Abstract
General control non-repressible 5 (GCN5)-related N-acetyltransferases (GNATs) catalyse the acetylation of a diverse range of substrates, thereby orchestrating a variety of biological processes within prokaryotes and eukaryotes. GNAT enzymes can catalyze the transfer of an acetyl group from acetyl coenzyme A to substrates such as aminoglycoside antibiotics, amino acids, polyamines, peptides, vitamins, catecholamines, and large macromolecules including proteins. Although GNATs generally exhibit low to moderate sequence identity, they share a conserved catalytic fold and conserved structural motifs. In this current study we characterize the high-resolution X-ray crystallographic structure of a GNAT enzyme bound with acetyl-CoA from Elizabethkingia anophelis, an important multi-drug resistant bacterium. The tertiary structure is comprised of six α-helices and nine β-strands, and is similar with other GNATs. We identify a new and uncharacterized GNAT dimer interface, which is conserved in at least two other unpublished GNAT structures. This suggests that GNAT enzymes can form at least five different types of dimers, in addition to a range of other oligomers including trimer, tetramer, hexamer, and dodecamer assemblies. The high-resolution structure presented in this study is suitable for future in-silico docking and structure–activity relationship studies.
Subject terms: Enzymes, Structural biology
Introduction
Elizabethkingia anophelis is a newly discovered gram negative bacterium, isolated from the midgut of Anopheles gambiae mosquito1,2. The bacterium is emerging as an important and opportunistic human pathogen, with capacity for causing neonatal meningitis and sepsis. Multi-drug resistance profiles against several antibiotics have been reported3, although the antimicrobial susceptibility and the resistance mechanism to antibiotics remain unresolved. Recent studies have reported antibiotic resistance to ampicillin, chloramphenicol, kanamycin, streptomycin and tetracycline4,5, while resistance against tetracycline, trimethoprim/sulfamethoxazole, and ciprofloxacin are of high concern since these antibiotics are first-line therapies for infections caused by this pathogen6. Infections caused by E. anophelis are generally hospital-acquired and include nosocomial pneumonia, bacteremia, sepsis, meningitis, skin and soft-tissue infection, urinary tract infection, and biliary tract infection in neonates and adults with underlying other diseases including malignancies and/or immunosuppression7–9. Several outbreaks of E. anophelis across many countries, has led to recent interest in discovering the underlying mechanisms of antibiotic resistance and possible new drug targets to overcome resistance10–12.
General control non-repressible 5 (GCN5)-related N-acetyltransferases (GNATs) are a large superfamily of enzymes, playing prominent roles across a large number of biological processes including aminoglycoside antibiotic resistance, transcription regulation, protein acetylation and stress reaction13,14. GNAT proteins possess a highly conserved catalytic fold, and can acetylate a wide range of substrates ranging from antibiotics to proteins15,16. The transfer of an acetyl group from acetyl-CoA (AcCoA) to substrates such as aminoglycoside antibiotics alter the fundamental characteristics of these molecules and can render them inactive17,18.
Despite the relatively low number of studies on bacterial protein acetylation, evidence is gathering to suggest that this post-translational modification occurs on many bacterial virulence factors and may play a key role in bacterial virulence14. Previous studies have demonstrated that GNAT enzymes of several bacteria are capable of acetylating different antibiotics16,19,20. E. anophelis strain 12012‐2PRCM has been reported to be resistant against aminoglycosides, β‐lactams, polypeptides, sulfonamides, chloramphenicols, quinolones, and tetracyclines5.
Previous structural characterizations of GNAT proteins have led to a consensus or conserved fold common to GNAT family members. Functional GNAT proteins generally contain six (or sometimes seven) β-strands and four α-helices arranged in a (β0)-β1-α1-α2-β2-β3-β4-α3-β5-α4-β6 topology21. These secondary structural elements comprise four conserved sequence motifs, motif C (β1, α1), motif D (β2-3-α2), motif A (β4-α3), B (β5-α4))21. The N-terminus is moderately well conserved while the C-terminal region varies considerably, in part, due to this region being responsible for substrate binding22. With the emergence of multi-drug resistance (MDR) in E. anophelis, here we determine the structure of an uncharacterized GNAT family acetyltransferase from this organism, which may provide a platform for in-silico fragment screening, drug-design, and/or structure–activity relationship studies.
Materials and methods
Cloning, expression, and purification
The gene for BAY10_3400 was amplified from genomic DNA and cloned into the expression vector BG1861 using ligation-independent cloning23. The expression vector provides a non-cleavable N-terminal His6-tag (SSGCID target ID ElanA.19303.a; UniProt ID A0A1T3E2H1). The enzyme was expressed in E. coli Rosetta BL21(DE3)R3 following standard SSGCID protocols, as described previously24. Purification was performed using Ni–NTA affinity and size exclusion chromatography following standard SSCID protocols25. Briefly, the bacterial cell pellet was suspended in a buffer comprised of 25 mM HEPES, 500 mM NaCl, 5% glycerol, 30 mM imidazole, 0.025% sodium azide, 0.5% CHAPS, 10 mM MgCl2, 1 mM TCEP, 250 µg/ml AEBSF, 0.05 µg/ml lysozyme pH 7.0). Cell lysis was undertaken by sonication, and the resulting extract clarified by centrifugation and passed over a HisTrap FF 5 ml column pre-equilibrated in 25 mM HEPES, 500 mM NaCl, 5% glycerol, 30 mM imidazole, 0.025% sodium azide, 1 mM TCEP pH 7.0. The column was washed with 20 column volumes of 25 mM HEPES, 500 mM NaCl, 5% glycerol, 30 mM imidazole, 0.025% sodium azide, 1 mM TCEP pH 7.0 to remove unbound proteins. The His-tagged protein was eluted with seven column volumes of a buffer comprised of 25 mM HEPES, 500 mM NaCl, 5% glycerol, 1 mM TCEP, 250 mM imidazole and 0.025% azide pH 7.0. The protein was further purified by size exclusion chromatography using a HiLoad 26/60 Superdex 75 preparative-grade column pre-equilibrated in 25 mM HEPES, 500 mM NaCl, 5% glycerol, 2 mM DTT, 0.025% azide pH 7.0, and calibrated using the GE Healthcare Calibration Kit (28-4038-41) with conalbumin, ovalbumin and ribonuclease A. Two protein peaks, corresponding to monomer and dimer species (molecular weights determined from the elution volume and a standard calibration curve) were pooled and analyzed by SDS–PAGE (see Supplementary Figure 1). The purified proteins were concentrated to 16 mg/ml (fractions from the dimer peak) and 36 mg/ml (fractions from the monomer peak) in 25 mM Hepes pH 7.5, 0.3 M NaCl, 1 mM DTT, 0.025% w/v sodium azide, 10% glycerol), flash frozen in liquid nitrogen and stored at − 80 °C.
Crystallization, data collection, and structure determination
Crystallization trials were performed with apo protein at 16 mg/ml (from the sample obtained from the first elution peak) (see Supplementary Figure 1), and 36 mg/ml (from the sample obtained from the second elution peak) (see Supplementary Figure 1), using 96-well XJR crystallization trays (Rigaku Reagents) with 0.4 μl protein mixed with 0.4 μl reservoir, equilibrating against 80 µl reservoir solution. Crystallization conditions were searched for with sparse matrix screens JCSG + (Rigaku Reagents), CrystalScreen HT, Index HT (Hampton Research), and PACT (Molecular Dimensions). Crystallization trays were incubated at 285 K. Crystals were observed in all trays. The same crystals were obtained from both protein preparations, and a crystal formed in 100 mM Tris/HCl pH 5.5, 2 M ammonium sulfate, and 5 mM acetyl-CoA) from the 16/ml sample was cryo-protected with a solution of reservoir with 25% ethylene glycerol, and vitrified in liquid nitrogen. Diffraction data were collected at the Advanced Photon Source Life Sciences Collaborative Access Team (APS LS-CAT) beamline 21 ID-G equipped with a Rayonic MX-300 CCD detector at a wavelength of 0.97856 Å. The same shaped crystals grown from the 36 ml/ml sample were not diffracted due to time limitations at the Synchrotron. Data sets were reduced with the XDS package26. Diffraction data are summarized in Table 1.
Table 1.
Data collection, refinement and structure quality.
| Parameter | Value |
|---|---|
| PDB ID | 6AO7 |
| Space group | P6522 |
| Unit-cell parameters | |
| a (Å) | 67.16 |
| b (Å) | 67.16 |
| c (Å) | 134.21 |
| α = β (°) | 90 |
| γ (°) | 120 |
| Matthews coefficient (Å3Da−1) | 2.33 |
| Solvent content (%) | 47.25 |
| Resolution range (Å) | 50–1.85 (1.90–1.85)a |
| Mean I/σ(I) | 35.45 (4.88) |
| No. of observed reflections | 15,998 (1150) |
| Completeness (%) | 99.6 (100) |
| Multiplicity | 13.8 (14.3) |
| Rmerge | 0.038 (0.515) |
| Refinement | |
| No. of used reflections | 15,975 |
| Rwork (%) | 0.200 |
| Rfree (%) | 0.244 |
| Mean B factor (Å2) | 48.0 |
| RMSD bonds (Å) | 0.007 |
| RMSD angles (°) | 0.901 |
| Model validationb | |
| MolProbity | |
| Clash score, all atoms | 2.84 |
| Ramachandran favored (%) | 98.7 |
| Ramachandran outliers (%) | 0 |
| MolProbity score | 1.23 |
aValue in parenthesis are statistics for the highest resolution shell.
bValues calculated using Molprobity.
The structure of the enzyme was solved by molecular replacement using the program Molrep27 from the CCP4 package28, with PDB entry 1YVK as the search model. The initial model was extended with ARP/wARP29. Manual model building was performed using Coot30, and the structure was refined in reciprocal space with Phenix31. The coordinates and structure factors of the apo structure were deposited in the PDB with accession code 6AO7.
Results and discussion
Protein expression and structure determination
To determine the crystallographic structure of an uncharacterized GNAT from E. anophelis and allow the identification of key motifs and structural hallmarks common to GNAT family members, we first cloned the gene and recombinantly expressed the protein as a 6-His fusion (see “Materials and methods”). Following purification of the His-tagged protein by affinity purification, the protein was further purified by size exclusion chromatography, yielding two peaks, corresponding to monomer and dimer species (Supplementary Figure 1). Analysis by SDS-PAGE confirmed that the same protein was present in both peaks (Supplementary Figure 1), and each peak was concentrated to 16 mg/ml (peak 1) and 36 mg/ml (peak 2) (Supplementary Figure 1). Crystallisation trials were performed with both protein preparations, and despite minor protein contaminations (Supplementary Figure 1), the same crystal morphology was obtained with both preparations in 100 mM Tris/HCl pH 5.5, 2 M ammonium sulfate, and 5 mM acetyl-CoA. Crystals grown from the protein preparation at 16 mg/ml (obtained from the dimer species) diffracted to 1.85 Å. Diffraction data were indexed and integrated in the space group P6522, with unit cell parameters a = 67.16 Å, b = 67.16 Å, c = 134.21 Å, α = β = 90°, γ = 120°. The structure was solved using molecular replacement in Molrep27 using chain A of PDB model 1YVK. One molecule was placed in the asymmetric unit and following model rebuilding in COOT30 and refinement in Phenix31, a final model was produced with an Rcryst and Rfree of 0.195 and 0.244 respectively. With the exception of the N-terminal methionine, all amino acid residues were modelled, and coordinates and associated data files were deposited to the Protein Data Bank and issued the PDB ID 6AO7. The reflection data, model, and refinement statistics are summarized in Table 1.
Structural characterization reveals the putative GNAT from Elizabethkingia anophelis contains the motifs and hallmarks for a GNAT
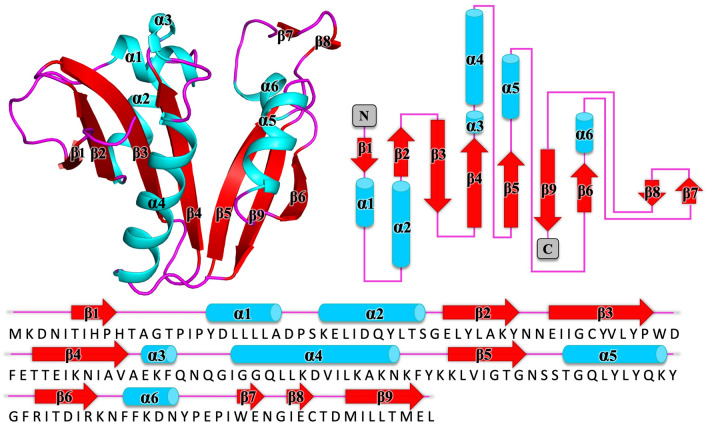
The protein was structured as an α/β protein and comprised of six α-helices and nine β-strands with a topology of β1-α1-α2-β2-β3-β4-α3-α4-β5-α5-β6-α6-β7-β8-β9 (Fig. 1). The core follows the conserved topology pattern of a typical GNAT family member, β1-α1-α2-β2-β3-β4-α3-β5-α4-β6, with additional secondary structural elements highlighted in italics. The structure exhibits two β-sheets comprised of β-strands 1-4, and β-strands 5, 6, and 9, splayed to create a V-shape (Fig. 1). Stabilization of the splayed β-sheets is mediated through H-bonding between Thr63 and Ile65 on β-strand 4, and Val100 on β-strand 5.
Figure 1.
Tertiary structure the GNAT acetyltransferase from Elizabethkingia anophelis. Top left, shown in cartoon format, α-helices, β-strands, and loops are shown in cyan, red, and magenta, respectively. Top right, topology map of the GNAT acetyltransferase from Elizabethkingia anophelis with α-helices shown as cylinders colored in cyan, β-strands are presented as red arrows, and loops are shown with magenta. Bottom panel, sequence of the GNAT acetyltransferase from Elizabethkingia anophelis spanning these structural elements.
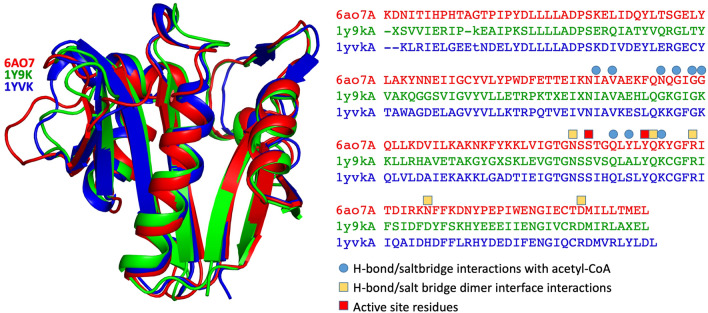
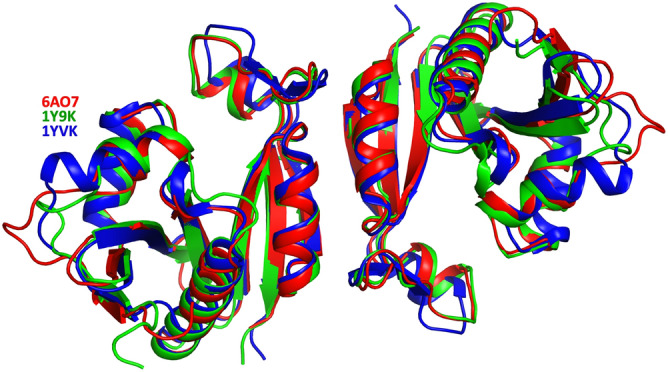
The GNAT has not been characterized previously, and we therefore performed both BLAST and DALI searches to determine proteins with similar sequence and structural features. A sequence homology search (excluding Elizabethkingia (taxid: 308865)), revealed the most closely related proteins to be GNAT family members from Runella zeae (55% sequence identity with 100% coverage), Microscillaceae bacterium (60% identity with 90% coverage), and Caenibacillus caldisaponilyticus (59% identity with 90% coverage). Within the top 20 Blast results, 3 GNAT proteins were aminoglycoside 6′-N-acetyltransferases: Chitinophaga eiseniae (53% identity with 98% coverage), Pedobacter nutrimenti (59% identity and 89% coverage), and Rhabdobacter roseus (50% identity and 98% coverage) (Supplementary Table 1). Moreover, docking of kanamycin produced a structural model within the active site and no steric clashes (Supplementary Figure 2), however further experimentation is required to confirm the substrate. Since GNAT proteins often exhibit low sequence identity but high structural homology15, we also performed a DALI search. We found only one protein with a rmsd value of less than 1 Å, PDB 1y9k (0.9 Cα rmsd, 48% seq id; no primary citation), solved by a structural genomics consortium that remains to be published. The next most similar protein was 1yvk (1.4 Cα rmsd, 49% seq id; no primary citation), and this was also solved by a structural genomics group and remains unpublished. The sequence identity between 1y9k and 1yvk is 59% (the top10 Dali results and a sequence based structural alignment are presented in Supplementary Table 2 and Supplementary Figure 3 respectively). Interestingly, 1y9k is predicted to be a dimer, while 1vyk is predicted to be a tetramer (discussed below). All other PDB entries have a sequence identity of less than 20%, and Cα rmsd greater than 2. Structural superposition of these structures, together with a structure-based sequence alignment is presented in Fig. 2.
Figure 2.
Tertiary structure the GNAT acetyltransferase from Elizabethkingia anophelis (shown in cartoon format in red), superimposed with the two most closely related structures identified from DALI (1Y9K in green, and 1YVK in blue). The alignment of these corresponding sequences are presented on the right, with active site residues highlighted with a red box, residues involved in dimer interface bonding in yellow boxes, and residues involved in acetyl-CoA binding in blue circles.
Structural analysis of the acetyl-CoA binding site
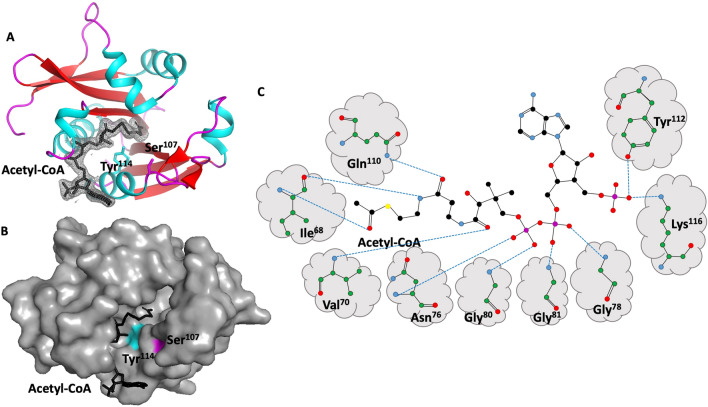
Our structure contained a well-ordered acetyl-CoA molecule bound in the active site (Fig. 3AB). The interface between acetyl-CoA buries 593 Å2 of surface area and mediated by 10 hydrogen bonds. Key residues involved in these hydrogen bond interactions include Ile68, Val70, Asn76, Gly78, Gly80, Gly81, Gln110, Tyr112, and Lys116 (Fig. 3C), and specific interactions are summarized in Table 2. Based on the position of the acetyl-CoA in the structure and well characterized kinetic studies of GNAT enzymes, the most likely active residues are Ser107 and Try114. Most GNATs contain either a Glu, Asp or Ser near the active site, which serves to act as a general base by extracting a proton from the substrate15,32–34. Nucleophilic attack on acetyl-CoA leads to the creation of a transient zwitterionic tetrahedral intermediate, and receival of a proton from a general acid (usually Tyr or Ser)15,35. Approximately 62% of GNATs have been reported to contain a conserved Tyr as a general acid to initiate the catalysis, and the positioning of Tyr114 from our structural analysis is consistent with this36. Both the acetyl-CoA residues involved in H-bonding, and active site residues are shown in Fig. 2.
Figure 3.
Acetyl-CoA binding site depicted within the tertiary structure the GNAT acetyltransferase from Elizabethkingia anophelis. Top left, shown in cartoon format, α-helices, β-strands, and loops are shown in cyan, red, and magenta, respectively. Acetyl-CoA is depicted in stick mode in black, with associated electron density map (2Fo-Fc) contoured at 1.0σ. The side chains of active site residues Tyr114 and Ser107 are depicted in stick mode. Bottom left, the same structure as above, but shown in surface view and coloured grey to depict the binding cleft. The location of Tyr114 and Ser107 are coloured cyan and magenta respectively. Right panel, schematic of hydrogen bond interactions between GNAT acetyltransferase residues and acetyl-CoA.
Table 2.
Hydrogen bond interactions between GNAT acetyltransferase from Elizabethkingia anophelis and acetyl-CoA.
| Bonding atom/residue | Distance | Acetyl-CoA (ACO) | ||||
|---|---|---|---|---|---|---|
| N | ILE | 68 | 3.12 | O | ACO | 201 |
| O | ILE | 68 | 2.81 | N4P | ACO | 201 |
| N | VAL | 70 | 2.76 | O9P | ACO | 201 |
| N | ASN | 76 | 3.13 | O5A | ACO | 201 |
| N | GLY | 78 | 2.79 | O1A | ACO | 201 |
| N | GLY | 80 | 2.86 | O4A | ACO | 201 |
| N | GLY | 81 | 3.04 | O2A | ACO | 201 |
| NE2 | GLN | 110 | 2.95 | O5P | ACO | 201 |
| OH | TYR | 112 | 2.56 | O9A | ACO | 201 |
| NZ | LYS | 116 | 2.51 | O9A | ACO | 201 |
Quaternary structure
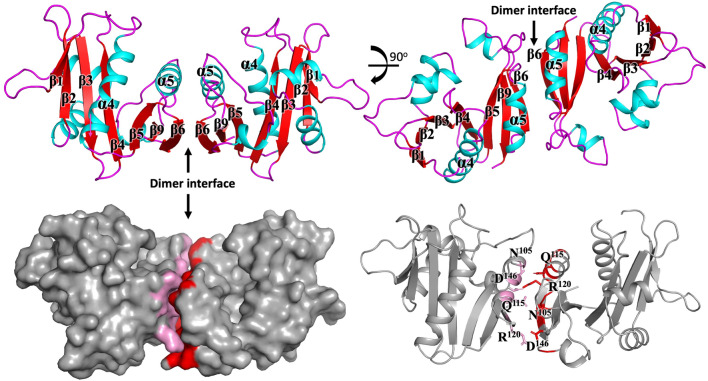
GNATs adopt a wide variety of assemblies, ranging from monomers, dimers, trimers, tetramers, hexamers, and dodecameric (double ring) structures37–44. Analysis of the interfaces present in the crystal using PISA45 suggests the protein is most likely to be a dimer (Fig. 4).
Figure 4.
The structure of the GNAT acetyltransferase from Elizabethkingia anophelis exhibits a significant dimer interface. The secondary structural elements mediating the dimer interface are presented in the left panels. Residues within α-helix five are depicted in the right panel and pack form an array of hydrogen bond and salt bridge interactions summarized in Tables 3 and 4.
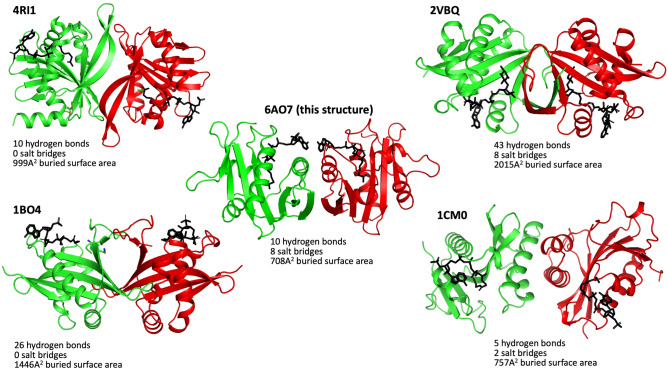
To test whether this interface is conserved in the most closely related GNATs, we examined whether the same dimer structures were present from proteins of different crystal space groups and distinctly different sequences. We found that the same dimer structure is present in both of the crystal structures of 1YVK and 1Y9K (Fig. 5). Since these sequences are distinctly different, and the crystal space groups are unrelated in all three structures, this suggests that these dimer interfaces are unlikely to be a crystal artefact. In this assembly configuration, the two monomers bury a surface area of 1464 Å, and mediated by ten hydrogen bonds (Table 3) and eight salt bridges (Table 4). Key interactions at the dimer interface include Arg120 forming a salt bridge interaction with Asp146, and Gln115 hydrogen bonding with Asn105. Dimers are the most common GNAT assembly, and there are four different types of dimers that have been described in the literature21. One form involves the C-terminal β-strands interacting to form a continuous β-sheet15,46–49. A second form involves two monomers exchanging their C-terminal β-strands (Eg ScHpa2, SeAAC(6′)-Iy and ScGNA139,50,51. A third form involves two monomers forming a β-barrel (eg SmAAC(3)-Ia52, while the fourth type is formed through a four-helical bundle (e.g. pPCAF)53. The dimer interface found in our structure is distinctly different to any other dimer described previously (Fig. 6), and is identical in at least two other GNATs suggesting this dimer interface is conserved.
Figure 5.
The dimer interface observed in the GNAT acetyltransferase from Elizabethkingia anophelis is conserved in two other unpublished structures. Superposition of the two structures (PDBs 1Y9K (green) and 1YVK (blue), both unpublished), demonstrate a conserved dimer interface.
Table 3.
Hydrogen bonding at the dimer interface.
| Interface on chain A | Distance (Å) | Interface on chain B |
|---|---|---|
| A:GLN 115[NE2] | 2.92 | B:ASN 105[O] |
| A:ASN 105[ND2] | 2.76 | B:GLN 115[OE1] |
| A:ARG 120[NH2] | 3.13 | B:ASN 127[OD1] |
| A:ARG 120[NE] | 2.98 | B:ASP 146[OD1] |
| A:ARG 120[NH2] | 2.75 | B:ASP 146[OD2] |
| A:ASN 105[O] | 2.92 | B:GLN 115[NE2] |
| A:GLN 115[OE1] | 2.76 | B:ASN 105[ND2] |
| A:ASN 127[OD1] | 3.13 | B:ARG 120[NH2] |
| A:ASP 146[OD1] | 2.98 | B:ARG 120[NE] |
| A:ASP 146[OD2] | 2.75 | B:ARG 120[NH2] |
Table 4.
Salt bridge interactions at the dimer interface.
| Interface on chain A | Distance (Å) | Interface on chain B |
|---|---|---|
| A:ARG 120[NH2] | 3.36 | B:ASP 146[OD1] |
| A:ARG 120[NE] | 2.98 | B:ASP 146[OD1] |
| A:ARG 120[NH2] | 2.75 | B:ASP 146[OD2] |
| A:ARG 120[NE ] | 3.84 | B:ASP 146[OD2] |
| A:ASP 146[OD1] | 2.98 | B:ARG 120[NE] |
| A:ASP 146[OD1] | 3.36 | B:ARG 120[NH2] |
| A:ASP 146[OD2] | 3.84 | B:ARG 120[NE] |
| A:ASP 146[OD2] | 2.75 | B:ARG 120[NH2] |
Figure 6.
GNATs form a range of different dimers. Presented here are the 5 (including this dimer) different dimer structures. The PDB codes 4RI1, 2VBQ, 1BO4 and 1CM0, representing each different dimer are shown below each dimeric structure.
Conclusion
Here, we describe the structure of a GNAT family member from E. anophelis. Structural analysis suggests that the protein exhibits the hallmarks typical of a GNAT fold. Structural and sequence-based alignments suggest that this acetyltransferase may be a possible aminoglycoside 6′-N-acetyltransferase, however this needs to be determined experimentally. Residues involved in acetyl-CoA binding have been identified and are conserved in closely related GNATs. We identify a new type of dimeric interface, and this is conserved in at least two other structures that have been deposited to the PDB, but remain unpublished. The oligomerization of GNATs vary range from dimers through to dodecaomers, and oligomerization is important for function15. Thus characterization of new Gcn5-related N-acetyltransferases has the potential to expand our understanding of how these enzymes carry out acetylation of a wide range of substrates, and high-resolution structural elucidation may offer new opportunities for biotechnological applications and drug design.
Supplementary Information
Acknowledgements
Structural work for this project was funded in part by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN272201700059C. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE‐AC02‐06CH11357. The use of the LS‐CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri‐Corridor (Grant No. 085P1000817).
Author contributions
R.M.I., J.A., S.J.M., D.D.L., T.E.E. performed the experiments and deposited the structure. P.S., S.M.G. and J.K.F. wrote the main manuscript text and prepared the figures. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Thomas E. Edwards, Email: tom.edwards@ucb.com
Jade K. Forwood, Email: jforwood@csu.edu.au
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-020-79649-5.
References
- 1.Janda JM, Lopez DL. Mini review: New pathogen profiles: Elizabethkingia anophelis. Diagn. Microbiol. Infect. Dis. 2017;88:201–205. doi: 10.1016/j.diagmicrobio.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Kukutla P, et al. Insights from the genome annotation of Elizabethkingia anophelis from the malaria vector Anopheles gambiae. PLoS ONE. 2014;9:e97715. doi: 10.1371/journal.pone.0097715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breurec S, et al. Genomic epidemiology and global diversity of the emerging bacterial pathogen Elizabethkingia anophelis. Sci. Rep. 2016;6:30379. doi: 10.1038/srep30379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kämpfer P, et al. Elizabethkingia anophelis sp. nov., isolated from the midgut of the mosquito Anopheles gambiae. Int. J. Syst. Evol. Microbiol. 2011;61:2670–2675. doi: 10.1099/ijs.0.026393-0. [DOI] [PubMed] [Google Scholar]
- 5.Wang M, et al. The antibiotic resistance and pathogenicity of a multidrug-resistant Elizabethkingia anophelis isolate. MicrobiologyOpen. 2019;8:e804. doi: 10.1002/mbo3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han M-S, et al. Relative prevalence and antimicrobial susceptibility of clinical isolates of Elizabethkingia species based on 16S rRNA gene sequencing. J. Clin. Microbiol. 2017;55:274–280. doi: 10.1128/JCM.01637-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau SK, et al. Elizabethkingia anophelis bacteremia is associated with clinically significant infections and high mortality. Sci. Rep. 2016;6:26045. doi: 10.1038/srep26045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frank T, et al. First case of Elizabethkingia anophelis meningitis in the Central African Republic. Lancet. 2013;381:1876. doi: 10.1016/S0140-6736(13)60318-9. [DOI] [PubMed] [Google Scholar]
- 9.Lin J-N, Lai C-H, Yang C-H, Huang Y-H. Elizabethkingia infections in humans: From genomics to clinics. Microorganisms. 2019;7:295. doi: 10.3390/microorganisms7090295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teo J, et al. First case of E. anophelis outbreak in an intensive-care unit. Lancet. 2013;382:855–856. doi: 10.1016/S0140-6736(13)61858-9. [DOI] [PubMed] [Google Scholar]
- 11.Perrin A, et al. Evolutionary dynamics and genomic features of the Elizabethkingia anophelis 2015 to 2016 Wisconsin outbreak strain. Nat. Commun. 2017;8:15483. doi: 10.1038/ncomms15483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McTaggart LR, et al. Application of whole genome sequencing to query a potential outbreak of Elizabethkingia anophelis in Ontario, Canada. Access Microbiol. 2019;1:e000017. doi: 10.1099/acmi.0.000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hentchel KL, Escalante-Semerena JC. Acylation of biomolecules in prokaryotes: A widespread strategy for the control of biological function and metabolic stress. Microbiol. Mol. Biol. Rev. 2015;79:321–346. doi: 10.1128/MMBR.00020-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren J, Sang Y, Lu J, Yao Y-F. Protein acetylation and its role in bacterial virulence. Trends Microbiol. 2017;25:768–779. doi: 10.1016/j.tim.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 15.Vetting MW, et al. Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 2005;433:212–226. doi: 10.1016/j.abb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 16.Favrot L, Blanchard JS, Vergnolle O. Bacterial GCN5-related N-acetyltransferases: From resistance to regulation. Biochemistry. 2016;55:989–1002. doi: 10.1021/acs.biochem.5b01269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carabetta VJ, Cristea IM. Regulation, function, and detection of protein acetylation in bacteria. J. Bacteriol. 2017;199:e00107–00117. doi: 10.1128/JB.00107-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christensen D, et al. Mechanisms, detection, and relevance of protein acetylation in prokaryotes. mBio. 2019;10:e02708–e02718. doi: 10.1128/mBio.02708-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Czub MP, et al. A Gcn5-related N-acetyltransferase (GNAT) capable of acetylating polymyxin B and colistin antibiotics in vitro. Biochemistry. 2018;57:7011–7020. doi: 10.1021/acs.biochem.8b00946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agrawal A, et al. Global proteome profiling reveals drug-resistant traits in Elizabethkingia meningoseptica: An opportunistic nosocomial pathogen. Omics J. Integr. Biol. 2019;23:318–326. doi: 10.1089/omi.2019.0039. [DOI] [PubMed] [Google Scholar]
- 21.Salah Ud-Din AIM, Tikhomirova A, Roujeinikova A. Structure and functional diversity of GCN5-related N-acetyltransferases (GNAT) Int. J. Mol. Sci. 2016;17:1018. doi: 10.3390/ijms17071018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dyda F, Klein DC, Hickman AB. GCN5-related N-acetyltransferases: A structural overview. Annu. Rev. Biophys. Biomol. Struct. 2000;29:81–103. doi: 10.1146/annurev.biophys.29.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pappas G, Akritidis N, Bosilkovski M, Tsianos E. Brucellosis. N. Engl. J. Med. 2005;352:2325–2336. doi: 10.1056/NEJMra050570. [DOI] [PubMed] [Google Scholar]
- 24.Choi R, et al. Immobilized metal-affinity chromatography protein-recovery screening is predictive of crystallographic structure success. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011;67:998–1005. doi: 10.1107/S1744309111017374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bryan CM, et al. High-throughput protein production and purification at the Seattle Structural Genomics Center for infectious disease. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011;67:1010–1014. doi: 10.1107/S1744309111018367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kabsch W. Xds. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. doi: 10.1107/S0021889897006766. [DOI] [Google Scholar]
- 28.Collaborative CP. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994;50:760. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 29.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 2008;3:1171. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng K-C, Liao J-N, Lyu P-C. Crystal structure of the dopamine N-acetyltransferase–acetyl-CoA complex provides insights into the catalytic mechanism. Biochem. J. 2012;446:395–404. doi: 10.1042/BJ20120520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schuetz A, et al. Crystal structure of a binary complex between human GCN5 histone acetyltransferase domain and acetyl coenzyme A. Proteins Struct. Funct. Bioinform. 2007;68:403–407. doi: 10.1002/prot.21407. [DOI] [PubMed] [Google Scholar]
- 34.Pourreza A, et al. Mutagenesis analysis of a conserved region involved in acetyl coenzyme A binding in the aminoglycoside 6′-N-acetyltransferase type Ib encoded by plasmid pJHCMW1. Antimicrob. Agents Chemother. 2005;49:2979–2982. doi: 10.1128/AAC.49.7.2979-2982.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Majorek KA, Kuhn ML, Chruszcz M, Anderson WF, Minor W. Structural, functional and inhibition studies of a GNAT superfamily protein PA4794: A new C-terminal lysine protein acetyltransferase from Pseudomonas aeruginosa. J. Biol. Chem. 2013;M113:501353. doi: 10.1074/jbc.M113.501353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He H, et al. Crystal structure of tabtoxin resistance protein complexed with acetyl coenzyme A reveals the mechanism for β-lactam acetylation. J. Mol. Biol. 2003;325:1019–1030. doi: 10.1016/S0022-2836(02)01284-6. [DOI] [PubMed] [Google Scholar]
- 37.Tsimbalyuk S, Shornikov A, Kuhn ML, Forwood JK. SpeG polyamine acetyltransferase enzyme from Bacillus thuringiensis forms a dodecameric structure and exhibits high catalytic efficiency. J. Struct. Biol. 2020;210:107506. doi: 10.1016/j.jsb.2020.107506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Srivastava P, et al. Structural characterization of a Gcn5-related N-acetyltransferase from Staphylococcus aureus. PLoS ONE. 2014;9:e102348. doi: 10.1371/journal.pone.0102348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Angus-Hill ML, Dutnall RN, Tafrov ST, Sternglanz R, Ramakrishnan V. Crystal structure of the histone acetyltransferase Hpa2: A tetrameric member of the Gcn5-related N-acetyltransferase superfamily. J. Mol. Biol. 1999;294:1311–1325. doi: 10.1006/jmbi.1999.3338. [DOI] [PubMed] [Google Scholar]
- 40.Vetting MW, Bareich DC, Yu M, Blanchard JS. Crystal structure of RimI from Salmonella typhimurium LT2, the GNAT responsible for Nα-acetylation of ribosomal protein S18. Protein Sci. 2008;17:1781–1790. doi: 10.1110/ps.035899.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen W, Biswas T, Porter VR, Tsodikov OV, Garneau-Tsodikova S. Unusual regioversatility of acetyltransferase Eis, a cause of drug resistance in XDR-TB. Proc. Natl. Acad. Sci. 2011;108:9804–9808. doi: 10.1073/pnas.1105379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maurice F, et al. Enzyme structural plasticity and the emergence of broad-spectrum antibiotic resistance. EMBO Rep. 2008;9:344–349. doi: 10.1038/embor.2008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vetting MW, Roderick SL, Yu M, Blanchard JS. Crystal structure of mycothiol synthase (Rv0819) from Mycobacterium tuberculosis shows structural homology to the GNAT family of N-acetyltransferases. Protein Sci. 2003;12:1954–1959. doi: 10.1110/ps.03153703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vetting MW, Yu M, Rendle PM, Blanchard JS. The substrate-induced conformational change of Mycobacterium tuberculosis mycothiol synthase. J. Biol. Chem. 2006;281:2795–2802. doi: 10.1074/jbc.M510798200. [DOI] [PubMed] [Google Scholar]
- 45.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 46.Ud-Din AI, Liu YC, Roujeinikova A. Crystal structure of Helicobacter pylori pseudaminic acid biosynthesis N-acetyltransferase PseH: Implications for substrate specificity and catalysis. PLoS ONE. 2015;10:e0115634. doi: 10.1371/journal.pone.0115634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vetting MW, De Carvalho LPS, Roderick SL, Blanchard JS. A novel dimeric structure of the RimL Nα-acetyltransferase from Salmonella typhimurium. J. Biol. Chem. 2005;280:22108–22114. doi: 10.1074/jbc.M502401200. [DOI] [PubMed] [Google Scholar]
- 48.Hung M-N, et al. Crystal structure of TDP-fucosamine acetyltransferase (WecD) from Escherichia coli, an enzyme required for enterobacterial common antigen synthesis. J. Bacteriol. 2006;188:5606–5617. doi: 10.1128/JB.00306-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Forouhar F, et al. Structural and functional evidence for Bacillus subtilis PaiA as a novel N1-spermidine/spermine acetyltransferase. J. Biol. Chem. 2005;280:40328–40336. doi: 10.1074/jbc.M505332200. [DOI] [PubMed] [Google Scholar]
- 50.Peneff C, Mengin-Lecreulx D, Bourne Y. The crystal structures of Apo and complexed Saccharomyces cerevisiae GNA1 shed light on the catalytic mechanism of an amino-sugar N-acetyltransferase. J. Biol. Chem. 2001;276:16328–16334. doi: 10.1074/jbc.M009988200. [DOI] [PubMed] [Google Scholar]
- 51.Magalhães ML, et al. Kinetic and structural analysis of bisubstrate inhibition of the Salmonella enterica aminoglycoside 6′-N-acetyltransferase. Biochemistry. 2008;47:579–584. doi: 10.1021/bi701957c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolf E, et al. Crystal structure of a GCN5-related N-acetyltransferase: Serratia marcescens aminoglycoside 3-N-acetyltransferase. Cell. 1998;94:439–449. doi: 10.1016/S0092-8674(00)81585-8. [DOI] [PubMed] [Google Scholar]
- 53.Shi S, et al. Dimeric structure of p300/CBP associated factor. BMC Struct. Biol. 2014;14:2. doi: 10.1186/1472-6807-14-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.