Abstract
Objectives
During gastrointestinal infection, dysbiosis can result in decreased production of microbially derived short‐chain fatty acids (SCFAs). In response to the presence of intestinal pathogens, we examined whether an engineered acetate‐ or butyrate‐releasing diet can rectify the deficiency of SCFAs and lead to the resolution of enteric infection.
Methods
We tested whether a high acetate‐ or butyrate‐producing diet (HAMSA or HAMSB, respectively) condition Citrobacter rodentium infection in mice and assess its impact on host‐microbiota interactions. We analysed the adaptive and innate immune responses, changes in gut microbiome function, epithelial barrier function and the molecular mechanism via metabolite sensing G protein‐coupled receptor 43 (GPR43) and IL‐22 expression.
Results
HAMSA diet rectified the deficiency in acetate production and protected against enteric infection. Increased SCFAs affect the expression of pathogen virulence genes. HAMSA diet promoted compositional and functional changes in the gut microbiota during infection similar to healthy microbiota from non‐infected mice. Bacterial changes were evidenced by the production of proteins involved in acetate utilisation, starch and sugar degradation, amino acid biosynthesis, carbohydrate transport and metabolism. HAMSA diet also induced changes in host proteins critical in glycolysis, wound healing such as GPX1 and epithelial architecture such as EZR1 and PFN1. Dietary acetate assisted in rapid epithelial repair, as shown by increased colonic Muc‐2, Il‐22, and anti‐microbial peptides. We found that acetate increased numbers of colonic IL‐22 producing TCRαβ+CD8αβ+ and TCRγδ+CD8αα+ intraepithelial lymphocytes expressing GPR43.
Conclusion
HAMSA diet may be an effective therapeutic approach for fighting inflammation and enteric infections and offer a safe alternative that may impact on human health.
Keywords: diet, GPR43, gut infection, IELs, IL‐22, short‐chain fatty acids
Specialised diets that produce high concentrations of acetate are a novel approach to protect against intestinal bacterial infections. We show that modified starches yielding short‐chain fatty acids delivered during gut infection mediate functional microbiota changes and intraepithelial lymphocyte expansion and activation via GPR43 central to aid in preserving epithelial barrier integrity and bacterial clearance.

Introduction
Alterations in microbial ecology or the imbalance of commensals favoring overgrowth of pathogens (dysbiosis) impacts gut homeostasis by decreasing protection from the intestinal barrier and promoting infection. Dysbiosis can lead the gut microbiota to lose its anti‐infectious/immunoregulatory role because of deficient production of short‐chain fatty acids (SCFAs), other beneficial metabolites and other regulatory molecules critical for controlling persistent infections and inflammatory responses. 1 Many inflammatory and autoimmune diseases such as type 1 diabetes (T1D) and metabolic diseases like type 2 diabetes (T2D) have been associated with a deficiency in SCFA production. 2 , 3 , 4 , 5
The gut microbiota produces the SCFAs acetate, propionate and butyrate, from fermentation of non‐digestible carbohydrates. SCFAs have anti‐inflammatory properties and help maintain a healthy gut barrier. 6 Resistant starches are an excellent source of fibre that can boost the production of SCFAs by the gut microbiota. 7 We and others have shown that specially designed diet based on high amylose resistant starches (HAMS), which are acetylated and butyrylated (HAMSA and HAMSB, respectively), enhance the production of microbial acetate and butyrate in the colon. 3 , 8 , 9 , 10 , 11 These specialised diet have many beneficial properties in restoring and maintaining gut health. The use of HAMS supplementation by itself in an oral rehydration solution decreases diarrhoea duration in both adults and children hospitalised for acute infectious diarrhoea. 12 A combination of HAMSA and HAMSB diets improved gut homeostasis correlating with 99% protection against autoimmune diabetes in mice. 3 Improved gut homeostasis achieved by HAMSA and HAMSB diets correlated with reduced serum LPS translocation, reduced pro‐inflammatory IL‐21, increased colonic Tregs, increased expression of junction markers and also the level of serum IL‐22, an important cytokine that maintains the commensal composition of the microbiota and gut integrity. 3 , 7 However, it is still unclear as to how dietary SCFAs affect the host‐microbiota interactions during enteric infections, particularly the intraepithelial lymphocyte (IELs) compartment.
To improve the clinical efficacy of dietary SCFAs to minimise gut infections, it is important to understand the fundamental changes in the gut microbial ecology and its interactions with the intestinal immune system. Likewise, efficient crosstalk between the gut microbiota and the mucosal immune system is critical to induce rapid responses from immune cells and to limit colonisation from opportunistic enteric bacteria. Intestinal epithelium possesses a large and diverse pool of innate and adaptive immune cells, 13 with protective functions against pathogenic bacteria and viral infections. 13 , 14 , 15
We hypothesise that HAMSA supplementation protects from enteric infections, by improving host‐microbiota interactions. This study shows that high SCFA‐producing diets dramatically affected C. rodentium infection in mice, and we reveal mechanisms by which dietary SCFAs directly affect the expression of pathogen virulence genes and thus influence infection dynamics including pathogen load, gut microbiota composition and function, as well mucosal architecture and immunity. Dietary SCFA offer an alternative therapy for the prevention or treatment of pathogenic gastrointestinal infections.
Results
Deficiency in SCFA acetate production increases the severity of C. rodentium infection
After being fed a diet supplemented with 15% HAMSA or HAMSB for 3 weeks prior and during induced infection, mice had twofold higher concentrations of faecal acetate (25 mM to 50 mM) and butyrate (15 mM to 30 mM) compared to control HAMS‐fed mice (Figure 1a). Similarly, the concentration of acetate was also significantly higher at 14 days post‐infection (DPI) in mice fed HAMSA and HAMSB compared to HAMS. In contrast, butyrate concentration was only significantly increased in uninfected but not infected HAMSB‐fed mice. Thus, these results suggest that enteric colonic infections affect the capacity of certain commensal bacteria to release butyrate from modified starch substrates. As such, mice fed diets containing zero dietary fibre (ZF) were more susceptible to C. rodentium infection compared to mice receiving a chow (non‐purified) diet containing standard fibre (SF) or a higher dietary fibre (HF) content (Supplementary figure 1a, b). Reduced susceptibility to infection in HF‐fed mice compared to ZF‐fed mice was consistent with a lower number of faecal C. rodentium bacteria 14 DPI (Supplementary figure 1b). Therefore, HAMSA diet, that exclusively boosts acetate, significantly ameliorated the clinical severity of infection at day 14 post‐C. rodentium infection evidenced by reduced weight loss and stool consistency scores at 14 DPI (Figure 1b and c). In contrast, HAMSB‐fed mice showed significantly increased weight loss and similarly severe stool consistency scores at 14 DPI compared to HAMS‐fed mice (Figure 1b and c). Colonisation of C57BL/6J fed on HAMS, HAMSA or HAMSB diet peaked between 8 and 10 DPI, and there was no significant difference in colonisation levels between groups of mice at 14 DPI (Figure 1d). Representative distal colon sections and data from histological colitis scores showed an increased inflammatory infiltrate in infected HAMSA‐fed mice at 14 DPI. Meanwhile, HAMSB‐fed mice presented increased epithelial damage compared to HAMS‐fed mice (Figure 1e‐g). No differences were found in the degree of enterocyte hyperplasia (Supplementary figure 1c).
Figure 1.
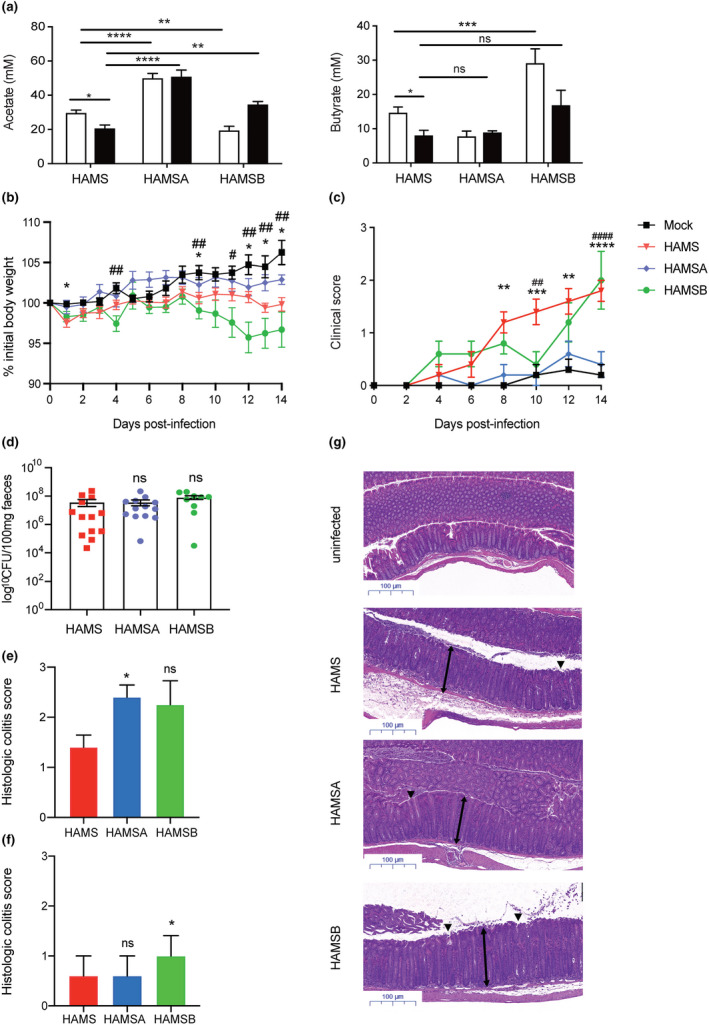
Restored concentrations of SCFA acetate by HAMSA diet is associated with reduced susceptibility to C. rodentium infection. (a) Acetate and butyrate concentrations in the faeces of C57Bl/6J mice fed high amylose starch (HAMS), acetylated HAMS (HAMSA) or butyrylated HAMS (HAMSB) supplement ad libitum from pre‐infection (3 weeks, white bars) and post‐infection (2 weeks, black bars) with C. rodentium. Pre‐infection faecal SCFA concentrations determined at day 0 of C. rodentium infection model, and 14 DPI (n = 5 per group). (b) Body weight changes and (c) stool scores in faeces (0 = normal stool; 1 = soft stool; 2 = diarrhoea; 3 = diarrhoea and anal bleeding) from C57Bl/6J mice fed HAMS, HAMSA or HAMSB diets at 14 DPI (n = 9 or 13). (d) Bacterial load of C. rodentium in faeces from mice at day 14 DPI. Histological colitis scores showing (e) inflammatory infiltrate and (f) epithelial damage of the distal colon as mean ± SEM (n > 4). The scoring system is described in the Methods. Uninfected mice (not represented) scored 0 in all categories (n = 4 or 5 mice per group). (g) Representative H&E slides from distal colon sections at 14 DPI. Single‐headed arrows indicate the degree of immune cell infiltration within the base of the mucosa, which appears to be greater in infected mice than uninfected mice. Double‐headed arrows indicate mucosal thickness, ▼ highlights noticeable tattering and erosion on the epithelial surface. Scale bar = 100 µm, (n = 5). Data are expressed as mean ± S.E.M. P‐values determined by one‐way ANOVA (a, d, e, f) or two‐way ANOVA (b, c) with Bonferroni’s correction. (b) *uninfected vs HAMS; #uninfected vs HAMSB. (c) *HAMSA vs HAMS (day 8, 12); #HAMSB vs HAMS. Graphs and disease incidence are representative of 2 independent experiments. ns = not significant. * or # P < 0.05, ** or ## P < 0.01, *** or ### P < 0.001, **** or #### P < 0.0001.
No changes were observed in energy or food intake between groups of mice fed the experimental diets, when compared to mice fed SF diets (Supplementary figure 1d), similar to what we found previously in the non‐obese diabetic (NOD) mouse model of autoimmune diabetes. 3 Clearance of C. rodentium from the colon was completely resolved at day 22 in HAMSA and HAMSB compared to control HAMS‐fed mice, indicating HAMSA and HAMSB diets are effective mediating colonisation resistance at later stages of infection (Supplementary figure 1e, f).
SCFAs inhibited C. rodentium growth in vitro
Given that HAMSA diet ameliorated the clinical severity at 14 DPI, we examined whether higher concentrations of acetate had a bacteriostatic effect and consequently reduced pathogenicity. We found under anaerobic conditions, physiological doses of acetate and butyrate found in the faeces of HAMSA‐ and HAMSB‐fed mice (50 mM and 35 mM, respectively) had no significant effect on the growth rate of C. rodentium over a 24‐h period, which is likely to reflect what occurs in vivo (Supplementary figure 2a). Assessment of a dose‐dependent response revealed that higher concentrations of butyrate significantly compromised C. rodentium growth under anaerobic conditions (Supplementary figure 2b–e), whereas under aerobic conditions, high concentrations of both acetate and butyrate significantly attenuated C. rodentium growth over 24 h (Supplementary figure 3a). Next, we pre‐cultured C. rodentium with 500 mM acetate or butyrate for 2 h and showed normal growth when returned to SCFA‐free media (Supplementary figure 2f), suggesting SCFA acetate and butyrate exhibit a bacteriostatic rather than a bactericidal effect on C. rodentium growth. Therefore, we examined changes in gene expression levels of C. rodentium virulence genes, Tir and EspB in the colon tissues of infected mice. We found both HAMSA‐ and HAMSB‐fed infected mice exhibited decreased expression of Tir and EspB (Supplementary figure 2g), which are essential for bacterial adhesion to host enterocytes. 16 , 17 , 18 , 19 , 20
HAMSA diet regulates early acute antibacterial response
Although butyrate displayed similar bacteriostatic effects as acetate in vitro and can reduce pathogenicity in vivo, we examined whether worse clinical and histological scores observed in HAMSB‐fed mice were because of defects in regulation of early acute antibacterial responses. Neutrophils, group 3 innate lymphoid cells (ILC3s), CD4+ T cells and IgA‐producing B cells in the lamina propria (LP) are essential for controlling C. rodentium infection. 21 , 22 , 23 We found that CD45+Ly6G+ neutrophils were significantly decreased in frequency and number in the LP of HAMSA‐ and HAMSB‐fed infected mice (Figure 2a), and no differences were found in IL‐22‐producing Ly6G+ neutrophils regardless of diet (data not depicted). ILC3s and CD4+T cells are the major producers of interleukin‐22 (IL‐22) and interleukin‐17 (IL‐17) and are key players in preserving gut homeostasis. 21 We found HAMSA diet significantly increased the frequency and numbers of total CD4+T cells and ILC3s in the colon (data not shown), which was explained by the increased number of IL‐22‐producing RORγt+ILC3s and IL‐22‐producing CD4+ T cells and RORγt+CD4+ T cells in the LP (Figure 2b, d). HAMSA diet slightly reduced the number of IL‐17‐producing CD4+ T cells but did not change the frequency or number of IL‐17‐producing RORγt+ILC3s and in the LP (Figure 2e, f). In contrast, infected HAMSB‐fed mice showed a significant decrease in frequency and number of CD4+FoxP3+ Tregs and no changes in the IL‐22‐ and IL‐17‐producing RORγt+ILC3s or CD4+ T cells in the LP (Figure 2g).
Figure 2.
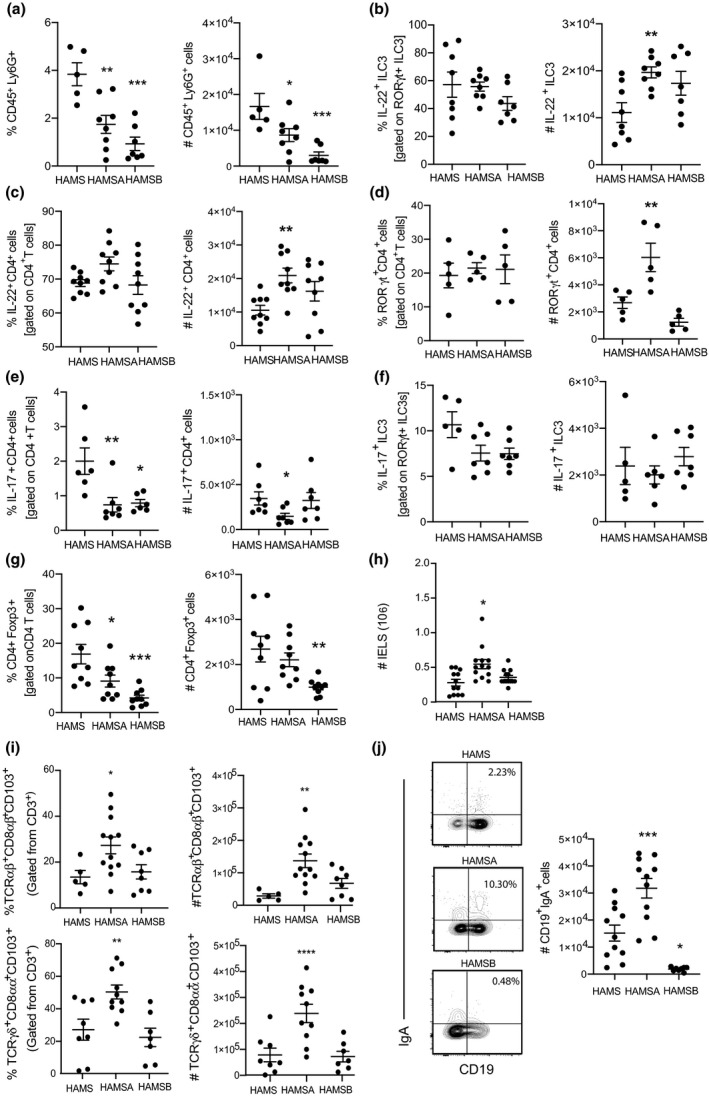
Increased regulatory IELs induced by HAMSA are associated with protection against C. rodentium. Frequency and numbers of (a) CD45+Ly6G+ neutrophils; (b) IL‐22+RORγt+ILC3s; (c) IL‐22+CD4+ T cells; (d) RORγt+CD4+ T cells; (e) IL‐17+CD4+ T cells; (f) IL‐17+RORγt+ILC3s; (g) CD4+FoxP3+ Tregs; (h) number of total IELs; (i) TCRαβ+CD8αβ+CD103+ and TCRγδ+CD8αα+CD103+ IELs and (j) CD19+IgA+ B cells analysed by flow cytometry. Data are expressed as mean ± S.E.M. P‐values were determined by one‐way ANOVA with Bonferroni’s correction. Each symbol represents data from an individual mouse. Graphs are representative of 2 or 3 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 vs HAMS.
We next examined the intraepithelial lymphocytes (IELs) compartment as these cells are the first line of immunological defence against bacteria and include fast responding effector innate immune cells and T cells that can mount strong immunity and affect the ability of C. rodentium to colonise IECs. 24 We found HAMSA but not HAMSB increased the number of total IELs (Figure 2h), which could be explained by the increased frequency and number of TCRαβ+CD8αβ+CD103+ and TCRγδ+CD8αα+CD103+ IELs (Figure 2i). Immunoglobulin A (IgA) is the most abundant antibody in the gut, which is induced towards bacteria. 25 IgA is a marker of the intense activity of the gut immunity‐microbiota interaction, as it has the capacity to bind bacteria and regulate the composition and function of gut microbiota and provide protection against enteric pathogens. 26 IgA‐producing B cells in the LP were significantly increased in HAMSA‐fed infected mice compared to control HAMS‐fed infected mice (Figure 2j). In contrast, the HAMSB diet decreased the numbers of IgA+CD19+B cells. These data suggest HAMSA‐ but not HAMSB‐modified diet is capable of regulating acute antibacterial immune responses, consistent with previous studies using specific agonists of SCFA. 21
An acetate‐yielding diet remodelled the gut microbial community associated with protection against C. rodentium
Next, we determined whether production of dietary SCFAs was linked to changes in the microbial community structure within the gut. Although feeding with HAMSA and HAMSB diets for three weeks increased the concentrations of acetate and butyrate in uninfected mice, only HAMSA supplement resulted in a marked change of the microbiota composition present in the faeces, as shown by a principal component analysis (PCA) plot (Figure 3a). In contrast, the gut microbiota of HAMSB‐fed mice was similar to control HAMS‐fed mice and did not show a strong separation from the non‐purified (NP) diet. Further characterisation of the faecal microbiota profile demonstrated that HAMSA supplement decreased the prevalence of the Bacteroides genus (P = 0.0002) and increased the population of an unknown genus of Alphaproteobacteria (P = 0.0187) compared to control HAMS supplement and the conventional NP diet (Figure 3b). Increased faecal acetate concentrations negatively correlated with the abundance of bacteria from the genera Lactococcus, Anaeroplasma, Akkermansia, Allobaculum, Oscillospira, Lactobacillus and Clostridium (Figure 3c). This indicates that HAMSA diet promotes the dominance of certain bacteria like Alphaproteobacteria, which includes E. coli species that have beneficial roles preventing the bloom of competitive pathogenic bacteria. 27 , 28 Correlation profiles between OTU and SCFA concentrations from the same samples were very distinctive (Figure 3d), especially between acetate and butyrate where there was little overlap in highly correlated OTUs. Altogether, these data suggest that the amount of acetate induced by HAMSA diet is a critical factor for protection against pathogenic bacteria by controlling the colonic microbiota colonisation.
Figure 3.
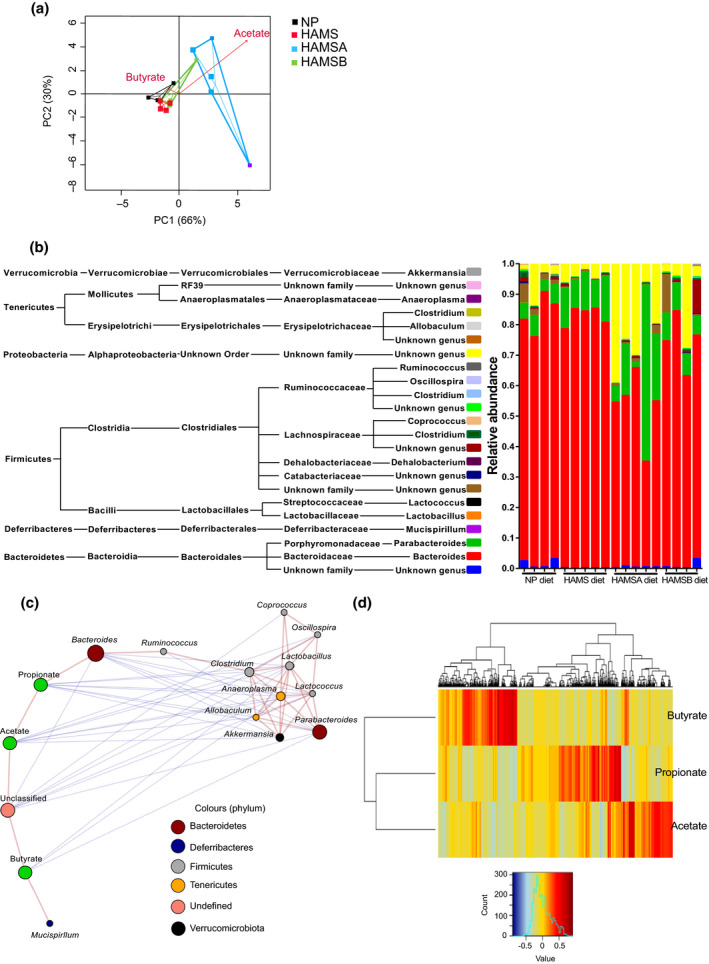
HAMSA and HAMSB diet alter gut microbiota. (a) A PCA plot showing variation between the faecal microbiota of uninfected mice fed for three weeks with NP, HAMS, HAMSA or HAMSB diets (n = 4 per diet group). Vectors indicate the influence of acetic and butyric acid, with length and direction of the arrows indicating influence of the SCFA on the data points. (b) A bar chart showing distribution of different genera detected in faeces from C57Bl/6J wild‐type mice after being fed different supplements. Each genus, picked at 97% sequence identity (QIIME), is represented by a different colour and is proportional to the relative abundance in each sample. The key text provides QIIME taxonomy classification of the different genera in the samples. (c) A Pearson correlation‐based network showing relationships between SCFAs acetate, butyrate and propionate measured in mouse faeces and bacterial genera in C57Bl/6J wild‐type mice fed NP, HAMS, HAMSA or HAMSB supplements. Genera‐depicting nodes are coloured by phylum that genus belongs to. The size of each genus node is proportional to the square root of the relative abundance of that genus; red lines connect positively, and blue lines connect negatively correlated nodes. (d) A heatmap showing Pearson correlations between OTUs (columns) and SCFA (rows).
An acetate‐yielding diet remodels the host and microbial faecal proteomic profile
After observing that HAMSA diet altered the composition of the gut microbiota, we investigated whether this diet impacted the abundance of either microbiota or mouse derived proteins found in faeces, which would indicate a functional change. Soluble faecal proteins were isolated from uninfected HAMS‐ and HAMSA‐fed mice or 14 DPI. Soluble faecal proteins were analysed as these are enriched for host derived proteins in addition to many microbial proteins which can provide insight into host‐microbiota interactions. 29 Spectra were searched against a database containing mouse and mouse gut microbial proteins resulting in identification of 814 mouse proteins and 2309 microbial protein clusters. When the combined mouse and microbial protein abundance was analysed by principal component analysis (PCA) (Figure 4a), the mice perfectly separated by both diet and infection status, indicating a profound remodelling of the microbial and gut proteome. Univariate analysis identified 362 proteins with an adj P < 0.05 that differed between the four treatment groups (Supplementary table 2). These included 84 microbial proteins and 18 mouse proteins significantly altered by diet in the infected groups (Supplementary tables 3 and 4).
Figure 4.
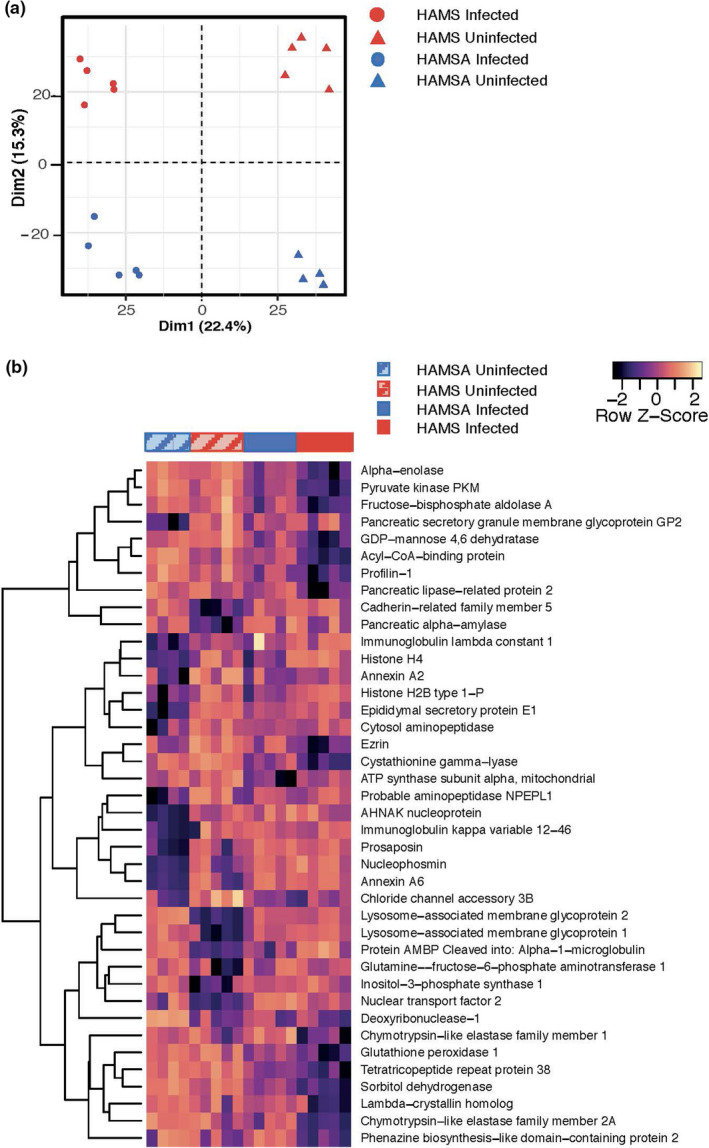
Acetylated diet and infection alter both host‐intestinal and microbiota derived protein abundance in mouse faeces. Soluble faecal proteins were isolated from HAMS‐ and HAMSA‐fed uninfected mice or 2‐weeks post‐infection and analysed by LC‐MS/MS. (a) Principal component analysis of identified faecal proteins separates subject groups by both infection and diet status. (b) All mouse proteins that significantly differed (FDR or adj P < 0.05) in HAMS‐ vs HAMSA‐fed mice (either in uninfected or infected groups) in a one‐way ANOVA model of diet and infection were used for unsupervised hierarchal clustering. A full list of the significant proteins is provided in Supplementary table 2.
The 84 microbial proteins that differed by diet in the infected mice are involved in amino acid biosynthesis (i.e. gdhA, asd, serA, dapdh), carbohydrate transport and metabolism (i.e. msmE, susA, Eno,Gpi), energy production (mdh, sdhA) and protein translation and turnover (trxA, tpx) as well as many proteins with unknown functions (Supplementary figure 4 and Supplementary tables 2 and 3). Enzymes involved in the biosynthesis of lysine, glycine, serine and threonine were increased in abundance by HAMSA feeding. The gut microbiota is a major source for systemic lysine availability 30 and arginine has many well‐described effects on immune regulation including myeloid cell function and nitric oxide synthase activity. 31 Amino acids including lysine, arginine and glycine are also substrates for SCFA production by the colonic bacteria. 32 From the total 192 proteins that were differentially abundant in bacteria, 77 proteins changed because of diet (HAMSA vs HAMS) in uninfected mice. The top changed proteins are involved in amino acid biosynthesis (asd, dapdh), metabolism and energy production (mdh), carbohydrate transport and metabolism (susA, susC), antioxidant defence (Sod2) and other involved in general functions signalling and cellular processes (ffrc and tonB) (Supplementary table 2). From these, 34 proteins were no longer different between diets in mice undergoing infection, which suggests that those provide benefits to mice in processes not related to infection. Noticeably, the top proteins that are no longer different are also involved in carbohydrate metabolism and energy production (gapdh, galK, mdh), protein translation and turnover (trxA), while others are involved in general functions including signalling and cellular processes (TonB hlpa, bmpA and ABC.PE.S) (Supplementary table 2). The most commonly assigned taxa amongst the proteins that differed by diet in either infected or uninfected mice was Bacteroides thetaiotaomicron of which 34 of 39 were upregulated, suggesting this species may increase in change functional state. The second most common taxa altered by HAMSA was a Parabacteroides distasonis. Both of these species are known starch degraders and acetate producers.
We then focused on proteins with potential relevance to infection resolution. Hence, from the bacterial proteome, we identified 75 proteins changed in HAMS because of infection (Supplementary table 2). The top upregulated proteins are involved in fatty acid and lipid metabolism (acpP, buk), amino acid metabolism (glsA, ilvC), carbohydrate transport and metabolism (pgmPTS‐Man‐EIIB, msmE), protein translation and turnover (trxA), and protein metabolism (pqqL). 36 proteins were no longer significantly different in HAMSA (infected vs non‐infected), meaning that HAMSA successfully reverted the changes caused by C. rodentium infection. The top 15 proteins that were no longer different include proteins involved in fatty acid and lipid metabolism acpP, cbh amino acid metabolism (ilvD) and carbohydrate transport and metabolism (Eno, Gapdh, rbsB) amongst others. The remaining 18 were significant in HAMS and HAMSA, which suggest that expression of these genes is not affected or regulated by HAMSA diet, rather by infection. Interestingly, the majority of the changed proteins were associated to bacteria from the phylum Bacteroidetes and Firmicutes. Complete and detailed information on the proteomic profile is shown in Supplementary table 2.
We found 18 mouse proteins that were significantly altered in HAMSA compared to HAMS diet during infection (Figure 4b and Supplementary table 4). The majority of these proteins had maintained high expression levels in HAMSA‐fed infected mice similar to what was found in uninfected mice while they were downregulated in the HAMS‐fed infected group. These included proteins involved in carbohydrate and protein digestion (Cela1a, Cela2a, Pnliprp2, Ambp), glycolysis and carbohydrate metabolism (Aldoa, Eno1, Pkm, Sord). Other proteins were involved in amino acids biosynthesis (Cth), cell adhesion (Ezr/Vil2, Pfn1), regulation of microvillus and wound healing (Gpx1). This suggests that dietary acetate helps to maintain a host glycolytic state, which has been linked to pro‐inflammatory mechanisms needed for a rapid response to infection or injury. 33
GPR43 plays a crucial role in the integral regulation of anti‐microbial responses
Given that HAMSA diet impacted on the susceptibility to C. rodentium infection, we examined whether the effect of high acetate‐yielding diets such as HAMSA operated through GPR43, a metabolite sensing receptor for acetate/propionate/butyrate. We found that Gpr43−/− mice were more susceptible to C. rodentium infection than C57.Gpr43+/+ littermates, which is similar to published studies. 21 , 34 , 35 As expected, control HAMSA‐fed C57.Gpr43+/+ mice showed reduced clinical severity of infection at 14 DPI based on stool consistency (Figure 5a) and bacteria load (Figure 5b), similar to the results in WT C57Bl/6J mice (see Figure 1b‐f). In contrast, HAMSA‐fed C57.Gpr43−/− mice had similarly high clinical scores to control HAMS‐fed C57.Gpr43+/+ littermates and no differences were found compared to HAMS‐fed C57.Gpr43−/− mice (Figure 5a). Histological analysis of mouse distal colons 14 DPI with C. rodentium showed an increased epithelial damage in infected C57.Gpr43−/− mice compared to C57.Gpr43+/+ mice (Figure 5c, d). There were no pathological changes induced by HAMSA supplementation (Figure 5c, d and Supplementary figure 5a). However, HAMS‐fed infected C57.Gpr43+/+ littermates (***P < 0.0001 non‐infected vs infected mice) had reduced colon length, which was significantly reverted when mice were fed HAMSA diet ($$ P = 0.0038 HAMSA vs HAMS‐fed mice) but not in infected C57.Gpr43−/− mice (Figure 5e). No differences were observed in the course of infection in C57.Gpr109−/− mice and C57.Gpr109+/+ littermates (Supplementary figure 5b, c), implying that butyrate via sensing GPR109A receptor is unlikely to contribute to C. rodentium protection, consistent with the HAMSB results shown in Figure 1.
Figure 5.
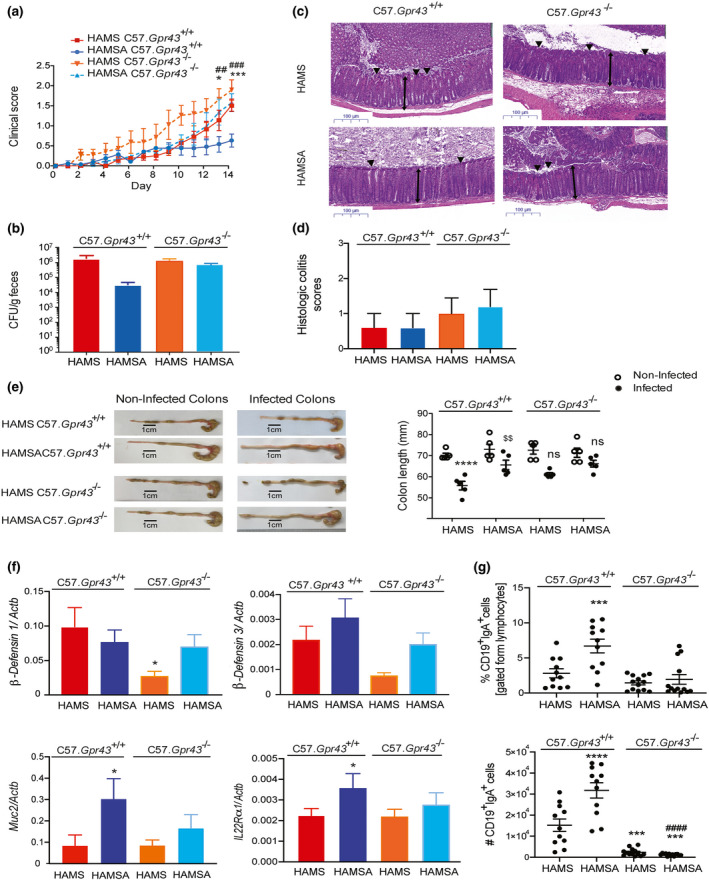
HAMSA‐mediated protection from C. rodentium infection is GPR43 dependent. (a) Stool scores in C. rodentium infected C57.Gpr43+/+ or C57.Gpr43−/− mice fed HAMS or HAMSA supplement ad libitum 3 weeks prior to inoculation and for the length of the experiment. (b) Bacterial load of C. rodentium in the faeces at the peak of disease, 14 DPI (n = 5). (c) Representative H&E slides from distal colon sections at 14 DPI. Single‐headed arrows indicate the degree of immune cell infiltration within the base of the mucosa, which appears to be greater in infected mice than uninfected mice. Double‐headed arrows indicate mucosal thickness, ▼ highlights noticeable tattering and erosion on the epithelial surface. Scale bar = 100 µm (n = 4 or 5). (d) Blinded histopathological scoring of infected C57.Gpr43+/+ or C57.Gpr43−/− mice fed HAMS or HAMSA supplement 14 DPI based on the indicated markers of tissue pathology (n = 5). The scoring system is described in the Methods. Uninfected mice (not represented) scored 0 in all categories. (e) Representative pictures and cumulative data of colon length comparing infected and non‐infected C57.Gpr43+/+ or C57.Gpr43−/− mice fed HAMS or HAMSA supplement 14 DPI. (f) Gene expression of anti‐microbial peptides (β‐defensin 1 and β‐defensin 3) in colon tissues, and Muc‐2 and Il‐22Rα1 in isolated IECs of infected mice 14 PDI determined by qPCR (n = 6 or 8). (g) Frequency and number of CD19+IgA+ B cells. Data are expressed as mean ± S.E.M. P‐values determined by one‐way ANOVA (b, d, f, g) or two‐way ANOVA (a, e) with Bonferroni’s correction. (a) *HAMSA‐fed C57.Gpr43 +/+ vs HAMS‐fed C57.Gpr43 +/+; #HAMSA‐fed C57.Gpr43 +/+ vs HAMSA‐fed C57.Gpr43−/−. (e) *HAMS‐fed C57.Gpr43+/+ non‐infected vs infected; $$infected HAMS‐fed C57.Gpr43 +/+ vs infected HAMSA‐fed C57.Gpr43 +/+. (f, g) *HAMS‐ vs HAMSA‐fed C57.Gpr43 +/+; $HAMS‐fed C57.Gpr43 +/+ vs HAMS‐fed C57.Gpr43−/−; #HAMSA‐fed C57.Gpr43 +/+ vs HAMSA‐fed C57.Gpr43−/−. Each symbol in (e, g) represents data from an individual mouse. Graphs are representative of 2 or 3 independent experiments. ns = not significant. * or $ or # P < 0.05, $$ or ## P < 0.01, *** or $$$ P < 0.001, **** or #### P < 0.0001.
The production of anti‐microbial peptides and mucus by enterocytes impair the ability of pathogens to colonise the gut and maintain a healthy epithelial barrier. 36 Infected HAMS‐fed Gpr43−/− mice presented impaired expression of β‐defensin 1 (*P = 0.0432 HAMS‐fed Gpr43−/− vs HAMS‐fed C57.Gpr43+/+), and a similar but to a lesser extent, the same was observed for β‐defensin 3 (Figure 5f). In contrast, no significant changes were observed in RegIIIγ gene expression and tight junction markers (data not depicted). HAMSA diet significantly increased the expression of both Muc‐2 (*P = 0.0295, HAMS vs HAMSA) and IL‐22rα1 in isolated IECs (*P = 0.0224, HAMS vs HAMSA (Figure 5f). During infection, the interaction between the gut bacteria and IECs induce a local cascade of mucosal immune responses including promoting effector T‐cell function and antibody production by plasma B cells. 37 Infected Gpr43−/− mice presented a significantly reduced number of IgA‐secreting CD19+ B cells (***P = 0.0007, HAMS‐fed Gpr43−/− vs HAMS‐fed C57.Gpr43+/+; ****P < 0.0001 HAMSA‐fed Gpr43−/− vs HAMS‐fed C57.Gpr43+/+) (Figure 5g), which is in line with previous studies. 38
Acetate enhances Gpr43 expression on colonic IELs
We next investigated whether the HAMSA diet might be stimulating IEL immune responses in a GPR43 dependent manner. IL‐22 and IL‐17 are reported to control C. rodentium infection by stimulating epithelial cells to recruit immune cells to the site of the infection. 39 , 40 HAMSA diet significantly elevated the expression of colonic Il‐22, Il‐17 and Il‐18 at 14‐days following infection (***P = 0.0005, ****P < 0.0001 and *P = 0.0168 for HAMSA‐fed mice vs HAMS‐fed mice, respectively) (Figure 6a). Increased IL‐22 and IL‐17 production correlated with the protective effects of HAMSA diet observed in WT C57Bl/6J mice and C57.Gpr43+/+ littermates, but not in the C57.Gpr43−/− mice as shown in Figures 1b and 5a. These cytokines work at different phases of the infection by maintaining gut mucosal integrity, mediating host‐microbial growth balance and regulating immune responses. 41 , 42 , 43 Neutrophils in the LP were reduced by HAMSA feeding and were also found to be reduced in GPR43 deficient mice, indicating a low recruitment (Supplementary Figure 6a). These results were supported by the increased expression of chemokines CcL3 (MIP‐1 alpha) and CcL4 (MIP‐1 beta) in control HAMS‐fed C57.Gpr43+/+ but not C57.Gpr43−/− infected mice compared to non‐infected mice (****P < 0.0001 HAMS‐fed mice infected vs non‐infected) (Supplementary Figure 6b). Only HAMSA‐fed C57.Gpr43+/+ but not C57.Gpr43−/− infected mice significantly decreased expression of these chemokines (**P = 0.0045 and **P = 0.0030 HAMSA‐fed vs HAMS‐fed mice, respectively), which impact on the migration and function of many different innate cells and T cells. 44 , 45 Our findings suggest that recruitment of neutrophils is dependent on SCFAs via activation of GPR43 receptor.
Figure 6.
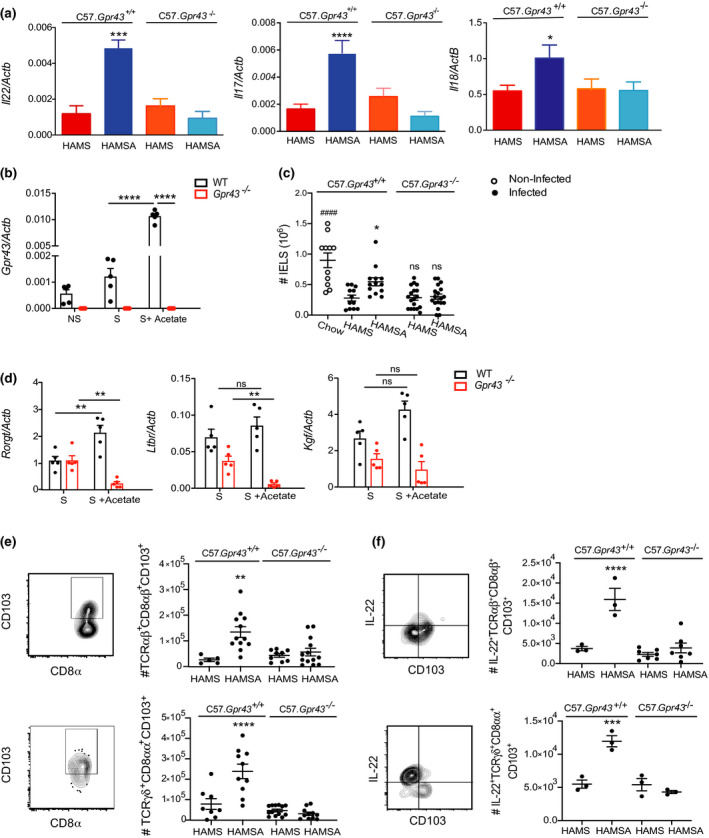
HAMSA diet enhances IL‐22, IL‐17 and number of IELs via GPR43. (a) Expression of Il‐22, Il‐17 and Il‐18 in the colon tissue of infected mice 14 DPI determined by qPCR.(b)Expression of Gpr43 in isolated colonic IELs from C57Bl/6J mice stimulated with PMA and with or without sodium acetate by qPCR. (c)Number of total colonic IELS from infected and non‐infected C57.Gpr43 +/+ or C57.Gpr43−/− mice fed HAMS and HAMSA diets analysed by flow cytometry. (d) Expression of Rorγt, Ltbr and Kgf in isolated colonic IELS from C57Bl/6J mice stimulated with PMA and with or without sodium acetate by qPCR (n = 5). (e) Number and FACS plots of TCRαβ+CD8αβ+CD103+ and TCRγδ+CD8αα+CD103+ IELs from C57.Gpr43 +/+ or C57.Gpr43−/− mice fed HAMS or HAMSA diet. (f) Number of IL‐22+TCRαβ+CD8αβ+CD103+ and IL‐22+TCRγδ+CD8αα+CD103+ IELs from C57.Gpr43 +/+ or C57.Gpr43−/− mice fed HAMS or HAMSA diet. All cellular proportions determined by flow cytometry at 14 DPI. Data are expressed as mean ± S.E.M. P‐values were determined by one‐way ANOVA (a, c, e) or two‐way ANOVA (b, d) with Bonferroni’s correction. (e, f) *HAMSA‐fed vs HAMS‐fed C57.Gpr43 +/+. Each symbol in b–f represents data from an individual mouse. Graphs are representative of 2 or 3 independent experiments. ns = not significant. *P < 0.05, **P < 0.01, ***P < 0.001 **** or #### P < 0.0001.
We sought to understand whether altered recruitment of potential IL‐17 or IL‐22 producing cells were coming from the draining lymph nodes. Neither HAMSA diets nor the absence of the GPR43 receptor change the frequencies of total CD4+ T cells, CD4+CD25+Foxp3+ Tregs nor CD4+IL17+IFNγ+ Th17 cells in the mesenteric lymph nodes (MLN) or draining lymph nodes from the colon (data not depicted) of infected mice (Supplementary figure 6c–e). Likewise, we found that irrespective of diet, the frequency and number of Tregs in the colon did not change in C57.Gpr43+/+ nor C57.Gpr43−/− infected mice (Supplementary Figure 6f). These data suggested that dietary acetate can moderate the severity of C. rodentium infection without potentiating an acute immune response and by promoting the resolution of inflammation in a faster manner.
To ensure intestinal homeostasis, IL‐22 acts on IECs to stimulate the mucosal immune system to respond to pathogens quickly while maintaining the state of tolerance to commensal microbes. 21 , 46 Although GPR43 is expressed by IECs (Supplementary figure 7a–c) and ILC3s, 21 , 47 we found that isolated colonic IELs from WT C57Bl/6J mice also expressed Gpr43 and was highly upregulated by sodium acetate under PMA stimulation (S + Acetate), compared to IELs stimulated without acetate (S) (~tenfold increase, ****P > 0.0001) (Figure 6b). IEL numbers remained reduced in C57.Gpr43−/− mice compared to control HAMS‐fed matched C57.Gpr43+/+ littermates when compared to non‐infected chow‐fed mice (P = 0.729, C57.Gpr43+/+vs C57.Gpr43−/− mice; #### P < 0.0001, non‐infected vs C57.Gpr43−/− mice) (Figure 6c). Only HAMSA increased the number of IELs similar to non‐infected mice.
Next, we determined whether HAMSA was affecting activation markers or the functional status of IELs, using real‐time PCR. To do this, isolated IELs were examined following stimulation with (S + Acetate) or without (S) sodium acetate. Particularly TCRγδ+ IELs produce a number of factors such as keratinocyte growth factor (KGF) to promote healing and protect intestinal integrity. 48 , 49 and lymphotoxin alpha receptor (LTβR), which previously has been shown to be essential for IL‐22 production by innate cells and protection against C. rodentium infection. 50 , 51 There was no impairment in Ltβr or Kgf expression in the absence of GPR43, and only expression of Rorγt significantly changed with acetate treatment in WT Gpr43+/+ IELs (**P = 0.0019 S vs S + Acetate) (Figure 6d). Likewise, IL‐17/IL‐22‐secreting cells are defined by expression of the transcription factor RORγt. 39 , 50 Our data are consistent with the expansion of RORγt+CD4+ T cells, IL‐22‐producing RORγt+ ILC3s in HAMSA‐fed C57.Gpr43+/+mice and numbers were significantly reduced in C57.Gpr43−/− mice or between diet groups (Supplementary figure 7d, e and example of gating strategy Supplementary figure 7g). No changes were observed in IL‐22‐producing CD4+ T cells and IL‐17‐producing RORγt+ ILC3s in C57.Gpr43−/− mice compared to HAMS‐fed C57.Gpr43+/+mice (data not depicted). However, in the absence of the GPR43 receptor, mice showed an increased frequency and number of IL‐17‐producing CD4+ T cells (Supplementary figure 7f).
Analysis of colonic IELs following infection showed that HAMSA‐fed C57.Gpr43+/+ mice presented a significantly increased frequency and numbers of TCRαβ+CD8αβ+CD103+ (~twofold increase **P < 0.0011, HAMSA vs HAMS‐fed mice) and TCRγδ+CD8αα+CD103+ IELs (****P < 0.0001, HAMSA vs HAMS‐fed mice) (Figure 6e and example of gating strategy Supplementary figure 7g). We established that both TCRαβ+CD8αβ+ and TCRγδ+CD8αα+ IELs were free of contaminating epithelial cells by expression of CD103, which marks lymphocytes but not epithelial cells. CD103 has been associated with gut homing and retention. Moreover, HAMSA increased the frequency and number of IL‐22+TCRαβ+CD8αβ+ and IL‐22+TCRγδ+CD8αα+ IELs in C57.Gpr43+/+ mice (Figure 6f). The majority of IELs express T‐cell receptors (TCR) that have cytotoxic and repairing properties that are required to fight off early microbial invasion and limit excessive tissue damage at the end of infection. 13 , 52 For the first time, our data suggest a potential role for HAMSA‐delivering acetate and its sensing receptor GPR43 for IEL function during intestinal infection. Taken together, these results identify a novel mechanism for SCFA acetate regulation, via GPR43, of IEL function and production of cytokines that are important in preserving epithelial barrier integrity and bacterial clearance.
Discussion
HAMSA diets that target the gut microbiota to produce high concentrations of acetate are a novel approach to protect against intestinal bacterial infections. We have shown that deficiency in microbial acetate and butyrate production occurs following C. rodentium infection. Although both acetate and butyrate have a bacteriostatic effect in vitro and can reduce pathogenicity in vivo, only increased concentrations of acetate delivered by HAMSA‐modified gut microbiota significantly ameliorated the severity of C. rodentium infection. We show HAMSA diet mediates functional microbiota changes during gut infection. Restoration of gut epithelial barrier integrity by HAMSA diet was evidenced by the increased expression of anti‐microbial peptides genes, IgA‐secreting B cells and production of IL‐22 associated with protection against infection. We demonstrate that SCFA acetate, through GPR43 signalling, not only induced the expansion of RORγt+CD4+ T cells, IL‐22+ILC3s, but also was required for the regulation of IELs, with significant increase of IL‐22+TCRαβ+CD8αβ+CD103+ and IL‐22+TCRγδ+CD8αα+CD103+ IELs cells to limit C. rodentium infection. Thus, HAMSA diet activates and expands IL‐22‐producing IELs via GPR43, which is central to aid in preserving epithelial barrier integrity and bacterial clearance.
While many studies have used oral administration of SCFAs in drinking water, that approach does not model physiological sustained release and absorption of SCFAs in the colon, 53 , 54 questioning their physiological role and translational application. As such, SCFA administration in drinking water at higher than physiological levels has been associated with dysregulated T‐cell responses and tissue inflammation in the renal system. 54 It is known that SCFAs acetate and particularly butyrate differentially affect bacterial growth and pathogenesis. 55 In addition to the beneficial effects of SCFAs by limiting bacterial growth and A/E lesions induced by pathogenic E. coli, when acetate or butyrate is given in the form of acylated resistant starch, it not only becomes an effective vehicle for increasing SCFAs in the gut 11 , 56 but also promotes fluid and electrolyte uptake to promote oral rehydration therapy in a rat model of cholera. 57 Furthermore, the composition of these acylated high amylose starches may also promote the adhesion of bacteria to starch in the gastrointestinal tract, as it has been shown that Vibrio cholerae, a gram‐negative pathogen that causes cholera, adhere to granular corn‐starches in vitro. 58 Likewise, acetate delivered to the large bowel of mice fed acetylated high amylose maize starch (HAMSA) improved survival against EHEC O157:H7 infection by promoting acetate‐producing bacteria. 9 The high amylose starches not only work as carriers of SCFAs, but also may work by ‘encapsulating’ the pathogenic bacteria and impeding their attachment 59 to the mucosal barrier, 58 which might explain the reduction in C. rodentium colonisation at 22 DPI in the faeces. Acetate diet can act synergistically by boosting the commensal microbiota growth, stability and their beneficial physiological effects, 60 and slowing down the growth of C. rodentium, when the access of glucose becomes limiting. 61 In line with this, the administration of SCFAs, via dietary form, constitutes an effective and physiological way to increase the concentration of SCFAs to achieve beneficial effects. 3 , 9 , 10
The fact that HAMSB‐delivered butyrate concentrations did not ameliorate the severity of C. rodentium infection, even though they had bacteriostatic effects, suggests that HAMSB diet may not be sufficient to modulate the innate immune response as effectively as HAMSA supplementation. Our results highlight the critical importance of contrasting the effects of high and low concentrations of acetate and butyrate in modulating the immune system and challenge the current notion that high concentrations of SCFAs are needed for a beneficial outcome. Our data also provide insights into new approaches to maximise the efficacy of delivery of SCFAs.
Perturbations in the composition of the gut bacteria communities result in a critical reduction in production of the SCFAs necessary to maintain a healthy gut homeostasis. We found that increased acetate was associated with changes in the activity of Bacteroides genus. Our proteomic data showed altered proteins involved in acetate production and starch degrading associated with Bacteroides thetaiotaomicron and Parabacteroides distasonis, a commensal member of the gut microbiota, supporting bacteria functional changes. As such B. thetaiotaomicron can degrade starches and produce butyrate. 62 Although Bacteroides can have beneficial effects in health, 63 it has been reported that interaction between B. thetaiotaomicron and the enteric pathogens can enhance virulence and infection progression. 64 , 65 Furthermore, HAMSA feeding increased the proportion of an unknown Alphaproteobacteria in the faecal microbiota, similar to what has been observed in pigs fed a resistant starch diet. 66 Some Alphaproteobacteria (e.g. the Escherichia genus of which E.coli is a member) are beneficial to human beings 27 , 28 and the environments in general, others are pathogenic and thus cause diseases. Reports show that Alphaproteobacteria has a particular interaction with the host induced during infection. 67 In sublethal influenza infection within the Proteobacteria phylum, the proportion of Alphaproteobacteria and Gammaproteobacteria (Escherichia genus) classes increased, whereas Betaproteobacteria (Sutterella genus) decreased. 68 However, little is known about the role of Alphaproteobacteria in immunity. In both uninfected and infected mice, substantial changes in the microbial protein profile were induced by HAMSA including proteins involved in acetate utilisation, starch and sugar degradation and amino acid metabolism.
In different natural ecosystems like soil and groundwater, from the oxidation of organic compounds to reduce iron, Alphaproteobacteria produce significant amounts of acetate and to lesser extent propionate but not butyrate. 69 , 70 Alphaproteobacteria uses the glyoxylate cycle in the conversion of Coenzyme A to succinate to grow in the presence of acetate. 71 These are central pathways in metabolism and catabolic routes like glycolysis, fatty acid beta‐oxidation, carbohydrate and amino acid degradation, 71 which were impacted in HAMSA‐fed mice as shown by our microbial proteomic data (Supplementary figure 4 and Supplementary table 2, Supplementary table 3). Other bacteria with an altered proteome in response to HAMSA included acetate‐producing and starch degrading bacteria P. distasonis. Other studies utilising resistant starch feeding have observed an increase in P. distasonis abundance 11 suggesting these taxa may utilise the HAMSA starch and release acetate. Within the microbiota as a whole, bacterial proteins upregulated by HAMSA in infected mice included proteins involved in bacterial metabolism of glycine, serine and threonine. Non‐essential amino acids such as glycine produced by the microbiota are thought to have beneficial effects on the host including protective effects on the intestinal epithelium from damage caused by oxidative stress or DSS induced colitis. 72 Likewise, dietary serine is required for T‐cell expansion and promote T‐cell activation and effector function. 73 Interestingly, we observed that increased dietary serine protein in HAMSA‐fed mice was associated with increased CD4+ T‐cell expansion and activation status as shown by increased expression of RORγt and IL‐22. Acetate utilisation by bacteria drives a metabolic ‘switch’, which drives bacteria into the TCA cycle and gluconeogenesis producing precursor molecules for biosynthesis of amino acids, nucleotides, co‐factors and vitamins, 74 consistent with the functional changes we observed in the proteome. This implies SCFAs, particularly acetate, may target bacterial metabolism critical for colonisation, growth and competition for nutrients. 75 , 76
In addition to directly acting on pathogenic bacteria, our studies demonstrate high concentrations of acetate in faeces represent a key factor in maintaining gut integrity during infection. Acetate and butyrate are also used by epithelial cells (ECs) as an energy source. 77 , 78 We found that acetate particularly has multifactorial properties that are effective at reducing the pathogenesis of C. rodentium infection by inducing IEC‐producing anti‐inflammatory cytokines and increasing the abundance of proteins linked to wound healing, cell adhesion and microvillus regulation, as shown in the proteomic data. Likewise, HAMSA diet was effective at preventing the shortening of the colon following infection, along with increased levels of Muc‐2 expression an important component of the mucous barrier that is reduced in colitis. 79 Mice deficient in MUC‐2 were lethally susceptible to C. rodentium, highlighting its essential role in gut health. 80 The Muc‐2 gene is required for the production of mucus, and anti‐microbial peptides such as β‐defensins expression. 81 Overall, this is consistent with the role of acetate assisting in epithelial repair by promoting mineral absorption, mucin production and expression of anti‐microbial peptides 82 and generally improved mucosal immune functions. 9 , 10 , 83 , 84
Both innate and adaptive arms of the immune response are required to resolve C. rodentium infection. Innate cells and CD4+ T cells can both secrete IL‐22. 17 , 85 , 86 , 87 During early infection with C. rodentium, ILC3s are the major producers of IL‐22. 88 , 89 Meanwhile, other innate cells such as TCRγδ cells secrete IL‐22 at later stages of the infection. 86 , 87 HAMSA did change the frequency and numbers of IL‐22+CD4+ T cells and IL‐22+ILC3s in the LP as shown previously. 21 Moreover, HAMSA diet increased IL‐22+TCRαβ+CD8αβ+CD103+ and IL‐22+TCRγδ+CD8αα+CD103+ T cells in infected mice, which correlated with ameliorated clinical scores. It has been shown that SCFAs increase the production of vitamin A, 90 which stimulates IL‐22 production from TCRγδ+CD8αα+ T cells. 91 , 92 On the other hand, mucosal CD8αβ+TCRαβ+ induced IELs maintain a long‐term effector phase with repeated challenges, as they are functionally more mature and show an enhanced and sustained cytotoxic effector phenotype than memory T cells from the spleen or other tissues. 93 Although it is known that GPR43 is expressed on the gut epithelium and a myriad of immune cell subsets, 7 for the first time we reveal that HAMSA‐delivering acetate via GPR43 stimulates IELs‐producing IL‐22.
In the colon, the IELs are highly specialised lymphoid cells that play an important immunoregulatory function and help to maintain tolerance to commensal bacterial and food antigens alongside anti‐microbial responses to pathogenic bacteria. 94 , 95 The remarkable effects of acetate/GPR43 on IL‐22‐producing IELs have not been studied. Just recently it has been shown that SCFAs impact on ILC3s via GPR43. 21 , 47 Acetate/GPR43 can modulate TCRγδ+ CD8αα+ CD103+ IELs evidenced by enhanced anti‐microbial effectors such as Rorγt, Ltβr and Kgf, which are required to control microbial load and bacterial composition within intestinal epithelial cells. 95 , 96 , 97 , 98 Indeed, IELs have been linked with a protective memory response against mycobacterial infection, 99 and TCRδ‐deficient mice are fatally compromised in their resistance to lung infection by the bacterium Nocardia asteroids. 100 TCRγδ+ and TCRαβ+ IELs have been shown to have dynamic and distinct migratory patterns and regulatory functions within the intestinal epithelium. 101 , 102 , 103 We have shown that high acetate‐yielding diets may influence the retention and function of both TCRγδ+CD8αα+CD103+ and TCRαβ+CD8αβ+CD103+ IELs in the gut. The HAMSA‐modified gut microbiota may mediate immunoregulatory mechanisms dependent on TCRγδ+CD8αα+ activation of cytotoxic properties within TCRαβ+CD8αβ+ IELs by inducing IL‐22, as they typically encounter commensal bacteria rather than invasive pathogenic microorganisms. Following infection, protein clusters that were altered by HAMSA diet included Npc2, which is involved in lysosomal acidification and all‐trans retinoic acid triggered anti‐microbial activity 104 (Supplementary table 2). Taken together, these findings demonstrated that changes in TCRαβ+CD8αβ+ and TCRγδ+CD8αα+ IEL function may be a major mechanism in breaking the homeostasis of IELs in Gpr43−/− mice.
Conclusion
We demonstrate the impact of an engineered acetate‐releasing diet in subverting access of a pathogen (C. rodentium) to the host. Our core data showed that dietary SCFAs modulate the gut microbiota function and composition, mucosal immune system and host proteins to promote protection against gut infection. This initial work lays the groundwork for future studies by asking if this dietary technology is efficient to shape mutualism and if SCFAs as dietary supplementation can be used as novel mucosal therapy for a number of human infections.
Methods
Mice
Pathogen‐free C57Bl/6J mice were obtained from the Monash Animal Research Platform, Melbourne Australia. Gpr43−/− and Gpr109−/− mice 105 were obtained from our own specific pathogen‐free breeding colony at Monash Animal Services. From weaning, 4‐week‐old mice were acclimatised and randomly mixed between cages three times per week to homogenise the gut microbiota, prior to C. rodentium inoculation. Mice were aged 8–10 weeks at the starting point of experiments. Control mice for knockout (KO) mice were age‐ and gender‐matched with conventional C57Bl/6J mice and Gpr43+/ + littermates. Same cohort of mice were used for SCFA and microbiome analysis. See Supplementary table 1 for a complete description and formulation of the rodent diets. All experimental procedures involving mice were carried out according to protocols approved by the relevant Animal Ethics Committee of Monash University, Melbourne, Australia and complied with the NHMRC Australian code of practice for the care and use of animals for scientific purposes, as well as the ARRIVE guidelines, 106 including sample randomisation and blinding.
Administration of diets and infection with C. rodentium
Mice were fed their respective diets ad libitum for 3 weeks prior and during induced infection and continued until the endpoint of the experiment. Mice were orally gavaged with 200 µL of nalidixic acid resistant C. rodentium suspension in PBS (strain ICC169) ~3‐5 × 108 colony forming units (CFU). Control groups were vehicle‐treated non‐infected mice. The number of viable bacteria used was determined by retrospective plating. Over the 14‐ or 21‐day monitoring periods, mice were weighed and monitored daily for stool appearance and consistency as clinical score. Stool scores: 0 = normal stool; 1 = soft stool; 2 = diarrhoea; 3 = diarrhoea and anal bleeding. Upon sacrifice, number of viable bacteria per gram of faeces and colon were determined by serial dilution and plating on LB agar plates supplemented with nalidixic acid.
SCFA analysis
SCFAs in faeces were analysed as previously described. 107 In brief, faecal samples were stored at –80 °C until processing and acetate and butyrate were measured by gas chromatography after liquid‐liquid extraction. Briefly, conventionally SCFA are directly measured without derivatisation after solvent extraction using specialised polar phase GC‐MS Phenomenex Zebron ZBFFAP column (Phenomenex, Torrance, California). 200 μL of supernatant was mixed with internal standards (50 µL of 200 µM heptanoic acid internal standard and 50 µL of 10% sulfosalicylic acid). Samples were transferred into the glass test tubes containing 30 µL of 0.2 M NaOH and mixed vigorously. Samples were centrifuged, and the upper ether layer was removed and dried at 40 °C. A volume of 30 µL of cold phosphoric acid (1 M) was pipetted and mixed with pellet to dissolve it properly and then transferred into the GC vials. The residue was re‐dissolved in 30 μL of 1 M phosphoric acid in a glass insert GC vial. Samples were analysed on an Agilent 7890A gas chromatograph (Agilent Technologies, Santa Clara, California). Peaks were detected with a flame ionisation detector at 210 °C and identified and quantitated against calibration standards over the range 0–400 mM. Intra‐assay CVs were 14.2%, 11.8% and 10.3% for acetate, propionate and butyrate, respectively.
Mouse infection and enumeration of bacterial load in faeces
Citrobacter rodentium strain ICC169 was cultured in Luria‐Bertani (LB) broth supplemented with 50 µg mL–1 Nalidixic acid overnight and then pelleted by centrifugation and resuspended in sterile PBS. Mice (6 to 8 weeks old, male and female) were inoculated by oral gavage with 200 µL of approximately 5 x 108 CFU of C. rodentium. The viable count of the inoculum was determined retrospectively by plating dilutions of the inoculum onto LB agar with 50 µg mL–1 Nalidixic acid. C57.Gpr43−/− mice were fed a diet of either HAMS or HAMSA only. Mice were weighed every day for 14 days and faecal samples collected at day 4, 8, 10 and 14 post‐infection for enumeration of CFU. The viable bacterial count per 100 mg of faeces was determined by plating serial dilutions of faeces onto media containing 50 µg mL–1 Nalidixic acid for selection.
Histopathological analysis of mouse colons
Colons from representative groups of infected and uninfected C57BL/6 or C57.Gpr43−/− mice fed on either HAMS, HAMSA or HAMSB were collected following euthanasia at 14 DPI. The tissues were fixed in 4% (wt/vol) paraformaldehyde (Sigma‐Aldrich, St. Louis, MO, USA), then sectioned for H&E staining (University of Melbourne Histology Department) and assessment of gut pathology (APN Network, University of Melbourne). The scoring system (0 to 3) was used by a blinded veterinary pathologist to quantitate the morphological changes in tissue damage represented as follows: 0 = no damage, 1 = discrete lesion, 2 = mucosal erosion and 3 = extensive mucosal damage/ulceration (extending into muscularis and deeper); inflammatory infiltrate as follows: 0 = occasional infiltration, 1 = increasing leucocyte in lamina propria, 2 = confluence of leucocytes extending into submucosa and 3 = transmural extension of inflammatory infiltrate; and epithelial hyperplasia represented as: 0 = none, 1 = mild, 2 = moderate and 3 = severe. Eight measurements were taken from each section of distal colon from at least three individual mice per group.
Growth curve of C. rodentium supplemented with sodium acetate or sodium butyrate
Citrobacter rodentium was tested for its ability to grow in Luria‐Bertani Broth (LB) supplemented with various concentrations of either sodium acetate (500 mM, 250 mM, 125 mM, 60, mM, 50 mM) or sodium butyrate (500 mM, 250 mM, 125 mM, 60, mM, 35 mM). C. rodentium ICC169 was streaked onto LBA supplemented with 50 µg mL–1 Nalidixic Acid and incubated overnight at 37 °C. The following day, a single colony was inoculated into 10 mL of LB and incubated overnight at 37 °C in a shaking incubator. The following day, a subculture was prepared using a 1:100 dilution, where 50 μL of overnight culture was added to 5 mL of LB and incubated in a shaking incubator at 37 °C for 3 h. The optical density (OD600) was measured to standardise the culture to 0.1 in each media type (LB, LB plus sodium acetate, or LB plus sodium butyrate). Samples were prepared in across two 96‐well trays, one for measuring growth under aerobic conditions and the other for measuring growth under anaerobic conditions. Samples were prepared in triplicate, each well containing 200 μL of bacteria culture, or media alone as a blank. A CLARIOstar plate reader (BMG Labtech, Offenburg, Germany) was used to read the aerobic plate at and OD600 every 30 min for 24 h. A second plate reader, a FLUOstar OMEGA (BMG Labtech) fitted with a nitrogen gas injector device, was used to measure anaerobic growth. For both instruments, programme settings were set to 50 cycles multiplied by 1800, with double‐orbital shaking set to 200 rpm and temperature at 37 °C. Each experiment was performed three times.
Sequencing and bioinformatics
Bacterial genomic DNA from faeces was extracted using QIAamp DNA stool mini kit (QIAGEN). DNA samples were amplified targeting the V1‐V3 region of bacterial 16S rRNA gene using forward primer 5′‐AGAGTTTGATCCTGG‐3′; and a reverse primer, 5′‐TTACCGCGGCTGCT‐3′ and sequenced using Roche 454 GS FLX + sequencer. Bioinformatics analysis was performed with Quantitative Insights into Microbial Ecology (QIIME) software. Chimeric sequences were detected and removed using the Pintail algorithm 108 and de‐noised and error‐corrected with Acacia. 109 OTUs were picked de novo at 97% sequence identity using the UCLUST algorithm in QIIME. Taxonomies were assigned in QIIME using BLAST against the Greengenes database. 110 The Pearson correlation‐based network showing relationships between SCFA acetate, butyrate and propionate concentrations detected in mouse faeces and bacterial genera was visualised in Calypso. 111 The data from the bacteria DNA sequencing are publicly available in MG‐RAST database (accession code mgl640446).
Real‐time quantitative PCR analysis
RNA from the colon was extracted and converted to cDNA using Tetro cDNA synthesis kit (Bioline, Cincinnati, OH, USA) using oligo (dT)18 primers to amplify mRNA. qPCR was performed using AccuPower® 2X Greenstar™ qPCR Master Mix (Bioneer, Daejeon, South Korea) or QuantiNova SYBR Green PCR Kit (Qiagen, Hilden, Germany) on the CFX384 Touch Real‐Time PCR Detection System following manufacturer’s instructions (Bio‐Rad, Hercules, CA, USA). All expression were standardised to the housekeeping gene β‐actin. Gene expression of virulence genes Tir and EspB were normalised to 16S. Primers used are shown in Supplementary table 5a.
Isolation of colonic epithelial cells and immune cells from the colon and MLNs
Draining lymph nodes within the mesenteric lymph nodes (MLNs) were carefully removed and dissociated through a 70‐μm filter into FACS buffer as previously shown. 112 Isolation of immune cells from colon lamina propria and intraepithelial layer was performed as previously described. 113 , 114 Colons were dissected and mesenteric fat was removed. Colons were cut open longitudinally, washed with PBS to remove faeces and debris, cut into 0.5–1 cm pieces and incubated in PBS containing 5 mM EDTA, 1 mM DTT and 5% FBS for 20 min on a shaking platform (37 °C, 200 rpm). After being vortexed vigorously for 10 s, the dissociated cells containing IECs and IELs were collected by filtering through a 40‐μm cell strainer and centrifuged at 1500 rpm at 4 °C for 5 min. Pelleted cells were layered on a 40%/70% Percoll gradient (Sigma‐Aldrich) and centrifuged at 1,000 x g at room temperature for 20 min. IECs were collected from the top layer and pelleted for RNA extraction. IELs were collected from the 40%/70% interphase, washed with PBS and resuspended in FACS buffer. For the isolation of lamina propria immune cells, the remaining colonic tissues were digested in HBSS containing 0.5 mg mL–1Collagenase IV (Gibco, Waltham, MA, USA), 40 µg mL–1 DNase I (Roche, Basel, Switzerland) and 10% FBS for 45 min on a shaking platform (37 °C, 200 rpm). Cells were pelleted by centrifuging for 5 min (4 °C, 1500 rpm) and resuspended in FACS buffer for analysis by flow cytometry.
Flow cytometric analysis
Single‐cell suspensions were stained with a combination of fluorescence conjugated monoclonal antibodies. For surface marker staining, antibodies against CD3, CD4, CD8a, CD8b.2, TCRγ/δ, TCRβ, CD103, CD25, CD90.2, CD19, CD45 and Ly6G were used. Intracellular staining was performed using Foxp3/Transcription Factor Staining Buffer Set (eBioscience, San Diego, CA, USA) using antibodies against FOXP3, IL‐17A, RORγt, IL‐22 and IgA. All antibodies (Supplementary table 5b) were purchased from BD Biosciences (San Jose, CA, USA), eBioscience or BioLegend (San Diego, CA, USA). Samples were acquired using BD LSRII or LSRFortessa flow cytometers (BD Biosciences) and analysed with FlowJo version 9.3.2 (Tree Star, Inc., OR, USA).
In vitro IELs stimulation assay
Freshly isolated IELs (5 x 104) from the colon of 8 to 12‐week‐old C57BL/6J and C57.Gpr43−/− mice were stimulated or not with 40 ng mL–1 PMA (Sigma‐Aldrich) and 4 μg mL–1 ionomycin (Sigma‐Aldrich) for 4 h in complete RPMI 1640 media. Sodium acetate (50 mM) or sodium butyrate (30 mM) was added where indicated. At the end of the stimulation period, cells were pelleted, immediately lysed with RLT buffer (RNeasy Mini kit, Qiagen) and stored at −80 °C until real‐time PCR analysis.
Proteomics and bioinformatics
Faecal samples were collected from the colon at time of cull, immediately frozen on dry ice, and stored at −80 °C. Soluble protein extracts, peptide digests and mass spectrometry were performed as previously described. 29 Peptide spectrum matching followed by protein inference, grouping and quantitation was performed using the MetaPro‐IQ strategy with exceptions as follows. 115 The X! Tandem algorithm was implemented using RTandem and the final database search was performed with Spectrum Mill (Agilent, Santa Clara, CA, USA). Briefly, each raw file was first converted to mgf format using msconvert (Proteowizard) and searched against a custom database containing protein predictions from mouse metagenomics experiments 116 and the reference mouse proteome from Uniprot. Proteins identified in this search were then extracted into sample‐specific databases, and the spectra were re‐searched for the corresponding sample. All proteins identified in this second search were combined into a final non‐redundant database and spectra were searched a final time using Spectrum Mill (Agilent).
Carbamidomethylation of cysteine was included as a fixed modification. Oxidation of methionine and deamidation of asparagine were considered as variable modifications. A maximum of two missed cleavages was allowed. A decoy database, prepared by reversing the search database, was searched to determine thresholds to achieve a false discovery rate of 0.01 Proteins which shared 1 or more peptide were grouped. Proteins with only a single detected peptide were discarded. Proteins detected in fewer than 9 of 19 mice were excluded from univariate and multi‐variate analysis. Microbial proteins were assigned to KEGG orthologs using BlastKOALA. Intensities for microbial and human proteins were each scaled to the sum of the total microbial and murine intensities, respectively, and normalised by constant log ratio transformation with an offset corresponding to the minimum detected intensity across the dataset. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE 117 partner repository with the dataset identifier PXD008149.
Statistical analysis
GraphPad Prism (San Diego, CA, USA) (version 8.3.1) was used to perform statistical analysis. For comparisons between more than two independent groups, one‐way ANOVA and two‐way ANOVA with Bonferroni’s multiple comparison test was performed. Data are shown as mean ± S.E.M. as noted. P‐values less than 0.05 were considered significant. Each data point represents a biological, not technical, replicate. Disease incidence studies were analysed using two‐way ANOVA with Bonferroni’s multiple comparison test. Exclusion criteria are described in each Methods section. For proteomics data, one‐way ANOVA, presented as false discovery rate (FDR < 0.05), was performed to identify proteins with significance threshold associated with infection or diet followed by Tukey post hoc tests. Correction for multiple hypotheses was performed according to the method of Benjamini and Hochberg approach using R package. 118 ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05.
Conflict of interest
The authors declare no conflict of interest.
Author contribution
Yu‐Anne Yap: Formal analysis; Investigation; Validation; Writing‐review & editing. Keiran McLeod: Formal analysis; Investigation; Writing‐original draft. Craig I McKenzie: Formal analysis; Investigation; Methodology; Writing‐original draft. Patrick G Gavin: Formal analysis; Visualization. Mercedes Davalos‐Salas: Formal analysis; Validation; Writing‐review & editing. James Richards: Formal analysis; Investigation. Robert Moore: Formal analysis; Funding acquisition; Investigation. Trevor J Lockett: Writing‐review & editing. Julie Clarke: Funding acquisition; Writing‐review & editing. Vik Eng: Formal analysis; Investigation; Methodology. Jaclyn Pearson: Formal analysis; Funding acquisition; Investigation; Methodology; Writing‐review & editing. Emma Estelle Hamilton‐Williams: Data curation; Formal analysis; Funding acquisition; Supervision; Writing‐original draft. Charles Mackay: Funding acquisition; Writing‐review & editing. Eliana Marino: Conceptualization; Data curation; Formal analysis; Funding acquisition; Investigation; Methodology; Pr
oject administration; Supervision; Validation; Writing‐original draft; Writing‐review & editing.
Supporting information
Acknowledgments
We thank Ben Scherer from CSIRO for making the AIN93‐G based diets and undertaking SCFA analyses, Dragana Stanley, Juan Pacheco, Sara Bordbar, Hoey Yein Goh, Caroline Ang Kim Lian and Medina Pell for technical assistance. We thank the Dorothy Loo and the Translational Research Institute’s Proteomic Core Facility for their assistance. EHW was supported by grants from the Juvenile Diabetes Research Foundation (2‐2013‐34, 2‐SRA‐2015‐306‐Q‐R) and Children’s Hospital Foundation (WIS0202018). Part of this research was carried out at the Translational Research Institute, Woolloongabba, QLD 4102, Australia. The Translational Research Institute is supported by a grant from the Australian Government.
References
- 1. Antharam VC, Li EC, Ishmael A et al Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J Clin Microbiol 2013; 51: 2884–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. de Groot PF, Belzer C, Aydin O et al Distinct fecal and oral microbiota composition in human type 1 diabetes, an observational study. PLoS One 2017; 12: e0188475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marino E, Richards JL, McLeod KH et al Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat Immunol 2017; 18: 552–562. [DOI] [PubMed] [Google Scholar]
- 4. Zhao L, Zhang F, Ding X et al Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 2018; 359: 1151–1156. [DOI] [PubMed] [Google Scholar]
- 5. Huang J, Pearson JA, Peng J et al Gut microbial metabolites alter IgA immunity in type 1 diabetes. JCI Insight 2020;5: e135718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wong JMWRD, de Souza RRD, Kendall CWCP, Emam AM, Jenkins DJAMD. Colonic Health: Fermentation and Short Chain Fatty Acids. J Clin Gastroenterol 2006; 40: 235–243. [DOI] [PubMed] [Google Scholar]
- 7. Richards JL, Yap YA, McLeod KH, Mackay CR, Marino E. Dietary metabolites and the gut microbiota: an alternative approach to control inflammatory and autoimmune diseases. Clin Trans Immunol 2016; 5: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cummings JH, Beatty ER, Kingman SM, Bingham SA, Englyst HN. Digestion and physiological properties of resistant starch in the human large bowel. Br J Nutr 1996; 75: 733–747. [DOI] [PubMed] [Google Scholar]
- 9. Fukuda S, Toh H, Hase K et al Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011; 469: 543–549. [DOI] [PubMed] [Google Scholar]
- 10. Furusawa Y, Obata Y, Fukuda S et al Commensal microbe‐derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013; 504: 446–450. [DOI] [PubMed] [Google Scholar]
- 11. Clarke JM, Topping DL, Christophersen CT et al Butyrate esterified to starch is released in the human gastrointestinal tract. Am J Clin Nutr 2011; 94: 1276–1283. [DOI] [PubMed] [Google Scholar]
- 12. Binder HJ, Brown I, Ramakrishna BS, Young GP. Oral rehydration therapy in the second decade of the twenty‐first century. Curr Gastroenterol Rep 2014; 16: 376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yap YA, Marino E. An insight into the intestinal web of mucosal immunity, microbiota, and diet in inflammation. Front Immunol 2018; 9: 2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lepage AC, Buzoni‐Gatel D, Bout DT, Kasper LH. Gut‐derived intraepithelial lymphocytes induce long term immunity against Toxoplasma gondii. J Immunol 1998; 161: 4902–4908. [PubMed] [Google Scholar]
- 15. Muller S, Buhler‐Jungo M, Mueller C. Intestinal intraepithelial lymphocytes exert potent protective cytotoxic activity during an acute virus infection. J Immunol 2000; 164: 1986–1994. [DOI] [PubMed] [Google Scholar]
- 16. Frankel G, Phillips AD. Attaching effacing Escherichia coli and paradigms of Tir‐triggered actin polymerization: getting off the pedestal. Cell Microbiol 2008; 10: 549–556. [DOI] [PubMed] [Google Scholar]
- 17. Berger CN, Crepin VF, Roumeliotis TI et al The Citrobacter rodentium type III secretion system effector EspO affects mucosal damage repair and antimicrobial responses. PLoS Pathog 2018; 14: e1007406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Deng W, de Hoog CL, Yu HB et al A comprehensive proteomic analysis of the type III secretome of Citrobacter rodentium . J Biol Chem 2010; 285: 6790–6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mundy R, Petrovska L, Smollett K et al Identification of a novel Citrobacter rodentium type III secreted protein, EspI, and roles of this and other secreted proteins in infection. Infect Immun 2004; 72: 2288–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arbeloa A, Blanco M, Moreira FC et al Distribution of espM and espT among enteropathogenic and enterohaemorrhagic Escherichia coli . J Med Microbiol 2009; 58: 988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chun E, Lavoie S, Fonseca‐Pereira D et al Metabolite‐sensing receptor Ffar2 regulates colonic group 3 innate lymphoid cells and gut immunity. Immunity 2019; 51: 871–884.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bry L, Brigl M, Brenner MB. CD4+‐T‐cell effector functions and costimulatory requirements essential for surviving mucosal infection with Citrobacter rodentium . Infect Immun 2006; 74: 673–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bry L, Brenner MB. Critical role of T cell‐dependent serum antibody, but not the gut‐associated lymphoid tissue, for surviving acute mucosal infection with Citrobacter rodentium, an attaching and effacing pathogen. J Immunol 2004; 172: 433–441. [DOI] [PubMed] [Google Scholar]
- 24. Cheroutre H, Lambolez F, Mucida D. The light and dark sides of intestinal intraepithelial lymphocytes. Nat Rev Immunol 2011; 11: 445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grootjans J, Krupka N, Hosomi S et al Epithelial endoplasmic reticulum stress orchestrates a protective IgA response. Science 2019; 363: 993–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bunker JJ, Erickson SA, Flynn TM et al Natural polyreactive IgA antibodies coat the intestinal microbiota. Science 2017; 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hudault S, Guignot J, Servin AL. Escherichia coli strains colonising the gastrointestinal tract protect germfree mice against Salmonella typhimurium infection. Gut 2001; 49: 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reid G, Howard J, Gan BS. Can bacterial interference prevent infection? Trends Microbiol 2001; 9: 424–428. [DOI] [PubMed] [Google Scholar]
- 29. Gavin PG, Mullaney JA, Loo D et al Intestinal metaproteomics reveals host‐microbiota interactions in subjects at risk for Type 1 diabetes. Diabetes Care 2018; 41: 2178–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Metges CC, El‐Khoury AE, Henneman L et al Availability of intestinal microbial lysine for whole body lysine homeostasis in human subjects. Am J Physiol 1999; 277: E597–607. [DOI] [PubMed] [Google Scholar]
- 31. Peranzoni E, Marigo I, Dolcetti L et al Role of arginine metabolism in immunity and immunopathology. Immunobiology 2007; 212: 795–812. [DOI] [PubMed] [Google Scholar]
- 32. Neis EP, Dejong CH, Rensen SS. The role of microbial amino acid metabolism in host metabolism. Nutrients 2015; 7: 2930–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mills EL, Kelly B, Logan A et al Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell 2016; 167: 457–470.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim MH, Kang SG, Park JH, Yanagisawa M, Kim CH. Short‐chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology 145: 396–406.e310. [DOI] [PubMed] [Google Scholar]
- 35. Yang W, Xiao Y, Huang X et al Microbiota metabolite short‐chain fatty acids facilitate mucosal adjuvant activity of cholera toxin through GPR43. J Immunol 2019;203: 282–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schreiber F, Arasteh JM, Lawley TD. Pathogen Resistance Mediated by IL‐22 Signaling at the Epithelial‐Microbiota Interface. J Mol Biol 2015; 427: 3676–3682. [DOI] [PubMed] [Google Scholar]
- 37. Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol 2014; 14: 141. [DOI] [PubMed] [Google Scholar]
- 38. Wu W, Sun M, Chen F et al Microbiota metabolite short‐chain fatty acid acetate promotes intestinal IgA response to microbiota which is mediated by GPR43. Mucosal Immunol 2017; 10: 946–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Valeri M, Raffatellu M. Cytokines IL‐17 and IL‐22 in the host response to infection. Pathog Dis 2016; 74: ftw111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wolk K, Kunz S, Witte E, Friedrich M, Asadullah K, Sabat R. IL‐22 increases the innate immunity of tissues. Immunity 2004; 21: 241–254. [DOI] [PubMed] [Google Scholar]
- 41. Liu Z, Zaki MH, Vogel P et al Role of inflammasomes in host defense against Citrobacter rodentium infection. J Biol Chem 2012; 287: 16955–16964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Song X, Zhu S, Shi P et al IL‐17RE is the functional receptor for IL‐17C and mediates mucosal immunity to infection with intestinal pathogens. Nat Immunol 2011; 12: 1151–1158. [DOI] [PubMed] [Google Scholar]
- 43. Zheng Y, Valdez PA, Danilenko DM et al Interleukin‐22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med 2008; 14: 282. [DOI] [PubMed] [Google Scholar]
- 44. Li L, Shi QG, Lin F et al Cytokine IL‐6 is required in Citrobacter rodentium infection‐induced intestinal Th17 responses and promotes IL‐22 expression in inflammatory bowel disease. Mol Med Rep 2014; 9: 831–836. [DOI] [PubMed] [Google Scholar]
- 45. Patterson SJ, Pesenacker AM, Wang AY et al T regulatory cell chemokine production mediates pathogenic T cell attraction and suppression. J Clin Invest 2016; 126: 1039–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sabat R, Ouyang W, Wolk K. Therapeutic opportunities of the IL‐22‐IL‐22R1 system. Nat Rev Drug Discov 2014; 13: 21–38. [DOI] [PubMed] [Google Scholar]
- 47. Fachi JL, Secca C, Rodrigues PB et al Acetate coordinates neutrophil and ILC3 responses against C. difficile through FFAR2. J Exp Med 2020; 217: jem.20190489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Finch PW, Rubin JS. Keratinocyte growth factor/fibroblast growth factor 7, a homeostatic factor with therapeutic potential for epithelial protection and repair. Adv Cancer Res 2004; 91: 69–136. [DOI] [PubMed] [Google Scholar]
- 49. Chen Y, Chou K, Fuchs E, Havran WL, Boismenu R. Protection of the intestinal mucosa by intraepithelial γδ T cells. Proc Natl Acad Sci USA 2002; 99: 14338–14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tumanov AV, Koroleva EP, Guo X et al Lymphotoxin controls the IL‐22 protection pathway in gut innate lymphoid cells during mucosal pathogen challenge. Cell Host Microbe 2011; 10: 44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Spahn TW, Maaser C, Eckmann L et al The lymphotoxin‐beta receptor is critical for control of murine Citrobacter rodentium‐induced colitis. Gastroenterology 2004; 127: 1463–1473. [DOI] [PubMed] [Google Scholar]
- 52. Hayday A, Tigelaar R. Immunoregulation in the tissues by γδ T cells. Nat Rev Immunol 2003; 3: 233–242. [DOI] [PubMed] [Google Scholar]
- 53. Kim MH, Kang SG, Park JH, Yanagisawa M, Kim CH. Short‐Chain Fatty Acids Activate GPR41 and GPR43 on Intestinal Epithelial Cells to Promote Inflammatory Responses in Mice. Gastroenterology 2013; 145: 396–406. e310. [DOI] [PubMed] [Google Scholar]
- 54. Park J, Goergen CJ, HogenEsch H, Kim CH. Chronically elevated levels of short‐chain fatty acids induce T Cell‐mediated ureteritis and hydronephrosis. J Immunol 2016; 196: 2388–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nakanishi N, Tashiro K, Kuhara S, Hayashi T, Sugimoto N, Tobe T. Regulation of virulence by butyrate sensing in enterohaemorrhagic Escherichia coli . Microbiology 2009; 155: 521–530. [DOI] [PubMed] [Google Scholar]
- 56. Clarke JM, Bird AR, Topping DL, Cobiac L. Excretion of starch and esterified short‐chain fatty acids by ileostomy subjects after the ingestion of acylated starches. Am J Clin Nutr 2007; 86: 1146–1151. [DOI] [PubMed] [Google Scholar]
- 57. Clarke JM, Dharmalingam T, Srinivasan P, Young GP, Lockett T, Ramakrishna BS. Acetylated high amylose maize starch improves the efficacy of oral rehydration solution in a rat model of cholera. Gastroenterology 2011; 140: S‐134. [Google Scholar]
- 58. Gancz H, Niderman‐Meyer O, Broza M, Kashi Y, Shimoni E. Adhesion of Vibrio cholerae to granular starches. Appl Environ Microbiol 2005; 71: 4850–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Collins JW, Keeney KM, Crepin VF et al Citrobacter rodentium: infection, inflammation and the microbiota. Nat Rev Microbiol 2014; 12: 612–623. [DOI] [PubMed] [Google Scholar]
- 60. Fuentes‐Zaragoza E, Sánchez‐Zapata E, Sendra E, et al Resistant starch as prebiotic: a review. Starch 2011; 63: 406–415. [Google Scholar]
- 61. Nataro JP, Kaper JB. Diarrheagenic Escherichia coli . Clin Microbiol Rev 1998; 11: 142–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rodriguez‐Castano GP, Dorris MR, Liu X, Bolling BW, Acosta‐Gonzalez A, Rey FE. Bacteroides thetaiotaomicron Starch Utilization Promotes Quercetin Degradation and Butyrate Production by Eubacterium ramulus. Front Microbiol 2019; 10: 1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wrzosek L, Miquel S, Noordine M‐L et al Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol 2013; 11: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Curtis MM, Hu Z, Klimko C, Narayanan S, Deberardinis R, Sperandio V. The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe 2014; 16: 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhu W, Winter MG, Spiga L et al Xenosiderophore Utilization Promotes Bacteroides thetaiotaomicron Resilience during Colitis. Cell Host Microbe 2020; 27: 376–388.e378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Haenen D, Zhang J, Souza da Silva C et al A diet high in resistant starch modulates microbiota composition, SCFA concentrations, and gene expression in pig intestine. J Nutr 2013; 143: 274–283. [DOI] [PubMed] [Google Scholar]
- 67. Batut J, Andersson SG, O'Callaghan D. The evolution of chronic infection strategies in the alpha‐proteobacteria. Nat Rev Microbiol 2004; 2: 933–945. [DOI] [PubMed] [Google Scholar]
- 68. Sencio V, Barthelemy A, Tavares LP et al Gut dysbiosis during influenza contributes to pulmonary pneumococcal superinfection through altered short‐chain fatty acid production. Cell Rep 2020; 30: 2934–2947.e2936. [DOI] [PubMed] [Google Scholar]
- 69. Gagen EJ, Zaugg J, Tyson GW, Southam G. Goethite Reduction by a Neutrophilic Member of the Alphaproteobacterial Genus Telmatospirillum . Front Microbiol 2019; 10: 2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Khater DZ, El‐Khatib KM, Hassan HM. Microbial diversity structure in acetate single chamber microbial fuel cell for electricity generation. J Genet Eng Biotechnol 2017; 15: 127–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Beeckmans S. Glyoxylate Cycle. Encyclopedia of Microbiology (volume 5), 3rd edn Schaechter M, ed. Oxford: Elsevier; 2009; 159–179. [Google Scholar]
- 72. Sugihara K, Morhardt TL, Kamada N. The role of dietary nutrients in inflammatory bowel disease. Front Immunol 2018; 9: 3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ma EH, Bantug G, Griss T et al Serine is an essential metabolite for effector T cell expansion. Cell Metab 2017; 25: 482. [DOI] [PubMed] [Google Scholar]
- 74. Wolfe AJ. The acetate switch. Microbiol Mol Biol Rev 2005; 69: 12–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Louis P, Scott KP, Duncan SH, Flint HJ. Understanding the effects of diet on bacterial metabolism in the large intestine. J Appl Microbiol 2007; 102: 1197–1208. [DOI] [PubMed] [Google Scholar]
- 76. Sigala JC, Flores S, Flores N et al Acetate metabolism in Escherichia coli strains lacking phosphoenolpyruvate: carbohydrate phosphotransferase system; evidence of carbon recycling strategies and futile cycles. J Mol Microbiol Biotechnol 2009; 16: 224–235. [DOI] [PubMed] [Google Scholar]
- 77. Koh A, De Vadder F, Kovatcheva‐Datchary P, Backhed F. From Dietary Fiber to Host Physiology: Short‐Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016; 165: 1332–1345. [DOI] [PubMed] [Google Scholar]
- 78. Louis P, Flint HJ. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol 2017; 19: 29–41. [DOI] [PubMed] [Google Scholar]
- 79. Okayasu I, Hatakeyama S, Yamada M, Ohkusa T, Inagaki Y, Nakaya R. A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology 1990; 98: 694–702. [DOI] [PubMed] [Google Scholar]
- 80. Bergstrom KS, Kissoon‐Singh V, Gibson DL et al Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog 2010; 6: e1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Cobo ER, Kissoon‐Singh V, Moreau F, Holani R, Chadee K. MUC2 mucin and butyrate contribute to the synthesis of the antimicrobial peptide cathelicidin in response to entamoeba histolytica‐ and dextran sodium sulfate‐induced colitis. Infect Immun 2017; 85: e00905‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ashida H, Ogawa M, Kim M, Mimuro H, Sasakawa C. Bacteria and host interactions in the gut epithelial barrier. Nat Chem Biol 2012; 8: 36–45. [DOI] [PubMed] [Google Scholar]
- 83. Ericsson A, Svensson M, Arya A, Agace WW. CCL25/CCR9 promotes the induction and function of CD103 on intestinal intraepithelial lymphocytes. Eur J Immunol 2004; 34: 2720–2729. [DOI] [PubMed] [Google Scholar]
- 84. Smith PM, Howitt MR, Panikov N et al The microbial metabolites, short‐chain fatty acids. Regulate colonic treg cell homeostasis. Science 2013; 341: 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lee YS, Yang H, Yang JY et al Interleukin‐1 (IL‐1) signaling in intestinal stromal cells controls KC/ CXCL1 secretion, which correlates with recruitment of IL‐22‐ secreting neutrophils at early stages of Citrobacter rodentium infection. Infect Immun 2015; 83: 3257–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Backert I, Koralov SB, Wirtz S et al STAT3 activation in Th17 and Th22 cells controls IL‐22‐mediated epithelial host defense during infectious colitis. J Immunol 2014; 193: 3779–3791. [DOI] [PubMed] [Google Scholar]
- 87. Steinbach S, Vordermeier HM, Jones GJ. CD4+ and γδ T Cells are the main producers of IL‐22 and IL‐17A in Lymphocytes from Mycobacterium bovis‐infected Cattle. Sci Rep 2016; 6: 29990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Guo X, Qiu J, Tu T et al Induction of innate lymphoid cell‐derived interleukin‐22 by the transcription factor STAT3 mediates protection against intestinal infection. Immunity 2014; 40: 25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Giacomin PR, Moy RH, Noti M et al Epithelial‐intrinsic IKKα expression regulates group 3 innate lymphoid cell responses and antibacterial immunity. J Exp Med 2015; 212: 1513–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Tan J, McKenzie C, Vuillermin PJ et al Dietary Fiber and Bacterial SCFA Enhance Oral Tolerance and Protect against Food Allergy through Diverse Cellular Pathways. Cell Rep 2016; 15: 2809–2824. [DOI] [PubMed] [Google Scholar]
- 91. Mielke LA, Jones SA, Raverdeau M et al Retinoic acid expression associates with enhanced IL‐22 production by γδ T cells and innate lymphoid cells and attenuation of intestinal inflammation. J Exp Med 2013; 210: 1117–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Raifer H, Mahiny AJ, Bollig N et al Unlike αβ T cells, γδ T cells, LTi cells and NKT cells do not require IRF4 for the production of IL‐17A and IL‐22. Eur J Immunol 2012; 42: 3189–3201. [DOI] [PubMed] [Google Scholar]
- 93. Masopust D, Vezys V, Wherry EJ, Barber DL, Ahmed R. Cutting edge: gut microenvironment promotes differentiation of a unique memory CD8 T cell population. J Immunol 2006; 176: 2079–2083. [DOI] [PubMed] [Google Scholar]
- 94. Cheroutre H, Lambolez F. Doubting the TCR coreceptor function of CD8αα. Immunity 2008; 28: 149–159. [DOI] [PubMed] [Google Scholar]
- 95. Ismail AS, Severson KM, Vaishnava S et al γδ intraepithelial lymphocytes are essential mediators of host–microbial homeostasis at the intestinal mucosal surface. Proc Natl Acad Sci USA 2011; 108: 8743–8748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ismail AS, Behrendt CL, Hooper LV. Reciprocal interactions between commensal bacteria and γδ intraepithelial lymphocytes during mucosal injury. J Immunol 2009; 182: 3047–3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Wang Y, Koroleva EP, Kruglov AA et al Lymphotoxin beta receptor signaling in intestinal epithelial cells orchestrates innate immune responses against mucosal bacterial infection. Immunity 2010; 32: 403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Yang H, Antony PA, Wildhaber BE, Teitelbaum DH. Intestinal intraepithelial lymphocyte γδ‐T cell‐derived keratinocyte growth factor modulates epithelial growth in the mouse. J Immunol 2004; 172: 4151–4158. [DOI] [PubMed] [Google Scholar]
- 99. Shen Y, Zhou D, Qiu L et al Adaptive Immune Response of Vγ2Vδ2+ T Cells During Mycobacterial Infections. Science 2002; 295: 2255–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. King DP, Hyde DM, Jackson KA et al Cutting edge: protective response to pulmonary injury requires γδ T lymphocytes. J Immunol 1999; 162: 5033–5036. [PubMed] [Google Scholar]
- 101. Edelblum KL, Shen L, Weber CR et al Dynamic migration of γδ intraepithelial lymphocytes requires occludin. Proc Natl Acad Sci USA 2012; 109: 7097–7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Edelblum KL, Singh G, Odenwald MA et al γδ intraepithelial lymphocyte migration limits transepithelial pathogen invasion and systemic disease in mice. Gastroenterology 2015; 148: 1417–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Shires J, Theodoridis E, Hayday AC. Biological insights into TCRγδ+ and TCRαβ+ intraepithelial lymphocytes provided by serial analysis of gene expression (SAGE). Immunity 2001; 15: 419–434. [DOI] [PubMed] [Google Scholar]
- 104. Wheelwright M, Kim EW, Inkeles MS et al All‐trans retinoic acid‐triggered antimicrobial activity against Mycobacterium tuberculosis is dependent on NPC2. J Immunol 2014; 192: 2280–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Maslowski KM, Vieira AT, Ng A et al Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 2009; 461: 1282–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kilkenny C, Browne WJ, Cuthi I, Emerson M, Altman DG. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. Vet Clin Pathol 2012; 41: 27–31. [DOI] [PubMed] [Google Scholar]
- 107. Bajka BH, Topping DL, Cobiac L, Clarke JM. Butyrylated starch is less susceptible to enzymic hydrolysis and increases large‐bowel butyrate more than high‐amylose maize starch in the rat. Br J Nutr 2006; 96: 276–282. [DOI] [PubMed] [Google Scholar]
- 108. Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. At Least 1 in 20 16S rRNA Sequence Records Currently Held in Public Repositories Is Estimated To Contain Substantial Anomalies. Appl Environ Microbiol 2005; 71: 7724–7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Bragg L, Stone G, Imelfort M, Hugenholtz P, Tyson GW. Fast, accurate error‐correction of amplicon pyrosequences using Acacia. Nat Methods 2012; 9: 425–426. [DOI] [PubMed] [Google Scholar]
- 110. DeSantis TZ, Hugenholtz P, Larsen N et al Greengenes, a chimera‐checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006; 72: 5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Zakrzewski M, Proietti C, Ellis JJ et al Calypso: a user‐friendly web‐server for mining and visualizing microbiome‐environment interactions. Bioinformatics 2016; 33: 782–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Houston SA, Cerovic V, Thomson C, Brewer J, Mowat AM, Milling S. The lymph nodes draining the small intestine and colon are anatomically separate and immunologically distinct. Mucosal Immunol 2016; 9: 468–478. [DOI] [PubMed] [Google Scholar]
- 113. Weigmann B, Tubbe I, Seidel D, Nicolaev A, Becker C, Neurath MF. Isolation and subsequent analysis of murine lamina propria mononuclear cells from colonic tissue. Nat Protoc 2007; 2: 2307–2311. [DOI] [PubMed] [Google Scholar]
- 114. Reißig S, Hackenbruch C, Hövelmeyer N. Isolation of T cells from the gut. Methods Mol Biol 2014; 1193: 21. [DOI] [PubMed] [Google Scholar]
- 115. Zhang X, Ning Z, Mayne J et al MetaPro‐IQ: A universal metaproteomic approach to studying human and mouse gut microbiota. Microbiome 2016; 4: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Xiao L, Feng Q, Liang S et al A catalog of the mouse gut metagenome. Nat Biotechnol 2015; 33: 1103–1108. [DOI] [PubMed] [Google Scholar]
- 117. Vizcaíno JA, Csordas A, Del‐Toro N et al 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 2016; 44: D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Rohart F, Gautier B, Singh A, Lê Cao KA. mixOmics: An R package for 'omics feature selection and multiple data integration. PLoS Comput Biol 2017; 13: e1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


