Abstract
Objective
The role of tumor-infiltrating lymphocytes (TILs) has not yet been characterized in sarcomas. The aim of this bioinformatics study was to explore the effect of TILs on sarcoma survival and genome alterations.
Methods
Whole-exome sequencing, transcriptome sequencing, and survival data of sarcoma were obtained from The Cancer Genome Atlas. Immune infiltration scores were calculated using the Tumor Immune Estimation Resource. Potential associations between abundance of infiltrating TILs and survival or genome alterations were examined.
Results
Levels of CD4+ T cell infiltration were associated with overall survival of patients with pan-sarcomas, and higher CD4+ T cell infiltration levels were associated with better survival. Somatic copy number alterations, rather than mutations, were found to correlate with CD4+ T cell infiltration levels.
Conclusions
This data mining study indicated that CD4+ T cell infiltration levels predicted from RNA sequencing could predict sarcoma prognosis, and higher levels of CD4+ T cells infiltration indicated a better chance of survival.
Keywords: Sarcoma, CD4+ T cells, survival, immune infiltration, copy number variation, histological subtype
Introduction
Sarcomas are rare malignant tumors of mesenchymal origin that are characterized by remarkable heterogeneity. Overall, sarcomas fall into two categories: soft tissue sarcomas (such as those from fat, muscle, nerve, nerve sheath, blood vessels, and other connective tissue) and bone sarcomas (including osteosarcoma, Ewing sarcoma, chondrosarcoma, and chordoma), according to the fourth edition of the World Health Organization (WHO) Classification of Tumors of Soft Tissue and Bone. Over 100 distinct histological sarcoma subtypes are recognized.1 Sarcomas are some of the deadliest cancers when considering refractory and metastatic disease, with a median overall survival (OS) of 12 to 18 months. Furthermore, their rarity and heterogeneity has made the development of novel therapies challenging,2 and a distant metastasis is noted after successful treatment for the primary tumor lesion in approximately 25% of sarcoma patients.
Tumor-infiltrating lymphocytes (TILs) have been implicated as an important prognostic factor with contradictory roles in sarcoma. In some studies examining Ewing sarcoma, gastrointestinal stromal tumor (GIST), cutaneous angiosarcoma (AS), leiomyosarcoma (LMS), synovial sarcoma (SS), and high-grade undifferentiated pleomorphic sarcoma (UPS), the presence of TIL has been associated with improved prognosis.3–5 However, in other studies examining multiple sarcoma histologies, T cell infiltration was associated with a worse survival or had no effect on survival.6–8 Overall, a better understanding of the particular T cell subgroup and the activation status is necessary.
In the present study, whole-exome sequencing, transcriptome sequencing, and survival data of sarcomas from The Cancer Genome Atlas (TCGA) were obtained. An in silico analysis method, Tumor IMmune Estimation Resource (TIMER), was used to construct an in-depth characterization of the immune cell subsets that infiltrate into sarcomas on the basis of the transcriptome sequencing data. We analyzed the potential association of immune infiltration with prognosis and with genome alterations.
Materials and methods
Ethical approval
This study used bioinformatics data from a public database and did not involve human participants; therefore, informed consent was not required. In addition, no approval was required from an institutional review board.
Data acquisition
TCGA sarcoma data, including mutations, somatic copy number alterations (SCNAs), gene expression, and clinical information, were obtained from cBioPortal (https://www.cbioportal.org/). In total, 255 samples, with both clinical and gene alteration data, were obtained and used in the analysis.
Survival analysis
The abundance of six immune infiltrates (B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells) was estimated using TIMER (https://cistrome.shinyapps.io/timer/) based on gene expression.9 Each sarcoma sample was then classified into a high (>median of all tumors) or low (≤median of all tumors) immune infiltration group according to the infiltration score in each immune cell group. Survival analysis was performed on the two immune infiltration groups. Kaplan–Meier curves were plotted to visualize survival differences, and the log-rank test was conducted to identify statistical significance.
Another method of estimating tumor immune infiltration, The Cancer Immunome Atlas (TCIA, https://tcia.at/), was used to validate the results from TIMER. Sub-groups of CD4+ T cells, including activated CD4+ T cells (Act CD4+), effector memory CD4+ T cells (Tem CD4+), and regulatory T cells (Tregs), were estimated in TCIA. Survival analysis was performed using the median cutoff of each cell type as mentioned above.
Identification of differentially expressed genes
To identify genes that may contribute to immune infiltration levels, we compared high and low CD4+ T cell infiltration groups using the Welch two-sample t-test based on log2-scaled FPKM (fragments per kilobase of transcript per million mapped reads) values. The p-values were adjusted using the false discovery rate (FDR) method for multiple comparisons. Differentially expressed genes (DEGs) were identified as those with an FDR < 0.05 and a log2 fold change (FC) ≥ 1 or ≤ −1.
Functional enrichment analysis
Functional enrichment analysis for Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways was performed using the R package clusterProfiler.10 A hypergeometric model was used to detect enriched gene categories with an FDR < 0.05.
Immune infiltration and gene alterations
We examined potential associations between immune infiltration and gene alterations for both mutations and SCNAs. Only frequently mutated genes were selected for the analysis. These included three genes (ATRX, RB1, and TP53) that predominantly harbored mutation and copy number losses, and four genes (CDK4, FRS2, HMGA, and MDM2) that predominantly harbored a copy number gain. For the gene mutation analysis, each tumor was categorized as mutated or wild-type (WT) on the basis of the occurrence of a given gene mutation. For the copy number variations, each tumor was categorized as loss (−1, −2), no change (0), or gain (1, 2) on the basis of results generated by GISTIC2.11 Boxplots were generated to visualize the distribution of immune infiltration score, and a two-sided Welch t-test was performed to determine statistical significance.
Results
Overall survival of pan-sarcoma patients
Six major histological sarcoma subtypes, including LMS, dedifferentiated liposarcoma (DDLPS), UPS, myxofibrosarcoma (MFS), SS, and malignant peripheral nerve sheath tumor (MPNST), from 255 individuals were obtained from TCGA. Two of these, SS and MPNST, were later excluded in the histological subtype survival analysis because of the limited number of individuals present.
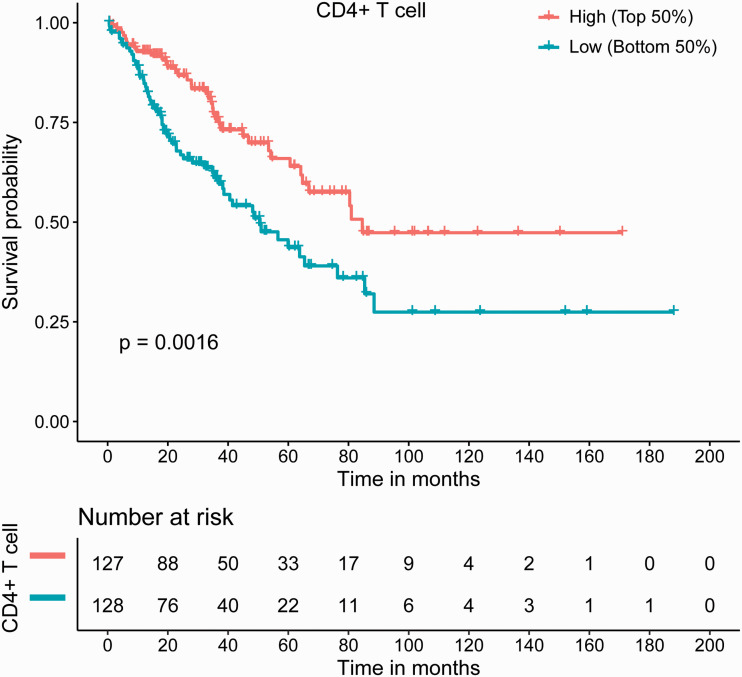
Immune infiltration scores of six distinct immune subsets, including B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells, were calculated by TIMER, with results based on transcriptome sequencing data from TCGA. Herein, we focused on lymphocytes, including B cells and T cells differentiated from a common lymphoid progenitor, with B cell, CD8+ T cell, and CD4+ T cell subsets selected for further analysis. Sarcoma prognosis in relation to the immune infiltration scores of the three cell subsets was assessed. The results showed that CD4+ T cell infiltration levels were significantly correlated with OS in pan-sarcomas (p = 0.0016), with higher infiltration levels indicating better prognosis (Figure 1). However, B cell and CD8+ T cell infiltration levels were not correlated with prognosis in pan-sarcomas (data not shown).
Figure 1.
CD4+ T cell infiltration levels were significantly correlated with sarcoma prognosis, with higher infiltration levels being associated with better survival. High and Low refer to the infiltration score.
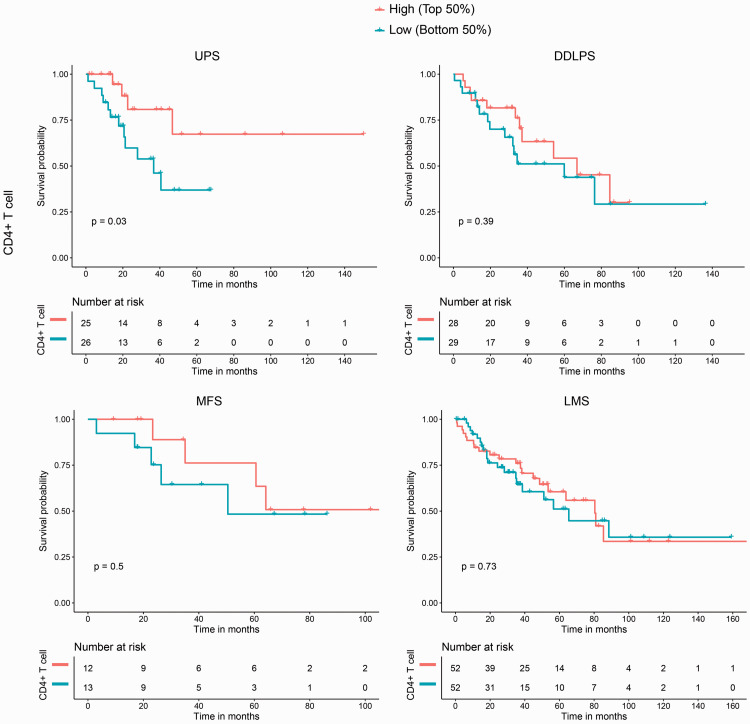
Considering the remarkable histological heterogeneity of sarcomas, we determined correlations between OS and immune infiltration scores within each sarcoma histological subtype. Four histological subtypes (UPS, DDLPS, MFS, and LMS) were investigated and the results showed that CD4+ T cell infiltration levels were significantly correlated with OS in UPS (p = 0.03), whereas no significant correlations were noted for the other subtypes; trends suggested that patients with higher survival rates also had higher CD4+ T cell infiltration scores (Figure 2). Compared with CD4+ T cell infiltration levels in sarcoma histological subtypes, levels of B cell and CD8+ T cell infiltration were not correlated with OS in the four histological subtypes, and the trends were irregular (data not shown).
Figure 2.
CD4+ T cell infiltration levels were significantly correlated with overall survival (OS) in undifferentiated pleomorphic sarcoma (UPS), but not in dedifferentiated liposarcoma (DDLPS), myxofibrosarcoma (MFS), or leiomyosarcoma (LMS). High and Low refer to the infiltration score in each histological subtype.
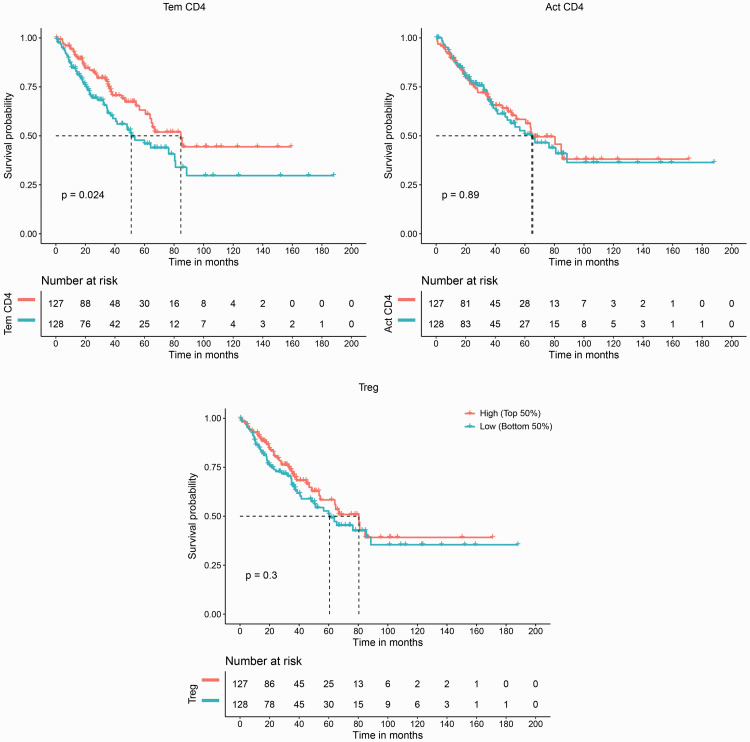
TIMER analyzes six major immune cell groups, which is useful in identifying clinical associations; however, higher resolution of the TIL landscape is required. Recently, several novel computational methods have attempted to enumerate tumor-infiltrating immune cells from transcriptomes. Therefore, we reanalyzed the TCGA RNA-Seq data of pan-sarcomas using an independent computational method, TCIA,12 to further validate our discovery. TCIA predicts immune-related factors classified into four categories: MHC molecules, immunomodulators, effector cells (EC), and suppressor cells (SC), and three CD4+ T cell-related parameters from EC and SC categories, activated (Act) CD4+, effector memory (Tem) CD4+, and regulatory T cells (Tregs),12 were further analyzed. The results showed that infiltration levels of Tem CD4+ cells were significantly related to OS (p = 0.024,), with higher infiltration indicating a better OS, whereas Act CD4+ T cells and Tregs were not significantly related to OS (Figure 3). The results showed that Tem CD4+ T cells infiltration may be associated with better survival in sarcomas.
Figure 3.
Validation of the association between CD4+ T cell infiltration and sarcoma survival. An independent computational method, The Cancer Immunome Atlas (TCIA, https://tcia.at/), was used to validate the association between CD4+ T cell infiltration and sarcoma survival discovered by Tumor IMmune Estimation Resource (TIMER). Infiltration of effector memory CD4+ T (Tem CD4+) cells was significantly correlated with overall survival (OS), but infiltration of activated CD4+ T cells (Act CD4+) and regulatory T cells (Treg) was not. High and Low refer to the infiltration score in each immune cell group.
Immune activation processes and pathways
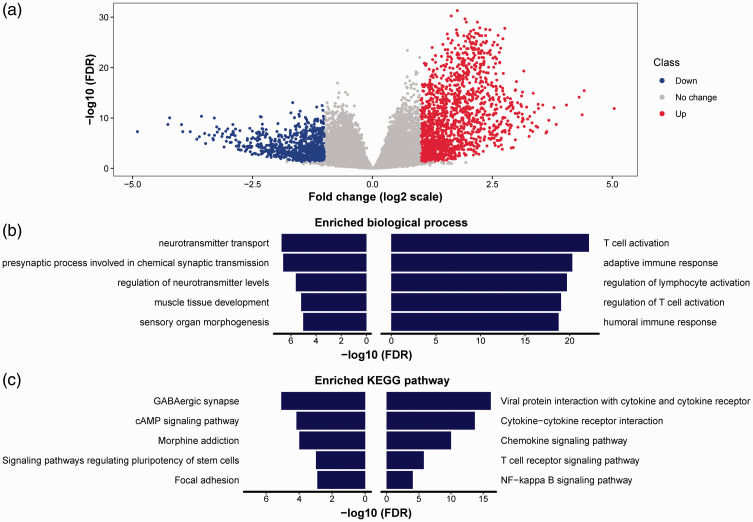
Next, we determined factors contributing to CD4+ T cell infiltration. We analyzed DEGs between high and low tumor infiltration groups. In total, we identified 2181 DEGs, including 1388 upregulated and 793 downregulated genes in the high CD4+ T cells infiltration group (Figure 4a). We then performed functional analysis for biological processes of GO and for KEGG pathways using both upregulated and downregulated genes (the top 200 DEGs were selected and ranked by fold change). The upregulated genes were mainly enriched in immune activation–related biological processes such as T cell activation, adaptive immune response, and regulation of lymphocyte activation (Figure 4b) and in immune-related KEGG pathways such as cytokine−cytokine receptor interaction, chemokine signaling pathway, and T cell receptor signaling pathway (Figure 4c). The biological processes and KEGG pathways enriched in downregulated genes were not related to the immune system. These results showed that immune activation may contribute to CD4+ T cell infiltration.
Figure 4.
Identification of differentially expressed genes (DEGs) between high and low CD4+ T cell infiltration groups and function enrichment analysis. (a) Volcano plot comparing each gene between the high and low CD4+ T cell infiltration groups. The red and blue dots represented upregulated and downregulated genes in high CD4+ T cell group, respectively. The gray dots represented genes without statistical significance. (b) The top 5 enriched biological processes from Gene Ontology using top 200 upregulated (right column) and downregulated (left column) genes. (c) The top 5 enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways using top 200 upregulated (right column) and downregulated (left column) genes.
FDR, false discovery rate.
Genomic aberrations and TIL infiltration levels
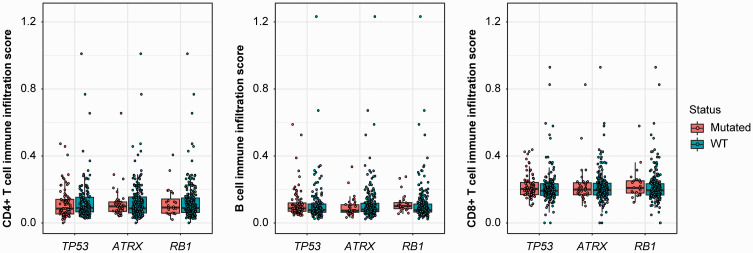
Tumor development and progression are associated with multiple genomic aberrations, which in turn may influence TILs. We analyzed potential correlations of TIL infiltration levels with gene mutations. Mutations of TP53, ATRX, and RB1 were not associated with infiltration of CD4+ T cells, B cells, or CD8+ T cells (Figure 5). When we examined the histological subtypes separately, the mutation rates of TP53, ATRX, and RB1 did not affect TIL infiltration levels in 4 subtypes, including DDLPS, LMS, MFS, and UPS (data not shown).
Figure 5.
CD4+ T cell infiltration levels were not associated with mutations in highly altered genes. Three highly mutated genes, TP53, ATRX, and RB1, were not associated with CD4+ T cell infiltration levels. The boxes span from the first to the third quartile, with the line inside the box representing the median value. The whiskers show the minimum and maximum values or values up to 1.5 times the interquartile range below or above the first or third quartile. The circles refer to the sarcoma patients.
WT, wild-type.
SCNAs and TIL infiltration levels
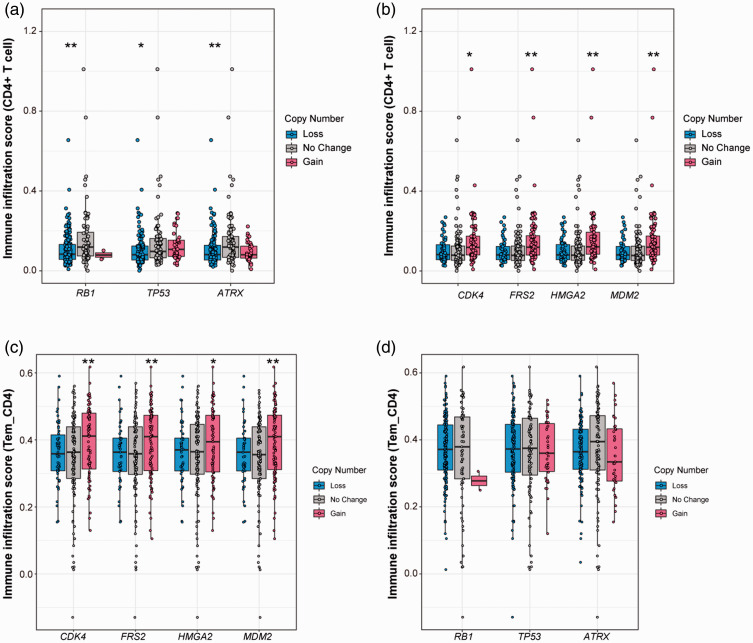
Next, we examined potential correlations between SCNAs and TIL infiltration levels. Unlike the gene mutations, SCNAs were found to be associated with TIL infiltration levels. Losses of TP53, RB1, and ATRX were found to be significantly correlated with decreased CD4+ T cell infiltration levels, and loss of ATRX was significantly correlated with a decrease in B cell infiltration (Figure 6a). Furthermore, gains of MDM2, FRS2, CDK4, and HMGA2 were significantly associated with increased CD4+ T cell infiltration levels but not with B cell or CD8+ T cell infiltration levels (Figure 6b). Overall, SCNAs rather than mutations were associated with TILs, especially CD4+ T cells.
Figure 6.
CD4+ T cell infiltration levels were correlated with somatic copy number alterations (SCNAs) (A, B) and the correlation was validated by The Cancer Immunome Atlas (TCIA, https://tcia.at/) (C, D). (a) The loss of TP53, ATRX, and RB1 was associated with a decrease of CD4+ T cell infiltration. (b) The gain of CDK4, FRS2, HMGA2, and MDM2 was correlated with an increase of CD4+ T cell infiltration. (C, D) Association between infiltration levels of Tem CD4+ and SCNAs of TP53, ATRX, and RB1 (c), and CDK4, FRS2, HMGA2 and MDM2 (d). The boxes span the first to the third quartile with the line inside the box representing the median value. The whiskers show the minimum and maximum values or values up to 1.5 times the interquartile range below or above the first or third quartile. The circles refer to the sarcoma patients. Tem CD4+, effector memory CD4+ T cells (one subtype of CD4+ lymphocytes). **p < 0.01; *p < 0.05.
We then examined SCNAs in relation to TIL infiltration levels in the histological subtypes. Loss of RB1 and ATRX was significantly associated with a decrease in CD4+ T cell infiltration levels in LMS (all p < 0.05), whereas loss of ATRX was associated with decreased B cell infiltration (p < 0.05). Moreover, gains of FRS2 and MDM2 were significantly correlated with an increase in CD4+ T cell infiltration in DDLPS (all p < 0.05). Although not significant, most individuals with gains of CDK4 and HMGA2 had higher CD4+ T cell infiltration scores in DDLPS. In the other 3 subtypes, individuals with gains of MDM2, FRS2, CDK4, and HMGA2 had higher CD4+ T cell infiltration levels, although not significant. Additionally, gains of MDM2, FRS2, CDK4, and HMGA2 were significantly correlated with decreased CD8+ T cell infiltration in UPS when separately analyzed (all p < 0.05).
Next, we validated the infiltration levels of Tem CD4+ calculated by TCIA in relation to sarcoma SCNAs. Increased Tem CD4+ infiltration levels were significantly correlated with gain of MDM2, FRS2, CDK4, and HMGA2 (all p < 0.05; Figure 6c). Although no significant correlations were noted, trends suggested that decreased Tem CD4+ infiltration levels were associated with individuals having loss of TP53, RB1, and ATRX (Figure 6D). These results may confirm the link between SCNAs and CD4+ T cells.
Discussion
Genomic landscape studies of sarcoma have provided large quantities of sequencing data that can aid in the analysis of immune cell infiltration by utilizing bioinformatics. The present study is the first to investigate the role of TILs in sarcoma and showed that TIL infiltration levels are correlated with prognosis and associated with SCNAs.
TILs play an important role in tumor development, with several studies indicating that they can serve a prognostic role in different cancer types.13–16 Furthermore, these TILs have been shown to have functional plasticity, with both pro- and antitumor properties. Although CD8+ cytotoxic T lymphocytes are generally antitumorigenic and restrain tumor development, CD4+ T and B lymphocytes have been shown to have dual roles. For example, in colon and lung carcinomas, CD4+ T cell infiltration correlates with a favorable prognosis, whereas in breast and renal cancers, CD4+ T cell infiltration correlates with poor survival.17–20
In this study, we found that CD4+ T cell infiltration levels were significantly correlated with OS in pan-sarcomas. An independent computational method, TCIA, was used to validate this discovery, and the degree of infiltration of the Tem CD4+ subgroup was found to be correlated with OS of sarcoma. When analyzing histological subtypes individually, CD4+ T cell infiltration levels were found to be significantly correlated with prognosis in UPS, with consistent trends noted in other histological subtypes (DDLPS, MFS, and LMS), although these were not significant (Figure 2). However, given the heterogeneity of sarcoma and the limited number of samples examined for each subtype, it is still possible that an association is present. The validation by TCIA helps strengthen the possibility of such an association. Thus, future studies could analyze more samples for each individual subtype. Unfortunately, an alternative appropriate dataset, in addition to the TCGA dataset, that could be used to validate and confirm the discoveries presented herein is not available, which is a major limitation of this study.
Another way to determine CD4+ T cell infiltration levels is by immunohistochemistry (IHC). Previous IHC studies have investigated the prognostic significance of CD4+ T lymphocyte infiltration in sarcomas. Sorbye et al.3 analyzed 249 soft tissue sarcomas, including 68 UPS, 67 LMS, and 34 LPS. The sarcoma subtypes were partially overlapped with the TCGA cohort used in our study, and were analyzed using IHC to evaluate the prognostic significance of CD3+, CD4+, CD8+, CD20+, and CD45+ lymphocytes infiltration. In univariate analyses, increased infiltrated CD4+ lymphocyte numbers correlated with a significantly improved disease-specific survival in patients with wide resection margins (p = 0.008). However, no such relationship was apparent for CD8+ lymphocytes,3 which is consistent with our findings. The consistency indicates that CD4+ lymphocytes may have a significant role in sarcoma survival. Nevertheless, when examining the peritumoral capsule of a sarcoma, CD4+ and CD8+ lymphocytes numbers had no relationship with disease-specific survival, and B cells were negatively associated with disease-specific survival.21 The above two studies3,21reveal that TILs have different roles in intratumor and peritumor tissues. In another study, predominantly examining GIST, which is absent from the TCGA cohort, low infiltration of CD3+ (p = 0.050) and CD4+ (p = 0.050) appeared to correlate with better OS,6 which is in contrast to our results, perhaps because of the histological heterogeneity of GIST. Unfortunately, the cited studies did not examine TIL infiltration levels and did not explore potential associations between TIL infiltration levels and prognosis within individual histological subtypes. No suitable IHC data were available to validate our findings. Furthermore, in the present study, IHC could not be performed due to a lack of sarcoma tissue samples.
The association between SCNAs of highly altered genes and CD4+ lymphocyte infiltration in sarcoma requires further study. We speculate that the infiltration of CD4+ lymphocytes into the tumor microenvironment is related to the signal pathways involved with these genes. Unfortunately, no research addressing related findings has been identified. Future studies should investigate the mechanism of CD4+ lymphocyte infiltration and its role in survival in sarcomas.
Conclusions
Our findings indicate that predicted CD4+ T cell infiltration levels may be correlated with sarcoma prognosis, and that sarcoma SCNAs may be associated with CD4+ T cell infiltration levels. Further studies focusing on CD4+ T cell infiltration and its role in prognosis are needed to improve our understanding of the mechanism of sarcoma pathogenesis. Additionally, the potential role of genomic alterations in relation to CD4+ T cell infiltration requires further investigation.
Acknowledgments
The results shown here are based on data generated by the TCGA Research Network (https://cancergenome.nih.gov/). We thank the patients who donated samples to the TCGA and consented to share the resulting data. We also thank TCGA for the unrestricted access provided to the data used in this work. We thank LetPub (www.letpub.com.cn) for English language editing.
Footnotes
Declaration of conflicting interest: The authors declare that there is no conflict of interest.
Funding: This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
ORCID iD: Jie Lin https://orcid.org/0000-0003-3784-8120
References
- 1.Jo VY, Fletcher CD. WHO classification of soft tissue tumours: an update based on the 2013 (4th) edition. Pathology 2014; 46: 95–104. doi: 10.1097/PAT.0000000000000050. [DOI] [PubMed] [Google Scholar]
- 2.Wilky BA, Goldberg JM. From vision to reality: deploying the immune system for treatment of sarcoma. Discov Med 2017; 23: 61–74. [PubMed] [Google Scholar]
- 3.Sorbye SW, Kilvaer T, Valkov A, et al. Prognostic impact of lymphocytes in soft tissue sarcomas. PLoS One 2011; 6: e14611. doi: 10.1371/journal.pone.0014611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berghuis D, Santos SJ, Baelde HJ, et al. Pro-inflammatory chemokine-chemokine receptor interactions within the Ewing sarcoma microenvironment determine CD8(+) T-lymphocyte infiltration and affect tumour progression. J Pathol 2011; 223: 347–357. doi: 10.1002/path.2819. [DOI] [PubMed] [Google Scholar]
- 5.Fujii H, Arakawa A, Utsumi D, et al. CD8(+) tumor-infiltrating lymphocytes at primary sites as a possible prognostic factor of cutaneous angiosarcoma. Int J Cancer 2014; 134: 2393–2402. doi: 10.1002/ijc.28581. [DOI] [PubMed] [Google Scholar]
- 6.D’Angelo SP, Shoushtari AN, Agaram NP, et al. Prevalence of tumor-infiltrating lymphocytes and PD-L1 expression in the soft tissue sarcoma microenvironment. Hum Pathol 2015; 46: 357–365. doi: 10.1016/j.humpath.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim C, Kim EK, Jung H, et al. Prognostic implications of PD-L1 expression in patients with soft tissue sarcoma. BMC Cancer 2016; 16: 434. doi: 10.1186/s12885-016-2451-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toulmonde M ,Adam J, Bessede A, et al. Integrative assessment of expression and prognostic value of PDL1, IDO, and kynurenine in 371 primary soft tissue sarcomas with genomic complexity. J Clin Oncol 2016; 34(15_suppl): 11008. (Abstr.) [Google Scholar]
- 9.Li T, Fan J, Wang B, et al. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res 2017; 77: e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012; 16: 284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mermel CH, Schumacher SE, Hill B, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 2011; 12: R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charoentong P, Finotello F, Angelova M, et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep 2017; 18: 248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 13.Shah W, Yan X, Jing L, et al. A reversed CD4/CD8 ratio of tumor-infiltrating lymphocytes and a high percentage of CD4(+)FOXP3(+) regulatory T cells are significantly associated with clinical outcome in squamous cell carcinoma of the cervix. Cell Mol Immunol 2011; 8: 59–66. doi: 10.1038/cmi.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han S, Zhang C, Li Q, et al. Tumour-infiltrating CD4(+) and CD8(+) lymphocytes as predictors of clinical outcome in glioma. Br J Cancer 2014; 110: 2560–2568. doi: 10.1038/bjc.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gooden MJ, De Bock GH, Leffers N, et al. The prognostic influence of tumour-infiltrating lymphocytes in cancer: a systematic review with meta-analysis. Br J Cancer 2011; 105: 93–103. doi: 10.1038/bjc.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azimi F, Scolyer RA, Rumcheva P, et al. Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J Clin Oncol 2012; 30: 2678–2683. doi: 10.1200/JCO.2011.37.8539. [DOI] [PubMed] [Google Scholar]
- 17.Ruffell B, DeNardo DG, Affara NI, et al. Lymphocytes in cancer development: polarization towards pro-tumor immunity. Cytokine Growth Factor Rev 2010; 21: 3–10. doi: 10.1016/j.cytogfr.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siddiqui SA, Frigola X, Bonne-Annee S, et al. Tumor-infiltrating Foxp3-CD4+CD25+ T cells predict poor survival in renal cell carcinoma. Clin Cancer Res 2007; 13: 2075–2081. doi: 10.1158/1078-0432.CCR-06-2139. [DOI] [PubMed] [Google Scholar]
- 19.Wakabayashi O, Yamazaki K, Oizumi S, et al. CD4+ T cells in cancer stroma, not CD8+ T cells in cancer cell nests, are associated with favorable prognosis in human non-small cell lung cancers. Cancer Sci 2003; 94: 1003–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matkowski R, Gisterek I, Halon A, et al. The prognostic role of tumor-infiltrating CD4 and CD8 T lymphocytes in breast cancer. Anticancer Res 2009; 29: 2445–2451. [PubMed] [Google Scholar]
- 21.Sorbye SW, Kilvaer TK, Valkov A, et al. Prognostic impact of peritumoral lymphocyte infiltration in soft tissue sarcomas. BMC Clin Pathol 2012; 12: 5. doi: 10.1186/1472-6890-12-5. [DOI] [PMC free article] [PubMed] [Google Scholar]