Abstract
Gastric cancer (GC) is one of the most common malignancies of the digestive system. In diffuse-type GC, differentiation is relatively poor, and the probability of distant metastasis and lymph node metastasis is high, resulting in poor clinical prognosis. The purpose of this study was to identify specific genes that can predict the prognosis of different types of GC. Differentially expressed genes (DEGs) were screened in the GSE62254 dataset obtained from the Gene Expression Omnibus using the ‘limma’ and ‘survival’ R packages. A total of 355 survival-related DEGs were selected according to specific screening criteria, of which 293 were associated with diffuse-type GC and 62 with intestinal-type GC. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes were used for functional annotation and pathway enrichment analysis of DEGs. Using protein-protein interaction networks and Cytoscape software, three hub genes were identified in diffuse-type GC-associated DEGs, including angiotensinogen (AGT), C-X-C motif chemokine ligand 12 (CXCL12) and adrenoceptor β2 (ADRB2). Immunohistochemical staining and reverse transcription-quantitative PCR revealed that the expression levels of the three genes in diffuse-type GC samples were upregulated compared with in intestinal-type GC samples. Kaplan Meier analysis indicated that a higher expression levels of these three hub genes were associated with a poorer prognosis of diffuse-type GC. In summary, the present findings suggested that AGT, CXCL12 and ADRB2 might contribute to the progression of diffuse-type GC, and could serve as potential biomarkers or therapeutic targets for this disease.
Keywords: gastric cancer, diffuse-type, bioinformatics, differentially expressed gene, hub gene
Introduction
Gastric cancer (GC) is one of the most common malignant tumors in the digestive system worldwide, accounting for ~720,000 GC-related deaths every year (1). According to Globocan 2018 data, the incidence and mortality rates of GC among all malignant tumors occupied fifth and third place respectively, and >70% of these cases occurred in developing countries, with 50% in East Asia (2). Early diagnosis and treatment of GC can often produce an improved therapeutic effect and longer overall survival time. However, most patients are diagnosed by endoscopy only when they present with symptoms, at which point the disease is in advanced stage, and the optimal opportunity for surgical intervention has been missed. Even after complete R0 resection, one-third of patients experience recurrence (3). Most GC cases are adenocarcinomas, representing a highly heterogeneous disease with differences in epidemiology and histopathology. There has been no evident breakthrough for the treatment of patients with advanced GC, and surgery is still the primary therapy, with the majority of cancer patients dying due to tumor recurrence and metastasis, and a median overall survival (OS) <1 year (4).
GC can be categorized using different classification systems, such as the Bormann, the Lauren, and the World Health Organization classifications (5–7). In addition, some scholars have attempted to classify GC using molecular and genetic features, such as The Cancer Genome Atlas (8) and Asian Cancer Research Group classifications (9). Since the Lauren classification was proposed in 1965, it has been widely recognized by both clinicians and pathologists and continues to be used. The Lauren classification primarily divides GC into intestinal, diffuse and mixed types based on cell morphology and histochemistry (6). Histologically, intestinal-type GC cells are large, clear in boundary, variable in morphology and closely arranged, exhibiting tubular and glandular differentiation. In contrast, diffuse-type GC cells are typically scattered and often appear as solitary cells or in small clusters due to lack of adhesion. Thus, gland formation is hard to observe in diffuse-type GC tumor tissue, and is easy to disseminate. Mixed-type GC presents all of the aforementioned characteristics (6). Epidemiologically, intestinal GC is the most common type, has the highest five-year survival rate, and is more common in men and the elderly. Diffuse-type GC is more likely in women and younger patients and has a lower 5-year survival rate (10–12). Mix-type has the highest degree of malignancy due to variable biological behavior (13).
With the development of medicine and bioinformatics, high-throughput sequencing has become a common tool for medical research (14). For instance, data from gene expression profiling studies can be uploaded to public repositories, such as, the Gene Expression Omnibus (GEO) within the National Center for Biotechnology Information. Reanalyzing and reintegrating such datasets often provides some meaningful insights for research. A number of microarray datasets of GC have been developed in recent years (15–17) and a large number of significant differentially expressed genes (DEGs) have already been identified.
In the present study, the GSE62254 dataset was downloaded from the GEO and screened for DEGs using the ‘limma’ and ‘survival’ R packages. Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was carried out on selected DEGs, identifying key biological features and signaling pathways. Moreover, protein-protein interaction (PPI) network of DEGs associated with diffuse-type was constructed, identifying three hub genes using the Cytoscape. Lastly, Kaplan-Meier analysis was used to evaluate overall survival (OS) in patients with different expression levels of these newly identified hub genes in GSE62254 and GSE15459 datasets.
Materials and methods
Data collection
The GSE62254 (9,18) microarray dataset was downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/geo/). GSE62254 was obtained using the GPL570 platform with an Affymetrix Human Genome U133 Plus 2.0 Array (Thermo Fisher Scientific, Inc.) and contains 300 different Lauren subtypes GC samples (128 diffuse-type samples, 137 intestinal-type samples and 35 mixed-type samples).
Analysis of differentially expressed genes
The Robust multi-array average (RMA) algorithm (19) in the R environment (v3.6.1) (20) was used to normalize and transform the raw data to expression values. DEGs between diffuse- and intestinal-type samples were screened using the ‘limma’ package in R (v3.6.1) (21), using the cut-off criteria of adjusted P<0.05 and |log2FC|>0.585. Accordingly, DEGs with log2FC>0.585 were considered associated with diffuse-type GC, whereas DEGs with log2FC<-0.585 were considered associated with intestinal-type GC. To identify genes associated with OS, the ‘survival’ package (22) in R (v3.6.1) was used for Cox regression analysis. The resuls are presented as hazard ratios (HRs) and P-values for all genes in the GSE62254 dataset. Genes with P<0.05 were identified as OS-related genes. Venny's (v2.1) online software (http://bioinfogp.cnb.csic.es/tools/venny/index.html) was used to draw Venn diagrams of OS-related genes associated with diffuse or intestinal GC.
GO and KEGG enrichment analysis
The Database for Annotation, Visualization and Integrated Discovery (DAVID, http://david.ncifcrf.gov/) provides a comprehensive set of functional annotation tools for investigators to examine biological meaning behind large lists of genes. DAVID (v6.8) was used for GO functional annotation and KEGG pathway analysis of DEGs associated with diffuse and intestinal GC. P<0.05 was the cut-off for statistically significant terms.
PPI network construction and screening of hub genes in diffuse-type GC
PPI data of diffuse-type GC DEGs were constructed using the Search Tool for the Retrieval of Interacting Genes (STRING) database (version 10.0; http://string-db.org) (23). Interactions with an interaction score of >0.700 were used to construct the PPI network. Cytoscape (https://cytoscape.org; v3.7.1) (24) was used to visualize the PPI network, and the cytoHubba (v1.6) (25) plug-in was used to identify hub genes of the PPI network via four different algorithms. The algorithms used for analysis included degree, Edge Percolated component, Closeness and EcCentricity. Genes overlapping in the four groups were deemed hub genes.
Patients and tissues samples
A total of 40 GC patients who received a gastrectomy in The Third Affiliated Hospital of Anhui Medical University between December 2016 to July 2018 were recruited in this study. This cohort included 27 male and 13 female patients aged 41–83 years (average, 56.35±9.60). The inclusion criteria were: i) The postoperative pathological diagnosis of diffuse- or intestinal-type GC was consistent by two pathologists; ii) patients were newly diagnosed with GC; and iii) patients had not received any radiotherapy, chemotherapy or biological therapy. The exclusion criteria were: i) Patients were diagnosed with other types of tumor; ii) postoperative recurrent patients; and iii) patients had severe functional diseases, such as heart, liver, kidney and immunological diseases; iv) patients were treated with other therapies before surgery; and v) patients were pregnant.
All specimens were handled and made anonymous according to ethical and legal standards. Tissue samples were collected during the surgery for GC and were confirmed by tissue pathology examination. The patients were divided into diffuse-type and intestinal-type GC groups (n=20 in each group) according to the postoperative pathology results. All tumor tissues specimens were collected from formalin-fixed paraffin-embedded tissues of resection surgical procedures.
Immunohistochemical analysis
Immunohistochemistry was performed to determine the expressions of angiotensinogen (AGT), C-X-C motif chemokine ligand 12 (CXCL12) and adrenoceptor β2 (ADRB2) in diffuse-type intestinal-type GC tissue samples. GC tissues were fixed in 10% neutral buffered formalin at room temperature for 48 h and embedded in paraffin at 62°C for 45 min. The sections were then cut into 4-µm thick sections and dried overnight at 56°C. Paraffin-embedded tissue were passed through dimethylbenzene and gradient ethanol solution to deparaffinize and rehydrate the sections. Antigen retrieval was performed by heating the sections in a microwave oven in 10 mM sodium citrate-hydrochloric acid buffer (pH 6.0) for ~15 min at 95°C. To block endogenous peroxidase activity, 0.3% peroxidase quenching solution was used at 37°C for 10 min. After blocking for 30 min with 10% skim milk at 37°C, each section was incubated with a rabbit anti-human AGT antibody (1:50; cat. no. ab108334; Abcam), rabbit anti-human CXCL12 antibody (1:200; cat. no. ab9797; Abcam) or rabbit anti-human ADRB2 antibody (1:100; cat. no. ab182136; Abcam) overnight at 4°C. The slides were then incubated with HRP-secondary antibodies (Abcam; cat. no. ab6721; 1:1,000) at room temperature for 30 min. The sections then were incubated with 3,3′-diaminobenzidine (DAB) solution at room temperature for 5 min, and every slide were counterstained with hematoxylin at room temperature for 1 min, dehydrated, and sealed with cover slips. The stained sections were examined under an optical microscope.
For quantitative analysis, images from each sample were analyzed using Image-Pro Plus 6.0 software (Media Cybernetics). For each slide, three randomly selected regions were examined under the same exposure time and white balance setting. The blank area of the images was selected for optical density correction. Positive expression was defined as brown-yellow granules in the cytoplasm. The expression intensity was calculated as MD=IOD/area of target region, where MD is the mean density and IOD is the accumulated value of integrated optical density. The MD of the three fields of view was used to quantify the expression levels of the hub genes.
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
RT-qPCR was used to examine the expression levels of AGT, CXCL12 and ADRB2 between diffuse- and intestinal-type GC tissue samples. Total RNA was extracted with the TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. RNA purity was detected using a microplate reader (Infinite M1000 PRO; Tecan Group, Ltd.). A PrimeScript™ RT reagent kit (Takara Bio, Inc.) was used for cDNA synthesis, according to the manufacturer's protocol, at 37°C for 15 min and 85°C for 5 sec. RT-qPCR was carried out using SYBR® Premix ExTaq™ (Takara Bio, Inc.) on an ABI Prism 7500 Sequence Detection System (Applied Biosystems; Thermo Fisher Scientific, Inc.). GAPDH was used as reference gene. The relative mRNA expression levels were quantified using the 2−ΔΔCq method (26). The following thermocycling conditions were used: Initial denaturation at 95°C for 7 min; followed by 40 cycles of 95°C for 15 sec and 60°C for 30 sec; and a final extension at 72°C for 30 sec. Primers were as follows: i) AGT forward, 5′-CCCTGGCTTTCAACACCTAC-3′ and reverse, 5′-CTGTGGGCTCTCTCTCATCC-3′; ii) CXCL12 forward, 5′-GATTGTAGCCCGGCTGAAGA-3′ and reverse, 5′-TTCGGGTCAATGCACACTTGT-3′; iii) ADRB2 forward, 5′-AACTGGTTGGGCTATGTCAA-3′ and reverse, 5′-GTTAGTGTCCTGTCAGGGAG-3′; and iv) GAPDH forward, 5′-TGTGGGCATCAATGGATTTGG-3′ and reverse 5′-ACACCATGTATTCCGGGTCAAT-3′.
Kaplan-Meier analysis of hub genes
Kaplan-Meier Plotter (https://kmplot.com/analysis/) was used to examine the effect of hub genes on OS of patients with diffuse-type GC. In order to improve the reliability of the results, the GEO GSE62254 (9,18) and GSE15459 (27,28) datasets were used as the research target. The three genes (AGT, CXCL12 and ADRB2) were uploaded into the database to obtain the Kaplan-Meier survival plots. Log rank P-value and hazard ratio (HR) with 95% confidence intervals were calculated. Patients with expression above the median (high expression) are indicated in the red line, and patients with expressions below the median (low expression) are indicated in the black line. P<0.05 was the cut-off criterion.
Statistical analysis
The differences between the groups were compared using an unpaired Student's t-test on Graph Prism 7.0 (GraphPad Software, Inc.). P<0.05 was considered to indicate a statistically significant difference.
Results
DEG identification
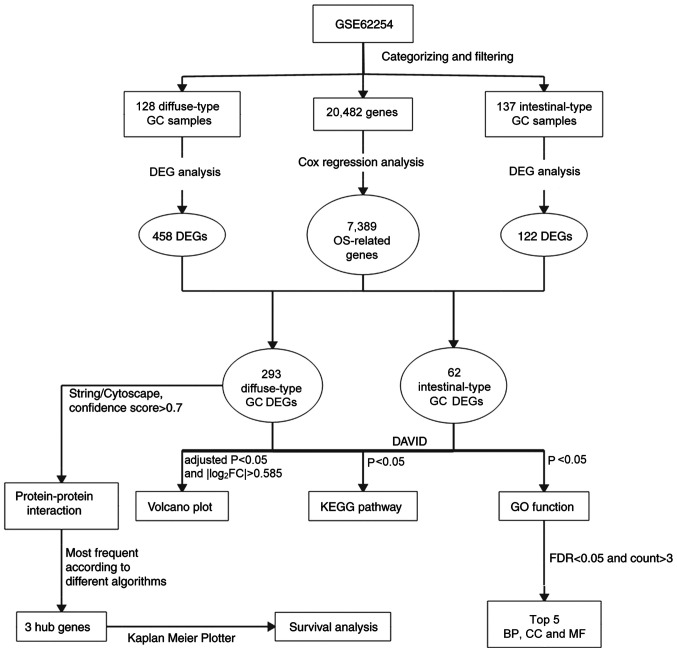
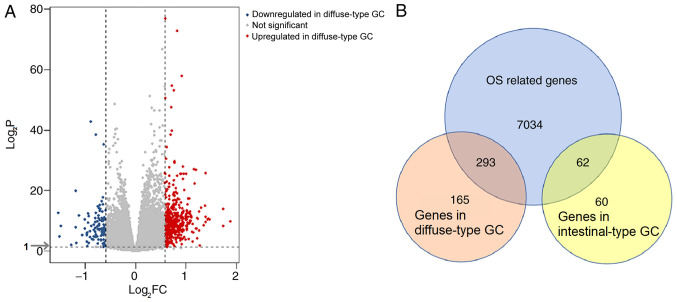
A flowchart of the bioinformatics analytical methods is presented in Fig. 1. The GSE62254 database includes 300 different Lauren-subtype GC samples. Of these, 265 genes with definite Lauren subtypes and survival data were singled out, including 128 diffuse-type and 137 intestinal-type GC samples. Detailed information regarding these samples is presented in Table SI. According to the screening criteria of |log2FC|>0.585 and adjusted P<0.05, 584 DEGs, including 458 genes associated with diffuse-type and 122 genes associates with intestinal-type GC, were identified (Fig. 2A). To identify genes with prognostic value, Cox regression analysis was carried out on the GSE62254 dataset. A total of 7,389 genes were identified as significantly associated with OS. Of these OS-related DEGs, 293 genes were identified in diffuse-type GC samples and 62 in intestinal-type GC (Fig. 2B).
Figure 1.
Flowchart of the bioinformatics analysis. GC, gastric cancer; OS, overall survival; DEG, differentially expressed gene; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; DAVID, Database for Annotation, Visualization and Integrated Discovery; FDR, false discovery rate; BP, biological process; CC, cellular component; MF, molecular function.
Figure 2.
Analysis of DEGs. (A) Volcano plot of DEGs identified in the GSE62254 dataset. Red symbols represent genes highly associated with diffuse-type GC, whereas blue symbols represent genes upregulated in intestinal-type GC. DEGs were screened according to |logFC|>0.585. (B) Venn diagrams of subtype-specific DEGs screened through the intersection of OS-related genes with intestinal- and diffuse-type GC genes. GC, gastric cancer; DEG, differentially expressed gene; OS, overall survival.
GO enrichment and KEGG pathway analyses of DEGs
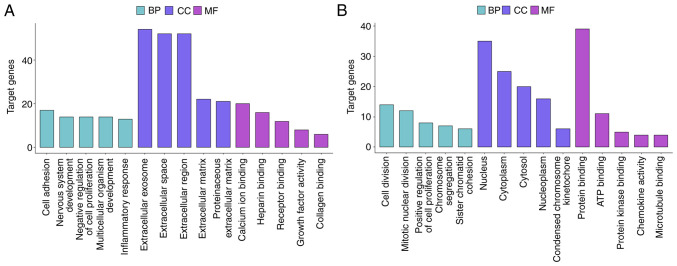
DEGs associated with diffuse-type and intestinal-type GC were functionally annotated using GO and analyzed using KEGG pathway analysis. GO analysis suggested that diffuse-type DEGs were primarily enriched in the ‘cell adhesion’ BP, the ‘extracellular exosome’ CC and the ‘calcium ion’ binding MF. By contrast, intestinal-type DEGs were primarily enriched in the ‘cell division’ BP, the ‘nucleus’ CC and the ‘protein binding’ MF (Figs. 3 and S1A and B). The top 5 BP, CC and MF results from the GO enrichment analysis of the subtype-specific DEGs are listed in Table I.
Figure 3.
Top 5 enriched GO terms. (A) Top 5 BPs, CCs and MFs in DEGs associated with diffuse-type GC. (B) Top 5 BPs, CCs and MFs in DEGs associated with intestinal-type GC. GC, gastric cancer; DEG, differentially expressed gene; GO, Gene Ontology; BP, biological process; CC, cellular component; MF, molecular function.
Table I.
Top 5 enriched GO terms associated with GC subtype-specific differentially expressed genes.
| A, Diffuse-type GC | ||||
|---|---|---|---|---|
| Category | Term | GO ID | Count | P-value |
| BP | Cell adhesion | GO:0007155 | 17 | 6×10−4 |
| BP | Nervous system development | GO:0007399 | 14 | 1×10−4 |
| BP | Negative regulation of cell proliferation | GO:0008285 | 14 | 3×10−3 |
| BP | Multicellular organism development | GO:0007275 | 14 | 2×10−2 |
| BP | Inflammatory response | GO:0006954 | 13 | 6×10−3 |
| CC | Extracellular exosome | GO:0070062 | 54 | 1×10−2 |
| CC | Extracellular space | GO:0005615 | 52 | 5×10−11 |
| CC | Extracellular region | GO:0005576 | 52 | 2×10−8 |
| CC | Extracellular matrix | GO:0031012 | 22 | 1×10−9 |
| CC | Proteinaceous extracellular matrix | GO:0005578 | 21 | 1×10−9 |
| MF | Calcium ion binding | GO:0005509 | 20 | 2×10−3 |
| MF | Heparin binding | GO:0008201 | 16 | 2×10−9 |
| MF | Receptor binding | GO:0005102 | 12 | 6×10−3 |
| MF | Growth factor activity | GO:0008083 | 8 | 5×10−3 |
| MF | Collagen binding | GO:0005518 | 6 | 1×10−3 |
| B, Intestinal-type GC | ||||
| Category | Term | GO ID | Count | P-value |
| BP | Cell division | GO:0051301 | 14 | 7×10−11 |
| BP | Mitotic nuclear division | GO:0007067 | 12 | 3×10−10 |
| BP | Positive regulation of cell proliferation | GO:0008284 | 8 | 7×10−4 |
| BP | Chromosome segregation | GO:0007059 | 7 | 8×10−8 |
| BP | Sister chromatid cohesion | GO:0007062 | 6 | 2×10−5 |
| CC | Nucleus | GO:0005634 | 35 | 2×10−6 |
| CC | Cytoplasm | GO:0005737 | 25 | 2×10−2 |
| CC | Cytosol | GO:0005829 | 20 | 4×10−3 |
| CC | Nucleoplasm | GO:0005654 | 16 | 2×10−2 |
| CC | Condensed chromosome kinetochore | GO:0000777 | 6 | 7×10−6 |
| MF | Protein binding | GO:0005515 | 39 | 2×10−3 |
| MF | ATP binding | GO:0005524 | 11 | 1×10−2 |
| MF | Protein kinase binding | GO:0019901 | 5 | 3×10−2 |
| MF | Chemokine activity | GO:0008009 | 4 | 4×10−4 |
| MF | Microtubule binding | GO:0008017 | 4 | 2×10−2 |
GO, Gene Ontology; GC, gastric cancer; BP, biological process; CC, cellular component; MF, molecular function.
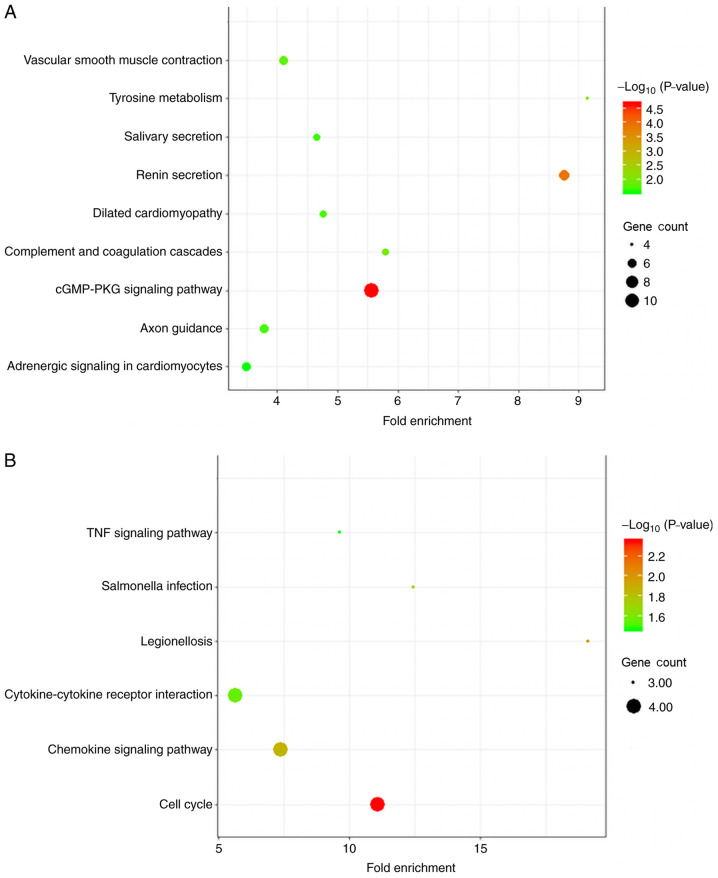
In the KEGG pathway analysis, diffuse-type GC was primarily enriched in DEGs associated with ‘cGMP-PKG signaling pathway’. Intestinal-type DEGs were primarily enriched in ‘cell cycle’ (Fig. 4 and Table II).
Figure 4.
KEGG pathway enrichment analysis of GC subtype-specific DEGs. (A) DEGs in diffuse-type GC. (B) DEGs in intestinal-type GC. GC, gastric cancer; DEG, differentially expressed gene; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Table II.
KEGG pathway analysis of GC subtype-specific differentially expressed genes.
| A, Diffuse-type GC | |||
|---|---|---|---|
| Term | KEGG ID | Count | P-value |
| cGMP-PKG signaling pathway | hsa04022 | 11 | 2.19E-05 |
| Renin secretion | hsa04924 | 7 | 1.26E-04 |
| Tyrosine metabolism | hsa00350 | 4 | 0.00896 |
| Complement and coagulation cascades | hsa04610 | 5 | 0.01021 |
| Vascular smooth muscle contraction | hsa04270 | 6 | 0.01456 |
| Dilated cardiomyopathy | hsa05414 | 5 | 0.01983 |
| Axon guidance | hsa04360 | 6 | 0.02008 |
| Salivary secretion | hsa04970 | 5 | 0.02142 |
| Adrenergic signaling in cardiomyocytes | hsa04261 | 6 | 0.02758 |
| B, Intestinal-type GC | |||
| Term | KEGG ID | Count | P-value |
| Cell cycle | hsa04110 | 4 | 0.00449 |
| Legionellosis | hsa05134 | 3 | 0.0095 |
| Chemokine signaling pathway | hsa04062 | 4 | 0.0137 |
| Salmonella infection | hsa05132 | 3 | 0.02153 |
| Cytokine-cytokine receptor interaction | hsa04060 | 4 | 0.02776 |
| TNF signaling pathway | hsa04668 | 3 | 0.03451 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; GC, gastric cancer.
Construction of the PPI network
To further examine subtype-specific genes in diffuse-type GC, a PPI network of the 293 DEGs associated with diffuse-type GC was constructed. A total of 112 nodes and 182 interactions were involved in the PPI network (Fig. S1C). The results were analyzed in Cytoscape software, and the top 15 hub genes were ranked using four different algorithms of the CytoHubba plugin according to predicted scores. Only those genes that overlapped in the results of all four ranking methods were used for further study, and three overlapping hub genes were identified for further analysis, namely, AGT, CXCL12 and ADRB2 (Table III).
Table III.
Hub genes for diffuse type DEGs ranked in cytoHubba plugin of Cytoscape.
| Rank methods in cytoHubba | ||||
|---|---|---|---|---|
| Rank | Degree | EPC | Closeness | EcCentricity |
| 1 | AGTa | AGTa | AGTa | PRKAR2B |
| 2 | CXCL12a | CXCL12a | CXCL12a | ITGA1 |
| 3 | IGF1 | S1PR1 | PRKAR2B | GRP |
| 4 | EDNRB | EDNRB | EDNRB | ACTG2 |
| 5 | FBN1 | CXCL13 | IGF1 | MYH11 |
| 6 | IGFBP5 | ARHGEF25 | ADRB2a | AGTa |
| 7 | CCL19 | GRP | ITGA1 | CXCL12a |
| 8 | MYH11 | CX3CR1 | FGF2 | LMOD1 |
| 9 | TAC1 | TAC1 | CCL19 | ADRB2a |
| 10 | GRP | EDNRA | CXCL13 | ITGA8 |
| 11 | ADRB2a | CCL19 | CX3CR1 | CAV1 |
| 12 | ITGA1 | P2RY12 | S1PR1 | THBS4 |
| 13 | SCG2 | HTR2B | P2RY12 | EDNRA |
| 14 | EDNRA | ADRB2a | FGF13 | SLIT2 |
| 15 | CXCL13 | PRKAR2B | TAC1 | ROBO1 |
Overlapping genes in the top 15 hub genes identified using four ranking methods in cytoHubba. EPC, edge percolated component.
AGT, CXCL12 and ADRB2 expression is increased in diffuse-type GC tissues
Immunohistochemical analysis indicated that the staining intensity of AGT, CXCL12 and ADRB2 was associated with GC Lauren subtypes. Indeed, MD values for AGT, CXCL12 and ADRB2 proteins in the diffuse-type GC samples were significantly increased, compared with intestinal-type GC samples (P=0.023 for AGT, P=0.011 for CXCL12 and P=0.007 for ADRB2, respectively; Table IV). Thus, compared with intestinal-type GC tissue samples, AGT, CXCL12 and ADRB2 stained more strongly in diffuse-type GC tissues (Fig. 5).
Table IV.
Immunohistochemical staining intensity results for AGT, CXCL12 and ADRB2.
| Gene | Intestinal-type GC | Diffuse-type GC | P-value |
|---|---|---|---|
| AGT | 2.36±1.29 | 6.87±4.37a | 0.023 |
| CXCL12 | 1.92±1.38 | 6.51±4.83a | 0.011 |
| ADRB2 | 2.32±0.95 | 7.68±5.13a | 0.007 |
P<0.05, vs. intestinal-type GC. GC, gastric cancer; AGT, angiotensinogen; CXCL12, C-X-C motif chemokine ligand 12; ADRB2, adrenoceptor β2.
Figure 5.
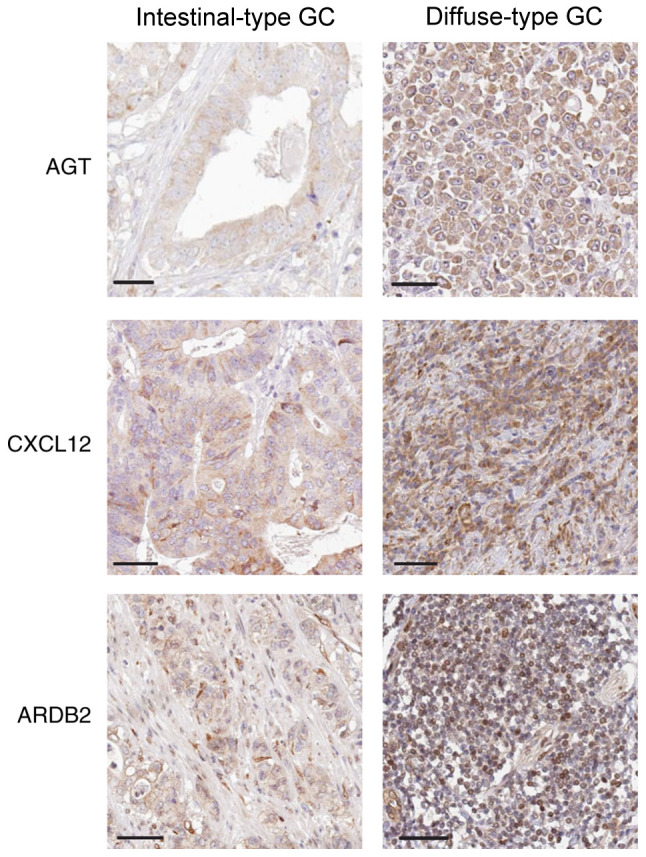
Immunohistochemical detection of AGT, CXCL12 and ADRB2 expression in intestinal and diffuse-type GC tissue samples. Magnification, ×400. Scale bar, 50 µm. GC, gastric cancer; AGT, angiotensinogen; CXCL12, C-X-C motif chemokine ligand 12; ADRB2, adrenoceptor β2.
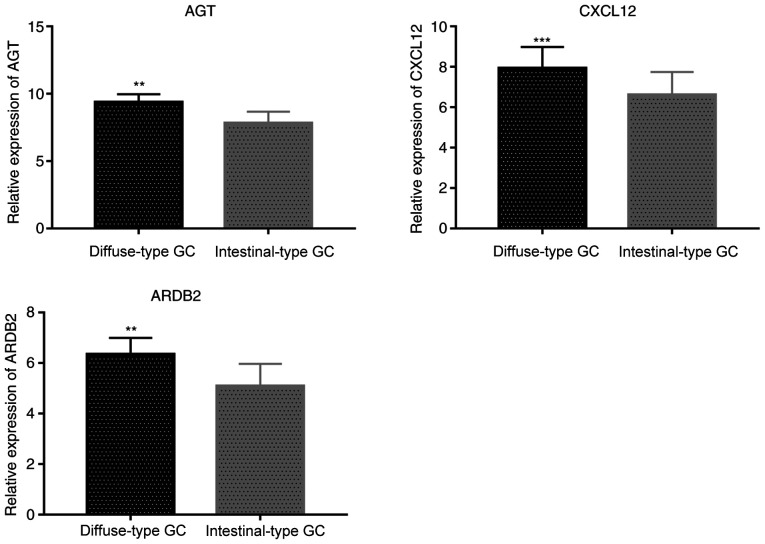
RT-qPCR results were consistent with immunohistochemistry analysis, showing that expression levels of AGT, CXCL12 and ADRB2 were significantly higher in diffuse-type GC than in intestinal-type GC tissues (P=0.0067 for AGT, P=0.00018 for CXCL12 and P=0.0043 for ADRB2, respectively; Fig. 6).
Figure 6.
Reverse transcription-quantitative PCR validation of AGT, CXCL12 and ADRB2 expression levels. **P<0.01, ***P<0.001. GC, gastric cancer; AGT, angiotensinogen; CXCL12, C-X-C motif chemokine ligand 12; ADRB2, adrenoceptor β2.
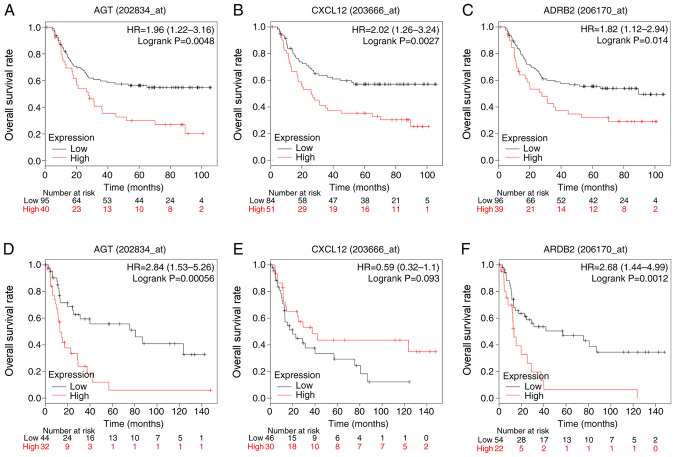
Association between hub genes and OS in patients with diffuse-type GC
To evaluate the prognostic value of the three identified hub genes in diffuse-type GC, Kaplan-Meier analysis was carried out. To improve the reliability of results, two datasets, GSE62254 and GSE15459, were used. In the GSE62254, high expression of AGT (P=0.0048), CXCL12 (P=0.0027) and ADRB2 (P=0.014) was associated with reduced overall survival rates in patients with diffuse-type GC patients. In GSE15459, high expression of AGT (P=0.00056) and ARDB2 (P=0.0012) presented similar results, indicating a poor prognosis for diffuse-type GC patients. However, expression of CXCL12 (P=0.093) was not associated with overall survival in this dataset (Fig. 7).
Figure 7.
Overall survival curves of hub genes in diffuse-type GC. The x-axis represents overall survival time and the y-axis represents survival rate. Patients with expression above the median are indicated in red line, and patients with expressions below the median in black line (A-C) Survival analysis evaluating the effects (A) AGT, (B) CXCL12 and (C) ADRB2 expression on survival in the GSE62254 dataset. (D-F) Survival analysis evaluating the effects of (D) AGT, (E) CXCL12 and (F) ADRB2 expression on survival in the GSE15459 dataset. GC, gastric cancer; AGT, angiotensinogen; CXCL12, C-X-C motif chemokine ligand 12; ADRB2, adrenoceptor β2, HR, hazard ratio.
Discussion
GC is a highly heterogeneous disease. Since the Lauren classification was first proposed in 1965, it has been widely recognized by clinicians and pathologists and is still currently used. For many years, the value of histopathological classification for evaluating the prognosis of GC has been very limited, and Lauren classification is considered the most valuable clinicopathological classification. There are significant differences between different Lauren subtypes (29–31), suggesting that some specific biomarkers might play an important role during the pathogenesis and development of GC. Although several studies have examined the mechanism of the occurrence and development of GC, few have evaluated specific GC subtypes.
To identify genes specifically associated with Lauren GC subtypes, the GSE62254 dataset was used, which contains 265 samples of diffuse or intestinal GC with survival data. A total of 598 DEGs identified, including 293 diffuse-type DEGs and 62 intestinal-type DEGs. To predict the biological pathways and functions involving these DEGs, GO and KEGG analyses were performed. To identify key genes for diffuse-type GC progression among the numerous DEGs, the top 15 hub genes in a PPI network were identified and screened using four different algorithms. Genes overlapping in the results of all four algorithms were then identified as hub genes and selected for further study, namely, AGT, CXCL12 and ADRB2. All three hub genes were associated with OS in patients with diffuse-type GC. Thus, these hub genes may represent novel diagnostic and prognostic biomarkers for diffuse-type GC.
The AGT gene encodes pre-angiotensinogen, also known as the angiotensinogen precursor protein, which is primarily expressed in the liver and cleaved by the renin enzyme in response to reduced blood pressure. The resulting product, angiotensin 1 (Ang I), is cleaved by the AGT-converting enzyme (ACE) to produce angiotensin 2 (Ang II). Thus, the product of AGT constitutes a key component of the renin-angiotensin system (RAS) (32). RAS has been implicated in arterial hypertension, kidney disease, and other cardiovascular conditions (33–35), as well as cancer growth and dissemination (36). RAS components are expressed in tumor microenvironments and directly or indirectly affect cell proliferation, invasion, migration, metastasis, apoptosis, angiogenesis, and cancer-associated inflammation and immunomodulation (36,37). RAS can also promote tumor growth indirectly, for instance, by regulating cancer-associated fibroblasts (38) and promoting VEGF-mediated angiogenesis (39,40) in solid tumors. Based on the results of the present study, it was hypothesized that AGT might be a potential indicator for the diagnosis and prognosis of diffuse-type GC.
The CXCL12 gene, also known as stromal cell-derived factor 1 (SDF1), encodes a chemokine of the intercrine family. The CXCL12 chemokine binds primarily to the CXC receptor 4 (CXCR4) receptor, playing an essential role in diverse cellular functions (41–43). CXCR4 is widely expressed on hematopoietic cells, embryonic pluripotent stem cells and several types of tissue-committed stem cells (44), which have direct or indirect proangiogenic properties. The CXCL12/CXCR4 axis is associated with tumor progression, angiogenesis, metastasis, and survival. For example, CXCL12 overexpression enhances the proliferation and invasion of colon cancer cells through the MAPK/PI3K/AP-1 signaling pathway (45). High CXCR4 expression may represent a biomarker indicating poor prognosis for hepatocellular carcinoma patients (46). However, in the present study, the survival curves from two datasets of patients with diffuse-type GC divided according to CXCL12 expression exhibited opposite trends. This discrepancy could stem from differential patient characteristics, such as ethnicity, sex and Helicobacter pylori infection. CXCL12 has been reported as a prognostic marker in a variety of cancers according to the literature, including GC (47). According to the present results, the relationship between the expression of CXCL12 and its prognosis in human diffuse type GC between GSE62254 and GSE15459 presented different results. To explain differences between the two datasets, more comprehensive and precise analysis is needed.
The ADRB2 gene encodes the β2-adrenergic receptor, which belongs to the G protein-coupled receptor superfamily. The ADRB2 protein increases cAMP, and downstream L-type calcium channel interaction via adenylate cyclase stimulation through trimeric G-proteins, thereby mediating physiological responses, such as bronchodilation and smooth muscle relaxation (48). Previous studies have indicated that ARDB2 also plays an important role in several cancer types. For instance, ADRB2 signaling negatively regulates autophagy, leading to hypoxia-inducible factor-1α stabilization and inducing sorafenib resistance in hepatocellular carcinoma (HCC) (49). Moreover, ADRB2 expression is associated with prognosis of patients with HCC (50). In prostate cancer, high ADRB2 expression levels activated an angiogenic switch, regulated angiogenesis and affected the phenotype of prostate cells and promoted their ability to migrate and invade (51). In GC, chronic stress caused by stress hormone-induced activation of the ADRB2 signaling pathway plays a crucial role in progression and metastasis (52). Additionally, ADRB2 signaling regulates GC progression (53). In the present study, ARDB2 was highly expressed in diffuse-type GC and was associated with OS.
There are certain limitations to the present study. Only using immunohistochemical and RT-qPCR methods to study the qualitative and semiquantitative expression of AGT, CXCL12 and ADRB2 in diffuse-type GC may yield results that are not sufficiently rigorous. These data should be supplemented with further experiments to validate expression levels of AGT, CXCL12 and ADRB2 in GC tissue. Furthermore, key signaling pathways related to the identified hub genes should also be examined in in vitro experiments.
In summary, the present study demonstrated that expression of AGT, CXCL12 and ADRB2 was increased in diffuse-type GC, relative to intestinal-type GC, and that expression levels of these proteins negatively correlated with disease prognosis.
Supplementary Material
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the GEO data repository, [https://www.ncbi.nlm.nih.gov/geo]. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
GW conceived and designed the present study. SL and CY collected, extracted and analyzed the data. SL and FD performed the experiments and interpreted the data. SL wrote the manuscript. YC, FD and GW contributed to the data collection and performed the statistical analysis. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by The Ethics Committee of The Third Affiliated Hospital of Anhui Medical University. Written informed consent was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Allemani C, Weir HK, Carreira H, Harewood R, Spika D, Wang XS, Bannon F, Ahn JV, Johnson CJ, Bonaventure A, et al. Global surveillance of cancer survival 1995–2009: Analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2) Lancet. 2015;385:977–1010. doi: 10.1016/S0140-6736(14)62038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Spolverato G, Ejaz A, Kim Y, Squires MH, Poultsides GA, Fields RC, Schmidt C, Weber SM, Votanopoulos K, Maithel SK, Pawlik TM. Rates and patterns of recurrence after curative intent resection for gastric cancer: A United States multi-institutional analysis. J Am Coll Surg. 2014;219:664–675. doi: 10.1016/j.jamcollsurg.2014.03.062. [DOI] [PubMed] [Google Scholar]
- 4.In H, Solsky I, Palis B, Langdon-Embry M, Ajani J, Sano T. Validation of the 8th edition of the AJCC TNM staging system for gastric cancer using the national cancer database. Ann Surg Oncol. 2017;24:3683–3691. doi: 10.1245/s10434-017-6078-x. [DOI] [PubMed] [Google Scholar]
- 5.Borrmann R. Geschwulste des magens und des duodenums. In: Henke F, Lubarsch O, editors. Handbuch spez pathol anat und histo. Springer Verlag; Berlin: 1926. pp. 812–1054. (In German) [Google Scholar]
- 6.Lauren P. The two histological main types of gastric carcinoma: Diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- 7.Fléjou JF. WHO Classification of digestive tumors: The fourth edition. Ann Pathol. 2011;31:S27–S31. doi: 10.1016/j.annpat.2011.08.001. (In French) [DOI] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research Network, corp-author. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS, Liu J, Yue YG, Wang J, Yu K, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015;21:449–456. doi: 10.1038/nm.3850. [DOI] [PubMed] [Google Scholar]
- 10.Zheng H, Takahashi H, Murai Y, Cui Z, Nomoto K, Miwa S, Tsuneyama K, Takano Y. Pathobiological characteristics of intestinal and diffuse-type gastric carcinoma in Japan: An immunostaining study on the tissue microarray. J Clin Pathol. 2007;60:273–277. doi: 10.1136/jcp.2006.038778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiu MZ, Cai MY, Zhang DS, Wang ZQ, Wang DS, Li YH, Xu RH. Clinicopathological characteristics and prognostic analysis of Lauren classification in gastric adenocarcinoma in China. J Transl Med. 2013;11:58. doi: 10.1186/1479-5876-11-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gong EJ, Lee JY, Bae SE, Park YS, Choi KD, Song HJ, Lee GH, Jung HY, Jeong WJ, Cheon GJ, et al. Characteristics of non-cardia gastric cancer with a high serum anti-Helicobacter pylori IgG titer and its association with diffuse-type histology. PLoS One. 2018;13:e0195264. doi: 10.1371/journal.pone.0195264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang CT, Zeng L, Yang J, Zeng C, Chen Y. Analysis of the incidence and survival of gastric cancer based on the lauren classification: A large population-based study using SEER. Front Oncol. 2020;10:1212. doi: 10.3389/fonc.2020.01212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 15.Yang G, Zhang Y, Yang J. Identification of potentially functional CircRNA-miRNA-mRNA regulatory network in gastric carcinoma using bioinformatics analysis. Med Sci Monit. 2019;25:8777–8796. doi: 10.12659/MSM.916902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen S, Pan S, Wu H, Zhou J, Huang Y, Wang S, Liu A. ICAM1 regulates the development of gastric cancer and may be a potential biomarker for the early diagnosis and prognosis of gastric cancer. Cancer Manag Res. 2020;12:1523–1534. doi: 10.2147/CMAR.S237443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li R, Jiang J, Shi H, Qian H, Zhang X, Xu W. CircRNA: A rising star in gastric cancer. Cell Mol Life Sci. 2020;77:1661–1680. doi: 10.1007/s00018-019-03345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh SC, Sohn BH, Cheong JH, Kim SB, Lee JE, Park KC, Lee SH, Park JL, Park YY, Lee HS, et al. Clinical and genomic landscape of gastric cancer with a mesenchymal phenotype. Nat Commun. 2018;9:1777. doi: 10.1038/s41467-018-04179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson CL, Miller CJ. Simpleaffy: A BioConductor package for Affymetrix quality control and data analysis. Bioinformatics. 2005;21:3683–3685. doi: 10.1093/bioinformatics/bti605. [DOI] [PubMed] [Google Scholar]
- 20.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 21.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Therneau TM, Grambsch PM. Modeling Survival Data: Extending the Cox Model. Springer; Berlin: 2000. The Cox model; pp. 39–77. [Google Scholar]
- 23.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Ooi CH, Ivanova T, Wu J, Lee M, Tan IB, Tao J, Ward L, Koo JH, Gopalakrishnan V, Zhu Y, et al. Oncogenic pathway combinations predict clinical prognosis in gastric cancer. PLoS Genet. 2009;5:e1000676. doi: 10.1371/journal.pgen.1000676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muratani M, Deng N, Ooi WF, Lin SJ, Xing M, Xu C, Qamra A, Tay ST, Malik S, Wu J, et al. Nanoscale chromatin profiling of gastric adenocarcinoma reveals cancer-associated cryptic promoters and somatically acquired regulatory elements. Nat Commun. 2014;5:4361. doi: 10.1038/ncomms5361. [DOI] [PubMed] [Google Scholar]
- 29.Qiu M, Zhou Y, Zhang X, Wang Z, Wang F, Shao J, Lu J, Jin Y, Wei X, Zhang D, et al. Lauren classification combined with HER2 status is a better prognostic factor in Chinese gastric cancer patients. BMC Cancer. 2014;14:823. doi: 10.1186/1471-2407-14-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen YC, Fang WL, Wang RF, Liu CA, Yang MH, Lo SS, Wu CW, Li AF, Shyr YM, Huang KH. Clinicopathological variation of lauren classification in gastric cancer. Pathol Oncol Res. 2016;22:197–202. doi: 10.1007/s12253-015-9996-6. [DOI] [PubMed] [Google Scholar]
- 31.Janjigian YY, Werner D, Pauligk C, Steinmetz K, Kelsen DP, Jäger E, Altmannsberger HM, Robinson E, Tafe LJ, Tang LH, et al. Prognosis of metastatic gastric and gastroesophageal junction cancer by HER2 status: A European and USA international collaborative analysis. Ann Oncol. 2012;23:2656–2662. doi: 10.1093/annonc/mds104. [DOI] [PubMed] [Google Scholar]
- 32.Lu H, Cassis LA, Kooi CW, Daugherty A. Structure and functions of angiotensinogen. Hypertens Res. 2016;39:492–500. doi: 10.1038/hr.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kobori H, Nangaku M, Navar LG, Nishiyama A. The intrarenal renin-angiotensin system: From physiology to the pathobiology of hypertension and kidney disease. Pharmacol Rev. 2007;59:251–287. doi: 10.1124/pr.59.3.3. [DOI] [PubMed] [Google Scholar]
- 34.Ferrario CM. Role of angiotensin II in cardiovascular disease therapeutic implications of more than a century of research. J Renin Angiotensin Aldosterone Syst. 2006;7:3–14. doi: 10.3317/jraas.2006.003. [DOI] [PubMed] [Google Scholar]
- 35.Paul M, Poyan Mehr A, Kreutz R. Physiology of local renin-angiotensin systems. Physiol Rev. 2006;86:747–803. doi: 10.1152/physrev.00036.2005. [DOI] [PubMed] [Google Scholar]
- 36.George AJ, Thomas WG, Hannan RD. The renin-angiotensin system and cancer: Old dog, new tricks. Nat Rev Cancer. 2010;10:745–759. doi: 10.1038/nrc2945. [DOI] [PubMed] [Google Scholar]
- 37.Ager EI, Neo J, Christophi C. The renin-angiotensin system and malignancy. Carcinogenesis. 2008;29:1675–1684. doi: 10.1093/carcin/bgn171. [DOI] [PubMed] [Google Scholar]
- 38.Chauhan VP, Martin JD, Liu H, Lacorre DA, Jain SR, Kozin SV, Stylianopoulos T, Mousa AS, Han X, Adstamongkonkul P, et al. Angiotensin inhibition enhances drug delivery and potentiates chemotherapy by decompressing tumour blood vessels. Nat Commun. 2013;4:2516. doi: 10.1038/ncomms3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ji Y, Wang Z, Li Z, Li K, Le X, Zhang T. Angiotensin II induces angiogenic factors production partly via AT1/JAK2/STAT3/SOCS3 signaling pathway in MHCC97H cells. Cell Physiol Biochem. 2012;29:863–874. doi: 10.1159/000171034. [DOI] [PubMed] [Google Scholar]
- 40.Anandanadesan R, Gong Q, Chipitsyna G, Witkiewicz A, Yeo CJ, Arafat HA. Angiotensin II induces vascular endothelial growth factor in pancreatic cancer cells through an angiotensin II type 1 receptor and ERK1/2 signaling. J Gastrointest Surg. 2008;12:57–66. doi: 10.1007/s11605-007-0403-9. [DOI] [PubMed] [Google Scholar]
- 41.Balkwill F. Cancer and the chemokine network. Nat Rev Cancer. 2004;4:540–550. doi: 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]
- 42.Murdoch C. CXCR4: Chemokine receptor extraordinaire. Immunol Rev. 2000;177:175–184. doi: 10.1034/j.1600-065X.2000.17715.x. [DOI] [PubMed] [Google Scholar]
- 43.Balabanian K, Lagane B, Infantino S, Chow KY, Harriague J, Moepps B, Arenzana-Seisdedos F, Thelen M, Bachelerie F. The chemokine SDF-1/CXCL12 binds to and signals through the orphan receptor RDC1 in T lymphocytes. J Biol Chem. 2005;280:35760–35766. doi: 10.1074/jbc.M508234200. [DOI] [PubMed] [Google Scholar]
- 44.Ratajczak MZ, Zuba-Surma E, Kucia M, Reca R, Wojakowski W, Ratajczak J. The pleiotropic effects of the SDF-1-CXCR4 axis in organogenesis, regeneration and tumorigenesis. Leukemia. 2006;20:1915–1924. doi: 10.1038/sj.leu.2404357. [DOI] [PubMed] [Google Scholar]
- 45.Ma J, Su H, Yu B, Guo T, Gong Z, Qi J, Zhao X, Du J. CXCL12 gene silencing down-regulates metastatic potential via blockage of MAPK/PI3K/AP-1 signaling pathway in colon cancer. Clin Transl Oncol. 2018;20:1035–1045. doi: 10.1007/s12094-017-1821-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liepelt A, Tacke F. Stromal cell-derived factor-1 (SDF-1) as a target in liver diseases. Am J Physiol Gastrointest Liver Physiol. 2016;311:G203–G209. doi: 10.1152/ajpgi.00193.2016. [DOI] [PubMed] [Google Scholar]
- 47.Ishigami S, Natsugoe S, Okumura H, Matsumoto M, Nakajo A, Uenosono Y, Arigami T, Uchikado Y, Setoyama T, Arima H, et al. Clinical implication of CXCL12 expression in gastric cancer. Ann Surg Oncol. 2007;14:3154–3158. doi: 10.1245/s10434-007-9521-6. [DOI] [PubMed] [Google Scholar]
- 48.Yang-Feng TL, Xue FY, Zhong WW, Cotecchia S, Frielle T, Caron MG, Lefkowitz RJ, Francke U. Chromosomal organization of adrenergic receptor genes. Proc Natl Acad Sci USA. 1990;87:1516–1520. doi: 10.1073/pnas.87.4.1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu FQ, Fang T, Yu LX, Lv GS, Lv HW, Liang D, Li T, Wang CZ, Tan YX, Ding J, et al. ADRB2 signaling promotes HCC progression and sorafenib resistance by inhibiting autophagic degradation of HIF1α. J Hepatol. 2016;65:314–324. doi: 10.1016/j.jhep.2016.04.019. [DOI] [PubMed] [Google Scholar]
- 50.Chen D, Xing W, Hong J, Wang M, Huang Y, Zhu C, Yuan Y, Zeng W. The beta2-adrenergic receptor is a potential prognostic biomarker for human hepatocellular carcinoma after curative resection. Ann Surg Oncol. 2012;19:3556–3565. doi: 10.1245/s10434-012-2396-1. [DOI] [PubMed] [Google Scholar]
- 51.Zahalka AH, Arnal-Estapé A, Maryanovich M, Nakahara F, Cruz CD, Finley LWS, Frenette PS. Adrenergic nerves activate an angio-metabolic switch in prostate cancer. Science. 2017;358:321–326. doi: 10.1126/science.aah5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang X, Zhang Y, He Z, Yin K, Li B, Zhang L, Xu Z. Chronic stress promotes gastric cancer progression and metastasis: An essential role for ADRB2. Cell Death Dis. 2019;10:788. doi: 10.1038/s41419-019-2030-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhi X, Li B, Li Z, Zhang J, Yu J, Zhang L, Xu Z. Adrenergic modulation of AMPK-dependent autophagy by chronic stress enhances cell proliferation and survival in gastric cancer. Int J Oncol. 2019;54:1625–1638. doi: 10.3892/ijo.2019.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the GEO data repository, [https://www.ncbi.nlm.nih.gov/geo]. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.