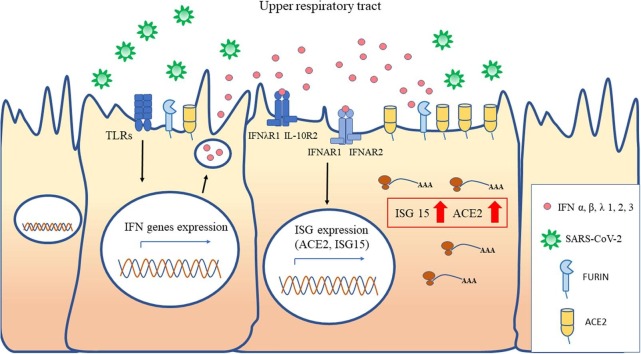
Graphical abstract
Keywords: ACE2, Interferon, Truncated ACE2 isoform, SARS-COV-2, Furin
Abstract
In vitro interferon (IFN)α treatment of primary human upper airway basal cells has been shown to drive ACE2 expression, the receptor of SARS-CoV-2. The protease furin is also involved in mediating SARS‐CoV‐2 and other viral infections, although its association with early IFN response has not been evaluated yet. In order to assess the in vivo relationship between ACE2 and furin expression and the IFN response in nasopharyngeal cells, we first examined ACE2 and furin levels and their correlation with the well-known marker of IFNs’ activation, ISG15, in children (n = 59) and adults (n = 48), during respiratory diseases not caused by SARS-CoV-2. A strong positive correlation was found between ACE2 expression, but not of furin, and ISG15 in all patients analyzed. In addition, type I and III IFN stimulation experiments were performed to examine the IFN-mediated activation of ACE2 isoforms (full-length and truncated) and furin in epithelial cell lines. Following all the IFNs treatments, only the truncated ACE2 levels, were upregulated significantly in the A549 and Calu3 cells, in particular by type I IFNs. If confirmed in vivo following IFNs’ activation, the induction of the truncated ACE2 isoform only would not enhance the risk of SARS‐CoV‐2 infection in the respiratory tract.
1. Introduction
Starting from December 2019 in China, a novel virus of the Coronaviridae family, named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is spreading worldwide [1]. SARS-CoV-2 infections range from completely asymptomatic to mild symptoms, up to a severe acute respiratory distress syndrome (ARDS) and death. SARS-CoV-2 entry into respiratory tract cells starts with the binding to its main receptor, the angiotensin-converting enzyme 2 (ACE2) [2], the same receptor used by SARS-CoV-1 [3]. Differently from SARS-CoV-1, the spike protein of the novel CoV is activated by the serine protease furin that enhances viral entry into human lung cells [4]. The efficiency of SARS-CoV-2 entry may be related to the number and type of cells programmed to express ACE2 and/or furin other than to the rate of their expression in the specific epithelial cells. Besides being SARS-CoV-2 binding/entry receptor, ACE2 exerts a protective role in acute lung injury through its involvement in renin-angiotensin system (RAS) signaling and its down regulation in SARS-CoV infections might also play a causal role in disease progression to ARDS [5].
ACE2 has been recently proposed to be an interferon (IFN)-stimulated gene (ISG) in epithelial tissues [6], [7] coherently with the notion that its upregulated expression may be functional in preventing ARDS during viral infections [5], [8]. Accordingly, we sought to verify this ISG-type induction of ACE2 expression in upper airways cells, collected during children’s and adult’s respiratory infections caused by pathogens other than SARS-CoV-2.
Subsequent studies [9], [10], [11] demonstrated the existence of truncated ACE2 isoforms (dACE2) of unknown functions, not able to serve as SARS-CoV-2 entry receptor. Moreover, the authors observed that IFNs and viruses induced dACE2, but not full-length ACE2, in several, but not all, the studied cell models [9], [10], [11]. Therefore, to clarify this issue, we performed experiments of in vitro stimulation of the A549 and Calu3 cell lines with type I and III IFNs, to characterize the activation of ACE2/dACE2 and to investigate the potential induction of the furin gene as well.
2. Materials and methods
2.1. Patients and samples
Fifty-nine previously healthy children admitted to the Paediatric Department of the University Hospital “Policlinico Umberto I” with a clinical diagnosis of bronchiolitis and pneumonia, and 48 adults with respiratory symptoms attending the same Hospital were enrolled over the 2019/2020 winter season. The local ethics committee approved the study protocol (Sapienza University of Rome, University Hospital “Policlinico Umberto I”); all adult patients and parents/guardians gave informed consent to participate in the study and patients’ data were anonymized. Respiratory specimens (nasopharyngeal washings for children, nasopharyngeal swabs for adults) were tested for 14 respiratory viruses and for SARS-CoV-2 [12]; remaining aliquots were centrifuged, and RNA extracted from cell pellet for gene expression analysis [13].
2.2. In vivo gene expression analysis
The ACE2 mRNAs levels, of the serine protease furin and that of the well-known marker of IFNs’ activation, ISG15, were measured in respiratory cells by quantitative RT-Real time PCR assays. ACE2 was tested with a commercial qPCR Assay (Hs.PT.58.27645939. IDT, Iowa, USA), targeting exons 14–15 that are common to ACE2 isoforms; furin was tested with a commercial assay (Hs.PT.58.1294962. IDT, Iowa, USA) and ISG15 with primers: F 5′-TGGCGGGCAACGAATT-3′, R 5′-TGATCTGCGCCTTCA-3′; Probe 5′-[6FAM]TGAGCAGCTCCATGTC[TAM]n-3′. The mRNA copy numbers of target genes were measured, in co-amplification with the beta-glucuronidase gene (GUS; F 5′-TCTGTCAAGGGCAGTAACCTG-3′, R 5′-GCCCACGACTTTGTTTTCTG-3′, Probe 5′-[6FAM]TATGTCTTTCGATATGCAGCCAAGTTTTACCG[TAM]n-3′) gene to normalize the amount of total RNA, using the threshold cycle relative quantification (the 2−ΔCt method) as previously reported [13]. Each raw Ct (threshold cycle) value was tagged as undetermined when fell between levels of 40 and 45. For mRNA copies that were undetectable by the RT-PCR assays, the Ct was set to 45, for calculating the fold changes to the GUS Ct of the same sample.
2.3. In vitro IFN induction experiments
The experiments of IFNs exogenous administration in A549 and Calu-3 (human lung carcinoma derived cell lines) were performed treating, for 24 h, 200.000 cells/well cultured in 24-well plates, with natural IFNα (Alfa Wassermann SpA, Bologna, Italy) at 5x104 IU/mL, IFNβ1a (Avonex Pen, Biogen Idec Ltd. Cambridge, Massachusetts, USA) and IFNλ1-3 (R&D systems, Minneapolis, MN, USA) all at 500 ng/mL. The comparative threshold cycle method was applied to calculate the fold changes, compared with non-stimulated cell (2-ΔΔC t) of the genes of interest: furin and ISG15 were tested as described above. In addition, the full-length ACE2 (f-lACE2) and dACE2 isoforms were quantified using specific primers and probes [9], and two other ISGs were tested: ISG56 (F 5′-TGAAGAAGCTCTAGCCAACATGTC-3′, R 5′-GAGCTTTATCCACAGAGCCTTTTC-3′, Probe 5′-[6FAM]TATGTCTTTCGATATGCAGCCAAGTTTTACCG[TAM]n-3′), IRF-7 (Hs.PT.58.24613215. IDT, Iowa, USA). All experiments were conducted in triplicates.
2.4. Statistical analysis
Differences between the level of gene expression among patients infected with different viruses, or not infected or in terms of demographic factors, were compared using the Mann–Whitney test or Kruskal-Wallis test. Spearman's ρ coefficient was calculated to assess the correlation between the level of ACE2, and furin with that of ISG15. Two-tailed paired-Samples t test was used to compare levels of ISGs, ACE2 and furin in cell lines treated in vitro with IFN. To assess the association of ACE2 with ISG15, a generalized linear model was constructed and coefficients, and standard errors (SE) were calculated for each covariate. Age, sex, and viral infection positivity were simultaneously included in the model. The significance was fixed at the 5% level. Analysis was performed with Stata 15 (2017, College Station, TX).
3. Results and discussion
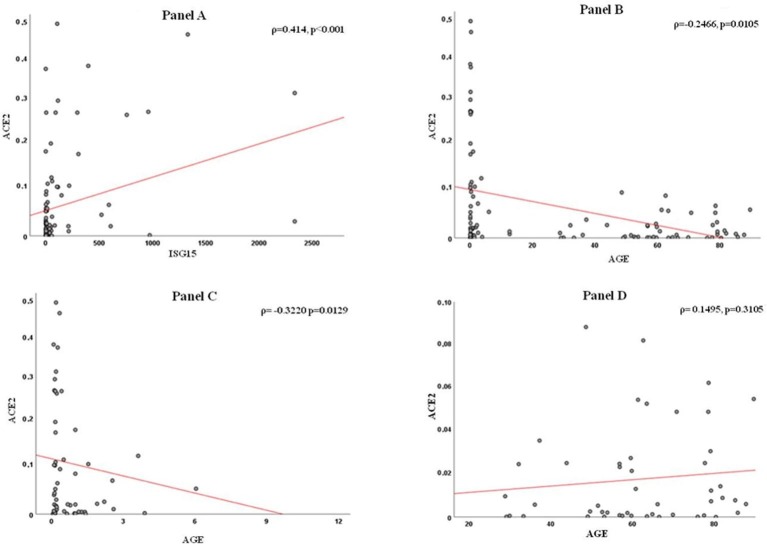
A total of 107 patients, 59 children and 48 adults, were tested for gene expression: demographic, clinical and virological data of these patients are summarized in Table 1 . To assess consistency of gene expression measurements, we first compared the GUS Ct values between nasopharyngeal washings (children) and swabs (adults), as a proxy for respiratory cells abundance: values were not significantly different between the two groups (p = 0.116, Table 1). ACE2 transcript levels, measured with an assay that does not discriminate isoforms, were higher in children compared with adults but the difference did not reach significance (p = 0.061). When comparing only samples positive to respiratory viruses, the difference in ACE2 expression reached significance (54 children, median value = 0.0238, vs 10 adults, median value = 0.0014, p = 0.021). Furin transcript levels did not differ between groups whereas ISG15 transcripts were augmented in children (p < 0.001, Table 1). To examine relationships of ACE2 and furin with the IFN response, we examined their levels with respect to that of ISG15. We found that a strict relation does exist for ACE2 (ρ = 0.414, p < 0.001; supplementary Figure, panel A) but not for furin (ρ = 0.141, p = 0.148). Furthermore, ACE2 expression levels were inversely correlated with patient’s age in years (ρ = -0.2466, p = 0.0105; supplementary Figure, panel 1 B). However, when analyzing the two age groups separately, the relationship with ACE2 remained significant for children (ρ = -0.3220, p = 0.0129) but not for adults (ρ = 0.1495, p = 0.3105). No significant difference was evident when testing ACE2 transcripts with respect to sex either in total or by age group (data not shown). These findings suggest that ACE2 levels in the respiratory tract are not consistently related to age, although its expression was higher in children (age < 13 years, median 1.21 ± 2.45 years) underlying the complex regulation of ACE2 expression. To overcome limitation due to the interdependency of age and infection status, a multivariate analysis was performed to minimize potential confounders (age and presence of a viral infection). This analysis confirmed that ACE2 levels were consistently associated with those of ISG15 (coef. 0.00046, SE 0.0019, p = 0.018). As far as furin is concerned, its expression was positively correlated with that of ACE2 (ρ = 0.344, p < 0.001), but not significantly related to patients’ age (ρ = -0.157, p = 0.109).
Table 1.
Demographic, clinical, virological features and gene expression data (GUS, ACE2, furin, ISG15) of study patients (n = 107).
| Items | Total patients n = 107 |
Children n = 59 |
Adults n = 48 |
|---|---|---|---|
| Age (years)* | 28.34 ± 32.29 | 1.21 ± 2.45 | 61.67 ± 16.91 |
| Male (%) | 69/107 (64.5%) | 38/59 (55.1%) | 31/48 (44.9%) |
| Virus positive | 64/107 (59.8%) | 54/59 (91.5%) | 10/48 (20.83%)● |
| RSV | 29/64 (45.3%) | 28/54 (51.9%) | 1/10 (10%) |
| HRV | 18/64 (28.1%) | 16/54 (29.6%) | 2/10 (20%) |
| HKU1 | 5/64 (7.8%) | 2/54 (3.7%) | 3/10 (30%) |
| FLU A | 6/64 (9.4%) | 5/54 (9.3%) | 1/10 (10%) |
| HMPV | 1/64 (1.6%) | 0/54 (0%) | 1/10 (10%) |
| Coinfections | 5/64 (7.8%) | 3/54 (5.5%) | 2/10 (20%) |
| Gene expression | |||
| GUS** | 34.62 ± 3.67 | 34.12 ± 3.83 | 35.24 ± 3.40 |
| ACE2*** | 0.01 (0.002–0.06) | 0.03 (0.006–0.14) | 0.006 (0.0008–0.02) |
| Furin*** | 0.006 (0.0006–0.02) | 0.006 (0.001–0.026) | 0.004 (0.0005–0.01) |
| ISG15*** | 7.01 (1.88–52.71) | 41.07 (7.49–213.79) | 2.03 (0.88–4.83)▾ |
*Age is expressed as mean ± SD. **Ct (threshold cycle) values of GUS (beta-glucuronidase gene) are indicated as mean ± SD. ***Transcript levels of ISG15, ACE2 and furin, calculated using 2-ΔCt, are indicated as median (interquartile range = IQR).
●p < 0.001 by Chi-Squared test; ▾p < 0.001 by Mann-Whitney U test.
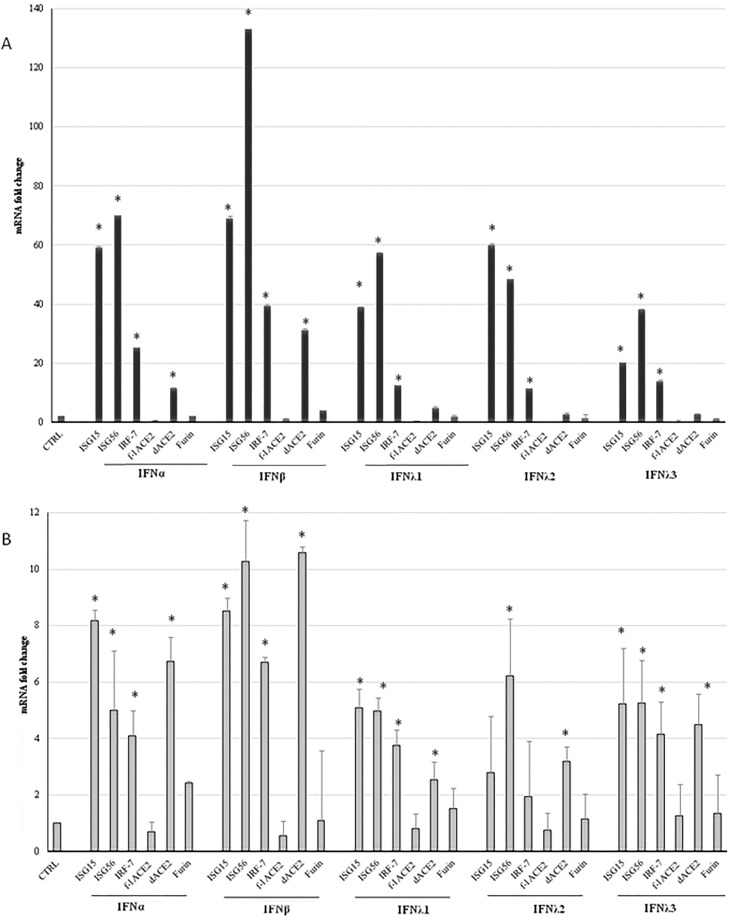
After we had measured ACE2 gene expression in patients, the existence of truncated ACE2 isoforms became known [9], [10], [11]. The commercial qPCR assay we used could not distinguish isoforms and, unfortunately, respiratory samples were not enough to be tested with isoform-specific assays. Hence, in order to discriminate which of the isoforms could be upregulated by IFNs, we performed experiments of IFNα, IFNβ, and IFNλs1-3 in vitro stimulation of the lung epithelial cell lines A549 and Calu-3, measuring ACE2 transcripts with tailored Real time PCR assays [9] for f-lACE2 and dACE2, before and after stimulations, in relation to the expression of well-known ISGs (ISG15, ISG56 and IRF7). Together with ACE2 isoforms, the induction of furin transcript was studied, as well. Constitutive expression levels of the study genes, i.e. 2−ΔCt values at baseline, were much higher in Calu-3 than in the A549: ISG15 = 1.8153, ISG56 = 1.0825, IRF7 = 0.1465, f-lACE2 = 0.0014, dACE2 = 0.0009, furin = 0.0007 in the A549; ISG15 = 12.4965, ISG56 = 5.1230, IRF-7 = 1.2226, f-lACE2 = 0.6378, dACE2 = 0.0078; furin = 0.0218 in the Calu-3). Following IFNs’ treatments, ISGs transcription levels were significantly elevated in both cell lines, with fold-changes about ten-times higher in A549 than in Calu-3 cells (Fig. 1 ).
Fig. 1.
Induction of the Interferon (IFN)-stimulated genes (ISG), IRF-7, ACE2 and furin after type I and III IFN stimulation of the A549 and Calu-3 epithelial cell lines. Expression of ISG15, ISG56, IRF7, full-length (f-l) dACE2 and furin in A549 (panel A) and Calu-3 (panel B) cells treated with IFNα (5x104 IU/mL), IFNβ1a (500 ng/mL), IFNλ1 (500 ng/mL), IFNλ2 (500 ng/mL) and IFNλ3 (500 ng/mL) for 24 h. Data are expressed as fold change (ΔΔCt method), from the equation 2-ΔΔCt used to calculate the mRNA levels of the stimulated genes, compared to baseline values in untreated cells (set to 1 and indicated in the graphs as CTRL). Differences in mRNA expression levels between untreated and treated cells were evaluated using paired T-test. *=p < 0.05.
Furin levels of expression were not augmented in either cell lines following stimulation with any IFN (Fig. 1); undoubtedly, IFN treatments could have altered furin intracellular trafficking, glycosylation and/or enzymatic activation, that we did not address. Previous studies observed that furin mRNA and protein expression is negatively regulated by IFNγ and that two ISGs (GBP2 and GBP5) are able to inhibit furin proteolytic activity, as summarized in a recent review [14].
After treatments by IFNs, the f-lACE2 levels were not increased, or were even diminished (in the A549 by type III IFNs) (Fig. 1). Differently, dACE2 transcripts were significantly elevated, especially after type I IFNs treatments (Fig. 1), in the A549 (IFNα = 11.4-fold and β = 31.1-fold) and Calu-3 cells (IFNα = 6.8 fold and β = 10.6-fold). Hence, these in vitro experiments further supported that the mRNA(s) for the truncated isoform dACE2, and not that for the full-length ACE2, is induced by IFNs [9], [10], [11]. Compared to ‘traditional’ ISGs, dACE2 regulation by IFNs appeared weaker and its expression might be also affected by different cell-specific factors. The latter aspect is not a novelty since the exact set and number of genes that are regulated by IFN is known to largely vary between different cell types, often depending on the constitutive level of expression of the IFN genes [15]. In this respect, Onabajo et al [9] reported that dACE2 transcript was not inducible by IFNs in Vero E6, and moderately stimulated in HEK293T cells. Notably, Ng et al [10] found that dACE2 is more abundant in the respiratory mucosa with respect to the gastrointestinal tract and that SARS-CoV-2 infected Calu3 cells induced dACE2 but not ACE2. Interestingly, Blume et al. reported that in airway primary cells, dACE2 is upregulated in response to IFN-β treatment and rhinovirus, but not by SARS-CoV-2 infection [11].
The function, if any, of the truncated ACE2 protein is not yet known and it is not clear whether dACE2 transcript might be activated as one of the antiviral weapons of the IFN response. It was also hypothesized that the ratio ACE2/dACE2 in different tissue and/or in individuals could explain the different rates of SARS-CoV-2 infection [10].
Collectively, our results in patients suggest that ACE2 transcripts’ level (all isoforms) in vivo were correlated with those of ISG15, a marker of type I and III IFNs’ activation. However, the ISG-like induction should be relevant only for dACE2 as we observed in lung epithelial cell lines. If confirmed in patients that the IFN response increase the truncated ACE2 isoform only, this activation would not enhance the risk of SARS‐CoV‐2 infection in the respiratory tract.
Further studies are needed to measure the specific expression of ACE2 isoforms in additional cell lines and primary tissues and in vivo studies should clarify their possible role in SARS-CoV-2 tissue tropism and relationship with host immunity.
CRediT authorship contribution statement
Carolina Scagnolari: Conceptualization, Methodology. Camilla Bitossi: . Agnese Viscido: . Federica Frasca: . Giuseppe Oliveto: . Mirko Scordio: . Laura Petrarca: . Enrica Mancino: . Raffaella Nenna: . Elisabetta Riva: . Corrado De Vito: . Fabio Midulla: . Guido Antonelli: . Alessandra Pierangeli: Conceptualization.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Funding: This work was supported by a Sapienza University grant to AP “Ricerca 2019” n. RM11916B893FC0D0 and by grant to GA from the Italian Ministry of Health: COVID-2020-12371817.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cyto.2021.155430.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Fig. 1.
References
- 1.Lu R., Zhao X., Li J., et al. Genomic characterisation and epidemiology of novel coronavirus: implications for virus origins and receptor binding. Lancet. 2019;395(2020):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wan Y., Shang J., Graham R., et al. Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020;94:e00127–e220. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamming I., Timens W., Bulthuis M., et al. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann M., Kleine-Weber H., Pöhlmann S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell. 2020;78:779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuba K., Imai Y., Rao S., et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ziegler C.G.K., Allon S.J., Nyquist S.K., et al. SARS-CoV-2 Receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith J.C., Sausville E.L., Girish V., et al. Cigarette Smoke Exposure and Inflammatory Signaling Increase the Expression of the SARS-CoV-2 Receptor ACE2 in the Respiratory Tract. Dev. Cell. 2020;8 doi: 10.1016/j.devcel.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu H., Xie Z., Li T., et al. Angiotensin-converting enzyme 2 inhibits lung injury induced by respiratory syncytial virus. Sci. Rep. 2016;6:19840. doi: 10.1038/srep19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.OO Onabajo, AR Banday, ML Stanifer, et al. Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor. Nat Genet. (2020) Oct 1doi: 10.1038/s41588-020-00731-Epub ahead of print. [DOI] [PMC free article] [PubMed]
- 10.KW. Ng, J. Attig, W. Bolland, et al. Tissue-specific and interferon-inducible expression of nonfunctional ACE2 through endogenous retroelement co-option. Nat Genet. (2020) Oct 19. 10.1038/s41588-020-00732-8. Epub ahead of print. [DOI] [PMC free article] [PubMed]
- 11.C Blume, CL Jackson, CM Spalluto, et al. A novel isoform of ACE2 is expressed in human nasal and bronchial respiratory epithelia and is upregulated in response to RNA respiratory virus infection bioRxiv 2020.07.31.230870; 10.1101/2020.07.31.230870.
- 12.Mancino E., Cristiani L., Pierangeli A., et al. A single centre study of viral community-acquired pneumonia in children: No evidence of SARS-CoV-2 from October 2019 to March 2020. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pierangeli A., Viscido A., Bitossi C., et al. Differential interferon gene expression in bronchiolitis caused by respiratory syncytial virus-A genotype ON1. Med. Microbiol. Immunol. 2020;209:23–28. doi: 10.1007/s00430-019-00633-6. [DOI] [PubMed] [Google Scholar]
- 14.Braun E., Sauter D. Furin-mediated protein processing in infectious diseases and cancer. Clin. Transl. Immunology. 2019;8:e1073. doi: 10.1002/cti2.107315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu A.C., Parsons K., Barr I., Lowther S., Middleton D., Hansbro P.M., Wark P.A. Critical role of constitutive type I interferon response in bronchial epithelial cell to influenza infection. PLoS ONE. 2012;7:e32947. doi: 10.1371/journal.pone.0032947. [DOI] [PMC free article] [PubMed] [Google Scholar]