Abstract
RAS (KRAS, NRAS and HRAS) is the most frequently mutated gene family in cancers, and, consequently, investigators have sought an effective RAS inhibitor for more than three decades. Even 10 years ago, RAS inhibitors were so elusive that RAS was termed ‘undruggable’. Now, with the success of allele-specific covalent inhibitors against the most frequently mutated version of RAS in non-small-cell lung cancer, KRASG12C, we have the opportunity to evaluate the best therapeutic strategies to treat RAS-driven cancers. Mutation-specific biochemical properties, as well as the tissue of origin, are likely to affect the effectiveness of such treatments. Currently, direct inhibition of mutant RAS through allele-specific inhibitors provides the best therapeutic approach. Therapies that target RAS-activating pathways or RAS effector pathways could be combined with these direct RAS inhibitors, immune checkpoint inhibitors or T cell-targeting approaches to treat RAS-mutant tumours. Here we review recent advances in therapies that target mutant RAS proteins and discuss the future challenges of these therapies, including combination strategies.
RAS (KRAS, NRAS and HRAS) is the most frequently mutated gene family in cancers. Mutations in KRAS are known drivers of three of the most lethal cancers (lung cancer, colorectal cancer (CRC) and pancreatic cancer). For more than three decades, development of effective therapeutics to inhibit RAS-driven oncogenesis has eluded the field and RAS was thought to be ‘undruggable’. However, a clinically approved mutant selective KRAS therapy is now within sight as the FDA has granted an allele-specific covalent inhibitor, AMG 510, Fast Track designation1. AMG 510 binds to KRAS-G12C, the RAS mutatant most commonly found in non-small-cell lung tumours. This successful inhibition of KRAS-G12C, has given hope that a range of mutant RAS allele-specific targeted therapies could become therapeutically tractable.
In normal cells, RAS is activated at the membrane downstream of growth factor receptors, including members of the epidermal growth factor receptor (EGFR) family (FIG. 1). This family contains EGFR itself as well as the related ERBB receptors (known as HER in humans). RAS is a small switch signalling GTPase that toggles between its GTP-bound active state and the GDP-bound inactive state. Although RAS proteins exhibit both intrinsic GTP hydrolysis and nucleotide exchange, their cellular signalling state results from activation by guanine exchange factors (GEFs) that catalyse the loading of GTP and deactivation by GTPase-activating proteins (GAPs) that increase hydrolysis of GTP. In its GTP-bound state, RAS directly interacts with and activates several downstream effector pathways including the mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase (PI3K) pathways. Mutations in RAS disrupt the guanine exchange cycle, typically by becoming GAP-independent and ‘locking’ RAS in the active, GTP-bound state, thereby activating downstream signalling pathways resulting in tumour cell growth.
Fig. 1 |. Clinical development of inhibitors for RAS-mutant tumours.
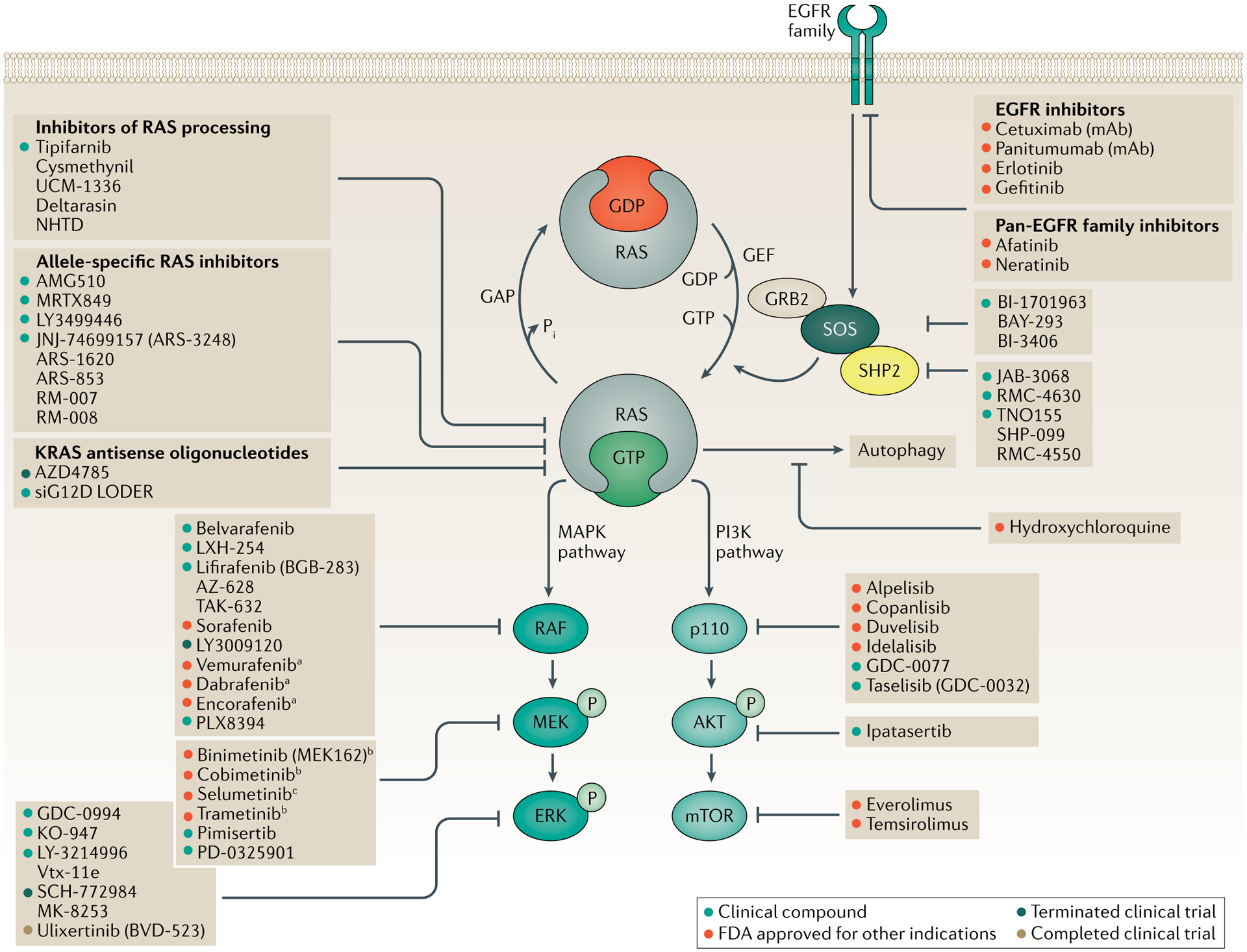
Activation of receptor tyrosine kinases, such as members of the epidermal growth factor receptor (EGFR) family, promotes the exchange of GDP for GTP in RAS, thereby activating RAS. Inhibition of EGFR can reduce this activation. Inhibition of SOS or SHP2 decreases the rate of GDP–GTP exchange and reduces the GTP-bound RAS population. Mutant RAS proteins accumulate in the GTP-bound state. A number of approaches have been developed to directly inhibit RAS, including covalent allele-specific inhibitors that bind to KRAS-G12C. GTP-bound RAS activates downstream signalling by binding to the RAS-binding domain of effector proteins, such as RAF and p110, to activate the MAPK and PI3K signalling cascades, respectively. Both the MAPK and PI3K signalling cascades can be inhibited at each kinase tier. Data compiled from ClinicalTrials.gov and AccessData.FDA.gov. aOnly effective against monomeric BRAF (BRAF-V600E/K). bApproved for the treatment of BRAF-mutant melanoma. cApproved for the treatment of paediatric patients with NF1 mutations.
In this Review, we describe recent advances in the development of therapies targeting mutant RAS proteins and discuss combination strategies that potentially increase the clinical benefit of RAS inhibition (FIG. 1). First, we discuss the prevalence of different RAS isoforms and differences in their biochemical properties. Then, we discuss recent strategies to directly or indirectly inhibit RAS, highlighting the breakthrough therapies that target KRAS-G12C and inhibitors that block Son of Sevenless homologue 1 (SOS1), SHP2 (PTPN11) and RAS membrane association. Third, we provide a clinical update on the use of inhibitors targeting RAS itself as well as RAS effector pathways, namely MAPK and PI3K. Last, we detail emerging therapeutic strategies for treating RAS-mutant tumours.
RAS mutations and splice variants
RAS mutations are genetic drivers in numerous cancer types including CRC, pancreatic ductal adenocarcinoma (PDAC), lung adenocarcinoma (LUAD; a subtype of non-small-cell lung cancer (NSCLC)), melanoma and certain haematological cancers2–6. Although these tumour types are driven by RAS mutations, the isoform (KRAS, NRAS or HRAS), codon and frequency of RAS mutations vary by tissue type (FIG. 2). For example, a large percentage of LUAD (32%), PDAC (86%) and CRC (41%) (FIG. 2a) are driven by KRAS mutations, which predominantly occur at codon 12 in these tumour types2,4,5 (FIG. 2b). By contrast, 29% of melanomas are driven by mutations in NRAS, and unlike KRAS, these mutations occur at codon 61 (REF.3) (FIG. 2). HRAS mutations occur less frequently than mutations in KRAS or NRAS, but a subset of head and neck squamous cell carcinoma (HNSCC; 5%) and bladder cancers (6%) are driven by HRAS mutations and these mutations occur at either codon 12 or 61 (REFS7,8) (FIG. 2a). Genetically engineered mouse models (GEMMs) recapitulate the isoform and codon mutational preferences observed in patient tumours. Expression of KrasG12D in colonic epithelial cells resulted in hyperproliferation, but the expression of NrasG12D in these cells did not alter cell proliferation9. Expression of NrasQ61R, but not NrasG12D, in melanocytes induced melanomas10. Approaches to target RAS-driven cancers must therefore account for the specific isoform and the specific codon mutation.
Fig. 2 |. Frequency and distribution of RAS mutations in human cancers.
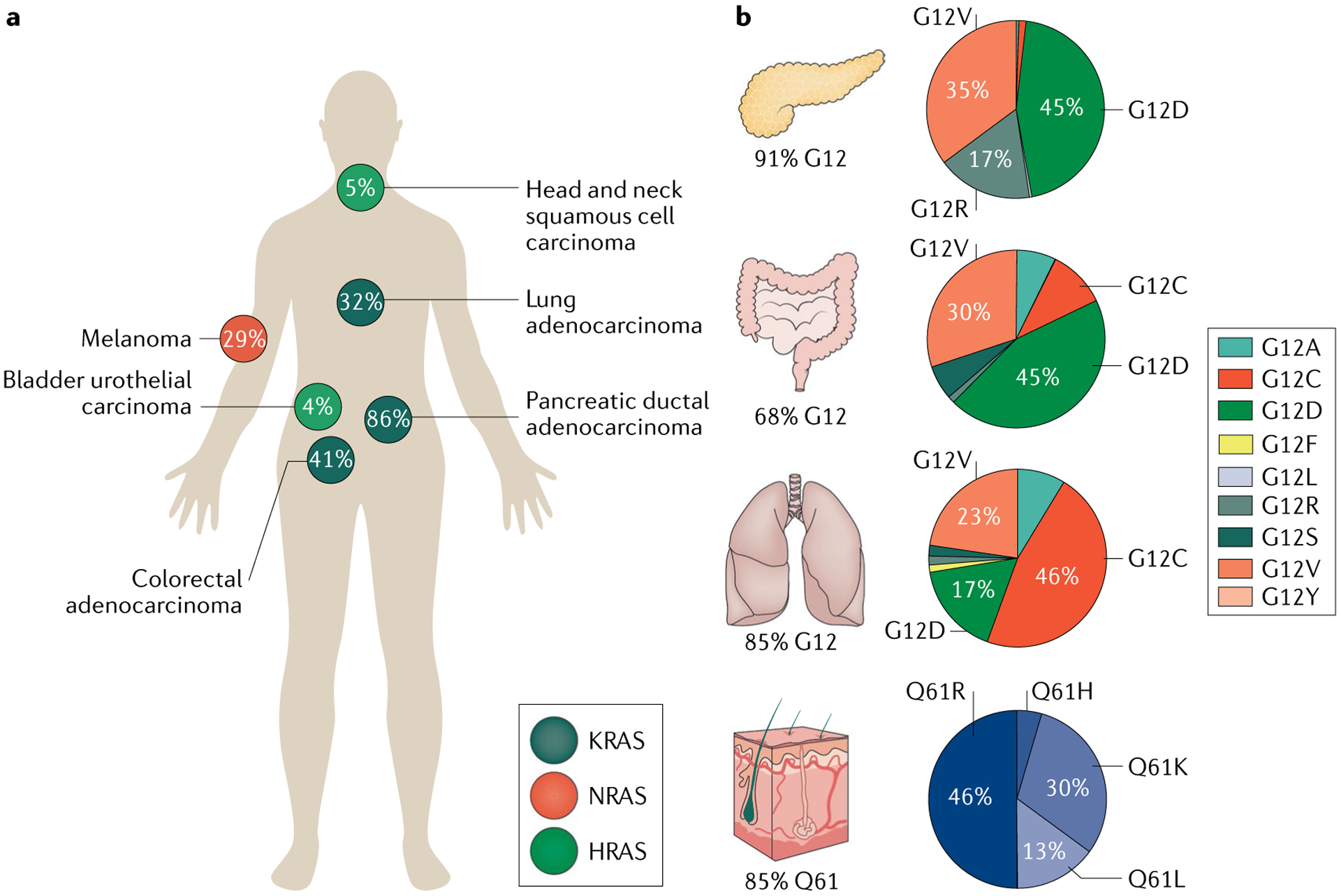
Human cancers differ in which has the most frequently mutated RAS isoform, codon and amino acid substitution. a | Distribution of RAS isoform (KRAS, NRAS and HRAS) mutations across tumour types and the frequency of the RAS mutation by isoform in each tumour type. b | Percentages of KRAS mutations that are in codon 12 by tissue type for pancreatic, colorectal and lung adenocarcinoma, and the percentage of NRAS mutations that are in codon 61 for melanoma. The distributions of amino acid substitutions at the mutated codon (12 or 61) for each tissue type are shown in pie charts beside the relevant organ. Data acquired from The Cancer Genome Atlas (pan-Cancer) from cBioPortal and from Project GENIE269 (GENIE v7.0 public).
The KRAS gene encodes two splice variants which use different exon 4s, resulting in KRAS4A and KRAS4B. KRAS4A contains an additional 22 or 23 amino acids in the C terminus and therefore has different post-translational modifications and membrane localization11–13. KRAS4B has long been viewed as the major isoform as it is ubiquitously and highly expressed in human cancers14,15. Recently, however, KRAS4A was shown to be widely expressed in cancer cell lines and expressed at equivalent levels to KRAS4B in colorectal tumours12. KRAS4A is dispensable in GEMMs: genetic deletion of exon 4A results in viable embryos, whereas deletion of Kras results in embryonic lethality16. Recently, Kras4B was also shown to be dispensable in mice, indicating that the two isoforms are functionally redundant during development17. However, deletion of either Kras4A or Kras4B resulted in resistance to lung tumour formation, suggesting that tumour initiation requires both isoforms17. The two isoforms may also have specific roles in the tumour microenvironment — KRAS4A expression increases adaptability to stress, such as hypoxia, and KRAS4B is expressed in stem cells and progenitor cells. Tumours can adapt, through splicing, to express KRAS4A in times of stress17. These recent studies have renewed the focus on the role of KRAS4A in tumorigenesis and have shifted the perspective for inhibiting KRAS, as KRAS4A now requires consideration.
Biochemical features of mutant RAS
Small GTPases, such as RAS, cycle between a GDP-bound inactive state and a GTP-bound active state (FIG. 3). GEFs, such as SOS or Ras guanyl nucleotide-releasing protein (RasGRP), promote the exchange of GDP for GTP18–20. To stimulate GTP hydrolysis and therefore return RAS to the inactive GDP state, RAS GAPs, such as neurofibromin (NF1) or p120GAP, mediate GTP hydrolysis21,22. Mutations in codons 12, 13 and 61 of RAS disrupt GAP-mediated GTP hydrolysis, allowing these mutants to accumulate in a persistently GTP-bound state (FIG. 3a,b). GTP-bound RAS activates downstream effector pathways to promote cell proliferation, most notably the MAPK and PI3K pathways.
Fig. 3 |. Biochemical features of mutant RAS proteins.
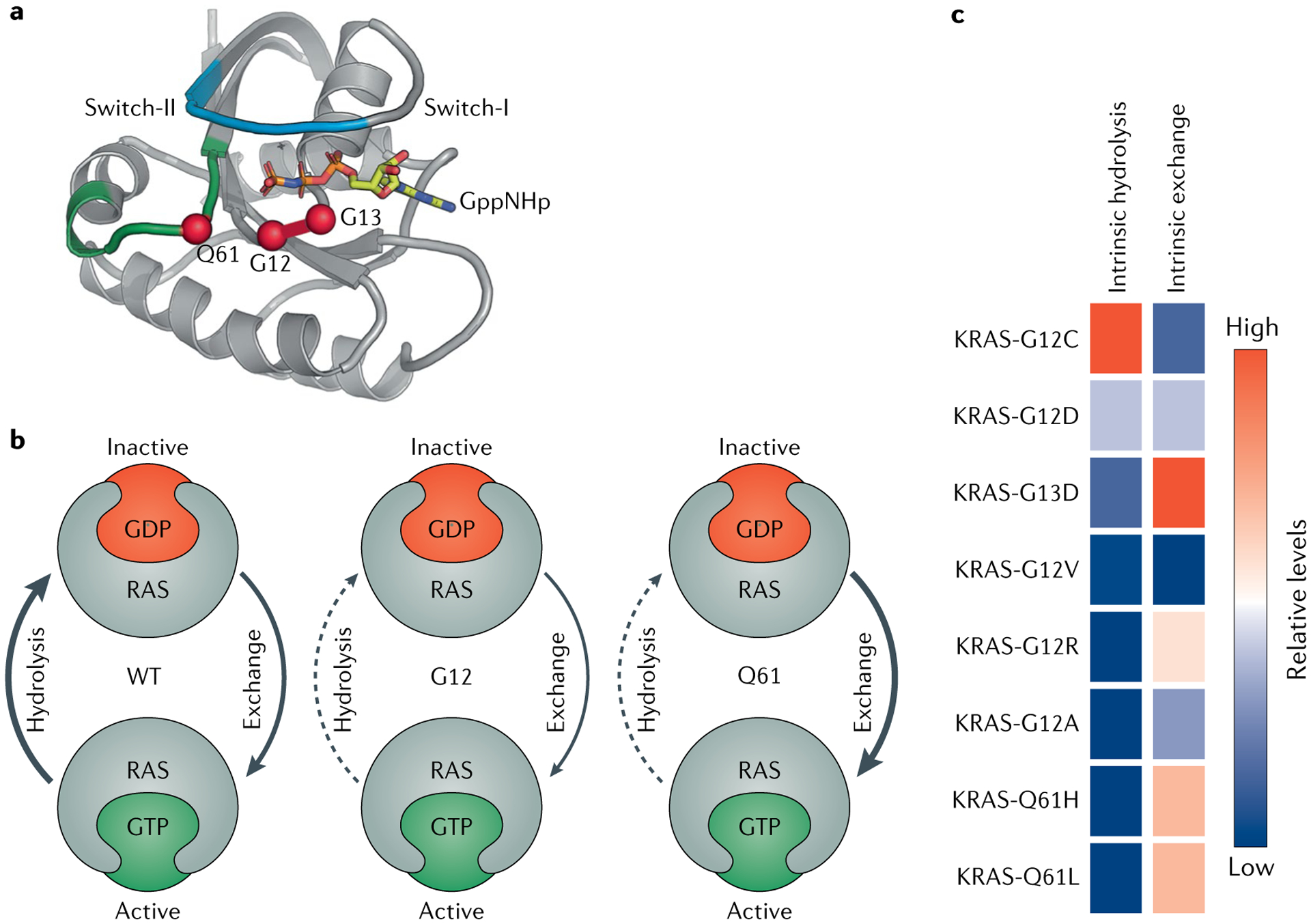
Mutations in codons 12, 13 and 61 disrupt the GTP hydrolysis and guanine exchange rates of RAS proteins. a | Ribbon diagram of HRAS. Switch-I is shown in light blue, switch-II is in green, GppNHp (a GTP analogue) is shown in yellow, and oncogenic hotspot residues G12, G13 and Q61 are shown as red spheres. b | Summary of generalized biochemical disruption of hydrolysis and guanine exchange upon mutations in codon 12 or 61. Generally, mutations in codon 12 disrupt the GTPase activity of RAS and thereby decrease the rate of GTP hydrolysis, so the mutant protein accumulates in the GTP-bound state. Mutations in codon 61 accelerate the rate of GDP–GTP exchange and simultaneously decrease the rate of GTP hydrolysis, so codon 61 RAS mutants also accumulate in the GTP-bound state. c | Biochemical properties of specific amino acid substitutions at KRAS codon 12, 13 or 61, rank ordered by intrinsic hydrolysis. Data acquired from results in Hunter et al.23. WT, wild type.
Intrinsic GTPase and GDP–GTP exchange rates can vary among the different RAS mutants and this observation may offer insight into how to best target each mutant23. For example, codon 12, 13 and 61 mutations generally have diminished intrinsic GTPase activity, except for KRAS-G12C, which exhibits near-wild-type intrinsic GTPase activity despite its reduced p120 GAP-mediated hydrolysis rate23 (FIG. 3c). Indeed, this unique biochemical property of KRAS-G12C was leveraged by Shokat and colleagues to target KRAS-G12C using covalent inhibitors that bind to the GDP-bound state of KRAS-G12C (REF.24). By contrast, KRAS-G13D has elevated intrinsic exchange activity relative to wild-type RAS, suggesting that the GDP-bound state is short-lived23 (FIG. 3c). KRAS with a mutation in codon 13 is partially sensitive to NF1 GAP-mediated hydro lysis, whereas KRAS isoforms with mutations in codon 12 or 61 are insensitive to NF1 (REF.25). Interestingly, KRAS codon 13 mutations are frequently co-mutated with NF1 mutations, further exemplifying the evolutionary pressure of a tumour to rid KRAS-G13 of NF1 GAP activity and thus become more biochemically active. The biochemical differences between mutant versions of RAS will determine which nucleotide-bound state of RAS would therefore be most appropriate to target with allele-specific inhibitors. Low levels of GTPase activity or high levels of guanine exchange could pose difficulties in targeting the GDP-bound state.
Approaches to target RAS directly
Directly inhibiting RAS is a desirable approach for treating RAS-mutant tumours. Here, we highlight the latest efforts in targeting RAS directly, including the development of switch-II mutant-selective covalent inhibitors and pan-RAS inhibitors. Intense efforts in developing mutant-specific RAS (KRAS-G12C) switch-II covalent inhibitors are underway, and progress is being made.
Covalent inhibitors targeting KRAS-G12C
Kras is essential for mouse development, whereas Nras and Hras are dispensable26,27. If humans are similar in this regard, this requirement for KRAS creates toxicity concerns when targeting the wild-type KRAS protein. However, when Kras is replaced with Hras, mice are viable, which reduces toxicity concerns28. Conditional deletion of Kras in adult mice would directly examine these concerns, but such studies have yet to be published. The field of RAS inhibitors, pioneered by Shokat and colleagues, has focused on covalent inhibition of KRAS-G12C, which would be expected to circumvent toxicity attributed to inhibiting all RAS isoforms.
The inherently reactive nature of cysteine, which is found at codon 12 of KRAS-G12C, can be exploited to create covalent small-molecule inhibitors. Covalently targeting active site cysteines is a widely used strategy in drug discovery29. Importantly, wild-type KRAS lacks cysteines in the active site so KRAS-G12C can be specifically inhibited with this covalent approach. As discussed above, the G12 codon is a mutational hotspot in KRAS and G12C is the third most common mutation at this position. G12C mutations predominantly occur in LUAD and are transversion mutations (G>T and G>C) associated with smoking (FIG. 2b).
Shokat and colleagues first identified a novel allosteric binding pocket behind switch-II, termed the switch-II pocket, in the mutant KRAS-G12C protein24 (Figs 4,5a,b). They developed the first series of compounds to irreversibly target KRAS-G12C. These compounds bound KRAS-G12C in the GDP-bound state, blocked SOS-catalysed nucleotide exchange and blocked KRAS-G12C association with RAF24. Notably, these compounds only bound to KRAS-G12C in the GDP-bound state and therefore required KRAS-G12C to first undergo GTP hydrolysis. About 75% of KRAS-G12C is GTP-bound in the steady state, but KRAS-G12C has the highest level of intrinsic GTPase activity among the common oncogenic mutations and so is vulnerable to covalent attack30 (FIG. 3c). Because mutations in KRAS other than KRASG12C have a lower rate of intrinsic GTP hydrolysis, it is unclear whether targeting the switch-II pocket via a similar approach in these other mutant forms will be successful23.
Fig. 4 |. Chemical structures of compounds that bind to KRAS-G12C.
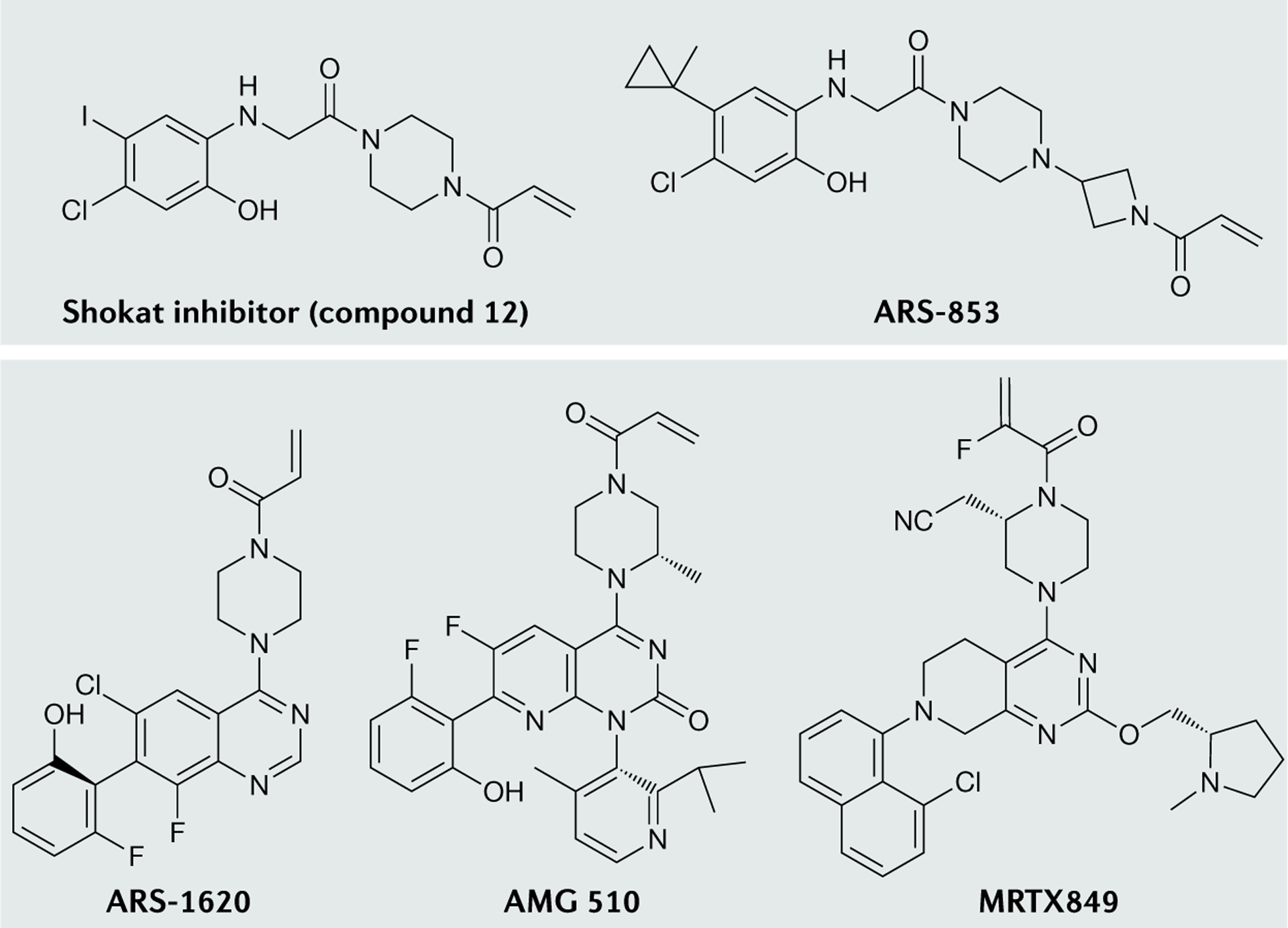
Shokat and colleagues developed the first series of small molecules to bind KRAS-G12C, the most potent of which is compound 12 (REF.24). Modification of the linker and hydrophobic binding pocket led to the development of a more potent and cellular active compound, ARS-853 (REF.36). Further improvements, such as the introduction of a quinazoline-based series and a fluorophenol hydrophobic binding moiety, enhanced the potency and pharmacological properties and led to the development of ARS-1620 (REF.30). Using an alternative orientation of His95 in the switch-II pocket allowed the addition of aromatic rings to enhance the protein–protein interactions with KRAS-G12C, leading to the development of AMG 510 (REF.32). Structure-based drug design approaches and optimization led to the development of MRTX849 (REF.34).
Fig. 5 |. Structures of RAS surfaces targeted by therapeutics.
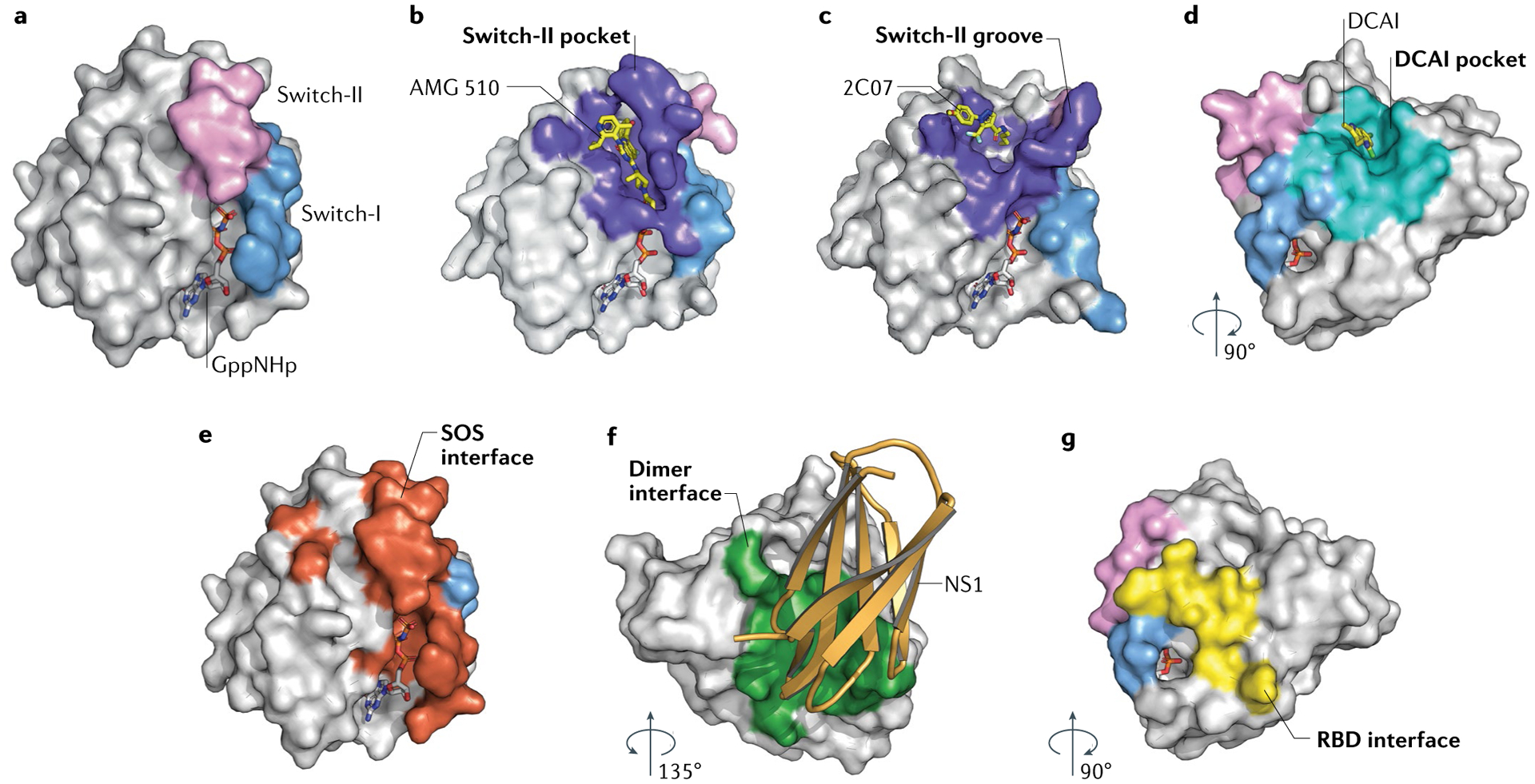
Proteins are depicted by surface representation, and compounds and nucleotides are shown as stick models. Each panel is coloured to highlight important surfaces, with switch-I in blue, switch-II in pink and relevant interfaces coloured uniquely. a | HRAS binds to GppNHp, a non-hydrolysable nucleotide, which can be used as a reference for orientation (Protein Data Bank (PDB): 5P21). b | Switch-II pocket (purple) of KRAS-G12C bound to AMG 510 (PDB: 6OIM; compound identifier: MOV). c | Switch-II groove (purple) bound to 2C07 (PDB: 5VBZ; compound identifier: 92V). d | DCAI pocket (teal) of KRAS-G12D bound to the DCAI compound (PDB: 4DST; compound identifier: 9LI). e | Son of Sevenless (SOS) binding interface (red) of HRAS (PDB: 1BKD). f | Proposed HRAS dimerization interface (green) bound by NS1 monobody (gold) (PDB: 5E95). g | RAF–RAS-binding domain (RBD) binding interface (yellow) of HRAS (PDB: 4G0N).
Numerous companies are developing more potent inhibitors against KRAS-G12C and some of these molecules are in clinical trials, although few details have been published. AMG 510 (FIG. 4) was the first molecule to enter clinical trials and the preliminary results of the phase I trial are promising, particularly in NSCLC: of 13 patients receiving the target dose of 960 mg, 7 patients had a partial response (PR) and 6 had stable disease (SD)1 (NCT03600883; TABLE 1). The activity in CRC is far less striking: only 1 of 12 patients had a PR and 10 patients had SD31. Notably, of the total of 34 patients in the study, none showed dose-limiting toxicities or adverse events causing discontinuation. In preclinical models, AMG 510 potently inhibited cellular viability exclusively in KRAS-G12C cell lines (half-maximal inhibitory concentration (IC50) ≈ 9 nM and IC50 ≈ 6 nM in the cell lines MiaPaCa-2 and NCI-H358, respectively) and induced tumour regression in xenograft models32. AMG 510 had synergistic growth inhibitory effects when combined with inhibitors of proteins that activate or are activated by RAS — such as MEK, AKT, PI3K, SHP2 and members of the EGFR family — or with immunotherapy32.
Table 1 |.
Single-agent inhibitors in clinical development
| Drug | Biomarker | Disease setting | Study phase | ClinicalTrials.gov registration |
|---|---|---|---|---|
| KRAS-G12C inhibitors | ||||
| AMG 510 | KRASG12C mutation | Advanced solid tumours | I/II | NCT03600883 |
| MRTX849 | KRASG12C mutation | Advanced solid tumours | I/II | NCT03785249 |
| JNJ-74699157 (ARS-3248) | KRASG12C mutation | Advanced solid tumours | I | NCT04006301 |
| LY3499446 | KRASG12C mutation | Advanced solid tumours | I/II | NCT04165031 |
| SOS inhibitors | ||||
| BI-1701963 | KRAS mutations | Advanced or metastatic solid tumours | I | NCT04111458 |
| SHP2 inhibitors | ||||
| RMC-4630 | Mutations that hyperactivate the MAPK pathway | Relapsed or refractory solid tumours | I | NCT03634982 |
| TNO155 | EGFR or KRASG12C mutations | Advanced solid tumours | I | NCT03114319 |
| Farnesyltransferase inhibitors | ||||
| Tipifarnib | HRAS mutations | Thyroid cancer, squamous head and neck cancer and squamous cell carcinoma | II | NCT02383927 |
| RAF inhibitors | ||||
| Belvarafenib (HM95573) | BRAF, KRAS or NRAS mutations | Advanced solid tumours | I | NCT03118817 |
| LXH-254 | MAPK pathway mutation | Advanced solid tumours | I | NCT02607813 |
| MEK inhibitors | ||||
| Binimetinib (MEK162) | NRAS mutation | Unresectable or metastatic melanoma | III | NCT01763164 |
| ERK inhibitors | ||||
| KO-947 | BRAF, KRAS, NRAS or HRAS mutation | Advanced solid tumours | I | NCT03051035 |
| LY-3214996 | BRAF or NRAS mutations | Metastatic melanoma and NSCLC | I | NCT02857270 |
| Adoptive cell therapies | ||||
| Anti-RAS-G12D mTCR | HLA-A11:01 RASG12D mutation | Advanced solid tumours | I/II | NCT03745326 |
| Anti-RAS-G12V mTCR | HLA-A11:01 RASG12V mutation | Advanced solid tumours | I/II | NCT03190941 |
| Cancer vaccines | ||||
| mRNA-5671 | HLA-A11:01 and/or HLA-C08:02; KRASG12C, KRASG12D, KRASG12V or KRASG13D mutation | NSCLC, non-MSI-H CRC, PDAC | I | NCT03948763 |
CRC, colorectal cancer; HLA, human leukocyte antigen; MSI-H, microsatellite instability-high; mTCR, murine T cell receptor; NSCLC, non-small-cell lung cancer; PDAC, pancreatic ductal adenocarcinoma; SOS, Son of Sevenless.
MRTX849 (FIG. 4) is also in phase I/II clinical trials (NCT03785249; TABLE 1). In early clinical studies (in seven patients at 600 mg twice daily), three of five patients with NSCLC achieved a PR and one of two patients with CRC achieved a PR33. In preclinical models, MRTX849 potently reduced cellular viability exclusively in KRAS-G12C cell lines (IC50 ≈ 94 nM and IC50 ≈ 107 nM in MiaPaCa-2 and NCI-H358 cells, respectively) and caused tumour regression in xenograft models34. MRTX849 exhibited synergistic effects, resulting in tumour regression when combined with inhibitors of the EGFR family, SHP2, mTOR, or cyclin-dependent kinase 4 (CDK4) and CDK6, even in MRTX849-refractory tumours34. In a CRISPR screen, loss of NRAS or KEAP1 resulted in resistance to MRTX849 whereas loss of SHP2, MYC or mTOR pathway genes further sensitized tumours to MRTX849 (REF.34).
A third KRAS-G12C covalent inhibitor, JNJ-74699157 (ARS-3248), is currently in a phase I clinical trial (NCT04006301; TABLE 1); results have not been published. Two previous compounds (ARS-853 and ARS-1620; FIG. 4) diminished cell growth and inhibited downstream signalling to MAPK exclusively in tumour cell lines with KRASG12C mutations30,35,36. A fourth KRAS-G12C covalent inhibitor, LY3499446, is currently in a phase I/II clinical trial (NCT04165031; TABLES 1,2) as a monotherapy and in combination with inhibitors of CDK4/CDK6 or EGFR or with chemotherapy (docetaxel). Results have not been published.
Table 2 |.
Combination therapies in clinical development
| Drugs | Biomarker | Disease setting | Study phase | ClinicalTrials.gov registration |
|---|---|---|---|---|
| KRAS-G12C combinations | ||||
| AMG510 and antibodies to PD1 or PDL1 | KRASG12C mutation | Advanced NSCLC | II | NCT03600883 |
| MRTX849 and TNO155 | KRASG12C mutation | Advanced or metastatic solid tumours | I/II | NCT04330664 |
| LY3499446 and abemaciclib | KRASG12C mutation | Advanced solid tumours | I/II | NCT04165031 |
| LY3499446 and cetuximab | KRASG12C mutation | Advanced solid tumours | I/II | NCT04165031 |
| LY3499446 and erlotinib | KRASG12C mutation | Advanced solid tumours | I/II | NCT04165031 |
| LY3499446 and docetaxel | KRASG12C mutation | Advanced solid tumours | I/II | NCT04165031 |
| SOS inhibitor combinations | ||||
| BI-3406 and trametinib | KRAS mutation | Advanced or metastatic solid tumours | I | NCT04111458 |
| SHP2 inhibitor combinations | ||||
| TNO155 and spartalizumab | EGFR or ALK WT NSCLC | Advanced solid tumours | Ib | NCT04000529 |
| TNO155 and ribociclib | WT EGFR or WT ALK NSCLC, KRAS-mutant CRC or NSCLC | Advanced solid tumours | Ib | NCT04000529 |
| RAF inhibitor combinations | ||||
| Belvarafenib and cobimetinib | RAS or RAF mutations | Locally advanced or metastatic tumours | Ib | NCT03284502 |
| LXH-254 and an antibody to PD1 | NRAS-mutant melanoma and KRAS-mutant NSCLC | Advanced solid tumours | I | NCT02607813 |
| LXH-254 and trametinib | KRAS-mutant or BRAF-mutant NSCLC or NRAS-mutant melanoma | Advanced or metastatic solid tumours | Ib | NCT02974725 |
| LXH-254 and LTT462 | KRAS-mutant or BRAF-mutant NSCLC or NRAS-mutant melanoma | Advanced or metastatic solid tumours | Ib | NCT02974725 |
| LXH-254 and ribociclib | KRAS-mutant or BRAF-mutant NSCLC or NRAS-mutant melanoma | Advanced or metastatic solid tumours | Ib | NCT02974725 |
| BGB-283 and PD-0325901 | KRAS-mutant NSCLC or endometrial cancer | Advanced or refractory solid tumours | Ib | NCT03905148 |
| MEK inhibitor combinations | ||||
| Cobimetinib and atezolizumab | KRAS mutation | Advanced and metastatic NSCLC | II | NCT03600701 |
| Cobimetinib and RMC-4630 | Mutations that hyperactivate the MAPK pathway | Relapsed or refractory solid tumours | Ib/II | NCT03989115 |
| Selumetinib and afatinib | KRAS mutation, WT PIK3CA | Advanced and metastatic NSCLC | I/II | NCT02450656 |
| Selumetinib and MK-8353 | RAS or RAF mutation | Advanced solid tumours | Ib | NCT03745989 |
| Trametinib and ponatinib | KRAS mutation | Advanced NSCLC | I | NCT03704688 |
| Trametinib and hydroxychloroquine | None | Advanced pancreatic cancer | I | NCT03825289 |
| ERK inhibitor combinations | ||||
| MK-8353 and pembrolizumab | None | Advanced solid tumours and CRC | Ib | NCT02972034 |
| Ulixertinib (BVD-523) and nab-paclitaxel + gemcitabine | None | Metastatic pancreatic cancer | Ib | NCT02608229 |
| Ulixertinib (BVD-523) and palbociclib | None | Advanced or metastatic pancreatic cancer or solid tumours | I | NCT03454035 |
| LY-3214996 and midazolam | BRAF or RAS mutations | Advanced or metastatic solid tumours | I | NCT02857270 |
| LY-3214996 and abemaciclib | BRAFor RAS mutations | Advanced or metastatic solid tumours | I | NCT02857270 |
| LY-3214996 and nab-paclitaxel + gemcitabine | KRAS mutations | Advanced or metastatic solid tumours | I | NCT02857270 |
| siRNA combinations | ||||
| siG12D-LODERand nab-paclitaxel + gemcitabine | KRASG12D mutation | Unresectable locally advanced pancreatic cancer | II | NCT01676259 |
| Cancer vaccine combinations | ||||
| mRNA-5671 and pembrolizumab | HLA-A11:01 and/or HLA-C08:02 KRASG12C, KRASG12D, KRASG12V or KRASG13D mutation | NSCLC, non-MSI-H CRC, PDAC | I | NCT03948763 |
| Tyrosine kinase inhibitor combinations | ||||
| Neratinib and divalproex sodium | KRAS or NRAS mutation | Advanced solid tumours | I/II | NCT03919292 |
| Cetuximab and irinotecan | RASG131 mutation | Advanced colorectal cancer | II | ACTRN12612 000901808 |
CRC, colorectal cancer; HLA, human leukocyte antigen; MSI-H, microsatellite instability-high; nab-paclitaxel, nanoparticle albumin-bound paclitaxel; NSCLC, non-small-cell lung cancer; PDAC, pancreatic ductal adenocarcinoma; SOS, Son of Sevenless; TBA, to be advised; WT, wild type.
As the covalent inhibitors discussed require KRAS-G12C to be in the GDP-bound state, resistant mutations could arise in KRASG12C that disable the GTPase activity or that promote the guanine exchange of GDP for GTP. Proposed resistance mechanisms to covalent KRAS-G12C inhibitors have been identified through CRISPR screens and include the loss of either NF1 or one of the other RAS isoforms (NRAS and HRAS)34,37.
Recently, molecules have been discovered that bind both the GDP-bound and GTP-bound state of KRAS38. These molecules bind a new groove (switch-II groove), which is adjacent to the switch-II pocket but away from the nucleotide-binding site (FIG. 5c). This discovery highlights the dynamic nature of the switch-II pocket and, importantly, provides proof-of-concept evidence that both nucleotide-bound states of RAS can be targeted with inhibitors.
Other mutation-specific approaches
As described above, KRASG12C mutations only account for a proportion of KRAS mutations and are primarily found in LUAD (FIG. 2b). To effectively inhibit the other common KRAS mutations, KRASG12D and KRASG12V, different approaches are needed as these mutants lack reactive cysteines in the active site. The development of inhibitors against these specific mutations will need to consider the biochemical differences and evaluate which state (bound to GDP or GTP) to target, as each mutation will differ in the relative prevalence of these states.
Revolution Medicines is developing a tri-complex platform, RAS(ON), in which a compound mediates a non-natural protein–protein interaction between different mutant KRAS proteins, including KRAS-G12C, and a chaperone, such as cyclophilin A39. Formation of such complexes prevents mutant KRAS from binding SOS and RAS-binding domain (RBD)-containing effector proteins. Interestingly, these molecules (RM-007 and RM-008) covalently bind KRAS-G12C and KRAS-G13C, respectively, in the GTP-bound state, and have anti-proliferative activity in cells39. As molecules that target the active GTP-bound state break the canonical GTP conformation or block effector interactions, approaches like this, which target the GTP-bound state, would impede the presumed mechanisms of resistance to GDP-bound KRAS-G12C covalent inhibitors.
RAS–effector interaction inhibitors
Although targeting mutation-specific states is effective for KRAS-G12C covalent inhibitors, it will be cumbersome to identify effective therapeutics for each mutant RAS protein. Directly targeting conserved ligand binding sites on all RAS proteins (KRAS4A, KRAS4B, NRAS and HRAS) could provide a single therapeutic approach to inhibit RAS across mutation and tumour types. One such compound, compound 3144, binds a conserved residue, Asp38, in switch-I and blocks RAS effector binding40. Compound 3144 bound wild-type KRAS, NRAS and HRAS in vitro and suppressed the growth of KRAS-G13D tumours in vivo; however, toxicity and off-target activity were reported. Indeed, pan-RAS inhibitors are unlikely to be tolerated because RAS is essential in normal cell signalling. Deletion of all three RAS isoforms results in embryonic lethality in mouse models and the absence of cellular proliferation in vitro41,42.
An early RAS-binding small molecule, DCAI, weakly inhibits SOS1-mediated nucleotide exchange on RAS43 (FIG. 5d). Cocrystallization with KRAS revealed that DCAI binds a pocket between the α2 helix and the core β-sheet, β1–β3, here referred to as the DCAI pocket, which blocks the interaction between RAS and SOS1. Small molecules that bind in the DCAI pocket prevent SOS1-mediated guanine exchange and so RAS proteins cannot adopt the GTP-bound active state, making this pocket a desirable drug target.
Two independent protein–protein interactor compound series, Abd- and Ch-, reversibly bound RAS in the DCAI pocket and inhibited the interactions of CRAF, RalGDS and the catalytic p110α or p110γ subunits of PI3K with mutant KRAS, NRAS or HRAS44,45. A third compound, BI-2852, also bound the DCAI pocket and reduced SOS1-mediated exchange, which reduced the phosphorylation of the downstream kinases ERK and AKT46.
These series function as pan-RAS inhibitors; the DCAI pocket is present in the wild-type RAS proteins and therefore these compounds are not mutant-selective inhibitors. Further studies are required to optimize parameters for mutant selectivity, as pan-RAS inhibition could pose toxicity issues.
Approaches to target RAS indirectly
Normal RAS activation requires nucleotide exchange, processing, membrane localization and effector binding. Altering one of these essential steps can be used to indirectly inhibit RAS activation.
Inhibitors of the nucleotide exchange cycle
SOS inhibitors.
Initial efforts to block GEF activity towards RAS identified a small molecule that bound KRAS between switch-I and switch-II (Kd = 190 μM), thereby inhibiting SOS binding and SOS-mediated nucleotide exchange47 (FIG. 5e). The next attempts to inhibit the SOS1–RAS interaction used a molecule designed to mimic an orthosteric SOS helix. Although the compound bound to SOS1 with nanomolar affinity, it had low cellular activity48,49. The field then shifted focus and tried to find small-molecule inhibitors of SOS1. Three groups identified small molecules that bound the CDC25 domain of SOS1 and a region adjacent to switch-II on RAS in the RAS–SOS1–RAS ternary complex50–52. Interestingly, binding of small molecules to this site could activate or inhibit of the SOS1–RAS interaction52. One group identified a small-molecule inhibitor (BAY-293; FIG. 1) that inhibited the SOS1–KRAS interaction at nanomolar levels52. However, BAY-293 weakly inhibited KRAS-mutant cell proliferation (IC50 = 3 μM) but more potently inhibited proliferation of wild-type KRAS cells (IC50 = 1 μM). Of note, BAY-293 and the KRAS-G12C covalent inhibitor ARS-853 showed synergistic growth-inhibitory effects in a KRAS-G12C cell model. This observation suggests that SOS1 inhibitors could be used in combination with KRAS-G12C inhibitors that bind to the GDP state, as SOS1 inhibition would increase the pool of GDP-bound KRAS-G12C. Currently, a SOS1 inhibitor, BI-1701963, is in a phase I clinical trial as a single agent and in combination with the MEK inhibitor trametinib (NCT04111458; FIG. 1; TABLES 1,2).
SHP2 inhibitors.
SHP2 is a non-receptor protein tyrosine phosphatase that is required for full activation of the MAPK pathway53. Mutations in PTPN11, which encodes SHP2, cause RASopathies and are found in about 50% of people with Noonan syndrome54–56. Although the biological function of SHP2 remains unclear, in the current model SHP2 functions as a scaffold protein, binds GRB2 and SOS1, and thereby increases RAS nucleotide exchange57–59. Inhibition of SHP2 would function similarly to a SOS1 inhibitor and block the loading of wild-type RAS with GTP. KRAS-mutant tumours depend on SHP2, as deletion of Ptpn11 in established tumours delays tumour progression, but does not induce tumour regression60.
SHP-099 (FIG. 1) was found, in a compound library screen for molecules that lock SHP2 in the auto-inhibited conformation, to potently and allosterically inhibit SHP2 (IC50 = 0.071 μM)61. SHP-099 synergistically reduces cell proliferation when combined with trametinib in KRAS-G12D patient-derived organoids and xenograft models of PDAC and NSCLC60,62. However, no sensitivity to SHP-099 was observed in a KRAS-G13D-mutant cell line, MDA-MB-213 (REF.61).
A potent and selective SHP2 allosteric inhibitor, RMC-4550 (FIG. 1), binds at the same site as SHP-099 and stabilizes the auto-inhibited conformation of SHP2 (REF.63). Treatment with RMC-4550 reduced cell proliferation in preclinical models but this effect was evident only in cells harbouring mutations in codon 12, not codon 13 or 61, of KRAS. Moreover, the greatest sensitivity was observed in KRASG12C-mutant cells; KRASG12D- or KRASG12V-mutant cells were modestly sensitive to the compound. This observation highlights that the biochemical properties of each mutation determine the extent to which RAS activity depends on guanine exchange, and that mutations with elevated intrinsic or GAP-mediated hydrolysis are particularly sensitive to SHP2 inhibition.
The clinical candidate derived from RMC-4550, RMC-4630 (FIG. 1), is currently in a phase I monotherapy clinical trial (NCT03634982; TABLE 1) and a phase Ib/II clinical trial in combination with another MEK inhibitor, cobimetinib (NCT03989115; TABLE 2). A second SHP2 allosteric inhibitor, JAB-3068 (FIG. 1), is currently in a phase I/II clinical trial and results have not been published (NCT03518554, NCT03565003; TABLE 1). A third SHP2 inhibitor, TNO155 (FIG. 1), is in a phase I monotherapy clinical trial (NCT03114319; TABLE 1) and a phase I/II clinical trial in combination with MRTX849 (REF.64) (NCT04330664; TABLE 2). Results from these studies have not been published.
Inhibitors of RAS processing
RAS is only active when localized to the cell membrane. To associate with the membrane, RAS requires three enzymatic post-translational processing steps: prenylation of the CAAX box by farnesyltransferase (FTase) or geranylgeranyltransferase (GGTase); cleavage of the terminal AAX residues by RAS-converting enzyme (RCE1); and methylation of the cysteine residue of the CAAX box by isoprenylcysteine carboxyl methyltransferase (ICMT). Inhibition of RAS processing should prevent membrane association and downstream RAS signalling.
Although FTase inhibitors (FTIs) were clinically disappointing in KRAS-mutant cancers, possibly because of the functional redundancy between FTase and GGTase, there is a renewed focus on the use of FTIs in HRAS-mutant cancers. Unlike KRAS and NRAS, HRAS is prenylated exclusively by FTase so FTIs could be useful for treating HRAS-mutant cancers65. Indeed, responses to the FTI tipifarnib were observed in patient-derived models of HRAS-mutant HNSCC and NSCLC66. Tipifarnib is currently in a phase II clinical trial for the treatment of HRAS-mutant HNSCC and thyroid cancer (NCT02383927; TABLE 1). In the reported results, six patients received tipifarnib of whom four had a PR and two had SD67.
A strategy to circumvent the compensation of FTIs by GGTase in KRAS-mutant and NRAS-mutant tumours is to target the downstream RAS processing enzymes, RCE1 and ICMT, which are essential in KRAS-mutant cells, but not wild-type KRAS cells68. By contrast, the β-subunit of FTase is essential in cells with either wild-type or mutant KRAS, so the complete loss of FTase activity is detrimental to all cells68. Inhibition of RCE1 or ICMT may provide mutant selectivity and reduce the toxicity with FTIs.
A small-molecule inhibitor of ICMT, cysmethynil (IC50 = 2.4 μM), impaired RAS membrane association and reduced cellular growth of RAS-mutant cell lines in vitro (half-maximal effective concentration (EC50) ≈ 20 μM) and in vivo69–71. Structural modification of cysmethynil improved the potency of ICMT inhibition but this compound did not inhibit cellular growth72. The most potent ICMT inhibitor to date (IC50 = 1.3 nM) only mildly reduces proliferation (EC50 = 0.3 to>10 μM)72. Recently, another ICMT inhibitor, UCM-1336 (IC50 = 2 μM), was found to impair membrane association of all four RAS isoforms, regardless of mutational status, and to reduce the cellular growth of RAS-mutant cell lines in vitro (EC50 = 2–12 μM) and in vivo73. UCM-1336 is structurally distinct from the cysmethynil-derived compounds, but still requires further enhancement for effective therapeutic use. No potent and selective inhibitors have been identified for RCE1 (REFS74,75).
After post-translational modification, RAS localization and trafficking is regulated by the prenyl-binding protein phosphodiesterase-δ (PDEδ), which binds to farnesylated RAS76. Deltarasin, a small molecule that binds to the farnesyl-binding pocket of PDEδ, prevents KRAS binding (Kd = 7.6 nM) and results in mislocalization of KRAS and reduced cell proliferation77. Computational docking later identified another compound, NHTD, which disrupts KRAS binding to PDEδ78. NHTD inhibited the growth of mutant KRAS tumour cells in vitro and in vivo (EC50 = 2–7 μM), but did not have the cytotoxic effects associated with deltarasin.
Notably, the enzymes discussed above also process other membrane-associated proteins, which could lead to off-target effects. Direct inhibition of RAS therefore has the most potential as a primary therapeutic option.
RAS oligomerization and effector binding
Once RAS is effectively processed, RAS proteins localize in the membrane where they undergo oligomerization or dimerization, which is required for effective RAS-driven signalling. RAS proteins were observed to organize into isoform-specific assemblies of five to nine proteins termed ‘nanoclusters’, which are important for the activation of downstream pathways79–85. More recently, RAS dimerization has emerged as another possible mechanism by which RAS can self-associate to enhance scaffolding and signalling activities86–89.
RAS dimers and oligomers are difficult to reconstitute in vitro and typically must be studied in the context of post-translationally modified RAS within reconstituted lipid membranes or nanodiscs. Owing to the absence of definitive structural and biochemical data, most efforts to define the molecular details of RAS–RAS interaction surfaces have been restricted to computational modelling combined with experimental validations, as well as leveraging information from crystal packing interactions across the hundreds of RAS structures deposited in the Protein Data Bank (PDB; see Related links)88,90–93. Although numerous interfaces have been identified and validated, many computational and experimental studies have converged on a single RAS dimerization interface, the α4–α5 interface, which is also prevalent as a crystal packing interaction in many of the HRAS, KRAS and NRAS structures.
The proposed α4–α5 interface can be targeted with a nanobody, NS1 (REF.92) (FIG. 5f). NS1 disrupts HRAS and KRAS self-association through directly binding the α4–α5 interface (Kd = 13 nM for HRAS and 65 nM for KRAS), reducing activation of downstream pathways and inhibiting cell proliferation, while leaving RAS localization and GTPase activity unperturbed92,94. Further work should define and validate these RAS–RAS interaction surfaces: a recent study demonstrated that fully processed KRAS remains monomeric in lipid membranes across a wide range of concentrations78.
A newly discovered mechanism of RAS auto-inhibition, membrane occlusion, has provided another potential avenue for targeting RAS protein–protein interactions. In membrane occlusion, direct interactions between RAS and the lipid membrane sequester the effector binding interface of RAS away from the cytosol95,96. A small molecule, Cmpd2, can bind to the interface between RAS and the lipid membrane (Kd = 1 μM), promoting membrane occlusion and reducing binding to the RBD domain of RAF95. Owing to the highly conserved nature of the RAS–effector interface (FIG. 5g), this type of strategy provides an opportunity to effectively inhibit all downstream signalling pathways driven by RAS.
Targeting the RAS pathways
Two distinct approaches exist to target the RAS pathways: identifying genes that are synthetically lethal with RAS mutations, or targeting the tyrosine kinase receptors (EGFR family) and RAS effector pathways, namely MAPK and PI3K.
Synthetic lethal screens.
The Broad Institute dependency map (DepMap) portal compiles gene essentiality scores from CRISPR screens in hundreds of cancer cell lines (see Related links). In KRAS-mutant and NRAS-mutant cell lines, besides RAS itself, the top essential genes are RAF1 (which encodes CRAF) and SHOC2. An independent study showed that RAF1 and SHOC2 are required in RAS-mutant human acute myeloid leukaemia cell lines68.
Ablation of Craf in a KrasG12V;Trp53−/− LUAD mouse model significantly reduced tumour size97. The authors observed no adverse systemic effects of CRAF ablation and the loss of CRAF did not alter MAPK signalling, suggesting that kinase-independent CRAF functions may play a role in this context. Furthermore, combined ablation of Egfr and Craf in KrasG12V;Trp53−/− PDAC mouse models induced complete regression of PDAC tumours98. Again, the dual ablation of Craf and Egfr was well tolerated in the GEMMs. In a second approach, using PDAC patient-derived xenografts (PDXs), the dual knockdown of EGFR and CRAF expression also reduced tumour growth. These findings highlight the therapeutic advantage of inhibiting CRAF and suggest the need for selective inhibitors against CRAF. However, novel approaches are likely to be required for selective CRAF inhibition because of the high homology between RAF family kinase domains and the fact that CRAF may have a kinase-independent function in this context.
A phosphatase complex containing SHOC2, MRAS and protein phosphatase 1 (PP1) dephosphorylates RAF at Ser259. This dephosphorylation alleviates the negative regulation of RAF by 14-3-3 and enables RAF dimerization99. Therefore, SHOC2 promotes RAF dimerization and, importantly, SHOC2 is required for maximal ERK activity. Ablation of Shoc2 in a KrasG12D;Trp53R172H LUAD mouse model reduced tumour burden and pro longed survival without toxicity100. In a second model, genetic deletion of SHOC2 in KRAS-mutant and EGFR-mutant NSCLC cell lines reduced growth in xenograft models and sensitized cells to MEK inhibition (using selumetinib, trametinib, PD0325901 or pimasertib). A CRISPR–Cas9 screen identified that SHOC2 loss is synthetically lethal with trametinib treatment101. These findings suggest that SHOC2 inhibitors could not only provide therapeutic benefit as a single agents, but potentially also in combination with MEK inhibitors.
EGFR family therapies.
There is also evidence of interplay between RAS and the EGFR family of receptor tyrosine kinases. As the EGFR family is upstream of RAS, inactivation of these receptor tyrosine kinases can reduce RAS activation36. Until recently, studies have focused on inhibition of EGFR in particular for the treatment of RAS-mutant tumours.
The use of cetuximab or panitumumab, monoclonal antibodies against EGFR, improved overall and progression-free survival in patients with metastatic CRC and wild-type KRAS alleles (also with no mutations in NRAS, BRAF or PI3KCA, which encodes the catalytic subunit of PI3Kα), but these antibodies had no effect in KRAS-mutant tumours, specifically those with mutations at codons 12 or 13 (REFS102,103). The results of these clinical trials led to the approval of the use of cetuximab and panitumumab in the treatment of metastatic CRCs that lack mutations in KRAS. Additionally, EGFR tyrosine kinase inhibitors (TKIs), such as erlotinib and gefitinib, which are approved for the treatment of EGFR-mutant NSCLC, are ineffective as a monotherapy in KRAS-mutant NSCLC104–107. Erlotinib is approved in combination with gemcitabine for the treatment of PDAC, albeit with limited benefit: over 90% of patients with PDAC have KRAS mutations108.
A retrospective study of the clinical trials of cetuximab and panitumumab in metastatic CRC analysed their clinical benefit in relation to the specific codon mutation. Patients with KRAS-G13D-mutated tumours (n = 32) had increased overall survival and progression-free survival compared with those with other KRAS mutations when receiving cetuximab109. However, in a second retrospective analysis of panitumumab, patients with KRAS-G13D-mutant tumours were deemed unlikely to benefit from this treatment110. A phase II clinical trial, ICECREAM, is underway to evaluate the efficacy of cetuximab treatment in patients with metastatic CRC with KRASG13D mutations (ACTRN12612000901808; TABLE 2)111.
Although wild-type KRAS CRCs respond to cetuximab treatment, ultimately metastatic CRCs become refractory to anti-EGFR treatment and acquire KRAS mutations as a mechanism of resistance. In one study, these mutations included KRAS amplification (1 of 11) or KRAS somatic mutations, most commonly G13D (5 of 11)112. However, it is unclear whether these mutations were present prior to cetuximab treatment in a KRAS-mutant clone, or arose as a result of treatment.
Genetic depletion of Egfr suppresses the development of KrasG12D-driven PDAC and NSCLC in GEMMs113–115. Upregulation of EGFR family members other than EGFR itself, such as ERBB2, ERBB3 and ERBB4, was found as an early event in tumours with either KRAS-G12D or erlotinib resistance115,116. Interestingly, patients with NSCLC, including those with mutations in EGFR or amplification of EGFR, HER2, HER3 or HER4, had poor survival117. Treatment of autochthonous KRAS-G12D tumours with the FDA-approved pan-ERBB inhibitors, afatinib or neratinib, reduced both tumour size and initiation compared with erlotinib or gefitinib treatment115,116. Currently, a phase I/II clinical trial is evaluating the efficacy of neratinib in RAS-mutant solid tumours in combination with the histone deacetylase inhibitor divalproex sodium118 (NCT03919292; TABLE 2).
Pan-ERBB and EGFR inhibitors have promise in combination with MEK or KRAS-G12C covalent inhibitors. ERBB3 suppression with afatinib sensitized KRAS-mutant CRC and NSCLC cell lines to MEK inhibition (by selumetinib)119. Afatinib or neratinib treatment, in combination with MEK inhibition (by trametinib), increased the survival of KRAS-G12D NSCLC mouse models116. The animals were treated daily for one week and monitored for survival, indicating that this combinatorial approach induces a durable response. Treatment of KRAS-G12C cells with ARS-853 and either erlotinib or gefitinib gave a synergistic effect and erlotinib decreased the fraction of KRAS-G12C bound to GTP35,36. As ARS-853 and other KRAS-G12C covalent inhibitors bind to KRAS-G12C in the GDP-bound state, the efficacy of EGFR TKIs suggests that EGFR is capable of activating mutant RAS and the inhibition of EGFR reduces the amount of GTP-bound KRAS-G12C, providing increased efficacy.
MAPK pathway: RAF inhibitors.
Active GTP-bound RAS promotes RAF dimerization and phosphorylation, which induces RAF kinase activity and results in phosphorylation of the RAF substrates MEK1 and MEK2. The phosphorylation cascade continues with MEK phosphorylation of the terminal kinases, ERK1 and ERK2. ERK kinase activity both activates growth-promoting transcription factors, including members of the ETS family, and participates in negative feedback loops120. This dynamic process controls the duration and amplitude of the MAPK signalling cascade. ERK can negatively suppress MAPK signalling by phosphorylating upstream components, SOS or CRAF, or by altering the transcription of the dual-specific phosphatase (DUSP) family and the Sprouty (SPRY) family121–124. DUSPs dephosphorylate ERK and SPRYs inhibit receptor tyrosine kinase signalling by sequestering SOS–GRB2 (REFS125,126). To effectively treat RAS-mutant tumours, the MAPK pathway must be almost completely suppressed. One strategy to achieve this is to use kinase inhibitors against components of the MAPK pathway, in combination with therapies that target other mechanisms discussed in this Review.
Currently, kinase inhibitors targeting BRAF-V600 and MEK are approved for BRAFV600-mutant metastatic melanoma but not for RAS-mutant tumours. Clinically approved BRAF-V600 inhibitors, such as vemurafenib and dabrafenib, induce an outward shift of the αC helix in the kinase domain of RAF127. These inhibitors effectively inhibit RAF monomers (BRAF-V600 signals as a monomer) but cannot be used for RAS-mutant tumours, which signal through BRAF and CRAF dimers. Furthermore, in RAS-mutant tumours, these inhibitors have been shown to paradoxically activate the MAPK pathway by binding to wild-type RAF, inducing RAF dimerization and the downstream phosphorylation of MEK and ERK128,129. Paradoxical activation depends on the mode of inhibitor binding: RAF dimer inhibitors exhibit far less paradoxical activation than approved BRAF-V600 inhibitors in RAS-mutant tumours130.
RAF dimer inhibitors, such as AZ-628, belvarafenib, LY3009120 and LXH-254, bind RAF in a DFG-motif ‘out’, αC helix ‘in’ active position131–133 (FIG. 1). These inhibitors are effective against RAF monomers and dimers, and although they can also promote dimerization, they have minimal paradoxical activation, as they can bind both dimer components. Many of these inhibitors act as pan-RAF inhibitors and have been reported to have efficacy in RAS-mutant and BRAF-mutant tumours131,132,134–139. A phase I clinical trial evaluating LY3009120 was terminated owing to lack of clinical efficacy as the best overall outcome was SD (observed in eight patients (15%))140.
Currently, two pan-RAF inhibitors are under phase I clinical evaluation for the treatment of RAS-mutant tumours: belvarafenib (NCT02405065, NCT03118817; TABLE 1) and LXH-254 (NCT02607813; TABLE 1). Belvarafenib has clinical activity as a monotherapy in subsets of patients with BRAF-mutant and NRAS-mutant tumours, with a PR observed in two of six individuals with BRAF-mutant melanoma, two of seven with BRAF-mutant CRC and two of nine with NRAS-mutant melanoma138. Treatment with belvarafenib, unlike BRAF-V600 inhibitors, did not induce squamous cell carcinomas, a known result of paradoxical activation138,141,142.
MAPK pathway: MEK inhibitors.
Currently, there are three allosteric kinase inhibitors targeting MEK — cobimetinib, trametinib and binimetinib — clinically approved for the treatment of BRAFV600-mutant melanoma. Clinical trials evaluating MEK inhibitors as monotherapies for RAS-mutant tumours have found no improvement and none of the MEK inhibitors are clinically approved for treating RAS-mutant tumours143–147. MEK inhibitors, like RAF inhibitors, induce pathway feedback loops in RAS-mutant tumours, resulting in relatively modest efficacy in these tumours123,148–150.
Although single-agent MEK inhibitors have largely failed in the clinic for RAS-mutant tumours, NRAS-mutant melanomas have some sensitivity151,152 (FIG. 1). For example, in a phase II trial of binimetinib, 20% of people with NRAS-mutant melanomas had a PR151. Based on these data, a phase III trial of binimetinib compared with dacarbazine is ongoing to evaluate the efficacy in treating patients with NRASQ61 mutations (NCT01763164; TABLE 1). Additionally, in a phase II trial, pimasertib improved progression-free survival compared with dacarbazine in patients with NRAS-mutant melanoma (NCT01693068)152. In the overall patient population, pimasertib failed to improve overall survival compared with dacarbazine and did not receive FDA approval153. Notably, similar clinical trials of MEK inhibitors for KRAS-mutant PDAC, CRC and NSCLC have shown no benefit over the standard of care147,154–156. In cell culture, NRAS-mutant melanoma cells have greater sensitivity to pan-RAF inhibition than KRAS-mutant cells and have comparable sensitivity to BRAFV600E cells130. The differential sensitivity for NRAS-mutant melanoma requires further exploration to determine whether the sensitivity is due to an intrinsic property of melanoma, mutant NRAS or the biochemical properties of codon 61 mutations.
As MEK or RAF inhibitor monotherapy failed to provide clinical benefit for KRAS-mutant tumours, the efficacy of these inhibitors used in combination is being explored in the clinic. Owing to the complexity of the feedback loops within the MAPK pathway, targeting multiple nodes of the pathway could lead to sustained and durable suppression of phosphorylated ERK (a measure of activated ERK and a direct target of MEK; phosphorylated ERK is commonly used to measure overall MAPK pathway activity). Indeed, the combination of MEK and RAF inhibitors exhibited synergy in preclinical models of RAS-mutant tumour cells and blocked pathway reactivation130. Furthermore, the induction of phosphorylated MEK and GTP-bound RAS upon MEK inhibitor treatment is required for the synergistic effect of MEK and RAF combination treatment130. The strongest synergistic effects of MEK and RAF inhibition were observed in cells with mutant KRAS with high levels of intrinsic nucleotide exchange, such as KRAS-G13D, which enhances GTP-bound RAS levels: this observation suggests that RAF dimerization, induced by MEK inhibition, probably contributes to this synergy. Currently, two phase I clinical trials combining RAF and MEK inhibitors are underway. One is investigating the combination of belvarafenib and cobimetinib (NCT03284502; TABLE 2) and the other is investigating the combination of trametinib and LXH-254 (NCT02974725; TABLE 2).
MAPK pathway: ERK inhibitors.
Inhibition of ERK, the culminating kinase in the cascade, could directly reduce the oncogenic transcriptional output and provide a valuable therapeutic option for tumours that are resistant to MEK or RAF inhibition157,158. The clinical development of ERK inhibitors lags behind the development of MEK, BRAF monomer and pan-RAF inhibitors. In addition, the early clinical trials of ERK inhibitors for the treatment of RAS-mutant tumours have been largely unsuccessful (FIG. 1).
The preclinical compound SCH-772984 (FIG. 1) acts as a dual mechanism ERK inhibitor by binding to and inhibiting ERK1/2 while also inducing a conformational shift that prevents ERK1/2 phosphorylation by upstream kinases159,160. Treatment of RAS-mutant cancer cell lines with SCH-772984 reduced levels of phosphorylated ERK and reduced cell proliferation161. The clinical compound, MK-8353, was developed by Merck to have improved pharmacokinetic properties compared with SCH-772984 (FIG. 1). Similar to SCH-772984, MK-8353 acts as a dual mechanism inhibitor and reduced cell proliferation of NRAS-mutant melanoma cell lines in preclinical models160,162. However, in a phase I monotherapy clinical trial for MK-8353, no antitumour responses were observed amongst the 26 patients with KRAS or NRAS mutations who were enrolled160. Three PRs, however, were observed in patients with BRAFV600-mutant melanoma. MK-8353 is under clinical evaluation in combination with selumetinib (NCT03745989; TABLE 2) and pembrolizumab (NCT02972034; TABLE 2) in trials that include patients with RAS-mutant tumours.
GDC-0994 (an ERK inhibitor, FIG. 1) exhibited efficacy in combination with cobimetinib in KRAS-mutant tumour models163. Although preclinical xenograft models showed a therapeutic dose leading to growth reduction could be achieved, the phase I clinical trial of cobimetinib and GDC-0994 was terminated because patients could not tolerate the combination163,164.
However, when GDC-0994 was evaluated as a monotherapy, in a phase I clinical trial, the recommended phase II dose of 400 mg daily on a 21-days-on/7-days-off schedule was tolerable165. In the trial, 14 patients with BRAF-mutant CRC or gastric cancer were treated with GDC-0994 and two patients had a PR, seven had SD and five had disease progression165. Of the 14 patients enrolled with KRAS-mutant tumours, four achieved SD and ten had progressive disease165. GDC-0994 induced MAPK pathway suppression (19–51%) in paired tumour biopsies using NanoString gene expression, and greater suppression was observed in patients with BRAF-mutant CRC (three of four) than in those with KRAS-mutant PDAC (one of four)165. Further evaluation of NRAS-mutant tumours is required in a phase II study as only one patient was evaluated at the lower dose of 100 mg daily165.
In a recently completed phase I clinical trial, ulixertinib (BVD-523; FIG. 1) showed antitumour effects in NRAS-mutant melanoma and BRAF-mutant solid tumours166. Ulixertinib is a selective, reversible, ATP-competitive ERK1/2 inhibitor166. In the trial, 17 patients with NRAS-mutant melanoma were treated with ulixertinib and three patients (18%) had a PR, six had SD and eight had disease progression166. Although these results are encouraging for NRAS-mutant melanomas, they suggest that ulixertinib may not work for KRAS-mutant tumours. Because NRAS-mutant tumours have historically responded better to MEK and pan-RAF inhibitors than KRAS-mutant tumours, KRAS-mutant tumours should continue to be evaluated. Ulixertinib is currently being evaluated in combination with chemotherapy, nanoparticle albumin-bound paclitaxel (nab-paclitaxel) and gemcitabine in patients with metastatic PDAC in a phase I clinical trial (NCT02608229; TABLE 2).
KO-947 (FIG. 1), currently in a phase I clinical trial for RAS-mutant and BRAF-mutant NSCLC, potently reduced levels of phosphorylated ERK in preclinical models167 (NCT03051035; TABLE 1). Sustained responses were observed; levels of phosphorylated ERK were suppressed for up to 5 days following a single dose in vitro167. These properties differ from those of the other ERK inhibitors discussed and suggest that KO-947 may provide a therapeutic advantage. However, this sustained suppression of phosphorylated ERK may not be well tolerated in patients.
LY-3214996 (FIG. 1), currently in a phase I clinical trial, is a potent and selective inhibitor in vitro (IC50 = 5 nM) of both ERK1 and ERK2 (REF.168) (NCT02857270; TABLE 1). In a dose-escalation arm, 33 patients with RAS-mutant and 16 with BRAF-mutant tumours were treated with LY-3214996 (REF.169). Seven patients with BRAF mutations had tumour regression, two had SD while one patient with RAS-mutant tumour had SD; the others had disease progression169.
Overall, single-agent MEK, RAF or ERK inhibitors show little efficacy in the treatment of RAS-mutant tumours. Therefore, these inhibitors will have to be used in combination with other inhibitors of the MAPK pathway or with other approaches discussed in this Review. Finding the optimal combination for each type of inhibitor will be a challenge. However, the advent of allele-specific RAS inhibitors has increased the number of potential combinations that can be used to achieve maximal pathway suppression.
PI3K pathway inhibitors.
Of all the RAS effectors, the MAPK pathway has been the main focus for inhibition of RAS-mutant tumours. However, a second effector pathway, PI3K, is also activated by RAS. The next sections describe in more detail the types of PI3K inhibitors and the efficacy of these inhibitors for treating RAS-driven tumours.
There are three classes (I–III) of PI3Ks. Class I PI3Ks, upon activation by GTP-bound RAS, phosphorylate phosphatidylinositol 4,5-bisphosphate (PIP2) to generate phosphatidylinositol (3,4,5)-trisphosphate (PIP3), which recruits AKT to the membrane and allows activation of mTOR. Class I PI3Ks are generally composed of a heterodimer consisting of a catalytic subunit, p110, which contains an RBD, and a regulatory subunit, p85. Three genes (PIK3CA, PIK3CB, PIK3CD and PIK3CG) encode the four p110 subunit isoforms — p110α, p110β, p110γ and p110δ, respectively170. The p110α and p110β isoforms are ubiquitously expressed, whereas p110γ and p110δ are generally only expressed in immune cells171,172. The p110β and p110γ isoforms can be activated by both G protein-coupled receptors and receptor tyrosine kinases173,174. Cancers upregulate the PI3K pathway through activating mutations in PIK3CA, amplification of AKT, or loss of PTEN, which encodes a terminal phosphatase that converts PIP3 to PIP2 (REFS175–177). Interestingly, PI3K pathway mutations can coexist with RAS mutations, whereas RAS mutations and MAPK pathway mutations are mutually exclusive, such as EGFR or BRAF4. This suggests that RAS mutations are sufficient to dysregulate the MAPK but not the PI3K pathway. Activation of the PI3K pathway, which can occur in response to chemotherapy or MAPK inhibition, confers resistance to these two therapies178. Additionally, this suggests that the combined inhibition of MAPK and PI3K could be efficacious in treating RAS-driven tumorigenesis.
For the purpose of this Review, we discuss p110α inhibitors, as p110α is ubiquitously expressed and exclusively activated by RAS, unlike p110γ and p110δ. Four class I PI3K inhibitors have received FDA approval, and one of these, alpelisib, is p110α-specific, and one, copanlisib, is pan-class I PI3K (FIG. 1). These inhibitors are not approved for the treatment of RAS-mutant tumours.
RAS activates both the PI3K and MAPK pathways, and there are overlapping feedback mechanisms that provide crosstalk. Inhibition of one pathway can lead to the compensatory activation of the other; therefore, the inhibition of both MAPK and PI3K is a compelling strategy179,180. Preclinical models suggested that the combined inhibition of PI3K and MEK is efficacious and clinically achievable for the treatment of RAS-mutant tumours181,182. However, in clinical trials, the combination of these inhibitors was not tolerated and had little efficacy, presumably as a result of dose reduction due to toxicity183–186.
To overcome this toxicity, studies focused on identifying receptor tyrosine kinases that could suppress PI3K, MAPK or both pathways. One such receptor, insulin-like growth factor 1 receptor (IGF1R), is required for mutant-RAS-mediated activation of PI3K187,188. Combined inhibition of IGF1R and MEK synergized effectively in CRC and NSCLC models187,188. Furthermore, replacing the MEK inhibitor with a KRAS-G12C covalent inhibitor in the combination with an IGF1R inhibitor, linsitinib, improved the efficacy and tolerability of this combination in mouse models189. The efficacy and tolerability of combinations of IGF1R and KRAS-G12C covalent inhibitors will need to be evaluated in a clinical setting.
No AKT inhibitors are currently approved for clinical use, in RAS-mutant tumours or otherwise (FIG. 1). The combination of AKT and MEK inhibitors is under clinical evaluation but toxicity issues similar to those observed in the PI3K–MAPK inhibitor studies have been reported190,191. The combination of mTOR inhibitors (everolimus or temsirolimus) with MEK inhibitors (trametinib or pimasertib) also had poor tolerability192,193 (FIG. 1). A recent review on targeting the PI3K pathway details the clinical status of AKT and mTOR inhibitors194.
As the tissue type dictates the expression of p110 isoforms and the prevalence of RAS or PI3K mutations, further studies are required to determine the best setting for combinatorial approaches. Isoform-specific p110 inhibitors are expected to have fewer off-target effects and better toxicity profiles as they should more specifically target malignant cells. As p110δ/p110γ inhibitors have been approved for the treatment of chronic lymphocytic leukaemia and small lymphocytic lymphoma, and these isoforms are exclusively expressed in leukocytes, the use of these inhibitors could be efficacious in treating RAS-driven leukaemias. PI3K therapies should also be evaluated in combination with other approaches discussed in this Review, such as allele-specific inhibitors, including those for KRAS-G12C.
Emerging therapeutics
Small interfering RNA therapies
Systemic delivery of nanoparticles containing KRAS-targeting small interfering RNA (siRNA) in mouse models suggests that these could be effective approaches for targeting KRAS and could be mutant-specific195. However, a chemically modified antisense oligonucleotide, AZD4785, which significantly reduced levels of KRAS following subcutaneous injection in preclinical models, failed to sufficiently reduce KRAS levels in patients in a clinical trial (NCT03101839). Work is ongoing to improve uptake and internalization and to improve our understanding of how to make this approach more effective. A mutant-specific siRNA against KRAS-G12D, siG12D-LODER, showed promise in a phase I trial in combination with chemotherapy in 12 patients with PDAC; two patients achieved a PR while ten achieved SD (NCT01188785)196. An ongoing phase II trial in patients with KRASG12D PDAC will evaluate siG12D-LODER in combination with gemcitabine and nab-paclitaxel (NCT01676259; TABLE 2).
Autophagy
Genetic suppression of KRAS with siRNA or short hairpin RNA or by pharmacological inhibition of the MAPK pathway using inhibitors against KRAS-G12C (ARS-853, ARS-1620), MEK (trametinib, cobimetinib) or ERK (SCH772984) increases autophagy197,198. PDAC cells have increased levels of autophagy and these cells require autophagy for growth, as pharmacological inhibition or genetic depletion of autophagy have induced tumour regression in preclinical models199,200. From these studies, a clinical study that inhibited autophagy with hydroxychloroquine, which is FDA-approved for the treatment of malaria, as a monotherapy was performed in 20 patients with PDAC and showed limited activity, as 18 of the 20 patients had disease progression201,202. Results from the use of hydroxychloroquine for preoperative care in combination with chemotherapy (gemcitabine plus nab-paclitaxel) were promising, as this treatment increased overall survival and decreased levels of the blood-borne tumour biomarker carbohydrate antigen 19–9 (CA 19–9)203.
Although the treatment of PDACs with hydroxychloroquine had limited efficacy, the combination of hydroxychloroquine and inhibitors of the MAPK pathway showed promising results in preclinical models of PDAC and NRAS-mutant melanoma197,198. Combined treatment of hydroxychloroquine and trametinib resulted in robust tumour regression in xenograft and PDX models197. Treatment of a single patient with metastatic PDAC, who was refractory to all standard-of-care therapeutic options, with a combination of trametinib and hydroxychloroquine profoundly reduced CA 19–9 levels and induced a PR with a 50% reduction in tumour voume197. With this promising result, a phase I clinical trial to investigate the efficacy of trametinib plus hydroxychloroquine for the treatment of patients with PDAC is currently under evaluation (NCT03825289; TABLE 2).
Immunotherapy
Immune checkpoint inhibitors.
Tumours evade detection of the immune system through negative regulatory antigens (checkpoints). The widely discussed immune checkpoints include cytotoxic T lymphocyte protein 4 (CTLA4), PD1 and PDL1. CTLA4 negatively regulates T cell activation. PD1 is expressed on T cells and generates an intracellular inhibitory signal when bound to PDL1 (REF.204). Tumours express PDL1 on the cell surface, so this interaction inhibits immune activity in the vicinity of the tumour.
Seven antibodies that target immune checkpoint proteins have received FDA approval: one anti-CTLA4 (ipilimumab), three anti-PD1 (nivolumab, pembrolizumab, cemiplimab) and three anti-PDL1 (atezolizumab, avelumab, durvalumab) antibodies. Immunotherapies are approved for the treatment of NSCLC and melanoma, two of the four main RAS-mutant tumour types. Ipilimumab is currently approved as a monotherapy in metastatic melanoma, as an adjuvant therapy in melanoma205,206 and in combination with nivolumab for the treatment of advanced NSCLC and melanoma207,208. The anti-PD1 antibodies nivolumab and pembrolizumab are currently approved for the treatment of unresectable or metastatic melanoma, as adjuvant therapy for melanoma (nivolumab) and for the treatment of advanced NSCLC209–216. The anti-PDL1 antibodies atezolizumab and durvalumab are also approved for the treatment of NSCLC (NCT02951767, NCT02108652)217–219. Clinical trials are underway to evaluate the efficacy of these anti-PDL1 antibodies for the treatment of melanoma (NCT02535078, NCT01772004)220,221.
High mutational burden, high expression levels of PDL1 and increased numbers of tumour-infiltrating lymphocytes (TILs) are predictive of a response to immunotherapy222–224. PDAC and CRCs have low immunogenicity because they have immunosuppressive microenvironments and low mutational burdens204,225–227. Unsurprisingly, immunotherapies are not therapeutically beneficial in these tumours228–230. However, a subset of patients with PDAC or CRC that show high levels of microsatellite instability (MSI-H) or deficient mismatch repair (dMMR) respond to immunotherapies; fewer than 15% of CRCs and 20% of PDACs are MSI-H231–234. Given this success, pembrolizumab is approved for the treatment of MSI-H solid tumours and nivolumab is approved for MSI-H or dMMR CRC.
The clinical response to anti-PD1 and anti-PDL1 antibodies in NSCLC is limited to a subset of patients; roughly 10–20% of patients have a clinical response to these antibodies when used as single agents235. The mutational landscape of NSCLC can dictate the response to checkpoint antibodies. In KRAS-mutant NSCLC, the presence of LKB1 mutations reduces the overall response rate (7.4%) to PD1 blockade, whereas TP53 mutations increase the response rate (35.7%) relative to tumours with KRAS mutations alone (28.6%)236.
Combination treatment of allele-specific RAS inhibitors or inhibitors of the MAPK pathway and immunotherapies could improve the response of RAS-mutant tumours to immunotherapy. Interestingly, a clinical trial showed that NSCLC tumours with KRAS mutations had increased levels of PDL1, which could provide these tumours with immunoresistance215. Mutant KRAS directly upregulates PDL1 expression; this upregulation is reversed with a MEK inhibitor (trametinib) or a selective KRAS-G12C inhibitor (ARS-853)237. Additionally, in immune-competent mouse models, treatment with AMG 510 induced a pro-inflammatory tumour microenvironment, with an increased number of TILs, and the drug synergized with anti-PD1 treatment32. The combination of AMG 510 and anti-PD1 or anti-PDL1 for the treatment of NSCLC is being investigated in a phase II clinical trial (NCT03600883; TABLE 2). MEK inhibition also increases the number of TILs and, when combined with anti-PDL1 treatment, synergistically induces tumour regression238. Indeed, there is a phase II clinical trial investigating the efficacy of the combination of atezolizumab and cobimetinib in the treatment of NSCLC (NCT03600701; TABLE 2), although this combination did not provide a clinical benefit in metastatic CRC145. Additionally, a phase I clinical trial is currently evaluating spartalizumab, an anti-PD1 antibody, in combination with TNO155, a SHP2 inhibitor (NCT04000529; TABLE 2).
Adoptive cell therapy.
A second immunotherapeutic approach to treat RAS-driven cancers involves engineering the immune system to recognize antigens specific to mutant RAS proteins. This approach uses adoptive cell therapy to transfer T cells (either TILs or transgenic T cells), which have been expanded ex vivo, into patients. TILs recognize specific antigens displayed on tumours and transferring expanded TILs to patients provides clinical benefit in melanoma239,240. Technical developments in engineering T cell receptors (TCRs) and expressing them in peripheral blood lymphocytes achieves similar clinical benefits and allows the expansion of this technology beyond individual patients241.
Given the successful identification of tumour-specific antigens in melanoma, a similar approach was used to identify KRAS-mutant specific antigens. From a patient with metastatic CRC, CD8+ TILs were identified that specifically recognized KRAS-G12D and, upon infusion of the expanded TILs, the patient had tumour regression in seven pulmonary metastatic lesions with a durable PR lasting 9 months242. Another study identified KRAS-G12V-mutant-specific TCRs in CD4+ T cells from a patient with NSCLC243. These studies showed that antigens from KRAS-G12D or KRAS-G12V are immunogenic in humans.
These antigens can then be used to engineer T cells. Wang and colleagues immunized mice bearing a transgene for a human leukocyte antigen, HLA-A*11:01, with mutant RAS peptides to generate T cells that recognized these mutations244. TCRs from the mouse T cells were identified, cloned and retrovirally transduced into peripheral blood lymphocytes to prime the T cells against RAS-G12D or RAS-G12V antigens244. In a xenograft model, these engineered mouse T cells detected human KRAS-mutant PDAC cells, leading to tumour reduction or complete regression244. Two clinical trials are currently using this technology to transduce human peripheral blood lymphocytes with the murine TCRs against RAS-G12D or RAS-G12V in patients with HLA-A*11:01 (NCT03745326, NCT03190941; TABLE 1).
Cancer vaccines.
A third immunotherapeutic approach to treat RAS-mutant tumours uses known RAS-mutant tumour antigens to elicit T cell responses through vaccination. One such approach involves intradermal injection of peptides from mutant RAS proteins in combination with granulocyte–macrophage colony-stimulating factor (GM–CSF)245. This stimulation activates dendritic cells and triggers a T cell response against the mutant pepetides245. In a phase I/II clinical trial in patients with PDAC treated with a mutant-RAS-specific vaccine, Targovax TG-01, patients had an increased immune response and increased overall survival (NCT02261714)246. A second-generation vaccine, TG-02, has been used to treat patients with CRC but results have not been published (NCT02933944).
A second approach uses an mRNA that encodes neo-epitopes for common KRAS mutations (G12C, G12D, G12V and G13D)247. A lipid nanoparticle-formulated mRNA vaccine is given intramuscularly and the mRNA nanoparticle is taken up by antigen-presenting cells and translated and presented on the cell surface, which leads to T cell responses to the mutant RAS neo-epitopes. A phase I clinical trial is underway to evaluate one such mRNA vaccine, mRNA-5671, in patients with KRAS mutations, in which the mRNA is used either as a single agent or in combination with pembrolizumab (NCT03948763; TABLES 1,2).
Future directions and conclusions
KRAS-G12C allele-specific inhibitors will change the treatment landscape for RAS-driven tumours. These inhibitors are expected to be the first FDA-approved therapeutics for RAS-mutant tumours and will be used to treat refractory cancers driven by mutant RAS, such as PDAC, CRC and LUAD. Although the development of these inhibitors is incredibly exciting, new challenges and questions will arise.
There will be continued development of inhibitors specific to other alleles, such as KRAS-G12D and KRAS-G12V. These alleles are the most common KRAS variants and are therefore associated with the largest patient populations. Eventually, specific inhibitors could be developed for all mutant RAS alleles, providing a personalized medicine approach. Targeting mutant RAS proteins is the best approach for RAS-mutant tumours; however, allele-specific inhibitors will probably have limited efficacy as monotherapies. The greatest antitumour effects will require combinations with other inhibitors.
Determining which combination strategies will work best in patients will be challenging for several reasons. First, each variation in RAS has distinct biochemical properties and these properties will determine the response to many of the therapeutic approaches discussed. For example, SHP2 inhibition using RMC-4550 was effective in treating cells with KRASG12 mutations, and was more effective against KRASG12C than against KRASG12D or KRASG12V (REF.63). This observation suggests that a combination of SHP2 and KRAS-G12C inhibitors would be an effective therapeutic strategy. Understanding both the requirements of specific mutant RAS codons and the response of inhibitors to these alleles will be necessary in developing strategic combination therapies.
Second, as discussed throughout this Review, the tumour type can dramatically impact the response rate. RAS-mutant CRC and PDAC have a minimal response to inhibitors of MAPK or immune checkpoint blockade. Early data from the AMG 510 trial showed that CRC is more refractory to treatment than LUAD, suggesting that CRC will require combination therapies1. CRC tumours are particularly challenging to treat. However, promising antitumour effects have been observed in BRAFV600E metastatic CRCs treated with the triple combination of binimetinib, encorafenib and cetuximab, suggesting that KRAS-mutant CRC will require an aggressive combination strategy to achieve a response248.
Third, historically, combination treatments have been toxic and have poor safety profiles. However, the lack of dose-limiting toxicities observed with AMG 510 is encouraging and mutant-specific therapies should have limited off-target effects. Allele-specific inhibitors (with low toxicity) could be combined with other inhibitors that have greater toxicity, without encountering the problems observed with combining two toxic compounds, as occurred with combinations of the PI3K and MEK inhibitors.
As the treatment of RAS-driven tumours becomes more personalized, potential resistance mechanisms to allele-specific inhibition need to be evaluated. The heterogeneity of tumours could provide intrinsic mechanisms of resistance. For example, one tumour may contain 95% KRASG12C and 0.1% KRASG12V cells. Upon treatment with an allele-specific KRAS-G12C inhibitor, the tumour will regress but ultimately the KRASG12V cells will be selected for and the tumour will relapse. Tumour heterogeneity also can lead to intrinsic resistance, as subpopulations of cells are resistant to KRAS-G12C inhibition249. Preclinical models have not accounted for tumour heterogeneity and responses in the clinic will clarify this effect. Currently, there is little evidence regarding the types of de novo mutations that will arise upon treatment with KRAS-G12C allele-specific inhibitors.
The mechanisms of resistance that have been described for EGFR and BRAF-V600 inhibitors may shed some light as to what de novo mutations we might expect to see clinically with allele-specific KRAS-G12C inhibitors. Cysteine mutations are one expected mechanism of resistance, as these have been observed for covalent EGFR inhibitors250,251; however, in the case of KRAS-G12C, the cysteine mutation is required for oncogenic activity and hence it is less likely that this will be observed clinically unless the residue is mutated to another oncogenic KRAS-G12-mutant allele (such as to an aspartate, valine or other residue). Gene amplification, including BRAF or RAS, has been described as a mechanism of resistance to BRAF-V600 inhibitors252–254, and MET amplification has been observed in the context of EGFR inhibitors255,256. In this case, amplification of EGFR or another upstream EGFR family member could drive elevated GTP-bound RAS levels in cells, conferring resistance to the KRAS-G12C inhibitors as these molecules bind to the GDP-bound form. Additionally, RAS amplification, either HRAS or NRAS, or amplification of KRAS itself could potentially drive RAS dimerization and elevated GTP-bound RAS levels, thereby conferring resistance to the KRAS-G12C inhibitors. The majority of acquired resistance mechanisms in BRAF-mutant tumours reactivate the MAPK pathway and, in addition to the above mechanisms, this can be achieved by mutations in NRAS and MEK1/2 (REFS257–259). Therefore, reactivation of the MAPK pathway poses numerous conceivable mechanisms of resistance to allele-specific inhibitors and could be achieved through either altering upstream signalling (through EGFR mutations or amplification or RAS amplification), or downstream mechanisms, such as RAF or MEK mutations. Reactivation of EGFR can also occur through autocrine and/or paracrine mechanisms such as secretion of proheparin-binding EGF-like growth factor249.
Cell state changes, such as a switch from NSCLC to SCLC260,261 or upregulation of the master regulator MITF262,263, in melanoma have been described as resistance mechanisms to EGFR and BRAF-V600 inhibitors. Additionally, epithelial–mesenchymal transition reprogramming has been associated with resistance to EGFR inhibition in NSCLC249,260,264,265 and with BRAF-V600 inhibition in melanoma266,267. Certainly, transcriptional mechanisms of resistance to KRAS-G12C inhibitors could ultimately render the tumour cell state independent of mutant KRAS. Each of these resistance mechanisms will likely require novel combination approaches in order to achieve more complete and durable suppression of RAS-driven tumours.
Mutations that disable the GTPase activity of RAS are another hypothetical mechanism of resistance. In agreement with this hypothesis, a CRISPR screen showed that loss of NF1 promotes resistance37. Importantly, loss of NF1 renders BRAFV600E tumours resistant to vemurafenib treatment, suggesting that RAS-mutant tumours may utilize a similar approach268. Additionally, when a mutation blocking GTPase activity (A59G) was introduced in conjunction with KRAS-G12C, allele-specific KRAS-G12C inhibitors had a diminished effect as KRAS-G12C was predominately in the GTP-bound state34,35. However, it is unclear whether these predicted mechanisms would be consistent across RAS mutations or tumour types.
This is an exciting time to be at the forefront of ‘drugging the undruggable’ as there is new optimism that RAS-mutant cancers can be successfully treated.
Glossary
- Allosteric
A site that is outside the active site of an enzyme.
- RASopathies
A group of clinically defined genetic syndromes caused by germline mutations of regulators or components of the MAPK pathway.
- Noonan syndrome
An autosomal dominant RASopathy characterized by distinctive craniofacial features. Frequently germline mutated genes in Noonan syndrome include PTPN11, SOS1, RAF1, KRAS, NRAS, MRAS, SHOC2, CBL and RIT1.
Footnotes
Competing interests
F.McC. is a consultant for the following companies: Amgen, Pfizer Inc., and Quanta Therapeutics; is a consultant and co-founder with ownership interest including stock options of BridgeBio Pharma, Inc; and is Scientific Director of the NCI Ras Initiative at Frederick National Laboratory for Cancer Research/Leidos Biomedical Research Inc. S.M. is an employee of Genentech/Roche. A.R.M. and S.C.R. are also post-doctoral fellows employed by Genentech/Roche.
References
- 1.Amgen. Amgen Announces New Clinical Data Evaluating Novel Investigational KRAS(G12C) Inhibitor in Larger Patient Group at WCLC 2019 www.amgen.com https://www.amgen.com/media/news-releases/2019/09/amgen-announces-new-clinical-data-evaluating-novel-investigational-krasg12c-inhibitor-in-larger-patient-group-at-wclc-2019/ (2019).
- 2.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Network. Genomic classification of cutaneous melanoma. Cell 161, 1681–1696 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Network. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell 32, 185–203.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox AD, Fesik SW, Kimmelman AC, Luo J & Der CJ Drugging the undruggable RAS: mission possible? Nat. Rev. Drug Discov 13, 828–851 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517, 576–582 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robertson AG et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell 171, 540–556 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haigis KM et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat. Genet 40, 600–608 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burd CE et al. Mutation-specific RAS oncogenicity explains NRAS codon 61 selection in melanoma. Cancer Discov. 4, 1418–1429 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laude AJ & Prior IA Palmitoylation and localisation of RAS isoforms are modulated by the hypervariable linker domain. J. Cell Sci 121, 421–427 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Tsai FD et al. K-Ras4A splice variant is widely expressed in cancer and uses a hybrid membrane-targeting motif. Proc. Natl Acad. Sci. USA 112, 779–784 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGrath JP et al. Structure and organization of the human Ki-ras proto-oncogene and a related processed pseudogene. Nature 304, 501–506 (1983). [DOI] [PubMed] [Google Scholar]
- 14.Pells S et al. Developmentally-regulated expression of murine K-ras isoforms. Oncogene 15, 1781–1786 (1997). [DOI] [PubMed] [Google Scholar]
- 15.Plowman SJ et al. K-ras 4A and 4B are co-expressed widely in human tissues, and their ratio is altered in sporadic colorectal cancer. J. Exp. Clin. Cancer Res 25, 259–267 (2006). [PubMed] [Google Scholar]
- 16.Plowman SJ et al. While K-ras is essential for mouse development, expression of the K-ras 4A splice variant is dispensable. Mol. Cell. Biol 23, 9245–9250 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen W-C et al. Regulation of KRAS4A/B splicing in cancer stem cells by the RBM39 splicing complex. Preprint at bioRxiv 10.1101/646125 (2019). [DOI] [Google Scholar]
- 18.Bonfini L, Karlovich CA, Dasgupta C & Banerjee U The Son of sevenless gene product: a putative activator of Ras. Science 255, 603–606 (1992). [DOI] [PubMed] [Google Scholar]
- 19.Buday LDJ Epidermal growth factor regulates p21ras through the formation of a complex of receptor, Grb2 adapter protein, and Sos nucleotide exchange factor. Cell 73, 611–620 (1993). [DOI] [PubMed] [Google Scholar]
- 20.Ebinu JO et al. RasGRP, a Ras guanyl nucleotide-releasing protein with calcium- and diacylglycerol-binding motifs. Science 280, 1082–1086 (1998). [DOI] [PubMed] [Google Scholar]
- 21.Xu GF et al. The neurofibromatosis type 1 gene encodes a protein related to GAP. Cell 62, 599–608 (1990). [DOI] [PubMed] [Google Scholar]
- 22.Trahey MMF A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science 238, 542–545 (1987). [DOI] [PubMed] [Google Scholar]
- 23.Hunter JC et al. Biochemical and structural analysis of common cancer-associated KRAS mutations. Mol. Cancer Res 13, 1325–1335 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Ostrem JM, Peters U, Sos ML, Wells JA & Shokat KM K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rabara D et al. KRAS G13D sensitivity to neurofibromin-mediated GTP hydrolysis. Proc. Natl Acad. Sci. USA 116, 22122–22131 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson L et al. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev. 11, 2468–2481 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura K et al. Partial functional overlap of the three ras genes in mouse embryonic development. Oncogene 27, 2961–2968 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Potenza N et al. Replacement of K-Ras with H-Ras supports normal embryonic development despite inducing cardiovascular pathology in adult mice. EMBO Rep. 6, 432–437 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gehringer M & Laufer SA Emerging and re-emerging warheads for targeted covalent inhibitors: applications in medicinal chemistry and chemical biology. J. Med. Chem 62, 5673–5724 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Janes MR et al. Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell 172, 578–589 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Amgen. Amgen Announces New Clinical Data Evaluating Novel Investigational KRAS(G12C) Inhibitor in Patients with Solid Tumors at ESMO 2019 www.amgen.com https://www.amgen.com/media/news-releases/2019/09/amgen-announces-new-clinical-data-evaluating-novel-investigational-krasg12c-inhibitor-in-patients-with-solid-tumors-at-esmo-2019/ (2019).
- 32.Canon J et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 575, 217–223 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Mirati Therapeutics. Mirati Therapeutics Presents First Clinical Data of Phase 1/2 Trial of MRTX849 at the 2019 AACR-NCI-EORTC International Conference on Molecular Targets and Cancer Therapeutics ir.mirati.com https://ir.mirati.com/news-releases/news-details/2019/Mirati-Therapeutics-Presents-First-Clinical-Data-Of-Phase-12-Trial-Of-MRTX849-At-The-2019-AACR-NCI-EORTC-International-Conference-On-Molecular-Targets-And-Cancer-Therapeutics/default.aspx (2019). [Google Scholar]
- 34.Hallin J et al. The KRASG12C inhibitor, MRTX849, provides insight toward therapeutic susceptibility of KRAS mutant cancers in mouse models and patients. Cancer Discov. 10, 54–71 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lito P, Solomon M, Li LS, Hansen R & Rosen N Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science 351, 604–608 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patricelli MP et al. Selective inhibition of oncogenic KRAS output with small molecules targeting the inactive state. Cancer Discov. 6, 316–329 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Lou K et al. KRAS(G12C) inhibition produces a driver-limited state revealing collateral dependencies. Sci. Signal 12, eaaw9450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gentile DR et al. Ras binder induces a modified switch-II pocket in GTP and GDP states. Cell Chem. Biol 24, 1455–1466 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Revolution Medicines. Revolution Medicines to Present Preclinical Data on Novel Inhibitors of Oncogenic RAS(ON) Mutants at AACR-NCI-EORTC International Conference on Molecular Targets and Cancer Therapeutics www.revmed.com https://www.revmed.com/media/revolution-medicines-present-preclinical-data-novel-inhibitors-oncogenic-rason-mutants-aacr (2019). [Google Scholar]
- 40.Welsch ME et al. Multivalent small-molecule pan-RAS inhibitors. Cell 168, 878–889 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drosten M, Lechuga CG & Barbacid M Ras signaling is essential for skin development. Oncogene 33, 2857–2865 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Drosten M et al. Genetic analysis of Ras signalling pathways in cell proliferation, migration and survival. EMBO J. 29, 1091–1104 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maurer T et al. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc. Natl Acad. Sci. USA 109, 5299–5304 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cruz-Migoni A et al. Structure-based development of new RAS-effector inhibitors from a combination of active and inactive RAS-binding compounds. Proc. Natl Acad. Sci. USA 116, 2545–2550 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quevedo CE et al. Small molecule inhibitors of RAS-effector protein interactions derived using an intracellular antibody fragment. Nat. Commun 9, 3169 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kessler D et al. Drugging an undruggable pocket on KRAS. Proc. Natl Acad. Sci. USA 116, 15823–15829 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun Q et al. Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew. Chem. Int. Ed. Engl 51, 6140–6143 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leshchiner ES et al. Direct inhibition of oncogenic KRAS by hydrocarbon-stapled SOS1 helices. Proc. Natl Acad. Sci. USA 112, 1761–1766 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patgiri A, Yadav KK, Arora PS & Bar-Sagi D An orthosteric inhibitor of the Ras-Sos interaction. Nat. Chem. Biol 7, 585–587 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winter JJ et al. Small molecule binding sites on the Ras:SOS complex can be exploited for inhibition of Ras activation. J. Med. Chem 58, 2265–2274 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Burns MC et al. Approach for targeting Ras with small molecules that activate SOS-mediated nucleotide exchange. Proc. Natl Acad. Sci. USA 111, 3401–3406 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hillig RC et al. Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS-SOS1 interaction. Proc. Natl Acad. Sci. USA 116, 2551–2560 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi ZQ, Yu DH, Park M, Marshall M & Feng GS Molecular mechanism for the Shp-2 tyrosine phosphatase function in promoting growth factor stimulation of Erk activity. Mol. Cell. Biol 20, 1526–1536 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tartaglia M et al. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat. Genet 29, 465–468 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Castel P et al. RIT1 oncoproteins escape LZTR1-mediated proteolysis. Science 363, 1226–1230 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rauen KA The RASopathies. Annu. Rev. Genomics Hum. Genet 14, 355–369 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dance M, Montagner A, Salles JP, Yart A & Raynal P The molecular functions of Shp2 in the Ras/mitogen-activated protein kinase (ERK1/2) pathway. Cell. Signal 20, 453–459 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Li W et al. A new function for a phosphotyrosine phosphatase: linking GRB2-Sos to a receptor tyrosine kinase. Mol. Cell. Biol 14, 509–517 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bennett AM, Tang TL, Sugimoto S, Walsh CT & Neel BG Protein-tyrosine-phosphatase SHPTP2 couples platelet-derived growth factor receptor beta to Ras. Proc. Natl Acad. Sci. USA 91, 7335–7339 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ruess DA et al. Mutant KRAS-driven cancers depend on PTPN11/SHP2 phosphatase. Nat. Med 24, 954–960 (2018). [DOI] [PubMed] [Google Scholar]
- 61.Chen YN et al. Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature 535, 148–152 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Lu H et al. SHP2 inhibition overcomes RTK-mediated pathway reactivation in KRAS-mutant tumors treated with MEK inhibitors. Mol. Cancer Ther 18, 1323–1334 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Nichols RJ et al. RAS nucleotide cycling underlies the SHP2 phosphatase dependence of mutant BRAF-, NF1- and RAS-driven cancers. Nat. Cell Biol 20, 1064–1073 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mirati Therapeutics. Mirati Announces Clinical Collaboration to Evaluate MRTX849 in Combination with SHP2 Inhibitor TNO155 ir.mirati.com https://ir.mirati.com/news-releases/news-details/2019/Mirati-Announces-Clinical-Collaboration-to-Evaluate-MRTX849-in-Combination-with-SHP2-Inhibitor-TNO155/default.aspx (2019).
- 65.Whyte DB et al. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J. Biol. Chem 272, 14459–14464 (1997). [DOI] [PubMed] [Google Scholar]
- 66.Kessler L, Scholz C, Gualberto A, Liu Y & Burrows F Tipifarnib is highly active in HRAS mutant lung squamous carcinoma tumor models [abstract]. Cancer Res. 78 (Suppl. 13), 4917 (2018). [Google Scholar]
- 67.Ho A et al. Preliminary results from a phase 2 trial of tipifarnib in HRAS-mutant head and neck squamous cell carcinomas [abstract]. Int. J. Radiat. Oncol. Biol. Phys 100, 1367 (2018). [Google Scholar]
- 68.Wang T et al. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic Ras. Cell 168, 890–903 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Winter-Vann AM et al. A small-molecule inhibitor of isoprenylcysteine carboxyl methyltransferase with antitumor activity in cancer cells. Proc. Natl Acad. Sci. USA 102, 4336–4341 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang M et al. Inhibition of isoprenylcysteine carboxylmethyltransferase induces autophagic-dependent apoptosis and impairs tumor growth. Oncogene 29, 4959–4970 (2010). [DOI] [PubMed] [Google Scholar]
- 71.Manu KA et al. Inhibition of isoprenylcysteine carboxylmethyltransferase induces cell-cycle arrest and apoptosis through p21 and p21-regulated BNIP3 induction in pancreatic cancer. Mol. Cancer Ther 16, 914–923 (2017). [DOI] [PubMed] [Google Scholar]
- 72.Judd WR et al. Discovery and SAR of methylated tetrahydropyranyl derivatives as inhibitors of isoprenylcysteine carboxyl methyltransferase (ICMT). J. Med. Chem 54, 5031–5047 (2011). [DOI] [PubMed] [Google Scholar]
- 73.Marin-Ramos NI et al. A potent isoprenylcysteine carboxylmethyltransferase (ICMT) inhibitor improves survival in Ras-driven acute myeloid leukemia. J. Med. Chem 62, 6035–6046 (2019). [DOI] [PubMed] [Google Scholar]
- 74.Hampton SE, Dore TM & Schmidt WK Rce1: mechanism and inhibition. Crit. Rev. Biochem. Mol. Biol 53, 157–174 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mohammed I et al. 8-Hydroxyquinoline-based inhibitors of the Rce1 protease disrupt Ras membrane localization in human cells. Bioorg. Med. Chem 24, 160–178 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chandra A et al. The GDI-like solubilizing factor PDEδ sustains the spatial organization and signalling of Ras family proteins. Nat. Cell Biol 14, 148–158 (2011). [DOI] [PubMed] [Google Scholar]
- 77.Zimmermann G et al. Small molecule inhibition of the KRAS-PDEδ interaction impairs oncogenic KRAS signalling. Nature 497, 638–642 (2013). [DOI] [PubMed] [Google Scholar]
- 78.Leung EL et al. Identification of a new inhibitor of KRAS-PDEδ interaction targeting KRAS mutant nonsmall cell lung cancer. Int. J. Cancer 145, 1334–1345 (2019). [DOI] [PubMed] [Google Scholar]
- 79.Inouye K, Mizutani S, Koide H & Kaziro Y Formation of the Ras dimer is essential for Raf-1 activation. J. Biol. Chem 275, 3737–3740 (2000). [DOI] [PubMed] [Google Scholar]
- 80.Prior IA, Muncke C, Parton RG & Hancock JF Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J. Cell Biol 160, 165–170 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Plowman SJ, Muncke C, Parton RG & Hancock JF H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc. Natl Acad. Sci. USA 102, 15500–15505 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Abankwa D, Gorfe AA, Inder K & Hancock JF Ras membrane orientation and nanodomain localization generate isoform diversity. Proc. Natl Acad. Sci. USA 107, 1130–1135 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Solman M et al. Specific cancer-associated mutations in the switch III region of Ras increase tumorigenicity by nanocluster augmentation. Elife 4, e08905 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tian T et al. Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nat. Cell Biol 9, 905–914 (2007). [DOI] [PubMed] [Google Scholar]
- 85.Zhou Y & Hancock JF Ras nanoclusters: versatile lipid-based signaling platforms. Biochim. Biophys. Acta 1853, 841–849 (2015). [DOI] [PubMed] [Google Scholar]
- 86.Guldenhaupt J et al. N-Ras forms dimers at POPC membranes. Biophys. J 103, 1585–1593 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lin WC et al. H-Ras forms dimers on membrane surfaces via a protein-protein interface. Proc. Natl Acad. Sci. USA 111, 2996–3001 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Muratcioglu S et al. GTP-dependent K-Ras dimerization. Structure 23, 1325–1335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nan X et al. Ras-GTP dimers activate the mitogen-activated protein kinase (MAPK) pathway. Proc. Natl Acad. Sci. USA 112, 7996–8001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Prakash P et al. Computational and biochemical characterization of two partially overlapping interfaces and multiple weak-affinity K-Ras dimers. Sci. Rep 7, 40109 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sarkar-Banerjee S et al. Spatiotemporal analysis of K-Ras plasma membrane interactions reveals multiple high order homo-oligomeric complexes. J. Am. Chem. Soc 139, 13466–13475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Spencer-Smith R et al. Inhibition of RAS function through targeting an allosteric regulatory site. Nat. Chem. Biol 13, 62–68 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ambrogio C et al. KRAS dimerization impacts MEK inhibitor sensitivity and oncogenic activity of mutant KRAS. Cell 172, 857–868 (2018). [DOI] [PubMed] [Google Scholar]
- 94.Khan I, Spencer-Smith R & O’Bryan JP Targeting the α4-α5 dimerization interface of K-RAS inhibits tumor formation in vivo. Oncogene 38, 2984–2993 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fang Z et al. Inhibition of K-RAS4B by a unique mechanism of action: stabilizing membrane-dependent occlusion of the effector-binding site. Cell Chem. Biol 25, 1327–1336 (2018). [DOI] [PubMed] [Google Scholar]
- 96.Mazhab-Jafari MT et al. Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site. Proc. Natl Acad. Sci. USA 112, 6625–6630 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sanclemente M et al. c-RAF ablation induces regression of advanced Kras/Trp53 mutant lung adenocarcinomas by a mechanism independent of MAPK signaling. Cancer Cell 33, 217–228 (2018). [DOI] [PubMed] [Google Scholar]
- 98.Blasco MT et al. Complete regression of advanced pancreatic ductal adenocarcinomas upon combined inhibition of EGFR and C-RAF. Cancer Cell 35, 573–587 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Young LC et al. SHOC2-MRAS-PP1 complex positively regulates RAF activity and contributes to Noonan syndrome pathogenesis. Proc. Natl Acad. Sci. USA 115, E10576–E10585 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jones GG et al. SHOC2 phosphatase-dependent RAF dimerization mediates resistance to MEK inhibition in RAS-mutant cancers. Nat. Commun 10, 2532 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sulahian R et al. Synthetic lethal interaction of SHOC2 depletion with MEK inhibition in RAS-driven cancers. Cell Rep. 29, 118–134 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Karapetis CS et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N. Engl. J. Med 359, 1757–1765 (2008). [DOI] [PubMed] [Google Scholar]
- 103.Amado RG et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J. Clin. Oncol 26, 1626–1634 (2008). [DOI] [PubMed] [Google Scholar]
- 104.Linardou H et al. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol. 9, 962–972 (2008). [DOI] [PubMed] [Google Scholar]
- 105.Mao C et al. KRAS mutations and resistance to EGFR-TKIs treatment in patients with non-small cell lung cancer: a meta-analysis of 22 studies. Lung Cancer 69, 272–278 (2010). [DOI] [PubMed] [Google Scholar]
- 106.Pao W et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl Acad. Sci. USA 101, 13306–13311 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kobayashi S et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N. Engl. J. Med 352, 786–792 (2005). [DOI] [PubMed] [Google Scholar]
- 108.Moore MJ et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J. Clin. Oncol 25, 1960–1966 (2007). [DOI] [PubMed] [Google Scholar]
- 109.De Roock W et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA 304, 1812–1820 (2010). [DOI] [PubMed] [Google Scholar]
- 110.Peeters M et al. Mutant KRAS codon 12 and 13 alleles in patients with metastatic colorectal cancer: assessment as prognostic and predictive biomarkers of response to panitumumab. J. Clin. Oncol 31, 759–765 (2013). [DOI] [PubMed] [Google Scholar]
- 111.Segelov E et al. ICECREAM: randomised phase II study of cetuximab alone or in combination with irinotecan in patients with metastatic colorectal cancer with either KRAS, NRAS, BRAF and PI3KCA wild type, or G13D mutated tumours. BMC Cancer 16, 339 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Misale S et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 486, 532–536 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ardito CM et al. EGF receptor is required for KRAS-induced pancreatic tumorigenesis. Cancer Cell 22, 304–317 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Navas C et al. EGF receptor signaling is essential for k-ras oncogene-driven pancreatic ductal adenocarcinoma. Cancer Cell 22, 318–330 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Moll HP et al. Afatinib restrains K-RAS-driven lung tumorigenesis. Sci. Transl Med 10, eaao2301 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kruspig B et al. The ERBB network facilitates KRAS-driven lung tumorigenesis. Sci. Transl Med 10, eaao2565 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chen HY et al. EGFR-activating mutations, DNA copy number abundance of ErbB family, and prognosis in lung adenocarcinoma. Oncotarget 7, 9017–9025 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dent P et al. Neratinib degrades MST4 via autophagy that reduces membrane stiffness and is essential for the inactivation of PI3K, ERK1/2, and YAP/TAZ signaling. J. Cell. Physiol 10.1002/jcp.29443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sun C et al. Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell Rep. 7, 86–93 (2014). [DOI] [PubMed] [Google Scholar]
- 120.Xie Y et al. COP1/DET1/ETS axis regulates ERK transcriptome and sensitivity to MAPK inhibitors. J. Clin. Invest 128, 1442–1457 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dougherty MK et al. Regulation of Raf-1 by direct feedback phosphorylation. Mol. Cell 17, 215–224 (2005). [DOI] [PubMed] [Google Scholar]
- 122.Owens DM & Keyse SM Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases. Oncogene 26, 3203–3213 (2007). [DOI] [PubMed] [Google Scholar]
- 123.Pratilas CA et al. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc. Natl Acad. Sci. USA 106, 4519–4524 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lake D, Correa SA & Muller J Negative feedback regulation of the ERK1/2 MAPK pathway. Cell. Mol. Life Sci 73, 4397–4413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kim HJ & Bar-Sagi D Modulation of signalling by Sprouty: a developing story. Nat. Rev. Mol. Cell Biol 5, 441–450 (2004). [DOI] [PubMed] [Google Scholar]
- 126.Mason JM, Morrison DJ, Basson MA & Licht JD Sprouty proteins: multifaceted negative-feedback regulators of receptor tyrosine kinase signaling. Trends Cell Biol. 16, 45–54 (2006). [DOI] [PubMed] [Google Scholar]
- 127.Karoulia Z et al. An integrated model of RAF inhibitor action predicts inhibitor activity against oncogenic BRAF signaling. Cancer Cell 30, 501–503 (2016). [DOI] [PubMed] [Google Scholar]
- 128.Hatzivassiliou G et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature 464, 431–435 (2010). [DOI] [PubMed] [Google Scholar]
- 129.Poulikakos PI, Zhang C, Bollag G, Shokat KM & Rosen N RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature 464, 427–430 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yen I et al. Pharmacological induction of RAS-GTP confers RAF inhibitor sensitivity in KRAS mutant tumors. Cancer Cell 34, 611–625 (2018). [DOI] [PubMed] [Google Scholar]
- 131.Peng SB et al. Inhibition of RAF isoforms and active dimers by LY3009120 leads to anti-tumor activities in RAS or BRAF mutant cancers. Cancer Cell 28, 384–398 (2015). [DOI] [PubMed] [Google Scholar]
- 132.Nakamura A et al. Antitumor activity of the selective pan-RAF inhibitor TAK-632 in BRAF inhibitor-resistant melanoma. Cancer Res. 73, 7043–7055 (2013). [DOI] [PubMed] [Google Scholar]
- 133.Wang X & Kim J Conformation-specific effects of Raf kinase inhibitors. J. Med. Chem 55, 7332–7341 (2012). [DOI] [PubMed] [Google Scholar]
- 134.Okaniwa M et al. Discovery of a selective kinase inhibitor (TAK-632) targeting pan-RAF inhibition: design, synthesis, and biological evaluation of C-7-substituted 1,3-benzothiazole derivatives. J. Med. Chem 56, 6478–6494 (2013). [DOI] [PubMed] [Google Scholar]
- 135.Sun Y et al. A brain-penetrant RAF dimer antagonist for the noncanonical BRAF oncoprotein of pediatric low-grade astrocytomas. Neuro Oncol. 19, 774–785 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Vakana E et al. LY3009120, a panRAF inhibitor, has significant anti-tumor activity in BRAF and KRAS mutant preclinical models of colorectal cancer. Oncotarget 8, 9251–9266 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yao YM et al. Mouse PDX trial suggests synergy of concurrent inhibition of RAF and EGFR in colorectal cancer with BRAF or KRAS mutations. Clin. Cancer Res 23, 5547–5560 (2017). [DOI] [PubMed] [Google Scholar]
- 138.Kim TW et al. Belvarafenib, a novel pan-RAF inhibitor, in solid tumor patients harboring BRAF, KRAS, or NRAS mutations: phase I study. J. Clin. Oncol 37, 3000–3000 (2019).31557067 [Google Scholar]
- 139.Monaco K-A et al. RAF inhibitor LXH254 effectively inhibits B-and-CRAF, but not ARAF [abstract]. Cancer Res. 79 (Suppl. 13), LB–144 (2019). [Google Scholar]
- 140.Sullivan RJ et al. A phase I study of LY3009120, a pan-RAF inhibitor, in patients with advanced or metastatic cancer. Mol. Cancer Ther 19, 460–467 (2020). [DOI] [PubMed] [Google Scholar]
- 141.Su F et al. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N. Engl. J. Med 366, 207–215 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Flaherty KT et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med 363, 809–819 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Carter CA et al. Selumetinib with and without erlotinib in KRAS mutant and KRAS wild-type advanced nonsmall-cell lung cancer. Ann. Oncol 27, 693–699 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Janne PA et al. Selumetinib plus docetaxel compared with docetaxel alone and progression-free survival in patients with KRAS-mutant advanced non-small cell lung cancer: the SELECT-1 randomized clinical trial. JAMA 317, 1844–1853 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hellmann MD et al. Phase Ib study of atezolizumab combined with cobimetinib in patients with solid tumors. Ann. Oncol 30, 1134–1142 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zimmer L et al. Phase I expansion and pharmacodynamic study of the oral MEK inhibitor RO4987655 (CH4987655) in selected patients with advanced cancer with RAS-RAF mutations. Clin. Cancer Res 20, 4251–4261 (2014). [DOI] [PubMed] [Google Scholar]
- 147.Blumenschein GR Jr. et al. A randomized phase II study of the MEK1/MEK2 inhibitor trametinib (GSK1120212) compared with docetaxel in KRAS-mutant advanced non-small-cell lung cancer (NSCLC). Ann. Oncol 26, 894–901 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Friday BB et al. BRAF V600E disrupts AZD6244-induced abrogation of negative feedback pathways between extracellular signal-regulated kinase and Raf proteins. Cancer Res. 68, 6145–6153 (2008). [DOI] [PubMed] [Google Scholar]
- 149.Hatzivassiliou G et al. Mechanism of MEK inhibition determines efficacy in mutant KRAS-versus BRAF-driven cancers. Nature 501, 232–236 (2013). [DOI] [PubMed] [Google Scholar]
- 150.Lito P et al. Disruption of CRAF-mediated MEK activation is required for effective MEK inhibition in KRAS mutant tumors. Cancer Cell 25, 697–710 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ascierto PA et al. MEK162 for patients with advanced melanoma harbouring NRAS or Val600 BRAF mutations: a non-randomised, open-label phase 2 study. Lancet Oncol. 14, 249–256 (2013). [DOI] [PubMed] [Google Scholar]
- 152.Lebbe C et al. Pimasertib (PIM) versus dacarbazine (DTIC) in patients (pts) with cutaneous NRAS melanoma: a controlled, open-label phase II trial with crossover [abstract 1136P]. Ann. Oncol 27 (Suppl. 6), vi389 (2016). [Google Scholar]
- 153.Van Cutsem E et al. Phase I/II trial of pimasertib plus gemcitabine in patients with metastatic pancreatic cancer. Int. J. Cancer 143, 2053–2064 (2018). [DOI] [PubMed] [Google Scholar]
- 154.Bodoky G et al. A phase II open-label randomized study to assess the efficacy and safety of selumetinib (AZD6244 [ARRY-142886]) versus capecitabine in patients with advanced or metastatic pancreatic cancer who have failed first-line gemcitabine therapy. Invest. N. Drugs 30, 1216–1223 (2012). [DOI] [PubMed] [Google Scholar]
- 155.Bennouna J et al. A phase II, open-label, randomised study to assess the efficacy and safety of the MEK1/2 inhibitor AZD6244 (ARRY-142886) versus capecitabine monotherapy in patients with colorectal cancer who have failed one or two prior chemotherapeutic regimens. Invest. N. Drugs 29, 1021–1028 (2011). [DOI] [PubMed] [Google Scholar]
- 156.Hainsworth JD et al. A phase II, open-label, randomized study to assess the efficacy and safety of AZD6244 (ARRY-142886) versus pemetrexed in patients with non-small cell lung cancer who have failed one or two prior chemotherapeutic regimens. J. Thorac. Oncol 5, 1630–1636 (2010). [DOI] [PubMed] [Google Scholar]
- 157.Morris EJ et al. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov. 3, 742–750 (2013). [DOI] [PubMed] [Google Scholar]
- 158.Hatzivassiliou G et al. ERK inhibition overcomes acquired resistance to MEK inhibitors. Mol. Cancer Ther 11, 1143–1154 (2012). [DOI] [PubMed] [Google Scholar]
- 159.Chaikuad A et al. A unique inhibitor binding site in ERK1/2 is associated with slow binding kinetics. Nat. Chem. Biol 10, 853–860 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Moschos SJ et al. Development of MK-8353, an orally administered ERK1/2 inhibitor, in patients with advanced solid tumors. JCI Insight 3, e92352 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.He Y et al. Identification and validation of PROM1 and CRTC2 mutations in lung cancer patients. Mol. Cancer 13, 19 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Boga SB et al. MK-8353: discovery of an orally bioavailable dual mechanism ERK inhibitor for oncology. ACS Med. Chem. Lett 9, 761–767 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Merchant M et al. Combined MEK and ERK inhibition overcomes therapy-mediated pathway reactivation in RAS mutant tumors. PLoS One 12, e0185862 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Weekes CD et al. A phase Ib study to evaluate the MEK inhibitor cobimetinib in combination with the ERK1/2 inhibitor GDC-0994 in patients with advanced solid tumors. Cancer Res. 77, CT107 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Varga A et al. A first-in-human phase I study to evaluate the ERK1/2 inhibitor GDC-0994 in patients with advanced solid tumors. Clin. Cancer Res 26, 1229–1236 (2020). [DOI] [PubMed] [Google Scholar]
- 166.Sullivan RJ et al. First-in-class ERK1/2 inhibitor ulixertinib (BVD-523) in patients with MAPK mutant advanced solid tumors: results of a phase I dose-escalation and expansion study. Cancer Discov. 8, 184–195 (2018). [DOI] [PubMed] [Google Scholar]
- 167.Burrows F et al. KO-947, a potent ERK inhibitor with robust preclinical single agent activity in MAPK pathway dysregulated tumors [abstract]. Cancer Res. 77 (Suppl. 13), 5168 (2017). [Google Scholar]
- 168.Bhagwat SV et al. Discovery of LY3214996, a selective and novel ERK1/2 inhibitor with potent antitumor activities in cancer models with MAPK pathway alterations [abstract]. Cancer Res. 77 (Suppl. 13), 4973 (2017).28754668 [Google Scholar]
- 169.Pant S et al. A phase I dose escalation (DE) study of ERK inhibitor, LY3214996, in advanced (adv) cancer (CA) patients (pts) [abstract]. J. Clin. Oncol 37 (Suppl. 15), 3001 (2019). [Google Scholar]
- 170.Fruman DA et al. The PI3K pathway in human disease. Cell 170, 605–635 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Vanhaesebroeck B et al. P110δ, a novel phosphoinositide 3-kinase in leukocytes. Proc. Natl Acad. Sci. USA 94, 4330–4335 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hirsch E et al. Central role for G protein-coupled phosphoinositide 3-kinase γ in inflammation. Science 287, 1049–1053 (2000). [DOI] [PubMed] [Google Scholar]
- 173.Houslay DM et al. Coincident signals from GPCRs and receptor tyrosine kinases are uniquely transduced by PI3Kβ in myeloid cells. Sci. Signal 9, ra82 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Schmid MC et al. Receptor tyrosine kinases and TLR/IL1Rs unexpectedly activate myeloid cell PI3kγ, a single convergent point promoting tumor inflammation and progression. Cancer Cell 19, 715–727 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Samuels Y et al. High frequency of mutations of the PIK3CA gene in human cancers. Science 304, 554 (2004). [DOI] [PubMed] [Google Scholar]
- 176.Li J et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 275, 1943–1947 (1997). [DOI] [PubMed] [Google Scholar]
- 177.Aoki M, Batista O, Bellacosa A, Tsichlis P & Vogt PK The Akt kinase: molecular determinants of oncogenicity. Proc. Natl Acad. Sci. USA 95, 14950–14955 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wang Q et al. PIK3CA mutations confer resistance to first-line chemotherapy in colorectal cancer. Cell Death Dis. 9, 739 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Mao M et al. Resistance to BRAF inhibition in BRAF-mutant colon cancer can be overcome with PI3K inhibition or demethylating agents. Clin. Cancer Res 19, 657–667 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wee S et al. PI3K pathway activation mediates resistance to MEK inhibitors in KRAS mutant cancers. Cancer Res. 69, 4286–4293 (2009). [DOI] [PubMed] [Google Scholar]
- 181.Engelman JA et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat. Med 14, 1351–1356 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hoeflich KP et al. Intermittent administration of MEK inhibitor GDC-0973 plus PI3K inhibitor GDC-0941 triggers robust apoptosis and tumor growth inhibition. Cancer Res. 72, 210–219 (2012). [DOI] [PubMed] [Google Scholar]
- 183.Shapiro GI et al. Phase Ib study of the MEK inhibitor cobimetinib (GDC-0973) in combination with the PI3K inhibitor pictilisib (GDC-0941) in patients with advanced solid tumors. Invest. New Drugs 38, 419–432 (2020). [DOI] [PubMed] [Google Scholar]
- 184.Bedard PL et al. A phase Ib dose-escalation study of the oral pan-PI3K inhibitor buparlisib (BKM120) in combination with the oral MEK1/2 inhibitor trametinib (GSK1120212) in patients with selected advanced solid tumors. Clin. Cancer Res 21, 730–738 (2015). [DOI] [PubMed] [Google Scholar]
- 185.Juric D et al. A phase 1b dose-escalation study of BYL719 plus binimetinib (MEK162) in patients with selected advanced solid tumors [abstract]. J. Clin. Oncol 32 (Suppl. 15), 9051 (2014). [Google Scholar]
- 186.Algazi AP et al. A dual pathway inhibition strategy using BKM120 combined with vemurafenib is poorly tolerated in BRAF V600(E/K) mutant advanced melanoma. Pigment. Cell Melanoma Res 32, 603–606 (2019). [DOI] [PubMed] [Google Scholar]
- 187.Ebi H et al. Receptor tyrosine kinases exert dominant control over PI3K signaling in human KRAS mutant colorectal cancers. J. Clin. Invest 121, 4311–4321 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Molina-Arcas M, Hancock DC, Sheridan C, Kumar MS & Downward J Coordinate direct input of both KRAS and IGF1 receptor to activation of PI3 kinase in KRAS-mutant lung cancer. Cancer Discov. 3, 548–563 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Molina-Arcas M et al. Development of combination therapies to maximize the impact of KRAS-G12C inhibitors in lung cancer. Sci. Transl Med 11, eaaw7999 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Tolcher AW et al. Phase I study of the MEK inhibitor trametinib in combination with the AKT inhibitor afuresertib in patients with solid tumors and multiple myeloma. Cancer Chemother. Pharmacol 75, 183–189 (2015). [DOI] [PubMed] [Google Scholar]
- 191.Shoushtari AN et al. A randomized phase 2 study of trametinib with or without GSK2141795 in patients with advanced uveal melanoma [abstract]. J. Clin. Oncol 34 (Suppl. 15), 9511 (2016). [Google Scholar]
- 192.Tolcher AW et al. A phase IB trial of the oral MEK inhibitor trametinib (GSK1120212) in combination with everolimus in patients with advanced solid tumors. Ann. Oncol 26, 58–64 (2015). [DOI] [PubMed] [Google Scholar]
- 193.Mita M et al. Phase I trial of MEK 1/2 inhibitor pimasertib combined with mTOR inhibitor temsirolimus in patients with advanced solid tumors. Invest. N. Drugs 35, 616–626 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Janku F, Yap TA & Meric-Bernstam F Targeting the PI3K pathway in cancer: are we making headway? Nat. Rev. Clin. Oncol 15, 273–291 (2018). [DOI] [PubMed] [Google Scholar]
- 195.Ross SJ et al. Targeting KRAS-dependent tumors with AZD4785, a high-affinity therapeutic antisense oligonucleotide inhibitor of KRAS. Sci. Transl Med 9, eaal5253 (2017). [DOI] [PubMed] [Google Scholar]
- 196.Golan T et al. RNAi therapy targeting KRAS in combination with chemotherapy for locally advanced pancreatic cancer patients. Oncotarget 6, 24560–24570 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kinsey CG et al. Protective autophagy elicited by RAF→MEK→ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat. Med 25, 620–627 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Bryant KL et al. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat. Med 25, 628–640 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Yang A et al. Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer Discov. 4, 905–913 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Yang S et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 25, 717–729 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wolpin BM et al. Phase II and pharmacodynamic study of autophagy inhibition using hydroxychloroquine in patients with metastatic pancreatic adenocarcinoma. Oncologist 19, 637–638 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.White NJ The treatment of malaria. N. Engl. J. Med 335, 800–806 (1996). [DOI] [PubMed] [Google Scholar]
- 203.Boone BA et al. Safety and biologic response of pre-operative autophagy inhibition in combination with gemcitabine in patients with pancreatic adenocarcinoma. Ann. Surg. Oncol 22, 4402–4410 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Chen DS & Mellman I Elements of cancer immunity and the cancer-immune set point. Nature 541, 321–330 (2017). [DOI] [PubMed] [Google Scholar]
- 205.Hodi FS et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med 363, 711–723 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Eggermont AM et al. Prolonged survival in stage III melanoma with ipilimumab adjuvant therapy. N. Engl. J. Med 375, 1845–1855 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hellmann MD et al. Nivolumab plus ipilimumab in advanced non-small-cell lung cancer. N. Engl. J. Med 381, 2020–2031 (2019). [DOI] [PubMed] [Google Scholar]
- 208.Wolchok JD et al. Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N. Engl. J. Med 377, 1345–1356 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Weber JS et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 16, 375–384 (2015). [DOI] [PubMed] [Google Scholar]
- 210.Robert C et al. Nivolumab in previously untreated melanoma without BRAF mutation. N. Engl. J. Med 372, 320–330 (2015). [DOI] [PubMed] [Google Scholar]
- 211.Weber J et al. Adjuvant nivolumab versus ipilimumab in resected stage III or IV melanoma. N. Engl. J. Med 377, 1824–1835 (2017). [DOI] [PubMed] [Google Scholar]
- 212.Brahmer J et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N. Engl. J. Med 373, 123–135 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Reck M et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N. Engl. J. Med 375, 1823–1833 (2016). [DOI] [PubMed] [Google Scholar]
- 214.Ribas A et al. Pembrolizumab versus investigator-choice chemotherapy for ipilimumab-refractory melanoma (KEYNOTE-002): a randomised, controlled, phase 2 trial. Lancet Oncol. 16, 908–918 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Garon EB et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N. Engl. J. Med 372, 2018–2028 (2015). [DOI] [PubMed] [Google Scholar]
- 216.Robert C et al. Pembrolizumab versus ipilimumab in advanced melanoma. N. Engl. J. Med 372, 2521–2532 (2015). [DOI] [PubMed] [Google Scholar]
- 217.Rittmeyer A et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet 389, 255–265 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Socinski MA et al. Atezolizumab for first-line treatment of metastatic nonsquamous NSCLC. N. Engl. J. Med 378, 2288–2301 (2018). [DOI] [PubMed] [Google Scholar]
- 219.Antonia SJ et al. Durvalumab after chemoradiotherapy in stage III non-small-cell lung cancer. N. Engl. J. Med 377, 1919–1929 (2017). [DOI] [PubMed] [Google Scholar]
- 220.Sullivan RJ et al. Atezolizumab plus cobimetinib and vemurafenib in BRAF-mutated melanoma patients. Nat. Med 25, 929–935 (2019). [DOI] [PubMed] [Google Scholar]
- 221.Keilholz U et al. Avelumab in patients with previously treated metastatic melanoma: phase 1b results from the JAVELIN solid tumor trial. J. Immunother. Cancer 7, 12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Van Allen EM et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350, 207–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Keenan TE, Burke KP & Van Allen EM Genomic correlates of response to immune checkpoint blockade. Nat. Med 25, 389–402 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Topalian SL et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med 366, 2443–2454 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Herbst RS et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 515, 563–567 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Alexandrov LB et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Vonderheide RH & Bayne LJ Inflammatory networks and immune surveillance of pancreatic carcinoma. Curr. Opin. Immunol 25, 200–205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Royal RE et al. Phase 2 trial of single agent ipilimumab (anti-CTLA-4) for locally advanced or metastatic pancreatic adenocarcinoma. J. Immunother 33, 828–833 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Patnaik A et al. Phase I study of pembrolizumab (MK-3475; anti-PD-1 monoclonal antibody) in patients with advanced solid tumors. Clin. Cancer Res 21, 4286–4293 (2015). [DOI] [PubMed] [Google Scholar]
- 230.O’Reilly EM et al. Durvalumab with or without tremelimumab for patients with metastatic pancreatic ductal adenocarcinoma: a phase 2 randomized clinical trial. JAMA Oncol. 5, 1431–1438 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Le DT et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Le DT et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med 372, 2509–2520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Overman MJ et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 18, 1182–1191 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Eatrides JM et al. Microsatellite instability in pancreatic cancer [abstract]. J. Clin. Oncol 34 (Suppl. 15), e15753 (2016). [Google Scholar]
- 235.Zou W, Wolchok JD & Chen L PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: mechanisms, response biomarkers, and combinations. Sci. Transl Med 8, 328rv324 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Skoulidis F et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov. 8, 822–835 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Coelho MA et al. Oncogenic RAS signaling promotes tumor immunoresistance by stabilizing PD-L1 mRNA. Immunity 47, 1083–1099 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Ebert PJR et al. MAP kinase inhibition promotes T cell and anti-tumor activity in combination with PD-L1 checkpoint blockade. Immunity 44, 609–621 (2016). [DOI] [PubMed] [Google Scholar]
- 239.Robbins PF et al. A mutated beta-catenin gene encodes a melanoma-specific antigen recognized by tumor infiltrating lymphocytes. J. Exp. Med 183, 1185–1192 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Dudley ME et al. Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science 298, 850–854 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Robbins PF et al. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J. Clin. Oncol 29, 917–924 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Tran E et al. T-cell transfer therapy targeting mutant KRAS in cancer. N. Engl. J. Med 375, 2255–2262 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Veatch JR et al. Endogenous CD4+ T cells recognize neoantigens in lung cancer patients, including recurrent oncogenic KRAS and ERBB2 (Her2) driver mutations. Cancer Immunol. Res 7, 910–922 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Wang QJ et al. Identification of T-cell receptors targeting KRAS-mutated human tumors. Cancer Immunol. Res 4, 204–214 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Gjertsen MK et al. Intradermal ras peptide vaccination with granulocyte-macrophage colony-stimulating factor as adjuvant: clinical and immunological responses in patients with pancreatic adenocarcinoma. Int. J. Cancer 92, 441–450 (2001). [DOI] [PubMed] [Google Scholar]
- 246.Palmer DH, Dueland S, Valle JW & Aksnes A-K A phase I/II trial of TG01/GM-CSF and gemcitabine as adjuvant therapy for treating patients with resected RAS-mutant adenocarcinoma of the pancreas [abstract]. J. Clin. Oncol 35 (Suppl. 15), 4119 (2017). [Google Scholar]
- 247.Merck. Moderna and Merck expand mRNA cancer vaccines collaboration merck.com https://investors.merck.com/news/press-release-details/2018/Moderna-and-Merck-Expand-mRNA-Cancer-Vaccines-Collaboration/default.aspx (Merck, 2018). [Google Scholar]
- 248.Van Cutsem E et al. Binimetinib, encorafenib, and cetuximab triplet therapy for patients with BRAF V600E-mutant metastatic colorectal cancer: safety lead-in results from the phase III BEACON colorectal cancer study. J. Clin. Oncol 37, 1460–1469 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Xue JY et al. Rapid non-uniform adaptation to conformation-specific KRAS(G12C) inhibition. Nature 577, 421–425 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Sharma SV, Bell DW, Settleman J & Haber DA Epidermal growth factor receptor mutations in lung cancer. Nat. Rev. Cancer 7, 169–181 (2007). [DOI] [PubMed] [Google Scholar]
- 251.Schwartz PA et al. Covalent EGFR inhibitor analysis reveals importance of reversible interactions to potency and mechanisms of drug resistance. Proc. Natl Acad. Sci. USA 111, 173–178 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Yaeger R et al. Mechanisms of acquired resistance to BRAF V600E inhibition in colon cancers converge on RAF dimerization and are sensitive to its inhibition. Cancer Res. 77, 6513–6523 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Johnson DB et al. Acquired BRAF inhibitor resistance: a multicenter meta-analysis of the spectrum and frequencies, clinical behaviour, and phenotypic associations of resistance mechanisms. Eur. J. Cancer 51, 2792–2799 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Van Allen EM et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 4, 94–109 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Engelman JA et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 316, 1039–1043 (2007). [DOI] [PubMed] [Google Scholar]
- 256.Bean J et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc. Natl Acad. Sci. USA 104, 20932–20937 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Nazarian R et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 468, 973–977 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Emery CM et al. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proc. Natl Acad. Sci. USA 106, 20411–20416 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Villanueva J et al. Concurrent MEK2 mutation and BRAF amplification confer resistance to BRAF and MEK inhibitors in melanoma. Cell Rep. 4, 1090–1099 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Sequist LV et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci. Transl Med 3, 75ra26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Yu HA et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin. Cancer Res 19, 2240–2247 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Johannessen CM et al. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature 504, 138–142 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Garraway LA et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 436, 117–122 (2005). [DOI] [PubMed] [Google Scholar]
- 264.Thomson S et al. Epithelial to mesenchymal transition is a determinant of sensitivity of non-small-cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer Res. 65, 9455–9462 (2005). [DOI] [PubMed] [Google Scholar]
- 265.Frederick BA et al. Epithelial to mesenchymal transition predicts gefitinib resistance in cell lines of head and neck squamous cell carcinoma and non-small cell lung carcinoma. Mol. Cancer Ther 6, 1683–1691 (2007). [DOI] [PubMed] [Google Scholar]
- 266.Caramel J et al. A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell 24, 466–480 (2013). [DOI] [PubMed] [Google Scholar]
- 267.Richard G et al. ZEB1-mediated melanoma cell plasticity enhances resistance to MAPK inhibitors. EMBO Mol. Med 8, 1143–1161 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Whittaker SR et al. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Discov. 3, 350–362 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.The AACR Project GENIE Consortium. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 7, 818–831 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]


