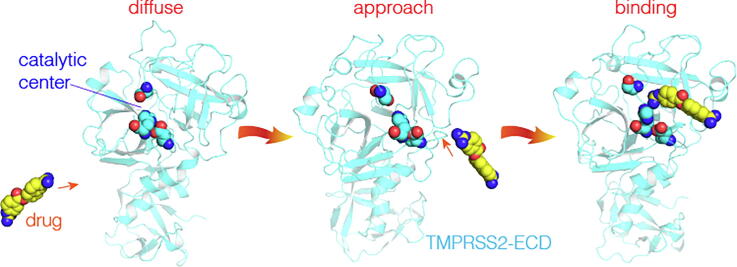
Graphical abstract
Keywords: COVID-19, SARS-CoV-2, TMPRSS2, Drug action, Spontaneous binding simulation
Abstract
Effective treatment or vaccine is not yet available for combating SARS coronavirus 2 (SARS-CoV-2) that caused the COVID-19 pandemic. Recent studies showed that two drugs, Camostat and Nafamostat, might be repurposed to treat COVID-19 by inhibiting human TMPRSS2 required for proteolytic activation of viral spike (S) glycoprotein. However, their molecular mechanisms of pharmacological action remain unclear. Here, we perform molecular dynamics simulations to investigate their native binding sites on TMPRSS2. We revealed that both drugs could spontaneously and stably bind to the TMPRSS2 catalytic center, and thereby inhibit its proteolytic processing of the S protein. Also, we found that Nafamostat is more specific than Camostat for binding to the catalytic center, consistent with reported observation that Nafamostat blocks the SARS-CoV-2 infection at a lower concentration. Thus, this study provides mechanistic insights into the Camostat and Nafamostat inhibition of the SARS-CoV-2 infection, and offers useful information for COVID-19 drug development.
1. Introduction
Coronaviruses (CoVs) are single-stranded RNA viruses that can spread in animals and humans, causing a variety of diseases, such as respiratory, intestinal, kidney, and nervous system diseases [1]. To date, three highly pathogenic human coronaviruses (hCoVs) have been identified, including severe acute respiratory syndrome coronavirus (SARS-CoV) [2], Middle East respiratory syndrome coronavirus (MERS-CoV) [3] and the 2019 novel coronavirus (SARS-CoV-2) that emerged in Wuhan in December 2019 [4]. All three viruses have the ability to infect human host cells and thus to transmit from humans to humans. However, according to the data released by WHO, the spread rate of SARS-CoV-2 in humans has significantly exceeded those of SARS-CoV and MERS-CoV [5], leading to a global pandemic of the coronavirus disease 2019 (COVID-19). Up to December 20, 2020, approximately 76.340 million COVID-19 cases were confirmed worldwide, including 1,686,590 deaths [6]. Genomic sequencing and phylogenetic analysis showed that SARS-CoV-2 shares 79.6% sequence identity to SARS-CoV, and belongs to β-coronavirus. Because the overall genome sequence reaches the highest similarity (96.2%) to the bat coronavirus bat-SL-CoV-RaTG13, it was thought that this novel virus might have been transmitted from bats to humans via unknown intermediate host animals [7]. Currently, there is no effective treatment or vaccine to combat SARS-CoV-2. Therefore, scientists and researchers all over the world are having a race against time to develop effective drugs and vaccines for preventing SARS-CoV-2 infection [8], including repurposing of existing drugs to target viral and host proteins [9], [10], [11], especially drugs that could block the SARS-CoV-2 entry into the human cells.
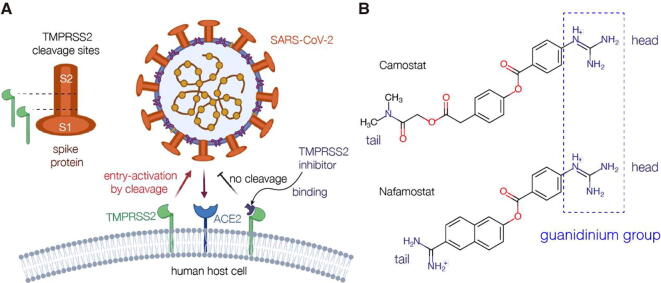
The hCoV entry into the human cells is mediated by its trimeric transmembrane spike (S) glycoprotein [12]. In general, to release the viral RNA genome into a host cell, the S protein binds to the host receptor angiotensin-converting enzyme 2 (ACE2) on the host cell surface, and upon endocytic uptake the viral membrane fuses with the endosomal membrane [13]. In addition to ACE2, human proteases are essential to prime and to activate the S-ectodomain for binding and fusion [14]. Two cleavage sites, S1/S2 and S2′, located at the boundary between the S1 and S2 subunits, have to be proteolytic processed by different proteases, such as furin, the transmembrane protease serine 2 (TMPRSS2), and cathepsin L and B [15]. Cleavage of the S1/S2 site by furin primes the spike protein for efficient binding of S1 to ACE2 through its receptor-binding domain (RBD) attaching the virus to the cell surface [16]. TMPRSS2 on the cell surface processes S2′, priming the S protein to mediate fusion with the endosomal membrane by its fusion peptide [17]. Although both endosomal cathepsins process S2′ as well, TMPRSS2 has been shown to be more essential for the S protein priming and thus infection by SARS-CoV-2 [18]. In consequence, the TMPRSS2 processing is one of the key steps to activate the membrane function of the SARS-CoV-2 S protein [19]. Hence, using drugs to inhibit the proteolytic activity of TMPRSS2 is likely to block the membrane fusion of SARS-CoV-2 (Fig. 1A). Interestingly, TMPRSS2 is a human protease, and as the drug target, will not cause the problem of developing drug resistance like the viral protein targets [20]. Indeed, TMPRSS2 is one of the most promising targets for the anti-SARS-CoV-2 drugs [21].
Fig. 1.
TMPRSS2 activation of SARS-CoV-2 entry into human host cell and two potential COVID-19 drugs targeting TMPRSS2. (A) The schematic diagram of TMPRSS2 activation of SARS-CoV-2 into the host cell. The spike protein of SARS-CoV2 is cleaved by human proteases at the S1/S2 boundary and/or within the S2 subunit with conserved Arginine residues. TMPRSS2 inhibitors could block the entry activating process. (B) The chemical structures of two potential COVID-19 drugs that inhibit the cleavage of the spike protein by TMPRSS2: Camostat and Nafamostat. The guanidinium group of the drug (Camostat or Nafamostat) is defined as its head, and the other terminus as its tail.
Previous studies have shown that several TMPRSS2 inhibitors could effectively block the hCoV infection [22]. Kawase et al. found that Camostat, a drug for treating chronic pancreatitis, can block the SARS-CoV and HCoV-NL63 infections by inhibiting the TMPRSS2 activity [23]. Later, Yamaoto et al. screened a library of 1017 FDA-approved drugs using Dual Split Protein (DSP) reporter fusion assay, and then found that Nafamostat, another related agent for pancreatitis and disseminated intravascular coagulation, can also block the MERS-CoV membrane fusion [24]. After the emergence of SARS-CoV-2, scientists investigated the ability of these two drugs to block the SARS-CoV-2 infection. Hoffmann et al. firstly showed that Camostat has inhibitory effects on the SARS-CoV-2 in TMPRSS2-expressing human cells [18]. More recently, two groups at the same time confirmed that Nafamostat can block the SARS-CoV-2 fusion at a concentration less than one-tenth that required for Camostat; they reported that 1 ~ 10 nM of Nafamostat can significantly inhibit the cell infection of SARS-CoV-2 [25], [26]. These results strongly supported that both Camostat and Nafamostat are potential drugs for treating COVID-19 [27], [28], [29]. Indeed, several clinical trials for evaluating their therapeutic effects against SARS-CoV-2 are now underway [8]. Recent studies have shown that clinical trials of these two drugs in the treatment of COVID-19 have achieved preliminary and obvious effects [30], [31]. Moreover, these two old drugs have been commercialized for many years; so, once they pass the clinical trials, they may be immediately applied to the COVID-19 treatment.
Although they are undergoing the COVID-19 clinical trials, the molecular mechanisms in which Camostat and Nafamostat inhibit the TMPRSS2 activity remain unclear. To provide guidance for their repurposing for treating COVID-19, it is urgent to answer the following questions: Can Camostat or Nafamostat bind to the catalytic center of TMPRSS2 and then inhibit its proteolytic activity? What are the key molecular interactions in binding process? What are the stable conformations of the drugs bound to TMPRSS2? Besides the on-target binding site (i.e., the catalytic center), are there other off-target binding hotspots on TMPRSS2? To address these questions, we performed atomic-level, unbiased molecular dynamics (MD) simulations to investigate the dynamic binding processes of Camostat and Nafamostat to TMPRSS2. In these simulations, the drug molecules were initially placed at random positions distant from the TMPRSS2 surface; then, without giving any prior knowledge of the drug binding sites, drugs diffused around TMPRSS2 to spontaneously “recognize” their native binding sites on TMPRSS2. Our simulations showed that both Camostat and Nafamostat could spontaneously and stably bind to the TMPRSS2 catalytic center, indicating that the catalytic center is their native binding site. We found that the main driving forces for the binding are the electrostatic attractions between the drug guanidinium group and the surface Asp/Glu residues around the catalytic center. As the drugs enter the catalytic center, van der Waals forces and hydrogen bonds between the drugs and TMPRSS2 stabilize them in the center in an induced-fit way. For this reason, the bound drugs occupy the space required for the substrate binding. This may inhibit the proteolytic activity of TMPRSS2.
2. Materials and methods
2.1. Homology modeling of the TMPRSS2-ECD structure
Because no experimental structure is available for TMPRSS2, an initial, all-atom model of TMPRSS2 extracellular domain (TMPRSS2-ECD, aa 146 ~ 492) was first generated by homology modeling via the Swiss-Model server (https://swissmodel.expasy.org/). The amino-acid sequence of TMPRSS2 was obtained from UniProt (UniProt ID: O15393; GenBank No: U75329). Then, by uploading the sequence of TMPRSS2-ECD, the 3D model was built using the fully automated modeling mode of the Swiss-Model server. The server searched the existing structure that shares the highest sequence identity to that of TMPRSS2-ECD as the template for building the model. The crystal structure of the extracellular region of the transmembrane serine protease hepsin with a resolution of 1.55 Å (PDB ID: 1Z8G) was found to share the best sequence coverage and the highest GMQE (Global Model Quality Estimation) to TMPRSS2-ECD. So, with this crystal structure as the template, a 3D atomistic model was eventually constructed for TMPRSS2-ECD (aa 146 ~ 492).
2.2. MD refinement of theTMPRSS2-ECD model
The homology all-atom model of TMPRSS2-ECD was then optimized by the MD simulation in solution state. The MD simulations were conducted using GROMACS (Ver. 5.1.4). The CHARMM27 force field [32] and the TIP3P water model [33] were employed to model the simulation system. In the simulation system, the all-atom structure of TMPRSS2-ECD was placed in the center of a rectangular water box with a minimal distance of 15 Å from the protein surface to its boundary. Certain numbers of Na+ and Cl− ions were added to the system for setting an ionic concentration of 150 mM and neutralizing the system.
To optimize the system, the energy minimization of the system was first carried out using the steepest descent algorithm for a maximum of 50,000 steps or until the maximum force <1000 kJ·mol−1·nm−1. Following the energy minimization, the system was heated by NVT equilibration at 320 K for 100 ps. Then, NPT equilibration for 1 ns was followed by setting the system pressure to 1 bar. In the simulations, the integration time step was 2 fs. The V-rescale method [34] was used to maintain the average temperature of 320 K; and the Parrinello-Rahman barostat [35] was used for maintaining the average pressure of 1 bar. The periodic boundary conditions (PBC) were applied in all three dimensions. The bond lengths and angles were constrained using the LINCS algorithm [36]. The Particle Mesh Ewald (PME) method [37] was used for the long-range electrostatics; and a cut-off distance of 10 Å was employed for both short-range electrostatic interactions and van der Waals interactions. Finally, MD refinement simulation was performed in the NVT ensemble for 200 ns. In the simulation, the coordinates of the system atoms were recorded per 10 ps for the analysis.
2.3. System set-up for spontaneous binding simulations
As in the above MD refinement, CHARMM27 force field was used to model the protein, and the TIP3P water model was employed for the solvent. The 2D chemical structures of Camostat and Nafamostat were downloaded from PubChem (https://pubchem.ncbi.nlm.nih.gov). The ionization state of the drugs at pH ~ 7.0 was predicted using ChemAxon (https://chemaxon.com/) and the final 3D structures of two drugs with protons were obtained using MolView (http://molview.org/). The drugs were modeled using the General Amber Force Field (GAFF) [38]. Their topology and parameter files were generated using tLEaP module of AmberTools17 and converted to GROMACS-compatible files with ACPYPE [39].
To build the initial system for simulating a drug association with TMPRSS2, the refined structure of TMPRSS2-ECD was placed in the center of a rectangular water box, with a distance at least 15 Å from protein surface to the box boundary. Then, a drug molecule (Camostat or Nafamostat) was placed at a random position in the solvent, at least 30 Å away from the TMPRSS2 catalytic center. The system was added certain numbers of Na+ and Cl− ions for setting the ionic concentration to 150 mM and neutralization. Next, the systems were minimized and equilibrated using the same control methods of temperature and pressure as those in the above MD refinement. Finally, for each drug 15 independent simulations were performed in the NVT ensemble, and each simulation lasted at least for 150 ns. In the simulations, the coordinates of the system atoms were recorded per 100 ps for the analysis.
2.4. Time-dependent drug distance to the catalytic center ()
We determined whether a drug (Camostat or Nafamostat) binds to the TMPRSS2 catalytic center in the simulations by defining a drug distance to the catalytic center (). In the simulations, the TMPRSS2 region within 3 Å of S441 and H296 is regarded as the catalytic center. Then, in a given frame of the MD trajectories, the minimum distance from any atom of the drug to any atom of the S441 and H296 is defined as . In a given simulation, if < 3 Å, the drug molecule is considered to enter the catalytic center in that MD frame. If this state lasts for more than 20 ns, the simulation will be considered as a successful trajectory to capture the spontaneous binding process of the drug to the catalytic center.
2.5. Binding free energy calculation
As in our previous studies [40], [41], we used AutoDockTools [42] to calculate the free energy of a drug (Camostat or Nafamostat) that bind to TMPRSS2-ECD in the simulation snapshots. AutoDockTools uses the AutoDock 4.1 semi-empirical free energy force field to estimate the free energy of a small-molecular drug binding to a protein receptor based on their drug-protein complex, With the given complex, the binding free energy () equals to the change in the free energy of the drug from the unbound state to the bound state:
| (1) |
where the first two terms are the energies of the drug-protein complex in the bound state, consisting of the intermolecular and intramolecular free energies. The third term is the conformational entropy change of the drug in the binding and is directly calculated from the sum of the torsional degrees of freedom. The fourth term is the reference energy of the drug in its unbound state and is defined as 0.0 kcal·mol−1. Of them, the intermolecular energy involves in van der Waals, hydrogen bonding, desolvation and electrostatic contributions, and is calculated by:
| (2) |
For the drug-TMPRSS2 complex in a given MD frame, Python programs in AutoDockTools were directly used to calculate the AutoDock 4.1 parameters of the drug and TMPRSS2 and the binding free energy according to standard procedures.
3. Results
3.1. Building the 3D model of the TMPRSS2 extracellular domain
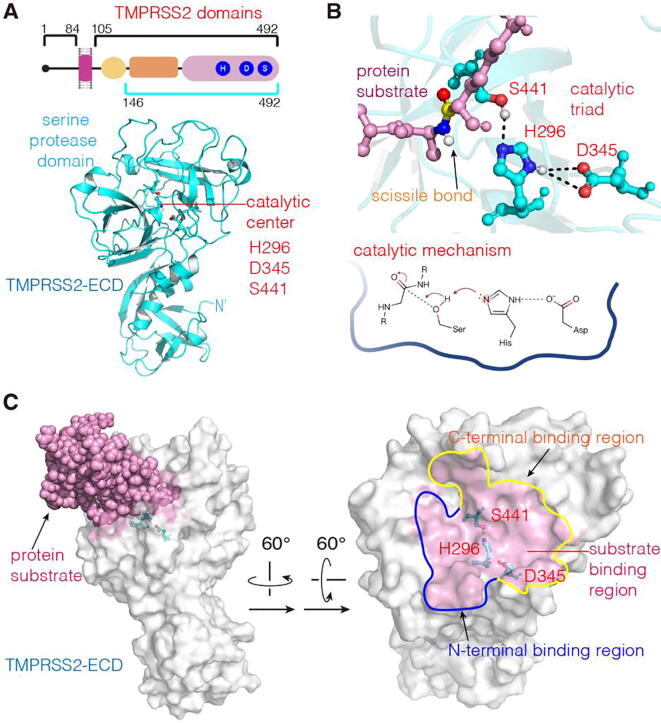
Human TMPRSS2 protein consists of 492 amino acids and is divided into three domains: the intracellular (aa 1–84), the transmembrane (aa 85–105) and the extracellular (aa 106–492) domain (Fig. 2A) [43]. The catalytic site for proteolysis is localized in the extracellular domain (ECD). So far, no three-dimensional (3D) structure of TMPRSS2 has been resolved. Thus, we predicted the atomic model of the TMPRSS2 extracellular domain (TMPRSS2-ECD) by computational methods.
Fig. 2.
The 3D structure of TMPRSS2-ECD and its catalytic center. (A) The 3D atomic model of TMPRSS2-ECD constructed via the Swiss-Model server. (B) The catalytic center with catalytic triad consisting of H296, A345 and S441, and corresponding catalytic mechanism for cleaving the protein substrate [49]. (C) The substrate-binding region. TMPRSS2-ECD is represented by the surface model (white) and the substrate is shown as pink spheres. The substrate-binding region in the right panel (pink) is defined as TMPRSS2 atoms being in a distance to the substrate < 3 Å. The N- and C-terminal binding regions are enclosed by the curves in blue and yellow, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To build the model, we used the Swiss-Model server (https://swissmodel.expasy.org/). Amino-acid sequence alignment indicated that the structure of a type II transmembrane trypsin-like serine protease hepsin (PDB ID: 1Z8G) shares the best sequence coverage (89%) and the highest GMQE (Global Model Quality Estimation, 0.64) to TMPRSS2-ECD (Supplementary Fig. S1). In fact, 1Z8G has been widely used as the modeling template of TMPRSS2 [44], [45], [46]. Thus, this crystal structure was employed as the template for our model construction. Because this structure lacks the N-terminal segment corresponding to aa 106–145 of TMPRSS2, a 3D structure for the aa 146–492 of TMPRSS2-ECD was built. The 3D model implies that the distance from the lacking N-terminal segment (aa 106–145) to the active site is greater than 37 Å. Next, to establish a TMPRSS2-ECD structure in the native state, we performed MD simulation with explicit solvent to further optimize the built TMPRSS2-ECD structure (see Section 2.2). As shown in Supplementary Fig. S2, the average root mean square deviation (RMSD) of the simulation system in the simulation time >60 ns is about 5 Å, and the ECD structure maintains a stable state in aqueous solution up to 200 ns. To verify this, we also examined whether the catalytic center of the TMPRSS2-ECD model possesses a conformation identical to that of a typical serine protease. So, we first compared the MD snapshot structure at 200 ns with the crystal structure of a serine protease of S1 family with a peptide inhibitor, bovine cationic trypsin (PDB ID: 2PTC) [47]. The catalytic centers of two structures are very similar and have an RMSD of 1.67 Å (Supplementary Fig. S3A). Since 2PTC is not a human protein, we also compared the MD structure with a recently published model of human TMPRSS2 (hTMPRSS2) [48], found that their RMSD of the catalytic centers is 1.88 Å (Supplementary Fig. S3B). Thus, the TMPRSS2-ECD structure obtained from the homology modeling and the MD refinement has a conserved catalytic center of serine proteases. Eventually, we chose the MD snapshot structure at 200 ns as the starting model of TMPRSS2-ECD for the following simulations (Fig. 2A).
Since the experimental structure of the TMPRSS2-spike protein complex is not yet available, to understand the binding mode of the substrate to the TMPRSS2 active site, we compared TMPRSS2-ECD with 2PTC in complex with a peptide inhibitor. Structural alignment indicated that the scissile peptide bond of the protein substrate is located in the catalytic center of TMPRSS2 (Fig. 2B), implying that the catalytic triad of TMPRSS2 likely uses the same proteolytic mechanism of serine proteases [49] to cleave its protein substrate. Based on the orientation of the scissile bond in Fig. 2B, we designated the TMPRSS2 areas occupied by the N-t- rminal and C-terminal substrate segments of the scissile bond as the N-terminal and C-terminal binding regions, respectively (Fig. 2C).
3.2. Two drugs spontaneously enter the TMPRSS2 catalytic center
Any drug molecule that effectively inhibits the proteolysis function of TMPRSS2 has to be bound to the catalytic amino-acids and/or the substrate-binding regions. To capture the dynamic association process of a given drug with the protein receptor, atomic-level, unbiased MD simulation is an effective tool [40], [41], [50]. To simulate those association processes for Camostat and Nafamostat in an aqueous environment, we used the mentioned TMPRSS2-ECD structure at 200 ns to establish the simulation systems. As described in Subsect. 2.3, the protein structure was placed at the center of the simulation box, while the drug molecule (Camostat or Nafamostat) was randomly placed around the protein, with initial position away at least 30 Å from the catalytic triad and at least 15 Å from the protein surface, ensuring that the dynamic binding of the drug to TMPRSS2 is not predetermined by the initial arrangement of components (Supplementary Fig. S4). In the simulations, no bias forces were added to the drug molecules; in other words, the drug movement from the initial position to the catalytic center is completely driven by the interplay of the drug with solvent molecules and TMPRSS2. For each drug, we performed at least 15 independent simulations starting from a random position of drug and lasting at least for 150 ns (Table 1).
Table 1.
Summary of spontaneous binding simulations.
| Drug | Number of independent simulations | Trajectory number of drugs bound to catalytic center | Number of binding from the N-terminal binding region | Number of binding from the C-terminal binding region | The lowest binding energy at catalytic center (kcal·mol−1) |
|---|---|---|---|---|---|
| Camostat | 15 | 6 | 4 | 2 | −9.66 |
| Nafamostat | 15 | 6 | 2 | 4 | −9.16 |
To determine whether a drug is bound to the TMPRSS2 catalytic center, we defined the minimum pairwise atomic distance between the drug and the catalytic residues S441 and H296 as “drug distance to the catalytic center” (). For < 3 Å lasting at least for 20 ns, the drug molecule is considered to be bound to the catalytic center. Within the simulation timescale of 150 ns both Camostat and Nafamostat were found to spontaneously bind to the catalytic center with a successful rate of ~40% (Table 1 and Supplementary Fig. S5). The 6 successful binding trajectories of 15 independent simulations showed that Camostat and Nafamostat follow two association pathways: the drug binds to the catalytic center from either the N-terminal binding region (Fig. 3A, B) or the C-terminal binding region (Fig. 3C, D). Among them, Camostat prefers to bind to the catalytic center from the N-terminal binding region, Nafamostat prefers to the C-terminal binding region (Table 1). As seen in Fig. 3, both drugs from distantly random positions could associate with TMPRSS2, and finally enter the catalytic center after short periods of conformational adjustment (see also Supplementary Movies S1 and S2). In the processes, even if the drugs diffused in the solvent to positions with > 60 Å, eventually they entered the catalytic center.
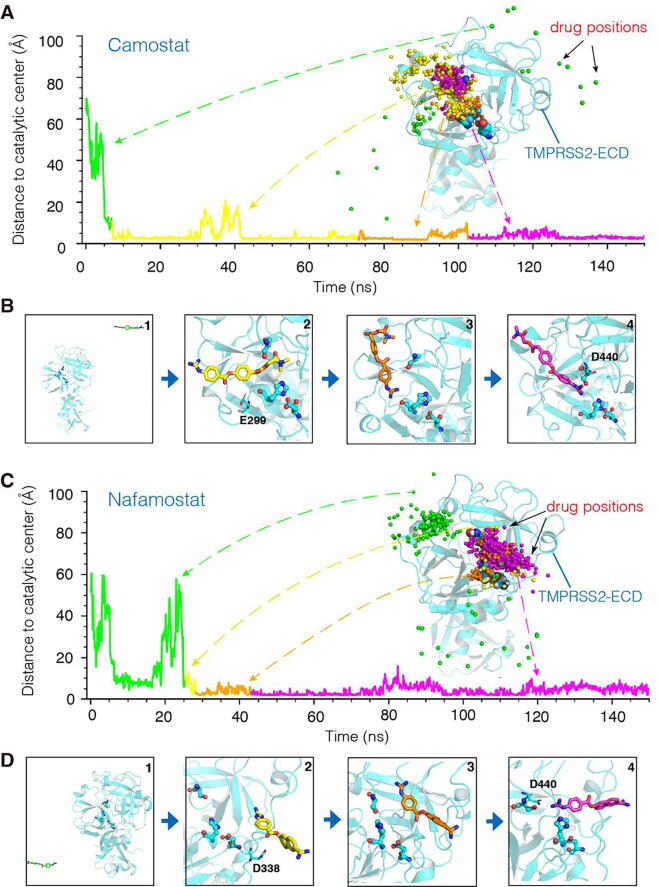
Fig. 3.
Typical spontaneous binding processes to the catalytic center of TMPRSS2. (A) Time-dependent Camostat distance to the catalytic center (). TMPRSS2-ECD is represented by the cartoon in cyan, and corresponding drug positions represented by the drug atoms closest to the catalytic center (spheres in colors). See also Movie S1 in Supplementary Materials. (B) Representative conformations of bound Camostat. (C) Time-dependent Nafamostat distance to the catalytic center (). TMPRSS2-ECD is represented by the cartoon in cyan, and corresponding drug positions represented by the drug atoms closest to the catalytic center (spheres in colors). See also Movie S2 in Supplementary Materials. (D) Representative conformations of bound Nafamostat. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To elucidate the driving forces of the binding, we analyzed the successful trajectories that capture the association processes of the drugs with the catalytic center (Supplementary Fig. S5). The simulations showed that whenever the drugs move toward the catalytic center either from the N-terminal binding region or from the C-terminal binding region, their association processes could be divided into two phases, as illustrated by the two typical trajectories in Fig. 3. Initially, driven by the thermal motions of the solvent molecules, the Camostat and Nafamostat drugs diffused randomly around the TMPRSS2 surface until they became attracted to the substrate-binding region (Fig. 3A, C). Here, as there are many negatively charged Asp/Glu residues in/around this region (Supplementary Fig. S6), the electrostatic interactions between the Asp/Glu oxygens and the positively charged guanidinium group of the drugs attracted the drugs to the TMPRSS2 surface in the vicinity of the substrate-binding region (e.g., Fig. 3B, D, panel 2). Next, by forming hydrogen bonds with amino acids in the catalytic center (Fig. 3B, D), the drugs continued to adjust their conformations and positions, and eventually achieved the final stable poses in the catalytic center. For example, as illustrated by the trajectory in Fig. 3A, Camostat even adjusted its head-tail orientation, so that its guanidinium group eventually contacted the catalytic center (Fig. 3B, panel 3). As shown by the trajectory in Fig. 3C, Nafamostat directly approached the catalytic center without any significant head-tail adjustment (Fig. 3D, panel 3). Finally, the hydrogen bonds formed by the drug guanidinium group with the amino acids in the catalytic center stabilized both drugs in the center (Fig. 3B, D, panel 4). As a result, the hydrogen bonding between S441 and H296 (Fig. 2B) is disrupted by the drug guanidinium group being positioned in the middle of H296 and S441. On-going simulations revealed that the values of both drugs are almost less than 3 Å, and the average RMSDs of both drugs are less than 2.1 Å (Camostat:2.01 Å; Nafamostat:1.30 Å) (Supplementary Fig. S7), indicating that they are in the stable binding states at least in the time windows of the simulations (Fig. 3A, C). Note that, besides the trajectories in Fig. 3, drug binding processes of other independent simulation trajectories in Supplementary Fig. S5 are similar. Because of the dynamic nature of the drug binding, all the trajectories are somehow different, but the results are identical: Camostat and Nafamostat become attracted to and spontaneously enter the substrate-binding cavity of TMPRSS2 from the N-terminal or C-terminal binding region, and finally bind to the TMPRSS2 catalytic center.
3.3. Both drugs are stably bound to the TMPRSS2 catalytic center
To elucidate the key interacting groups between the drugs and TMPRSS2 at the stable state, we used AutoDock 4.1 semi-empirical free energy function [51] to calculate the free energies of drug binding to the catalytic center using the snapshots of the drug-TMPRSS2 complexes in the trajectories in Fig. 3. As supported by our previous studies [40], [41], this calculation method of binding free-energy is fast and reliable for analyzing large numbers of inhibitor-receptor snapshot complexes in the MD simulations. The calculations for the trajectories in Fig. 3 showed that the lowest binding free energy of the drug-TMPRSS2 complex conformation is −9.16 kcal·mol−1 (Camostat) and −9.16 kcal·mol−1 (Nafamostat), respectively (see also Supplementary Fig. S8). So, although experimental values are not yet available, according to , the equilibrium dissociation constants of the two drugs are predicted in the order of nanomoles (Camostat: ~83 nM; Nafamostat: ~190 nM). And these predictions need to be validated by further in vitro or in vivo experiments.
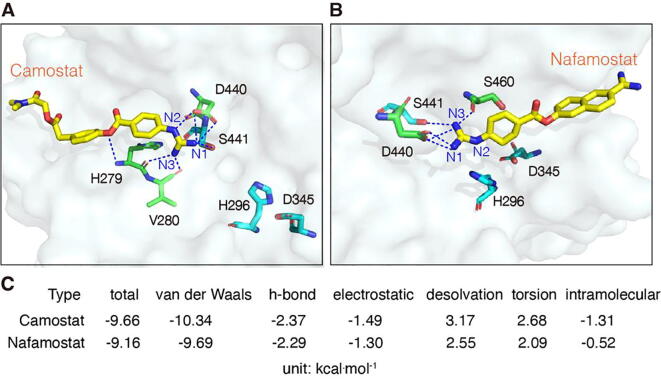
As shown in Fig. 4, Camostat and Nafamostat embed either in the N-terminal or C-terminal regions of the binding cavity near the catalytic center. The guanidinium groups of the drugs are located between S441 and H296. All three N atoms of the Camostat guanidinium group participate in hydrogen bonding: N1 and N2 form four hydrogen bonds with D440 and S441, and N3 forms a bidentate hydrogen bond with H279 and V280 (Fig. 4A). These hydrogen bonds fix the position of the guanidinium group. In addition, the oxygen atom between two aromatic rings also forms a hydrogen bond with H279, stabilizing the molecular center in the binding cavity. For Nafamostat, the two N atoms of the guanidinium group participate in bonding: N1 and D440 form a bidentate hydrogen bond, and N3 simultaneously interacts with D440, S441 and S460 (Fig. 4B). These hydrogen bonds highly restrict the structural flexibility of the guanidinium group. In addition, compared to the catalytically active conformation in Fig. 2B, the distance between H296 and S441 exceeds 5 Å. Such a distance enlargement is attributed to the steric effects of the bound guanidinium group on the H296 side chain.
Fig. 4.
The binding poses of drugs at the catalytic center with the lowest binding free energy. (A and B) The binding poses of Camostat and Nafamostat, respectively. Hydrogen bonds are shown as dotted lines. (C) Intramolecular and intermolecular terms of the lowest binding free energies corresponding to the binding poses in A (Camostat) and B (Nafamostat).
To understand the intermolecular forces that maintain the drug-TMPRSS2 complex in the stable binding state, we further analyzed the energy terms contributing to the binding free energies. As shown in Fig. 4C, the main intermolecular forces involved in binding are van der Waals forces, hydrogen bonds, and electrostatic forces. Among them, the van der Waals forces contribute the most. This may be attributed to the matching of the drug shapes to that of the binding cavity, which results in a close contact between the drug atoms and the amino acids in the catalytic center. As mentioned, the drugs also form multiple hydrogen bonds with the amino acids in the catalytic center (Fig. 4A, B). This restricts their orientation in the catalytic center, and therefore enhances the binding stability. Finally, the electrostatic forces between the positively charged guanidinium group of the drugs and the oxygen atoms in the binding cavity may further strengthen the drug-TMPRSS2 binding stability (Supplementary Fig. S6).
In conclusion, the drugs interact with the catalytic amino acids via their guanidinium groups, and form stably bound conformations in the catalytic center, thereby occupy the space required for the substrate binding. Then, Camostat and Nafamostat may inhibit the catalytic activity of TMPRSS2. The guanidinium group appears to be the key pharmacophore of the drugs. Indeed, the conserved arginine (Arg) at the cleavage site of the TMPRSS2 substrate also fully supports this, because the Arg side-chain possesses a guanidinium group. This molecular similarity implies that both drugs binds to the TMPRSS2 in the same way as the substrate does.
3.4. Nafamostat is more specific for the on-target binding
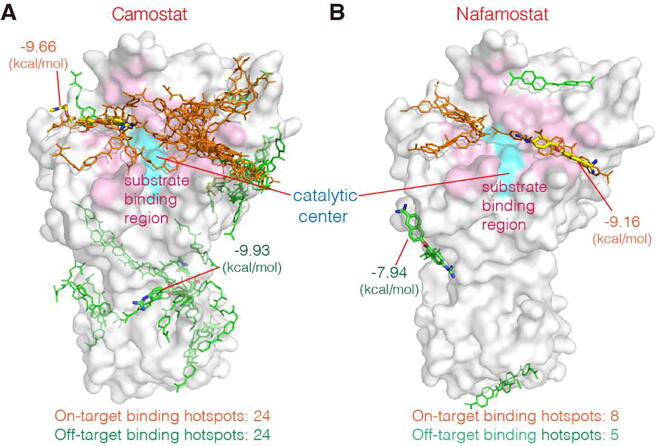
To find out all potential high-affinity binding hotspots of the drugs on TMPRSS2, we carried out statistical analysis of the transient complex conformations of the drugs bound to TMPRSS2 in all 30 independent MD simulations in Table 1. To this end, we firstly sorted out the high-affinity complex conformations with the binding free energy < 6.82 kcal·mol−1 (corresponding to < 10 μM). Next, we calculated their pairwise RMSDs, and then classified any two conformations with a pairwise RMSD < 4 Å into a conformational cluster. We used the conformation with the lowest binding free energy in the cluster as the representative conformation, and regarded its binding site as the TMPRSS2 binding hotspot of the cluster. In Fig. 5, all high-affinity binding hotspots of both drugs are shown. As seen, both Camostat and Nafamostat have certain numbers of high-affinity binding hotspots. Since the high-affinity hotspots outside the substrate-binding region may affect the association efficiency of the drugs with the catalytic center (i.e., the target binding site), we defined the hotspots contacting the substrate-binding region as on-target hotspots, while the others as off-target hotspots. The distribution of the on-target hotspots in Fig. 5 demonstrates that both drugs could occupy the substrate-binding region in multiple orientations, suggesting that they are able to efficiently bind to TMPRSS2 and thereby inhibit its activity.
Fig. 5.
The high-affinity binding hotspots of two drugs on TMPRSS2 revealed in the MD simulations. (A) The binding hotspots of Camostat. (B) The binding hotspots of Nafamostat. Each hotspot is represented by line model of the lowest-energy pose of corresponding conformational cluster of the drug (Camostat or Nafamostat). We defined the hotspots contacting the substrate-binding region as the on-target binding hotspots (orange), while the others as the off-target binding hotspots (green). Corresponding hotspots with the lowest binding energies are indicated by stick models with the energy values. The catalytic center is shown as cyan surface. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
However, both the total numbers of the on-target and off-target hotspots of Camostat are higher than those of Nafamostat (Fig. 5A, B, panel bottom). Very likely, this is attributed to the structural difference between two drug molecules. Although the structures of Camostat and Nafamostat are similar, Nafamostat is slightly shorter in size and possess more aromatic rings, resulting in a more rigid structure. Thus, less conformational clusters are possible when binding to TMPRSS2. In contrast, Camostat is slightly longer and more flexible, which may give rise to form more binding poses on TMPRSS2 and thus more binding clusters. As a result, Camostat has more off-target hotspots than Nafamostat. Obviously, the off-target hotspots could decrease the binding efficiency of Camostat to the substrate-binding region. Thus, if the numbers of the drug molecules are equal, the proportion of Camostat bound to the substrate-binding region is lower than that of Nafamostat, because of the stronger competition of the off-target hotspots for the binding. In other words, to target the same number of the TMPRSS2 molecules, Nafamostat requires lower concentration than Camostat. This is fully consistent with experimental observations on SARS-CoV-2 membrane fusion showing inhibition by Nafamostat in the range of 1 ~ 10 nM while Camostat requires 10 ~ 100 nM to achieve a similar extent of inhibition [25], [26]. Note that, as mentioned, the used TMPRSS2-ECD model lacks 40 N-terminal amino acids. However, compared to the whole sequence of TMPRSS2-ECD, the missing N-terminal segment is relatively short, so it is reasonable to consider that the number of the off-target binding hotspots on this segment is small and negligible.
Moreover, our analysis of the binding free-energy revealed that the lowest binding free energy of Camostat was found for an off-target hotspot with −9.93 kcal·mol−1, which is lower than all those of the identified on-target hotspots (Fig. 5A). This implies that Camostat could bind to this off-target hotspot with a greater affinity than to the on-target hotspots. This may further reduce the probability of Camostat to bind to the substrate-binding region. In contrast, the binding energy of Nafamostat to the substrate-binding region is lower than those for any off-target site (−9.16 kcal·mol−1 in Fig. 5B), indicating that the catalytic center provides the higher-affinity for Nafamostat. To further characterize these, we employed the method in our previous study [40] to construct the binding energy landscapes of two drugs using all the simulation trajectories, as shown in Supplementary Fig. S9. Consistent with the results in Fig. 5, the catalytic center of TMPRSS2 is the high-affinity/high-probability binding region for both drugs; also, Camostat has more off-target binding hotspots than Nafamostat. So, Camostat and Nafamostat are effective inhibitors of TMPRSS2; and Nafamostat is more specific for the on-target binding, implying that the antiviral effect of Nafamostat against SARS-CoV-2 is likely better than that of Camostat [25], [26].
4. Discussion and conclusion
During the current challenging period of accelerated development of COVID-19 therapeutic vaccines and drugs around the world, repurposing of existing drugs could accelerate the discovery of effective treatments for COVID-19, e.g., by screening drugs that target TMPRSS2 catalytic site with virtual screening method [52]. Here we have performed the spontaneous binding simulations to investigate the molecular mechanisms of pharmacological action of two old drugs (Camostat and Nafamostat), which were repurposed against SARS-CoV-2 and are undergoing clinical trials [8]. To avoid simulation artifacts, our study did not assign any drug binding sites before the simulations, and therefore the drug associations with the binding sites were completely driven by the native intermolecular forces between the drugs and the TMPRSS2 residues, without any biased forces. Our simulations successfully captured their dynamic association processes with the TMPRSS2 catalytic center (Fig. 3). This demonstrates that both drugs diffusing from randomly initial positions in the solvent, approach the TMPRSS2 surface by attractions of the Asp/Glu residues at the substrate binding site, and finally bind to the catalytic center by the van der Waals forces and hydrogen bonds (Fig. 4). In addition, by mapping the high-affinity hotspots of both drugs on TMPRSS2 (Fig. 5), we found that Nafamostat is more specific for the binding to the TMPRSS2 catalytic center. This provides a molecular explanation for the observation that Nafamostat possesses better inhibition effects than Camostat [25], [26].
Our study implies that Camostat and Nafamostat are effective inhibitors to block the TMPRSS2-mediated cleavage of the S protein required for the SARS-CoV-2 infection in humans. Moreover, we revealed that the guanidinium group of the drugs is critical for driving the drug associations with the TMPRSS2 catalytic center. When developing new drugs targeting TMPRSS2, one should pay specific attention to this group. No doubt, there are still open questions to be addressed for deeply understanding the molecular mechanisms of drug action. For example, experimental determination of the full-length TMPRSS2 structure in complex with Camostat or Nafamostat will be very helpful in confirming our simulations. Meanwhile, in all the simulations of 150 ns (Table 1), we did not observe drug binding modes similar to that of Camostat in the protease prostasin [53]. Because so far no experimental evidence for a covalent binding of the two drugs to TMPRSS2 is available, further studies are needed to clarify this issue.
In summary, this study not only provides mechanistic insights into the Camostat and Nafamostat inhibition of the SARS-CoV-2 infection, but also offers useful information for the repurposing of Camostat and Nafamostat for treating COVID-19.
CRediT authorship contribution statement
Haixia Zhu: Investigation, Formal analysis, Writing - original draft, Writing - review & editing. Wenhao Du: Investigation, Formal analysis, Visualization, Writing - original draft, Writing - review & editing. Menghua Song: Formal analysis, Visualization. Qing Liu: Formal analysis, Visualization. Andreas Herrmann: Conceptualization, Writing - original draft, Writing - review & editing, Funding acquisition. Qiang Huang: Supervision, Conceptualization, Writing - original draft, Writing - review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was partially supported by the grants from the National Major Scientific and Technological Special Project for “Significant New Drugs Development” [2018ZX09J18112], the National Natural Science Foundation of China [31671386, 31971377, 91430112], the Shanghai Municipal Science and Technology Major Project [2018SHZDZX01] and ZJLab, and the Corona Virus Pre Exploration Project by Berlin University Alliance GC2 Global Health (to A.H.). Computational resources were provided by the Shanghai Supercomputer Center. We thank Dr. Yi Wang for his discussion in the initial phase of this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.12.035.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhong N.S., Zheng B.J., Li Y.M., Poon LLM, Xie Z.H., Chan K.H., Li P.H., Tan S.Y., Chang Q., Xie J.P., Liu X.Q., Xu J., Li D.X., Yuen K.Y., Peiris JSM, Guan Y. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 4.Zhu N.a., Zhang D., Wang W., Li X., Yang B.o., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coronavirus disease 2019 (COVID-19) situation report – 45. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200305-sitrep-45-covid-19.pdf?sfvrsn=ed2ba78b_2. [accessed on 20 March 2020]
- 6.World Map. https://coronavirus.jhu.edu/map.html. [accessed on 20 December 2020].
- 7.Zhou Peng, Yang Xing-Lou, Wang Xian-Guang, Hu Ben, Zhang Lei, Zhang Wei, Si Hao-Rui, Zhu Yan, Li Bei, Huang Chao-Lin, Chen Hui-Dong, Chen Jing, Luo Yun, Guo Hua, Jiang Ren-Di, Liu Mei-Qin, Chen Ying, Shen Xu-Rui, Wang Xi, Zheng Xiao-Shuang, Zhao Kai, Chen Quan-Jiao, Deng Fei, Liu Lin-Lin, Yan Bing, Zhan Fa-Xian, Wang Yan-Yi, Xiao Geng-Fu, Shi Zheng-Li. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.COVID-19 vaccine & therapeutics tracker. https://biorender.com/covid-vaccine-tracker. [accessed on 20 December 2020].
- 9.Meyer-Almes Franz-Josef. Repurposing approved drugs as potential inhibitors of 3CL-protease of SARS-CoV-2: virtual screening and structure based drug design. Comput Biol Chem. 2020;88:107351. doi: 10.1016/j.compbiolchem.2020.107351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Touret Franck, Gilles Magali, Barral Karine, Nougairède Antoine, van Helden Jacques, Decroly Etienne, de Lamballerie Xavier, Coutard Bruno. In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication. Sci Rep. 2020;10:13093. doi: 10.1038/s41598-020-70143-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shyr Zeenat A., Gorshkov Kirill, Chen Catherine Z., Zheng Wei. Drug discovery strategies for SARS-CoV-2. J Pharmacol Exp Ther. 2020;375:127–138. doi: 10.1124/jpet.120.000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walls Alexandra C., Park Young-Jun, Tortorici M. Alejandra, Wall Abigail, McGuire Andrew T., Veesler David. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Wenhui, Moore Michael J., Vasilieva Natalya, Sui Jianhua, Wong Swee Kee, Berne Michael A., Somasundaran Mohan, Sullivan John L., Luzuriaga Katherine, Greenough Thomas C., Choe Hyeryun, Farzan Michael. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simmons Graham, Zmora Pawel, Gierer Stefanie, Heurich Adeline, Pöhlmann Stefan. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antiviral Res. 2013;100:605–614. doi: 10.1016/j.antiviral.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaimes Javier A., Millet Jean K., Whittaker Gary R. Proteolytic cleavage of the SARS-CoV-2 spike protein and the role of the novel S1/S2 Site. iScience. 2020;23:101212. doi: 10.1016/j.isci.2020.101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tai W., He L., Zhang X., Pu J., Voronin D., Jiang S. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. 2020;17:613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ou Xiuyuan, Zheng Wangliang, Shan Yiwei, Mu Zhixia, Dominguez Samuel R., Holmes Kathryn V., Qian Zhaohui, Perlman S. Identification of the fusion peptide-containing region in betacoronavirus spike glycoproteins. J Virol. 2016;90:5586–5600. doi: 10.1128/JVI.00015-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffmann Markus, Kleine-Weber Hannah, Schroeder Simon, Krüger Nadine, Herrler Tanja, Erichsen Sandra, Schiergens Tobias S., Herrler Georg, Wu Nai-Huei, Nitsche Andreas, Müller Marcel A., Drosten Christian, Pöhlmann Stefan. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuyama Shutoku, Nao Naganori, Shirato Kazuya, Kawase Miyuki, Saito Shinji, Takayama Ikuyo, Nagata Noriyo, Sekizuka Tsuyoshi, Katoh Hiroshi, Kato Fumihiro, Sakata Masafumi, Tahara Maino, Kutsuna Satoshi, Ohmagari Norio, Kuroda Makoto, Suzuki Tadaki, Kageyama Tsutomu, Takeda Makoto. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci USA. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuritzkes Daniel R. Drug resistance in HIV-1. Curr Opin Virol. 2011;1:582–589. doi: 10.1016/j.coviro.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou H., Fang Y., Xu T., Ni W.J., Shen A.Z. Potential therapeutic targets and promising drugs for combating SARS-CoV-2. Br J Pharmacol. 2020;177:3147–3161. doi: 10.1111/bph.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen Li Wen, Mao Hui Juan, Wu Yan Ling, Tanaka Yoshimasa, Zhang Wen. TMPRSS2: a potential target for treatment of influenza virus and coronavirus infections. Biochimie. 2017;142:1–10. doi: 10.1016/j.biochi.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawase M., Shirato K., van der Hoek L., Taguchi F., Matsuyama S. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J Virol. 2012;86:6537–6545. doi: 10.1128/JVI.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamamoto Mizuki, Matsuyama Shutoku, Li Xiao, Takeda Makoto, Kawaguchi Yasushi, Inoue Jun-ichiro, Matsuda Zene. Identification of nafamostat as a potent inhibitor of middle east respiratory syndrome coronavirus S protein-mediated membrane fusion using the split-protein-based cell-cell fusion assay. Antimicrob Agents Chemother. 2016;60:6532–6539. doi: 10.1128/AAC.01043-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamoto M, Kiso M, Sakai-Tagawa Y, Iwatsuki-Horimoto K, Imai M et al. The anticoagulant nafamostat potently inhibits SARS-CoV-2 S protein-mediated fusion in a cell fusion assay system and viral infection in vitro in a cell-type-dependent manner. Viruses (2020) 12: 629. [DOI] [PMC free article] [PubMed]
- 26.Hoffmann M, Schroeder S, Kleine-Weber H, Müller MA, Drosten C et al. Nafamostat mesylate blocks activation of SARS-CoV-2: new treatment option for COVID-19. Antimicrob Agents Chemother (2020) 64: e00754-00720. [DOI] [PMC free article] [PubMed]
- 27.McKee Dwight L., Sternberg Ariane, Stange Ulrike, Laufer Stefan, Naujokat Cord. Candidate drugs against SARS-CoV-2 and COVID-19. Pharmacol Res. 2020;157:104859. doi: 10.1016/j.phrs.2020.104859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guy R. Kiplin, DiPaola Robert S., Romanelli Frank, Dutch Rebecca E. Rapid repurposing of drugs for COVID-19. Science. 2020;368:829–830. doi: 10.1126/science.abb9332. [DOI] [PubMed] [Google Scholar]
- 29.Yamaya Mutsuo, Nishimura Hidekazu, Deng Xue, Kikuchi Akiko, Nagatomi Ryoichi. Protease inhibitors: candidate drugs to inhibit severe acute respiratory syndrome coronavirus 2 replication. Tohoku J Exp Med. 2020;251:27–30. doi: 10.1620/tjem.251.27. [DOI] [PubMed] [Google Scholar]
- 30.Hofmann-Winkler H, Moerer O, Alt-Epping S, Bräuer A, Büttner B et al. Camostat mesylate may reduce severity of coronavirus disease 2019 sepsis: a first observation. Crit Care Explor (2020) 2: e0284. [DOI] [PMC free article] [PubMed]
- 31.Jang Sukbin, Rhee Ji-Young. Three cases of treatment with nafamostat in elderly patients with COVID-19 pneumonia who need oxygen therapy. Int J Infectious Dis. 2020;96:500–502. doi: 10.1016/j.ijid.2020.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacKerell A.D., Jr., Bashford D., Bellott M., Dunbrack R.L., Jr., Evanseck J.D., Field M.J., Fischer S., Gao J., Guo H., Ha S., Joseph-McCarthy D., Kuchnir L., Kuczera K., Lau F.T.K., Mattos C., Michnick S., Ngo T., Nguyen D.T., Prodhom B., Reiher W.E., Roux B., Schlenkrich M., Smith J.C., Stote R., Straub J., Watanabe M., Wiórkiewicz-Kuczera J., Yin D., Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 33.Price Daniel J., Brooks Charles L., III A modified TIP3P water potential for simulation with Ewald summation. J Chem Phys. 2004;121:10096–10103. doi: 10.1063/1.1808117. [DOI] [PubMed] [Google Scholar]
- 34.Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F., DiNola A., Haak J.R. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 35.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys. 1981;52:7182–7190. [Google Scholar]
- 36.Hess Berk, Bekker Henk, Berendsen Herman J.C., Fraaije Johannes G.E.M. LINCS: a linear constraint solver for molecular simulations. J Comput Chem. 1997;18:1463–1472. [Google Scholar]
- 37.Darden Tom, York Darrin, Pedersen Lee. Particle mesh Ewald: an N ⋅log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 38.Wang Junmei, Wolf Romain M., Caldwell James W., Kollman Peter A., Case David A. Development and testing of a general amber force field. J Comput Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 39.Sousa da Silva Alan W, Vranken Wim F. ACPYPE - AnteChamber PYthon Parser interfacE. BMC Res Notes. 2012;5:367. doi: 10.1186/1756-0500-5-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Qing, Herrmann Andreas, Huang Qiang. Surface binding energy landscapes affect phosphodiesterase isoform-specific inhibitor selectivity. Comput Struct Biotechnol J. 2019;17:101–109. doi: 10.1016/j.csbj.2018.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song Menghua, Li Gan, Zhang Qi, Liu Jianping, Huang Qiang. De novo post-SELEX optimization of a G-quadruplex DNA aptamer binding to marine toxin gonyautoxin 1/4. Comput Struct Biotechnol J. 2020;18:3425–3433. doi: 10.1016/j.csbj.2020.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morris Garrett M., Huey Ruth, Lindstrom William, Sanner Michel F., Belew Richard K., Goodsell David S., Olson Arthur J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin B., Ferguson C., White J.T., Wang S., Vessella R. Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Res. 1999;59:4180–4184. [PubMed] [Google Scholar]
- 44.Vivek-Ananth R.P., Rana A., Rajan N., Biswal H.S., Samal A. In silico identification of potential natural product inhibitors of human proteases key to SARS-CoV-2 infection. Molecules. 2020;25:3822. doi: 10.3390/molecules25173822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hussain M., Jabeen N., Amanullah A., Baig A.A., Aziz B. Molecular docking between human TMPRSS2 and SARS-CoV-2 spike protein: conformation and intermolecular interactions. AIMS Microbiol. 2020;6:350–360. doi: 10.3934/microbiol.2020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chikhale R.V., Gupta V.K., Eldesoky G.E., Wabaidur S.M., Patil S.A. Identification of potential anti-TMPRSS2 natural products through homology modelling, virtual screening and molecular dynamics simulation studies. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1798813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marquart M., Walter J., Deisenhofer J., Bode W., Huber R. The geometry of the reactive site and of the peptide groups in trypsin, trypsinogen and its complexes with inhibitors. Acta Crystallogr B Struct Sci. 1983;39:480–490. [Google Scholar]
- 48.Brooke Greg N., Prischi Filippo. Structural and functional modelling of SARS-CoV-2 entry in animal models. Sci Rep. 2020;10:15917. doi: 10.1038/s41598-020-72528-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Antalis T.M., Buzza M.S., Hodge K.M., Hooper J.D., Netzel-Arnett S. The cutting edge: membrane-anchored serine protease activities in the pericellular microenvironment. Biochem J. 2010;428:325–346. doi: 10.1042/BJ20100046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dror Ron O., Pan Albert C., Arlow Daniel H., Borhani David W., Maragakis Paul, Shan Yibing, Xu Huafeng, Shaw David E. Pathway and mechanism of drug binding to G-protein-coupled receptors. Proc Natl Acad Sci USA. 2011;108:13118–13123. doi: 10.1073/pnas.1104614108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huey Ruth, Morris Garrett M., Olson Arthur J., Goodsell David S. A semiempirical free energy force field with charge-based desolvation. J Comput Chem. 2007;28:1145–1152. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- 52.Singh Natesh, Decroly Etienne, Khatib Abdel-Majid, Villoutreix Bruno O. Structure-based drug repositioning over the human TMPRSS2 protease domain: search for chemical probes able to repress SARS-CoV-2 Spike protein cleavages. Eur J Pharm Sci. 2020;153:105495. doi: 10.1016/j.ejps.2020.105495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spraggon Glen, Hornsby Michael, Shipway Aaron, Tully David C., Bursulaya Badry, Danahay Henry, Harris Jennifer L., Lesley Scott A. Active site conformational changes of prostasin provide a new mechanism of protease regulation by divalent cations. Protein Sci. 2009;18:1081–1094. doi: 10.1002/pro.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.