Abstract
Leptospirosis is a worldwide infectious and zoonotic disease. The incidence of this disease is high in temperate regions, especially in northern Iran. The aim of this study was to investigate the effects of temperature, pH, and Phyllanthus amarus plant extract on the lipL32 gene expression in pathogenic Leptospira spp. Fifty water samples were collected. Culture and PCR technique were used to isolate and identify the bacterium and the presence of the lipL32 gene. The samples were exposed to different temperatures and pH levels for one day and the Ph. amarus plant extract at different concentrations for one and seven days. RNA was extracted, and cDNA synthesis was performed for all the samples. All cDNAs were evaluated by the real-time PCR (SYBR green) technique. Out of the 50 samples, ten samples (20%), using PCR were determined to contain the pathogenic Leptospira. Fold change of the expression of the lipL32 gene associated with stresses was as follows: temperature stress of 40°C, 35°C, and 25°C reduced the lipL32 gene expression in all three isolates, especially in the isolates type 1. The pH stress, i.e., pH values equal to 8 or 9 reduced the gene expression in three types of isolates, and pH = 6 stress increases the lipL32 gene expression in the isolates of type 1. Ph. amarus plant extract stress reduced the mentioned gene expression only in isolates of type 2. Temperature and pH stresses could lead to differences in the expression level and cause the lipL32 gene expression decrease in three pathogenic isolates. The MIC results showed anti-leptospiral effect of Ph. amarus plant extract.
Key words: Leptospira, Phyllanthus amarus, lipL32 gene expression, real-time PCR, stress
Introduction
Leptospirosis is a contagious zoonotic disease spread worldwide and is caused by the bacterium Lepto spira (Zakeri et al. 2010). The genus Leptospira includes pathogenic, intermediate, and non-pathogenic species with the pathogenic species causing infection in humans and animals (Ko et al. 2009). Leptospirosis is widespread throughout the world except in Antarctica, and it is endemic in tropical regions with high rainfall (Costa et al. 2015). The World Health Organization recognizes leptospirosis, also known as Weil’s disease, as a neglected tropical disease with a significant global health burden. Globally, it has been estimated to be 1.03 million cases annually with 58,900 deaths (Fann et al. 2020) The highest estimates of disease morbidity and mortality were observed in Global Burden of Disease (GBD) regions of South and Southeast Asia, Oceania, Caribbean, Andean, Central, Tropical Latin America, and East Sub-Saharan Africa) Costa et al. 2015).
The natural reservoirs of Leptospira are rodents, but reservoirs include a variety of wild and domestic animals, livestock, and insectivores (De Vries et al. 2014). The exposure may occur through direct contact with an infected animal or indirect contact via soil or water contaminated with urine from an infected animal. Individuals with occupations at risk for direct contact with potentially infected animals include veterinarians, farmworkers (particularly in dairy milking situations), hunters, animal shelter workers, scientists, and technologists handling animals in laboratories or during fieldwork. Agricultural workers at risk for leptospirosis include rice field workers, banana farmers, sugar cane, and pineapple field harvesters (Haake and Levett 2015).
Many virulence factors are involved in the pathogenesis and infection of this bacterium, including lipopolysaccharide (LPS), hemolysin, surface proteins, adhesion molecules, and the ability to move and swim in liquid environments (Fraga et al. 2011). The 32-kDa lipL32 lipoprotein, a type of outer membrane protein of Leptospira, which is highly conserved in pathogenic species, exists only in pathogenic strains and is expressed during infection (Podgorsek et al. 2020). LipL32 binds to collagens V, VI, and I as well as to lamin. It also binds to fibronectin via the calcium-dependent pathway (Shen-Hsing et al. 2017).
Bacteria rely on the ability to sense and respond to environmental stressors, including changes in temperature, pH, osmolarity, oxygen availability, and nutrient conditions. Environmental signals, such as increased oxidative stress or temperature, can stimulate the response of regulators of virulence genes to pathogenic Leptospira during infection (Fraser and Brown 2017). Phyllanthus amarus is a plant widely used for the treatment of human diseases. The aqueous and methanolic extracts of this plant possess an excellent and useful inhibitory activity against Leptospira (Verma et al. 2014).
The aim of this research was to isolate and identify the leptospires from the farming fields of Tonekabon town situated in northern Iran as well as to study the effect of different environmental stresses, including pH, temperature, and Ph. amarus plant extract on the survival capability and the lipL32 gene expression in the pathogenic leptospires. Study of the influence of Ph. amarus plant extract on these bacteria could reveal the important role of this plant in the prevention, control, and treatment of leptospirosis. So far, no research has been performed to investigate the effect of pH on the lipL32 gene expression in pathogenic Leptospira spp. and our study is a part of new investigations.
Experimental
Material and Methods
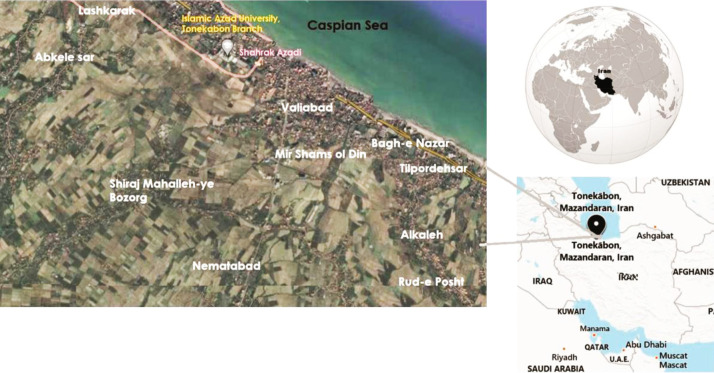
Samples collection, cultivation, and isolation. 50 samples from the stagnant waters of the rice field were collected under the aseptic conditions, including wearing gloves and the boot in Tonekabon city (Iran, Mazandaran province). Sampling was performed in four seasons of the year from the autumn of 2017 to the end of the summer of 2018 on days with stable weather conditions (sunny weather and a few days after rain). A sampling of creeks and water canals of agricultural fields was done from ten places, including: Valiabad, Mir Shams-ol Din, Bagh-e Nazar, Tilpordehsar, Alkaleh, Rud-e Posht, Nematabad, Shiraj Mahalleh-ye Bozorg, Abkele Sar, and Lashkarak (Fig. 1). The samples were placed in on ice after sampling each time and transferred to Tonekabon University’s Microbiology Research Laboratory as soon as possible.
Fig. 1.
Localization of ten environmental sites investigated.
Samples were centrifuged at 10,000 × g for 10 min. The supernatant was discarded, and the remaining contents were passed through 0.45 µm and 0.2 µm filters, respectively, and transferred to liquid EMJH medium (Ellinghausen and Mccullough 1965; Johnson and Harris 1967). All the samples were incubated at 30°C for 30 days. Aliquots of the medium were transferred to the solid EMJH medium and cultured. Plates were incubated at 30°C for 30 days. Leptospira interrogans serovar Icterohaemorrhagiae (RTCC2823) was used as a reference strain. The mentioned strain was from a Leptospira Reference Laboratory, Razi Vaccine and Serum Research Institute, Karaj, Iran.
DNA extraction. DNA was extracted from all Leptospira isolates along with the reference strain using the Purification Kia Spin polymerase chain reaction (PCR) Kit (Kiagen, Iran). The purity of the extracted DNA was evaluated by optical absorption at 260 and 280 nm wavelengths using a Biophotometer (Eppendorf, Germany).
Amplification of the lipL32 and 16S rRNA genes by PCR method. At this stage, the presence of the 16S rRNA and lipL32 genes was evaluated using specific primers (Table I). The primers were produced by TAG Kopenhagen (Denmark) and used to amplify the mentioned genes (Vital-Brazil et al. 2010). Each reaction was performed in a total volume of 25 μl and included 14 μl dd water, 2.5 μl 10 × PCR buffer, 1 μl of 10 pmol of each primer, 0.5 μl of 10 mM dNTPs, 0.75 μl of 50 mM MgCl2, 0.25 μl Taq polymerase enzyme (CinnaGen, Iran) and 5 μl of DNA. Escherichia coli was used as a negative control sample, and L. interrogans serovar Icterohaemorrhagiae (RTCC2823) as a positive control sample. The mentioned control samples were amplified along with the other samples.
Table I.
The specific primers of Leptospira.
| Name of primer | Type | Sequence of primer |
|---|---|---|
| 16S rRNA | Forward | 5’-GAACTGAGACACGGTCCAT-3’ |
| 16S rRNA | Reverse | 5’-GCCTCAGCGTCAGTTTTAGG-3’ |
| LipL32 | Forward | 5’-ATCTCCGTTGCACTCTTTGC-3’ |
| LipL32 | Reverse | 5’-ACCATCATCATCATCGTCCA-3’ |
PCR amplification conditions using a thermocycler (Sensoquest, Germany) were as follows: one denaturation cycle at 95°C for 5 min, 40 cycles of denaturation at 95°C for 1 min, annealing at 55°C for 30 sec, extension at 72°C for 1 min, and a final extension at 72°C for 7 min.
Agarose gel electrophoresis. PCR products were run on 1.5% agarose gel with 100 bp DNA ladder (Fermentas, Russia). Five microliters of the PCR products were electrophoresed at 75 V for 40 min. DNA fragments were visualized using Safe view-II Nucleic Acid stain (Kiagen, Iran) and UVDoc (England) imaging.
The 16S rRNA gene sequencing. 16S rRNA PCR products were sent to Macrogen in South Korea (http://www.macrogen.com/) for DNA sequencing.
Bioinformatics applications. All sequence data were subjected to BLAST analysis (http://www.ncbi.nlm.nih.gov/BLAST/) to definitively identify each respective amplicon of the 16S rRNA gene.
Thermal and pH stress conditions. Three isolates were selected from Leptospira colonies whose PCR confirmed the presence of the lipL32 gene. A similar number of colonies was cultured in liquid EMJH medium for seven days until the bacteria reached the log phase (Larson et al. 1959). One ml of the above culture was added to 4 ml of liquid EMJH medium and incubated at 25, 30, 35, and 40°C (Parker and Walker 2011) for one day. To apply the pH stress, liquid EMJH media were prepared at the different pH of 6, 7, 8, and 9 (pH-meter, Euteoh, Malaysia). Then, 1 ml of the mentioned culture was added to 4 ml of liquid EMJH medium with the above-mentioned pHs and incubated at 30°C for one day. For each of the stressors, the reference strain was also considered. Experiments were performed in three biological replicates. The reference strain, as mentioned isolates, was affected by different temperature and pH stresses.
Plant extract stress conditions. Plant collection. Ph. amarus plant was obtained from the ABS medicinal plant research center, Karippatti, Salem Tamilnadu, India, during December 2018. Then, it was washed several times with sterile distilled water and again sterilized, dried, and powdered.
Preparation of an aqueous extract. Ten grams of the plant powder were added to sterile distilled water and heated for 2 h. The liquid contents of the extract were centrifuged at 10,000 × g for 10 min. The supernatant was heated (2 h) and centrifuged, and the cycle was repeated again (Mohan et al. 2016). The liquid contents were first passed through the Whatman 1 filter paper and then filtered through a 0.2 µm filter and heated for 2 h. It was then dried in an oven for 6 h at 70°C (Fiaz et al. 2013).
Anti-leptospiral sensitivity test. The effect of aqueous extracts of Ph. amarus against pathogenic Leptospira species and reference Leptospira strain was investigated using a minimum inhibitory concentration (MIC) and a tube dilution technique (TDT) (Mohan et al. 2016). One ml of 5, 10, 20, 40, 80, 160, 320, 640, 1,280, 2,560, 5,120, 10,240 µg/ml concentrations was added to 1 ml of liquid EMJH medium, and the work was continued according to the above techniques for three isolates. Tubes were evaluated for anti-leptospiral activity, and the effect of plant extract stress on three selected isolates and a reference strain was investigated at 1- and 7-day intervals. Experiments were repeated in triplicate to evaluate the plant extract stress.
RNA extraction and cDNA synthesis. The RNA of three isolates of Leptospira and the reference strain was isolated and purified with and without shock using a hybrid-RTM (Gene ALL Korea-South Seoul) RNA extraction kit. cDNA synthesis was performed by the TwoStep kit HyperScriptTM first-strand synthesis kit (Gene ALL Korea, South Seoul). cDNA was used as a template for PCR and real-time PCR.
Real-Time PCR. The real-time PCR reaction was performed on Step One and Step One Plus Real-Time PCR systems (Applied Biosystems-Thermo Fisher Scientific). Twenty μl of real-time PCR mixture was assayed, including 10 µl of 2 × real-time PCR SYBR Green Master mix (Amplicon, Denmark), 0.2 µl of forward primer, 0.2 µl of reverse primer, lipL32 primer (Table I), 2 µl of cDNA as a sample and 7.6 µl of dd water. Real-time PCR temperature program consisted of holding stage at 95°C for 15 min, cycling stage comprising 33 cycles (95°C for 40 s, 57°C for 1 min, 72°C for 30 s), melt curve stage at 72°C for 1 min, and 97°C for 5 s. All the experiments were repeated at least three times. The raw data was removed from the system as a CT. Analysis of relative gene expression data was conducted using real-time PCR (SYBR Green) and the 2–ΔΔCT method (Livak and Schmittgen 2001).
Statistical Analysis. SPSS (22) was used for statistical analysis. Variation among the groups was analyzed using the ANOVA test. A minimum range of variance p < 0.05 was reflected as significant.
Results
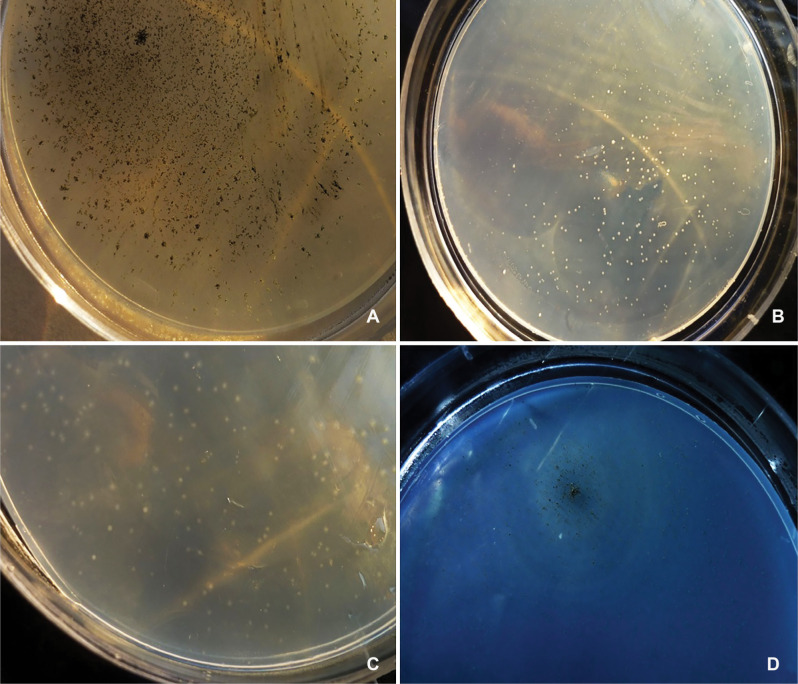
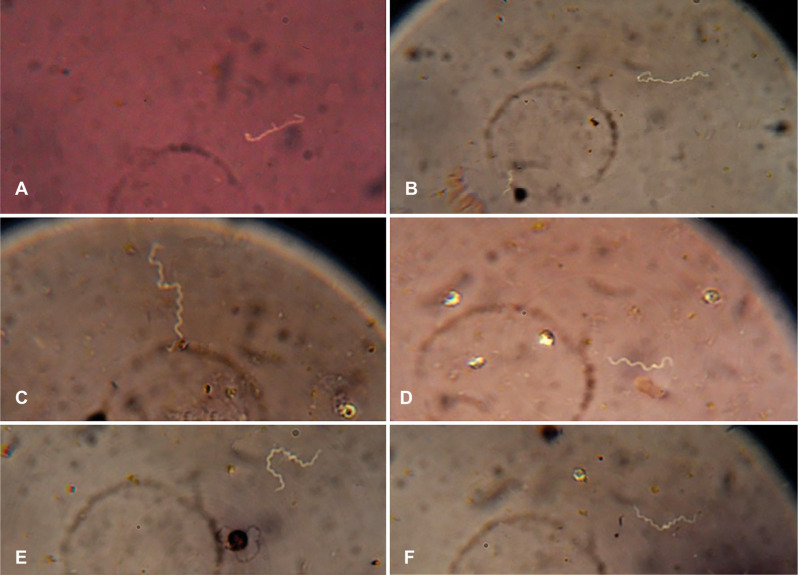
Results of macroscopic and microscopic observations of the samples. Different colony morphology of leptospiral growth was observed (Fig. 2). During negative staining, the observation of thin, helical organisms by optical microscopy at 100 × magnification revealed Leptospira’s presence in the samples Leptospira are usually curved in one or two ends (Fig. 3). Overall, of the 50 samples, 21 (42%) contained Leptospira based on colony formation and microscopic observation.
Fig. 2.
Leptospira colonies.
(a) Formation of surface’s black colonies, with round shape, needle tip size (at 30 days of incubation on EMJH agar), wooly margins, and hilly elevation related to Leptospira pathogen species. (b) Formation of sub surface’s white colonies with round shape, small size, and some smaller colonies, size 1–2 mm (after 30 days of incubation on EMJH agar), smooth (entire) margins, flat elevation related to Leptospira perdikensis. (c) Formation of sub surface’s very bright cream colonies with round shape, small size, 1–3 mm, (after 30 days of incubation on EMJH agar), smooth (entire) margins, with small to medium halo, flat elevation related to Leptospira ryugenii. (d) It had the characteristics of a colony (a), except that in this case, rings were seen to the same distance around the surface black colony related to Leptospira pathogen species.
Fig. 3.
Microscopic observation of Leptospira with a 100× magnification optical microscope
A: Multiplying Leptospira; the image relates to pathogenic Leptospira spp., B: The Leptospira bacterium that is twisted over itself is located on the right. In the lower-left a shorter Leptospira is visible; the image related to Leptospira ryugenii, C and E: Long Leptospira, D: The hook area of the Leptospira bacterium was clearly observed in a microscopic view; the image related to Leptospira perdikensis, F: Two Leptospira bacteria that are interconnected from one end and form a long separable leptospiral chain; the image related to Leptospira sp.
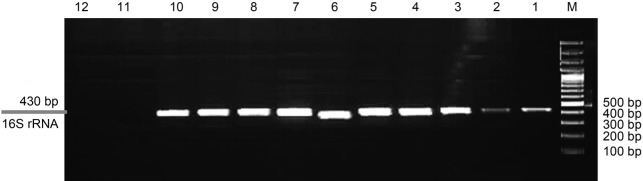
Detection of pathogenic and saprophytic Leptospira spp. by PCR. Of the 50 samples examined, 10 (20%) contained pathogenic species that were positive by PCR and specific primers for the 16S rRNA and lipL32 genes, respectively. PCR amplification of the samples with the aforementioned primers was performed separately in both 430 bp and 474 bp regions, respectively. Amplification of a 474 bp fragment confirmed the presence of pathogenic Leptospira (Fig. 4). Eleven (22%) samples contained saprophytic species that were positive by PCR using the 16S rRNA gene-specific primer, and amplification of 430 bp-gene fragments was observed (Fig. 5).
Fig .4.
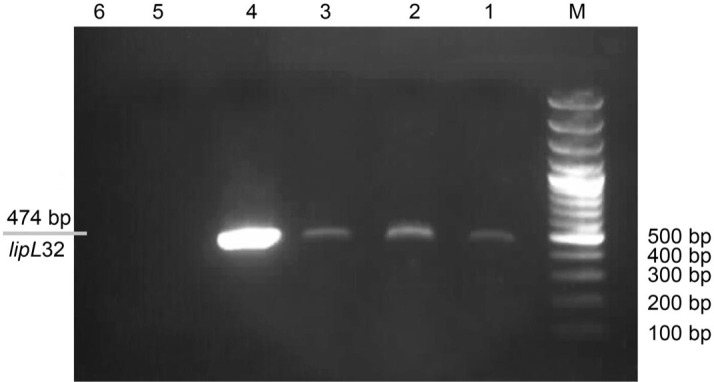
Agarose gel electrophoresis (1.5%) of PCR products of environmental samples.
Columns 1, 2, and 3, pathogenic Leptospira spp.; Column 4 – a reference strain; Column 5 – a negative control; Column 6 – E. coli; Column M – 100 bp DNA ladder.
Fig. 5.
Agarose gel (1.5%) analysis of a PCR products of environmental samples; Columns 1–9 – species of Leptospira; Column 10 – a reference strain; column 11 – a negative control; Column 12 – E. coli E. coli; Column M – 100 bp DNA ladder. Sampling sites that were positive for the presence of pathogenic Leptospira by both culture and PCR were reported in Fig. 1 and Table II.
Table II.
The sampling sites and positive cases reported in the study.
| Number of positive cases of saprophytic Leptospira | Number of positive cases of pathogenic Leptospira | Number of samples | Site |
|---|---|---|---|
| 3 | 2 | 5 | Valiabad |
| 2 | 2 | 5 | Bagh e nazar |
| 0 | 1 | 5 | Mir shams-ol |
| 1 | 1 | 5 | Rud e posht |
| 1 | 0 | 5 | Alkaleh |
| 1 | 0 | 5 | Abkele sar |
| 0 | 1 | 5 | Lashkarak |
| 1 | 1 | 5 | Nematabad |
| 1 | 1 | 5 | Shiraj mahalleh-ye bozorg |
| 1 | 1 | 5 | Tilpordehsar |
| 11 (22%) | 10 (20%) | 50 | Total |
16S rRNA PCR and sequencing analysis. Of the six samples selected randomly and sent for sequencing; three could not be sequenced. Elements of the nucleotide sequence of the other three samples in the NCBI site confirmed the existence of Leptospira (Table III).
Table III.
The results of the sequencing.
| Molecular detection | Max Score | Total Score | Query Cover | E Value | Per Ident | Accession |
|---|---|---|---|---|---|---|
| Leptospira perdikensis strain CES 16S ribosomal RNA gene | 612 | 612 | 93% | 3e-171 | 99.41% | MN086353.1 |
| Leptospira ryugenii strain CES 16S ribosomal RNA gene | 176 | 176 | 26% | 5e-40 | 100% | MN086356.1 |
| Leptospira sp. MS336 gene for 16S ribosomal RNA gene | 593 | 593 | 100% | 2e-166 | 100% | AB758752.1 |
The anti-leptospiral activity of the aqueous extract of Ph. amarus. Stress exerted by the aqueous extract of Ph. amarus plant was acceptable for all three isolates of pathogenic Leptospira and L. interrogans serovar Icterohaemorrhagiae and their growth was inhibited for seven days. A complete inhibition (100%) of aqueous extract of Ph. amarus plant was observed against isolates of pathogenic Leptospira No. 1, 2, and 3 at the concentrations of 10,240, 5,120, 2,560, 1,280, 640 µg/ml. For the reference strain, a similar inhibition level was visible at the concentrations of 10,240, 5,120, 2,560, 1,280, 640, 320 µg/ml, respectively. But the aqueous extract of Ph. amarus plant did not show the full inhibitory effect for three isolates of pathogenic Leptospira and reference strain of Leptospira on the first day of experiments.
The lipL32 gene expression in Leptospira isolates by the real-time PCR and data analysis. The lipL32 gene expression were successfully observed using real-time PCR, and the curve showed the lipL32 gene amplification, indicating proper proliferation and function without any impediments. Samples without cDNA (i.e., negative) and those containing cDNA (i.e., positive) were assayed with other samples by RT-PCR.
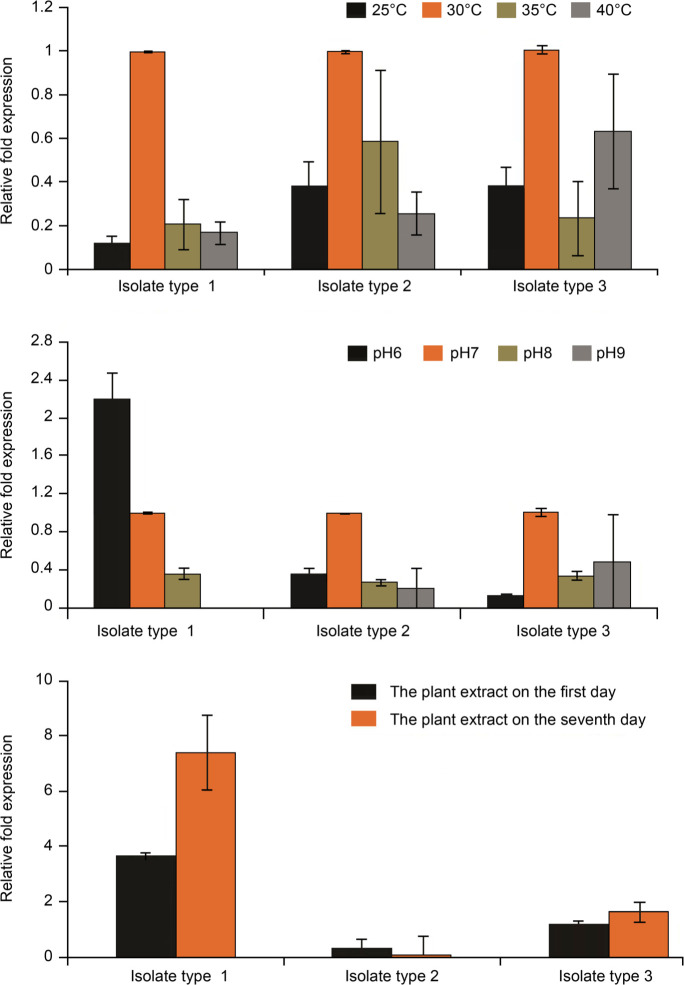
Fold change associated to gene lipL32, according to the results of 2–ΔΔCT affected by temperature stress, pH, and Ph. amarus plant extract includes the following.
Fold change associated with the lipL32 gene was in all three types of isolates affected by temperature stress of 30°C (equal to 1), 40°C, 35°C, and 25°C (less than 1). Thus, temperature stresses of 40°C, 35°C, and 25°C reduced the lipL32 gene expression in the isolates and this reduction significantly led to a further downward trend in type 1 isolates (25°C = 0.12-fold; 35°C = 0.21-fold; 40°C = 0.17-fold) when compared to type 2 and 3 isolates (Fig. 6). In all three types of Leptospira, there was a significant difference in the CT values for the lipL32 gene in control and the samples investigated under different thermal stresses (p < 0.05).
Fig. 6.
Comparison of the lipL32 gene expression in pathogenic Leptospira under the influence of temperature, pH, and Ph. amarus plant extract.
Fold change associated with the lipL32 gene affected by pH stress was equal to 1 to type 2 and 3 isolates at pH = 7, and lower than 1 at pH = 8 and 9. Therefore, the stress of pH 8 and 9 reduced the lipL32 gene expression in type 2 and 3 isolates, and this decrease was higher in type 2 isolates than in type 3 isolates. Isolates of type 1 did not show the expression at pH = 9 and demonstrated a decrease in the lipL32 gene expression at pH 8. Isolates of type 1 were the only isolates that showed an increase in the lipL32 gene expression (2.21-fold) at pH 6 (Fig. 6). In all the three isolates, there was a significant difference between the control lipL32 gene CT and the lipL32 gene CT under different pH conditions (p < 0.05).
Fold change associated with the lipL32 gene expression affected by Ph. amarus plant extract was 0.31-fold on day one and 0.05-fold on day 7 in isolates of type 2. These results indicated a reduction in the lipL32 gene expression under Ph. amarus plant extract’s stress. A further decrease of the gene expression in isolates type 2 was observed on the seventh day; however, the fold change associated to the lipL32 gene expression treated with Ph. amarus plant extract showed an increase in type 1 and 3 isolates. Fold change of this gene for isolates type 1 was 3.68-fold on first day and 7.41-fold of the seventh day, and for isolates of type 3 – 1.18-fold on first day and 7.41 of the seventh day (Fig. 6). Effect of stress imposed by the extract of Ph. amarus on the first and seventh days was significantly different in the three isolates (p < 0.05).
Ph. amarus plant extract, according to the MIC test, had strong anti-leptospiral activity and decreased the lipL32 gene expression in isolates type 2.
Discussion
Due to the temperate climate of the northern regions of Iran, the prevalence of leptospirosis is high in these areas. In the study of the pollution status of stagnant waters and wetlands, more than 7% of the evaluated wetlands in Mazandaran were contaminated with pathogenic Leptospira, and leptospirosis is an endemic and occupational disease in these regions (Rafiei et al. 2012).
Studies have shown that pathogenic Leptospira species may be detected by real-time PCR because the lipL32 gene is not present in non-pathogenic Leptospira species. Real-time PCR based on the lipL32 gene is a tool used in the rapid diagnosis of acute leptospirosis, especially in cases with the potential for rapid mortality before a diagnosis could be performed by serology or culture (Stoddard et al. 2009).
In our study, a real-time PCR technique was used to evaluate the lipL32 gene expression. The expression of the lipL32 gene was affected by temperature, pH, or Ph. amarus plant extract. In the previous study by Fraser and Brown (2017), the effects of temperature and oxidative stress on the expression of virulence genes in Leptospira borgpeterseneii Jules and Leptospira interrogans Portlandvere species were investigated. Bacteria were grown in EMJH medium at 30°C before the transition to 37°C. A total of 14 virulence-related genes (i.e., fliY, invA, lenA, ligB, lipL32, lipL36, lipL41, lipL45, loa22, lsa21, mce, ompL1, sph2, and tlyC) were evaluated using Endpoint PCR. Transcriptional analysis of the lenA, lipL32, lipL41, loa22, and sph2 genes was performed by quantitative real-time PCR. Temperature had reduced the lipL32 gene expression in 30°C and 37°C for both Portlandvere and Jules serovars in the study mentioned above, and 30°C and 37°C were temperatures that also reduced the lipL32 gene expression in our study.
Another study was conducted on the viability of Leptospira isolated from a human outbreak in Thailand in waters with different thermal and pH conditions (Stoddard et al. 2014). Lepospira spp. isolated from patients was genotyped using multilocus sequence typing during a multi-year outbreak in Thailand. The survival of ST34 isolates at different pH values, temperatures, and water sources was investigated. Most isolates survived until the end of the study, except for those subjected to temperatures higher than 37°C and pH = 8.65. In our study, the lipL32 gene expression was assessed at different pH values, and the isolates of type 1 did not show any growth in pH = 9. The results of the electrophoresis of PCR products were negative and real-time PCR results did not show the lipL32 gene expression when the isolates grew at pH = 9.
Today, due to the side effects of chemical drugs, herbal remedies gain interest. An important feature of medicinal plants is that treatment takes longer. In our study, the effect of Ph. amarus plant extract was higher on the lipL32 gene expression on the seventh day than on the first day, especially in the isolates type 2, confirming that herbal drugs take time.
In 2016, a study using Eclipta alba and Ph. amarus was performed, and their activity against L. interrogans was evaluated (Mohan et al. 2016). In this study, the anti-leptospiral activity of plant extracts against L. interrogans, expressed as the MIC value following standard TDT was evaluated. The Ph. amarus extract (at the concentration of 160 µg/ml) had superior antileptospiral activity than the E. alba extract (at the concentration of 320 µg/ml). When compared with our study, the Ph. amarus extract at the concentration of ≥ 640 µg/ml completely inhibited the growth of the three selected Leptospira isolates on the seventh day. Methanolic and aqueous extracts of E. alba and Ph. amarus had anti-leptospiral activity and proved to be the best anti-leptospiral drugs in vitro (Chandan et al. 2012). Since no research has been conducted to evaluate the Leptospira lipL32 gene expression under different physicochemical conditions, this study can be considered an innovative research in Iran examining the expression of the lipL32 gene in pathogenic Leptospira under different pH, temperature, or Ph. amarus extract stress conditions.
In this study, an increase or decrease in the lipL32 gene expression can be attributed to the influence of a single factor or a combination of factors such as growth temperature, strain variation, pH conditions, the plant extract, and other unknown factors. On the other hand, it is well known that Leptospira bacteria favor neutral to slightly alkaline pH. The results of our study showed a reduction in the lipL32 gene expression in pH = 9. The reason for choosing temperature 40°C in vitro was that we aimed to determine whether the lipL32 gene expression might occur in patients with leptospirosis who had a fever of 40°C.
The decrease in the lipL32 gene expression was associated with a 40°C thermal stress in isolates types 1, 2, and 3. According to our results and the findings of Fraser and Brown (2017) study, it can be stated that the lipL32 gene expression may depend on various thermal conditions, genus and strain of Leptospira, and different pH levels.
The results of this study showed the inhibitory anti-Leptospira effect of Ph. amarus plant extract expressed as the MIC value, and the reduction of the lipL32 gene expression in Leptospira isolates (Type 2) as it was demonstrated with real-time PCR technique. Therefore, it can be concluded that Ph. amarus plant can play an integral role in the prevention, control, and treatment of leptospirosis after examining its non-toxicity in vivo at the concentration used in this study.
Acknowledgements
Thesis advisors Masood Ghane and Monir Doudi are acknowledged and appreciated extremely in the research process as well as all employees of the Microbiology Research Laboratory ward in the Islamic Azad University of Tonekabon Branch.
Footnotes
Conflict of interest
The authors do not report any financial or personal connections with other persons or organizations, which might negatively affect the contents of this publication and/or claim authorship rights to this publication.
ORCID
Monir Doudi https://orcid.org/0000-0002-0895-1586
Literature
- Chandan S, Umesha S, Balamurugan V. Antileptospiral, antioxidant and DNA damaging properties of Eclipta alba and Phyllanthus amarus. Open Access Sci Rep. 2012;1(4):231 10.4172/scientificreports.231 [DOI] [Google Scholar]
- Costa F, Hagan JE, Calcagno J, Kane M, Torgerson P, Martinez-Silveira MS, Stein C, Abela-Ridder B, Ko AI. Global morbidity and mortality of Leptospirosis: A systematic review. PLoS Negl Trop Dis. 2015. September 17;9(9):e0003898 10.1371/journal.pntd.0003898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries SG, Visser BJ, Nagel IM, Goris MGA, Hartskeerl RA, Grobusch MP. Leptospirosis in Sub-Saharan Africa: a systematic review. Int J Infect Dis. 2014. November;28:47–64. 10.1016/j.ijid.2014.06.013 [DOI] [PubMed] [Google Scholar]
- Ellinghausen HC Jr, McCullough WG. Nutrition of Leptospira pomona and growth of 13 other serotypes: fractionation of oleic albumin complex (OAC) and a medium of bovine albumin and polysorbate 80. Am J Vet Res. 1965. January;26:45–51. [PubMed] [Google Scholar]
- Fann RJ, Vidya RR, Chong HE, Indralingam V, Christopher Chan WS. Clinical presentations and predictors of mortality for leptospirosis – A study from suburban area in Malaysia. Med J Malaysia. 2020. January;75(1):52–56. [PubMed] [Google Scholar]
- Fiaz AM, Rehaman H, Ansar Y, Zahid IA, Naeem A. Antimicrobial activities of the leaves and roots of Elaeagnus umbellata Thunb. Afr J Biotechnol. 2013. November 27;12(48):6754–6760. 10.5897/AJB11.4181 [DOI] [Google Scholar]
- Fraga TR, Barbosa AS, Isaac L. Leptospirosis: aspects of innate immunity, immunopathogenesis and immune evasion from the complement system. Scand J Immunol. 2011. May;73(5):408–419. 10.1111/j.1365-3083.2010.02505.x [DOI] [PubMed] [Google Scholar]
- Fraser T, Brown PD. Temperature and oxidative stress as triggers for virulence gene expression in pathogenic Leptospira spp. Front Microbiol. 2017. May 09;8:783 10.3389/fmicb.2017.00783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake DA, Levett PN. Leptospira species (Leptospirosis) In: Bennett JE, Dolin R, Blaser MJ, editors. Principles and practice of infectious diseases. 8th ed Philadelphia (USA): Saunders; 2015. p. 2714–2720. [Google Scholar]
- Johnson RC, Harris VG. Differentiation of pathogenic and saprophytic letospires. I. Growth at low temperatures. J Bacteriol. 1967; 94(1):27–31. 10.1128/JB.94.1.27-31.1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko AI, Goarant C, Picardeau M. Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat Rev Microbiol. 2009. October;7(10):736–747. 10.1038/nrmicro2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson AD, Treick RW, Edwards CL, Cox CD. Growth studies and plate counting of leptospires. J Bacteriol. 1959;77(3):361–366. 10.1128/JB.77.3.361-366.1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 2001. December;25(4):402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Mohan A, Pandurangan S, Ramalingam S. In vitro screening of Phyllanthus amarus and Eclipta alba against Leptospira autumnalis. J Med Sci Clin Res. 2016. May 26;4(5):10620–10627. 10.18535/jmscr/v4i5.38 [DOI] [Google Scholar]
- Parker J, Walker M. Survival of a pathogenic Leptospira serovar in response to combined in vitro pH and temperature stresses. Vet Microbiol. 2011. August;152(1–2):146–150. 10.1016/j.vetmic.2011.04.028 [DOI] [PubMed] [Google Scholar]
- Podgoršek D, Ružić-Sabljić E, Logar M, Pavlović A, Remec T, Baklan Z, Pal E, Cerar T. Evaluation of real-time PCR targeting the lipL32 gene for diagnosis of Leptospira infection. BMC Microbiol. 2020. December;20(1):59 10.1186/s12866-020-01744-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiei A, Hedayati Zadeh-Omran A, Babamahmoodi F, Alizadeh Navaei R, Valadan R. Review of leptospirosis in Iran. Majallah-i Danishgah-i Ulum-i Pizishki-i Mazandaran. 2012;22(94):102–110. [Google Scholar]
- Shen-Hsing H, Cheng-Chieh H, Ming-Yang Ch, Yi-Ching K, Huang-Yu Y, Hsiang-Hao H. Ya-Chung Ti, Li-Fang Ch, Rong-Long P, Fan-Gang T, Chih-Wei Y. Active components of Leptospira outer membrane protein LipL32 to Toll-Like Receptor 2. Sci Rep. 2017;7(1). 10.1038/s41598-017-08743-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard RA, Bui D, Wuthiekanun V, Haberling DL, Thaipadungpanit J, Hoffmaster AR. Viability of Leptospira isolates from a human outbreak in Thailand in various water types, pH, and temperature conditions. Am J Trop Med Hyg. 2014. November 05;91(5): 1020–1022. 10.4269/ajtmh.13-0748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard RA, Gee JE, Wilkins PP, McCaustland K, Hoffmaster AR. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn Microbiol Infect Dis. 2009. July;64(3):247–255. 10.1016/j.diagmicrobio.2009.03.014 [DOI] [PubMed] [Google Scholar]
- Verma S, Sharma H, Garg M. Phyllanthus amarus: A review. J Pharmacogn Phytochem. 2014;3(2):18–22. [Google Scholar]
- Vital-Brazil JM, Balassiano IT, Oliveira FS, Costa ADS, Hillen L, Pereira MM. Multiplex PCR-based detection of Leptospira in environmental water samples obtained from a slum settlement. Mem Inst Oswaldo Cruz. 2010. May;105(3):353–355. 10.1590/S0074-02762010000300020 [DOI] [PubMed] [Google Scholar]
- Zakeri S, Khorami N, Ganji ZF, Sepahian N, Malmasi AA, Gouya MM, Djadid ND. Leptospira wolffii, a potential new pathogenic Leptospira species detected in human, sheep and dog. Infect Genet Evol. 2010. March;10(2):273–277. 10.1016/j.meegid.2010.01.001 [DOI] [PubMed] [Google Scholar]