBS 133580(T) = CPC 19792 | KC005798 | KC005777 | MF951730 |
| Zasmidium thailandicum | CBS 145027(T) | NG_066342.1 | NR_164463.1 | _ |
| Zasmidium tsugae | ratstk | EF114705 | EF114688 | – |
| Zasmidium velutinum | CBS 101948(T) = CPC 2262 | EU041838 | EU041781 | MF951731 |
| Zasmidium xenoparkii | CBS 111185(T) = CPC 1300 | JF700966 | DQ303028 | MF951732 |
| Nothopericoniella perseamacranthae | CBS 122097 = RoKi 2995 | GU452682 | MF951354 | MF951583 |
Phylogenetic analyses
Our newly-generated sequences were aligned by locus with ex-type and other representative sequences of Zasmidium species, which were downloaded from GenBank (Table 1). Alignments were made using the online version of MAFFT v. 7.307 (Katoh and Standley 2016) and manually improved, where necessary, using MEGA v. 6.06 (Tamura et al. 2013). Mesquite v. 2.75 (Maddison 2008) was used to concatenate the aligned sequences of the different loci. Ambiguous regions were excluded from analyses using AliView (Larsson 2014) and gaps were treated as missing data. DNA base differences of different gene loci between our strains and ex-type or representative strains of relative taxa are shown in Table 2. The alignment document is available in TreeBASE under the study ID 27250.
Table 2.
The DNA base differences between our Chinese strains and related taxa in the three gene regions. Asterisks (*) denote our material.
| Species | Strain number | ITS (1-480 bp) |
LSU
(481-1254 bp) |
RPB2
(1255-2355 bp) |
| Zasmidium citri-griseum * | GUCC 1507.3 | - | - | - |
| Zasmidium citri-griseum | CBS 146.36 | 8 | 1 | 0 |
| Zasmidium suregadae | P36 | 6 | 0 | / |
| Zasmidium anthuriicola | CBS 118742T | 6 | 0 | 52 |
| Species | Strain number |
ITS
(1-475 bp) |
LSU
(476-1297 bp) |
RPB2
(1298-2141 bp) |
| Zasmidium liboense sp. nov.* | GUCC 1720.2 | - | - | - |
| Zasmidium cellare | CBS 122455NT | 23 | 19 | 72 |
Maximum Parsimony (MP) analyses were performed in PAUP v. 4.0b10 (Swofford 2002), using the heuristic search option with 1,000 random sequence addition replicates and tree bisection-reconnection (TBR) as the branch swapping algorithm. Maxtrees was set at 10,000. The Tree Length (TL), Consistency Indices (CI), Retention Indices (RI), Rescaled Consistency Indices (RC) and Homoplasy Index (HI) were calculated for each tree generated.
Maximum Likelihood (ML) was inferred using the IQ-tree (Nguyen et al. 2015, Chernomor et al. 2016) under the Edge-linked partition model. Bootstrapping was done under 10000 ultrafast replicates (Hoang et al. 2018). ModelFinder (Kalyaanamoorthy et al. 2017) was used to select the best-fit partition model (Edge-linked) using BIC criterion. The best-fit model, according to BIC was: ITS: TNe+I+G4, LSU: TN+F+I+G4, rpb2: TN+F+I+G4.
For Bayesian Inference (BI), GTR+I+G was selected as the best model for all three loci (ITS, LSU and rpb2) as determined by MrModeltest v2 (Nylander 2004). BI analysis was undertaken using MrBayes v. 3.2.6 (Ronquist et al. 2012). Six Markov Chain Monte Carlo runs were launched with random starting trees for 1,000,000 generations and sampling every 1,000 generations. The first 25% resulting trees were discarded as burn-in.
Taxon treatments
Zasmidium liboense
Y.Y. An, Yong Wang bis & K.D. Hyde sp. nov.
D97D94FD-E802-51FF-B7FC-2455428769D8
Materials
Type status: Holotype. Occurrence: catalogNumber: HGUP 1720.2; recordedBy: Wang Yong; Taxon: scientificName: Zasmidium liboense; kingdom: Fungi; class: Dothideomycetes; order: Capnodiales; family: Mycosphaerellaceae; genus: Zasmidium; Location: country: China; stateProvince: Guizhou; locality: Gold Thread Cave of Libo county; Identification: identifiedBy: Yuan-Yan An; dateIdentified: 2020; Record Level: type: ex-type cultures GUCC 1070.2; MFLUCC 20-0139; language: en
Description
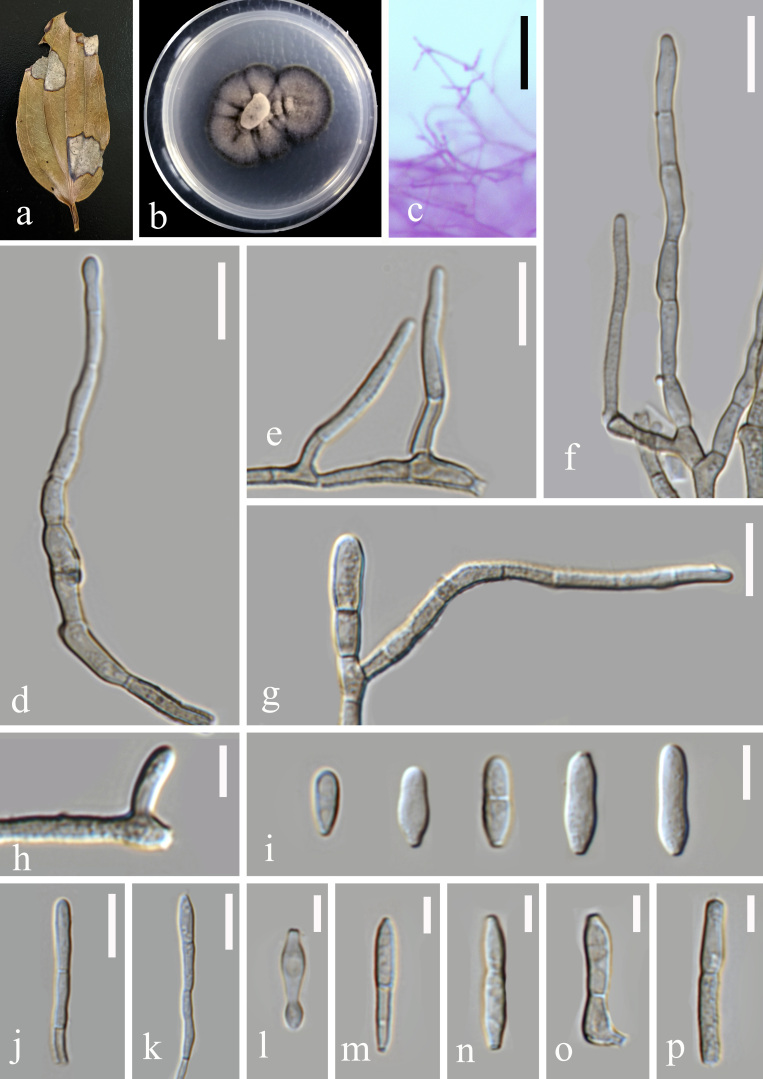
Pathogenic on the leaf spot of Smilax china (Liliales, Smilacaceae). Lesions on the upper leaf surface, scattered, distinct, irregular, rather large, the maximum length of the spot more than 20 mm, the edges of the disease reddish-brown, the centre dead to greyish-white, on the lower leaf surface similar. Colonies on PDA 10–15 mm diam. in 2 weeks, with an even, dark coffee margin. Mycelium composed of hyaline and pale brown to dark blackish-brown hyphae, verruculose, septate, branching, uniform in width, 2.5 μm. Conidiophores arising from hyphae, pale olivaceous brown to pale blackish-brown, finely verruculose, straight or slightly curved, dendritic rugged or rugose on the surface 20–350 × 1.5–3.5 μm. Conidiogenous cells integrated, apical, polyblastic, proliferating sympodially, with rim-like conidiogenous loci, thickened and darkened, located apically and lateraly as in a short rachis 1.5–2.5 μm diam. Conidia solitary, occasionally catenate, pale blackish-brown to pale olivaceous brown, verruculose, ellipsoidal, cylindrical to obclavate, base obconically truncate and apex rounded, straight or curved, 6–21 × 2–4 μm, 0–1-septate, sometimes constricted at septa, with hila thickened and darkened, 1–1.5 μm diam. (Fig. 1)
Figure 1.
Zasmidium liboense (GUCC 1720.2). a. Leaf spot symptoms on the host; b. Culture on PDA; c, d. hyphae and conidiophores on PDA; e–m. Conidiophores and conidia; n, o. Conidiogenous cells and conidia; p. Conidia. Scale bars: c, d = 50 µm, e–j = 10 µm, k = 5 µm, l–p = 10 µm.
MycoBank Number
MB836278
Etymology
In reference to the location (Libo county, Guizhou Province), where the holotype was isolated.
Zasmidium citri-griseum
(F.E. Fisher) U. Braun & Crous, IMA Fungus 5 (2): 337 (2014)
6940A98C-A2F7-5387-B8E6-56FC1C1650A1
Materials
Type status: Other material. Occurrence: catalogNumber: HGUP 1507.3; recordedBy: An Yuan-Yan; occurrenceID: living culture GUCC 1507.3 and MFLUCC 20-0138; Taxon: scientificName: Zasmidium citri-griseum; kingdom: Fungi; class: Dothideomycetes; order: Capnodiales; family: Mycosphaerellaceae; genus: Zasmidium; Location: country: China; stateProvince: YunNan; locality: Xishuangbanna Dai Autonomous Prefecture; Identification: identifiedBy: Yuan-Yan An; dateIdentified: 2020; Record Level: language: en
Description
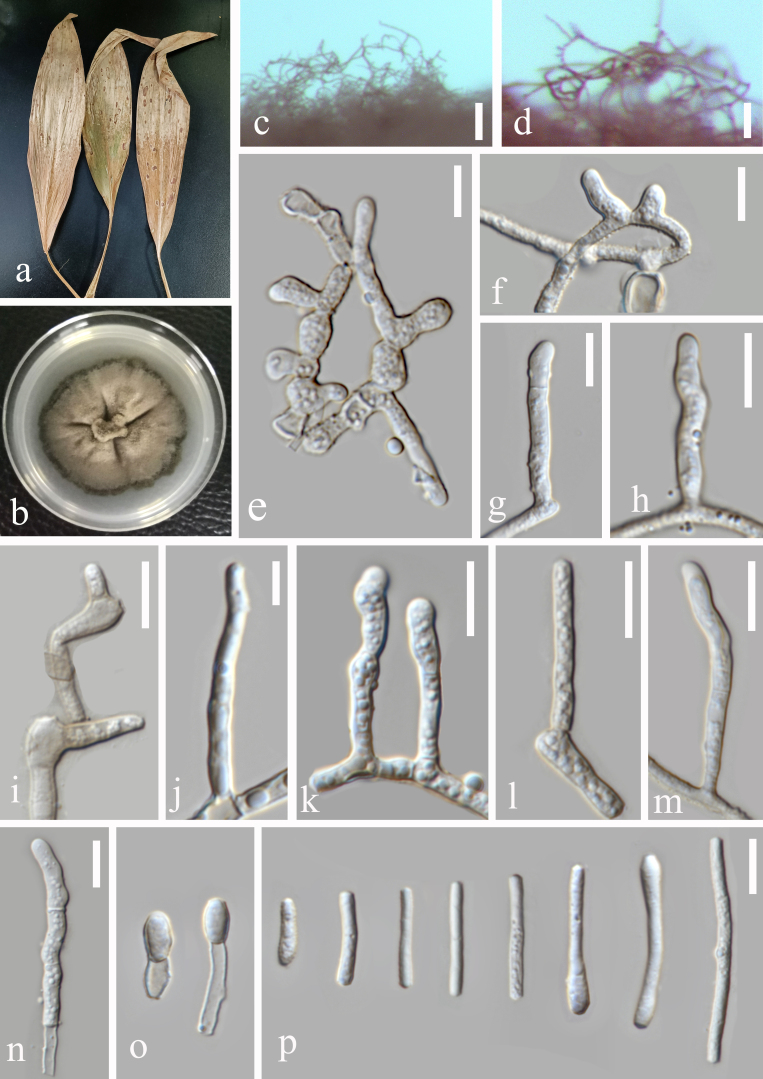
Pathogenic on leaf spot of Cremastra appendiculata (Asparagales, Orchidaceae). Lesions on the upper leaf surface, scattered to confluent, distinct, angular, spots elliptic to suborbicular, reddish-brown to dark brown, 2–5 mm, on the lower leaf surface similar. Colonies on PDA 10–15 mm diam. in 2 weeks, with an even, dark-brown margin. Surface fold, with gully shape. Conidiophores arising singly as lateral branches of superficial hyphae, semi-macronematous to macronematous, mononematous, erect to flexuous, sometimes curved, unbranched, thick-walled, non-smooth surface, 1–3-septate, cylindrical, geniculate, brown, 35–135 × 5–7 µm. Conidiogenous cells integrated, terminal, 5–10 µm long, sympodial, polyblastic, cylindrical, geniculate, scars slightly thickened and darkened, 0.5–1.5 µm in diam. Conidia solitary in simple or occasionally branched chains, short to long cylindrical, some ends swollen straight to somewhat curved, 8–41 × 2–5 µm, unseptate, light brown, thin-walled, verruculose, 2–3.5 µm wide. (Fig. 2)
Figure 2.
Zasmidium citri-griseum (GUCC 1507.3) a. Leaf spot symptoms on host; b. Culture on PDA; c. Hyphae and conidiophores on PDA culture; d–h. Conidiophores, conidiogenous cells and conidia; i. Conidia; j, k. Conidiogenous cells and conidia; l–p. Conidia. Scale bars: c = 100 µm, d–g =10 µm, h=5 µm, i, j = 10 µm, k–p = 5 µm.
Analysis
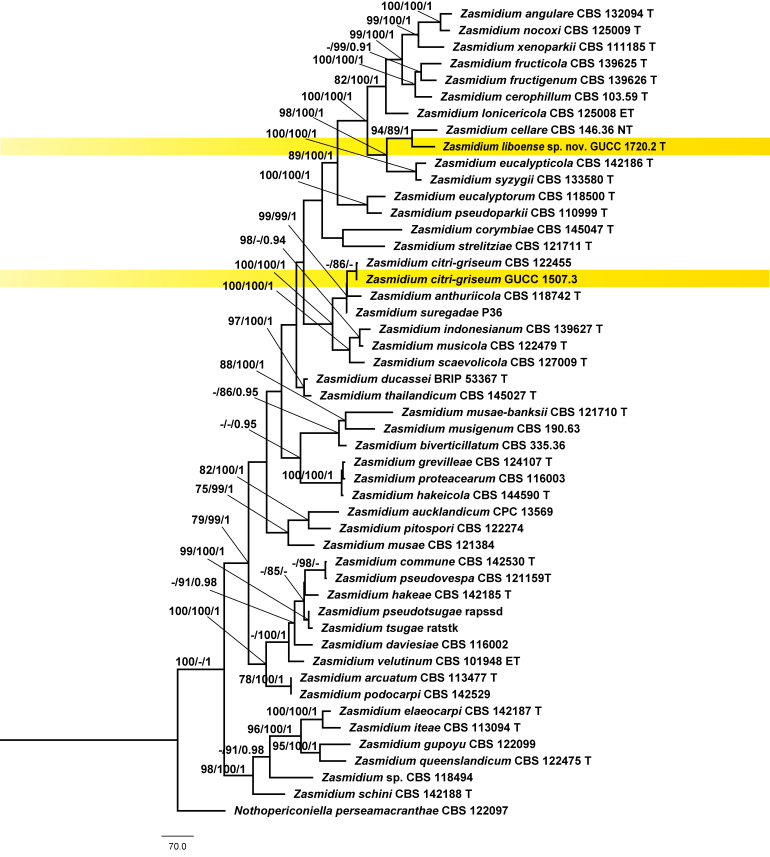
Our final concatenated alignment included 2407 characters , viz. (ITS: 1–537, LSU: 538–1354, rpb2: 1355–2407), of which 645 were parsimony-informative characters. MP inference resulted in two equally-parsimonious trees (TL = 3705, CI = 0.35, RC = 0.59, HI = 0.20, RC = 0.65) and one of them was selected to show the topology (Fig. 3). Forty-eight Zasmidium strains clustered together as a clade, which received maximum support from both ML and BI analyses. Zasmidium liboense sp. nov. (GUCC 1720.2) was retrieved as sister taxon of Z. cellare (Pers.) Fr. (CBS 146.36) with high support (94 MP/89 ML/1.00 PP). Strain GUCC1507.3 grouped with a lineage consisting of Z. anthuriicola (U. Braun & C.F. Hill) Crous & U. Braun, Z. citri-griseum (F.E. Fisher) U. Braun & Crous and Z. suregadae Phengs., K.D. Hyde & U. Braun, with high support (99 MP/99 ML/1.00 PP). It was placed sister to Z. citri-griseum (CBS 122455) (86 ML).
Figure 3.
MP phylogeny of Zasmidium reconstructed from a three-locus dataset (ITS, LSU, rpb2). MP and ML bootstraps > 70 and BI posterior probabilities (PP) > 0.9 are placed close to topological nodes and separated by “/”. Nothopericoniella perseamacranthae (CBS 122097) was selected as outgroup.
A comparison of DNA bases (Table 2) demonstrated that, between Z. liboense (GUCC 1720.2) and Z. cellara (CBS 146.36), there were 23 bp differences in the ITS region, 19 in the LSU and 72 in the rpb2. The ITS sequence of GUCC 1507.3 differed in 8 bp from Z. citri-griseum (CBS 122455). Their LSU sequences were identical, whereas a single base pair difference was found in the rpb2.
Discussion
Our phylogenetic analyses pointed out that Zasmidium liboense is different from Z. cellare. Morphologically, both species can be separated as well (Arzanlou et al. 2007). Zasmidium liboense produces conidia with 0-1 septa, whereas those of Z. cellare possess 0–1(–4) septa. Conidia of Z. cellare arise terminally or laterally and are subhyaline, whereas those of Z. liboense arise terminally and are pale blackish-brown to pale olivaceous brown. In addition, conidia of Z. liboense (6–21 × 2–4 μm) generally have larger dimensions compared to those of Z. cellare (6–9 × 1.8–2.5 µm). We also compared micro-morphology of our new taxon with 74 Zasmidium species, for which no sequence data are available. Most of those, 64 species, produce larger conidia with more septa compared to Z. liboense. In addition, Z. araliae, Z. cerophilum and Z. litseae all possess smaller conidia than Z. liboense. The conidia of Z. capparacearum, Z. eriolobi, Z. gahniicola and Z. mitellae have more septa compared to those of Z. liboense. Zasmidium clusiae produces conidia with basal unthickened hilum, whereas the conidia of Z. liboense, Z. deightonianum and Z. oxycocci have slightly or obviously thickened conidia. For these three species, the length of conidiophores can be used to distinguish amongst species: Z. liboense (20–350 μm), Z. deightonianum (10–30 μm) and Z. oxycocci (50–100 μm).
For strain GUCC 1507.3, our molecular phylogenetic data (Fig. 3, Table 2) showed that it was very close to Z. anthuriicola, Z. citri-griseum and Z. suregadae. Rbp2 sequences of strain GUCC 1507.3 and Z. citri-griseum (CBS 122455) are identical, but different from Z. anthuriicola (CBS 118742). Currently, no rpb2 sequences are available for Z. suregadae. Morphological comparison shows that conidia of our material (GUCC 1507.3) agree with the description of Z. citri-griseum; they are somewhat shorter although still in the range of measurements [Z. citri-griseum: 6–70(–120) × 2–4.5 μm] and they are aseptate [Z. citri-griseum: 0–1–(–6)-septate] (Braun et al. 2014). Zasmidium citri-griseum is one of the few species of Zasmidium with a wider host range. Braun et al. (2014) listed hosts in the Fabaceae, Musaceae and Rutaceae. The host of our Chinese material, Cremastra appendiculata (Orchidaceae), represents a new host family for this fungus.
Supplementary Material
Acknowledgements
This research was supported by the following sources: National Natural Science Foundation of China (No. 31972222, 31560489), Program of Introducing Talents of Discipline to Universities of China (111 Program, D20023), Talent Project of Guizhou Science and Technology Cooperation Platform ([2017]5788-5 and [2019]5641), Guizhou Science, Technology Department of International Cooperation Base Project ([2018]5806) and Guizhou Science and Technology Innovation Talent Team Project ([2020]5001).
References
- Arzanlou M, Groenewald J Z, Gams W, Braun U, Shin H D, Crous Pedro W. Phylogenetic and morphotaxonomic revision of Ramichloridium and allied genera. Studies in Mycology. 2007;58:57–93. doi: 10.3114/sim.2007.58.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensch Konstanze, Braun Uwe, Groenewald Johannes Zacharias, Crous Pedro W. The genus Cladosporium. Studies in Mycology. 2012;72:1–401. doi: 10.3114/sim0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun Uwe, Crous Pedro W, Schubert Konstanze, Shin Hyeon Dong. Some reallocations of Stenella species to Zasmidium. Schlechtendalia . 2010;20:99–104. [Google Scholar]
- Braun Uwe, Crous Pedro W, Nakashima Chiharu. Cercosporoid fungi (Mycosphaerellaceae) 2. Species on monocots (Acoraceae to Xyridaceae, excluding Poaceae) IMA Fungus. 2014;5(2):203–390. doi: 10.5598/imafungus.2014.05.02.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheek, Martin, Lughadha Eimear Nic, Niskanen Paul M Kirk,Heather Lindon,Tuula, et al. New scientific discoveries: Plants and fungi. Plants People Planet. 2020;5 doi: 10.1002/ppp3.10148. [DOI] [Google Scholar]
- Chernomor Olga, von Haeseler Arndt, Minh Bui Quang, et al. Terrace Aware Data Structure for Phylogenomic Inference from Supermatrices. Systematic Biology. 2016;65(6):997–1008. doi: 10.1093/sysbio/syw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomnunti Putarak, Hongsanan Sinang, Aguirre-Hudson Begoña, Tian Qing, Peršoh Derek, Dhami Manpreet K, Alias Aisyah S, Xu Jian Chu, Liu Xing Zhong, Stadler Marc, Hyde Kevin D. The sooty moulds. Fungal Diversity. 2014;66:1–36. doi: 10.1007/s13225-014-0278-5. [DOI] [Google Scholar]
- Crous Pedro W, Summerell Brett A, Carnegie Angus J, Wingfield Michael J, Groenewald Johannes Z. Novel species of Mycosphaerellaceae and Teratosphaeriaceae. Persoonia. 2009;23:119–146. doi: 10.3767/003158509X479531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoog G S, van den Ende A H G G. Molecular diagnostics of clinical strains of filamentous Basidiomycetes. MYCOSES-BERLIN- 1998;41:183–190. doi: 10.1111/j.1439-0507.1998.tb00321.x. [DOI] [PubMed] [Google Scholar]
- Fries E M. Sectio posterior, Uppsala; 1849. Summa vegetabilium Scandinaviae. [Google Scholar]
- Hoang Diep Thi, Chernomor Olga, Von Haeseler Arndt, Minh Bui Quang, Vinh Le Sy. UFBoot2: improving the ultrafast bootstrap approximation. Molecular Biology and Evolution. 2018;35(2):518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongsanan S, Hyde K D, Phookamsak R, Wanasinghe D N, McKenzie E H C, Sarma V V, Boonmee S, Lücking R, Bhat D J, Liu N G, Tennakoon D S, Pem D, Karunarathna A, Jiang S H, Jones E B G, Phillips A J L, Manawasinghe I S, Tibpromma S, Jayasiri S C, Sandamali D S, Jayawardena R S, Wijayawardene N N, Ekanayaka A H, Jeewon R, Lu Y Z, Dissanayake A J, Zeng X Y, Luo Z L, Tian Q, Phukhamsakda C, Thambugala K M, Dai D Q, Chethana K W T, Samarakoon M C, Ertz D, Bao D F, Doilom M, Liu J K, Pérez-Ortega S, Suija A, Senwanna C, Wijesinghe S N, Konta S, Niranjan M, Zhang S N, Ariyawansa H A, Jiang H B, Zhang J F, Norphanphoun C, de Silva N I, Thiyagaraja V, Zhang H, Bezerra J D P, Miranda-González R, Aptroot A, Kashiwadani H, Harishchandra D, Sérusiaux E, Aluthmuhandiram J V S, Abeywickrama P D, Devadatha B, Wu H X, Moon K H, Gueidan C, Schumm F, Bundhun D, Mapook Ausana, Monkai J, Chomnunti P, Suetrong S, Chaiwan N, Dayarathne M C, Yang J, Rathnayaka A R, Bhunjun C S, Xu J C, Zheng J S, Liu G, Feng Y, Xie N. Refined families of Dothideomycetes: Dothideomycetidae and Pleosporomycetidae. Mycosphere. 2020;11(1) doi: 10.5943/mycosphere/11/1/13. [DOI] [Google Scholar]
- Hyde Kevin D, Norphanphoun Chada, Chen Jie, Dissanayake Asha J, Doilom Mingkwan, Hongsanan Sinang, Jayawardena Ruvishika S, Jeewon Rajesh, Perera Rekhani H, Thongbai Benjarong, Wanasinghe D N, Wisitrassameewong Komsit, Tibpromma S, Stadler M. Thailand’s amazing diversity: up to 96% of fungi in northern Thailand may be novel. Fungal Diversity. 2018;93(1):215–239. doi: 10.1007/s13225-018-0415-7. [DOI] [Google Scholar]
- Kalyaanamoorthy S., Minh B. Q., Wong T. K.F., Haeseler A., Jermiin L. S., et al. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14 doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal . Cercosporoid fungi of India. Bishen Singh Mahendra Pal Singh; Dehradun: 2010. [Google Scholar]
- Katoh Kazutaka, Standley Daron M. A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics. 2016;32(13):1933–1942. doi: 10.1093/bioinformatics/btw108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson A. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics. 2014;30(22) doi: 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Yin, Ran Shuang Fei, Bhat Jayarama, Hyde Kevin D, Wang Yong, Zhao De Gang. Curvularia microspora sp. nov. associated with leaf diseases of Hippeastrum striatum in China. MycoKeys. 2018;(29) doi: 10.3897/mycokeys.29.21122. [DOI] [PMC free article] [PubMed]
- Liu Yajuan J, Whelen Sally, Hall Benjamin D. Phylogenetic relationships among Ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution. 1999;16(12):1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- Long Hui, Zhang Qian, Hao Yuan Yuan, Shao Xian Qiang, Wei Xiao Xing, Hyde Kevin D, Wang Yong, Zhao De Gang. Diaporthe species in south-western China. MycoKeys. 2019;57 doi: 10.3897/mycokeys.57.35448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison Wayne P. Mesquite: a modular system for evolutionary analysis. Evolution. 2008;62:1103–1118. [Google Scholar]
- Nguyen L., Schmidt H. A., Von Haeseler A., Minh B. Q., et al. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution. 2015;32(1) doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander J A A. MrModeltest v2. Evolutionary Biology Centre, Uppsala University, Uppsala 2004
- Ronquist Fredrik, Teslenko Maxim, Van Der Mark Paul, Ayres Daniel L, Darling Aaron, Höhna Sebastian, Larget Bret, Liu Liang, Suchard Marc A, Huelsenbeck John P. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Ya Ru, Goonasekara Ishani, Thambugala Kasun M, Jayawardena Ruvishika S, Wang Yong, Hyde Kevin D. Distoseptispora bambusae sp. nov.(Distoseptisporaceae) on bamboo from China and Thailand. Biodiversity Data Journal. 2020;8 doi: 10.3897/BDJ.8.e53678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D L. PAUP*: Phylogenetic analysis using parsimony (and other methods), version 4.0 b10. MA: Sinauer Associates, Sunderland, UK 2002
- Tamura Koichiro, Stecher Glen, Peterson Daniel, Filipski Alan, Kumar Sudhir. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgalys Rytas, Hester Mark. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology. 1990;172(8):4238–4246. doi: 10.1128/JB.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White Thomas J, Bruns Thomas, Lee S J W T, Taylor John. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. Vol. 18. Academic Press; 1990. 315-322. [DOI] [Google Scholar]
- Wijayawardene N N, Hyde K D, Al-Ani L K T, Tedersoo L, Haelewaters Danny, Rajeshkumar K C, Zhao R L, Aptroot A, Leontyev D, Saxena R K, Tokarev Y S, Dai D Q, Letcher P M, Stephenson S L, Ertz D, Lumbsch H T, Kukwa M, Issi I V, Madrid H, Phillips A J L, Selbmann L, Pfliegler W P, Horváth E, Bensch K, Kirk P M, Kolaříková K, Raja H A, Radek R, Papp V, Dima V, Ma J, Malosso E, Takamatsu S, Rambold G, Gannibal P B, Triebel D, Gautam A K, Avasthi S, Suetrong S, Timdal,E, Fryar S C, Delgado G, Réblová M, Doilom M, Dolatabadi S, Pawłowska JundefinedZ, Humber RundefinedA, Kodsueb R, Sánchez-Castro I, Goto B T, Silva D K A, de Souza F A, Oehl F, da Silva G A, Silva I R, Błaszkowski J, Jobim K, Maia LundefinedC, Barbosa FundefinedR, Fiuza PundefinedO, Divakar PundefinedK, Shenoy BundefinedD, Castañeda-Ruiz R F, Somrithipol S, Lateef A A, Karunarathna S C, Tibpromma S, Mortimer P E, Wanasinghe D N, Phookamsak R, Xu J, Wang Y, Tian F, Alvarado P, Li D W, Kušan I, Matočec N, Mešić A, Tkalčec Z, Maharachchikumbura SundefinedSundefinedN, Papizadeh M, Heredia G, Wartchow F, Bakhshi M, Boehm E, Youssef N, Hustad VundefinedP, Lawrey JundefinedD, Santiago AundefinedLundefinedCundefinedMundefinedA, Bezerra JundefinedDundefinedP, Souza-Motta CundefinedM, Firmino AundefinedL, Tian Q, Houbraken J, Hongsanan S, Tanaka K, Dissanayake AundefinedJ, Monteiro JundefinedS, Grossart HundefinedP, Suija A, Weerakoon G, Etayo J, Tsurykau A, Vázquez V, Mungai P, Damm U, Li Q R, Zhang H, Boonmee S, Lu Y Z, Becerra A G, Kendrick B, Brearley F Q, Motiejūnaitė J, Sharma B, Khare R, Gaikwad S, Wijesundara DundefinedSundefinedA, Tang LundefinedZ, He MundefinedQ, Flakus A, Rodriguez-Flakus P, Zhurbenko MundefinedP, McKenzie EundefinedHundefinedC, Stadler M, Bhat DundefinedJ, Liu JundefinedK, Raza M, Jeewon R, Nassonova EundefinedS, Prieto M, Jayalal RundefinedGundefinedU, Erdoğdu M, Yurkov A, Schnittler M, Shchepin OundefinedN, Novozhilov YundefinedK, Silva-Filho AundefinedGundefinedS, Gentekaki E, Liu P, Cavender JundefinedC, Kang Y, Mohammad S, Zhang LundefinedF, Xu RundefinedF, Li YundefinedM, Dayarathne MundefinedC, Ekanayaka AundefinedH, Wen TundefinedC, Deng CundefinedY, Pereira OundefinedL, Navathe S, Hawksworth DundefinedL, Fan XundefinedL, Dissanayake LundefinedS, Kuhnert E, Grossart HundefinedP, Thines M. Outline of fungi and fungus-like taxa. Mycosphere. 2020;11(1):1060–1456. doi: 10.5943/mycosphere/11/1/8. [DOI] [Google Scholar]
- Wijesinghe Subodini N, Wang Yong, Camporesi Erio, Wanasinghe Dhanushka N, Boonmee Saranyaphat, Hyde Kevin D. A new genus of Bambusicolaceae (Pleosporales) on Corylus avellana (Fagales) from Italy. Biodiversity Data Journal. 2020;8 doi: 10.3897/BDJ.8.e55957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Qian, Yang Zai Fu, Cheng Wei, Wijayawardene Nalin N, Hyde Kevin D, Chen Zhuo, Wang Yong. Diseases of Cymbopogon citratus (Poaceae) in China: Curvularia nanningensis sp. nov. MycoKeys. 2020;63 doi: 10.3897/mycokeys.63.49264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.