Abstract
Purpose
To investigate the efficacy of radiomics in diagnosing patients with coronavirus disease (COVID-19) and other types of viral pneumonia with clinical symptoms and CT signs similar to those of COVID-19.
Methods
Between 18 January 2020 and 20 May 2020, 110 SARS-CoV-2 positive and 108 SARS-CoV-2 negative patients were retrospectively recruited from three hospitals based on the inclusion criteria. Manual segmentation of pneumonia lesions on CT scans was performed by four radiologists. The latest version of Pyradiomics was used for feature extraction. Four classifiers (linear classifier, k-nearest neighbour, least absolute shrinkage and selection operator [LASSO], and random forest) were used to differentiate SARS-CoV-2 positive and SARS-CoV-2 negative patients. Comparison of the performance of the classifiers and radiologists was evaluated by ROC curve and Kappa score.
Results
We manually segmented 16,053 CT slices, comprising 32,625 pneumonia lesions, from the CT scans of all patients. Using Pyradiomics, 120 radiomic features were extracted from each image. The key radiomic features screened by different classifiers varied and lead to significant differences in classification accuracy. The LASSO achieved the best performance (sensitivity: 72.2%, specificity: 75.1%, and AUC: 0.81) on the external validation dataset and attained excellent agreement (Kappa score: 0.89) with radiologists (average sensitivity: 75.6%, specificity: 78.2%, and AUC: 0.81). All classifiers indicated that "Original_Firstorder_RootMeanSquared" and "Original_Firstorder_Uniformity" were significant features for this task.
Conclusions
We identified radiomic features that were significantly associated with the classification of COVID-19 pneumonia using multiple classifiers. The quantifiable interpretation of the differences in features between the two groups extends our understanding of CT imaging characteristics of COVID-19 pneumonia.
Abbreviations: AI, artificial intelligence; COVID-19, coronavirus disease; CT, Computed tomography; KNN, k-nearest neighbour; LASSO, least absolute shrinkage and selection operator; RF, random forest; RMS, original_firstorder_RootMeanSquared; ROI, region of interest; RT-PCR, Reverse transcriptase polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2
Keywords: Radiology; Pneumonia; Coronavirus infections; Tomography, X-Ray computed; Machine learning
1. Introduction
The number of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections is rapidly increasing. As of 1 December 2020, over 62.1 million cases and 1.4 million deaths were reported globally since the start of the pandemic [1]. The coronavirus strain SARS-CoV-2 causes the 2019 coronavirus disease (COVID-19), resulting in impaired lung function and decreased blood oxygen saturation in affected patients. The main symptoms of COVID-19 include fever, cough, fatigue, and body aches. In addition to the recommended viral nucleic acid real-time reverse transcriptase-polymerase chain reaction (RT-PCR) test, the value of radiological images for diagnosis of COVID-19 has been verified [2,3]. Based on radiologists' evaluations of the signs of pneumonia on chest computed tomography (CT), an average sensitivity of 80 % and specificity of 83 % of clinical diagnosis have been achieved [4].
The distinct characteristics of COVID-19 pneumonia on CT images include bilateral involvement, peripheral distribution, multifocality, mixed ground-glass opacity, consolidation, and vascular thickening [5,6]. However, as radiologists' evaluations of these specific characteristics may be affected by subjective experience, the accuracy of COVID-19 diagnosis varies [4], especially in cases of viral pneumonia with clinical symptoms and CT signs similar to those of COVID-19. Previous studies have confirmed the value of a pre-designed artificial intelligence (AI) framework for task-oriented COVID-19 analysis based on radiological images [7,8]. However, this method is currently hampered by insufficient training data. Recent studies underscore the need for further verification of the sensitivity and stability of current AI-based solutions [9] and for more evidence to establish AI as a production-ready solution for COVID-19 diagnosis [10,11].
Based on the region of interest (ROI) for pneumonia lesions delineated by radiologists, radiomics may provide additional knowledge for survival prognosis and classification of illness severity for COVID-19 pneumonia [[12], [13], [14], [15]]. The latest analytical tool of radiomics, Pyradiomics, has paved the way for standardised radiomics analysis [16]. As an open-source Python package for extracting radiomic features from the ROI on medical images, the standardised radiomic feature extraction program provided by Pyradiomics can reduce programming bias in researchers, improve the reproducibility of radiomic features, and thus enhance the credibility of radiomics results. We hypothesised that radiomics features could be used to differentiate COVID-19 from other types of viral pneumonia with clinical symptoms and CT signs similar to those of COVID-19. We evaluated the performance of radiomics features by using different feature selection and prediction classifiers and investigated standardised high-throughput CT image features extracted by Pyradiomics to clarify the value of radiomics features for the diagnosis of COVID-19.
2. Materials and methods
Between 18 January 2020 and 20 May 2020, patients with viral pneumonia were initially recruited from two hospitals in China and one hospital in the United States. All procedures performed in studies that involved human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration. Our institutional review board approved this retrospective study (IRB: no.2020-P-036 and no. 51059) and waived the need for written informed consent. There are no conflicts of interest to declare.
2.1. Patients
All patients included in this study were admitted during the COVID-19 pandemic. RT-PCR tests for COVID-19 were performed using respiratory secretions obtained by bronchoalveolar lavage, endotracheal aspirate, nasopharyngeal swab, or oropharyngeal swab. These COVID-19 RT-PCR tests were performed at least twice for each patient, to reduce the potential false-negative RT-PCR test.
For SARS-CoV-2 positive patients, the inclusion criteria were as follows: 1) patient over 18 years of age, 2) complete clinical data and CT scan available at admission, and 3) all RT-PCR test results positive. CT scans were reviewed by at least two radiologists to identify the outline of each pneumonia lesion. Patients whose pneumonia lesions could not be visually detected by radiologists were excluded from this study because the ROI of the pneumonia abnormalities could not be delineated for radiomics analysis. For SARS-CoV-2 negative patients, in addition to the points 1) and 2) of the above-mentioned inclusion criteria, 3) all RT-PCR test results negative, 4) patients with viral pneumonia with clinical symptoms and CT signs similar to those of COVID-19, as reviewed by radiologists, and 5) relevant exposure history, was selected as an inclusion criterion for negative patients from China because these patients were at a high risk of COVID-19 infection during the outbreak. The related exposure histories were as follows: H1) history of travel to Wuhan within 14 days before admission, H2) history of contact with a confirmed COVID-19 patient and H3) history of contact with a dense crowd. All SARS-CoV-2 positive patients recruited from China were classified as having mild, common, severe, or critical illness according to the National Health Commission of the People's Republic of China, diagnosis and treatment of COVID-19 pneumonia (Tentative Standard 7).
2.2. CT scans
All patients underwent a chest CT scan at admission. The detailed CT scanning parameters of the three hospitals included in this study are listed in Supplementary Appendix A. We tabulated the number of days between symptom onset and date of the CT scan. The time from symptom onset to admission for CT examination was defined as early (0–2 days), intermediate (3–5 days), or late (6–12 days).
2.3. Manual segmentation for pneumonia lesions
Four radiologists from our local hospital, each with >5 years of radiological experience, completed the manual segmentation of all pneumonia lesions layer-by-layer using ITK-Snap software (v.3.6.0) [17,18]. Any potential signs of COVID-19 on chest CT were reviewed by radiologists, including bilateral involvement, peripheral distribution, multifocality, mixed ground-glass opacity, consolidation, and vascular thickening, as reported previously [5,6]. Examples of pneumonia lesion segmentation are provided in Supplementary Appendix B. All four radiologists underwent training to identify the characteristics of COVID-19 pneumonia on CT images, to distinguish the CT signs of COVID-19 pneumonia and other types of pneumonia, and to detect the boundary of the pneumonia lesions on CT images. All training sessions were completed online, and approximately eight lessons were provided by radiologists/respiratory experts with experience in the clinical diagnosis of COVID-19 pneumonia. All segmentations were summarised and reviewed by an expert with >10 years of radiological experience. Based on the expert evaluation, the four radiologists held an online consensus meeting to resolve disagreements as to lesion segmentation and to determine the boundaries of these lesions on the CT images.
To verify the credibility of the manual segmentation and feature extraction in this study, ten patients' CT scans were randomly selected and segmented by one radiologist from a different hospital to correct for the potential experience-related bias in feature extraction. Analysis of variance (ANOVA) and Mann-Whitney U tests were performed on the features extracted from the two rounds of segmenting the pneumonia lesion images to verify the inter-observer agreement.
2.4. Pyradiomics for feature extraction
The recommended standardised radiomics analysis workflow [19] was used in this study. Pyradiomics was used to extract high-throughput image features of the ROI of pneumonia lesions on CT images. The latest Pyradiomics version available online includes 120-dimensional features, including first-order statistics (19 features), 3D shape-based (16 features), 2D shape-based (10 features), Grey Level Co-occurrence Matrix (24 features), Grey Level Run Length Matrix (16 features), Grey Level Size Zone Matrix (16 features), Neighbouring Grey Tone Difference Matrix (5 features), and Grey Level Dependence Matrix (14 features). A detailed description has been presented previously [16,20].
2.5. Radiomics classifier
To verify the performance of radiomics features for the classification of COVID-19 pneumonia, the following classifiers were used in this study: linear classifier, k-nearest neighbour (KNN), least absolute shrinkage and selection operator (LASSO), and random forest (RF).
The pneumonia lesion images from China were randomly divided into training and test datasets with an 80:20 ratio based on the related radiomics studies [[21], [22], [23]]. After obtaining the classification model from the training dataset which executed on the server in China, the model was then provided to the dataset from the United States. The images from the United States were used as an external validation dataset to independently verify the method proposed in this study. The pneumonia lesion images manually delineated by the radiologists will be published for open-access after the review process.
2.6. Clinical use
To compare the diagnostic results of radiomics with radiologists, CT images in the two test datasets were additionally diagnosed by three radiologists from another hospital in China with 3, 5, and 10 years of radiological experience, respectively. The radiologists were blinded to the patients' RT-PCR results. We compared the sensitivity and specificity of the diagnosis obtained by radiologists and the radiomics methods. Also, a Kappa consistency analysis was used to evaluate the agreement of the classification result in the classifiers and radiologists on the external validation dataset.
2.7. Statistical analysis
The ROC curve, the area under the curve (AUC), sensitivity, and specificity were used to evaluate the diagnostic accuracy for COVID-19. All statistical analyses were performed using the R language (version 3.4.3, Vienna, Austria). The linear classifier, KNN, LASSO, and RF were implemented by the "lm", "kknn", "glmnet", and "randomforest" functions, respectively. Chi-square tests and ANOVA were used to evaluate the differences in demographics between the two groups. For Kappa analysis, the classification results of the two methods were considered to be excellent agreement when the Kappa score ≥0.85, and [0.6, 0.85) indicated good agreement, and [0.45, 0.6) indicated moderate agreement, and Kappa score <0.45 was considered poor agreement. P<0.05 was considered statistically significant.
3. Results
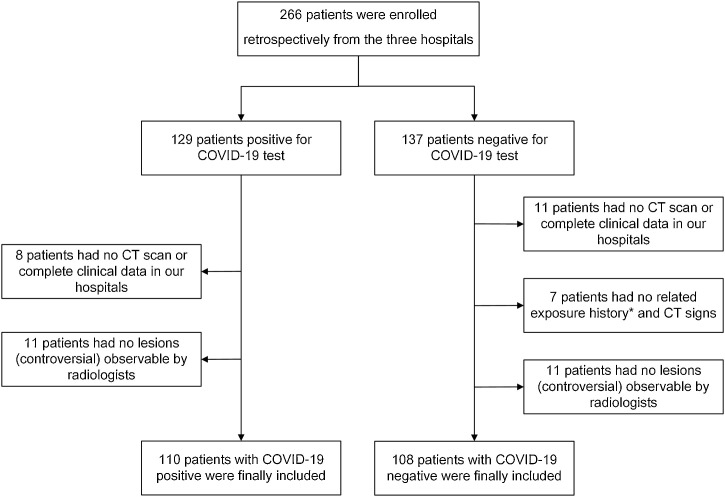
In total, 266 patients were initially recruited from the three hospitals. Based on the inclusion criteria, 218 patients were enrolled in this study, including 110 patients who were SARS-CoV-2 positive and 108 patients who were SARS-CoV-2 negative, but who had other types of viral pneumonia. There were 184 patients (112 males and 72 females) from China and 34 patients (18 males and 16 females) from the United States, as shown in Table 1 . No significant differences in demographic data were observed between male and female patients (P > 0.05). A flowchart of patient enrolment is shown in Fig. 1 . Regarding the time interval from initial symptom onset to admission CT scan, the ratio of individuals classified as early, intermediate, and late in the SARS-CoV-2 positive group was 21:30:49, and that in the SARS-CoV-2 negative group was 30:20:50, respectively. For all patients the second test was performed one day after the first test. In 26% patients the first RT-PCR test was performed immediately after (within the same day) the suspected pneumonia lesions were found on CT scan. In the other patients, the first RT-PCR test and CT scan were performed at the same time on admission. Statistics regarding the prevalence of the main characteristics of CT presentation in SARS-CoV-2 positive and SARS-CoV-2 negative patients in this study are shown in Table 2 .
Table 1.
Demographics of the patients enrolled in this study. Hospital 1 and 2 are in China, and Hospital 3 is in the United States of America.
| Hospital 1 |
Hospital 2 |
Hospital 3 |
||||
|---|---|---|---|---|---|---|
| COVID-19 | Non-COVID-19 | COVID-19 | Non-COVID-19 | COVID-19 | Non-COVID-19 | |
| Number | 73 | 71 | 20 | 20 | 17 | 17 |
| Age | 44.1 (15.7) | 39.8 (16.5) | 39.1 (12.4) | 35.5 (12.2) | 56.6 (20.3) | 56.9 (12.1) |
| Sex | ||||||
| Male | 44 | 40 | 14 | 14 | 10 | 8 |
| Female | 29 | 31 | 6 | 6 | 7 | 9 |
| Time Interval | 6.1 | 4.9 | 7.4 | 4.2 | 10.5 | 11.3 |
| History | ||||||
| H1 | 28 | 17 | 13 | 2 | NA | NA |
| H2 | 11 | 10 | 6 | 5 | NA | NA |
| H3 | 25 | 45 | 11 | 19 | NA | NA |
Note. Age is presented with mean and standard deviation. History denotes the related exposure history (H1-H3) defined for included patients in China in this study. NA: not applicable.
Fig. 1.
Flowchart of patient enrolment in this study.
Table 2.
Statistics of the prevalence of the main characteristics of CT presentation in SARS-CoV-2 positive and SARS-CoV-2 negative patients in this study.
| Main CT characteristics |
|||||
|---|---|---|---|---|---|
| GGO | Crazy paving | Consolidation | Combined (2) | Combined (3) | |
| Positive (%) | 45.8 | 65.0 | 16.9 | 62.2 | 14.5 |
| Negative (%) | 32.6 | 6.7 | 88.8 | 7.0 | 2.2 |
Note. GGO: ground-glass opacity; Combined (2): two main characteristics that significantly appeared in a patient; Combined (3): three main characteristics that significantly appeared in a patient.
For manual segmentation, 12,001 images containing 24,216 pneumonia lesion ROIs were obtained from the Chinese datasets and were randomly divided into the training dataset (9601) and test dataset (2400). A total of 4052 images containing 8409 pneumonia lesion ROIs were manually delineated from the United States dataset. For a case with approximately 20 CT layers containing pneumonia lesions (single or multiple infection foci), the manual segmentation of the ROIs by one radiologist averaged approximately 10 min. Using Pyradiomics, 120 radiomic features were extracted from the original pneumonia lesion images. The statistical results of the ANOVA and Mann-Whitney U test revealed no significant differences between the features extracted from the two rounds of the segmented pneumonia images (P > 0.05).
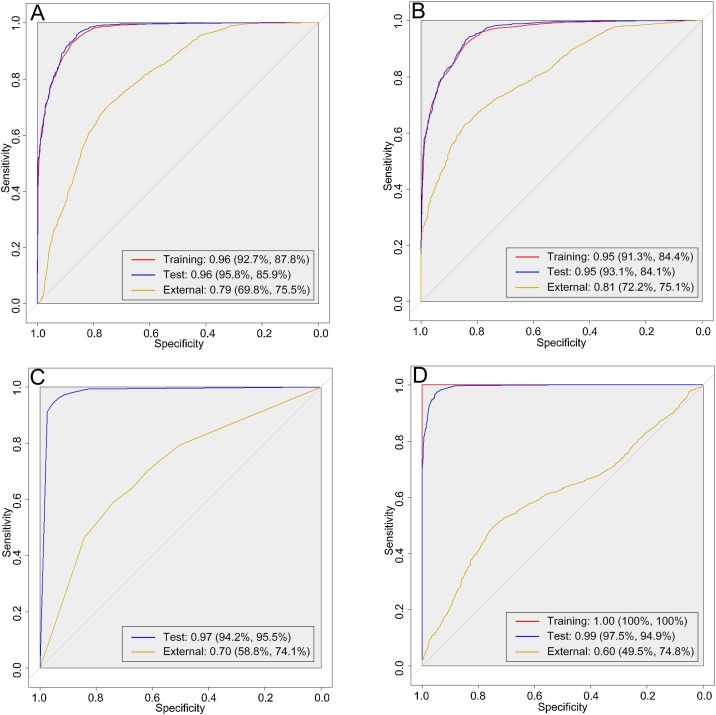
For the training, test, and external validation datasets, the AUCs (sensitivity and specificity) of the linear classifier were 0.96 (92.7% and 87.8%), 0.96 (95.8% and 85.9%), and 0.79 (69.8% and 75.5%), respectively. The results of LASSO indicated that AUCs of 0.95 (93.1% and 84.1%) and 0.81 (72.2% and 75.1%) were obtained for the test and external validation datasets, respectively. The AUCs for RF were 0.99 (97.5% and 94.9%) and 0.60 (49.5% and 74.8%) for the test and external validation datasets, respectively. AUCs of 0.97 (94.2% and 95.5%) and 0.70 (58.8% and 74.1%) were obtained by the KNN for the test and external validation datasets, respectively. The ROC curves of the four classifiers are presented in Fig. 2 . The results of the radiologists' diagnosis indicated an average sensitivity of 75.6% and specificity of 78.2% for the images in the two test datasets, the detailed comparison is presented in Table 3 . For Kappa analysis, the Kappa score between the radiologists was 0.93, and results indicated that excellent agreement score was obtained between the LASSO and the radiologists (Kappa = 0.89). In contrast, the agreement between the radiologists and linear classifier (Kappa = 0.73), RF (Kappa = 0.40), and KNN (Kappa = 0.55) were lower. A significant difference was found when compared the classification results by the classifiers (P < 0.05).
Fig. 2.
The receiver-operating characteristic curves of the four classifiers for the datasets in this study. Area under the curve with sensitivity and specificity are presented for each classifier. A: linear classifier, B: least absolute shrinkage and selection operator, C: k-nearest neighbour, and D: random forest.
Table 3.
Comparison of sensitivity (%) and specificity (%) of classification among the four machine learning classifiers and radiologists on different datasets.
| Linear |
LASSO |
RF |
KNN |
Radiologists |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sen (%) | Spe (%) | Sen (%) | Spe (%) | Sen (%) | Spe (%) | Sen (%) | Spe (%) | Sen (%) | Spe (%) | |
| Training | 92.7 | 87.8 | 91.3 | 84.4 | 100 | 100 | NA | NA | NA | NA |
| Test | 95.8 | 85.9 | 93.1 | 84.1 | 97.5 | 94.9 | 94.2 | 95.5 | 75.6 | 78.2 |
| External test | 69.8 | 75.5 | 72.2 | 75.1 | 49.5 | 74.8 | 58.8 | 74.1 | ||
NA: not applicable. Sen: sensitivity, Spe: specificity.
Different radiomics features were selected as the key features for classification when using different machine learning classifiers. For the linear classifier, a total of 26 features were identified as statistically significant for the classification task in this study (P < 0.0001). For the LASSO, 17 features were identified as the key features for the classification when the minimum objective function was obtained. For the RF, the top-20 features of importance for the classification were acquired. Of the four classifiers, LASSO had the best classification performance (highest AUC) for external validation dataset. A detailed list of the significant features selected by the three classifiers (ranking of significant features was not available in KNN) and the classifier constructed by the LASSO are shown in Supplementary Appendix C. Also, the source code of model training and the structure of the model is shown in Supplementary Appendix D.
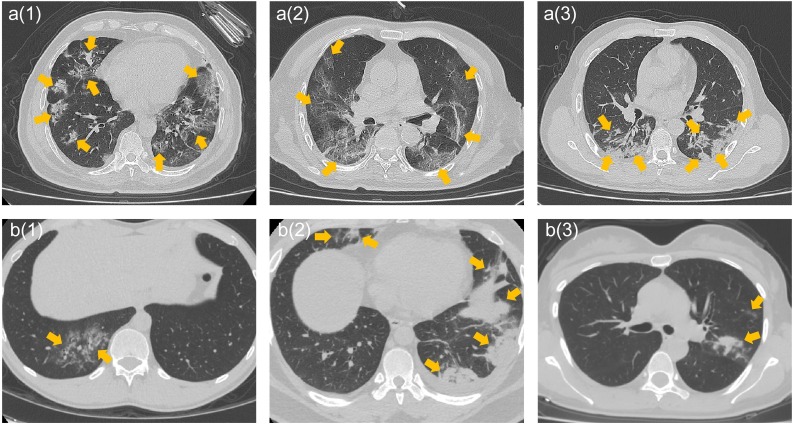
The features of "original_firstorder_RootMeanSquared" (RMS) and "original_firstorder_Uniformity" were significant features validated by all classifiers. Specifically, the values of the RMS feature in COVID-19 lesion images were significantly lower than those in non−COVID-19 lesion images in all datasets (P < 0.05, t-test) in this study (Table 4 ). The values of the "original_firstorder_Uniformity" feature in the COVID-19 lesion images were significantly lower than those in the non−COVID-19 lesion images in the three datasets (P < 0.05, t-test). Evaluation of the differences in this feature reflected in the CT images revealed that the structure of COVID-19 lesion images was more chaotic than that of non−COVID-19 lesion images with high feature values. Specifically, the internal structure of COVID-19 lesions and boundary structure were more likely to demonstrate a sharp contrast, and the internal structure tended to be more heterogeneous than that of non−COVID-19 lesions. Further, the uniformity of COVID-19 pneumonia lesion images with low feature values tended to be worse (Fig. 3 ). Additionally, although the discrimination ability of the "diagnostics_Image-original_Mean" feature was significant in both the training and test datasets, the performance was decreased in the external validation dataset.
Table 4.
The mean and standard deviation of the three significant features selected by the classifiers based on the three datasets.
| Mean |
RMS |
Uniformity |
||||
|---|---|---|---|---|---|---|
| COVID-19 | Non-COVID-19 | COVID-19 | Non-COVID-19 | COVID-19 | Non-COVID-19 | |
| Training | −707.98 ± 138.98 | −994.61 ± 170.22 | 584.00 ± 100.45 | 662.61 ± 127.87 | 0.03 ± 0.01 | 0.06 ± 0.03 |
| Test | −706.98 ± 135.03 | −991.52 ± 170.70 | 586.28 ± 99.27 | 662.99 ± 127.02 | 0.03 ± 0.01 | 0.06 ± 0.03 |
| External | −513.18 ± 105.32 | −561.83 ± 77.57 | 552.90 ± 90.07 | 638.77 ± 125.23 | 0.03 ± 0.01 | 0.06 ± 0.02 |
Note: Mean: diagnostics_Image-original_Mean. RMS: original_firstorder_RootMeanSquared. Uniformity: original_firstorder_Uniformity.
Fig. 3.
The expression of the differences in COVID-19 (a1-a3) and non−COVID-19 (b1-b3) images by the feature of "original_firstorder_Uniformity". Figure a(1) represents the lung of a 63-year-old male with fever, chest tightness, and anorexia for 9 days. CT manifested as bilateral involvement and multifocality. Figure a(2) represents the lungs of a 88-year-old male with anorexia for 6 days and fever for 2 days. CT manifestation indicates peripheral distribution, diffuseness, and mixed ground-glass opacity. Figure a(3) represents the lungs of a 53-year-old male with weakness and muscle aches for more than 10 days. CT manifestation included bilateral involvement, consolidation, and vascular thickening. Figure b(1) represents the lungs of a 29-year-old male with cough for 6 days. CT manifested as pneumonia lesions similar to the visual characteristics of Figure a(1) in the right lung. Figure b(2) represents the lungs of a 30-year-old male with cough and chest pain for 2 days. CT manifestation included bilateral involvement with multiple consolidation lesions. Figure b(3) represents the lungs of a 37-year-old female with a cough for 3 days. CT manifested as mixed ground-glass opacity, consolidation, and vascular thickening in the left lung.
4. Discussion
We verified the value of radiomics for differentiating COVID-19 from other types of viral pneumonia with clinical symptoms and CT signs similar to those of COVID-19 based on the standardised radiomics workflow. Using the features extracted by Pyradiomics, our study identified the radiomics features that were significantly associated with the classification of pneumonia patients with and without COVID-19 using multiple classifiers. We also clarified the differences in CT images reflected by different key features.
Image recognition technologies have been demonstrated to be effective methods for the clinical diagnosis of COVID-19 [24,25]. Deep learning-based methods have developed rapidly due to the unnecessary manual segmentation of pneumonia lesions on CT images [26,27]. However, owing to the black-box nature of AI [28,29], neither the operator nor the clinician can intuitively understand the working process of AI for distinguishing patients with COVID-19 pneumonia from those with non−COVID-19 pneumonia. Although high diagnostic accuracy was reported by AI, we require a deeper understanding in clinical practice to perceive the differences in imaging characteristics between COVID-19 and non−COVID-19 pneumonia. Our study demonstrated that the radiomics method can provide feasible solutions. No significant statistical differences in radiomics features obtained by manual segmentation between radiologists ensured the accuracy of the analysis of pneumonia lesions in this study. Based on the features extracted by Pyradiomics (and although the results of screening for distinctive features were different in the four classifiers), there were three measurements on the evaluation of the intensity and homogeneity of pneumonia lesions that were significantly associated with the classification task and were demonstrated cross multiple classifiers. As a measure of image homogeneity, the feature of "original_firstorder_Uniformity" demonstrated that higher feature values were observed in non−COVID-19 pneumonia lesion images. In addition, based on the measurement of the CT grey level magnitude, RMS indicated that the range of grey level of COVID-19 pneumonia was higher. These findings suggested that COVID-19 pneumonia lesions were more likely to be heterogenous, and the alveoli fluid of COVID-19 pneumonia contributed to the more-chaotic structure of infections foci on images. Furthermore, the significantly worse uniformity and the sharper contrast of peripheral and internal structure of COVID-19 pneumonia lesion on CT images indicated that the proposed radiomics features could be used to assist radiologists in improving the performance of visually differentiating CT manifestations of COVID-19 from other types of viral pneumonia. On the other hand, Pyradiomics-based analysis of the pneumonia lesions could further quantify the difference between COVID-19 and other types of viral pneumonia more intuitively, which provide a reference for more accurate clinical diagnosis.
Results of our study indicated that the performance of different classifiers varied substantially in the discrimination of pneumonia lesion features. Other classifiers, such as linear classifier, KNN, and RF achieved good performance in the training and test datasets. However, their performance in the external validation dataset was poorer (P < 0.05) than that of the LASSO. Therefore, studies that use a single machine learning classifier for radiomics feature selection and signature construction may have potential defects. The use of multiple classifiers to evaluate radiomic features in future radiomics studies may play an important role to ensure the credibility of radiomics results. Although the main purpose of this study was not to propose a method beyond the radiologists' diagnosis, we found that the diagnostic performance and Kappa score of the LASSO were closest to that of the radiologists' based on the diagnosis of CT slices. This finding supports the superiority of the LASSO in radiomics workflow, which has been reported in previous studies such as human oncological diseases and immunotherapy-induced pneumonitis [18,[30], [31], [32], [33], [34]].
Our results support the application of radiomics to assist with diagnosis of COVID-19 pneumonia. The recent development of radiomics has provided a new research paradigm in clinical studies [35,36], and there have already been radiomics studies published for COVID-19 survival prognosis and illness severity identification [[12], [13], [14], [15]]. However, current radiomics studies on the differentiation of COVID-19 from other types of viral pneumonia with clinical symptoms and CT signs similar to those of COVID-19, and the evaluation of radiomic feature among different classifiers on COVID-19, are scarce. Our study differed from previous studies [[37], [38], [39]] in several ways. First, we used a variety of machine learning classifiers to prove the effectiveness of radiomics for COVID-19 lesion feature analysis, and the performance of the current mainstream classifiers for COVID-19 classification was determined. Next, the radiomic features that were significant for COVID-19 classification were identified by multi-classifiers. This further strengthened the conclusion that radiomics can be used to assist in the diagnosis of COVID-19 pneumonia in previous studies, which provides a reference for more accurate COVID-19 diagnosis. Finally, we found that different classifiers produced significantly different classification results for the same radiomic features. This finding is of significance for the future radiomics studies to validate multiple machine learning classifiers to improve the credibility of the radiomics results.
In addition to the current specific CT manifestations of COVID-19 observed by radiologists, there is a clinical need for interpretable features that reflect the differential expression of COVID-19 and non−COVID-19 pneumonia on CT images. Radiomic features can be used as bridges to identify differences in pneumonia lesions on CT images in a manner that can be quantitatively interpreted and demonstrated the performance of different classifiers to this task. Based on the proposed significant radiomics features, the experiment could be reproduced using the open access Pyradiomics tool alongside other COVID-19 datasets. Furthermore, use of multiple machine learning classifiers in future radiomics studies will help identify more interpretable and reproducible radiomic features for clinical diagnosis of COVID-19 pneumonia.
This study has several limitations. First, only patients from China and the United States were included. Future studies should employ datasets from other countries to provide a more detailed classification of pneumonia subtypes, such as eosinophilic pneumonia, and other interstitial pneumonia to improve the robustness of the radiomics results. Next, four current mainstream classifiers were used in this study to differentiate COVID-19 and non−COVID-19 pneumonia. Still, the results indicated that the key radiomics features among the classifiers varied, and the results from the external validation dataset indicated that all four classifiers produced overfitting. More approaches should be tested to determine the optimal classifier. Also, even though each patient underwent at least two RT-PCR tests, the possibility of double false-negative RT-PCR tests cannot be eliminated. Finally, this study did not limit CT scanning parameters. Although robustness was improved, the accuracy of the external validation dataset was reduced. Future studies should design an optimised image normalisation method to mitigate the decrease in accuracy caused by different CT scanning parameters.
In conclusion, this study provides new evidence supporting the use of radiomics for the diagnosis of COVID-19 pneumonia. Our study identified the CT imaging features with significant discriminatory potential for patients with COVID-19 pneumonia and non−COVID-19 viral pneumonia using multiple classifiers. We also clarified the differences in CT images reflected by these key features. Our results expand knowledge regarding CT characteristics of COVID-19 pneumonia, which will improve the clinical diagnosis of COVID-19 pneumonia in the current radiological workflow.
Funding
This study received funding from China Postdoctoral Science Foundation (2018M630310), and China Scholarship Council (201908210051), and Irish Clinical Academic Training (ICAT) Programme, supported by the Wellcome Trust and the Health Research Board (Grant Number 203930/B/16/Z), the Health Service Executive National Doctors Training and Planning and the Health and Social Care, Research and Development Division, Northern Ireland and the Faculty of Radiologists, Royal College of Surgeons in Ireland. This work was completed as part of a Fulbright-HRB Health Impact award 2020. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Declaration of Competing Interest
None.
Acknowledgements
The dataset link: https://doi.org/10.17632/yn7vpd7bxk.1.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.ejrad.2021.109552.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.WHO . World Health Organization; 2020. Coronavirus Disease (COVID-2019) Situation Reports, Coronavirus Disease (COVID-2019) Situation Reports.https://www.who.int/emergencies/diseases/novel-coronavirus-2019 [Google Scholar]
- 2.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020:200343. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi H., Han X., Jiang N., Cao Y., Alwalid O., Gu J., Fan Y., Zheng C. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect. Dis. 2020;20(4):425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harrison B.H., Bai X., Xiong Zeng, Halsey Kasey, Ji Whae Choi, Tran Thi My Linh, Pan Ian, Shi Lin-Bo, Wang Dong-Cui, Ji Mei, Jiang Xiao-Long, Zeng Qiu-Hua, Egglin Thomas K., Ping-Feng Hu, Agarwal Saurabh, Xie Fangfang, Li Sha, Healey Terrance, Atalay Michael K., Liao Wei-Hua. Performance of radiologists in differentiating COVID-19 from viral pneumonia on chest CT. Radiology. 2020:200823. doi: 10.1148/radiol.2020200823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernheim A., Mei X., Huang M., Yang Y., Fayad Z.A., Zhang N., Diao K., Lin B., Zhu X., Li K. Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiology. 2020:200463. doi: 10.1148/radiol.2020200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zu Z.Y., Jiang M.D., Xu P.P., Chen W., Ni Q.Q., Lu G.M., Zhang L.J. Coronavirus disease 2019 (COVID-19): a perspective from China. Radiology. 2020:200490. doi: 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ting D.S.W., Carin L., Dzau V., Wong T.Y. Digital technology and COVID-19. Nat. Med. 2020;26(4):459–461. doi: 10.1038/s41591-020-0824-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mei X., Lee H.C., Diao K.Y., Huang M., Lin B., Liu C., Xie Z., Ma Y., Robson P.M., Chung M., Bernheim A., Mani V., Calcagno C., Li K., Li S., Shan H., Lv J., Zhao T., Xia J., Long Q., Steinberger S., Jacobi A., Deyer T., Luksza M., Liu F., Little B.P., Fayad Z.A., Yang Y. Artificial intelligence-enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020;26:1224–1228. doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naude W. Artificial intelligence vs COVID-19: limitations, constraints and pitfalls. AI Soc. 2020;(1):1–5. doi: 10.1007/s00146-020-00978-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rawat W., Wang Z. Deep convolutional neural networks for image classification: a comprehensive review. Neural Comput. 2017;29(9):2352–2449. doi: 10.1162/NECO_a_00990. [DOI] [PubMed] [Google Scholar]
- 11.Wang L., Lin Z.Q., Wong A. Covid-net: A tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. Sci. Rep. 2020;10(1):1–12. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei W., Hu X.W., Cheng Q., Zhao Y.M., Ge Y.Q. Identification of common and severe COVID-19: the value of CT texture analysis and correlation with clinical characteristics. Eur. Radiol. 2020;30(12):6788–6796. doi: 10.1007/s00330-020-07012-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang X., Li X., Bian Y., Ji X., Lu J. Radiomics nomogram for the prediction of 2019 novel coronavirus pneumonia caused by SARS-CoV-2. Eur. Radiol. 2020;30(12):6888–6901. doi: 10.1007/s00330-020-07032-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y., Wang Y., Zhang Y., Zhang N., Zhao S., Zeng H., Deng W., Huang Z., Liu S., Song B. A Quantitative and Radiomics approach to monitoring ARDS in COVID-19 patients based on chest CT: a retrospective cohort study. Int. J. Med. Sci. 2020;17(12):1773–1782. doi: 10.7150/ijms.48432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Q., Wang S., Li L., Wu Q., Qian W., Hu Y., Li L., Zhou X., Ma H., Li H., Wang M., Qiu X., Zha Y., Tian J. Radiomics analysis of computed tomography helps predict poor prognostic outcome in COVID-19. Theranostics. 2020;10(16):7231–7244. doi: 10.7150/thno.46428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Griethuysen J.J.M., Fedorov A., Parmar C., Hosny A., Aucoin N., Narayan V., Beets-Tan R.G.H., Fillion-Robin J.C., Pieper S., Aerts H. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(21):e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Becker A.S., Chaitanya K., Schawkat K., Muehlematter U.J., Hötker A.M., Konukoglu E., Donati O.F. Variability of manual segmentation of the prostate in axial T2-weighted MRI: a multi-reader study. Eur. J. Radiol. 2019;121:108716. doi: 10.1016/j.ejrad.2019.108716. [DOI] [PubMed] [Google Scholar]
- 18.Song J., Tian J., Zhang L., Qu X., Qian W., Zheng B., Zhang L., Zhao J., Niu M., Zhou M., Cui L., Liu Y., Zhao M. Development and validation of a prognostic index for efficacy evaluation and prognosis of first-line chemotherapy in stage III-IV lung squamous cell carcinoma. Eur. Radiol. 2019;29(5):2388–2398. doi: 10.1007/s00330-018-5912-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lambin P., Leijenaar R.T., Deist T.M., Peerlings J., De Jong E.E., Van Timmeren J., Sanduleanu S., Larue R.T., Even A.J., Jochems A. Radiomics: the bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017;14(12):749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 20.Joost van Griethuysen A.F., Aucoin Nicole, Fillion-Robin Jean-Christophe, Hosny Ahmed, Pieper Steve, Aerts Hugo. 2020. PyRadiomics: Radiomic Features.https://pyradiomics.readthedocs.io/en/latest/features.html [Google Scholar]
- 21.Zhou J., Tan H., Bai Y., Li J., Lu Q., Chen R., Zhang M., Feng Q., Wang M. Evaluating the HER-2 status of breast cancer using mammography radiomics features. Eur. J. Radiol. 2019;121:108718. doi: 10.1016/j.ejrad.2019.108718. [DOI] [PubMed] [Google Scholar]
- 22.Leijenaar R.T., Bogowicz M., Jochems A., Hoebers F.J., Wesseling F.W., Huang S.H., Chan B., Waldron J.N., O’Sullivan B., Rietveld D. Development and validation of a radiomic signature to predict HPV (p16) status from standard CT imaging: a multicenter study. Br. J. Radiol. 2018;91(1086):20170498. doi: 10.1259/bjr.20170498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsao H.-Y., Chan P.-Y., Su E.C.-Y. Predicting diabetic retinopathy and identifying interpretable biomedical features using machine learning algorithms. BMC Bioinformatics. 2018;19(9):283. doi: 10.1186/s12859-018-2277-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q., Cao K., Liu D., Wang G., Xu Q., Fang X., Zhang S., Xia J., Xia J. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020:200905. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang S., Zha Y., Li W., Wu Q., Li X., Niu M., Wang M., Qiu X., Li H., Yu H., Gong W., Bai Y., Li L., Zhu Y., Wang L., Tian J. A fully automatic deep learning system for COVID-19 diagnostic and prognostic analysis. Eur. Respir. J. 2020;56(5) doi: 10.1183/13993003.00775-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roy S., Menapace W., Oei S., Luijten B., Fini E., Saltori C., Huijben I., Chennakeshava N., Mento F., Sentelli A., Peschiera E., Trevisan R., Maschietto G., Torri E., Inchingolo R., Smargiassi A., Soldati G., Rota P., Passerini A., Van Sloun R.J.G., Ricci E., Demi L. Deep learning for classification and localization of COVID-19 markers in point-of-care lung ultrasound. IEEE Trans. Med. Imaging. 2020;39(8):2676–2687. doi: 10.1109/TMI.2020.2994459. [DOI] [PubMed] [Google Scholar]
- 27.Li D., Wang D., Dong J., Wang N., Huang H., Xu H., Xia C. False-negative results of real-time reverse-transcriptase polymerase chain reaction for severe acute respiratory syndrome coronavirus 2: role of deep-learning-based CT diagnosis and insights from two cases. Korean J. Radiol. 2020;21(4):505–508. doi: 10.3348/kjr.2020.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miotto R., Wang F., Wang S., Jiang X., Dudley J.T. Deep learning for healthcare: review, opportunities and challenges. Brief Bioinform. 2018;19(6):1236–1246. doi: 10.1093/bib/bbx044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ching T., Himmelstein D.S., Beaulieu-Jones B.K., Kalinin A.A., Do B.T., Way G.P., Ferrero E., Agapow P.M., Zietz M., Hoffman M.M., Xie W., Rosen G.L., Lengerich B.J., Israeli J., Lanchantin J., Woloszynek S., Carpenter A.E., Shrikumar A., Xu J., Cofer E.M., Lavender C.A., Turaga S.C., Alexandari A.M., Lu Z., Harris D.J., DeCaprio D., Qi Y., Kundaje A., Peng Y., Wiley L.K., Segler M.H.S., Boca S.M., Swamidass S.J., Huang A., Gitter A., Greene C.S. Opportunities and obstacles for deep learning in biology and medicine. J. R. Soc. Interface. 2018;15(141) doi: 10.1098/rsif.2017.0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Y.Q., Liang C.H., He L., Tian J., Liang C.S., Chen X., Ma Z.L., Liu Z.Y. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal Cancer. J. Clin. Oncol. 2016;34(18):2157–2164. doi: 10.1200/JCO.2015.65.9128. [DOI] [PubMed] [Google Scholar]
- 31.Zhang B., Tian J., Dong D., Gu D., Dong Y., Zhang L., Lian Z., Liu J., Luo X., Pei S., Mo X., Huang W., Ouyang F., Guo B., Liang L., Chen W., Liang C., Zhang S. Radiomics features of multiparametric MRI as novel prognostic factors in advanced nasopharyngeal carcinoma. Clin. Cancer Res. 2017;23(15):4259–4269. doi: 10.1158/1078-0432.CCR-16-2910. [DOI] [PubMed] [Google Scholar]
- 32.Jiang Y., Chen C., Xie J., Wang W., Zha X., Lv W., Chen H., Hu Y., Li T., Yu J., Zhou Z., Xu Y., Li G. Radiomics signature of computed tomography imaging for prediction of survival and chemotherapeutic benefits in gastric cancer. EBioMedicine. 2018;36:171–182. doi: 10.1016/j.ebiom.2018.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arshad M.A., Thornton A., Lu H., Tam H., Wallitt K., Rodgers N., Scarsbrook A., McDermott G., Cook G.J., Landau D., Chua S., O’Connor R., Dickson J., Power D.A., Barwick T.D., Rockall A., Aboagye E.O. Discovery of pre-therapy 2-deoxy-2-(18)F-fluoro-D-glucose positron emission tomography-based radiomics classifiers of survival outcome in non-small-cell lung cancer patients. Eur. J. Nucl. Med. Mol. Imaging. 2019;46(2):455–466. doi: 10.1007/s00259-018-4139-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colen R.R., Fujii T., Bilen M.A., Kotrotsou A., Abrol S., Hess K.R., Hajjar J., Suarez-Almazor M.E., Alshawa A., Hong D.S., Giniebra-Camejo D., Stephen B., Subbiah V., Sheshadri A., Mendoza T., Fu S., Sharma P., Meric-Bernstam F., Naing A. Radiomics to predict immunotherapy-induced pneumonitis: proof of concept. Invest. New Drugs. 2018;36(4):601–607. doi: 10.1007/s10637-017-0524-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song J., Yin Y., Wang H., Chang Z., Liu Z., Cui L. A review of original articles published in the emerging field of radiomics. Eur. J. Radiol. 2020;127:108991. doi: 10.1016/j.ejrad.2020.108991. [DOI] [PubMed] [Google Scholar]
- 36.Avanzo M., Stancanello J., El Naqa I. Beyond imaging: the promise of radiomics. Phys. Med. 2017;38:122–139. doi: 10.1016/j.ejmp.2017.05.071. [DOI] [PubMed] [Google Scholar]
- 37.Fu L., Li Y., Cheng A., Pang P., Shu Z. A novel machine learning-derived radiomic signature of the whole lung differentiates stable from progressive COVID-19 infection: a retrospective cohort study. J. Thorac. Imaging. 2020;35(6):361–368. doi: 10.1097/rti.0000000000000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H., Wang L., Lee E.H., Zheng J., Zhang W., Halabi S., Liu C., Deng K., Song J., Yeom K.W. Decoding COVID-19 pneumonia: comparison of deep learning and radiomics CT image signatures. Eur. J. Nucl. Med. Mol. Imaging. 2020:1–9. doi: 10.1007/s00259-020-05075-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie C., Ng M.-Y., Ding J., Leung S.T., Lo C.S.Y., Wong H.Y.F., Vardhanabhuti V. Discrimination of pulmonary ground-glass opacity changes in COVID‐19 and non-COVID-19 patients using CT radiomics analysis. Eur. J. Radiol. Open. 2020;7:100271. doi: 10.1016/j.ejro.2020.100271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.