Abstract
The present study was conducted to evaluate the infection rates of Entamoeba histolytica, Entamoeba dispar, and Entamoeba moshkovskii among asymptomatic individuals in Erbil City, northern Iraq. The research intent was to discover whether pathogenic or nonpathogenic species cause a high rate of symptomless Entamoeba infections. Stool samples were microscopically examined, and the 18S-rRNA gene was targeted utilizing the nested PCR technique in the positive specimens. Initial results based on morphological features showed that the Entamoeba prevalence rate was 7.4%. Significantly higher rates of infections were seen in females than in males and in low-income people than in moderate-income people. The incidence rates among the asymptomatic individuals, as determined by molecular analysis, were as follows: E. histolytica – 6%, E. dispar – 4.3%, and E. moshkovskii – 0.3%. Of all the Entamoeba positive samples, a single infection with E. histolytica was identified in 41.4% samples; the single infection with E. dispar in 18.6% samples, 35.7% samples had mixed infections with two Entamoeba species, and 4.3% had mixed infections with three species. The current study concluded that 7.4% of healthy people, who live in the endemic area under investigation, carry Entamoeba species asymptomatically. Additionally, the majority of asymptomatic Entamoeba infections were caused by the pathogenic E. histolytica (81.4%) compared to E. dispar (58.6%), and E. moshkovskii with the lowest rate of infection. Single and co-infections with E. histolytica and E. dispar were noted. E. moshkovskii, which was identified for the first time in the region, was only seen in mixed infections.
Key words: Entamoeba histolytica, Entamoeba dispar, Entamoeba moshkovskii, epidemiology, asymptomatic infections
Introduction
Parasitic infections are endemic to most tropical and subtropical regions of developing countries (WHO 1997). Entamoeba histolytica, a protozoan parasite that inhabits the human gastrointestinal tract, causes asymptomatic infections in about 90% of infected people playing a significant role in spreading the parasite. Prolonged asymptomatic infection can lead to invasive amoebiasis, whose symptoms may include bloody diarrhea, abdominal pain, flatulence, nausea, and vomiting. In some cases, the amebae may spread from the gastrointestinal tract to the liver and cause the formation of ulcerations and abscesses, resulting in amoebic liver abscesses (Haque et al. 2003).
Entamoeba dispar and Entamoeba moshkovskii are nonpathogenic intestinal protozoa that are morphologically identical to E. histolytica but are genetically and biochemically different (Clark and Diamond 1991; Diamond and Clark 1993). Previous studies showed that the infection rate of E. dispar in developed countries is much higher than E. histolytica (Pillai et al. 1999; Fotedar et al. 2007b). High levels of E. moshkovskii infection were reported on the Indian subcontinent. However, fewer studies have been conducted into the prevalence of this species. Human isolates have been reported in South Africa, North America, Italy, and Bangladesh (Ali 2003; Singh et al. 2009).
Amoebiasis develops in 50 million individuals globally, with an annual mortality rate of 40,000 to 100,000 (WHO 1997). This high infection rate is likely inflated as a result of false positives caused by the morphologically indistinguishable, nonpathogenic E. dispar/moshkovskii, and/or polymorphic nuclear leukocytes and macrophages with similar morphology in the stool samples (Walsh 1986; Tanyuksel and Petri 2003). New methods have been developed that are better in distinguishing between the pathogenic E. histolytica and nonpathogenic amoebae in the stool sample. Emerging molecular-based techniques, such as polymerase chain reaction (PCR), have improved test specificity, or true positive rate of the target E. histolytica DNA (Tanyuksel and Petri 2003; Paul et al. 2007). Therefore, the most-recent epidemiological studies of E. histolytica use molecular methods to provide accurate data (Santos et al. 2010). To date, the PCR technique has never been used for assessing the prevalence rate of E. histolytica in Erbil City. Several studies have reported the infections with E. histolytica in almost all Iraqi cities, but only a few applied molecular methods; most relied on microscopic examination (Hamad and Ahmed 2011; Al-Sorchee et al. 2013; Saqur et al. 2017). To date, no research has been conducted on asymptomatic individuals in Iraq, the least-studied group globally. Moreover, it is mostly unknown whether the asymptomatic individuals have been infected with E. histolytica or the nonpathogenic E. dispar and/or E. moshkovskii. This study was conducted to fill this gap in research and strives to determine the prevalence rate of Entamoeba in Erbil City, first using microscopic examination and then molecular techniques, to confirm the presence of and differentiate between pathogenic and nonpathogenic amoebae.
Experimental
Materials and Methods
A total of 950 random stool samples (524 male and 426 female) were collected from asymptomatic healthy adults in a cross-sectional study. The Central Laboratory of Erbil Province provided specimens from asymptomatic individuals. Specimen donors filled out a structured questionnaire about personal status, residency, and source of water supply. The collected fresh stool samples were microscopically examined using the iodine and saline wet mount microscopy to detect Entamoeba trophozoites and/or cysts. About 0.2 g of each specimen was preserved at –80°C for molecular analysis.
DNA was extracted from specimens using the QiaAmp Fast DNA Stool Mini Kit (Qiagen, Hilden, Germany) following the manufacturer protocol. Finally, the purified DNA concentrate was eluted from the silica membrane spin column with a low salt buffer. DNA concentration was measured with a nanospectrophotometer; then, each sample was labeled and stored at –20°C. A nested PCR was performed. The first PCR targeted the Entamoeba genus by amplifying the 897 bp of the 18S rRNA gene, while the second PCR primers targeted E. histolytica, E. dispar, and E. moshkovskii by amplifying the 439 bp, 174 bp, and 553 bp respectively. This method was previously described by Khairnar and Parija (2007). The primers targeting the 18S-ribosomal RNA gene were confirmed for specificity by the Basal Local Alignment Search Tool (BLAST), the genome database of all organisms from the National Center for Biotechnology Information (NCBI). PCR amplification was performed using a thermal cycler (Techne Ltd., Cambridge, UK) with 20 µl reaction volumes that consisted of 10 µl Hot Start Master Mix (containing Taq DNA polymerase 1 unit/10 µl, 2 × reaction buffer, enzyme stabilizer, 4 mM MgCl2, sediment, 0.5 mM each of dATP, dCTP, dGTP, dTTP, pH 9, and loading dye) (GeNet Bio, Daejeon, South Korea); 2 µl of both the forward and reverse primers (10 ρmole for each), 2 µl of DNA template, and 6 µl of water. The PCR cycling and running parameters were defined as one cycle of initial denaturation at 95°C for 10 min followed by 30 cycles of 94°C for 30 sec, 58°C for 30 sec, and 72°C for 30 sec with a final extension of 72°C for 5 min. The second PCR used the same cycling and running parameters except that the first step used 35 cycles, and the annealing temperature changed to 52°C. Negative and positive controls were used in both PCR rounds. Positive control DNA for E. histolytica HM-1:IMSS, E. dispar SAW760, and Laredo strains of E. moshkovskii were obtained from Kurdistan Biomedical Science University, Sanandaj, Iran. The PCR products were electrophoresed in 1%, 1.5%, and 2% agarose gels with a 1X Tris-boric acid-EDTA buffer (TBE) and stained with 0.2 mg/ml of ethidium bromide (Sigma-Aldrich, St. Louis, Missouri, USA), with a 100-bp DNA marker ladder (Promega Corp., Madison, Wisconsin, USA).
Sequencing of PCR products. A single sample of each species was randomly selected and sequenced with species-specific primers in both forward and reverse directions using BigDye terminators and an ABI 3730XL sequencer (Macrogen® Corp., Seoul, South Korea). The nucleotide sequences of forward and reverse reactions were manually edited, and the sequences for each identified species were submitted to the GenBank.
Statistical analysis. The data was analyzed using the IBM SPSS Statistics Server Version 23. Results expressed using descriptive statistics: frequencies, percentages, Fisher exact test, and chi-square. P value < 0.05 was regarded as statically significant.
Results
Sociodemographic factors associated with Entamoeba infection. Our simple random sample consisted of 55.2% male and 44.8% female individuals (Table I). As determined by the microscopic examination, 7.4%, or 70 out of 950 stool samples from asymptomatic individuals, tested positive for Entamoeba species cysts and/or characteristic features of the trophozoite. Quadrinucleated spherical cysts and amoebic trophozoites with multiple pseudopodia of Entamoeba were observed using light microscopy and identified based on their morphology. A significantly higher (p < 0.05) rate of infection was detected in females (9.6%) than in males (5.5%). Significantly higher (p < 0.05) rates were also recorded in low-income participants (11%) than in moderate-income individuals (5.2%).
Table I.
Statistical analysis of the risk factors associated with microscopic positives for Entamoeba species and PCR positives for E. histolytica in asymptomatic subjects.
| Variants | Total frequency | Frequency and percentages of positive by microscopy | Percentage within each group | 95%CI | p-value | |
|---|---|---|---|---|---|---|
| Lower | upper | |||||
| Gender | ||||||
| Male | 524 (55.2%) | 29 (41.4%) | 524 (5.5%) | 0.036 | 0.075 | 0.018* |
| Female | 426 (44.8%) | 41 (58.6%) | 426 (9.6%) | 0.069 | 0.125 | |
| Residency | ||||||
| Urban | 702 (73.9%) | 48 (68.6%) | 702 (6.8%) | 0.049 | 0.090 | 0.322 |
| Rural | 248 (26.1%) | 22 (31.4%) | 248 (8.9%) | 0.054 | 0.125 | |
| Age group | ||||||
| 15–18 | 87 (9.2%) | 4 (5.7%) | 87 (4.6%) | 0.009 | 0.090 | 0.76 |
| 19–25 | 311 (32.7%) | 24 (34.3%) | 311 (7.7%) | 0.048 | 0.108 | |
| 26–35 | 328 (34.5%) | 26 (37.1%) | 328 (7.9%) | 0.050 | 0.110 | |
| 36–45 | 157 (16.5%) | 13 (18.6%) | 157 (8.3%) | 0.00 | 0.100 | |
| > 45 | 67 (7.1%) | 3 (4.3%) | 67 (4.5%) | 0.00 | 0.308 | |
| Educational level | ||||||
| Primary school | 298 (31.4%) | 25 (35.7%) | 298 (8.4%) | 0.053 | 0.115 | 0.73 |
| Secondary and high school | 448 (47.2%) | 31 (44.3%) | 448 (6.9%) | 0.044 | 0.096 | |
| Bachelor | 204 (21.5%) | 14 (20%) | 204 (6.9%) | 0.036 | 0.107 | |
| Family size | ||||||
| 1–2 | 92 (9.7%) | 5 (7.1%) | 92 (5.4%) | 0.011 | 0.105 | 0.563 |
| 3–4 | 228 (24%) | 13 (18.6%) | 228 (5.7%) | 0.028 | 0.086 | |
| 5–6 | 301 (31.7%) | 24 (34.3%) | 301 (8%) | 0.050 | 0.112 | |
| > 6 | 329 (34.6%) | 28 (40%) | 329 (8.5%) | 0.055 | 0.116 | |
| Income status | ||||||
| Poor | 355 (37.4%) | 39 (55.7%) | 355 (11%) | 0.078 | 0.142 | 0.004* |
| Middle class | 594 (62.5%) | 31 (44.3%) | 594 (5.2%) | 0.033 | 0.069 | |
| Wealthy | 1 (0.1%) | 0 (0%) | 1 (0%) | 0.00 | 0.00 | |
| Source of water supply | ||||||
| Chlorinated water | 646 (68%) | 39 (55.7%) | 646 (6%) | 0.042 | 0.079 | 0.062 |
| Well water | 302 (31.8%) | 31 (44.3%) | 302 (10.3%) | 0.068 | 0.138 | |
| Others | 2 (0.2%) | 0 (0%) | 2 (0%) | 0.00 | 0.00 | |
| Eating out of home | ||||||
| Never | 259 (27.3%) | 17 (24.3%) | 259 (6.6%) | 0.039 | 0.096 | 0.857 |
| Sometimes | 310 (32.6%) | 24 (34.3%) | 310 (7.7%) | 0.045 | 0.110 | |
| Always | 381 (40.1%) | 29 (41.4%) | 381 (7.6%) | 0.050 | 0.104 | |
| History of taking medications | ||||||
| In the last 2 weeks | 140 (14.7%) | 6 (8.6%) | 140 (4.3%) | 0.013 | 0.075 | 0.161 |
| More than 2 weeks | 810 (85.3) | 64 (91.4%) | 810 (7.9%) | 0.059 | 0.098 | |
| Hygiene practice | ||||||
| Washing vegetables and fruits | 920 (96.8%) | 66 (94.3%) | 920 (7.2%) | 0.055 | 0.088 | 0.204 |
| Eating raw unwashed vegetables and fruits | 30 (3.2%) | 4 (5.7%) | 30 (13.3%) | 0.029 | 0.263 | |
presenting statistically significant differences < 0.05
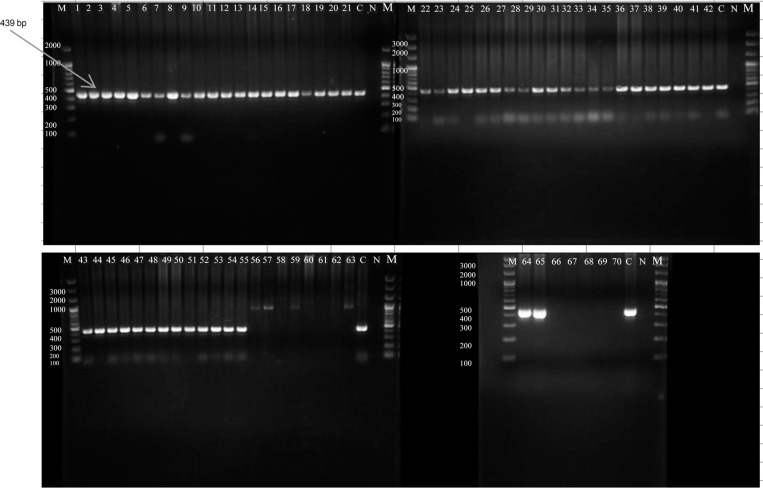
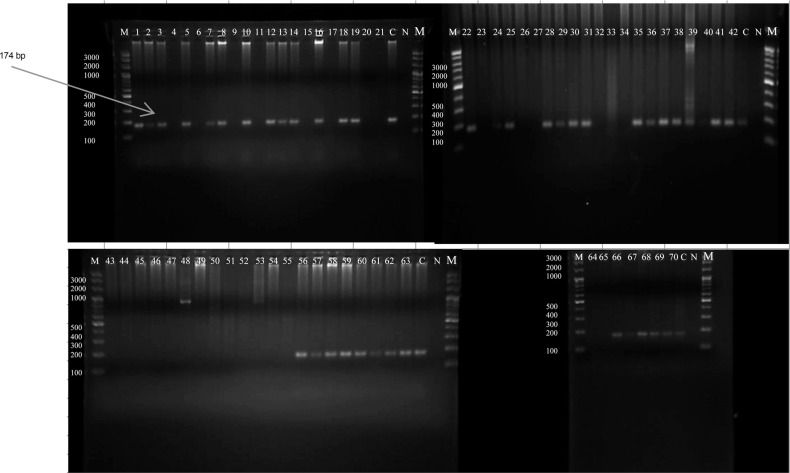
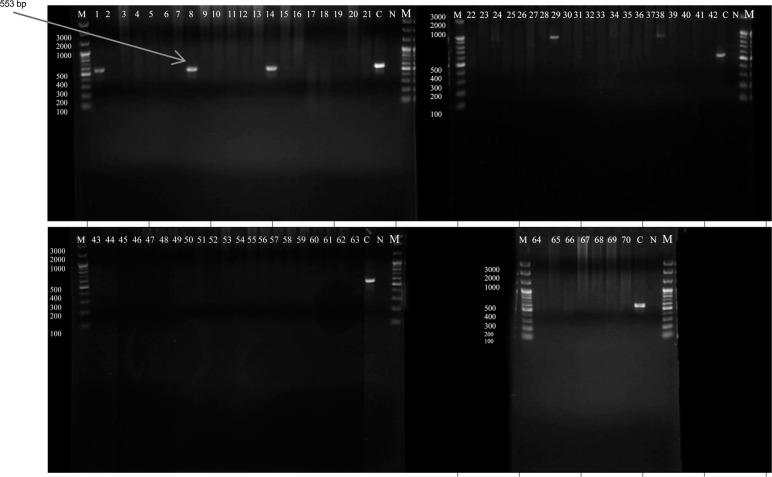
Nested PCR analysis. DNA was extracted from the 70 positive stool samples; their concentrations ranged from 5 µg/ml to 217 µg/ml, and purity ranged from 2.2–2.8, as measured by a nanospectrophotometer. Nested PCR results indicated that 57 samples tested positive for the 439 bp band for E. histolytica (Fig. 1), which is equivalent to 81.4% of the positive samples and 6% of the total number of samples (Table II). However, out of 57 positives, 29 carried a single infection, and 28 carried E. histolytica in combination with either E. dispar or E. dispar and E. moshkovskii. E. dispar accounted for 4.3% of the Entamoeba infections in the Erbil population (41 positives or 58.6% of the 70 positive samples, as revealed by the 174 bp band in the microscopic analysis (Fig. 2). Of samples testing positive for E. dispar, 13 carried E. dispar only, and 28 carried mixed infections with either E. histolytica or E. moshkovskii. Only three samples (4.3%) tested positive for the 553 bp band for E. moshkovskii (Fig. 3) as determined by microscopy, which indicated a 0.3% prevalence in Erbil City; all were mixed infections. The negative PCR results for E. histolytica (13 samples) represented 18.6% of the positive results for E. dispar as a single infection. However, the mixed infection rate for E. dispar with E. histolytica was 40% of the positive samples as determined by microscopy.
Fig. 1.
Agarose gel electrophoresis analysis for nested PCR products, using primers specific for E. histolytica, positive samples reveal 439 bp bands. Samples 1 to 55, 64,and 65 showed 439 bands; samples 56 to 63 and 66 to 70 tested negative for E. histolytica. Some of these negatives showed 900 bp bands, which are the product of the first PCR and an indicator for the presence of other Entamoeba species. C represented positive control, N represented negative control and M the 100 bp DNA marker.
Table II.
Frequency and percentages of positive and negative results for E. histolytica, E. dispar and E. moshkovskii as single and mixed infections.
| Entamoeba species | Frequency & percentage of Positives by PCR per total (microscopic) positives | Frequency & percentage of Negatives by PCR per total (microscopic) positives | Frequency & percentage of positives per population | Frequency and percentage of single infection/positives | Frequency and percentage of mixed infection/positives | Frequency and percentage of single infection/total positives | Frequency and percentage of mixed infection/total positives |
|---|---|---|---|---|---|---|---|
| E. histolytica | 57 /70 (81.4%) | 13/70 (18.6%) | 57/950 (6%) | 29/57 (50.9%) | 28/57 (49.1%) | 29/70 (41.4%) | 28/70 (40%) |
| E. dispar | 41/70 (58.6%) | 29/70 (41.4%) | 41/950 (4.3%) | 13/41 (31.7%) | 28/41 (68.3%) | 13/70 (18.6%) | 28/70 (40%) |
| E. moshkovskii | 3/70 (4.3%) | 67/70 (95.7%) | 3/950 (0.3%) | 0/3 (0%) | 3/3 (100%) | 0/70 (0%) | 3/70 (4.3%) |
Fig. 2.
Detection of E. dispar after using species specific primers in the second round of the nested PCR; amplification products were analyzed by agarose gel electrophoresis and the stained gels were visualized under UV light. Positive samples exhibited 174 bp bands, which appeared in sample numbers (1, 2, 3, 5, 7, 8, 10, 12, 13, 14, 16, 18, 19,22, 24, 25, 28, 29, 30, 31), (35 to 42), (56 to 63), and (66 to 70). Samples (4, 6, 9, 11, 15, 17, 20, 21, 23, 26, 27, 32, 33, 34, 64, 65) and (43 to 55) tested negative for E. dispar. C represented positive control, N represented negative control and M was the 100 bp DNA marker.
Fig. 3.
Nested PCR for identification of E. moshkovskii, determined with specific primers for each species, and analyzed by agarose gel electrophoresis. Positive samples amplifying 553 bp amplicon appeared in only three samples (1, 8, and 14); the remaining samples were negative. C represented positive control, N represented negativecontrol, and M was the 100 bp DNA marker.
Overall PCR results showed that, out of 70 positive samples, 25 (35.7%) carried mixed infection with both E. histolytica and E. dispar; 3 (4.3%) samples carried mixed infections with E. histolytica, E. dispar, and E. moshkovskii; 29 (41.4%) samples carried a single infection with E. histolytica; 13 (18.6%) samples carried a single infection with E. dispar, and none carried a single infection with E. moshkovskii.
Sequencing analysis of PCR products. The BLAST sequence analysis tool (NCBI) showed that the sequence of E. histolytica amplicon under accession number MT250837 was 99.7% identical to the available E. histolytica GenBank sequence, accession number KY884295.1.1. In comparison, E. dispar under accession number MT250839 sequence was 100% identical to the E. dispar GenBank sequence, accession number KP722600.1 and the E. moshkovskii under accession number sequence MT250838 showed 100% homology to the sequence of E. moshkovskii GenBank, accession number KY823428.1.
Discussion
Determining prevalence rates for E. histolytica in endemic regions using molecular techniques is a radical solution to light microscopy’s shortcomings (Haque et al. 1998; Tanyuksel and Petri 2003). For the first time in Erbil City lying in the north of Iraq, molecular methods were used to estimate the prevalence rates of the pathogenic E. histolytica, and nonpathogenic E. dispar and E. moshkovskii in asymptomatic populations.
The results of the present study, as determined by microscopic examination, showed that 7.4% of individuals residing in Erbil province are asymptomatic carriers of at least one Entamoeba species. Several previous studies have recorded the prevalence rate of Entamoeba in Erbil City using microscopy. For example, in a study that included 500 diarrheal stool samples from infants and children, Entamoeba infections were found in 35% of samples (Al-Sorchee et al. 2013). In another study, the infection rate was 51.7%, but this study did not exclude the commensal protozoa Entamoeba coli (Hamad and Ahmed 2011). Unlike the present study, all research that has previously been done in Erbil City was based on samples from symptomatic subjects only, and this may be a reason for the differences in the rate of infections. Additionally, polymorphic leukocytes and macrophages in diarrheal stool samples could be misidentified as Entamoeba species and results in false positives.
Statistical analysis of the present study showed a significant difference (p < 0.05) in infection by Entamoeba species between males and females, revealing a higher rate of infection in females than in males. Similar results were reported in rural Malaysian communities (Ngui et al. 2012). Furthermore, significantly (p < 0.05) higher rates of infections were detected in low-income people, who often reside in poor living conditions and have a lower quality of life. These results are consistent with research reported in northeastern India (Nath et al. 2015).
Nested PCR revealed that E. histolytica infection was the most common (6%), followed by infection with E. dispar (4.3%), and E. moshkovskii, which had the lowest infection rate (0.3%) within the Erbil population. These results indicate that around 6% of individuals living in endemic regions are at risk of acquiring an asymptomatic infection caused by pathogenic amoeba. Asymptomatic carriers of E. histolytica play a significant role in spreading the parasite, and a prolonged asymptomatic infection can lead to invasive amoebiasis and amoebic liver abscesses (Fotedar et al. 2007a).
Previously, there have only been four molecular-based studies that have reported the prevalence of E. histolytica in Iraq. Only one study targeted nonpathogenic E. dispar and E. moshkovskii. The studies that detected E. histolytica by molecular methods were conducted in Diwanyha (south-central), Baghdad (central), and Al-Najaf (southwest of Iraq) provinces. Reported prevalence rates were 44.3%, 7%, and 24%, respectively, among symptomatic patients (Al-Hameedawi 2014; Hussein et al. 2015; Al-Khalidi 2016). The high rates of infections reported in Diwanyha and Al-Najaf cities, which share internal boundaries, could be due to the small sample sizes of their respective studies, the differences in the study design (they studied the symptomatic population whereas the present work studied the asymptomatic population), the differences in environmental conditions and hygienic practices in these regions, and the higher population density in Al Najaf city (whose shrine receives thousands of visitors). Amoebiasis is regarded as one of the primary food and water-borne diseases; the high rates of infections could be attributed to poor nutrition and sanitation and contaminated water supply (Jackson 2000). It has been documented that about 0.5 million tons of sewage a day are dumped into Iraqi rivers, resulting in water supply contamination. This especially concerns southern cities that use the rivers as their primary water sources (Korzeniewski 2006).
The prevalence rate of the pathogenic E. histolytica is higher than the non-pathogenic E. dispar and E. moshkovskii in the present study. Similar results were reported in asymptomatic individuals in Yemen, Mexico, and Japan (Tachibana et al. 2000; Ramos et al. 2005; Al-Areeqi et al. 2017); the latter two studies did not estimate the rate of E. moshkovskii infection. Similarly, the prevalence rate of E. histolytica was higher than the infection rate with nonpathogenic species in symptomatic subjects in the United Arab Emirates, Malaysia, and northeast India. Additionally, the studies conducted in populations living in south-west Iran, Cairo, Gaza Strip, and Barcelona did not determine the rate of E. moshkovskii infection (Al-Hindi et al. 2005; Pestehchian et al. 2011; Ngui et al. 2012; Rodulfo et al. 2012; Anuar et al. 2013; Elbakri et al. 2013; Nath et al. 2015; Roshdy et al. 2017).
The only study which discriminated among the three species of Entamoeba in Iraq was conducted by D’asheesh (2016) in Diwanyha city, south-central Iraq; the study involved symptomatic diarrheal patients. D’asheesh’s results differed from the present study by reporting the higher prevalence rates of E. dispar than E. histolytica. Similar results were recorded in the central and north-west regions and the Kurdistan province of Iran; Izmir, Turkey; Australia; and north--west Ethiopia (Dagci et al. 2007; Fotedar et al. 2007b; Mojarad et al. 2010; Fallah et al. 2014; Yimer et al. 2017; Bahrami et al. 2019).
The present study reported the lowest rate of infections by E. moshkovskii; similar results were documented in western Iran, northeast India, Malaysia, Diwanyha, and south-central Iraq (Ngui et al. 2012; Nath et al. 2015; D’asheesh 2016; Bahrami et al. 2019).
In conclusion, the current study finds that 7.4% of individuals who live in Erbil City, where amoeba infections are endemic, carry intestinal Entamoeba species, asymptomatically. The incidence rate of E. histolytica was higher than the incidence rate of E. dispar or E. moshkovskii among asymptomatic carriers. In the present study, E. histolytica and E. dispar were reported as single or mixed infections; only three cases of E. moshkovskii were documented as mixed infections with both E. histolytica and E. dispar.
Acknowledgments
We gratefully thank Dr. Banaz Omer Kareem (Leicester University, UK), Dr. Zana Hameed (Liverpool University, UK) and Dr. Hareem (Sulaimani University, Iraq) for providing professional, technical, and general advice throughout this study. We would like to extend our gratitude to Dr. Bahrami (Kurdistan Biomedical Science University, Iran) for providing us with control positive DNAs.
Footnotes
Conflict of interest
The authors do not report any financial or personal connections with other persons or organizations, which might negatively affect the contents of this publication and/or claim authorship rights to this publication.
ORCID
Shler Akram Faqe Mahmood https://orcid.org/0000-0001-5635-1422
Ethical approval
The study was conducted in accordance with the Declaration of Helsinki – Ethical Principles for Medical Research, revised in 2008, and was approved by the Ethics Committee of Hawler Medical University.
Literature
- Al-Areeqi MA, Sady H, Al-Mekhlafi HM, Anuar TS, Al-Adhroey AH, Atroosh WM, Dawaki S, Elyana FN, Nasr NA, Ithoi I, et al. . First molecular epidemiology of Entamoeba histolytica, E. dispar and E. moshkovskii infections in Yemen: different species-specific associated risk factors. Trop Med Int Health. 2017. April;22(4):493–504. 10.1111/tmi.12848 [DOI] [PubMed] [Google Scholar]
- Al-Hameedawi JJY. Molecular identification of Entamoeba histolytica parasite by using actin and amebapore-a genes Kufa. J Nurs Sci. 2014; 4(2):1–8. [Google Scholar]
- Al-Hindi A, Shubair ME, Marshall I, Ashford RW, Sharif FA, Abed AA, Kamel EG. Entamoeba histolytica or Entamoeba dispar among children in Gaza, Gaza Strip? J Egypt Soc Parasitol. 2005. April;35(1):59–68. [PubMed] [Google Scholar]
- Ali IKM, Hossain MB, Roy S, Ayeh-Kumi PF, Petri WA Jr, Haque R, Clark CG. Entamoeba moshkovskii infections in children, Bangladesh. Emerg Infect Dis. 2003. May;9(5):580–584. 10.3201/eid0905.020548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khalidi KaH. Detection of Entamoeba histolytica in patients an infected infants with diarrhea in born and children’s hospital by classic methods and real-time polymerase chain reaction. J Al Qadisiyah Pure Sci. 2016;21(2):27–35. [Google Scholar]
- Al-Sorchee SMA, Rabat AA, Juma IM. Microbial causatives of diarrhea in children in Erbil city. Journal of Al-Nahrain University Science. 2013a. September 1;16(3):19–29. 10.22401/JNUS.16.3.03 [DOI] [Google Scholar]
- Al-Sorchee SMA, Rabat AA, Juma IM. Microbial causatives of diarrhea in children in Erbil city. Journal of Al-Nahrain University Science. 2013b. September 1;16(3):19–29. 10.22401/JNUS.16.3.03 [DOI] [Google Scholar]
- Anuar TS, Al-Mekhlafi HM, Abdul Ghani MK, Azreen SN, Salleh FM, Ghazali N, Bernadus M, Moktar N. Different clinical outcomes of Entamoeba histolytica in Malaysia: does genetic diversity exist? Korean J Parasitol. 2013. April 25;51(2):231–236. 10.3347/kjp.2013.51.2.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrami F, Haghighi A, Zamini G, Khademerfan M. Differential detection of Entamoeba histolytica, Entamoeba dispar and Entamoeba moshkovskii in faecal samples using nested multiplex PCR in west of Iran. Epidemiol Infect. 2019;147 e96:e96 10.1017/S0950268819000141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark CG, Diamond LS. The Laredo strain and other ‘Entamoeba histolytica-like’ amoebae are Entamoeba moshkovskii. Mol Biochem Parasitol. 1991. May;46(1):11–18. 10.1016/0166-6851(91)90194-B [DOI] [PubMed] [Google Scholar]
- D’asheesh TIA. Molecular identification of some species of Entamoeba isolated from patients with diarrhea in Afak City/Al-qadisiyah governorate using real-time PCR technique. Int J Recent Sci Res. 2016;7(5):11207–11211. [Google Scholar]
- Dagci H, Erdogan DD, Toz SO, Kurt O, Ustun S, Akarca U. Differentiation of Entamoeba histolytica and Entamoeba dispar by PCR: a preliminary study in Izmir, Turkey. New Microbiol. 2007. January;30(1):45–48. [PubMed] [Google Scholar]
- Diamond LS, Clark CG. A redescription of Entamoeba histolytica Schaudinn, 1903 (Emended Walker, 1911) separating it from Entamoeba dispar Brumpt, 1925. J Eukaryot Microbiol. 1993. May;40(3): 340–344. 10.1111/j.1550-7408.1993.tb04926.x [DOI] [PubMed] [Google Scholar]
- El Bakri A, Samie A, Ezzedine S, Odeh R. Differential detection of Entamoeba histolytica, Entamoeba dispar and Entamoeba moshkovskii in fecal samples by nested PCR in the United Arab Emirates (UAE). Acta Parasitol. 2013. January 1;58(2):185–190. 10.2478/s11686-013-0128-8 [DOI] [PubMed] [Google Scholar]
- Fallah E, Shahbazi A, Yazdanjoii M, Rahimi-Esboei B. Differential detection of Entamoeba histolytica from Entamoeba dispar by parasitological and nested multiplex polymerase chain reaction methods J Anal Res. Clin Med (Lond). 2014;2(1):25–29. [Google Scholar]
- Fotedar R, Stark D, Beebe N, Marriott D, Ellis J, Harkness J. Laboratory diagnostic techniques for Entamoeba species. Clin Microbiol Rev. 2007a. July;20(3):511–532. 10.1128/CMR.00004-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotedar R, Stark D, Beebe N, Marriott D, Ellis J, Harkness J. PCR detection of Entamoeba histolytica, Entamoeba dispar, and Entamoeba moshkovskii in stool samples from Sydney, Australia. J Clin Microbiol. 2007b. March 01;45(3):1035–1037. 10.1128/JCM.02144-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamad NR, Ahmed RK. Intestinal parasites among patients attending general central public health laboratory in Erbil City-Iraq during 1998–2004, J Edu Sci. 2011a;24(4):79–86. [Google Scholar]
- Haque R, Ali IKM, Akther S, Petri WA Jr. Comparison of PCR, isoenzyme analysis, and antigen detection for diagnosis of Entamoeba histolytica infection. J Clin Microbiol. 1998;36(2):449–452. 10.1128/JCM.36.2.449-452.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque R, Huston CD, Hughes M, Houpt E, Petri WA Jr. Amebiasis. N Engl J Med. 2003. April 17;348(16):1565–1573. 10.1056/NEJMra022710 [DOI] [PubMed] [Google Scholar]
- Hussein RA, Al-Mayah QS, Merdaw MA-z, Al-Bashier NT, Al-Abbas AA, Jasem IA. Evaluation of multiplex real-time PCR and ELISA in detection of intestinal protozoan parasites from children with diarrheal disease. Int J Adv Res (Indore). 2015;3(9):782–788. [Google Scholar]
- Jackson TFHG. Amebiasis. London (United Kingdom): Imperial College Press; 2000. p. 47–63. [Google Scholar]
- Khairnar K, Parija SC. A novel nested multiplex polymerase chain reaction (PCR) assay for differential detection of Entamoeba histolytica, E. moshkovskii and E. dispar DNA in stool samples. BMC Microbiol. 2007;7(1):47–56. 10.1186/1471-2180-7-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzeniewski K. The epidemiological situation in Iraq. Przegl Epidemiol. 2006;60(4):845–855. [PubMed] [Google Scholar]
- Mojarad EN, Zahra Nochi NS, Nejad MR, Dabiri H, Haghighi A. Characterization of Entamoeba histolytica and Entamoeba dispar in fresh stool by PCR. Gastroenterol Hepatol Bed Bench. 2010; 3(1):37–41. [Google Scholar]
- Nath J, Ghosh SK, Singha B, Paul J. Molecular epidemiology of amoebiasis: A cross-sectional study among north east Indian population. PLoS Negl Trop Dis. 2015. December 3;9(12):e0004225 10.1371/journal.pntd.0004225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngui R, Angal L, Fakhrurrazi S, Lian YL, Ling L, Ibrahim J, Mahmud R. Differentiating Entamoeba histolytica, Entamoeba dispar and Entamoeba moshkovskii using nested polymerase chain reaction (PCR) in rural communities in Malaysia. Parasit Vectors. 2012;5(1):187–193. 10.1186/1756-3305-5-187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J, Srivastava S, Bhattacharya S. Molecular methods for diagnosis of Entamoeba histolytica in a clinical setting: an overview. Exp Parasitol. 2007. May;116(1):35–43. 10.1016/j.exppara.2006.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestehchian N, Nazary M, Haghighi A, Salehi M, Yosefi H. Frequency of Entamoeba histolytica and Entamoeba dispar prevalence among patients with gastrointestinal complaints in Chelgerd city, southwest of Iran. J Res Med Sci. 2011. November;16(11):1436–1440. [PMC free article] [PubMed] [Google Scholar]
- Pillai DR, Keystone JS, Sheppard DC, MacLean JD, MacPherson DW, Kain KC. Entamoeba histolytica and Entamoeba dispar: epidemiology and comparison of diagnostic methods in a setting of nonendemicity. Clin Infect Dis. 1999. November 01;29(5):1315–1318. 10.1086/313433 [DOI] [PubMed] [Google Scholar]
- Ramos F, Ramiro M, Gómez A, Melendro E, García G, González E, Ximénez C, De León MDCG, Valadez A, Morán P. High prevalence rate of Entamoeba histolytica asymptomatic infection in a rural Mexican community. Am J Trop Med Hyg. 2005. July 01; 73(1):87–91. 10.4269/ajtmh.2005.73.87 [DOI] [PubMed] [Google Scholar]
- Rodulfo H, Ahmar B, Rodríguez ME, Mora L, De Donato M. Nested PCR reveals elevated over-diagnosis of E. histolytica in Barcelona, Venezuela. Invest Clin. 2012. December;53(4):365–377. [PubMed] [Google Scholar]
- Roshdy MH, Abd El-Kader NM, Ali-Tammam M, Fuentes I, Mohamed MM, El-Sheikh NA, Rubio JM. Molecular diagnosis of Entamoeba spp. versus microscopy in the Great Cairo. Acta Parasitol. 2017. January 1;62(1):188–191. 10.1515/ap-2017-0022 [DOI] [PubMed] [Google Scholar]
- Santos HLC, Bandea R, Martins LAF, de Macedo HW, Peralta RHS, Peralta JM, Ndubuisi MI, da Silva AJ. Differential identification of Entamoeba spp. based on the analysis of 18S rRNA. Parasitol Res. 2010. March;106(4):883–888. 10.1007/s00436-010-1728-y [DOI] [PubMed] [Google Scholar]
- Saqur I, Al-Warid H, Albahadely H. The prevalence of Giardia lamblia and Entamoeba histolytica/dispar among Iraqi provinces. Karbala Int J Mod Sci. 2017;3:93–96. [Google Scholar]
- Singh A, Houpt E, Petri WA. Rapid diagnosis of intestinal parasitic protozoa, with a focus on Entamoeba histolytica. Interdiscip Perspect Infect Dis. 2009;2009(547090):547090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana H, Kobayashi S, Nagakura K, Kaneda Y, Takeuchi T. Asymptomatic cyst passers of Entamoeba histolytica but not Entamoeba dispar in institutions for the mentally retarded in Japan. Parasitol Int. 2000. March;49(1):31–35. 10.1016/S1383-5769(99)00032-X [DOI] [PubMed] [Google Scholar]
- Tanyuksel M, Petri WA Jr. Laboratory diagnosis of amebiasis. Clin Microbiol Rev. 2003. October;16(4):713–729. 10.1128/CMR.16.4.713-729.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh JA. Problems in recognition and diagnosis of amebiasis: estimation of the global magnitude of morbidity and mortality. Clin Infect Dis. 1986. March 01;8(2):228–238. 10.1093/clinids/8.2.228 [DOI] [PubMed] [Google Scholar]
- WHO Amoebiasis. Weekly Epidemiological Record. Geneva (Switzerland): World Health Organization; 1997. April 4;72(14): 97–100.9100475 [Google Scholar]
- Yimer M, Zenebe Y, Mulu W, Abera B, Saugar JM. Molecular prevalence of Entamoeba histolytica/dispar infection among patients attending four health centres in north-west Ethiopia. Trop Doct. 2017. January;47(1):11–15. 10.1177/0049475515627236 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.