Abstract
To date, there is no representative in vitro model for liver sinusoidal endothelial cells (LSECs), as primary LSECs dedifferentiate very fast in culture and no combination of cytokines or growth factors can induce an LSEC fate in (pluripotent stem cell (PSC)-derived) endothelial cells (ECs). Furthermore, the transcriptional programmes driving an LSEC fate have not yet been described. Here, we first present a computational workflow (CenTFinder) that can identify transcription factors (TFs) that are crucial for modulating pathways involved in cell lineage specification. Using CenTFinder, we identified several novel LSEC-specific protein markers, such as FCN2 and FCN3, which were validated by analysis of previously published single-cell RNAseq data. We also identified PU.1 (encoded by the SPI1 gene) as a major regulator of LSEC-specific immune functions. We show that SPI1 overexpression (combined with the general EC TF ETV2) in human PSCs induces ECs with an LSEC-like phenotype. The ETV2-SPI1-ECs display increased expression of LSEC markers, such as CD32B and MRC1, as well as several of the proposed novel markers. More importantly, ETV2-SPI1-ECs acquire LSEC functions, including uptake of FSA-FITC, as well as labelled IgG. In conclusion, we present the CenTFinder computational tool to identify key regulatory TFs within specific pathways, in this work pathways of lineage specification, and we demonstrate its use by the identification and validation of PU.1 as a master regulator for LSEC fating.
Subject terms: Differentiation, Reprogramming, Stem-cell differentiation
Introduction
Liver sinusoidal endothelial cells (LSECs) are highly specialised endothelial cells (ECs) lining the sinusoidal capillaries of the liver, where they reside along the space of Disse separating them from hepatocytes. LSECs have five main functions. First, LSEC fenestrations actively regulate the flow of macromolecules, such as lipids and chylomicron remnants towards the hepatocytes1,2. Second, LSECs endocytose larger lipid complexes, soluble macromolecules, hyaluronan, and glycosylation end products via the mannose receptor (MR or CD206), CD32B, stabilin-1 (STAB1), and stabilin-2 (STAB2)3,4. Third, in response to hepatic injury, LSECs regulate liver regeneration by releasing hepatocyte growth factor, WNT2, and angiopoietin-25,6. Fourth, LSECs can clear a multitude of viruses7–12. Furthermore, LSECs clear bacterial compounds via the MR, STAB1, and STAB2 receptors. LSECs also express Toll-like receptors, as well as several proteins involved in inflammasome formation (such as NLRP-1, NLRP-3, and AIM2)13. Expression of CD32B allows uptake of IgG immunocomplexes, while STAB2 is involved in endocytosis of Gram-positive and -negative bacteria. Finally, LSECs have a major role in adaptive immunity. LSECs are antigen-presenting cells through MR-mediated antigen uptake and expression of MHC-I14. Upon antigen presentation by MHC-I, LSECs can activate CD8+ T cells. In addition, LSECs are the main cells in the liver responsible for the conversion of CD4+ T cells into CD4+ CD25+ Foxp3+ regulatory T cells15.
In pathological conditions, LSECs become dysfunctional and play a role in non-alcoholic fatty liver disease and fibrosis16–19. In liver fibrosis, LSECs dedifferentiate, lose their fenestrations, and deposit a basement membrane (a process called capillarisation). As a result, when chronic fibrosis or cirrhosis ensues, lack of blood flow towards hepatocytes aggravates hepatotoxicity and inflammation, leading to portal hypertension20.
It should therefore be clear that LSECs, that play important roles in crosstalk with other (non-) parenchymal liver cell types in health and disease, should be included in liver co-culture models. However, primary LSECs very quickly lose their characteristics (e.g., fenestrae and endocytic capacity) and undergo apoptosis within 4–6 days of in vitro culture21–23. Furthermore, limited availability of human primary LSECs is a major obstacle in the progression of LSEC research. A few immortalised LSEC lines exist, such as SK-HEP-1, that retain some LSEC characteristics24. However, SK-HEP-1 cells do not take up FITC-ALB24 and immortalised lines may not respond similarly in co-culture and disease models as primary LSECs do.
A number of protocols have been described to differentiate PSCs towards ECs25–27. However, only a few studies have attempted to differentiate PSCs into LSECs, either through culturing PSC-derived ECs in hypoxic conditions with a TGFβ inhibitor28, or the combination of cAMP and a TGFβ inhibitor29. Classically, differentiation protocols for a given cell type deploy sequential growth factor and cytokine cocktails to mimic in vivo differentiation. However, relatively little is known regarding LSEC development and, hence, signals required for LSEC differentiation in vitro. It has been hypothesised that LSECs derive from the septum transversum mesenchyme30–32. However, recent lineage tracing experiments suggest that a large portion of the liver vasculature may derive from the sinus venosus endocardium33. It is yet to be elucidated, though, whether the endocardium contributes to LSECs rather than macrovascular ECs. As an alternative, transcription factors (TFs) have been overexpressed to guide differentiation of PSCs towards cell types of all germ layers34–37. However, the transcriptional programmes or TFs that drive LSEC development are not well known.
Here, we designed a novel integrative meta-analysis pipeline (available as the CenTFinder R package) to facilitate analysis of published EC microarray data, and identified pathways and TFs that might be crucial in fating LSECs. We combined several complementary analysis methods, including gene co-expression analysis (using weighted gene correlation network analysis (WGCNA)38), TF motif binding enrichment analysis (using RcisTarget39), and differential expression analysis on bulk RNAseq data (using DeSeq240), and identified several candidate TFs that may play a role in LSEC differentiation. As SPI1, encoding for PU.1, was identified as one of the top candidate TFs central to transcriptional networks for LSECs, we overexpressed SPI1 in PSCs, and demonstrated that this drives specification of PSC progeny to an LSEC-like phenotype.
Materials and methods
Microarray selection and retrieval
We defined query keywords, i.e., ‘EC AND Homo sapiens’ and ‘LSEC AND Homo sapiens’, to construct URLs with base https://www.ebi.ac.uk/arrayexpress/xml/v3/experiments?keywords = to query the ArrayExpress database, using the RCurl R package. ArrayExpress study accession codes were extracted from the HTTP responses. Subsequently, the respective SDRF files were downloaded to select relevant samples within each study. Only Affymetrix samples that were of human non-cancerous origin, and that were biotin-labelled were kept for analysis. Subsequently, new URLs were constructed to download the selected CEL files.
Microarray raw analysis
Platform information was extracted from all downloaded CEL files using the affyio R package. Microarray platforms with only one downloaded CEL file were excluded from analysis. Subsequently, for each platform type, the respective CEL files were RMA normalised, using the oligo R package. Probe intensity values were aggregated per gene by their geometric mean.
Platform selection and platform effect removal
As microarray platforms differed in the number of genes for which probes were coated, we weighed the benefits of analysing more samples and hence fewer genes in common or analysing fewer samples with more genes in common. For standard analyses, we selected as a rule of thumb those platforms for which the product of the number of samples and the number of common genes was maximised. However, the LSEC arrays of platform type HG-U219 contained probes for a slightly smaller than optimal number of genes. Nevertheless, as these were the only retrieved LSEC arrays, we included these arrays as well and, hence, the downstream analysis was performed with 279 EC arrays and 16,248 genes. Subsequently, we applied the Combat algorithm of the sva R package to remove platform and batch confounding effects.
Gene filtering
We excluded TFs that did not change by at least twofold across all EC microarray samples, as such minimal expression changes are not likely to be biologically meaningful. A list of human TFs was downloaded from http://humantfs.ccbr.utoronto.ca. Furthermore, to save on computational time, we restricted the number of genes that were not TFs to the 7,551 most variable ones (log2 fold change standard deviation > 0.5961016). The total number of genes for downstream analysis was thus set to 9,000.
Weighted gene correlation network analysis
WGCNA was performed using the WGCNA R package. We constructed a signed network and tested soft-thresholding powers ranging from 1 to 30. A cluster dendrogram was created based on the topological overlap matrix with a minimum cluster size of 30. Subsequently, modules with a dissimilarity <0.3 were merged.
Gene Ontology and KEGG pathway enrichment analysis
Gene Ontology (GO) enrichment was calculated for all identified WGCNA modules, using Fisher’s exact test from the topGO R package. The top 15 GO terms were retained for each GO category (i.e., biological process, cellular component, and molecular function).
RcisTarget analysis
We imported motif rankings from the ‘h19-tss-centred-10kb-7species.mc9nr’ database (https://resources.aertslab.org/cistarget/). Overrepresented DNA-binding motifs were identified within each of the WGCNA modules, using the RcisTarget R package according to the creators’ guidelines.
Bulk RNA-sequencing analysis
Sample acquisition
PSCs, wherein the coding region of the TF ETV2 (controlled by a TET-ON system) was recombined in the AAVS1 locus (as described in Ordovas et al.41), were differentiated until day 10. RNA samples (N = 3) were prepared and sequenced on an Illumina NextSeq 500 platform by the VIB Nucleomics Core (KULeuven, Belgium). Three RNAseq samples annotated as primary human LSECs were obtained from ArrayExpress (E-GEOD-40291 and E-GEOD-43984). However, only in one of these samples (E-GEOD-43984) LSEC-specific genes, FCGR2B, CLEC4M, STAB2, and CLEC4G, were expressed. The other samples were excluded from analysis.
Read trimming
Cutadapt was used to trim the adapter sequence from all reads. Bases with Phred-scores <20 were trimmed. Poly A {10} tails were removed and reads shorter than 20 bases were removed.
Genome index generation and read alignment
The human genome fasta files (GRCh38.92) and GTF file were downloaded from Ensemble, and assembled into a genome index using STAR v020201. Subsequently, STAR was used to align the filtered reads to the genome index. The R implementation of FeatureCounts (Rsubread) was used to generate raw read count matrices. Subsequently, we used the GTFtools Python package to calculate merged exon lengths, i.e., the union of all exons. Finally, we calculated the TPM matrix.
Differential gene expression analysis
The DESeq2 R package was used to calculate differential gene expression (Benjamini–Hochberg-corrected p value < 0.05). An absolute fold change of less than two was considered not biologically meaningful.
Module gene set variant analysis
We used the modules identified by WGCNA as gene sets in gene set variation analysis (GSVA), using the GSVA R package. Module gene sets were scored for each individual sample. Subsequently, mean GSVA scores were calculated per cell type to identify which modules were more prominently expressed for each cell type.
Transcription factor ranking and visualisation
TFs were considered for overexpression if they were higher expressed in LSECs compared to ECs derived from PSCs through overexpression of ETV2 (referred to as ETV2-ECs). In addition, TFs were only retained if their kME centrality measure was >0.5, and if their respective clusters were more active (GSVA) in LSECs than in other ECs. Next, the TFs were ranked based on the fraction of genes in their respective modules they regulated (inferred by RcisTarget). The modules of interest were visualised using Cytoscape. Regulons of the modules of interest were extracted and reformatted into a dataframe with Source and Target column, suitable for Cytoscape network import. TFs of the modules of interest were visualised when differentially expressed, regardless of whether they were higher or lower expressed in LSECs. For visualisation purposes we only retained the 20 most differentially expressed downstream targets, as evaluated by bulk RNAseq data. TFs assigned by WGCNA to other modules than the modules of interest were given the colour of the module of interest if they had a high cluster centrality in the respective module of interest (kME > 0.5). Node size was increased with increasing number of downstream targets.
Single-cell RNA-sequencing analysis
Fastq files from a recent human liver single-cell RNA-sequencing study (GSE12439542) were downloaded from the European Nucleotide Archive. Cellular barcode and UMI handling was done with UMI tools. We used Cutadapt to trim bases with Phred-score < 20 and to remove reads shorter than 20 bases. Read quality was assessed with FastQC. STAR was used to create a human genome index based on the GRCh38.92 release of Ensemble. STAR was used as well for alignment. The count matrix was generated with UMI tools. Cell quality control was performed using the Scater and Seurat R packages. Briefly, we removed cells with <2500 UMIs or <1500 detected genes. Only cells with a mitochondrial read content <15% were included for analysis. Only genes with a count higher than one in at least two cells were included for analysis. ENHANCE43 was used for imputation and denoising. The scran implementation (cyclone) of the method described by Scialdone et al.44. was used for cell cycle inference. SCENIC (with GRNBoost2 for ensemble learning) was used for detection of active gene regulatory networks (GRNs)39. Seurat was used to correct for confounding variables, i.e., number of detected genes, number of total counts, cell cycle status, and percentage of mitochondrial counts.
The count matrix of a second study (GSE11546945) was retrieved from the Gene Expression Omnibus. For each study, clusters were identified by Seurat and cell types were inferred by manually evaluating marker genes within the different clusters.
ECs were defined based on higher levels of CDH5 and KDR expression, and LSECs based on higher expression of CLEC4G, STAB1, and PECAM1. FCGR2B-low and FCGR2B-high LSEC subsets could be distinguished. Differential expression analysis was performed with a Wilcoxon rank-sum test followed by a Benjamini–Hochberg multiple test correction.
Stem cell culture
H9 embryonic stem cells (ESCs; WA09) were cultured in E8 Flex medium (cat. A2858501, ThermoFisher Scientific). Cells were passaged at 75% confluency using 0.1% EDTA in PBS (and tested for mycoplasma).
Genome engineering of ESC lines for endothelial differentiation
Two million H9 ESCs, engineered with an FRT and FRT3 site in the AAVS1 safe harbour locus41, were nucleoporated with a plasmid mix containing 2 µg of pCAGGS-FLPe (MES4488, Open Biosystems;without the puromycin resistance gene) and 8 µg of the donor plasmid containing the gene of interest. Donor plasmids also contained FRT and FRT3 sites in identical orientation, a promotorless puromycin cassette for gene trapping, and an inducible TETon system for overexpression of the gene of interest. The coding sequences of NM_014209.3 and of NM_001080547.1 were cloned into the donor plasmid template for overexpression of ETV2 and SPI1, respectively. ETV2 and SPI1 coding sequences were linked by a P2A autocleavage sequence. Nucleoporation and subsequent selection was performed, as described by Ordovas et al.41.
Differentiation of ESCs into endothelial cells
H9 ESCs were passaged at a 1:6 ratio and grown for 1–2 days in E8 Flex medium to ~40% confluency. On day 0 of endothelial differentiation, medium was changed to liver differentiation medium (LDM; composition as described in ref. 37) and 5 µg/ml doxycycline. On day 2 of differentiation, cells were grown in LDM with 5 µg/ml doxycycline and 2% FBS. Cells were passaged on days 4, 8, and 12 in a 1:3 ratio. After day 12, no further passaging was performed. When 24-well plate formats were required, ECs were passaged on day 4 from 6-well plates into 24-well plates (1 well into 10 wells, equivalent to a 1:2 split), and no further passaging was done until the time of read-out on day 12 of differentiation. To passage ECs, cells were washed with PBS and detached with StemPro™ Accutase™ Cell Dissociation Reagent (Gibco™, A1110501) for 35 s at 37 °C. Subsequently, accutase was removed, fresh medium added, and cells gently detached with a cell scraper. ECs derived from H9-ETV2 PSCs and H9-ETV2-SPI1 PSCs are further referred to as ETV2-ECs and ETV2-SPI1-ECs respectively. In the conditions where cells were supplemented with VEGFA (50 ng/ml; cat. 100-20, Peprotech), it was added to the culture medium as of day 6 of differentiation (Supplementary Fig. 1; N = 7 for samples without VEGFA, N = 4 for samples with VEGFA; statistical differences assessed by two-sided Student’s T test).
Isolation of mouse LSECs and hepatic stellate cells
Primary liver cells were isolated from 10- to 25-week-old BALB/c mice, (N = 3), as previously described46. LSECs and hepatic stellate cells (HSCs) were plated on collagen-coated wells in a 24-well plate (200,000 cells/well) in 0% FBS-DMEM for LSECs and 10% FBS-DMEM for HSCs. All methods, experimental protocols and animal experimentation ethics were carried out in accordance with the approved guidelines of the Vrije Universiteit Brussel (VUB, Belgium) and according to European Guidelines for the Care and Use of Laboratory Animals. All animal experimentation protocols were approved by the Ethical Committee of Animal Experimentation of the Vrije Universiteit Brussel (VUB, Belgium; LA 123 02 12, project 18-212-1).
Lentiviral overexpression of SPI1, ERG, and FLI1
H9-ETV2 PSCs were differentiated into ECs and passaged into 24-well plates on day 4. On day 6, cells were transduced with lentiviral vectors encoding for SPI1, FLI1, and ERG1 (for ERG overexpressions: N = 2; all other conditions: N = 3) in the presence of 4 µg/ml polybrene (cat. TR-1003-G, Merck; Supplementary Fig. 2). Where ETV2 overexpression was substituted for lentiviral overexpression of FLI1 and/or ERG, doxycycline was omitted from culture media from day 8 of differentiation onwards. Statistical differences were assessed by a two-sided Student’s T test.
Reverse-transcriptase quantitative polymerase chain reaction
Cells were lysed and RNA extracted with the GenEluteTM Mammalian RNA Extraction Kit (cat. RTN70, Sigma Aldrich) and reverse transcribed into cDNA, using SuperScript® III First-Strand Synthesis SuperMix (cat. 11752050, InvitrogenTM), according to the manufacturer’s protocol. Reverse-transcriptase quantitative polymerase chain reaction (RT-qPCR) reactions were made using the Platinum® SYBR® Green RT-qPCR SuperMix-UDG kit (cat. 11733046, ThermoFisher Scientific). Reactions were denatured at 95 °C for 20 s, followed by 40 cycles of a 1 s denaturation step at 95 °C and a 20 s annealing and elongation step at 60 °C. Gene expression values were normalised for GAPDH, RPL19, RPS23, and EEF1A1. Primer sequences are listed in Supplementary Table 1.
Immunostaining
Culture medium was removed and cells (on coverslips) washed with PBS. Cells were fixed with 4% paraformaldehyde for 10 min, washed with PBS and permeabilised with 0.2% Triton X-100 in PBS. For CD32B staining, cells were not permeabilised. Blocking was performed using 5% donkey serum in 0.2% Triton X-100 in PBS. Primary antibodies (Supplementary Table 2) were diluted according to manufacturer’s guidelines in DAKO Antibody Diluent (cat. S202230, Agilent). Cells were incubated with the primary antibody mixes overnight at 4 °C, washed with 0.2% Triton X-100 in PBS, and incubated for 30 min at room temperature with the secondary antibody (Supplementary Table 2; 1:500 diluted in DAKO Antibody Diluent). Cells were washed with 0.2% Triton X-100 in PBS, mounted with Prolong Gold (cat. P-36931, ThermoFisher Scientific), and visualised using a Zeiss Axioimager microscope. Stainings were replicated three times.
Flow cytometry
Day 12 PSC progeny were detached with accutase for 35 s at 37 °C. Accutase was inactivated by incubating cells in LDM + 10% FBS for 6 min at room temperature. Cells were washed with PBS and resuspended in PBS with 1% BSA (cat. A7979-50 ml, Sigma Aldrich). Cells were incubated with primary antibodies (Supplementary Table 2) for 30 min at 4 °C, washed with PBS with 1% BSA and incubated with secondary antibodies (Supplementary Table 2) for 30 min at 4 °C. Viability was assessed by propidium iodide (cat. 81845-25MG, Sigma Aldrich). Flow cytometry was performed using a BD FACSCantoTM II High-Throughput Sampler. Cells were gated according to the strategy depicted in Supplementary Fig. 3. Two-sided paired Student’s T tests were applied on population median intensities (replicates: N = 15 for CD32B and N = 14 for MRC1).
Tube formation assay
Wells of 24-well plates were coated with undiluted Matrigel® hESC-Qualified Matrix (protein concentration approximates 10 mg/ml; cat. 354277, Corning®). Matrigel was solidified by incubation for at least 10 min at 37 °C. Day 12-ECs (H9-ETV2 and H9-ETV2-SPI1) were seeded on the matrigel at a density of 150,000 cells per well. After 20 h, cells were fixed with 4% paraformaldehyde, washed with PBS, and permeabilised with 0.2% Triton X-100 in PBS for 30 min. Cells were washed with PBS and blocked with 0.1 M glycine in PBS for 30 min. Subsequently, cells were incubated with 2 U/ml phalloidin (Supplementary Table 2), 2.5 µg/ml DAPI (Supplementary Table 2), and 1% BSA in PBS for 90 min in the dark at room temperature. Cells were washed with PBS and tubes visualised by confocal imaging with a Zeiss LSM 880—Airyscan.
Scanning electron microscopy
ETV2-ECs and ETV2-SPI1-ECs were cultured on coverslips until day 12 of differentiation with VEGFA (50 ng/ml; cat. 100-20, Peprotech). BALB/c LSECs were cultured for 1.5–2 h after isolation, with (40 ng/ml, cat. V4512-10UG, Sigma Aldrich). Cells were washed thoroughly with PBS to avoid protein and serum remnants. Next, cells were immersed in 500 µl PBS and fixed for 10 min with 500 µl of 5% glutaraldehyde in 66 mM cacodylate buffer. The fixative solution was removed and cells maintained overnight at 4 °C in 2.5% glutaraldehyde in 66 mM cacodylate buffer. Cells were washed with 0.1 M cacodylate buffer at room temperature and post-fixed for 2 h at room temperature with 1% OsO4 + 1.5% K4Fe(CN)6 in 0.1 M cacodylate buffer. After post-fixation cells were washed with 0.1 M cacodylate buffer and dehydrated with increasing ethanol concentrations (30–50–70–90–100–100%) with 5 min incubation per concentration. Finally, the samples were critical point dried in a Leica CPD300 critical point dryer and the coverslips with cells mounted on pin stubs with carbon stickers. Cells were observed and imaged in a Zeiss Sigma scanning electron microscope at an accelerating voltage of 5 kV. Pore diameters were counted and measured using ImageJ and the EBImage R package.
FSA-FITC uptake assay
ETV2-ECs and ETV2-SPI1-ECs were cultured until day 12 of differentiation on Matrigel-coated wells in a 24-well plate. BALB/c LSECs were isolated and plated as well on collagen at 200,000 cells per well in a 24-well plate in 0% FBS-DMEM. After 1 h of LSEC plating, ETV2-ECs, ETV2-SPI1-ECs, and LSECs were supplemented with 10 µg/ml FSA-FITC (prepared as described in ref. 47) and 1:1000 IncuCyte® NucLight Rapid Red Reagent (cat. 4717, Sartorius). Cells were imaged hourly over a period of 24 h using the IncuCyte Cell analysis system (Essen Bioscience). Quantification was performed with the IncuCyte Cell analysis software. This experiment was repeated twice. Statistical differences were assessed by mixed ANOVA.
IgG-AF555 uptake assay
H9-ETV2 and H9-ETV2-SPI1 PSCs were differentiated to ECs as described above. On day 12 of differentiation cells were washed with PBS and incubated for 2 h at 37 °C with 100 µg/ml IgG (conjugated with Alexa Fluor 555; Supplementary Table 2) in LDM + 2% FBS + 5 µg/ml doxycycline. Cells were washed with PBS and fixed with 4% paraformaldehyde. For imaging intended for statistical assessment of uptake, samples were additionally stained with DAPI. CellProfiler version 3.1.948 was used for segmentation of the red channels using the Watershed algorithm. Mean intensities were calculated per object. For confocal imaging, samples were additionally stained with DAPI, phalloidin, and an anti-RAB5 antibody to detect endosomes (Details in Supplementary Table 2). This experiment was repeated twice. Co-localisation of RAB5 and IgG-AF555 was quantified using the JaCoP ImageJ plug-in, as described49. The Pearson’s correlation coefficient was computed after thresholding.
Statistical analysis of RT-qPCR
For all pair-wise comparisons (e.g., in the FLI1 and ERG overexpression and VEGFA addition experiments), we applied a Student’s T test. Statistical significance of the SPI1 lentiviral titre gradient on FCGR2B and LYVE1 expression was assessed using linear modelling. Time-course comparisons of RT-qPCR data and FITC-FSA were assessed by mixed ANOVA. Sample sizes were a priori decided through power analysis simulations in R, given a minimal effect size of interest of two cycle thresholds, an average standard deviation of one cycle threshold, a significance level of 0.05, and a minimally required power of 0.8.
Results
ETV2 overexpression fates pluripotent stem cells to the endothelial lineage
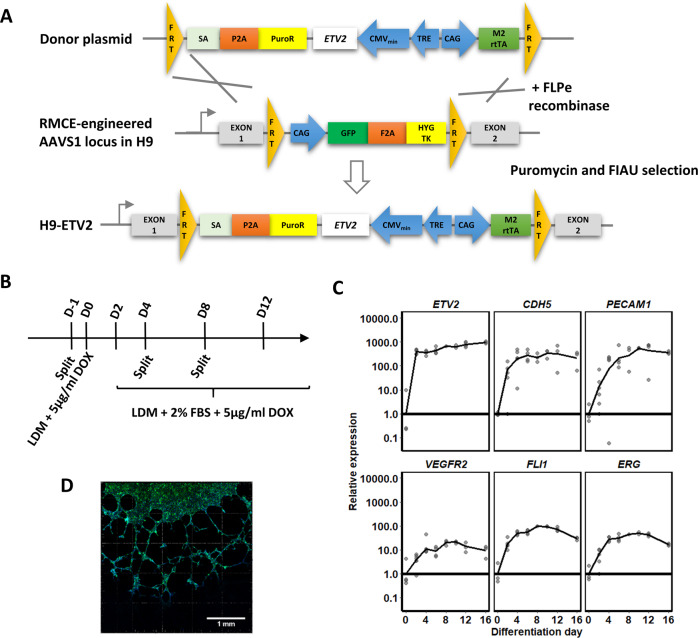
To effectively differentiate PSCs into ECs, we recombined a doxycycline-inducible ETV2 overexpression cassette in H9 ESC already containing an FRT-flanked exchange cassette (Fig. 1A), as described by Ordovas et al.41. Resulting ECs are further referred to as ETV2-ECs. We assessed the endothelial differentiation (Fig. 1B) by RT-qPCR (Fig. 1C). ETV2 was more than a 1000-fold overexpressed from day 2 of differentiation. As expected, expression levels of the pluripotency markers NANOG and POU5F1 decreased by >90% within 4 days of differentiation (Supplementary Fig. 4A). On day 6 of differentiation expression levels of the endothelial markers, i.e., PECAM1, CDH5, and VEGFR2, and endothelial TFs, i.e., FLI1 and ERG, were induced and remained stable (Fig. 1C). We confirmed expression of CD31 by immunostaining (Supplementary Fig. 4B). Low to no staining was observed for the LSEC markers CD32B and MRC1. Furthermore, tube formation, a key feature of ECs, was observed after seeding ETV2-ECs on matrigel (Fig. 1D).
Fig. 1. Generation and characterisation of ETV2-ECs.
A H9 human embryonic stem cells engineered with an RMCE cassette were recombined with a doxycycline-inducible ETV2 overexpression cassette (method described in and figure adapted from Ordovas et al.41). (FRT Flippase recognition target, SA splice acceptor, PuroR puromycin resistance gene, TRE tetracycline response element, M2rtTA M2 reverse tetracycline transactivator.) B Schematic representation for PSC differentiation into endothelial cells. C Gene expression (RT-qPCR) profile of ETV2, the endothelial markers CDH5, PECAM1, and VEGFR2, and the endothelial TFs FLI1 and ERG. Samples were collected every 2–4 days of differentiation until day 16 with N = 3 biological replicates per time point. Expression is relative to that of day 0 of differentiation (i.e., stem cells). D Confocal image of phalloidin-stained ETV2-ECs in Matrigel showing tube formation.
CenTFinder workflow
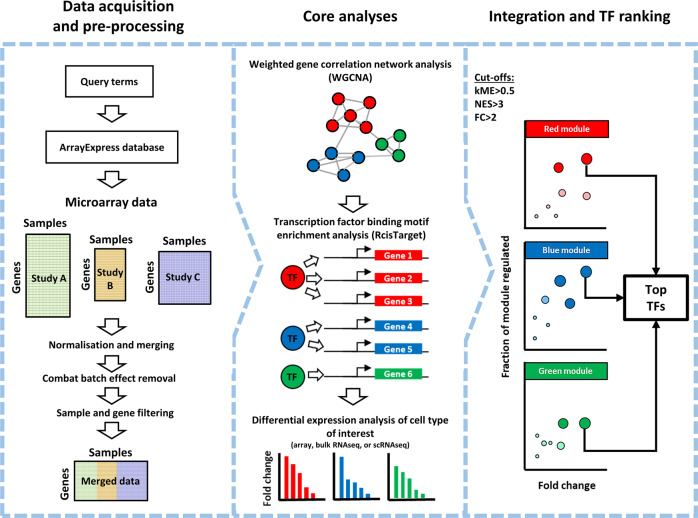
To identify and rank TFs required for fating ETV2-ECs towards LSECs, we designed a computational strategy (Fig. 2) to perform a meta-analysis on publicly available or in-house transcriptional data from various sources of ECs and LSECs (Supplementary Table 3). To ensure that the identified TFs play a ‘master regulator’ role in fating ECs to LSECs, we combined various computational methods, such as gene co-expression, TF binding motif enrichment, as well as differential expression analyses. Aside from TF rankings for LSECs, we also constructed TF rankings (Supplementary Table 4) for all other EC types that were included in the meta-analysis (Supplementary Table 3). The entire workflow is available as the CenTFinder R package on Github (https://github.com/jonathandesmedt92/CenTFinder).
Fig. 2. CenTFinder computational workflow.
Schematic overview of the CenTFinder workflow for identification of TFs (central in differentiation and specification). Left panel: data acquisition and pre-processing. The analysis starts by defining queries that are used for filtering and retrieving relevant expression data from the ArrayExpress database. Retrieved microarray data (of different formats) are combined and normalised and batch corrections are applied. Alternatively, a pre-processed expression matrix can be provided as input. Middle panel: core analyses. The expression matrix is subsequently subjected to gene co-expression analysis (WGCNA), TF binding motif enrichment analysis (RcisTarget), and the results are overlapped with differential expression analysis. Right panel: integration and TF ranking. The last step in the workflow is to combine the output of WGCNA, RcisTarget, and differential expression analyses to prioritise and rank TFs. Additional downstream analyses, e.g., Gene Ontology enrichment, and Cytoscape network visualisations are performed at this step.
Identification of LSEC markers by combination of microarray and single-cell RNAseq analyses
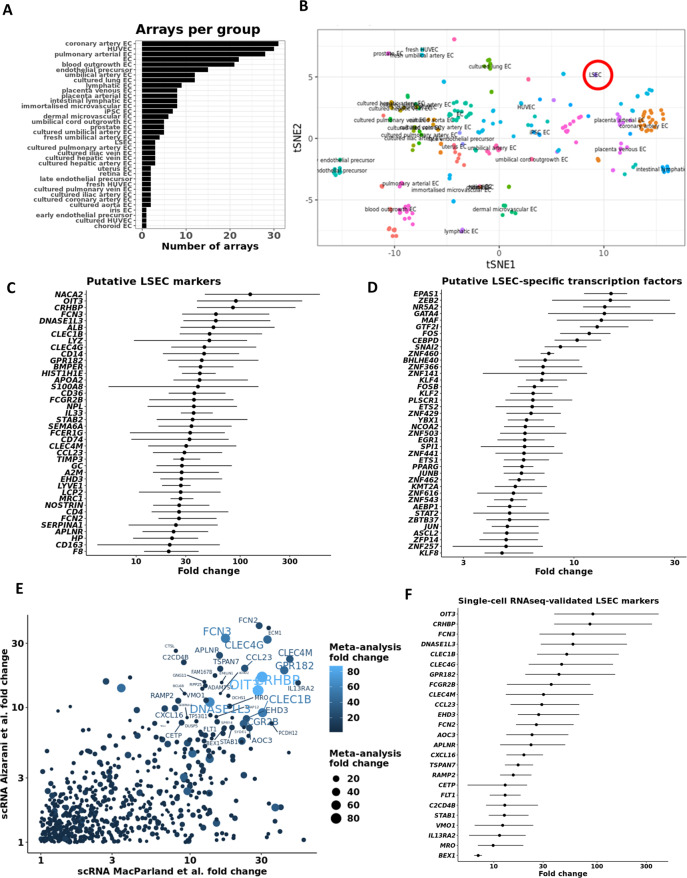
We downloaded, filtered, and normalised relevant microarray data (Fig. 2. left panel and Fig. 3A, and metadata in Supplementary Table 3). Next, we represented the gene expression profiles of all arrays (N = 279) by t-distributed Stochastic Neighbour Embedding (t-SNE), which shows great heterogeneity and sets the human primary LSECs apart from all other EC subtypes (Fig. 3B). Differential expression analysis between LSECs and all other EC types (Fig. 3C, D) confirmed established, as well as only recently identified LSEC marker genes, such as CLEC4G, FCGR2B, STAB2, MRC1, CLEC4M, F8, LYVE1, FCN2, FCN3, OIT3, CLEC1B, DNASE1L3, and GPR18245,50,51. In addition, we could identify putative novel LSEC marker candidates of which we validated LSEC-specific expression in two recently published liver scRNAseq datasets42,45 (Fig. 3E). Several putative markers were only expressed in the LSEC population, i.e., CDH5+ KDR+ CLEC4G+ cells42,51. In addition, some candidate markers, such as ALB and APOA2, were expressed in hepatocytes rather than LSECs. By merging fold changes of the CenTFinder and scRNAseq datasets, we generated a consensus list of scRNAseq-validated LSEC markers that were at least sevenfold higher expressed in LSECs compared to the other cells in the respective datasets42,45 (Fig. 3F).
Fig. 3. Intersection between microarray meta-analysis and single-cell RNAseq identifies putative LSEC markers.
A Bar chart indicating the number of microarrays per endothelial cell type that was included in the CenTFinder analysis (total number of arrays = 279). B t-SNE representation of all endothelial cells used in the analysis (LSECs circled in red). LSEC microarray data were contrasted to the expression data of all other endothelial cells used in the meta-analysis. Line charts show putative markers (C) and TFs (D) with increased expression in LSECs compared to all other endothelial cells (assessed by Wilcoxon rank-sum test). Putative markers and TFs were sorted by fold change. Dots indicate the mean fold change between LSECs and all other ECs. Lines span the interquartile range. E Overlap of genes that are specifically expressed in LSECs in the single-cell RNA-sequencing studies and in the microarray meta-analysis. The identified genes are more expressed in LSECs compared to all other endothelial cells, as well as compared to all other parenchymal and non-parenchymal liver cells. X- and Y-axes denote the two single-cell RNAseq studies42,45. F LSEC marker ranking by fold change compared to all other ECs in the microarray meta-analysis. For this analysis, genes were ranked only if in each of the studies they were more than sevenfold higher expressed in LSECs compared to the other cells. Dots indicate the mean fold change between LSECs and all other ECs. Lines span the interquartile range.
WGCNA and RcisTarget analysis identified SPI1 and IRFs as central to immune response transcriptional modules specific for LSECs
TFs were ranked by fold change (LSECs compared to other ECs in the microarray meta-analysis (Fig. 3D). However, such ranking identifies both putative TFs required for fating ECs to LSECs, as well as broadly acting TFs (e.g., JUN and FOS). Furthermore, transcripts of cell types other than LSECs may also bias this ranking as assessed by analysis of published single-cell RNAseq data42 (Supplementary Fig. 5). We subsequently used a gene regulatory network (GRN) approach (WGCNA) to select more specific TFs that might guide LSEC differentiation.
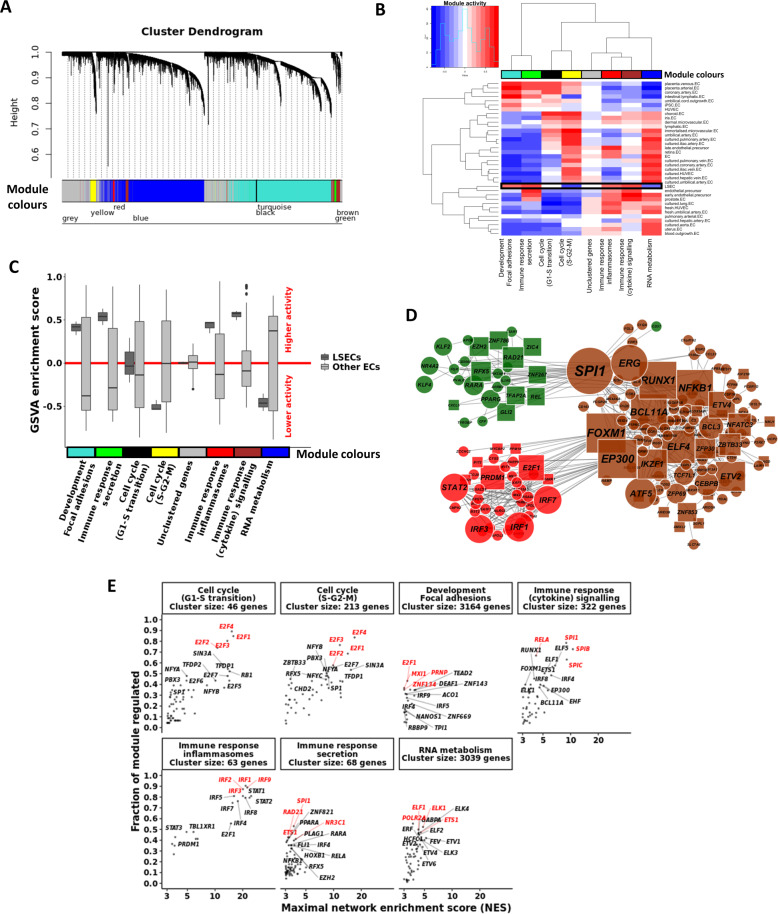
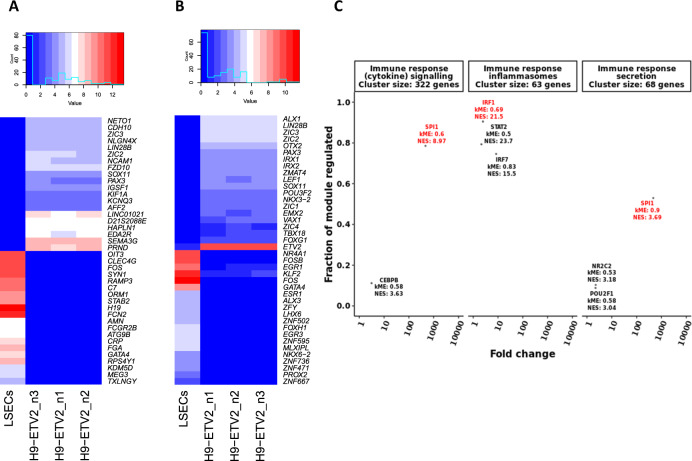
WGCNA is a tool that clusters genes with correlated expression patterns into modules that are, as a result, highly enriched for specific GO terms and pathways. We subjected the expression matrix to WGCNA38, to identify transcriptional modules, their associated GO terms and central TFs (i.e., TFs with a high module eigengene). We identified seven transcriptional modules (Fig. 4A) that were each active in at least one of the EC types included in the meta-analysis (Fig. 4B; excluding the grey module containing unassigned genes). By GO analysis, three of the modules were related to immune response genes (i.e., red, green, and brown), two to cell cycle genes (i.e., (S-G2-M) yellow and (G1-S transition) black), one to RNA metabolism and translation (i.e., blue), and one to development, gene expression regulation, and focal adhesions (i.e., turquoise; Supplementary Fig. 6). Comparison of module activities (calculated with gene set variation analysis, GSVA)52 demonstrated that the modules related to immune response, development, and focal adhesions were more active in LSECs compared to other ECs. In contrast, the (S-G2-M) cell cycle and the RNA metabolism modules were less active in LSECS (Fig. 4B, C). Interestingly, the immune clusters contained several C-type lectin receptors, such as CLEC10A, CLEC2D, CLEC4A, CLEC4E, CLEC5A, and CLEC7A, as well as several immunoglobulin receptors, such as FCER1G, FCGR1A, FCGR2A, FCGR2B, and FCGR3B (immune clusters represented in Fig. 4D).
Fig. 4. Gene co-expression (WGCNA) and transcription factor binding motif (RcisTarget) analyses.
A WGCNA cluster dendrogram visualising the gene co-expression modules identified in the microarray meta-analysis. These modules are active in at least one of the endothelial cell types. The grey module contains genes unassigned to any module due to low correlative values. B Heatmap and clustering of the gene set variation analysis (GSVA) activity scores for each of the modules and for each of the cell types. Module colours correspond with those from the cluster dendrogram in A. Black box identifies module activities in LSECs. C Boxplots of the GSVA enrichment scores for each of the modules, comparing LSECs to all other endothelial cell types. D Cytoscape visualisation of the LSEC-specific immune response modules. Edges indicate a regulatory link identified by RcisTarget. TF node size relates to the number of downstream targets. For the sake of simplicity, only nodes of the 20 most differentially expressed downstream targets are shown for each TF. Nodes were coloured according to their module colour. TFs belonging to other modules but with a kME > 0.5 for either the brown, red, or green modules were included as well and coloured accordingly. Circular (square) nodes represent genes that are higher (lower) expressed in LSECs compared to ETV2-ECs. E Enrichment of TF binding motifs was calculated for each of the WGCNA gene co-expression modules. Each dot represents an enriched TF binding motif with the x-axis indicating the RcisTarget network enrichment score (NES) and the y-axis indicating the fraction of genes in the respective module that has binding motifs for the respective gene. For genes with multiple enriched motifs, only the motif with the highest NES is shown. Top TFs are highlighted in red.
To avoid interference of spurious correlations due to technical or biological noise, we applied RcisTarget39 to each of the WGCNA modules (Fig. 4E). RcisTarget identifies TFs for which there is an enriched binding motif presence in the cis-regulatory regions of the genes in a given gene set. Only TFs with a network enrichment score (NES) > 3 were considered and TFs that regulate higher fractions of their respective modules were deemed more important or influential. In the immune response modules related to cytokine signalling and secretion, SPI1 had binding motifs in most of the genes in each of the respective clusters. In the immune response module related to inflammasomes, most genes were regulated by IRF TFs.
To further narrow down more promising TFs, we used bulk RNA-sequencing data to identify which of these TFs are differentially expressed between ETV2-ECs and LSECs (Fig. 5A, B). Next, we made an overlap between the cluster central and RcisTarget-enriched TFs that were expressed more in LSECs compared to ETV2-ECs (Fig. 5C). All TFs in the development and focal adhesions module were less expressed in LSECs compared to ETV2-ECs. In the immune reponse (cytokine signalling and secretion) modules SPI1 was the most highly differentially expressed (469-fold, p < 0.001; cluster centrality kME = 0.6 and kME = 0.9, respectively, and NES = 8.97 and NES = 3.69 respectively). In the immune response (inflammasomes) module, IRF1, IRF7, and STAT2 passed the filtering criteria. An analysis of the number of downstream targets for the different TFs demonstrated that PU.1 regulates considerably more genes than the other TFs (Supplementary Fig. 7). Furthermore, inspection of RcisTarget’s gene-motif database (hg19-tss-centred-10kb-7-species.mc9nr.feather) showed that, of all selected TFs, only PU.1 has binding motifs for which some of the LSEC markers are highly ranking targets (Supplementary Fig. 8). This was confirmed by in silico analysis of available ChiP-Seq data (downloaded from ChiP-Atlas https://chip-atlas.org/; Supplementary Fig. 9).
Fig. 5. CenTFinder analysis identifies SPI1 and IRF TFs as major LSEC regulators.
A Heatmap of the top 20 most upregulated and downregulated genes in LSECs compared to ETV2-ECs. B Heatmap of the top 20 most upregulated and downregulated TFs in LSECs compared to ETV2-ECs. C CenTFinder core analysis results were combined. Each dot represents a TF that was differentially and higher expressed in LSECs compared to ETV2-ECs. In addition, only TFs with kME > 0.5 and NES > 3 were considered. SPI1 was identified as the central TF in the immune response clusters related to cytokine signalling and secretion. IRF TFs were central to the immune response cluster related to inflammasomes. Top TFs are highlighted in red.
In conclusion, the microarray EC and LSEC meta-analysis identified SPI1 and IRFs to be candidate master regulators of LSEC fating. As the comparison of LSECs and ETV2-ECs also identified SPI1 to be the highest ranking TF in LSECs, subsequent studies were done to validate SPI1 as a master regulator of LSEC fating.
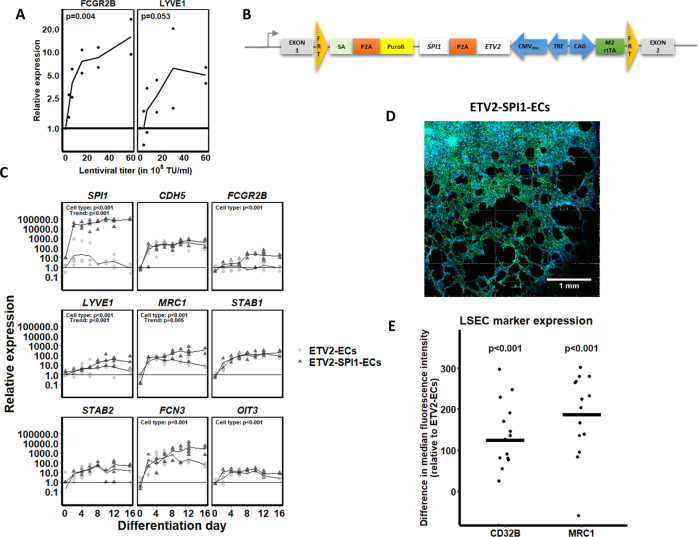
SPI1 overexpression induces LSEC marker expression
To evaluate if SPI1 can drive LSEC specification and maturation in ETV2-ECs, we cloned the SPI1 coding sequence into a pLVX backbone and transduced ETV2-ECs on day 6 of differentiation with the resulting lentiviral vector. Clear upregulation of the LSEC markers FCGR2B and LYVE1 was observed (Fig. 6A). To avoid the toxicity associated with lentiviral transduction, and to ensure that all cells contained the SPI1 TF, we next recombined a doxycycline-inducible cassette for both ETV2 and SPI1 in the AAVS1 safe harbour locus, to generate ETV2-SPI1-ECs (Fig. 6B). Combined induction of ETV2 and SPI1 did not impact general EC differentiation (Supplementary Fig. 10). Specifically, expression of NANOG and POU5F1 decreased by >90% on day 4 (Supplementary Fig. 10A) and no differences in expression of the EC markers PECAM1 and CDH5, and the EC TFs ERG, ETV2, and FLI1, were observed between ETV2- and ETV2-SPI1-ECs (Supplementary Fig. 10B). Consistently, immunostaining for CD31 was similar in ETV2 and ETV2-SPI-ECs (Supplementary Fig. 11). Transcript levels for SPI1 were >1000-fold higher expressed in ETV2-SPI1-ECs compared to ETV2-ECs (p < 0.001; Fig. 6C). In addition, tube formation by ETV2-ECs and ETV2-SPI1-ECs was similar (Fig. 6D).
Fig. 6. Characterisation of (liver sinusoidal) endothelial cell markers in ETV2-ECs and ETV2-SPI1-ECs.
A ETV2-ECs were transduced on day 6 of differentiation with increasing titres of the SPI1-encoding lentiviral vector. The effect of SPI1 overexpression on the LSEC markers FCGR2B and LYVE1 is shown. Statistical analysis by linear modelling (expression ~ titre); N = 2 biological replicates per concentration. B ETV2-SPI1 construct engineered into the AAVS1 safe harbour locus. Recombination was performed analogously to Fig. 1A. (FRT Flippase recognition target, SA splice acceptor, PuroR puromycin resistance gene, TRE tetracycline response element, M2rtTA M2 reverse tetracycline transactivator). C Time-course comparison between ETV2-ECs and ETV2-SPI1-ECs for the overexpressed SPI1, for the endothelial cell-specific gene CDH5, for the known LSEC markers FCGR2B, LYVE1, MRC1, STAB1, and STAB2, and for novel LSEC markers, such as FCN3, and OIT3. Statistical significances were assessed by mixed ANOVA. Undetected mRNA values were represented with a relative expression of 1. The x-axes denote the days of differentiation. The differentiation protocols for ETV2-ECs and ETV2-SPI1-ECs are identical. D Tube formation assay on day 12 of differentiation of ETV2-SPI1-ECs. E LSEC marker expression (CD32B and MRC1) in ETV2-SPI1-ECs compared to ETV2-ECs. Chart indicates the median fluorescence intensity shift for CD32B and MRC1 of ETV2-ECs and ETV2-SPI1-ECs on day 12 of differentiation (replicates: N = 15 for CD32B and N = 14 for MRC1). The CD32 and MRC1 positive cells were gated from single-cell, PI-negative, and VE-cadherin-positive populations (gating strategy in Supplementary Fig. 3).
Next, we compared the expression over time of the known and the proposed novel LSEC markers (as described in Fig. 3F). RT-qPCR demonstrated that the LSEC markers FCGR2B (p < 0.001), LYVE1 (p < 0.001), MRC1 (p = 0.002), CRHBP (p = 0.025), FCN3 (p = 0.011), and OIT3 (p = 0.018) were significantly higher expressed in ETV2-SPI1-ECs compared to ETV2-ECs (Fig. 6C and Supplementary Fig. 10C). This was further substantiated by immunostaining (Supplementary Fig. 11) and flow cytometry analysis (Fig. 6E and Supplementary Fig. 3), which showed a higher expression of CD32B and MRC1 in the ETV2-SPI1-ECs compared to the ETV2-ECs. CLEC4G, a highly specific LSEC marker, was however not expressed in either ETV2-ECs or ETV2-SPI1-ECs (data not shown).
As ETV2 is only expressed until E10.5 in the mouse53, we tested if persistent induction of ETV2 might prohibit differentiation of ETV2-ECs into ECs with a more mature, LSEC-like phenotype. Therefore, to replace ETV2 overexpression with TFs that are expressed in adult ECs, we cotransduced lentiviral vectors encoding the SPI1, FLI1, and/or ERG genes in ETV2-ECs on day 6 of differentiation, followed by doxycycline omission, and hence loss of ETV2 expression, as of day 8. Loss of ETV2 expression caused a significant drop (>90%, p < 0.01) in PECAM1 expression unless cells were cotransduced with FLI1 and/or ERG (Supplementary Fig. 2A). Combined induction of ETV2, ERG, and SPI1 led to the increased expression of MRC1 and OIT3 (p < 0.05). In contrast, combined induction of FLI1 and SPI1 decreased expression of FCN3 and DNASE1L3 (p < 0.05; Supplementary Fig. 2B, C). Overall, replacement of ETV2 overexpression with FLI1 or ERG overexpression could maintain the endothelial phenotype, but did not further enhance LSEC marker expression.
As the differentiation assay did not include VEGFA, and VEGFA may be beneficial for the LSEC phenotype17,54, we repeated differentiations with inclusion of VEGFA from day 6 onwards. Expression of LYVE1 in ETV2-SPI1-ECs was significantly induced by VEGFA (19.71-fold; p = 0.005; Supplementary Fig. 1). However, VEGFA did not significantly induce other LSEC markers, such as FCGR2B, STAB1, and STAB2.
Doxycycline-inducible SPI1 overexpression improves LSEC features in ETV2-ECs
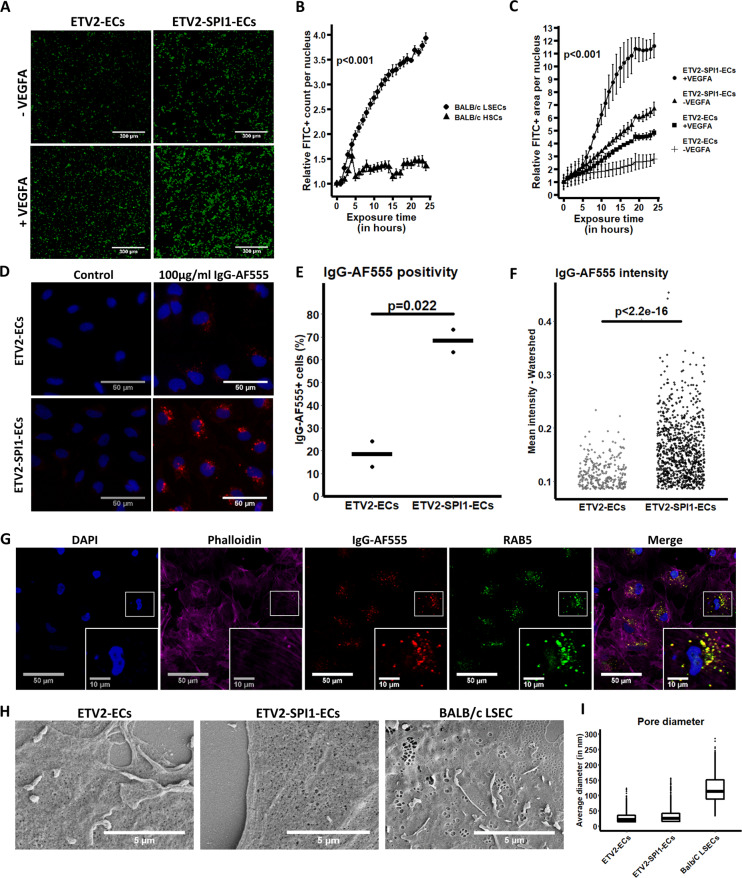
To assess LSEC functionality of ETV2-SPI1-ECs, we performed assays for the uptake of formaldehyde-treated FITC-coupled albumin (FSA-FITC; Fig. 7A–C), as well as IgG-AF555 (Fig. 7D–F). FSA-FITC uptake, which is scavenger receptor-mediated, is efficient in primary mouse LSEC (Fig. 7B). In PSC-derived ETV2-ECs, FSA-FITC uptake was enhanced in ETV2-SPI1-ECs when compared with ETV2-ECs (p < 0.001). In addition, when VEGFA was supplemented to the culture media, the uptake of FSA-FITC was increased in both cell populations (p < 0.001; Fig. 7C). Furthermore, CD32B-mediated IgG uptake was observed in only 22.21% (301/1355) of ETV2-ECs (Fig. 7D, E), while we could detect IgG-AF555 uptake in 68.60% (933/1360) of ETV2-SPI1-ECs (p = 0.022), with the IgG-AF555 fluorochrome co-localised with endosomes identified by a RAB5 antibody (Pearson correlation for co-localisation: 0.733 ± 0.055; Fig. 7G). In addition, the IgG-AF555-positive ETV2-SPI1-ECs were significantly brighter than the IgG-AF555-positive ETV2-ECs (p < 0.001; Fig. 7F).
Fig. 7. LSEC-specific functional characterisations of ETV2-ECs and ETV2-SPI1-ECs.
A–C FSA-FITC uptake assay (N = 2 biological replicates) on ETV2-ECs and ETV2-SPI1-ECs with and without addition of VEGFA, as well as on BALB/c LSECs and hepatic stellate cells (HSCs). Images are representative for 24 h exposure to FSA-FITC. Scale bars are 300 µm. FITC fluorescence was measured every hour. Statistical differences were assessed by mixed ANOVA. D IgG-AF555 uptake assay. ETV2-ECs and ETV2-SPI1-ECs were exposed for 2 h to 100 µg/ml IgG-AF555 to measure the clathrin-mediated endocytosis of IgGs upon their binding to CD32B. Scale bars are 50 µm (representative example of N = 3 biological replicates). E Comparison of the percentage of IgG-AF555-positive cells in ETV2-ECs and ETV2-SPI1-ECs at day 12 of differentiation. Statistical assessment by Student’s T test. F Comparison of the AF555 intensity of IgG-AF555-positive cells in ETV2-ECs and ETV2-SPI1-ECs at day 12 of differentiation. Statistical assessment by Wilcoxon rank-sum test. G Confocal imaging of ETV2-SPI1-ECs exposed to IgG-AF555. Cells were co-stained with an anti-RAB5 antibody to evaluate co-localisation of IgG-AF555 and endosomes. H Scanning electron microscopy of ETV2-ECs, ETV2-SPI1-ECs, and BALB/c LSECs. Scale bars are 5 µm (representative example of all cells per coverslip). Average pore diameters are shown in I.
We also performed scanning electron microscopy on ETV2-ECs and ETV2-SPI1-ECs (with mouse LSECs as control) to determine if induction of SPI1 induced fenestrae formation (Fig. 7H). Mouse LSECs showed clear fenestrations grouped in sieve plates. LSEC fenestration diameter was 122.73 ± 48.26 nm (Fig. 7I). However, neither ETV2- or ETV2-SPI1-ECs displayed fenestrations. Some transcytoplasmic holes were observed in ETV2-ECs and ETV2-SPI1-ECs, but these holes were much smaller, i.e., 28.38 ± 18.02 and 33.55 ± 22.79 nm, respectively, and likely due to SEM preparation artefacts, as previously indicated by Elvevold et al.55. We did note a clear morphological difference; ETV2-SPI1-ECs appeared more rounded with a few large cellular protrusions radiating from the main cell body, while ETV2-ECs appeared as flat cells with no or small and thin protrusions.
Discussion
To identify TFs that are crucial for LSEC fating, we created a computational workflow (CenTFinder) that applies several bioinformatic tools on (a meta-analysis of) transcriptomics datasets. Our analysis identified SPI1 as a key TF in a number of immune response pathways characteristic for LSECs. We demonstrated that forced expression of SPI1 in PSC-derived ECs can partially fate PSC-ECs to an LSEC-like phenotype.
Classically, in differential expression analyses genes or TFs are ranked by fold change and/or p value. However, such ranking may not necessarily yield (all) relevant TFs, as is exemplified by SPI1, which did not rank in the top 20 of differentially expressed TFs in the CenTFinder analysis. TFs might be differentially expressed due to differences between in vivo and in vitro environments, or due to impurities in primary cell populations. In addition, the fold change of a given TF does not provide knowledge regarding its number of dowstream targets, regarding its relevance to the cell-of-interest’s phenotype, or regarding the presence and complexity of its interactions with other proteins or signalling molecules.
To address these issues and to identify TFs that are crucial for LSEC fating, CenTFinder combines gene co-expression analysis (WGCNA), TF binding motif enrichment analysis (RcisTarget), and differential gene expression analysis. Several studies have been published that combine microarray meta-analysis with gene co-expression methods56,57; however, to date only a single study has combined this with TF binding motif enrichment as well58. Furthermore, the SCENIC workflow39, a well-established analysis tool for single-cell RNA-sequencing data, is in essence based on similar premises.
The underlying rationale for gene co-expression analyses is that genes that are associated with the same biological process or pathway tend to be coregulated, thereby increasing the correlation between their expression patterns. Expression coregulation implies the existence of a subset of responsible upstream TFs, which hence would display higher connectivity and cluster centrality (kME). Applying TF binding motif enrichment analysis on gene co-expression modules instead of the full gene set decreased the likelihood of prioritising a TF that was assigned to a module due to mere spurious correlation (e.g., correlation due to array probe position59). Simultaneously, it increased the likelihood of prioritising relevant TFs, as these TFs would not only be highly co-expressed with their module members, but would also have enriched binding motifs in the cis-regulatory elements of these very same module members. Furthermore, TFs with enriched binding motifs in relatively smaller clusters (such as SPI1 in our analysis) might not be detected by binding motif enrichment applied on the full list of differentially expressed genes. TFs were constrained by their centrality in a functional co-expression module and by the presence of enriched binding motifs. As TFs are assigned to a functional co-expression module, one can disregard TFs of modules that are not specifically related to the cell type of interest. For instance, in our analysis, we found that only three modules (immune response) out of seven were relevant for the LSEC phenotype, and hence we excluded TFs from other modules. Finally, we prioritised TFs that were differentially expressed between ETV2-ECs and LSECs, as we reasoned that correcting the respective module activities might suffice by mere overexpression of these TFs.
We note that such strategy could be of interest, as well in any other research aiming at improved differentiation and specification of PSC progeny towards a cell type of interest. In addition, the developed package can also identify TFs central to response to chemicals or different culturing conditions. The only limiting step in the described strategy is the number of arrays required for gene co-expression inference. As per their FAQ page (https://horvath.genetics.ucla.edu/html/CoexpressionNetwork/Rpackages/WGCNA/faq.html), the WGCNA authors do not recommend applying WGCNA on <20 samples. However, to counter noise, false–positive associations, and hence incorrect TF binding site enrichments, we would use at least 100 arrays. Extensive tutorials have been made available for WGCNA and RcisTarget analysis by their respective authors. Similarly, we made a tutorial available at https://github.com/jonathandesmedt92/CenTFinder.
We identified seven WGCNA modules, of which six were differentially active in LSECs compared to other ECs. Consistent with the known roles of LSECs in innate and immune functions, three modules contained genes related to immune response pathways. Interestingly, SPI1 was identified as one of the key regulators of two of these immune clusters. SPI1, the gene encoding for the PU.1 protein, is commonly believed to play a pivotal role in B cells, NK cells, granulocytes, dendritic cells, and macrophages60–63. Although PU.1 and IRF TFs were recently identified as TFs that are potentially relevant for the LSEC phenotype64, to our knowledge no group has functionally validated its role in fating ECs to LSEC-like cells.
We here provide conclusive evidence for PU.1 to be a master regulator for LSEC fating and as a key regulatory role in LSEC functionality. Over a hundred transciptional targets for PU.1 in haematopoietic progeny had already been identified by various groups65, including CLEC4G, FCGR2B, and MRC1. Although CLEC4G expression was not induced upon SPI1 overexpression in PSC-ECs, other established LSEC markers, such as FCGR2B and MRC1, were significantly induced. Furthermore, novel putative LSEC markers identified in this study (and in others42,45,50), such as FCN2, FCN3, CRHBP, and OIT3 were also induced by PU.1. Overexpression of PU.1 induced not only increased expression of FCGR2B, but the activity of the CD32B receptor was also significantly higher, as was evidenced by IgG-AF555 uptake.
Interestingly, not all LSEC markers were induced. We noted that these marker genes were not present in the immune response modules, but in the turquoise module related to development and focal adhesions (CLEC4G, FCN2, and STAB1), or in the grey module containing unclustered transcripts (CLEC4M, CRHBP, DNASE1L3, and STAB2). In addition, of the induced LSEC markers, FCN3 and OIT3 belonged to the turquoise module, and LYVE1 and MRC1 to the grey module. This could be caused by several factors. First, the Pearson correlation used for construction of the co-expression networks does not detect non-linear gene interactions. Combinatorial regulation of the LSEC markers may hence reduce the Pearson correlation of these markers with their respective regulator TFs. Expression of some of the induced and non-induced LSEC markers might thus be dependent on the interaction of PU.1, and one or more yet to be identified other TFs. Second, variables such the epigenetic status or chromatin conformation may effect correlation values as well. Third, interestingly, several LSEC markers clustered in the turquoise module, which contains genes related to substrate adhesion. Future research may address (co-)culturing in 3D cultures, and assess transcriptional changes in this module and the LSEC markers in this modules.
Finally, uptake of FSA-FITC, a commonly used functional assay for LSECs, was more evident in ETV2-SPI1-ECs, but not in ETV2-ECs, and this effect was enhanced with VEGFA supplementation, which is known to support in vivo LSEC differentiation66,67, as well as in vitro culture17. However, as STAB1/2 expression remained unchanged upon VEGFA addition, the mechanism could be post-translational, for instance at the level of receptor recycling.
While PU.1 is already known to regulate expression of cytokines, antibodies, and antibody receptors in a variety of cell types, to date no group, to our knowledge, described this in LSECs. Even more, it has been described that LSECs express CD45, a common haematopoietic marker68. Considering this, the fact that PU.1 is a critical TF for haemotopoietic stem cells, and given the unique immune roles of LSECs, it raises the question whether the LSEC might descend from an intermediate cell type in the haemogenic endothelium. Future lineage tracing experiments during development will be required to help define LSEC identity and origin. Based on the very recent single-cell RNAseq data of Wang et al.69 on human developing liver, it might also be of interest to test if specific TFs that are for instance present in endocardium during development, may aid in generating LSECs from PSCs.
Further research could also address the role of other TFs that are central in the immune response modules. Moreover, TFs that appear cluster central and have enriched binding motifs within their respective modules, but did not pass stringent differential expression criteria, could also be evaluated. The activity (or mere nuclear translocation) of such TFs could be modulated by interacting small molecules or other signalling factors. Furthermore, modules with lower activity in LSECs might contain TFs that interfere with immune response modules, or TFs inhibiting the formation of fenestrae. Hence, knockdown of such TFs might further improve the LSEC phenotype.
In summary, our findings combine complementary transcriptomics analysis methods in order to maximise knowledge extraction from transcriptome datasets. For this purpose, we developed the CenTFinder R package which bundles meta-analysis, gene co-expression analysis, TF binding motif enrichment analysis, and differential expression analysis in one integrated pipeline, providing also a framework for identifying TFs involved in, for instance, other endothelial lineage fating of stem cells. Based on this approach, we identified SPI1 as the most highly ranked LSEC-specific TF, and demonstrated the importance of PU.1 in LSEC differentiation from PSCs.
Supplementary information
Acknowledgements
We would like to thank Prof. Dr. Stein Aerts for his valuable input, advice, and discussions on bioinformatic strategies, Dr. Sara Aibar for her advice on the usage of RcisTarget, Dr. Cristina Øie for her critical assessment of and advice on the IgG uptake assays, and Dr. Aernout Luttun for his well-valued advice. Furthermore, we would like to acknowledge the VIB Nucleomics Core for the RNA-sequencing, related quality control, and fruitful discussions. Confocal Images were recorded on a Zeiss LSM 880—Airyscan (Cell and Tissue Imaging Cluster (CIC)), supported by Hercules AKUL/15/37_GOH1816N and FWO G.0929.15 to Pieter Vanden Berghe, University of Leuven.
Funding
This study was funded by grants to J.D.S (Fonds Wetenschappelijk Onderzoek 1S33916N) and R.B. (Agentschap voor Innovatie door Wetenschap en Technologie SB-121393), IWT-140045 and HILIM-3D to C.M.V. and L.A.v.G, H2020-766181-DeLIVER-ITN to L.A.v.G., FWO-G0D9917N to C.M.V., and funding from the EU-ToxRisk, a project running under the European Union’s Horizon 2020 research and innovation programme, grant agreement no. 681002.
Author contributions
J.D.S. wrote all code for analysis and developed the CenTFinder package. J.D.S. and I.T. analysed and interpreted the CenTFinder analysis, and performed the lentiviral transductions. J.D.S. and S.G. performed the RT-qPCR, immunostainings, and cell culture. J.D.S., S.Z., B.T., S.G., and S.V. all contributed to (the setup of) the flow cytometry readings. Confocal imaging and colocalisation analysis was entirely performed by B.T., M.A.V., and R.B. generated the H9-ETV2 stem cell line, and performed initial characterisations. J.D.S. generated the H9-ETV2-SPI1 stem cell line. R.F.M.D.C. performed the computational image analysis on immunostainings and the IgG uptake assays. J.D.S. and P.B. performed the SEM preparation, and P.B. did the SEM imaging. Primary mouse cells were isolated by E.A.v.O., FSA-FITC uptake assays were performed by E.A.v.O. and A.S., and IgG-AF555 uptake assays were performed by J.D.S., all statistical analyses were performed by J.D.S., J.D.S., L.A.v.G., and C.V. conceived the main concept of the presented study. J.D.S. and C.V. wrote majority of the manuscript. All authors contributed to the final version of the manuscript, as well as to discussions about and interpretations of the presented results.
Data availability
RNA-sequencing data is available from GEO (GSE156009).
Code availability
All code is reproducible and can be accessed via https://github.com/jonathandesmedt92/CenTFinder.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval and consent to participate
No ethical approval was required for this study.
Footnotes
Edited by I. Amelio
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Leo A. van Grunsven, Catherine M. Verfaillie
Contributor Information
Jonathan De Smedt, Email: jonathandesmedt92@gmail.com.
Catherine M. Verfaillie, Email: catherine.verfaillie@kuleuven.be
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41419-020-03356-2).
References
- 1.Fraser R, Bosanquet AG, Day WA. Filtration of chylomicrons by the liver may influence cholesterol metabolism and atherosclerosis. Atherosclerosis. 1978;29:113–123. doi: 10.1016/0021-9150(78)90001-1. [DOI] [PubMed] [Google Scholar]
- 2.Wisse E, de Zanger RB, Charels K, van der Smissen P, McCuskey RS. The liver sieve: considerations concerning the structure and function of endothelial fenestrae, the sinusoidal wall and the space of disse. Hepatology. 1985;5:683–692. doi: 10.1002/hep.1840050427. [DOI] [PubMed] [Google Scholar]
- 3.Smedsrød B, et al. Hepatic sinusoidal cells in health and disease: update from the 14th International Symposium. Liver Int. 2009;29:490–501. doi: 10.1111/j.1478-3231.2009.01979.x. [DOI] [PubMed] [Google Scholar]
- 4.Sørensen KK, et al. The scavenger endothelial cell: a new player in homeostasis and immunity. Am. J. Physiol. Integr. Comp. Physiol. 2012;303:R1217–R1230. doi: 10.1152/ajpregu.00686.2011. [DOI] [PubMed] [Google Scholar]
- 5.Ding BSen, et al. Inductive angiocrine signals from sinusoidal endothelium are required for liver regeneration. Nature. 2010;468:310–315. doi: 10.1038/nature09493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu J, et al. Endothelial cell-derived Angiopoietin-2 controls liver regeneration as a spatiotemporal rheostat. Science. 2014;343:416–419. doi: 10.1126/science.1244880. [DOI] [PubMed] [Google Scholar]
- 7.Lai WK, et al. Expression of DC-SIGN and DC-SIGNR on human sinusoidal endothelium: a role for capturing hepatitis C virus particles. Am. J. Pathol. 2006;169:200–208. doi: 10.2353/ajpath.2006.051191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Powlesland AS, et al. A novel mechanism for LSECtin binding to Ebola virus surface glycoprotein through truncated glycans. J. Biol. Chem. 2008;283:593–602. doi: 10.1074/jbc.M706292200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ganesan LP, et al. Rapid and efficient clearance of blood-borne virus by liver sinusoidal endothelium. PLoS Pathog. 2011;7:e1002281. doi: 10.1371/journal.ppat.1002281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simon-Santamaria J, et al. Efficient uptake of blood-borne BK and JC polyomavirus-like particles in endothelial cells of liver sinusoids and renal Vasa recta. PLoS ONE. 2014;9:e111762. doi: 10.1371/journal.pone.0111762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mates JM, et al. Mouse liver sinusoidal endothelium eliminates HIV-like particles from blood at a rate of 100 million per minute by a second-order kineticprocess. Front. Immunol. 2017;8:35. doi: 10.3389/fimmu.2017.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steffan AM, et al. Primary cultures of endothelial cells from the human liver sinusoid are permissive for human immunodeficiency virus type 1. Proc. Natl Acad. Sci. USA. 1992;89:1582–1586. doi: 10.1073/pnas.89.5.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boaru SG, Borkham-Kamphorst E, Tihaa L, Haas U, Weiskirchen R. Expression analysis of inflammasomes in experimental models of inflammatory and fibrotic liver disease. J. Inflamm. 2012;9:49. doi: 10.1186/1476-9255-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Limmer A, et al. Efficient presentation of exogenous antigen by liver endothelial cells to CD8+ T cells results in antigen-specific T-cell tolerance. Nat. Med. 2000;6:1348–1354. doi: 10.1038/82161. [DOI] [PubMed] [Google Scholar]
- 15.Carambia A, et al. TGF-β-dependent induction of CD4+CD25+Foxp3 + Tregs by liver sinusoidal endothelial cells. J. Hepatol. 2014;61:594–599. doi: 10.1016/j.jhep.2014.04.027. [DOI] [PubMed] [Google Scholar]
- 16.Pasarín M, et al. Sinusoidal endothelial dysfunction precedes inflammation and fibrosis in a model of NAFLD. PLoS ONE. 2012;7:e32785. doi: 10.1371/journal.pone.0032785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie G, et al. Role of differentiation of liver sinusoidal endothelial cells in progression and regression of hepatic fibrosis in rats. Gastroenterology. 2012;142:918. doi: 10.1053/j.gastro.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeLeve LD, Wang X, Guo Y. Sinusoidal endothelial cells prevent rat stellate cell activation and promote reversion to quiescence. Hepatology. 2008;48:920–930. doi: 10.1002/hep.22351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyao M, et al. Pivotal role of liver sinusoidal endothelial cells in NAFLD/NASH progression. Lab. Investig. 2015;95:1130–1144. doi: 10.1038/labinvest.2015.95. [DOI] [PubMed] [Google Scholar]
- 20.Rockey DC, Chung JJ. Reduced nitric oxide production by endothelial cells in cirrhotic rat liver: endothelial dysfunction in portal hypertension. Gastroenterology. 1998;114:344–351. doi: 10.1016/S0016-5085(98)70487-1. [DOI] [PubMed] [Google Scholar]
- 21.Elvevold K, Nedredal GI, Revhaug A, Bertheussen K, Smesrød B. Long-term preservation of high endocytic activity in primary cultures of pig liver sinusoidal endothelial cells. Eur. J. Cell Biol. 2005;84:749–764. doi: 10.1016/j.ejcb.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Tokairin T, et al. A highly specific isolation of rat sinusoidal endothelial cells by the immunomagnetic bead method using SE-1 monoclonal antibody. J. Hepatol. 2002;36:725–733. doi: 10.1016/S0168-8278(02)00048-X. [DOI] [PubMed] [Google Scholar]
- 23.Krause P, et al. Hepatocyte-supported serum-free culture of rat liver sinusoidal endothelial cells. J. Hepatol. 2000;32:718–726. doi: 10.1016/S0168-8278(00)80239-1. [DOI] [PubMed] [Google Scholar]
- 24.Tai Y, et al. SK-Hep1: not hepatocellular carcinoma cells but a cell model for liver sinusoidal endothelial cells. Int. J. Clin. Exp. Pathol. 2018;11:2931–2938. [PMC free article] [PubMed] [Google Scholar]
- 25.Levenberg S, Golub JS, Amit M, Itskovitz-Eldor J, Langer R. Endothelial cells derived from human embryonic stem cells. Proc. Natl Acad. Sci. USA. 2002;99:4391–4396. doi: 10.1073/pnas.032074999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sahara M, et al. Manipulation of a VEGF-Notch signaling circuit drives formation of functional vascular endothelial progenitors from human pluripotent stem cells. Cell Res. 2014;24:820–841. doi: 10.1038/cr.2014.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elcheva I, et al. Direct induction of haematoendothelial programs in human pluripotent stem cells by transcriptional regulators. Nat. Commun. 2014;5:1–11. doi: 10.1038/ncomms5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koui Y, et al. An in vitro human liver model by iPSC-derived parenchymal and non-parenchymal cells. Stem Cell Rep. 2017;9:490–498. doi: 10.1016/j.stemcr.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gage BK, et al. Generation of functional liver sinusoidal endothelial cells from human pluripotent stem-cell-derived venous angioblasts. Cell Stem Cell. 2020;27:254–269.e9. doi: 10.1016/j.stem.2020.06.007. [DOI] [PubMed] [Google Scholar]
- 30.Couvelard A, et al. Structural and functional differentiation of sinusoidal endothelial cells during liver organogenesis in humans. Blood. 1996;87:4568–4580. doi: 10.1182/blood.V87.11.4568.bloodjournal87114568. [DOI] [PubMed] [Google Scholar]
- 31.Gouysse G, et al. Relationship between vascular development and vascular differentiation during liver organogenesis in humans. J. Hepatol. 2002;37:730–740. doi: 10.1016/S0168-8278(02)00282-9. [DOI] [PubMed] [Google Scholar]
- 32.Shiojiri N, Sugiyama Y. Immunolocalization of extracellular matrix components and integrins during mouse liver development. Hepatology. 2004;40:346–355. doi: 10.1002/hep.20303. [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, et al. Genetic lineage tracing identifies endocardial origin of liver vasculature. Nat. Genet. 2016;48:537–543. doi: 10.1038/ng.3536. [DOI] [PubMed] [Google Scholar]
- 34.Tedesco FS, et al. Transplantation of genetically corrected human iPSC-derived progenitors in mice with limb-girdle muscular dystrophy. Sci. Transl. Med. 2012;4:140ra189. doi: 10.1126/scitranslmed.3003541. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.García-León JA, et al. SOX10 single transcription factor-based fast and efficient generation of oligodendrocytes from human pluripotent stem cells. Stem Cell Rep. 2018;10:655–672. doi: 10.1016/j.stemcr.2017.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boon R, et al. Amino acid levels determine metabolism and CYP450 function of hepatocytes and hepatoma cell lines. Nat. Commun. 2020;11:1–16. doi: 10.1038/s41467-020-15058-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aibar S, et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ordovás L, et al. Efficient recombinase-mediated cassette exchange in hPSCs to study the hepatocyte lineage reveals AAVS1 locus-mediated transgene inhibition. Stem Cell Rep. 2015;5:918–931. doi: 10.1016/j.stemcr.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aizarani N, et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature. 2019;572:199–204. doi: 10.1038/s41586-019-1373-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagner, F., Barkley, D. & Yanai, I. ENHANCE: accurate denoising of single-cell RNA-Seq data. Preprint at bioRxiv10.1101/655365 (2019).
- 44.Scialdone A, et al. Computational assignment of cell-cycle stage from single-cell transcriptome data. Methods. 2015;85:54–61. doi: 10.1016/j.ymeth.2015.06.021. [DOI] [PubMed] [Google Scholar]
- 45.MacParland SA, et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 2018;9:1–21. doi: 10.1038/s41467-018-06318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stradiot L, et al. Functionality based method for simultaneous isolation of rodent hepatic sinusoidal cells. Biomaterials. 2017;139:91–101. doi: 10.1016/j.biomaterials.2017.05.047. [DOI] [PubMed] [Google Scholar]
- 47.Li R, et al. Endocytosis of advanced glycation end-products in bovine choriocapillaris endothelial cells. Microcirculation. 2009;16:640–655. doi: 10.1080/10739680903133185. [DOI] [PubMed] [Google Scholar]
- 48.Lamprecht MR, Sabatini DM, Carpenter AE. CellProfilerTM: free, versatile software for automated biological image analysis. Biotechniques. 2007;42:71–75. doi: 10.2144/000112257. [DOI] [PubMed] [Google Scholar]
- 49.Bolte S, Cordelières F. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 50.Schmid CD, et al. GPR182 is a novel marker for sinusoidal endothelial differentiation with distinct GPCR signaling activity in vitro. Biochem. Biophys. Res. Commun. 2018;497:32–38. doi: 10.1016/j.bbrc.2018.01.185. [DOI] [PubMed] [Google Scholar]
- 51.Ramachandran P, et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature. 2019;575:512–518. doi: 10.1038/s41586-019-1631-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koyano-Nakagawa N, et al. Etv2 is expressed in the yolk sac hematopoietic and endothelial progenitors and regulates Lmo2 gene expression. Stem Cells. 2012;30:1611–1623. doi: 10.1002/stem.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.DeLeve LD, Wang X, Hu L, McCuskey MK, McCuskey RS. Rat liver sinusoidal endothelial cell phenotype is maintained by paracrine andautocrine regulation. Am. J. Physiol. Gastrointest. Liver Physiol. 2004;287:G757–G763. doi: 10.1152/ajpgi.00017.2004. [DOI] [PubMed] [Google Scholar]
- 55.Elvevold K, Smedsrød B, Martinez I. The liver sinusoidal endothelial cell: a cell type of controversial and confusing identity. Am. J. Physiol. Gastrointest. Liver Physiol. 2007;294:G391–G400. doi: 10.1152/ajpgi.00167.2007. [DOI] [PubMed] [Google Scholar]
- 56.Mistry M, Gillis J, Pavlidis P. Meta-analysis of gene coexpression networks in the post-mortem prefrontal cortex of patients with schizophrenia and unaffected controls. BMC Neurosci. 2013;14:1–16. doi: 10.1186/1471-2202-14-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun J, Xu H, Qi M, Zhang C, Jianxiang SHI. Identification of key genes in osteosarcoma by meta-analysis of gene expression microarray. Mol. Med. Rep. 2019;20:3075–3084. doi: 10.3892/mmr.2019.10543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Triska, M., Ivliev, A., Nikolsky, Y. & Tatarinova, T. V. Analysis of cis-regulatory elements in gene co-expression networks in cancer. Methods Mol. Biol.1613, 291–310 (2017). [DOI] [PubMed]
- 59.Qian J, Kluger Y, Yu H, Gerstein M. Identification and correction of spurious spatial correlations in microarray data. Biotechniques. 2003;35:42–48. doi: 10.2144/03351bm03. [DOI] [PubMed] [Google Scholar]
- 60.DeKoter RP, et al. Regulation of follicular B cell differentiation by the related E26 transformation-specific transcription factors PU.1, Spi-B, and Spi-C. J. Immunol. 2010;185:7374–7384. doi: 10.4049/jimmunol.1001413. [DOI] [PubMed] [Google Scholar]
- 61.Houston IB, Kamath MB, Schweitzer BL, Chlon TM, DeKoter RP. Reduction in PU.1 activity results in a block to B-cell development, abnormal myeloid proliferation, and neonatal lethality. Exp. Hematol. 2007;35:1056–1068. doi: 10.1016/j.exphem.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kamath MB, et al. Dose-dependent repression of T-cell and natural killer cell genes by PU.1 enforces myeloid and B-cell identity. Leukemia. 2008;22:1214–1225. doi: 10.1038/leu.2008.67. [DOI] [PubMed] [Google Scholar]
- 63.Colucci F, et al. Differential requirement for the transcription factor PU.1 in the generation of natural killer cells versus B and T cells. Blood. 2001;97:2625–2632. doi: 10.1182/blood.V97.9.2625. [DOI] [PubMed] [Google Scholar]
- 64.Danoy M, et al. Transcriptome profiling of hiPSC-derived LSECs with nanoCAGE. Mol. Omics. 2020;16:138–146. doi: 10.1039/C9MO00135B. [DOI] [PubMed] [Google Scholar]
- 65.Turkistany SA, Dekoter RP. The transcription factor PU.1 is a critical regulator of cellular communication in the immune system. Arch. Immunol. Ther. Exp. 2011;59:431–440. doi: 10.1007/s00005-011-0147-9. [DOI] [PubMed] [Google Scholar]
- 66.Walter TJ, Cast AE, Huppert KA, Huppert SS. Epithelial VEGF signaling is required in the mouse liver for proper sinusoid endothelial cell identity and hepatocyte zonation in vivo. Am. J. Physiol. Liver Physiol. 2014;306:G849–G862. doi: 10.1152/ajpgi.00426.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carpenter B, et al. VEGF is crucial for the hepatic vascular development required for lipoprotein uptake. Development. 2005;132:3293–3303. doi: 10.1242/dev.01902. [DOI] [PubMed] [Google Scholar]
- 68.Xie G, Wang L, Wang X, Wang L, DeLeve LD. Isolation of periportal, midlobular, and centrilobular rat liver sinusoidal endothelial cells enables study of zonated drug toxicity. Am. J. Physiol. Gastrointest. Liver Physiol. 2010;299:G1204. doi: 10.1152/ajpgi.00302.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang, X. et al. Comparative analysis of cell lineage differentiation during hepatogenesis in humans and mice at the single-cell transcriptome level. Cell Res.30, 1109–1126 (2020). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-sequencing data is available from GEO (GSE156009).
All code is reproducible and can be accessed via https://github.com/jonathandesmedt92/CenTFinder.