Abstract
Purpose:
Idiopathic normal pressure hydrocephalus (iNPH) and Alzheimer’s disease (AD) are geriatric diseases and common causes of dementia. Recently, many studies on the segmentation, disease detection, or classification of MRI using deep learning have been conducted. The aim of this study was to differentiate iNPH and AD using a residual extraction approach in the deep learning method.
Methods:
Twenty-three patients with iNPH, 23 patients with AD and 23 healthy controls were included in this study. All patients and volunteers underwent brain MRI with a 3T unit, and we used only whole-brain three-dimensional (3D) T1-weighted images. We designed a fully automated, end-to-end 3D deep learning classifier to differentiate iNPH, AD and control. We evaluated the performance of our model using a leave-one-out cross-validation test. We also evaluated the validity of the result by visualizing important areas in the process of differentiating AD and iNPH on the original input image using the Gradient-weighted Class Activation Mapping (Grad-CAM) technique.
Results:
Twenty-one out of 23 iNPH cases, 19 out of 23 AD cases and 22 out of 23 controls were correctly diagnosed. The accuracy was 0.90. In the Grad-CAM heat map, brain parenchyma surrounding the lateral ventricle was highlighted in about half of the iNPH cases that were successfully diagnosed. The medial temporal lobe or inferior horn of the lateral ventricle was highlighted in many successfully diagnosed cases of AD. About half of the successful cases showed nonspecific heat maps.
Conclusion:
Residual extraction approach in a deep learning method achieved a high accuracy for the differential diagnosis of iNPH, AD, and healthy controls trained with a small number of cases.
Keywords: artificial intelligence, deep learning, computer-aided diagnosis, idiopathic normal pressure hydrocephalus, Alzheimer’s disease
Introduction
Idiopathic normal pressure hydrocephalus (iNPH) and Alzheimer’s disease (AD) are geriatric diseases and common causes of dementia.1–3 iNPH is treated by surgical intervention of cerebrospinal fluid (CSF) shunting,4–6 and known as a treatable dementia. AD is mainly treated by medication such as an acetylcholinesterase inhibitor.7 Their treatment approaches are quite different, so it is necessary to differentiate iNPH and AD at an early stage. MRI is often used for morphological evaluation of the brain in iNPH and AD. In the diagnosis, selective atrophy of the medial temporal lobe is an important feature of AD.8 iNPH is characterized by ventricular enlargement with a disproportionately enlarged subarachnoid-space hydrocephalus (DESH),9 which includes dilated lateral ventricle and Sylvian fissure, narrow high-convexity sulci and a small callosal angle. However, cognitive decline and ventriculomegaly are seen in both iNPH and AD, it is difficult to make correct diagnosis in some cases.
Computer-aided diagnosis could be helpful in a daily clinical practice. There have been some reports that aimed to distinguish between iNPH and AD using three-dimensional (3D) T1-weighted images by measuring the hippocampal volume, lateral ventricle size, the corpus callosum angle, or the whole gray matter and white matter volumes.10–12 Even though segmental volume measurements can now be performed automatically, these techniques still require manual steps and some expertise. Brain deformation is severe in iNPH patients, so standardization or segmentation failure may frequently occur. We need other approach to achieve an automatic diagnosis.
Recently, many studies on deep learning for medical images have been conducted. Deep learning recognizes patterns in images well and makes a diagnosis immediately (usually less than several seconds). It can be applied to the diagnostic imaging of dementia.13–16 In general, we need thousands of data samples to construct a deep learning model, though few samples are available in actual clinical practice. So, it is waited to develop a model based on a small number of samples. Herein, we use a novel “residual extraction approach” in the deep learning method to effectively train a model even with a limited number of subjects. “Residual” is based on anomaly detection techniques, where the image is compressed and reconstructed. Difference between original image and reconstructed image is defined as a residual. This method has been applied to a detection of abnormal lesion in brain MRI of relatively small datasets.17 In the residual extraction approach, first we extract the residual, and then input that to a convolutional neural network. Extracted residual image is the essence of anomaly, so we expected that it would enhance model training and increase the diagnostic accuracy between iNPH and AD. The aim of this study was to automatically differentiate iNPH and AD using deep learning by a residual extraction approach.
Materials and Methods
Subjects
This study was approved by the Institutional Review Board of our hospital. This was a retrospective study and all persons gave their written informed consent prior to the MRI scan.
Twenty-three patients with iNPH (11 male and 12 female: mean age 74.6 years), 23 patients with AD (11 male and 12 female: mean age 75.0 years) and 23 age-matched healthy controls (11 male and 12 female: mean age 74.2 years) were included in this study. The patients with iNPH were consecutive cases from November 2010 to February 2012 suspected of iNPH and performed a CSF tap test, and thereafter diagnosed with probable iNPH. AD patients were randomly selected to match the age of iNPH patients whose MR images were taken during the same period. Diagnosis of iNPH was made according to the criteria of probable iNPH proposed by the Japanese Clinical Guidelines for Idiopathic Normal Pressure Hydrocephalus,18 and that of AD was made according to the criteria of probable AD dementia by the National Institute on Aging and Alzheimer’s Association workgroups.19
MRI data acquisition
All patients and volunteers underwent brain MRI with a 3T unit (Achieva; Philips Healthcare, Best, The Netherlands) and we used only whole-brain 3D T1-weighted images in this study. The images were obtained using a 3D magnetization-prepared rapid gradient-echo sequence with the following parameters: effective TR = 15 ms; effective TE = 3.4 ms; resolution = 0.8125 × 0.8125 × 0.86 mm3; field of view = 260 × 260 mm2; and total scan time = 400 s.
Case characteristics
The mean Mini-Mental State Examination score of iNPH and AD patients was 23.2 ± 4.1 and 19.8 ± 3.4, respectively. All iNPH patients underwent MRI before treatment intervention. Although it was difficult to clearly define the onset of AD, the mean time from symptom recognition to MRI was 2.6 ± 1.5 years.
Regarding image findings, ventricle enlargement (Evans index > 0.3) was observed in all iNPH cases and four AD cases, and not in healthy controls (HCs). Moderate to severe temporal lobe atrophy was seen in 13 AD patients and two NPH patients. The mean Fazekas grade of the periventricular white matter lesions evaluated with T1-weighted images was 2.57 ± 0.66 in iNPH, 2.13 ± 0.76 in AD, and 1.22 ± 0.67 in HC, respectively.
Interpretation experiments by radiologists
As a pilot study, two radiologists (experience of 9 and 5 years) conducted interpretation experiments. They were given only 3D T1-weighted images and diagnosed whether the case was iNPH, AD, or HC without referring to clinical symptoms. Images were shuffled between patients and the radiologists were blinded to the diagnosis of the patients.
Idiopathic normal pressure hydrocephalus was comprehensively evaluated with emphasis on the findings of dilated lateral ventricle and Sylvian fissure, narrow high-convexity sulci and a small callosal angle.18 Although morphological changes are not essential in the diagnostic criteria of Alzheimer’s disease, it is widely known that disproportionate atrophy of the medial temporal lobe is useful for diagnosis,8 so that was used as index findings. Since iNPH is a disease in which diagnostic imaging can be a clue for diagnosis, radiologists prioritized iNPH diagnosis when both iNPH and AD findings coexisted in a case.
The diagnostic accuracy of the two readers were 0.79 and 0.83 and the Cohen’s kappa coefficient between them was 0.80. We compared the sensitivity and specificity of the deep learning model and each radiologist simultaneously by McNemar χ2-test.20
Deep learning procedure
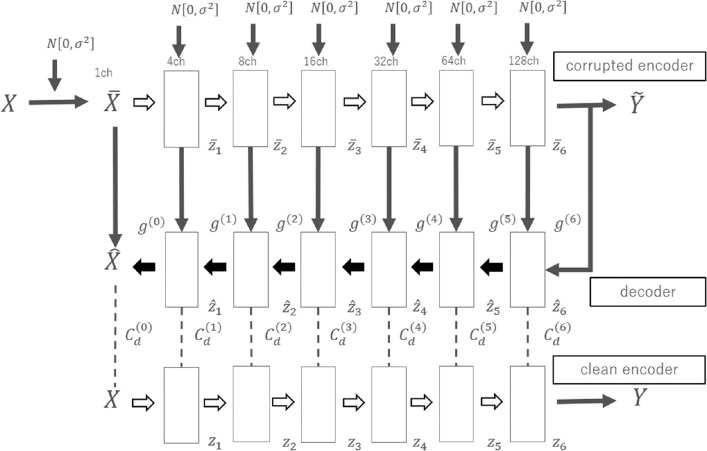
We designed a fully automated, end-to-end 3D deep learning classifier to differentiate iNPH, AD and controls. The number of samples (a total of 69) was too small to build and train a convolutional neural network using a general setup because of the risk of overfitting. Hence, we introduced a residual extraction phase followed by a neural network classifier. For the residual extraction phase, we built a 3D convolutional ladder network to reconstruct the 3D volume to be extracted from the original 3D volume. The residual volume was then fed again into the encoder to obtain a residual feature map. In the original study proposing the ladder network,21 the network was designed using neural networks without convolution layers. In the present study, the original ladder network was naturally extended to 3D convolutional networks. The feedforward path (X → z1 → ⋯ → Y) or “clean encoder” shared the mappings with the corrupted feedforward path () or “corrupted encoder”. The decoder () consisted of de-noising function g, which was the same function as used in the previous study,21 and had cost function Cd on each layer to minimize the absolute difference between ẑ and z (Fig. 1). Model training for each neural network classifier was performed separately from the following neural network classifier using the same training dataset for each trial of a leave-one-out test. The hyper-parameter settings for the training were as follows: the mini batch size was 23; the Adam optimization parameters α, β1, and β2 were 0.002, 0.9, and 0.999, respectively; and the number of training epochs was 1000.22 Because the loss function is calculated during the training, the model is expected to learn “common features” preferentially to minimize the mean absolute error between the original and reconstructed images generated through the encoder and decoder. After the 3D convolutional ladder network learned common features in the training datasets, the model extracted reconstructed common features from the input image. In brief, X is input to the clean encoder, Y is generated, and Y is input to the decoder, which outputs Z (= reconstructed X). We then created a residual image by subtracting Z from X, and the X − Z image was fed into the clean encoder to generate R, which we call the residual feature map (128ch × 5 × 3 × 5). The R map is input to the following fully connected neural network classifier. The residual images theoretically indicate specific features in each patient’s volume.
Fig. 1.
Architecture of the 3D convolutional ladder network. The first line is the “corrupted encoder”, the second line is the decoder, and the third line is the “clean encoder”. Each arrow in the encoder/decoder consists of 3 × 3 × 3 convolution/deconvolution with stride 2, batch normalization, and an exponential linear unit. g: de-noising function, Cd: cost function, N[0, σ2]: random sample from normal distribution with mean = 0 and variance = σ2.
In the neural network classifier part, we designed a relatively old-fashioned simple fully connected neural network classifier that has one hidden layer consisting of seven nodes, whose activation functions are all logistic sigmoids. The output layer includes three nodes activated by softmax function that represents the probability of iNPH, AD and controls respectively ranging from 0 to 1. Model training was performed separately from the above ladder network, i.e., we treated the residual feature map (128ch × 5 × 3 × 5) as a fixed input. The hyper-parameter settings for the training were as follows: the mini batch size was 34 (= a half of total number of training data); Adam parameters α, β1, and β2 were 0.0005, 0.9, and 0.999, respectively; and the number of training epochs was 200. The classification loss was defined to be hard-labelled softmax cross-entropy.
We evaluated the accuracy of our model at differentiating iNPH, AD and controls using a leave-one-out cross-validation test and presented the confusion matrix and the probability charts.
All model training was performed on a computer with 64 GB of CPU memory, a Xeon E5-2670 v3 CPU (Intel, Santa Clara, CA, USA), and a TITAN Xp graphics processing unit (NVIDIA, Santa Clara, CA, USA). The computer program was coded with Python 3.6 and the deep learning framework of Chainer 5.1.0 (http://chainer.org/).
Visual validation of deep learning diagnosis
We evaluated the validity of the result by visualizing important areas in the process of differentiating iNPH, AD and controls on the original input image using the Gradient-weighted Class Activation Mapping (Grad-CAM) technique.23 Grad-CAM uses the class-specific gradient information flowing into the final convolutional layer of a convolutional neural network to produce a coarse localization map of the important regions in the image. The present study focused on the third convolutional layer, which includes the 16 channels of the feature map needed to produce the Grad-CAM heat map image. The precise procedure was as follows: First, we performed backpropagation from true class activation (i.e., if the true class of the patient is AD, we start backpropagation from class AD). Then, we obtained the mean gradient of each channel and multiplied each of them by the post-activated values on the feature map, the absolute values of the gradient-weighted feature map are summed, and finally we obtained a coarse localization map. The map was thresholded and resized back to its original image resolution. To demonstrate the important regions in the differential diagnosis, we translated this localization map into a heat map by applying a color map that appears in the plot as a color bar.
Results
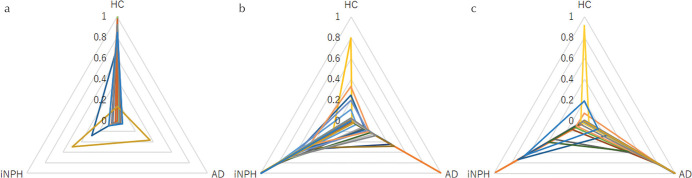
Twenty-one out of 23 iNPH cases, 19 out of 23 AD cases and 22 out of 23 controls were correctly diagnosed (Table 1). The accuracy was 0.90. The probability of the deep learning diagnosis in each subject was shown in the triangular radar charts (Fig. 2).
Table 1.
Confusion matrix of deep learning diagnosis
| Deep learning diagnosis | ||||
|---|---|---|---|---|
| HC | iNPH | AD | ||
| Clinical diagnosis | HC | 22 | 1 | 0 |
| iNPH | 1 | 21 | 1 | |
| AD | 1 | 3 | 19 | |
AD, Alzheimer’s disease; HC, healthy controls; iNPH, idiopathic normal pressure hydrocephalus.
Fig. 2.
Probability charts of the deep learning diagnosis. The triangular radar graph shows the probability of healthy controls (HC, a), idiopathic normal pressure hydrocephalus (iNPH, b) and Alzheimer’s disease (AD, c) in each subject ranging from 0 to 1.
The sensitivity of our deep learning model for iNPH was 0.91 and the specificity was 0.91. Meanwhile, the sensitivity of the first radiologist for iNPH was 0.96 and the specificity was 0.96, and the sensitivity of the second radiologist for iNPH was 0.96 and the specificity was 0.98. There were no significant differences between the deep learning model and two radiologists in sensitivity or specificity (McNemar χ2 = 1.7, P = 0.43 for radiologist 1 and χ2 = 2.1, P = 0.34 for radiologist 2). The sensitivity of the deep learning model for AD was 0.83 and the specificity was 0.98. The sensitivity of the first radiologist for AD was 0.61 and the specificity was 0.89, and the sensitivity of the second radiologist for AD was 0.74 and the specificity was 0.87. There were no significant differences between the deep learning model and two radiologists in sensitivity or specificity (McNemar χ2 = 5.9, P = 0.052 for radiologist 1 and χ2 = 4.0, P = 0.14 for radiologist 2).
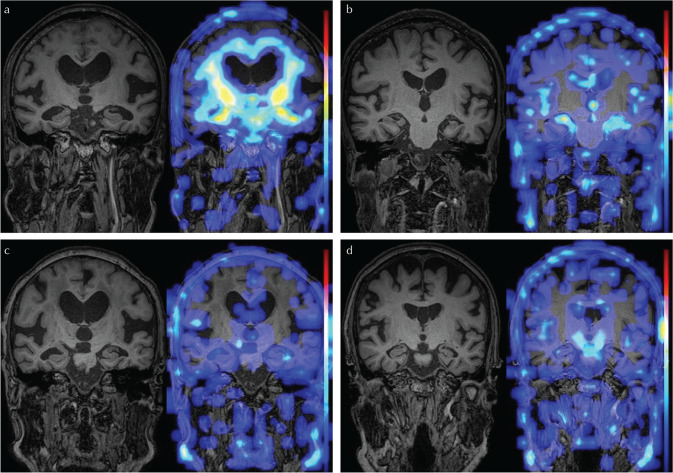
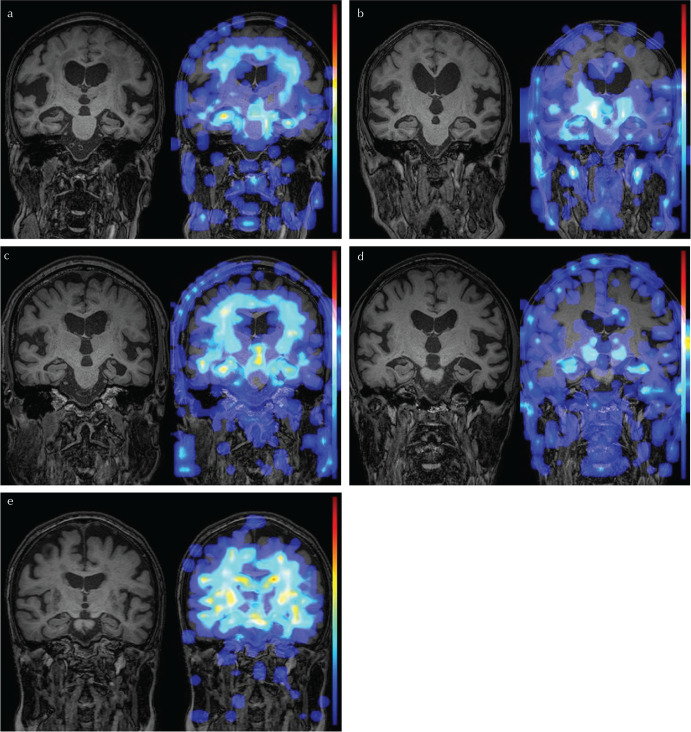
In the Grad-CAM heat map, the brain parenchyma surrounding the lateral ventricle was highlighted in the 10 cases of iNPH that were successfully diagnosed (Fig. 3a). The medial temporal lobe or inferior horn of the lateral ventricle was highlighted in 12 of the successfully diagnosed cases of AD (Fig. 3b). However, about half of the successful cases showed nonspecific heat maps (Fig. 3c and 3d). One iNPH case had enlarged Sylvian fissure and strong atrophy of the hippocampus and was misdiagnosed as AD, it is possible that AD coexisted in that patient (Fig. 4a). Another iNPH case was thought to be a typical iNPH with DESH but misdiagnosed as HC (Fig. 4b). The three cases misdiagnosed as iNPH (Fig. 4c) and the one case misdiagnosed as HC (Fig. 4d) in the AD group visually seemed to be typical AD with strong hippocampal atrophy. One HC case without hydrocephalus was misdiagnosed as iNPH (Fig. 4e).
Fig. 3.
Representative images of successfully diagnosed cases. The 3D T1-weighted image is on the left and the Gradient-weighted Class Activation Mapping heat map overlaid on the 3D T1-weighted image is on the right in each case. Brain parenchyma surrounding the lateral ventricle is highlighted in an idiopathic normal pressure hydrocephalus (iNPH) case (a). Medial temporal lobe or inferior horn of the lateral ventricle is highlighted in an AD case (b). About half of the successful cases show nonspecific heat maps (c: iNPH, d: AD).
Fig. 4.
Representative images of misdiagnosed cases. One idiopathic normal pressure hydrocephalus (iNPH) case with enlarged Sylvian fissure and strong atrophy of the hippocampus was misdiagnosed as AD (a). Another iNPH case showing typical disproportionately enlarged subarachnoid-space hydrocephalus was misdiagnosed as HC (b). Three apparently typical AD cases with strong hippocampal atrophy were misdiagnosed as iNPH (c) and one typical AD case was misdiagnosed as HC (d). One HC case without hydrocephalus was misdiagnosed as iNPH (e).
Discussion
In this study, we proposed the usage of residual extraction approach for deep learning based on a small number of subjects, and successfully developed a model that differentiates iNPH and AD with high accuracy. One of the main problems of image classification using deep learning is overfitting. The cause of overfitting is data noise, which can be categorized into pure noise and any features that are unrelated or marginally related to the estimation. For example, the skull or eyeball features are assumed to be unrelated to the classification of iNPH and AD. Such unrelated features are further categorized into a common feature and a non-related feature of the individual patient. The residual extraction phase of our model subtracts this common feature from the original image using bottleneck-shaped convolutional networks such as a ladder network to reconstruct the original image. Because the model was trained to minimize reconstruction loss, the model should have learned a way to efficiently minimize that loss. In other words, common features should have been preferentially learned. In the present study, although the basis was not clear in some cases, approximately 90% of iNPH and AD were correctly diagnosed. The residual extraction approach seems to be an effective method in a small number of samples.
In the current study, optimization of hyperparameters including model design was not comprehensively performed. However, some important hyperparameters were determined as follows. Adam parameter is an important parameter that determines the length of the optimization step. The default value was α = 0.001, but it had been confirmed that it did not significantly affect the convergence of the loss function that was in the range of 0.0001–0.002 in the preliminary model setup phase. In the optimization of the ladder network, since many voxels were involved in the loss function and hence the convergence was relatively stable, Adam α was set to 0.002. On the other hand, for the training of neural network classifier, we set it to 0.0005 to prioritize the stability of convergence. Number of epochs is also an important hyperparameter. In training the ladder network, the number of epochs was set to 1000 as the loss function (= least square error) did not become any smaller when monitoring loss-epoch curve. In training the neural network classifier, the number of epochs was set to 200 after obtaining the loss-epoch curve that gave the maximum prediction performance for the test data in the preliminary model setup phase. In order to eliminate arbitrariness, the number of epochs was fixed in all leave-one-out tests.
In this study, some cases were misdiagnosed, even though the image was apparently characteristic. Deep learning may identify various factors such as edges, texture, and signal intensity as well as brain atrophy. It is fundamentally a black box. To reveal the basis of diagnosis by deep learning, we used the Grad-CAM approach. There is no specific rule as to which convolutional layer should be used for Grad-CAM to produce heat map images. Each voxel in the feature maps of each layer basically reflects simple image features in the shallower layers and complex features in the deeper layers based on the range of the original image, the number of processes performed so far, and the number of convolutions. Therefore, it is necessary to arbitrarily select one layer to produce heat map images that is visually or scientifically meaningful. In this model, the real-world coordinate range in the input image corresponding to one voxel from first layer to sixth layer was as follows: first, 2.4 × 2.4 × 2.6 mm; second, 5.7 × 5.7 × 6.0 mm; third, 12.2 × 12.2 × 12.9 mm; fourth, 25.2 × 25.2 × 26.7 mm; fifth, 51.2 × 51.2 × 54.2 mm; and sixth, 103.2 × 103.2 × 109.2 mm. Based on the typical size of tissue which was likely to be conspicuous in the Grad-CAM output, we first determined third, fourth, and fifth layer as primary candidates for Grad-CAM heat map. On the fourth and fifth layer, the resolution of the heat map was too low to identify the hippocampus. Therefore, we selected the third layer for producing Grad-CAM heat map images.
The findings of Grad-CAM heat map suggested that atrophy of the medial temporal lobe is useful in distinguishing between iNPH and AD in our model. In contrast, the high convexity area in iNPH was not as emphasized as medial temporal lobe in AD. It is possible that our deep learning model was trained to diagnose AD cases by their characteristic medial temporal lobe atrophy. This model gave a high correct diagnosis rate without referring to the high-convexity area, which is usually useful in the diagnosis of iNPH. That suggests that in the future, it may be possible to find unknown diagnostic features from the analysis results of deep learning using Grad-CAM approach.
In previous studies, comparison of the volumes of brain gray matter and white matter by automated segmentation was able to differentiate between NPH and AD with accuracy of 94–96%,11,12 and comparison of the callosal angle and Evans index demonstrated the accuracy of 90–93%.12 Although the result of the present study was slightly inferior to those of the previous reports, our model based on the deep learning has the advantage that post-processing is unnecessary and the diagnosis can be obtained in 1–2 s per case by simply inputting 3D T1-weighted images. Although there was no statistically significant difference between the results of our model and the interpretation of the radiologists, the diagnostic accuracy of our model was higher than those of the radiologists who conducted the reading study: the radiologists tended to be more accurate for iNPH diagnosis, whereas the deep learning-based model tended to be more accurate for AD. Deep learning may be more accurate than radiologists for diagnosis of AD, by capturing information that is invisible to human’s eyes.
There are some limitations in our study. First, the sample size was small. To overcome this limitation, we used residual extraction approach. Even though our model achieved high accuracy, the results should be validated using another cohort in the future study. Another limitation is the uncertainty of the clinical diagnosis of iNPH. iNPH is a clinical entity without a basis of histology, and there are some cases in which iNPH and AD coexist.10,24 Amyloid-positron emission tomography can reliably exclude the diagnosis of AD,25 and will be helpful to create a highly reliable dataset for deep learning in the future study.
Conclusion
Residual extraction approach in a deep learning method achieved a high accuracy for the differential diagnosis of iNPH, AD, and HC. This method may help differential diagnosis even when a small number of cases are trained.
Acknowledgments
This work was supported by AMED under grant number JP19lk1010025h9902, by JSPS KAKENHI grant number JP16H06280, and by JSPS KAKENHI grant number 17K16486.
Footnotes
Conflicts of Interest
Yujiro Otsuka is an employee of Milliman Inc.; Yumiko Motoi is an endowed chair of Department of Diagnosis, Prevention and Treatment of Dementia, Juntendo University Graduate School of Medicine invested by Nihon Medi-Physics Co., Ltd. and Eisai Co., Ltd.; the other authors have no conflicts of interest.
References
- 1.Adams RD, Fisher CM, Hakim S, Ojemann RG, Sweet WH. Symptomatic occult hydrocephalus with “normal” cerebrospinal-fluid pressure. A treatable syndrome. N Engl J Med 1965; 273:117–126. [DOI] [PubMed] [Google Scholar]
- 2.Hakim S, Adams RD. The special clinical problem of symptomatic hydrocephalus with normal cerebrospinal fluid pressure. Observations on cerebrospinal fluid hydrodynamics. J Neurol Sci 1965; 2:307–327. [DOI] [PubMed] [Google Scholar]
- 3.Mathias JL, Burke J. Cognitive functioning in Alzheimer’s and vascular dementia: a meta-analysis. Neuropsychology 2009; 23:411–423. [DOI] [PubMed] [Google Scholar]
- 4.Black PM, Ojemann RG, Tzouras A. CSF shunts for dementia, incontinence, and gait disturbance. Clin Neurosurg 1985; 32:632–651. [PubMed] [Google Scholar]
- 5.Krauss JK, Droste DW, Vach W, et al. Cerebrospinal fluid shunting in idiopathic normal-pressure hydrocephalus of the elderly: effect of periventricular and deep white matter lesions. Neurosurgery 1996; 39:292–299; discussion 299–300. [DOI] [PubMed] [Google Scholar]
- 6.Kazui H, Miyajima M, Mori E, Ishikawa M, SINPHONI-2 Investigators Lumboperitoneal shunt surgery for idiopathic normal pressure hydrocephalus (SINPHONI-2): an open-label randomised trial. Lancet Neurol 2015; 14:585–594. [DOI] [PubMed] [Google Scholar]
- 7.Birks J. Cholinesterase inhibitors for Alzheimer’s disease. Cochrane Database Syst Rev 2006:CD005593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheltens P, Fox N, Barkhof F, De Carli C. Structural magnetic resonance imaging in the practical assessment of dementia: beyond exclusion. Lancet Neurol 2002; 1:13–21. [DOI] [PubMed] [Google Scholar]
- 9.Hashimoto M, Ishikawa M, Mori E, Kuwana N, Study of INPH on neurological improvement (SINPHONI) Diagnosis of idiopathic normal pressure hydrocephalus is supported by MRI-based scheme: a prospective cohort study. Cerebrospinal Fluid Res 2010; 7:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Savolainen S, Laakso MP, Paljärvi L, et al. MR imaging of the hippocampus in normal pressure hydrocephalus: correlations with cortical Alzheimer’s disease confirmed by pathologic analysis. AJNR Am J Neuroradiol 2000; 21:409–414. [PMC free article] [PubMed] [Google Scholar]
- 11.Serulle Y, Rusinek H, Kirov II, et al. Differentiating shunt-responsive normal pressure hydrocephalus from Alzheimer disease and normal aging: pilot study using automated MRI brain tissue segmentation. J Neurol 2014; 261:1994–2002. [DOI] [PubMed] [Google Scholar]
- 12.Miskin N, Patel H, Franceschi AM, et al. Diagnosis of normal-pressure hydrocephalus: use of traditional measures in the era of volumetric MR imaging. Radiology 2017; 285:197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suk HI, Shen D. Deep learning-based feature representation for AD/MCI classification. Med Image Comput Comput Assist Interv 2013; 16:583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu S, Liu S, Cai W, et al. Multimodal neuroimaging feature learning for multiclass diagnosis of Alzheimer’s disease. IEEE Trans Biomed Eng 2015; 62:1132–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ortiz A, Munilla J, Górriz JM, Ramírez J. Ensembles of Deep learning architectures for the early diagnosis of the Alzheimer’s disease. Int J Neural Syst 2016; 26:1650025. [DOI] [PubMed] [Google Scholar]
- 16.Ambastha AK, Leong TY, Alzheimer’s Disease Neuroimaging Initiative A deep learning approach to neuroanatomical characterisation of Alzheimer’s disease. Stud Health Technol Inform. 2017; 245:1249. [PubMed] [Google Scholar]
- 17.Chen X, Konukoglu E. Unsupervised detection of lesions in brain MRI using constrained adversarial auto-encoders. arXiv e-prints [Internet]. 2018. Available from: https://ui.adsabs.harvard.edu/\#abs/2018arXiv180604972C.
- 18.Mori E, Ishikawa M, Kato T, et al. Guidelines for management of idiopathic normal pressure hydrocephalus: second edition. Neurol Med Chir (Tokyo) 2012; 52:775–809. [DOI] [PubMed] [Google Scholar]
- 19.McKhann GM, Knopman DS, Chertkow H, et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement 2011; 7:263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hawass NE. Comparing the sensitivities and specificities of two diagnostic procedures performed on the same group of patients. Br J Radiol 1997; 70:360–366. [DOI] [PubMed] [Google Scholar]
- 21.Rasmus A, Valpola H, Honkala M, Berglund M, Raiko T. Semi-supervised learning with ladder networks. arXiv:1507.02672, 2015.
- 22.Kingma DP, Ba J. Adam: a method for stochastic optimization. arXiv:1412.6980v9, 2017.
- 23.Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-CAM: Visual explanations from deep networks via gradient-based localization. 2017 IEEE International Conference on Computer Vision (ICCV), Venice, 2017; 618–626. [Google Scholar]
- 24.Golomb J, Wisoff J, Miller DC, et al. Alzheimer’s disease comorbidity in normal pressure hydrocephalus: prevalence and shunt response. J Neurol Neurosurg Psychiatry 2000; 68:778–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Witte MM, Foster NL, Fleisher AS, et al. Clinical use of amyloid-positron emission tomography neuroimaging: practical and bioethical considerations. Alzheimers Dement (Amst) 2015; 1:358–367. [DOI] [PMC free article] [PubMed] [Google Scholar]