Transfer RNAs (tRNAs) are central adaptors that decode genetic information during translation and have been long considered static cellular components. However, whether dynamic changes in tRNAs and tRNA‐derived fragments actively contribute to gene regulation remains debated. In this issue, Huh et al (2020) highlight tyrosine tRNAGUA fragmentation at the nexus of an evolutionarily conserved adaptive codon‐based stress response that fine‐tunes translation to restrain growth in human cells.
Subject Categories: Cancer, RNA Biology, Protein Biosynthesis & Quality Control
Abstract
Recent work reports selective tRNA fragmentation as a conserved post‐transcriptional response mechanism to adjust protein synthesis and mRNA stability.
Protein synthesis is a fundamental process that relies on tRNAs for decoding the genetic information in mRNAs. Their central role as universal adaptor molecules in translation contributed to the prevailing notion that tRNAs are static cellular components with limited or no regulatory function. This view is changing due to increasing evidence that tRNA abundance and modifications are rewired to direct cellular translatomes during physiological and stress conditions in different types of normal and malignant cells (Gingold et al, 2014; Goodarzi et al, 2016; Chou et al, 2017). A renewed interest in tRNA biology was further sparked by the discovery of evolutionarily conserved stable tRNA‐derived fragments (tRFs) with pleiotropic roles in gene expression control across different cell types and upon stress in organisms from all kingdoms of life (Oberbauer & Schaefer, 2018). Findings that only a small portion of tRNA is cleaved suggested that the functional effects of tRFs on gene expression are mostly uncoupled from the translational impairments caused by the depletion of precursor or mature tRNAs (Thompson et al, 2008). However, an outstanding question is to what extent tRNA fragmentation coordinates the tRNA and tRF cellular pools at the codon level in order to molecularly adapt the stress response.
In this issue of The EMBO journal, Huh et al (2020) set out to address this question in human epithelial cells undergoing oxidative stress, which was known to induce robust tRF accumulation in multiple cell systems (Thompson et al, 2008). The authors employed a highly sensitive probe‐based sequencing approach to uncover reductions within a limited subset of tRNAs following stress. Depletion of these tRNAs overlapped, at least in part, with the generation of isodecoder‐specific fragments, highlighting tyrosine tRNAGUA (tRNATyr GUA) fragmentation as one of the most striking oxidative‐stress‐induced event. In an attempt to delineate the consequences of tRNATyr GUA depletion, authors performed elegant loss‐of‐function studies and revealed specific translation impairments in distinct tyrosine codon‐enriched mRNAs encoding metabolic and growth‐promoting factors. These defects were consistent with the cytostatic effects observed following oxidative stress and tRNATyr GUA downregulation. Findings that ribosome stalling was increased on tyrosine codons upon depletion of tRNATyr GUA strongly support that the availability of this cognate tRNA needs to meet the demand imposed by the mRNA codon usage during stress. This is concordant with previous reports illustrating a direct correspondence between tRNA abundance and codon usage that selectively impacts ribosome elongation rates and gene expression in response to stress and various biological cues (Gingold et al, 2014; Goodarzi et al, 2016).
Emerging evidence further suggests that reprogramming of tRNA modifications may provide an additional layer of translation control that impacts codon optimality in cells exposed to different environmental signals (Chan et al, 2012; Chou et al, 2017). Previous work by the Dedon laboratory demonstrated that increased levels of Trm4 methyltransferase‐induced 5‐methylcytosine (m5C) modification at the wobble position of tRNALeu CAA promoted selective translation of UUG codon‐enriched mRNAs involved in survival upon oxidative stress in yeast (Chan et al, 2012). Additional research indicates that the expression of tRNA modifying enzymes may be regulated and influence tRNA fragmentation across different types of normal and malignant cells and following stress (Blanco et al, 2016; Guzzi et al, 2018). These results raise a key question: do epitranscriptomic modifications impact processing and function of tRNATyr GUA and other stress‐sensitive isoacceptor tRNAs identified by Huh and colleagues (2020)? This may provide further mechanistic explanations for differences in selectivity of tRNA modulation observed between mammalian cells and unicellular organisms such as yeast and bacteria, which display profound rearrangements of their tRNA pools upon exposure to oxidative stress (Torrent et al, 2018).
One of most prominent mechanism of tRF generation relies on angiogenin (ANG), a member of the RNase A superfamily, which is activated under stress to cleave multiple mature cytoplasmic tRNAs into halves (Ivanov et al, 2011). Focusing on the accumulation of tRFTyr GUA, Huh et al (2020) report a new processing pathway that involves DIS3L2, a Wilms tumor‐associated exoribonuclease. Interestingly, tRFTyr GUA induction occurs rapidly following oxidative stress and is concomitant with a drastic reduction of pre‐tRNATyr GUA levels without changes in the mature tRNA pool. That DIS3L2 depletion does not fully restore pre‐tRNA levels upon oxidative stress points to an intermediate precursor tRFTyr GUA form that is processed. Evidence that this DIS3L2‐mediated processing event is evolutionarily conserved in C. elegans suggests that it may have widespread physiological implications for organismal growth. Thus, a future challenge is to determine how DIS3L2‐mediated tRF processing is regulated to generate mature tRFTyr GUA and possibly other tRFs.
There is a growing realization that tRFs may provide additional means to direct genetic information upon exposure to stress, during development and in disease (Oberbauer & Schaefer, 2018). Although still little is known about the function of tRFs, exciting studies suggest that specific fragments perturb mRNA stability and repress translation through critical interactions with RNA binding proteins (RBPs) and translation initiation factors (Ivanov et al, 2011; Goodarzi et al, 2015; Guzzi et al, 2018). Huh et al (2020) expand the tRF‐protein interactome by uncovering selective binding between tRFTyr GUA and two RBPs with multiple roles in RNA regulation, namely hnRNPA1 and La/SSB, in response to oxidative stress. Intriguingly, tRFTyr GUA may directly compete for hnRNPA1 binding with specific growth‐associated mRNA subsets, promoting their destabilization under stress conditions. This effect consolidates the translational repression of growth‐promoting transcripts imposed by the subsequent depletion of mature tRNATyr GUA. As such, these findings highlight divergent control programs that converge on pre‐tRNATyr GUA to adapt the cellular transcriptome and translatome during oxidative stress (Fig 1).
Figure 1. Stress‐induced tyrosine tRNA fragmentation controls cell growth.
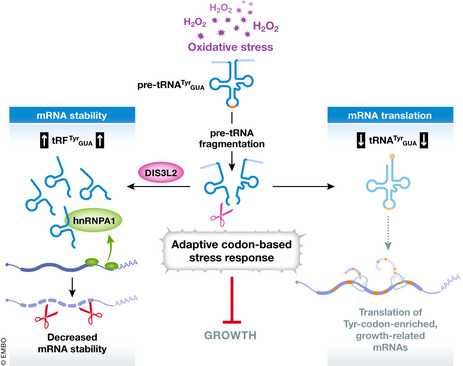
Schematic model depicts the adaptive codon‐based stress response governed by tyrosine tRNATyr GUA fragmentation in human cells. Through an unknown mechanism, oxidative stress induces rapid depletion of pre‐tRNATyr GUA and accumulation of tRNA fragments (tRFTyr GUA) in an exoribonuclease DIS3L2‐dependent manner. Left, tRFTyr GUA competes for hnRNPA1 binding with specific growth‐associated mRNA subsets promoting their destabilization. Right, pre‐tRNATyr GUA fragmentation subsequently depletes mature tRNATyr GUA, thereby hampering the translation of growth‐related mRNAs (e.g., USP3, EPCAM, and SCD) that are enriched in tyrosine codons. The synergistic effects on the transcriptome and translatome induced by the accumulation of tRFTyr GUA and depletion of tRNATyr GUA drive an evolutionarily conserved program that limits cell growth upon oxidative stress.
In conclusion, the recent study by Huh et al (2020) supports evidence that multifaceted gene regulatory mechanisms intrinsic to the genetic code are integral to the cellular stress response. While new exciting horizons are opening within this emerging research field, future work is needed to delineate the signaling cascades that coordinate tRNA fragmentation and reprogram protein synthesis during critical cell state transitions in development and tumorigenesis.
Acknowledgements
Research in the Bellodi laboratory is supported by the Swedish Foundations’ Starting Grant, Stem Therapy, Swedish Research Council (Vetenskapsrådet) and Swedish Cancer Society (Cancerfonden). C.B. is a Ragnar Söderberg Fellow in Medicine and Cancerfonden Young Investigator.
The EMBO Journal (2021) 40: e107097.
See also: D Huh et al (2020)
References
- Blanco S, Bandiera R, Popis M, Hussain S, Lombard P, Aleksic J, Sajini A, Tanna H, Cortes‐Garrido R, Gkatza N et al (2016) Stem cell function and stress response are controlled by protein synthesis. Nature 534: 335–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CTY, Pang YLJ, Deng WJ, Babu IR, Dyavaiah M, Begley TJ, Dedon PC (2012) Reprogramming of tRNA modifications controls the oxidative stress response by codon‐biased translation of proteins. Nat Commun 3: 937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou HJ, Donnard E, Gustafsson HT, Garber M, Rando OJ (2017) Transcriptome‐ wide analysis of roles for tRNA modifications in translational regulation. Mol Cell 68: 978–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingold H, Tehler D, Christoffersen NR, Nielsen MM, Asmar F, Kooistra SM, Christophersen NS, Christensen LL, Borre M, Sorensen KD et al (2014) A dual program for translation regulation in cellular proliferation and differentiation. Cell 158: 1281–1292 [DOI] [PubMed] [Google Scholar]
- Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF (2015) Endogenous tRNA‐derived fragments suppress breast cancer progression via YBX1 displacement. Cell 161: 790–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi H, Nguyen HCB, Zhang S, Dill BD, Molina H, Tavazoie SF (2016) Modulated expression of specific tRNAs drives gene expression and cancer progression. Cell 165: 1416–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzi N, Ciesla M, Ngoc PCT, Lang S, Arora S, Dimitriou M, Pimkova K, Sommarin MNE, Munita R, Lubas M et al (2018) Pseudouridylation of tRNA‐derived fragments steers translational control in stem cells. Cell 173: 1204–1216 [DOI] [PubMed] [Google Scholar]
- Huh D, Passarelli MC, Gao J, Dusmatova SN, Goin C, Fish L, Pinzaru AM, Molina H, McMillan EA, Asgharian H et al (2020) A stress‐induced Tyrosine tRNA depletion response mediates codon‐based translational repression and growth suppression. EMBO J 39: e106696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P (2011) Angiogenin‐induced tRNA fragments inhibit translation initiation. Mol Cell 43: 613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberbauer V, Schaefer MR (2018) tRNA‐derived small RNAs: biogenesis, modification, function and potential impact on human disease development. Genes‐Basel 9: 607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Lu C, Green PJ, Parker R (2008) tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 14: 2095–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrent M, Chalancon G, de Groot NS, Wuster A, Babu MM (2018) Cells alter their tRNA abundance to selectively regulate protein synthesis during stress conditions. Sci Signal 11: eaat6409 [DOI] [PMC free article] [PubMed] [Google Scholar]


