Abstract
Objective
To investigate the association between microRNA-3615 (miR-3615) expression and the prognosis and clinicopathological features in patients with hepatocellular carcinoma (HCC).
Methods
We obtained clinicopathological and genomic data and prognostic information on HCC patients from The Cancer Genome Atlas (TCGA) database. We then analyzed differences in miR-3615 expression levels between HCC and adjacent tissues using SPSS software, and examined the relationships between miR-3615 expression levels and clinicopathological characteristics. We also explored the influence of miR-3615 expression levels on the prognosis of HCC patients using Kaplan–Meier survival curve analysis.
Results
Based on data for 345 HCC and 50 adjacent normal tissue samples, expression levels of miR-3615 were significantly higher in HCC tissues compared with adjacent tissues. MiR-3615 expression levels in HCC patients were negatively correlated with overall survival time and positively correlated with high TNM stage, serum Ki-67 expression level, and serum alpha-fetoprotein level. There were no significant correlations between miR-3615 expression and age, sex, and pathological grade.
Conclusion
MiR-3615 may be a promising new biomarker and prognostic factor for HCC.
Keywords: MicroRNA-3615, hepatocellular carcinoma, prognosis, The Cancer Genome Atlas, survival, gene expression
Introduction
Hepatocellular carcinoma (HCC) is the main pathological type of primary liver cancer. HCC has the fifth highest annual incidence and third highest mortality of all cancers worldwide. China accounts for more than half of all cases of liver cancer globally each year.1 A lack of early sensitive diagnostic markers and methods means that HCC is usually diagnosed at a later stage, with a consequently poor prognosis and difficult clinical treatment, leading to a 5-year survival rate of less than 7%.2 There is thus an urgent need to identify biomarkers and therapeutic targets for the early detection of HCC, to improve its early diagnosis and survival rates.
MicroRNAs (miRNAs) are a class of endogenous single-stranded non-coding RNAs with a length of about 22 nucleotides, which have been confirmed to be involved in all types of tested cancers to date. MiRNAs can inhibit mRNA translation by binding to a target mRNA or long non-coding RNA (lncRNA), or can degrade bound mRNAs and lncRNAs.3 An imbalance in miRNA regulation can lead to the development of malignant tumors.2 MiR-3615 is an miRNA with regulatory roles in Barrett’s esophagus,4 follicular lymphoma,5 colorectal adenocarcinoma,6 and other diseases. However, the expression of miR-3615 in HCC and its relationship with clinicopathological features have not been well studied.
In this study, we investigated the expression of miR-3615 in HCC using an online database, and analyzed the relationships between its expression levels and clinical characteristics. We also plotted survival curves to determine the relationship between miR-3615 expression level and overall survival (OS), to determine its influence on the prognosis of HCC. The results of this study may provide new diagnostic and prognostic targets and thus improve the clinical outcomes of patients with HCC.
Patients and methods
Genomic and clinicopathological data
The Cancer Genome Atlas (TCGA, https: //tcga-data.nci.nih.gov/tcga/) is a public project using high-throughput genomic analysis techniques to improve the prevention, diagnosis, and treatment of cancer.7 We downloaded data from TCGA, as described previously.8 We compared miR-3615 expression levels in HCC patients between tumor and adjacent normal tissues from the same patients. We also examined the correlations between expression levels of miR-3615 in HCC tissues and various clinical features and prognosis in HCC patients. The data for this study were derived from a public database and ethical review was therefore deemed unnecessary.
Classification
Patients were classified before treatment according to sex, age, serum alpha fetoprotein (AFP) level (cutoff: 8.366 [median]), Ki-67 expression level (cutoff: 10.000 [median]), TNM stage, and pathological grade. Patients were also divided into high-expression and low-expression groups according to the median expression level of miR-3615 in the cancer tissues.
Statistical analysis
Statistical analysis was carried out using SPSS Statistics for Windows, Version 23.0 (IBM Corp., Armonk, New York, USA). Quantitative TCGA data consistent with a normal distribution were expressed as mean ± standard deviation. Differences in miR-3615 expression levels between HCC tissues and adjacent tissues were analyzed by t-tests, and relationships between miR-3615 expression levels and clinical characteristics were analyzed by χ2 tests. Kaplan–Meier survival curves were drawn to determine the relationship between miR-3615 expression level and prognosis of HCC patients. The significance probability threshold was set at 0.05.
Results
Patients and miR-3615 expression levels in HCC tissues and adjacent tissues
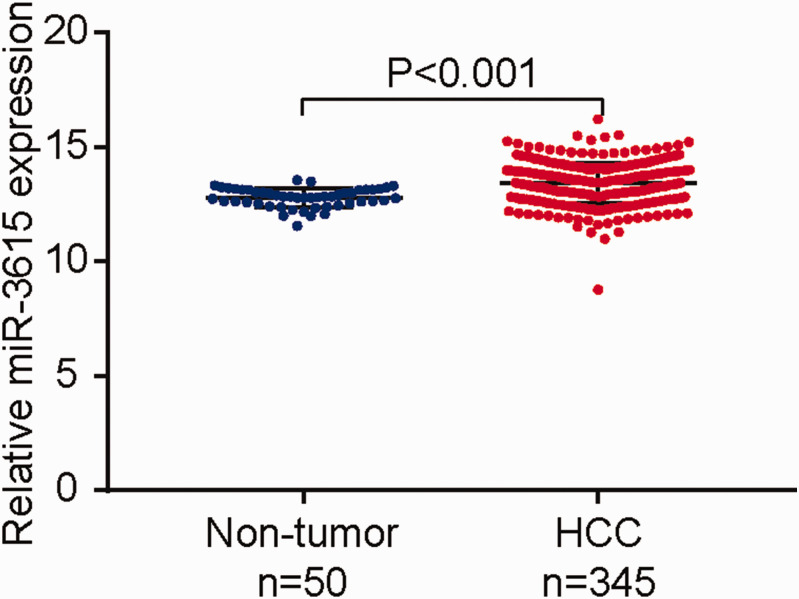
Data for 345 HCC samples (including 50 paired non-tumor tissues from the same patients) were obtained by screening the genomic data from TCGA database and excluding individuals with incomplete data on gene expression, clinicopathological features, and prognosis. Expression levels of miR-3615 were significantly higher in HCC tissues (relative expression: 13.41 ± 0.87) compared with adjacent tissues (12.78 ± 0.41) (P < 0.001) (Figure 1). These findings suggested that miR-3615 was significantly up-regulated in HCC tissues compared with non-tumor tissues.
Figure 1.
Expression of miR-3615 in 345 hepatocellular carcinoma (HCC) tissues and 50 adjacent non-tumor tissues. MiR-3615 relative expression levels (expressed in transcripts per million, TPM) were significantly higher in HCC tissues (13.41 ± 0.87) compared with adjacent non-tumor tissues (12.78 ± 0.41) (P < 0.001).
Relationships between miR-3615 expression level and clinical characteristics
MiR-3615 levels were positively correlated with TNM stage, serum Ki-67 expression level, and AFP level. Patients were divided into high-expression (n=173) and low-expression groups (n=172) according to the median expression level of miR-3615 in the cancer tissues (10.00)). Patients with high expression of miR-3615 expression had high TNM stage, and high serum Ki-67 and AFP levels. However, age, sex, and pathological grade were not correlated with miR-3615 (Table 1). Collectively, these findings suggested that elevated miR-3615 expression was associated with an unfavorable prognosis among patients with HCC.
Table 1.
Relationships between miR-3615 expression level and clinicopathological characteristics.
| General information and clinicopathological features |
Expression level of miR-3615 |
χ2 | P | |
|---|---|---|---|---|
| Low (n = 172) | High (n = 173) | |||
| Sex | ||||
| Female | 48 | 62 | 2.498 | 0.114 |
| Male | 124 | 111 | ||
| Age, years | ||||
| ≤60 | 93 | 78 | 2.784 | 0.095 |
| >60 | 79 | 95 | ||
| Pathological grade | ||||
| I–II | 111 | 104 | 0.717 | 0.391 |
| III–IV | 61 | 69 | ||
| TNM stage | ||||
| I–II | 139 | 116 | 8.472 | 0.004 |
| III–IV | 33 | 57 | ||
| Serum AFP level | ||||
| Low | 96 | 76 | 4.872 | 0.027 |
| High | 76 | 97 | ||
| Ki-67 level | ||||
| Low | 116 | 57 | 41.049 | < 0.001 |
| High | 56 | 116 | ||
TNM, tumor-node-metastasis; AFP, alpha fetoprotein.
Relationship between miR-3615 expression level and prognosis
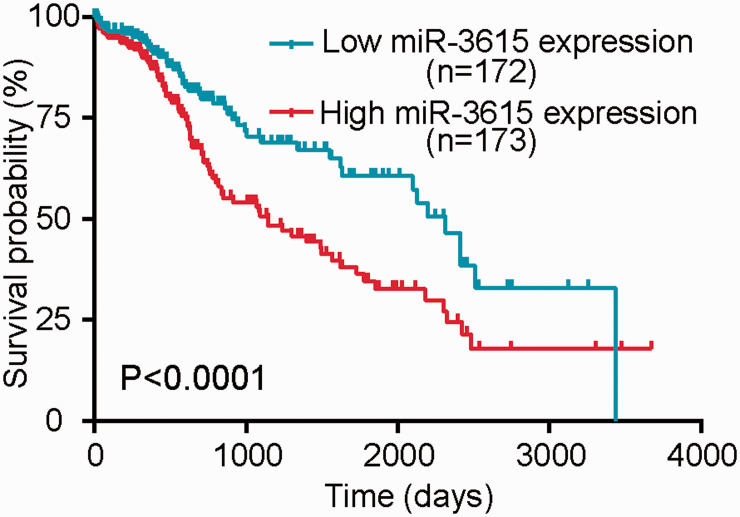
Kaplan–Meier survival curves indicated that HCC patients with low miR-3615 expression had significantly longer OS than those with high miR-3615 expression (P < 0.001) (Figure 2). These results suggested that miR-3615 could be a novel prognostic biomarker in HCC.
Figure 2.
Relationship between miR-3615 expression level and prognosis. Hepatocellular carcinoma patients with low miR-3615 expression had significantly longer overall survival than those with high miR-3615 expression (P < 0.0001).
Discussion
MiRNAs regulate the cell cycle, and cell differentiation and migration. They may also act as tumor suppressor genes or oncogenes during tumorigenesis and tumor development. Members of miRNA families in the miR17/18/19a/20/19b/92 cluster have previously demonstrated a strong embryonic signature, associated with proliferation and cancer.9 MiRNAs are also involved in the development of a variety of cancers and have been widely reported in HCC. Rashad et al.10 found that expression levels of miRNA-27a and miRNA-18b were related to the development of HCC and could be used as prognostic factors in HCC, indicating its distant metastasis, Child–Pugh grade, and degree of lymph node metastasis. Kong et al.11 proved that miR4435-2HG was up-regulated in liver cancer and promoted the proliferation of cancer cells through up-regulated miRNA-487A. In addition, Aye et al.12 reported that reducing miR-200b-3P in cancer cells promoted angiogenesis in HCC tissues by enhancing the expression of ERG (erythroblast transformation-specific [ETS]-related gene) in endothelial cells.
MiR-3615 was significantly down-regulated in high-grade intra-epithelial neoplasia/Barrett’s adenocarcinoma (HG-IEN/BAc) samples in patients with Barrett’s esophagus.4 In addition, miR-3615 was down-regulated in Chinese patients with follicular lymphoma, as demonstrated by quantitative reverse transcription-polymerase chain reaction.5 MiR-3615 was also identified as one target of five altered miRNAs in retinitis pigmentosa,13 suggesting an association between oxidative stress and the occurrence and development of retinitis pigmentosa.
In the current study, miR-3615 was highly expressed in liver cancer, and patients with high miR-3615 expression had poorer OS. MiR-3615 was also positively correlated with TNM stage, AFP level, and Ki-67 level. AFP level is widely used as a diagnostic marker of HCC, while high levels of Ki-67 can indicate tumor invasiveness, such as advanced tumor stage, portal vein invasion, and intrahepatic metastasis, which are in turn associated with high early recurrence rates and poor prognosis of HCC.14 Expression levels of miR-3615 were positively correlated with serum AFP and Ki-67 levels, suggesting that miR-3615 could be a useful biomarker for HCC. It could potentially make up for deficiencies in the early detection of serum AFP and imaging, and could indicate a poor prognosis. Detection of miR-3615 is thus an ideal candidate method for improving the early diagnosis of HCC.
This study was limited by restricting its analysis to HCC tissues from TCGA database, and further studies including serological and clinical data, as well as cell experiments, are needed to verify the results.
Conclusion
MiR-3615 is highly expressed in HCC tissues and its expression levels correlate with clinicopathological features and patient survival. MiR-3615 may thus represent a novel biomarker and therapeutic target in HCC.
Footnotes
Availability of data and materials: The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declaration of conflicting interest: The authors declare that there is no conflict of interest.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Natural Science Foundation of China [No. U1904164].
ORCID iD: Xin Yuan https://orcid.org/0000-0002-4355-0331
References
- 1.Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010; 127: 2893–2917. [DOI] [PubMed] [Google Scholar]
- 2.Li Q, Zou C, Zou C, et al. MicroRNA-25 functions as a potential tumor suppressor in colon cancer by targeting Smad7. Cancer Lett 2013; 335: 168–174. [DOI] [PubMed] [Google Scholar]
- 3.Zhao T, Xu J, Liu L, et al. Computational identification of epigenetically regulated lncRNAs and their associated genes based on integrating genomic data. FEBS Lett 2015; 589: 521–531. [DOI] [PubMed] [Google Scholar]
- 4.Fassan M, Realdon S, Cascione L, et al. Circulating microRNA expression profiling revealed miR-92a-3p as a novel biomarker of Barrett’s carcinogenesis. Pathol Res Pract 2020; 216: 152907. [DOI] [PubMed] [Google Scholar]
- 5.Pan Y, Guo Y, Luo Y, et al. MicroRNA expression profiling of Chinese follicular lymphoma by microarray: A preliminary study. Int Immunopharmacol 2016; 39: 41–47. [DOI] [PubMed] [Google Scholar]
- 6.Zheng G, Wang H, Zhang X, et al. Identification and validation of reference genes for qPCR detection of serum microRNAs in colorectal adenocarcinoma patients. PLoS One 2013; 8: e83025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015; 19: A68–A77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Liu X, Liu L, et al. Upregulation of FEN1 Is Associated with the tumor progression and prognosis of hepatocellular carcinoma. Dis Markers 2020; 2020: 2514090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farh KK, Grimson A, Jan C, et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science 2005; 310: 1817–1821. [DOI] [PubMed] [Google Scholar]
- 10.Rashad NM, El-Shal AS, Shalaby SM, et al. Serum miRNA-27a and miRNA-18b as potential predictive biomarkers of hepatitis C virus-associated hepatocellular carcinoma. Mol Cell Biochem 2018; 447: 125–136. [DOI] [PubMed] [Google Scholar]
- 11.Kong Q, Liang C, Jin Y, et al. The lncRNA MIR4435-2HG is upregulated in hepatocellular carcinoma and promotes cancer cell proliferation by upregulating miRNA-487a. Cell Mol Biol Lett 2019; 24: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moh-Moh-Aung A, Fujisawa M, Ito S, et al. Decreased miR-200b-3p in cancer cells leads to angiogenesis in HCC by enhancing endothelial ERG expression. Sci Rep 2020; 10: 10418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donato L, Bramanti P, Scimone C, et al. miRNA expression profile of retinal pigment epithelial cells under oxidative stress conditions. FEBS Open Bio 2018; 8: 219–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niu ZS, Niu XJ, Wang M. Management of hepatocellular carcinoma: Predictive value of immunohistochemical markers for postoperative survival. World J Hepatol 2015; 7: 7–27. [DOI] [PMC free article] [PubMed] [Google Scholar]