Abstract
Many studies on SARS-CoV-2 have been performed over short-time scale, but few have focused on the ultrastructural characteristics of infected cells. We used TEM to perform kinetic analysis of the ultrastructure of SARS-CoV-2-infected cells. Early infection events were characterized by the presence of clusters of single-membrane vesicles and stacks of membrane containing nuclear pores called annulate lamellae (AL). A large network of host cell-derived organelles transformed into virus factories was subsequently observed in the cells. As previously described for other RNA viruses, these replication factories consisted of double-membrane vesicles (DMVs) located close to the nucleus. Viruses released at the cell surface by exocytosis harbored the typical crown of spike proteins, but viral particles without spikes were also observed in intracellular compartments, possibly reflecting incorrect assembly or a cell degradation process.
Keywords: Covid-19, SARS-CoV-2, Virus/cell interactions, Electron microscopy
Introduction
Coronaviruses are infectious agents that infect many species of mammals and birds, and have high zoonotic potential. Some, such as HKU1, OC43, 229E, and NL63, circulate seasonally and cause benign respiratory illnesses in children or adults [1–3]. However, others, such as the severe acute respiratory syndrome coronavirus (SARS-CoV) in 2003 [4] and Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012 [5] have emerged more recently, through cross-species transmission between one or more mammals and humans. These emerging viruses can cause severe respiratory disease in adults. At the end of 2019, the identification, in China, of SARS-CoV-2 as the causal agent of atypical pneumopathies, provided the latest example of these emerging coronaviruses [6, 7]. SARS-CoV-2 is less virulent than SARS-CoV [8], and infected patients seem to be contagious before the onset of symptoms and even asymptomatic individuals appear to be able to transmit the disease [9]. This renders the identification of infected patients and chains of transmission much more complex and has resulted in the massive spread of this new virus worldwide, and the declaration of a pandemic by the WHO on March 11, 2020. The associated disease was named COVID-19 (coronavirus disease 2019). The pathophysiology of SARS-CoV-2 seems to differ in several ways from those of SARS-CoV and the benign human coronaviruses that generally circulate. However, it remains difficult to determine the exact contributions of viral and host factors to these differences.
Coronaviruses, with a single-stranded positive RNA genome of 26–32 kb, have the largest genomes of RNA viruses. The first two-thirds of the genome encodes two polyproteins that are subsequently cleaved by proteases to give rise to non-structural proteins involved in viral replication [10]. The last third of the genome encodes four structural proteins: spike (S), envelope (E), membrane (M), and nucleocapsid (N). Some coronavirus virions contain an additional membrane protein, a hemagglutinin esterase (HE). These proteins assemble to form enveloped virions of about 120 nm in diameter. The first stages of the viral cycle occur on virus-induced double-membrane structures derived from the endoplasmic reticulum (ER), commonly referred to as “double-membrane vesicles” (DMVs) [11]. These structures are mostly located in the perinuclear zone, and are also generated by other RNA viruses, such as arteriviruses, picornaviruses, and the flaviviruses of genus Hepacivirus [12, 13]. In coronaviruses, the nsp3 and nsp4 proteins appear to be sufficient to induce the formation of these structures. The viral genome replication complexes are anchored in the DMV membranes, and their fibrous content consists mostly of de novo synthesized double-stranded viral RNA. In SARS-CoV, these DMVs are not isolated, but integrated into an interconnected reticulovesicular network derived from the ER [14]. Later in the viral life cycle, DMVs fuse into larger cytoplasmic vacuoles containing numerous assembled virions that have budded, this budding probably occurring in the ERGIC (ER-Golgi intermediate compartment) [15].
There are many examples of the usefulness of electron microscopy for improving our understanding of the infectious cycle of viruses [16]. The Vero cell line has been shown to be a relevant model for studies of SARS-CoV-2/cell interactions [17]. We therefore used these cells for a kinetic study of virus-induced DMVs, viral morphogenesis and release for this new pathogen, based on immunofluorescence on confocal microscopy and transmission electron microscopy.
Materials and methods
Viral strain and cells
The SARS-CoV-2 strain used in this study was isolated with a nasal swab sample collected from one of the first COVID-19 cases confirmed in France—a 47 y-o female patient hospitalized in January 2020 in the Department of Infectious and Tropical Diseases, Bichat Claude Bernard Hospital, Paris [18]. This sample was collected according to the declaration of Helsinki and received approval from local ethics commission. The complete viral genome sequence obtained using Illumina MiSeq sequencing technology was then deposited after assembly on the GISAID EpiCoV platform (Accession ID EPI_ISL_411218) under the name BetaCoV/France/IDF0571/2020. Vero-81 cells (ATCC, CCL-81) were maintained in DMEM supplemented with 10% FBS at 37 °C, under an atmosphere containing 5% CO2. To generate virus stocks, we facilitated SARS-CoV-2 infection by transducing these cells with a lentiviral vector expressing transmembrane protease serine 2 (TMPRSS2). TMPRSS2 was inserted into a pTRIP vector, and lentiviral vectors were produced by the transfection of HEK293T cells with pTRIP-TMPRSS2, phCMV-VSVG and HIV gag–pol in the presence of Turbofect (Life Technologies) according to the manufacturer’s instruction. Supernatants containing lentiviral vectors were used to transduce cells twice, 48 h apart. SARS-CoV-2 was then propagated in these Vero-81 cells expressing TMPRSS2. The kinetic study following infection with this strain was performed on regular Vero-81 cells.
Kinetics of SARS-CoV-2 infection
Cells were plated in 24 wells plates (with or without glass coverslips) and infected the next day at a MOI of 0.25 for 1 h. Then the cells were rinsed twice with PBS and incubated at 37 °C. At various time point, post-infection cell supernatants were collected to quantify the secretion of virus and the cells were processed for different analyses (immunostaining, immunoblot, and qRT-PCR).
Immunofluorescence
Infected cells were fixed by incubation in 3% paraformaldehyde for 20 min, and stored in PBS at 4 °C until required for immunolabeling. Cells were permeabilized by incubation with 0.1% Triton X-100 for 5 min, and were then blocked by incubation for 30 min with 5% goat serum (GS) in PBS. Infected cells were labeled with a mixture of the mouse J2 monoclonal antibody against dsRNA (Scicons, diluted 1:1000) and a human monoclonal antibody directed against the spike protein (Sanyoubio #AHA003, diluted 1:250). The cells were incubated with primary antibodies in PBS supplemented with 5% GS for 30 min. They were washed three times with PBS and then incubated for 30 min with Alexa-488-conjugated goat anti-human IgG and cyanin-3-conjugated goat anti-mouse IgG secondary antibodies (Jackson Immunoresearch) in 5% goat serum in PS supplemented with 1 μg/ml DAPI (4′,6-diamidino-2-phenylindole). Coverslips were then rinsed four times with PBS, once in water and mounted on microscope slides in Mowiol 4-88-containing medium. Images were acquired on an Evos M5000 imaging system (Thermo Fisher Scientific) equipped with light cubes for DAPI, GFP, and RFP, and a 10× objective. For each coverslip, six 8-bit images of each channel were acquired. The total number of cells was determined by counting the nuclei. Infected cells, defined as positive for dsRNA and S immunolabeling, were counted, and the percentage of infected cells was calculated. The experiment was performed three times in duplicates. About 7,000–11,000 cells were counted per time point in each experiment using a homemade macro running in ImageJ.
Immunoblot
Infected cells were lysed in non-reducing Laemmli loading buffer. Lysates were incubated at 95 °C for 25 min before exit of the BSL3 facility. Then the samples were reduced by addition of 40 mM dithiothreitol and incubated for 10 min at 70 °C before separation of the proteins on a 10% SDS–polyacrylamide gel electrophoresis. Proteins were transferred on a nitrocellulose membrane (Amersham). Membrane-bound N and S proteins were detected using a rabbit polyclonal antibodiy to SARS nucleocapsid (Novus) and a mouse polyclonal antibody to SARS-CoV-2 S protein produced by Biotem, as primary antibodies, and horseradish peroxidase-conjugated secondary antibody (Jackson Immunoresearch). Detection was carried out by chemoluminescence (SuperSignal™ West Pico PLUS, Thermo Scientific). The signals were recorded using a LAS 3000 apparatus (Fujifilm).
Genome quantification
Total RNA of infected cells was extracted using the Nucleospin RNA kit (Macherey-Nagel) as recommended by the manufacturer. Reverse transcription was performed using the high-capacity cDNA reverse transcription kit (Life Technologies) according to the manufacturer’s instructions. Then RNA were subjected to qRT-PCR using the SYBR green PCR master mix (Life Technologies) with primers (GTGARATGGTCATGTGTGGCGG and CARATGTTAAASACACTATTAGCATA) for RdRp fragment amplification. qRT-PCR was performed with a Quantstudio 3 (Life Technologie). A standard curve was generated with in vitro transcribed RNA corresponding to the RdRp fragment.
Viral secretion
The quantity of virus secreted in cell supernatant was assessed by the TCID50 method.
Transmission electron microscopy
Cells were infected at a MOI of 0.25 for 1 h. Infected and mock-infected cells were then fixed at various time points post-infection, by incubating for 24 h in 1% glutaraldehyde, 4% paraformaldehyde, (Sigma, St-Louis, MO) in 0.1 M phosphate buffer (pH 7.2). Samples were then washed in phosphate-buffered saline (PBS) and post-fixed by incubation for 1 h with 2% osmium tetroxide (Agar Scientific, Stansted, UK). Cells were then fully dehydrated in a graded series of ethanol solutions and propylene oxide. They were impregnated with a mixture of (1∶1) propylene oxide/Epon resin (Sigma) and left overnight in pure resin. Samples were then embedded in Epon resin (Sigma), which was allowed to polymerize for 48 h at 60 °C. Ultra-thin sections (90 nm) of these blocks were obtained with a Leica EM UC7 ultramicrotome (Wetzlar, Germany). Sections were stained with 2% uranyl acetate (Agar Scientific), 5% lead citrate (Sigma), and observations were made with a transmission electron microscope (JEOL 1011, Tokyo, Japan). Infected cells were observed 4, 6, 8, 10, 12, and 24 h post-infection, and compared to mock-infected cells. For quantitative analysis, ultrastructural features were monitored in 100 cells (100 consecutive sections on the EM grid, except those with no nucleus section), for each cell pellet.
Results
Kinetics of SARS-CoV-2 infection in Vero cells
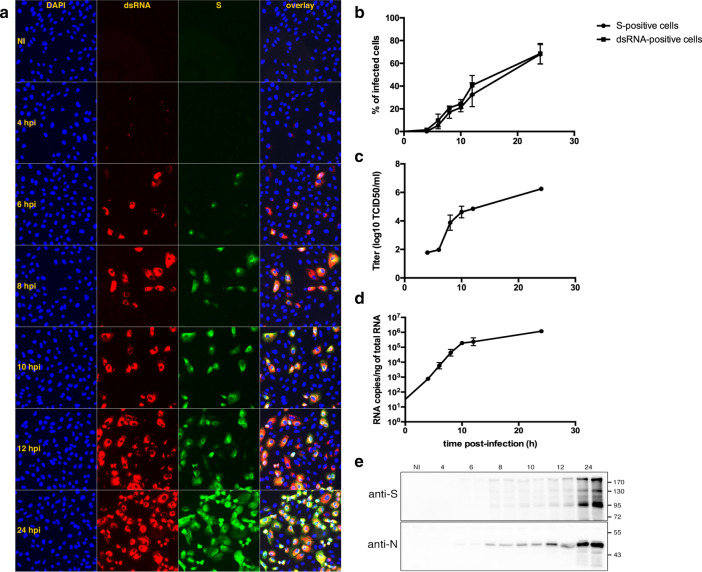
We analyzed the kinetics of SARS-CoV-2 infection. First, we quantified the number of infected cells based on the expression of the spike protein S and the presence of dsRNA, a replication complex marker, by evaluating immunofluorescence over time in infected Vero cells (Fig. 1a). At 4 h post-infection (hpi), a few cells displayed a weak dsRNA signal but no S staining. The first clear sign of infection was observed at 6 hpi, and a gradual increase in the number of infected cells and in the intensity of labeling was observed up to 8–10 hpi. At 12 hpi, the number of infected cells increased again, and groups of cells corresponding to infection foci appeared. These foci contained dsRNA-positive S-negative cells as observed in newly infected cells at 6 hpi, indicating that a new round of infection had occurred. We counted the number of dsRNA-positive cells and of S-positive cells, to measure the progression of the infection (Fig. 1b). Our results suggest that the replication cycle of SARS-CoV-2 has a periodicity of about 8 h in Vero cells. To further confirm this result, we measured the secretion of virus at each time point (Fig. 1c). Viral secretion was detectable at 8 h post-infection and increased over time. In parallel, we monitored viral replication by quantifying intracellular viral RNA (Fig. 1d). Viral replication was detected at 4hpi and increases until 10hpi to reach a plateau at 10–12 hpi. From 12 to 24 hpi, a 0.7 Log moderate increase of the quantity of viral RNA was observed. Finally, the gradual increase of N and S viral proteins in infected cells was confirmed by immunoblot (Fig. 1e).
Fig. 1.
Kinetics of infection analyzed by different methods. (a) Vero cells grown on glass coverslips were infected, fixed at the indicated time points and processed for immunofluorescence labeling of S (green) and dsRNA (red). Nuclei were stained with DAPI (blue). (b) Cells positive for S and dsRNA were counted and the percentage of infected cells was plotted against time. (c) The quantity of virus secreted in the cell supernatant was assessed by the TCID50 method. (d) Intracellular genome quantification was performed by qRT-PCR. (e) Intracellular N and S viral proteins were detected by immunoblot
Ultrastructural features of SARS-CoV-2 infected cells
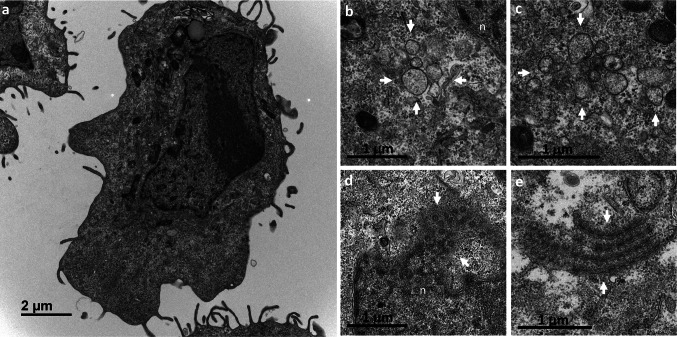
At 4 hpi, we observed no perceptible difference between infected and mock-infected cells (Fig. 2a). The first virus-induced modifications were observed at 6 hpi, when discrete clusters of single-membrane vesicles formed in the cytoplasm (Fig. 2b,c). Another change detectable as early as 6 hpi was the frequent presence of annulate lamellae (AL) in the cell sections (Fig. 2d,e). AL are a particular type of intracellular membrane, consisting of stacks of highly ordered ER-derived membranes, arranged in parallel, and containing nuclear pore complexes (NPCs) [19]. In these SARS-CoV-2-infected Vero cells, they appeared in continuity with the nuclear envelope (Fig. 2d) or within the cytoplasm (Fig. 2e).
Fig. 2.
Early ultrastructural changes encountered in Vero cells at 6 h post-infection with SARS-CoV-2. (a) At 4 hpi, no ultrastructural modifications relative to mock-infected cells, shown here, were visible. (b,c) At 6 hpi, the first discrete virus-induced structures appeared, consisting of clusters (delimited by the white arrows) of several single-membrane vesicles. These clusters contained 5–10 single-membrane vesicles per cell section, and were frequently observed in the perinuclear area (n, nucleus). (d,e) At 6 hpi, annulate lamellae (AL) were frequently observed in infected cells, either as nuclear (n) expansions (d, white arrows) or isolated in the cytoplasm (e, white arrows), suggesting that the formation of these structures may have been promoted by virus infection. AL consist of stacks of highly ordered endoplasmic reticulum-derived membranes, arranged in parallel and containing nuclear pore complexes, that can be visualized in tangential (e) or cross (d) sections
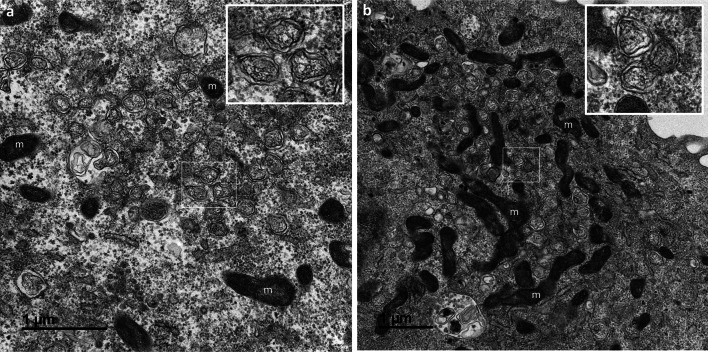
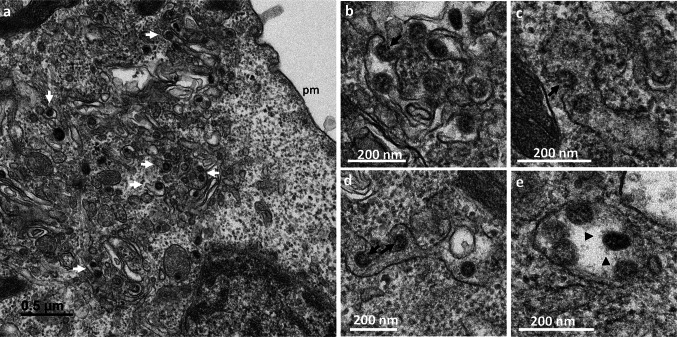
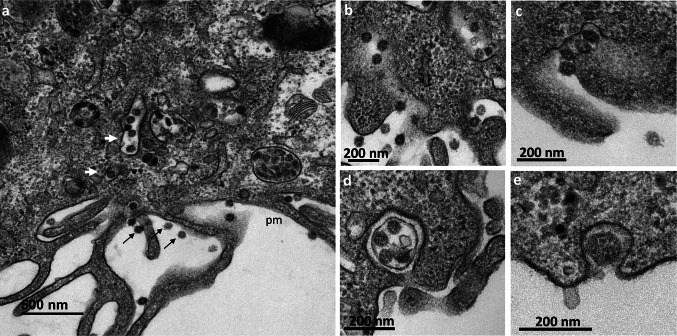
At 8 hpi, massive changes were observed, with the presence of numerous DMVs forming a replication network, frequently surrounded by mitochondria (Fig. 3a,b). At this time point, the first virus particles assembled within the cells were detected in intracellular vesicles probably related to the Golgi apparatus or ERGIC (Fig. 4a). In some sections, the budding of viruses toward the lumen of the ERGIC/Golgi vesicles could be seen (Fig. 4b–d). These budding viral particles or those fully assembled in the vesicle lumen (Fig. 4e) had prominent surface projections, or spikes. These intracellular spiky viruses were detected in small vesicles, each containing only a few particles, at 8 hpi. At 10 and 12 hpi, numerous extracellular particles could be seen at the cell surface (Fig. 5a). Images of virus-carrying vesicles that were presumably moving toward the plasma membrane (Fig. 5a), or merging with the plasma membrane (Fig. 5b–e), suggested that the virions were released by exocytosis mechanisms. The fact that these vesicles often contained several viral particles suggested that they represent viral exit rather than cellular entry.
Fig. 3.
Replication network established in Vero cells at 8 h post-infection with the SARS-CoV-2. (a,b) A massive replication network consisting of large clusters of numerous double-membrane vesicles (DMVs, presented at high magnification in the insets) was observed. These clusters of DMVs were frequently surrounded by mitochondria (m)
Fig. 4.
Morphogenesis of intracellular SARS-CoV-2 virions in Vero cells at 8 h post-infection. (a) Virus particles (white arrows) were observed in intracellular vesicles related to the Golgi apparatus or the endoplasmic reticulum/Golgi intermediate compartment (ERGIC). No virus was released at the plasma membrane (pm) at this time point. (b–d) The budding of the viruses (white arrows) toward the lumen of the ERGIC/Golgi vesicles was visualized in some cell sections. (e) At this time point, the intracellular viruses were detected in small vesicles, each containing only a few viral particles. These viral particles displayed prominent spikes, clearly visible at the surface (black arrowhead)
Fig. 5.
Release of the SARS-CoV-2 virions by exocytosis from the Vero cells at 10 h post-infection. (a) Virions (thin black arrows) were frequently observed at the plasma membrane (pm). Numerous virus-carrying vesicles (white arrows) presumably in transit to the plasma membrane were also visualized. (b–e) These virus-carrying vesicles fused with the plasma membrane to release their contents into the extracellular space by exocytosis. Similar ultrastructural events were observed at 12 hpi
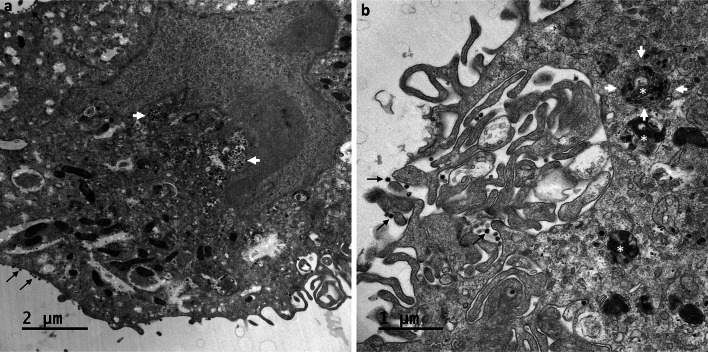
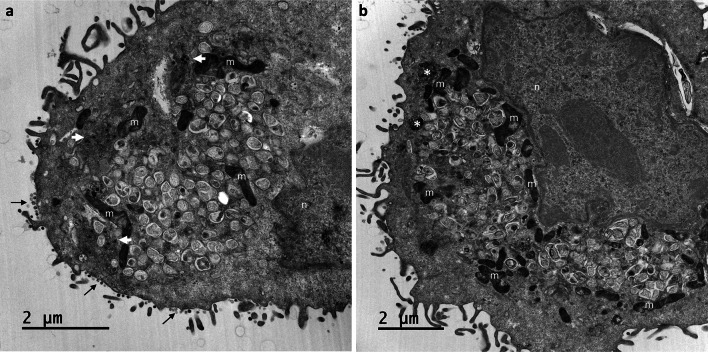
At 24 hpi, viruses were still detectable at the cell surfaces and were present in high amount. Although these viruses could represent virions released by the imaged cells, we could not exclude that some of them originate from adjacent cells before to be trapped at the plasma membrane of these cells. Also, large numbers of viral particles were found to have accumulated in very large intracellular vacuoles (Fig. 6a,b). Surprisingly, the viral particles present in these large intracellular compartments had a smooth surface and bore no spikes (Fig. 7a,b). Their appearance was clearly different from that of viruses released at the cell surface, which carried many spikes (Fig. 8a,b). At these later stages of the infection, most of the Vero cells infected with SARS-CoV-2 contained an abundant reticulovesicular network of DMVs, occupying an entire pole of the cytoplasm, in which viral replication and the assembly of new virions occurred simultaneously (Fig. 9a). DMVs were still the main component of this network, sometimes associated with other virus-induced structures, such as myelin-like membrane whorls or autophagic-like packaged membranes (Fig. 9b). These membrane whorls were sometimes also associated with the spike-less viral particles in the large intracellular vacuoles (Figs. 6b, 7a).
Fig. 6.
Intracellular accumulation of SARS-CoV-2 particles in large vacuoles within Vero cells at 24 hpi. (a,b) Viruses were still detected at the cell surface (thin black arrows), but large numbers of viral particles were observed accumulated in very large intracellular vacuoles (white arrows) of various sizes, predominantly located in the perinuclear region. At this time point, an accumulation of myelin-like membrane whorls or autophagic-like packaged membranes (white asterisk in b) was also observed in the cells, separated from or associated with the viral particles
Fig. 7.
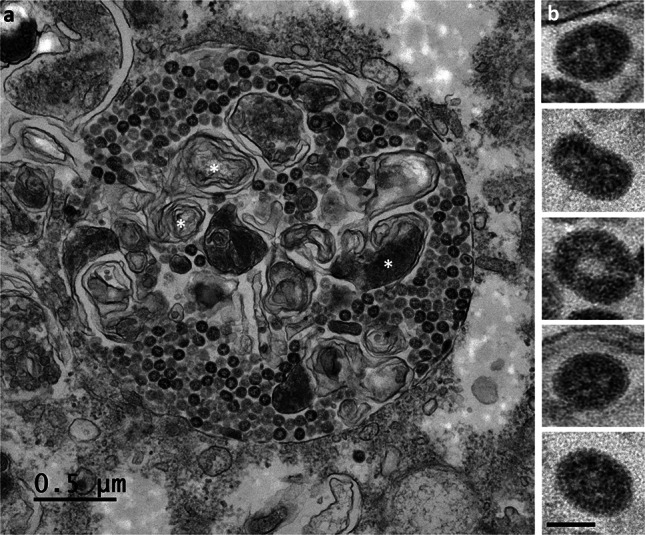
The SARS-CoV-2 particles accumulating in large intracellular vacuoles in Vero cells at 24 hpi have no spikes. (a) The numerous viral particles accumulating in these large intracellular compartments were often electron-dense and presented a smooth surface. Membrane whorls (white asterisks) were sometimes also associated with the viral particles within these large intracellular vacuoles. (b) At high magnification, the viral particles present in these large intracellular vacuoles were clearly observed to have no spikes (bar = 50 nm)
Fig. 8.
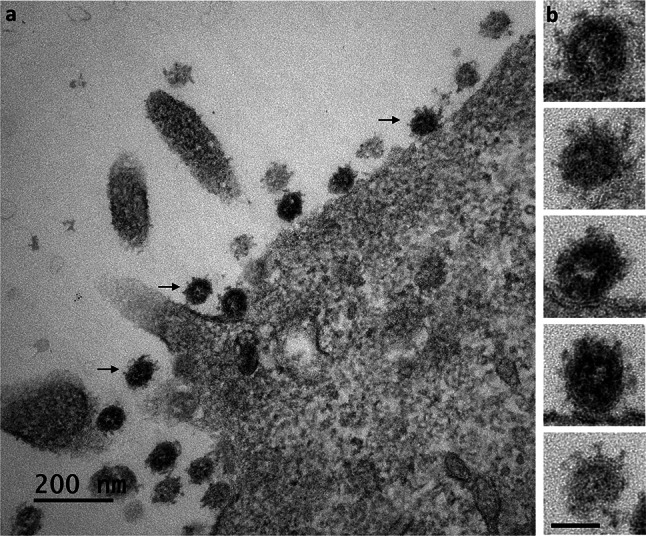
The SARS-CoV-2 particles released from the Vero cells at 24 hpi carry spikes. (a) All the viral particles released at the cell surface (thin black arrows) were surrounded by characteristic, “club-shaped”, spikes. (b) This was confirmed by an analysis of these viral particles at high magnification, revealing their distinctive crown-like appearance (bar = 50 nm)
Fig. 9.
Invasive replication network in Vero cells at 24 h post-infection with SARS-CoV-2. (a,b) At this time point, SARS-CoV-2-infected cells displayed an intense reticulovesicular network consisting mostly of large numbers of DMVs, which occupied almost all the cytoplasm and seemed to push against the nuclear compartment (n), with many mitochondria (m) recruited at the edge of this network. Virion assembly continued next to this network (white arrows in a), leading to release at the plasma membrane (thin black arrows in a). DMVs were often associated with myelin-like membrane whorls or autophagic-like packaged membranes (white asterisk in b)
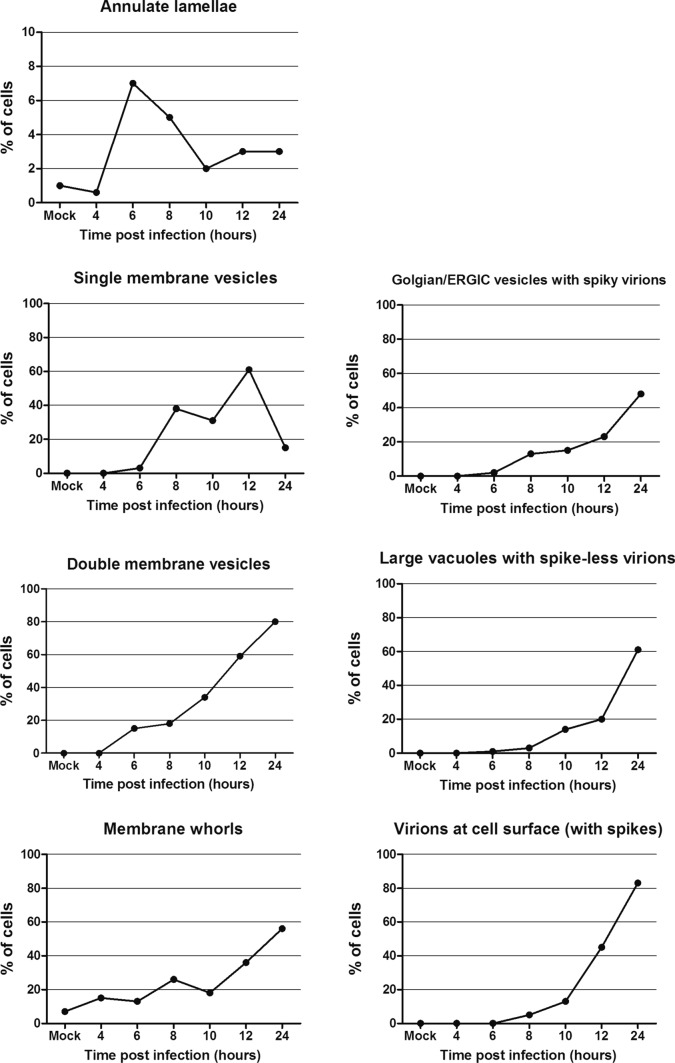
The quantification of these ultrastructural features in 100 consecutive cell sections showed that AL were present in only 1% of mock-infected cells, but increased up to 7% at 6 hpi, before to slowly decrease until 10 hpi and finally stabilize around 3% at 12 hpi (Fig. 10). Clusters of single-membrane vesicles that appeared between 6 and 8 hpi were observed in 40% of the cells at 8 hpi, before reaching 60% at 12 hpi and then dropping to <20% at 24 hpi. DMVs were observed in a constantly increasing number of cells between 4 and 24 hpi, reaching 80% of the cells at 24 hpi (Fig. 10). Membrane whorls and autophagic-like structures, observed in 7% of mock-infected cells, increased slowly during the first 10 hpi and then more rapidly to reach almost 60% of the cells at 24 hpi. Concerning the viral particles, spiky virions found in golgian/ERGIC vesicles were first observed at 8 hpi, and the number of cells with this feature regularly increased to reach 48% at 24 hpi. Large vacuoles containing spike-less virions started to accumulate in cells from 10 hpi, and their number increased to also reach a maximum at 24 hpi, being present in about 60% of the cells. Finally, the number of cells surrounded by extracellular spiky virions increases exponentially from 10 hpi, to reach 80% of the cells at 24 hpi (Fig. 10).
Fig. 10.
Quantitative analysis of the ultrastructural features in infected Vero cells, compared to mock-infected cells. For each point of the kinetics, 100 cell sections have been analyzed. Identification of at least one element in a cell section counted as one in the total account. For the annulate lamellae graph, due to the rarity of these structures and therefore the low associated cell percentage, Y axis displays only a 10% value. * At 4 hpi, 170 cell sections have been analyzed to find one cell containing annulate lamellae, leading to a <1% result
Discussion
As previously reported for coronaviruses and, more recently, for SARS-CoV-2 [20–23], our findings confirm that the SARS-CoV-2 viral cycle is supported by a reticulovesicular network derived from endoplasmic reticulum (ER) membranes and consisting mostly of double-membrane vesicles (DMVs). After the budding of the virions, presumably in the Golgi apparatus stacks or the ER-Golgi intermediate compartment (ERGIC), the virus is released by exocytosis at the plasma membrane. The kinetics of appearance of the DMVs and virions inside and on the surface of the cells are globally similar to those observed in a recent study performed with human pulmonary epithelial Calu-3-infected cells [21]. Thus, our findings confirm previous results, but they also shed new light on early and late stages of the viral infectious cycle.
We show that, at early stages of infection, the cells contained only discrete single-membrane vesicle clusters (Fig. 2b,c), potentially acting as the precursors of DMVs, as previously suggested for other viral models. This hypothesis is consistent with our quantitative analysis (Fig. 10), showing that the number of positive cells for these single-membrane vesicle clusters was decreasing in the late phase of the infection while the number of DMVs positive cells increased (Fig. 10). In hepatitis C virus (HCV), for example, it has been suggested that single-membrane vesicle clusters act as the precursors of the DMVs characterizing the membranous web induced by this virus [24, 25]. A similar process has also been described for poliovirus, for which early viral organelles are gradually transformed into double-membrane structures by the extension of membranous walls and/or the collapse of the luminal cavity of the single-membrane structure [26]. Recent investigations by high-resolution electron tomography analysis of SARS-CoV-2-infected Calu-3 cells have confirmed that DMVs are closely associated to the ER, being linked by smooth ER connectors or sometimes embedded into the rough ER such as the DMV outer membrane was contiguous to the ER membrane [21]. However, the mechanism underlying DMV biogenesis is still poorly understood and the potential role of these single-membrane vesicle clusters in their formation will certainly need further investigation.
We also report the presence of annulate lamellae (AL) in Vero cells infected with SARS-CoV-2 (Fig. 2d,e). These structures are not specific of the viral infection as they were observed in mock-infected cells in few cells (around 1%), but their presence increased following infection (peak at 7% 6 hpi) (Fig. 10). Similar observations were reported by another group, but little comment was made on their possible meaning [27]. AL have no obvious function, other than as a reservoir of nuclear envelope components. AL were initially thought to be an ultrastructural feature characteristic of rapidly growing germ and tumor cells, but they have since been characterized as a marker of infection for various viruses, including hepatitis A virus [28], Japanese encephalitis virus [29], and human herpes virus 6 [30]. However, their role in the infection process is unknown. Interestingly, in the HCV model, infected cells display an increase in the amount of cytoplasmic nuclear pore complex proteins (Nups), which accumulate in or close to cytoplasmic membranes enriched in HCV proteins [31]. It has, therefore, been suggested that cytoplasmic nuclear pore complexes (NPCs), or potentially derivatives of these structures, such as AL, may facilitate establishment of the HCV-induced membranous web [31, 32]. It will be interesting to investigate, whether such a mechanism is also involved in the biogenesis of the SARS-CoV-2 replication network.
As for AL, membrane whorls and autophagic-like structures were not specific of the viral infection as they were observed in mock-infected cells. However, if they were detected in 7% of uninfected cells, the number of cells containing these structures is slowly increased during the first 10 hpi, and then more rapidly to reach almost 60% of the cells at 24 hpi (Fig. 10). Once again, these data are in favor of virus-induced mechanisms stimulating the formation of these host cell membrane-derived structures.
At later stages of infection, we observed large intracellular vacuoles containing viral particles that appeared to be completely devoid of spikes. Such images have been reported by other groups, for SARS-CoV-2 propagated in cell culture [22, 23, 27, 33] or detected in the lung tissue of infected patients [34]. Similar findings were also reported in the very first electron microscopy investigations of SARS-CoV in cultured Vero cells [35, 36]. In the present study, a comparative analysis, at high EM magnifications, of the viral particles present on the cell surface and those present in these large intracellular vacuoles at these time points late in infection clearly showed differences in terms of the presence or absence of spikes (Figs. 7, 8). The reason for the absence of spikes on these intracellular viral particles is currently unknown. This does not seem to be related to a technical problem leading to a loss of the spikes of the intracellular viruses since spikes can be well visualized on the viruses present in the smaller vesicles of ERGIC or Golgi origin (Fig. 4). These large vacuoles containing spike-less viral particles may be intracellular compartments in which degradation through an autophagolysosomal process occurs. This hypothesis is supported by the presence of myelin-like membrane whorls in these compartments (Fig. 7a), as also previously reported for SARS-CoV [36]. However, these spike-less virions did not appear to be degraded in these compartments, although they were sometimes somewhat distorted (Fig. 7b), presenting a “doughnut shape” and a more pronounced electron-dense edge, as previously discussed by another group [27]. Alternatively, these spike-less viral particles may be formed by a defective virion assembly pathway, leading to immature and, thus, non-infectious particles. This would be consistent with previous findings showing that S protein is dispensable for the budding and assembly of coronaviruses [37, 38], whereas N protein seems to be essential for these mechanisms and to drive them [39]. Further investigations will be required to determine the potential role of these spike-free viral particles in the pathophysiology of SARS-CoV-2 infection, particularly given the huge numbers of such particles in infected cells.
A recent study by Ogando et al., in which Vero cells were also used to propagate SARS-CoV-2, suggested that this virus may carry fewer spikes than SARS-CoV [20]. We did not directly compare the two viruses in our study, but our EM images show that SARS-CoV-2 has many “club-shaped” projections at its surface (Fig. 8), as also previously reported for SARS-CoV. The underlying reasons for this remain unclear, but may reflect the ultimate release of spike-less viral particles from the intracellular vacuoles at very late stages of the infectious cycle in the study conducted by Ogando et al. [20], possibly due to cell lysis. This may explain why SARS-CoV-2 was found to be less infectious than SARS-CoV in this study, despite generating larger amounts of intracellular viral RNA.
It will certainly be important to continue this type of investigation with other relevant cellular models. Recent studies have shown the possibility of propagating the virus in polarized organoids of human airway epithelial [40]. Here also, although this has not been particularly commented by the authors, it seems that spike-free virus particles may accumulate intracellularly. In any case, our ultrastructural study provides new insight into the early and late steps of the SARS-CoV-2 infectious cycle, which merit further investigation in future studies of this important new pathogen.
Author contributions
SE: investigation, formal analysis, and writing original draft; YR: investigation, formal analysis, and writing original draft; OT: resources; EB: investigation and formal analysis; KS: investigation and formal analysis; MRC: resources; JD: conceptualization, formal analysis and manuscript editing; SB: conceptualization, investigation, formal analysis, writing original draft, manuscript editing and supervision; PR: conceptualization, investigation, formal analysis, writing original draft, manuscript editing and supervision.
Acknowledgments
S. Eymieux, E. Blanchard and P. Roingeard were supported by INSERM and the University of Tours; O. Terrier and M. Rosa-Calatrava were supported by a special COVID-19 grants from the REACTIng consortium (INSERM), the CNRS and the Institut Mérieux; Y. Rouillé, K. Seron, J. Dubuisson and S. Belouzard were supported by a special COVID-19 grant from the CNRS. We thank Andrès Pizzorno, Adeline Danneels, Fabienne Arcanger and Christine Rey for technical assistance. Our data were obtained with the assistance of the IBiSA Electron Microscopy Facility of the University of Tours.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Sandrine Belouzard and Philippe Roingeard contributed equally to this work.
References
- 1.van der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJM, Wolthers KC, Wertheim-van Dillen PM, Kaandorp J, Spaargaren J, Berkhout B. Identification of a new human coronavirus. Nat Med. 2009;10:368–773. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woo PCY, Lau SKP, Tsoi H-W, Huang Y, Poon RWS, Chu C-M, Lee RA, Luk WK, Wong GK, Wong BH, Cheng VC, Tang BS, Wu AK, Yung RW, Chen H, Guan Y, Chan KH, Yuen KY. Clinical and molecular epidemiological features of coronavirus HKU1-associated community-acquired pneumonia. J Infect Dis. 2005;192:1898–1907. doi: 10.1086/497151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edridge AWD, Kaczorowska J, Hoste ACR, Bakker M, Klein M, Loens K, Jebbink MF, Matser A, Kinsella CM, Rueda P, Ieven M, Goossens H, Prins M, Sastre P, Deijs M, van der Hoek L (2020) Seasonal coronavirus protective immunity is short-lasting. Nat Med. doi: 10.1038/s41591-020-1083-1 (online ahead of print) [DOI] [PubMed]
- 4.Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Ong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling AE, Humphrey CD, Shieh WJ, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang JY, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 5.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus ADME, Fouchier RAM. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 6.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W. A novel coronavirus from patients with pneumonia in China. N Engl J Med. 2019;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Zhou P, Yang X-L, Wang X-G. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verity R, Okell LC, Dorigatti I, Winskill P, Whittaker C, Imai N, Cuomo-Dannenburg G, Thompson H, Walker PGT, Fu H, Dighe A, Griffin JT, Baguelin M, Bhatia S, Boonyasiri A, Cori A, Cucunubá Z, FitzJohn R, Gaythorpe K, Green W, Hamlet A, Hinsley W, Laydon D, Nedjati-Gilani G, Riley S, van Elsland S, Volz E, Wang H, Wang Y, Xi X, Donnelly CA, Ghani AC, Ferguson NM. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect Dis. 2020;20:669–677. doi: 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He X, Lau EHY, Wu P, Deng X, Wang J, Hao X, Lau YC, Wong JY, Guan Y, Tan X, Mo X, Chen Y, Liao B, Chen W, Hu F, Zhang Q, Zhong M, Wu Y, Zhao L, Zhang F, Cowling BJ, Li F, Leung GM. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Liu Q, Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maier HJ, Neuman BW, Bickerton E, Keep SM, Alrashedi H, Hall R, Britton P. Extensive coronavirus-induced membrane rearrangements are not a determinant of pathogenicity. Sci Rep. 2016;6:1–12. doi: 10.1038/srep27126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Netherton CL, Wileman T. Virus factories, double membrane vesicles and viroplasm generated in animal cells. Curr Opin Virol. 2011;1:381–387. doi: 10.1016/j.coviro.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blanchard E, Roingeard P. Virus-induced double-membrane vesicles. Cell Microbiol. 2015;17:45–50. doi: 10.1111/cmi.12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knoops K, Kikkert M, van den Worm SHE, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, Mommaas AM, Snijder EJ. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stertz S, Reichelt M, Spiegel M, Kuri T, Martínez-Sobrido L, García-Sastre A, Weber F, Kochs G. The intracellular sites of early replication and budding of SARS-coronavirus. Virology. 2007;361:304–315. doi: 10.1016/j.virol.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roingeard P. Viral detection by electron microscopy: past, present and future. Biol Cell. 2008;100:491–501. doi: 10.1042/BC20070173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuyama S, Nao N, Shirato K, Kawase M, Saito S, Takayama I, Nagata N, Sekizuka T, Katoh H, Kato F, Sakata M, Tahara M, Kutsuna S, Ohmagari N, Kuroda M, Suzuki T, Kageyama T, Takeda M. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci USA. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pizzorno A, Padey B, Julien T, Trouillet-Assant S, Traversier A, Errazuriz-Cerda E, Fouret J, Dubois J, Gaymard A, Lescure FX, Dulière V, Brun P, Constant S, Poissy J, Lina B, Yazdanpanah Y, Terrier O, Rosa-Calatrava M. Characterization and treatment of SARS-CoV-2 in nasal and bronchial human airway epithelia. Cell Rep Med. 2020;1:100059. doi: 10.1016/j.xcrm.2020.100059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walther TC, Askjaer P, Gentzel M, Habermann A, Griffiths G, Wilm M, Mattaj IW, Hetzer M. RanGTP mediates nuclear pore complex assembly. Nature. 2003;424:689–694. doi: 10.1038/nature01898. [DOI] [PubMed] [Google Scholar]
- 20.Ogando NS, Dalebout TJ, Zevenhoven-Dobbe JC, Limpens RW, van der Meer Y, Caly L, Druce J, de Vries JJC, Kikkert M, Bárcena M, Sidorov I, Snijder EJ. SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J Gen Virol. 2020;101:925–940. doi: 10.1099/jgv.0.001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cortese M, Lee JY, Cerikan B, Neufeldt CJ, Oorschot VMJ, Köhrer S, Hennies J, Schieber NL, Ronchi P, Mizzon G, Romero-Brey I, Santarella-Mellwig R, Schorb M, Boermel M, Mocaer K, Beckwith MS, Templin RM, Gross V, Pape C, Tischer C, Frankish J, Horvat NK, Laketa V, Stanifer M, Boulant S, Ruggieri A, Chatel-Chaix L, Schwab Y, Bartenschlager R. Integrative imaging reveals SARS-CoV-2-induced reshaping of subcellular morphologies. Cell Host Microbe, in press. 2020 doi: 10.1016/j.chom.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendonça L, Howe A, Gilchrist JB, Sun D, Knight ML, Zanetti-Domingues LC, Bateman B, Krebs AS, Chen L, Radecke J, Sheng Y, Li VD, Ni T, Kounatidis I, Koronfel MA, Szynkiewicz M, Harkiolaki M, Martin-Fernandez ML, James W, Zhang P. SARS-CoV-2 assembly and egress pathway revealed by correlative multi-modal multi-scale Cryo-imaging. BioRxiv, preprint. 2020 doi: 10.1101/2020.11.05.370239.Preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neil D, Moran L, Horsfield C, Curtis E, Swann O, Barclay W, Hanley B, Hollinshead M, Roufosse C. Ultrastructure of cell trafficking pathways and coronavirus: how to recognise the wolf amongst the sheep. J Pathol, in press. 2020 doi: 10.1002/path.5547. [DOI] [PubMed] [Google Scholar]
- 24.Ferraris P, Blanchard E, Roingeard P. Ultrastructural and biochemical analyses of hepatitis C virus-associated host cell membranes. J Gen Virol. 2010;91:2230–2237. doi: 10.1099/vir.0.022186-0. [DOI] [PubMed] [Google Scholar]
- 25.Ferraris P, Beaumont E, Uzbekov R, Brand D, Gaillard J, Blanchard E, Roingeard P. Sequential biogenesis of host cell membrane rearrangements induced by hepatitis C virus infection. Cell Mol Life Sci. 2013;70:1297–1306. doi: 10.1007/s00018-012-1213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belov GA, Nair V, Hansen BT, Hoyt FH, Fischer ER, Ehrenfeld E. Complex dynamic development of poliovirus membranous replication complexes. J Virol. 2012;86:302–312. doi: 10.1128/JVI.05937-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brahim Belhaouari D, Fontanini A, Baudoin J-P, Haddad G, Le Bideau M, Yaacoub Bou Khali J, Raoult D, La Scola B. The strengths of scanning electron microscopy in deciphering SARS-CoV-2 infectious cycle. Front Microbiol. 2020;11:2014. doi: 10.3389/fmicb.2020.02014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marshall JA, Borg J, Coulepis AG, Anderson DA. Annulate lamellae and lytic HAV infection in vitro. Tissue Cell. 1996;28:205–214. doi: 10.1016/S0040-8166(96)80008-5. [DOI] [PubMed] [Google Scholar]
- 29.Wang JJ, Liao CL, Chiou YW, Chiou CT, Huang YL, Chen LK. Ultrastructure and localization of E proteins in cultured neuron cells infected with Japanese encephalitis virus. Virology. 1997;238:30–39. doi: 10.1006/viro.1997.8791. [DOI] [PubMed] [Google Scholar]
- 30.Cardinali G, Gentile M, Cirone M, Zompetta C, Frati L, Faggioni A, Torrisi MR. Viral glycoproteins accumulate in newly formed annulate lamellae following infection of lymphoid cells by human herpesvirus 6. J Virol. 1998;72:9738–9746. doi: 10.1128/JVI.72.12.9738-9746.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neufeldt CJ, Joyce MA, Levin A, Steenbergen RH, Pang D, Shields J, Tyrrell DL, Wozniak RW. Hepatitis C virus-induced cytoplasmic organelles use the nuclear transport machinery to establish an environment conducive to virus replication. PLoS Pathog. 2013;9:e1003744. doi: 10.1371/journal.ppat.1003744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonamassa B, Ciccarese F, Antonio VD, Contarini A, Palù G, Alvisi G. Hepatitis C virus and host cell nuclear transport machinery: a clandestine affair. Front Microbiol. 2015;6:e619. doi: 10.3389/fmicb.2015.00619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldsmith CS, Miller SE, Martines RB, Bullock HA, Zaki SR. Electron microscopy of SARS-CoV-2: a challenging task. Lancet. 2020;395:e99. doi: 10.1016/S0140-6736(20)31188-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martines RB, Ritter JM, Matkovic E, Gary J, Bollweg BC, Bullock H, Goldsmith CS, Silva-Flannery L, Seixas JN, Reagan-Steiner S, Uyeki T, Denison A, Bhatnagar J, Shieh WJ, Zaki SR (2020) Pathology and pathogenesis of SARS-CoV-2 associated with fatal coronavirus disease, United States. Emerg Infect Dis, in press. doi: 10.3201/eid2609.202095 [DOI] [PMC free article] [PubMed]
- 35.Ng ML, Tan SH, See EE, Ooi EE, Ling AE. Early events of SARS coronavirus infection in Vero cells. J Med Virol. 2003;71:323–331. doi: 10.1002/jmv.10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng ML, Tan SH, See EE, Ooi EE, Ling AE. Proliferative growth of SARS coronavirus in Vero E6 cells. J Gen Virol. 2003;84:3291–3303. doi: 10.1099/vir.0.19505-0. [DOI] [PubMed] [Google Scholar]
- 37.Rottier PJ, Horzinek MC, van der Zeijst BA. Viral protein synthesis in mouse hepatitis virus strain A59-infected cells: effect of tunicamycin. J Virol. 1981;40:350–357. doi: 10.1128/jvi.40.2.350-357.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ujike M, Taguchi F. Incorporation of spike and membrane glycoproteins into coronavirus virions. Viruses. 2015;7:1700–1725. doi: 10.3390/v7041700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McBride R, van Zyl M, Fielding BC. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu N, Wang W, Liu Z, Liang C, Wang W, Ye F, Huang B, Zhao L, Wang H, Zhou W, Deng Y, Mao L, Su C, Qiang G, Jiang T, Zhao J, Wu G, Song J, Tan W. Morphogenesis and cytopathic effect of SARS-CoV-2 infection in human airway epithelial cells. Nat Commun. 2020;11:3910. doi: 10.1038/s41467-020-17796-z. [DOI] [PMC free article] [PubMed] [Google Scholar]