Abstract
Background
Studies of the gut microbiome are becoming increasingly important. Such studies require stool collections that can be processed or frozen in a timely manner so as not to alter the microbial content. Due to the logistical difficulties of home-based stool collection, there has been a challenge in selecting the appropriate sample collection technique and comparing results from different microbiome studies. Thus, we compared stool collection and two alternative clinic-based fecal microbiome collection techniques, including a newer glove-based collection method.
Results
We prospectively enrolled 22 adult men from our prostate cancer screening cohort SABOR (San Antonio Biomarkers of Risk for prostate cancer) in San Antonio, TX, from 8/2018 to 4/2019. A rectal swab and glove tip sample were collected from each participant during a one-time visit to our clinics. A single stool sample was collected at the participant’s home. DNA was isolated from the fecal material and 16 s rRNA sequencing of the V1-V2 and V3-V4 regions was performed. We found the gut microbiome to be similar in richness and evenness, noting no differences in alpha diversity among the collection methods. The stool collection method, which remains the gold-standard method for the gut microbiome, proved to have different community composition compared to swab and glove tip techniques (p< 0.001) as measured by Bray-Curtis and unifrac distances. There were no significant differences in between the swab and glove tip samples with regard to beta diversity (p> 0.05). Despite differences between home-based stool and office-based fecal collection methods, we noted that the distance metrics for the three methods cluster by participant indicating within-person similarities. Additionally, no taxa differed among the methods in a Linear Discriminant Analysis Effect Size (LEfSe) analysis comparing all-against-all sampling methods.
Conclusion
The glove tip method provides similar gut microbiome results as rectal swab and stool microbiome collection techniques. The addition of a new office-based collection technique could help easy and practical implementation of gut microbiome research studies and clinical practice.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-020-02080-3.
Keywords: Gut microbiome, Stool collection, Rectal swabs, Digital rectal examination, 16 s rRNA gene sequencing
Background
The intestinal microbiome may play a role in the pathogenesis of many types of cancer and other diseases, thus providing a modifiable biomarker with potential for treatment interventions, which can complement standard screening programs [1–4]. The current gold standard for fecal microbiome collection is to obtain participant-collected stool with a home-based collection kit [5–7]. Unfortunately, participants may not adhere to the directions or send in the sample. For example, only 60% of participants returned their at-home collected stool specimens in a cohort at high risk for colorectal cancer [8]. Stool sample collection can be challenging due to participants’ comfort level, inconsistent sample collection, and increased collection cost [5, 6, 9–12]. Capturing samples during a clinic visit would save time and potentially increase the number of participants in a study, while reducing bias due to systematic differences between participants who do and do not send in samples or follow the directions for proper collection.
Studies have found that microbiome sample collection via rectal swabs versus participant-collected stool identify similar microbial composition [9, 10, 12]. We have recently published a study comparing exam gloves after a digital rectal exam (DRE) to swab methods for fecal material collection for microbiome studies [13]. We noted similar DNA yield and quality using glove tips compared to rectal swab techniques in different cohorts, yet did not perform both tests on the same participant. Herein, we used the 16S rRNA gene sequencing method to test the collection of fecal specimens with the office-based post-DRE glove tip and rectal swab, and participant-collected stool techniques, using the same participants to perform a paired investigation regarding the similarities and differences in microbial communities between these techniques.
Results
Population
We enrolled 22 individuals from whom we obtained three gut microbiome samples collected by different methods (participant-collected stool, rectal swab, post-DRE glove tip) from 8/2018 to 4/2019. All participants were men previously enrolled in a longitudinal observational prostate cancer screening study, with a median age of 74.5 years. We display the demographics for the cohort in Table 1.
Table 1.
Demographics Table
| Variable | N (%) |
|---|---|
| Race | |
| White | 20 (90.9) |
| Black | 2 (9.1) |
| Ethnicity | |
| Hispanic or Latino | 7 (31.8) |
| Not Hispanic or Latino | 15 (68.2) |
| Age | |
| 58–69 | 5 (22.7) |
| 70–79 | 14 (63.6) |
| 80–89 | 3 (13.6) |
| Median | 74.5 |
| BMI | |
| < 25.0 | 5 (22.7) |
| 25.0–35.0 | 12 (54.5) |
| > 35.0 | 5 (22.7) |
| Median | 30.26 |
| Smoking History | |
| Never smoked | 14 (63.6) |
| Former smoker | 8 (36.4) |
| Total | 22 |
Alpha and Beta diversity
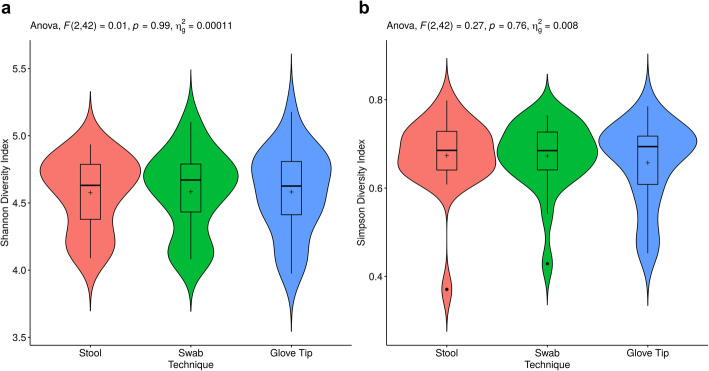
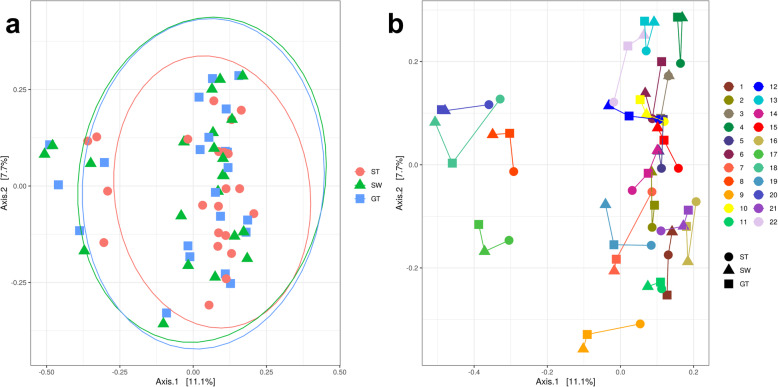
At the operational taxonomic unit (OTU) level, we found no difference in the Shannon (mean [SD] for participant-collected stool: 4.58 [0.27], swab: 4.58 [0.30], glove tip: 4.58 [0.31], p=0.99) or Simpson (mean [SD] for participant-collected stool: 0.67 [0.08], swab: 0.67 [0.08], glove tip: 0.66 [0.09], p=0.76) index of alpha diversity among the collection methods (Fig. 1) [14]. Regarding the beta diversity, there were differences among the collection methods in Bray-Curtis distances (p< 0.001; Fig. 2). In particular, in pairwise comparisons with p-values adjusted using the Benjamini-Hochberg approach, stool samples differed from swab (p< 0.001) and glove tip (p< 0.001); swab and glove tip samples did not differ significantly from one another (p=0.59). We also identified this pattern in tests of unifrac distances, both unweighted (global p< 0.001, swab vs. stool p< 0.001, glove tip vs. stool p< 0.001, glove tip vs. swab p=0.69) and weighted (global p< 0.001, swab vs. stool p< 0.001, glove tip vs. stool p< 0.001, glove tip vs. swab p=0.26). Secondary analyses examining the V1-V2 and V3-V4 regions separately had similar results. For both the V1-V2 region and the V3-V4 region, there were significant differences among the methods globally (p< 0.001 in each region), again driven by differences of stool compared with swab (p< 0.001 in each region) and glove tip (p< 0.001 in each region) methods, with no differences between the swab and glove tip methods (p=0.36 for V1-V2 region, p=0.20 for V3-V4 region). We present a PCoA plot with Bray-Curtis distances by technique (Fig. 2a) and by sampling technique and participant (Fig. 2b) for the mixed V1-V2 and V3-V4 region data, where there is proximity among samples from the same person, despite statistically significant differences among the methods.
Fig. 1.
Simpson and Shannon alpha diversity index assessment. Box denotes the first and third quartiles, whiskers denote the most extreme observations within 1.5 times the inter-quartile range distance from the box; outliers are marked as points. The horizontal bar within the box represents the median, and the “+” denotes the mean. The red, green, and blue areas represent kernel probability densities for the data. Repeated measures ANOVA found no significant differences in Shannon (p=0.99) or Simpson (p=0.76) diversity indices among the sample collection methods
Fig. 2.
Principal Coordinates Analysis (PCoA) using Bray-Curtis distances. (a) Principal coordinates analysis (PCoA) plot based on Bray-Curtis distances by sampling method with data ellipses based on multivariate t distributions (b) Principal coordinates analysis (PCoA) plot based on Bray-Curtis distances by person and method
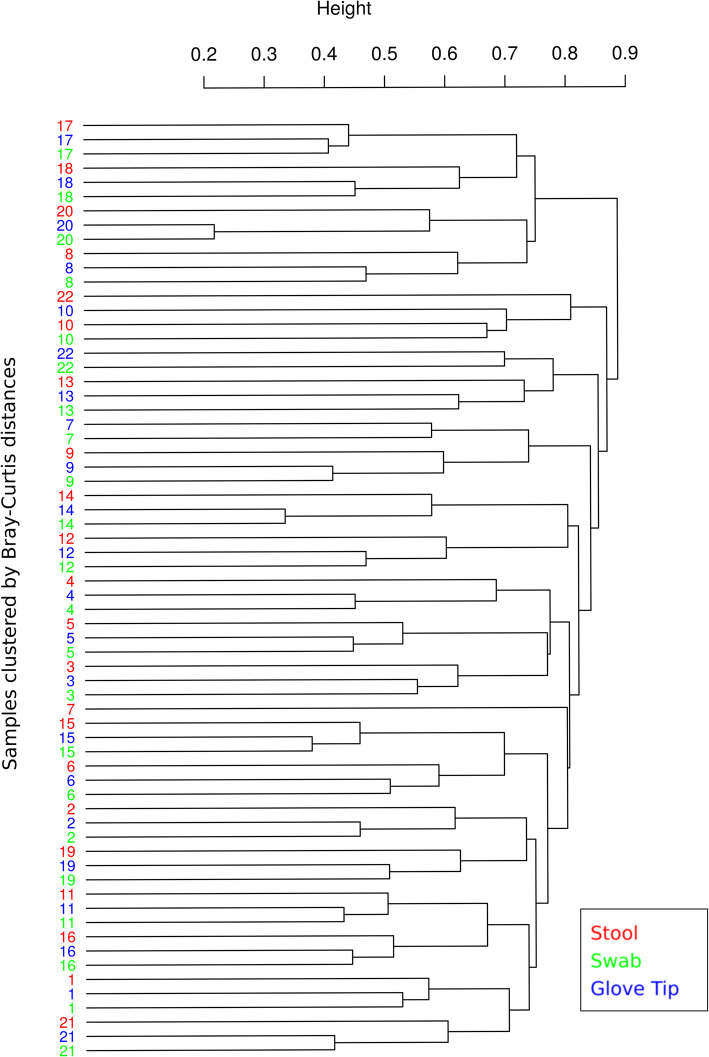
Most samples clustered together based on the person sampled from a hierarchical clustering procedure based on Bray-Curtis distances, as shown in the dendrogram (Fig. 3). If two participants (9% of the overall sample) were excluded (IDs 7, 22), the remaining 20 would each have their three measurements cluster together within a clade. Overall, 21 out of 22 people (95%) showed closer grouping between swab and glove tip samples relative to stool collection; the remaining person had stool and swab measurements more closely related.
Fig. 3.
Dendrogram of samples clustered based on Bray-Curtis distances. Bray-Curtis distances were calculated at the OTU level and clustered using the unweighted pair group method with arithmetic mean (UPGMA) algorithm. The numbers represent the individual patients and the sample is represented by color (Red = Stool, Green = Swab, and Blue = Glove tip)
Bacterial abundance analysis
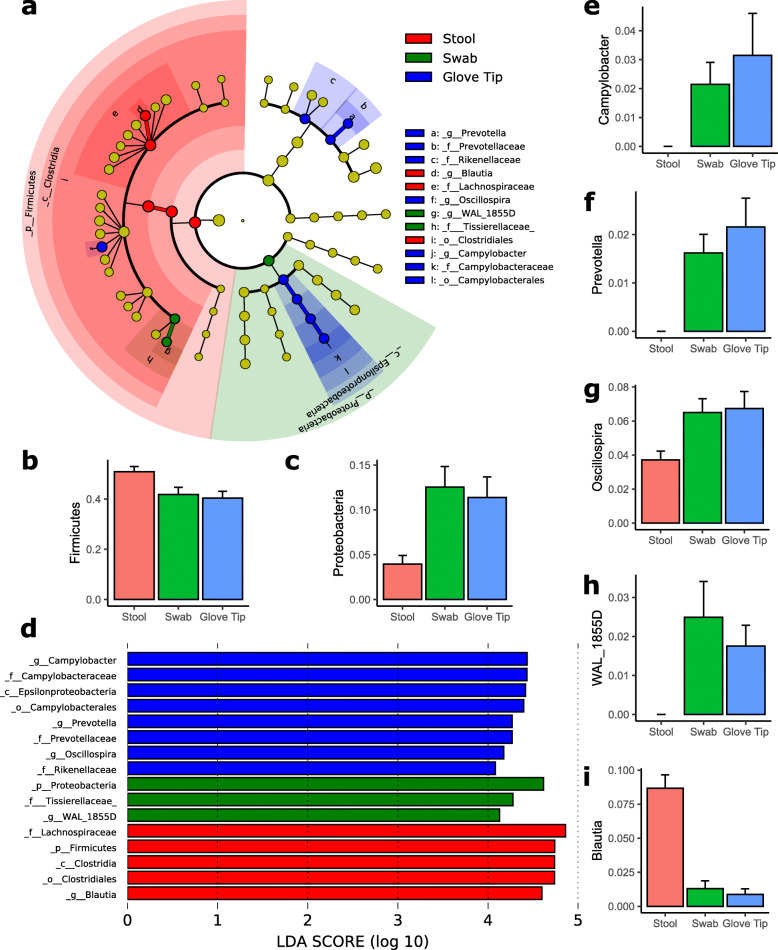
Analysis of differentially abundant taxa using Linear Discriminant Effect Size (LEfSe) analysis [15] found no significantly differently abundant taxa using the all-against-all algorithm, which identifies differences among all pairwise comparisons and therefore employs a more stringent multiple comparisons adjustment. We performed a secondary analysis with a lower effect size threshold of 2.0, and there were still no significantly differently abundant taxa using the all-against-all algorithm. Using the one-against-all algorithm, which compares abundance in each category to abundance in the other categories collectively and therefore requires a less stringent multiple testing correction, we identified the major phylum components unique to each technique. We found an enrichment of Firmicutes in the stool sample collection and Proteobacteria in the swab collection technique (Fig. 4). Within Firmicutes, the genus Blautia was enriched in stool samples, the genus Oscillospira was enriched in glove tip samples, and the genus WAL 1855D, a Sporobacterium, was enriched in swab samples. A cladogram, a plot of linear discriminant analysis (LDA) scores, and relative abundances of phylum and genus signatures of each method are displayed in Fig. 4. Absolute abundances of genera by phylum and sampling method are presented in Supplementary Fig. 1.
Fig. 4.
Summary of taxa which were differentially expressed. (a) Cladogram of taxa which were differentially expressed based on Linear discriminant analysis Effect Size (LEfSe), comparing each collection method against the other two combined (“one against all”). There were no significantly different taxa identified when testing “all against all” sampling methods. (b)-(c) Relative abundance of Firmicutes and Proteobacteria by sampling method. (d) Linear discriminant analysis (LDA) scores indicating the effect size of each differentially expressed taxon. (e)-(i): Relative abundances of differentially abundant genera by sampling method
Discussion
Our prospective, observational study, in which we obtained gut microbiome samples from healthy men using three techniques, showed an overall similarity between collection methods, especially at the participant level. Alpha diversity measured by Shannon’s and Simpson’s index did not significantly differ among the techniques examined. Beta diversity differed between stool samples and each of the other two collection methods; we did not observe differences between swab and glove tip collection techniques. LEfSe analysis showed that some taxa were differentially abundant among sampling methods when using a less stringent one-against-all testing scheme, but not with a more stringent all-against-all testing scheme. While examining different regions can lead to differences in abundance and taxa identified [16–18], we found similar results when examining the V1-V2 and V3-V4 regions separately in secondary analyses of beta diversity. Studies employing swab or glove tip collection methods should be aware that community composition may differ between these methods and stool sample collection.
Our study is consistent with previous literature noting similarities in microbiome analysis between the stool and rectal swabs [9, 19]. A hospital-based study of eight participants measured one stool and several swab samples taken over the course of a day, and found that between-person variation in beta diversity significantly exceeded within-person variation [9]. Our study found similar results, evidenced by the clustering of samples within participants based on Bray-Curtis distances. Another study by Jones et al. [20] compared stool with swab samples and mucosa biopsies, finding some taxa to vary between stool and swab samples, including Campylobacteracea and Prevotellaceae, which we observed to be over-represented in glove tip samples (which correlated highly with swab samples) and which the previous study found over-represented in swab samples. The study additionally found other families whose prevalence varied between stool and swab samples, which our study did not identify, and one family (Rikenellaceae) which had an opposite direction of effect. However, the Jones et al. study sample comprised people with a history of colon polyps, and overall gut microbial community structures may differ between their participants and ours. The Jones et al. study, and others, suggest that rectal swabs may represent more mucosal taxa than stool samples do, which may explain some differences between the sampling methods [21–23]. Mucosal taxa are not generally captured in stool, and can only be fully sampled using invasive mucosal biopsies. Given the similarities in our current study between glove tip and swab samples, it may be that glove tip samples capture mucosal species, possibly avoiding the need for mucosal biopsies in some cases. Our work is the first comparative study to include the glove tip technique, and we do not compare directly with mucosal biopsies, so further studies are needed to test this hypothesis.
The glove tip gut microbiome collection technique performed at the time of a clinical rectal exam is simple, requires no participant preparation, and samples can be transported easily from the clinic to the laboratory. The glove tip technique attempts to improve the implementation of microbiome studies and microbiome-based testing as compared to the more cumbersome at-home stool collection, which is complicated to collect and is participant-dependent. In some clinical settings, providers perform a standard-of-care digital rectal exam as part of cancer screening; use of the glove tip acquisition method means no other acquisition procedures, such as a rectal swab, are needed to obtain a sample for gut microbiome assessment. While stool samples remain the “gold standard” of gut microbiome assessment, our results suggest that inter-individual differences are still adequately captured using the glove tip technique.
There are limitations to our study. The sample size was relatively modest, leaving the possibility that there were differences in microbial communities among the collection methods that we did not detect. Specifically, stool collections were home based and swab and glove tips were collected in clinic, with differences being the swabs and glove tips were transferred to the lab within 4 h, but stool was placed immediately into the omigene kits to preserve DNA. All subjects were healthy, so we were unable to assess whether microbiome biomarkers for prostate cancer risk are as useful when measured using the glove tip method compared to other methods. However, similarities in diversity and lack of differentially expressed taxa under strict multiple testing criteria suggest that the methods’ results will be similar overall. Another limitation is that the material obtained using the glove collection method is small and may not be able to be used for metabolomic analysis; however, it provided enough sample for 16 s rRNA evaluation. Glove tips and swab samples were obtained in the same clinical visit, whereas the stool samples were collected at home, such that some differences may be attributed to timing of collection. Finally, providers may perform rectal exams less frequently in women’s health, and so the glove tip method may not be as useful in women’s health. Providers do use fecal occult testing for colorectal screening in both sexes; however, we did not test this particular technique.
Conclusions
We compare a new glove-based microbiome sample collection method to existing rectal swab and participant-collected stool sample collection methods. Concerning microbial diversity and taxonomic abundance, the glove tip collection is similar to the swab collection technique and generally similar to home-based stool collection. This new collection method, which can be conducted during clinic visits, has the potential to reduce barriers to gut microbiome collection and help implement microbiome sampling in clinical research and practice.
Methods
Study population
We attained approval by the Institutional Review Board at the University of Texas Health San Antonio (HSC20000030H). After approval, we prospectively enrolled men from our San Antonio Biomarkers of Risk (SABOR) prostate cancer screening cohort. In this sub-study, we collected rectal swabs, exam glove tips used during digital rectal exam (DRE), and participant-collected stool samples for comparison during a one-time collection period.
Sample collection
DRE glove collection
We used a double-glove technique where the urology provider wore two non-sterile latex gloves during DRE as described in Besasie et al. [13]. Briefly, we cut the glove tip from the outer glove and placed it in phosphate-buffered saline (PBS) in a 5 mL conical tube. We then stored the glove specimen at 4 °C and transported it to the laboratory. The glove tip was then placed in a 2 mL microfuge tube; the PBS solution from the collection tube was transferred to the 2 mL tube and used to rinse the glove tip to remove all collected fecal material. We then removed the glove tip to store the fecal material in the PBS at − 20 °C until DNA isolation.
Rectal swab collection
The provided used a swab at the time of DRE with an individual packet of sterile lubricating jelly. The provider placed the rectal swab in a 15 mL sterile centrifuge tube containing 1 mL of PBS. Personnel stored the swab specimen at 4 °C during transport to the laboratory. After delivery to the lab, the fecal swab was removed from the collection tube and the PBS solution was transferred into a 2 mL microfuge tube. The cotton portion of the swab was scraped into the microfuge tube using a sterilized scalpel blade. We stored the swab and PBS material at − 20 °C until DNA isolation.
Stool collection
We provided participants with the OMNIgene®-GUT stool collection kit (DNA Genotek, Ottawa, Canada) and instructions for home collection of stools. Stool collections occurred within median of 3 days of the clinic visit with 6 subjects within 24 h and two subjects at the latest of 5 days. The OMNIgene kits allow for DNA preservation at room temperature. The stool was either shipped to the lab or the patient brought the sample to one of our research coordinators. Once in the lab, the specimens were processed identically.
DNA isolation and quantification
For sample input, we attempted to provide a visible amount of stool on the glove tips and swabs. DNA was isolated from fecal samples using our standard operating procedure (see Appendix of Besasie et al. [13]). We performed a purification of genomic DNA from these respective fecal samples using the QIAamp® Fast DNA Stool Mini Kit according to the kit protocol (Qiagen, Germantown, MD). The DNA concentration was measured using the Thermo Scientific NanoDrop.
16S rRNA sequencing
Genomic DNA was used for amplification of V1-V2 variable region of the 16S rRNA genes with custom-designed primers (F27/R534, Youssef 2009, Applied and Environmental Microbiology), and V3-V4 variable region of the 16S rRNA genes following the Illumina 16S metagenomic library preparation guide. Final libraries were quantified, normalized, pooled together, and sequenced by Paired-end sequencing (2 × 300 bp) using Illumina Miseq platform. The average of 264,727 raw pair reads per sample were generated with read length of 300 bp. The sequences were exported as FASTQ files.
Statistical analysis
Sequence processing was performed using QIIME2 software, and statistical analysis was done using R (version 3.6). Dada2 was used to trim and join paired-end sequences, denoise, dereplicate, and remove chimeras from the sequence data. Forward and reverse reads were truncated to preserve Phred quality scores of 28 in 75% of reads at each base. Taxonomic units (OTUs) were assigned using the classify-sklearn feature classifier, which was trained against the Greengenes database with a 97% identity threshold. We removed mitochondria from our sample, and we removed OTUs with fewer than four reads in less than 10% of samples, and samples were rarefied to 2409, the minimum library size after filtering. We compared the groups using the Shannon index and Simpson’s index measures of alpha diversity at the OTU level by conducting a repeated-measures analysis of variance (ANOVA) accounting for differences between individuals [14]. If a global test result was significant, we used paired t-tests to make pairwise comparisons and adjusted p-values for multiple testing using the Benjamini-Hochberg procedure. We performed a beta diversity assessment with principal coordinates analysis using Bray-Curtis and weighted and unweighted unifrac distances at the OTU level. We then used permutational analysis of variance (PERMANOVA) implemented in the vegan package to test for differences in community composition among the collection techniques, stratified by each participant [24]. If the global test result was significant, we conducted pairwise tests between groups, and p-values were adjusted using the Benjamini-Hochberg procedure. We used Linear discriminant analysis Effect Size (LEfSe) to identify taxa, which were differentially abundant among the collection technique groups, using LDA score threshold of 4.0 and an alpha level of 0.05, at the genus level and above [25]. Both the all-against-one and all-against-all algorithms within the LEfSe package were employed. Our team used the Huttenhower Lab implementation of LEfSe on the Galaxy platform [15].
The UPGMA clustering algorithm was performed based on Bray-Curtis distances at the OTU level among the samples and was used to plot a dendrogram of the samples.
Supplementary Information
Additional file 1. Bar plot of genera within phyla by sampling method
Acknowledgments
The authors thank the research staff, investigators, and subjects who have contributed to the San Antonio Biomarkers of Risk for Prostate Cancer (SABOR) study and the laboratory staff of the Biobanking and Genome Sequencing Facility.
Abbreviations
- SABOR
San Antonio Biomarkers of Risk for prostate cancer
- DRE
Digital Rectal Exam
- PCoA
Principal Coordinates Analysis
- DNA
Deoxyribonucleic Acid
- rRNA
Ribosomal Ribonucleic Acid
- QIIME
Quantitative Insights Into Microbial Ecology
- ANOVA
Analysis of Variance
- OTU
Operational Taxonomic Unit
- PERMANOVA
Permutational Analysis of Variance
- LEfSe
Linear Discriminant analysis Effect Size
Authors’ contributions
MIS analyzed the microbiome data, made the figures, and drafted the bulk of the manuscript. RH and BDB coordinated patient visits, maintained subject data, and analyzed demographic data. DPS aided in the experimental design and interpretation of the data and made substantive edits to the manuscript. KRR, KW, RJL, SN, and TLJP provided substantive edits to the manuscript. ZL performed the microbial sequencing and drafted part of the manuscript. BF performed quality filtering, taxonomic classification and initial analysis of the microbiome data. MAL conceived the study design, collected the microbiome data, and drafted and edited the manuscript for important content. All authors read and approved the final manuscript.
Funding
The Los Padres Foundation based in San Antonio, Texas, supported this research with a generous gift. This research was supported in part by a grant from the UT Health San Antonio Center for Biomedical Neuroscience awarded to MS, DS, and BF as well as the SABOR grant from the Early Detection Network (U01 CA086402) The Mays Cancer Center grant provides support for the Biobanking and Genome Sequencing Facility (P30 CA054174).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The protocol for this study was approved by the Institutional Review Board at the University of Texas Health San Antonio (HSC20000030H), and all participants provided written informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Amirian ES, Petrosino JF, Ajami NJ, Liu Y, Mims MP, Scheurer ME. Potential role of gastrointestinal microbiota composition in prostate cancer risk. Infectious agents and cancer. 2013;8(1):42. doi: 10.1186/1750-9378-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Golombos DM, Ayangbesan A, O'Malley P, Lewicki P, Barlow L, Barbieri CE, et al. The role of gut microbiome in the pathogenesis of prostate cancer: a prospective, pilot study. Urology. 2018;111:122–128. doi: 10.1016/j.urology.2017.08.039. [DOI] [PubMed] [Google Scholar]
- 3.Sfanos KS, Yegnasubramanian S, Nelson WG, De Marzo AM. The inflammatory microenvironment and microbiome in prostate cancer development. Nature Reviews Urology. 2018;15(1):11–24. doi: 10.1038/nrurol.2017.167. [DOI] [PubMed] [Google Scholar]
- 4.Shrestha E, White JR, Yu S-H, Kulac I, Ertunc O, De Marzo AM, et al. Profiling the urinary microbiome in men with positive versus negative biopsies for prostate cancer. J Urol. 2018;199(1):161–171. doi: 10.1016/j.juro.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han M, Hao L, Lin Y, Li F, Wang J, Yang H, et al. A novel affordable reagent for room temperature storage and transport of fecal samples for metagenomic analyses. Microbiome. 2018;6(1):43. doi: 10.1186/s40168-018-0429-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu W-K, Chen C-C, Panyod S, Chen R-A, Wu M-S, Sheen L-Y, et al. Optimization of fecal sample processing for microbiome study—the journey from bathroom to bench. J Formos Med Assoc. 2019;118(2):545–555. doi: 10.1016/j.jfma.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Gevers D, Knight R, Petrosino JF, Huang K, McGuire AL, Birren BW, et al. The Human Microbiome Project: a community resource for the healthy human microbiome. PLoS biology. 2012;10(8). [DOI] [PMC free article] [PubMed]
- 8.von Euler-Chelpin M, Brasso K, Lynge E. Determinants of participation in colorectal cancer screening with faecal occult blood testing. J Public Health. 2010;32(3):395–405. doi: 10.1093/pubmed/fdp115. [DOI] [PubMed] [Google Scholar]
- 9.Bassis CM, Moore NM, Lolans K, Seekatz AM, Weinstein RA, Young VB, et al. Comparison of stool versus rectal swab samples and storage conditions on bacterial community profiles. BMC Microbiol. 2017;17(1):78. doi: 10.1186/s12866-017-0983-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Budding AE, Grasman ME, Eck A, Bogaards JA, Vandenbroucke-Grauls CM, van Bodegraven AA, et al. Rectal swabs for analysis of the intestinal microbiota. PloS one. 2014;9(7). [DOI] [PMC free article] [PubMed]
- 11.Coronado GD, Schneider JL, Sanchez JJ, Petrik AF, Green B. Reasons for non-response to a direct-mailed FIT kit program: lessons learned from a pragmatic colorectal-cancer screening study in a federally sponsored health center. Transl Behav Med. 2015;5(1):60–67. doi: 10.1007/s13142-014-0276-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones RM, Devers KJ, Kuzel AJ, Woolf SH. Patient-reported barriers to colorectal cancer screening: a mixed-methods analysis. Am J Prev Med. 2010;38(5):508–516. doi: 10.1016/j.amepre.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Besasie BD, Shah DP, Leach RJ, Liss MA. Comparison of in clinic-based fecal microbiome collection techniques for increase in study participation and utilization of microbiome analysis. Open Journal of Urology. 2019;9(03):51. doi: 10.4236/oju.2019.93006. [DOI] [Google Scholar]
- 14.Knight R, Vrbanac A, Taylor BC, Aksenov A, Callewaert C, Debelius J, et al. Best practices for analysing microbiomes. Nat Rev Microbiol. 2018;16(7):410–422. doi: 10.1038/s41579-018-0029-9. [DOI] [PubMed] [Google Scholar]
- 15.Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, et al. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46(W1):W537–WW44. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim M, Morrison M, Yu Z. Evaluation of different partial 16S rRNA gene sequence regions for phylogenetic analysis of microbiomes. J Microbiol Methods. 2011;84(1):81–87. doi: 10.1016/j.mimet.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 17.Fouhy F, Clooney AG, Stanton C, Claesson MJ, Cotter PD. 16S rRNA gene sequencing of mock microbial populations-impact of DNA extraction method, primer choice and sequencing platform. BMC Microbiol. 2016;16(1):123. doi: 10.1186/s12866-016-0738-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rintala A, Pietilä S, Munukka E, Eerola E, Pursiheimo J-P, Laiho A, et al. Gut microbiota analysis results are highly dependent on the 16S rRNA gene target region, whereas the impact of DNA extraction is minor. Journal of biomolecular techniques: JBT. 2017;28(1):19. doi: 10.7171/jbt.17-2801-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones RB, Zhu X, Moan E, Murff HJ, Ness RM, Seidner DL, et al. Inter-niche and inter-individual variation in gut microbial community assessment using stool, rectal swab, and mucosal samples. Sci Rep. 2018;8(1):1–12. doi: 10.1038/s41598-017-17765-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones RB, Zhu X, Moan E, Murff HJ, Ness RM, Seidner DL, et al. Inter-niche and inter-individual variation in gut microbial community assessment using stool, rectal swab, and mucosal samples. Sci Rep. 2018;8(1):4139. doi: 10.1038/s41598-018-22408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Budding AE, Grasman ME, Eck A, Bogaards JA, Vandenbroucke-Grauls CM, van Bodegraven AA, et al. Rectal swabs for analysis of the intestinal microbiota. PLoS One. 2014;9(7):e101344. doi: 10.1371/journal.pone.0101344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Albenberg L, Esipova TV, Judge CP, Bittinger K, Chen J, Laughlin A, et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology. 2014;147(5):1055–1063. doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Araujo-Perez F, McCoy AN, Okechukwu C, Carroll IM, Smith KM, Jeremiah K, et al. Differences in microbial signatures between rectal mucosal biopsies and rectal swabs. Gut Microbes. 2012;3(6):530–535. doi: 10.4161/gmic.22157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oksanen FGB, Michael Friendly, Roeland Kindt, Pierre Legendre, Dan McGlinn, Peter R. Minchin, R. B. O’Hara, Gavin L. Simpson, Peter Solymos, M. Henry H. Stevens, Eduard Szoecs and Helene Wagner Vegan: Community Ecology Package. 2.5–6 ed: R package; 2019.
- 25.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Bar plot of genera within phyla by sampling method
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.