Abstract
Modification of host hormone biology is a common strategy used by plant pathogens to promote disease. For example, the bacterial pathogen strain Pseudomonas syringae DC3000 (PtoDC3000) produces the plant hormone auxin (indole-3-acetic acid [IAA]) to promote PtoDC3000 growth in plant tissue. Previous studies suggest that auxin may promote PtoDC3000 pathogenesis through multiple mechanisms, including both suppression of salicylic acid (SA)-mediated host defenses and via an unknown mechanism that appears to be independent of SA. To test if host auxin signaling is important during pathogenesis, we took advantage of Arabidopsis thaliana lines impaired in either auxin signaling or perception. We found that disruption of auxin signaling in plants expressing an inducible dominant axr2–1 mutation resulted in decreased bacterial growth and that this phenotype was suppressed by introducing the sid2–2 mutation, which impairs SA synthesis. Thus, host auxin signaling is required for normal susceptibility to PtoDC3000 and is involved in suppressing SA-mediated defenses. Unexpectedly, tir1 afb1 afb4 afb5 quadruple-mutant plants lacking four of the six known auxin coreceptors that exhibit decreased auxin perception, supported increased levels of bacterial growth. This mutant exhibited elevated IAA levels and reduced SA-mediated defenses, providing additional evidence that auxin promotes disease by suppressing host defense. We also investigated the hypothesis that IAA promotes PtoDC3000 virulence through a direct effect on the pathogen and found that IAA modulates expression of virulence genes, both in culture and in planta. Thus, in addition to suppressing host defenses, IAA acts as a microbial signaling molecule that regulates bacterial virulence gene expression.
Keywords: auxin signaling, host defense, indole-3-acetic acid, pathogenesis, Pseudomonas syringae, virulence gene expression
Pseudomonas syringae is a gram-negative bacterium and a causal agent of leaf spot, leaf blight, leaf speck, and bacterial canker disease of tomato, Arabidopsis, and many cultivated crops and ornamental plant species all over the world (Agrios 1997). P. syringae is an extracellular hemibiotrophic pathogen that colonizes the surface of host plants as an epiphyte and, later, the intercellular space (apoplast) of the infected plant as a pathogen. Once in the apoplast, P. syringae suppresses basal defense responses by using the type III protein secretion system (T3SS), that delivers effector proteins directly into host cells. These effector proteins suppress host defenses and presumably alter other aspects of host physiology to elicit the release of nutrients and water from plant cells (Büttner 2016; Xin and He 2013). P. syringae then takes up nutrients and water, multiplies to high levels, and causes development of disease. During P. syringae pathogenesis, the levels of several plant hormones, including the auxin indole-3-acetic acid (IAA), increase in infected host tissue (Chen et al. 2007; O’Donnell et al. 2003; Schmelz et al. 2004; Spaepen and Vanderleyden 2011).
Although auxin has long been known to be an important virulence factor for gall-forming pathogens and root-associated bacteria (Spaepen and Vanderleyden 2011), it has more recently been discovered to be important during infection by leaf spotting pathogens such as the Pseudomonas syringae DC3000 (PtoDC3000) and Pseudomonas syringae pv. maculicola ES4326 (PmaES4326) strains (Chen et al. 2007; Mutka et al. 2013; Navarro et al. 2006; Wang et al. 2007). For example, treatment with exogenous auxin suppresses salicylic acid (SA)-mediated defenses in Arabidopsis thaliana, and application of auxin at the time of inoculation increases disease symptom severity (Chen et al. 2007; Navarro et al. 2006; Wang et al. 2007). Further, we observed that plants infected with the PtoDC3000 aldA mutant, which is impaired for IAA synthesis (McClerklin et al. 2018), supported reduced growth of the pathogen and that this was correlated with elevated expression of the defense gene PR1 (PATHOGENESIS RELATED PROTEIN 1). The reduced growth of the aldA mutant was restored to normal levels in sid2–1 mutant plants, which have impaired SA biosynthesis, suggesting that IAA promotes pathogen virulence by suppressing SA-mediated defenses (McClerklin et al. 2018). However, in a separate study, we found that elevated auxin levels in transgenic plants overexpressing the YUCCA1 auxin biosynthesis gene do not promote susceptibility simply by suppressing SA-mediated defenses (Mutka et al. 2013). This suggested that auxin also promotes susceptibility to PtoDC3000 by acting independently of SA.
An additional mechanism by which auxin may promote disease susceptibility is by altering host auxin signaling and physiology. Generally, auxin induces transcriptional changes by promoting ubiquitin-mediated degradation of AUX/IAA transcriptional repressors. The degradation of AUX/IAAs leads to activation of auxin response factors and expression of auxin-responsive genes (Mockaitis and Estelle 2008). Previous studies showed that P. syringae promotes pathogen growth and disease development through the action of the type III secreted effector protein AvrRpt2, which interferes with plant auxin signaling by promoting the degradation of AUX/IAA proteins, thereby increasing auxin sensitivity in the host (Chen et al. 2007;Cui et al. 2013). Further, Navarro et al. (2006) reported that basal defense responses induced against P. syringae results in stabilization of the AUX/IAA proteins and downregulation of auxin signaling, suggesting that inhibition of auxin signaling may be an important aspect of plant defense. If auxin-induced changes in the host are important during pathogenesis, we predict that host auxin signaling should contribute to host susceptibility, and host auxin responsiveness may be required for suppression of SA-mediated defenses upon infection by PtoDC3000.
In addition to a role for auxin in modulating basal host defenses, previous findings suggested that IAA promotes PtoDC3000 pathogenesis through one or more mechanisms that function independently of suppression of SA-mediated defenses (Mutka et al. 2013). One possible mechanism is by directly impacting the pathogen. IAA has been shown to regulate gene expression in several plant-associated bacteria (Aragón et al. 2014; Donati et al. 2013; Kunkel and Harper 2018; Van Puyvelde et al. 2011; Yang et al. 2007), however, the biological significance of auxin as a microbial signal during plant-microbe associations has not been explored. The objective of this study is to elucidate the one or more roles that auxin plays during PtoDC3000 pathogenesis. Here, we demonstrate that host auxin signaling is required to promote susceptibility to PtoDC3000 by suppressing SA-mediated defense. Secondly, we show that IAA acts as a microbial signaling molecule that regulates expression of bacterial virulence genes, both in culture and in plant tissue.
RESULTS
Plants expressing the dominant axr2–1 mutation exhibit impaired auxin responses.
Modulation of plant hormone physiology is an important virulence strategy for many plant pathogens, and several have been shown to target different aspects of host auxin biology, suggesting a role for auxin signaling in pathogenesis. Consistent with this hypothesis, a previous report by Wang et al. (2007) suggested that host auxin signaling may play a role in susceptibility to P. syringae, as the A. thaliana axr2–1 mutant, which is impaired in auxin responses (Timpte et al. 1994), had slightly reduced susceptibility to P. syringae pv. maculicola (PmaES4326) when inoculated with 1 × 106 CFU of bacteria per milliliter (Wang et al. 2007). However, as axr2–1 mutant plants are developmentally abnormal and severely dwarfed, it is difficult to interpret this observation. The dominant axr2–1 allele encodes a mutant form of Axr2, an Aux/IAA protein, that is not degraded upon auxin treatment and, therefore, auxin responses are not normally induced in this mutant. To further investigate the contribution of host auxin signaling to P. syringae pathogenesis, we took advantage of an A. thaliana transgenic line expressing a form of the Axr2–1 mutant protein that is translationally fused to the glucocorticoid receptor (Aoyama and Chua 1997), which allows us to induce nuclear localization of Axr2–1 upon dexamethasone (Dex) treatment. Use of this line provides us with plants that grow fairly normally to maturity and, then, allows us to disrupt auxin signaling with Dex treatment at the time of infection.
To confirm that expression of the axr2–1 mutation in these plants disrupts auxin signaling, we treated the adult GR-axr2–1 transgenic plants with Dex and then monitored auxin-responsive gene expression. Mature, four-to five-week-old GR-axr2–1 plants and WT Col-0 plants were sprayed with 10 μM Dex (suspended in 0.1% ethanol) or 0.1% ethanol (mock) 24 h prior to auxin treatment. Before the Dex treatment, the GR-axr2–1 transgenic plants looked normal, although they were slightly smaller than WT Col-0 plants (Supplementary Fig. S1). However, 1 day after Dex treatment, the plants exhibited abnormal morphology, in which the younger and newly emerged leaves did not expand normally and exhibited a downward curled shape (Supplementary Fig. S1). Col-0 WT plants treated with Dex and mock-treated GR-axr2–1 plants did not show this response, suggesting that axr2–1 impairs auxin-mediated events, such as expansion of young leaves (Woodward and Bartel 2005).
We monitored expression of the auxin-responsive genes GH3.3 and IAA19 in these plants in response to application of 1 μM of the synthetic auxin naphthaleneacetic acid (NAA) or a control (0.01% dimethyl sulfoxide [DMSO]), using quantitative reverse transcription-PCR (RT-qPCR). Transcripts of both IAA19 and GH3.3 accumulated to high levels in WT Col-0 plants treated with NAA compared with the control (Fig. 1A and B). In contrast, in plants carrying the axr2–1 mutation pretreated either with Dex or ethanol (mock), the expression of both GH3.3 and IAA19 were not induced in response to NAA (Fig. 1A and B). The observation that the GR-axr2–1 plants exhibited impaired auxin responses even in the absence of Dex treatment suggests that some nuclear localization of the GR-Axr2–1 protein occurs even in the absence of Dex. The observation that these plants were smaller than WT prior to Dex treatment (Supplementary Fig. S1) is consistent with this hypothesis. Our results indicate that plants expressing the axr2–1 mutation exhibit impaired auxin responses and, thus, these plants can be used to investigate if host auxin signaling is involved during pathogenesis by PtoDC3000.
Fig. 1.
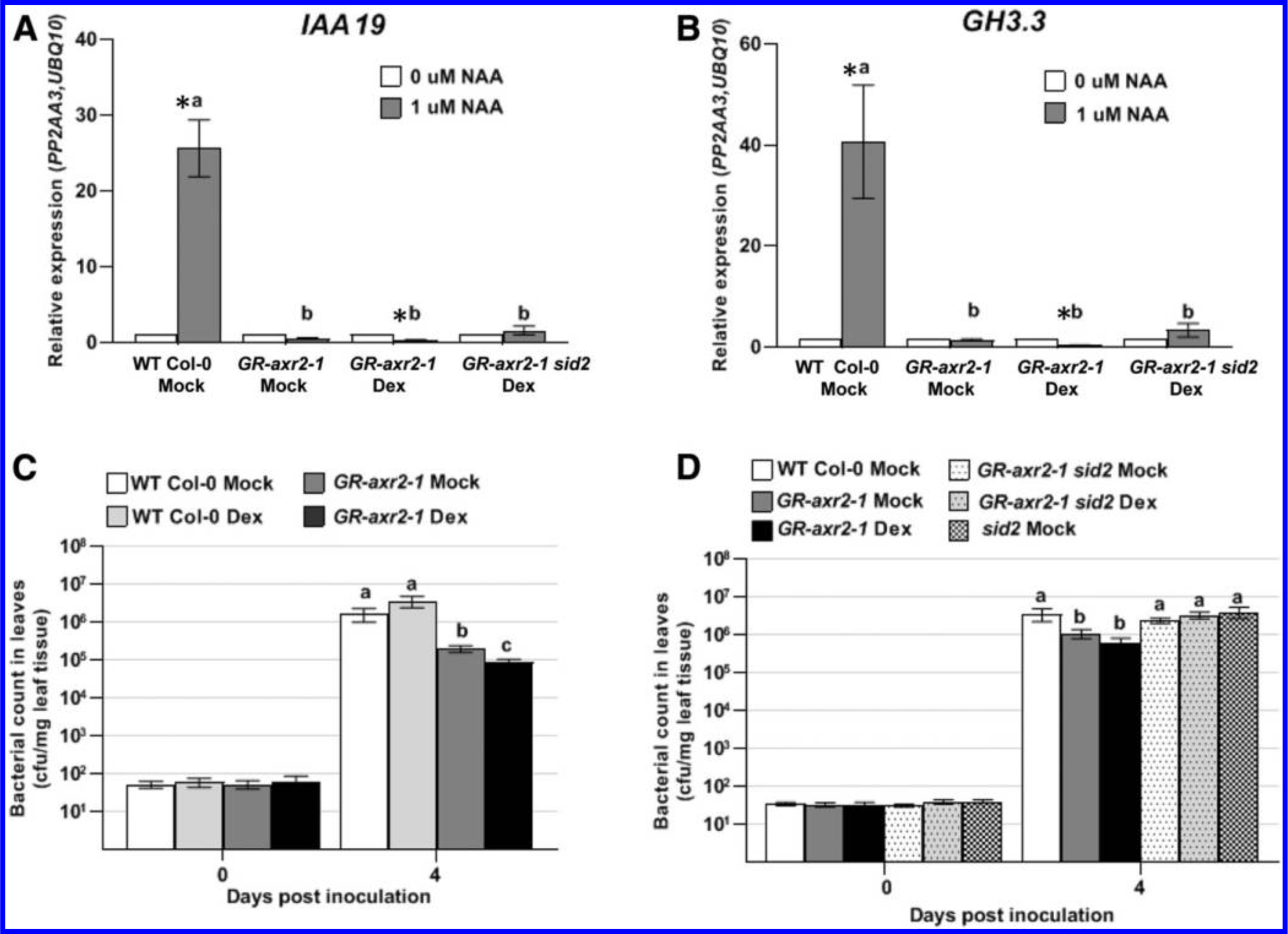
Expression of auxin-responsive genes IAA19 and GH3.3 and growth of PtoDC3000 in plants carrying the dominant axr2–1 mutation. A, Quantitative reverse transcription-PCR analysis of auxin-responsive gene expression IAA19 and B, GH3.3, 3 h after treatment with 1 μM naphthaleneacetic acid (NAA) or 0.01% dimethyl sulfoxide (0 μM NAA). Plants were pretreated with dexamethasone (Dex) or a buffer control (Mock) 24 h prior to NAA treatment. The expression data were normalized using PROTEIN PHOSPHATASE 2A SUBUNIT A3 (PP2AA3) and POLYUBIQUITIN 10 (UBQ10) as reference genes and the mock-treated sample was used as a calibrator of relative expression. Values are an average ± standard error of the mean (SEM) for data compiled from three independent experiments for Col-0 wild-type (WT) Mock (n = 9) and for two independent experiments for GR-axr2–1 and GR-axr2–1 sid2 (n = 6). Error bars are too small to see for several datapoints. C and D, Quantification of growth of PtoDC3000 in WT and GR-axr2–1 transgenic plants that were pretreated with Dex or a buffer control (Mock) 24 h prior to inoculation. In C, values are an average ± SEM for data from four independent experiments, carried out on different days, combined to generate composite growth curves, resulting in a total of 12 to 16 biological replicates for day 0 and 20 to 24 replicates for day 4. In D, values are an average ± SEM for data from three independent experiments, carried out on different days, combined to generate the composite growth curves, resulting in a total of 12 biological replicates for day 0 and 20 replicates for day 4. Statistical significance between plant genotypes determined by one-way analysis of variation, followed by a Tukey’s post hoc test. Samples indicated by different lower case letters are significantly different (P < 0.05). An asterisk (*) indicates significant difference between treatments (0 μM NAA vs. 1 μM NAA) with P < 0.05.
Plants expressing the axr2–1 mutation exhibit reduced susceptibility to P. syringae strain PtoDC3000.
To study the contribution of host auxin signaling to PtoDC3000 pathogenesis, we assayed the GR-axr2–1 transgenic plants for altered susceptibility to PtoDC3000. PtoDC3000 grew to high levels in WT Col-0 plants, regardless of whether they were treated with Dex (Fig. 1C). In contrast, plants carrying the axr2–1 mutation reproducibly supported significantly lower levels of PtoDC3000 growth compared with the WT Col-0 (Fig. 1C). We also observed that growth of PtoDC3000 in the GR-axr2–1 plants treated with Dex was slightly lower than in mock-treated GR-axr2–1 plants. Thus, although some GR-Axr2–1 protein appears to enter the nucleus in the absence of Dex, nuclear localization of additional Axr2–1 protein resulted in a further reduction in susceptibility to PtoDC3000. These results indicate that host auxin signaling is important for normal susceptibility to PtoDC3000.
Normal disease susceptibility in GR-axr2–1 plants is restored by introducing the sid2 mutation.
To test if the reduced susceptibility phenotype in GR-axr2–1 plants is dependent on SA-mediated defenses, we crossed the salicylic acid induction deficient 2 (sid2–2) mutation, which carries a deletion in ISOCHORISMATE SYNTHASE 1 (ICS1) and thus abolishes pathogen-induced SA accumulation (Wildermuth et al. 2001), into the GR-axr2–1 transgenic line. Introduction of sid2–2 did not substantially alter the reduced auxin-responsiveness in plants expressing the axr2–1 mutation (Fig. 1A and B). However, during infection, the GR-axr2–1 sid2–2 plants supported similar levels of PtoDC3000 as WT Col-0 and sid2–2 plants (Fig. 1D). Thus, introduction of the sid2–2 mutation restores normal levels of susceptibility to PtoDC3000 in GR-axr2–1 plants, suggesting that the reduced susceptibility in these plants with impaired auxin-signaling is due to elevated SA-mediated defenses. This is consistent with previous observations suggesting that SA and auxin signaling are mutually antagonistic in P. syringae/Arabidopsis interactions (McClerklin et al. 2018; Navarro et al. 2006; Wang et al. 2007).
TIR1/AFB auxin coreceptor mutants retain susceptibility to PtoDC3000 infection.
Given our observation that host auxin signaling contributes to disease susceptibility, we were interested in determining if any specific auxin coreceptors play a role in PtoDC3000 infection. The genome of A. thaliana encodes six auxin coreceptors (TIR1 and AFB1 through AFB5) that mediate diverse responses to the plant hormone auxin (Dharmasiri et al. 2005; Prigge et al. 2016). We took advantage of existing auxin coreceptor mutants to investigate the contributions of various combinations of the six known TIR1/AFB family proteins during PtoDC3000 pathogenesis. We tested two higher order mutants that lack four TIR1/AFB proteins but are still able to develop into mature plants: the tir1–1 afb1–3 afb2–3 afb3–4 (tir1 afb1 afb2 afb3) quadruple mutant (Parry et al. 2009) and the tir1–1 afb1–3 afb4–8 afb5–5 (tir1 afb1 afb4 afb5) quadruple mutant (Prigge et al. 2020). Although tir1 afb1 afb2 afb3 mutant plants are severely impaired developmentally as seedlings and are dwarfed as adult plants, they supported normal levels of PtoDC3000 growth, similar to that observed in WT Col-0 (Supplementary Fig. S2A). We also observed that tir1 afb1 afb4 afb5 mutants did not exhibit reduced disease susceptibility to PtoDC3000 (Fig. 2A). Unexpectedly, the tir1 afb1 afb4 afb5 mutant actually supported significantly higher levels of pathogen growth compared with the WT Col-0 plants (Fig. 2A; Supplementary Fig. S2B). These results indicate that inactivation of four of the six auxin coreceptors does not compromise susceptibility to PtoDC3000.
Fig. 2.
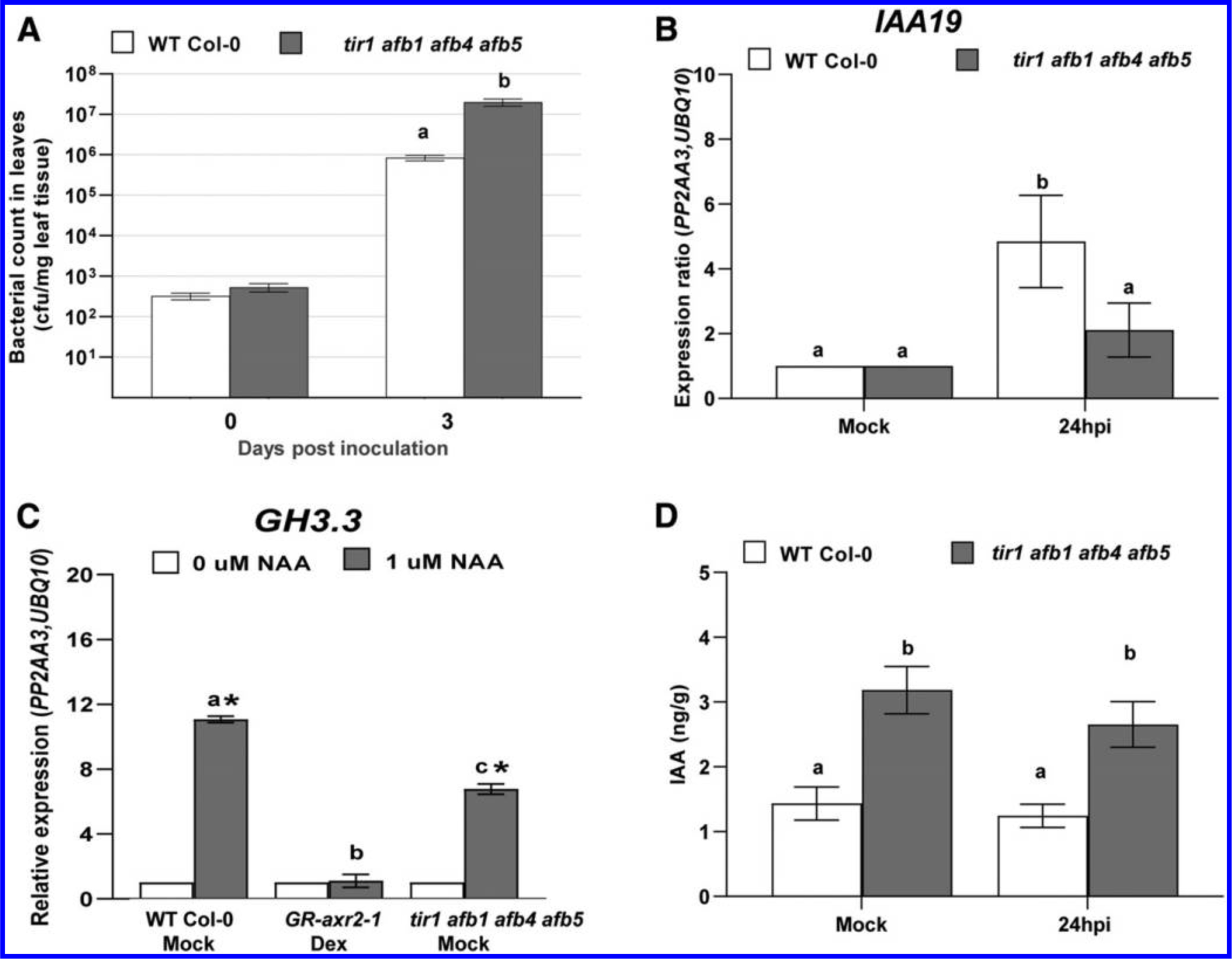
Growth of PtoDC3000, indole-3-acetic acid (IAA)-responsive gene expression, and quantification of IAA levels in auxin receptor mutants. A, Growth of PtoDC3000 in the tir1 afb1 afb4 afb5 mutant. Values are an average ± standard error of the mean (SEM) for data from four independent experiments, carried out on different days, combined to generate composite growth curves, resulting in a total of four to six biological replicates for day 0 and eight replicates for day 3. Statistical significance between plant genotypes was analyzed using the Student’s t test. B, Expression of the auxin-responsive gene IAA19 in wild type (WT) and tir1 afb1 afb4 afb5 mutant plants at 24 h after PtoDC3000 inoculation. Values are an average ± SEM for data compiled from two independent experiments with six biological replicates in total. C, Auxin-responsive GH3.3 gene expression in WT, GR-axr2–1, and tir1 afb1 afb4 afb5 mutant plants 3 h after treatment with 1 μM naphthaleneacetic acid (NAA) or 0.01% dimethyl sulfoxide (0 μM NAA). Plants were pretreated with dexamethasone (Dex) or a buffer control (Mock) 24 h prior to NAA treatment. Values are an average ± SEM for three biological replicates for Col-0 WT Mock, GR-axr2–1, and tir1 afb1 afb4 afb5. An asterisk (*) indicates significant difference between treatment (0 and 1 μM NAA) with P < 0.05. Similar results were observed for tir1 afb1 afb4 afb5 in a second independent experiment. For B and C, the expression data were normalized using PROTEIN PHOSPHATASE 2A SUBUNIT A3 (PP2AA3) and POLYUBIQUITIN 10 (UBQ10) and the mock-treated sample was used as a calibrator of relative expression. D, Free IAA levels in WT Col-0 and tir1 afb1 afb4 afb5 mutant plants, 24 h after Mock treatment (10 mM MgCl2) or inoculation with PtoDC3000. Values are an average ± SEM for data compiled from two independent experiments with six biological replicates in total. Results were analyzed using one-way analysis of variation, followed by a Tukey’s post hoc test. Similar results were obtained in a third independent experiment. Samples indicated by different lower case letters are significantly different (P < 0.05).
We originally expected that these TIR1/AFB coreceptor mutants would exhibit decreased susceptibility to PtoDC3000, because we hypothesized that they would have impaired auxin perception and signaling, like the GR-axr2–1 line. However, since the TIR1/AFB coreceptor mutants did not have decreased susceptibility to PtoDC3000, we hypothesized that the remaining two AFB family members (AFB4 and AFB5 in the tir1 afb1 afb2 afb3 mutant and AFB2 and AFB3 in the tir1 afb1 afb4 afb5 mutant) are sufficient to mediate normal auxin perception and signaling during infection. Therefore, to test if the tir1 afb1 afb4 afb5 mutant plants exhibit altered auxin perception during PtoDC3000 infection, we infected WT Col-0 and tir1 afb1 afb4 afb5 plants with PtoDC3000 and collected total RNA 24 h after inoculation, to monitor expression of the auxin-responsive gene IAA19. In WT Col-0 infected plants, we found that, as expected, the expression of IAA19 was induced by 24 h after infection, consistent with previous data that PtoDC3000 infection results in elevated IAA levels (Chen et al. 2007; O’Donnell et al. 2003; Schmelz et al. 2004). In contrast, expression of IAA19 was not significantly induced in tir1 afb1 afb4 afb5 plants at 24 h after infection (Fig. 2B), suggesting that this quadruple mutant exhibits reduced auxin perception, signaling, or both during PtoDC3000 infection.
However, since the tir1 afb1 afb4 afb5 mutant still harbors intact AFB2 and AFB3 auxin coreceptors, we predicted that the tir1 afb1 afb4 afb5 mutant plants may not be as severely compromised in auxin signaling as the GR-axr2–1 plants. To directly compare the auxin responsiveness of these two lines, we treated them with NAA and monitored expression of GH3.3 and IAA19. As observed previously, GH3.3 and IAA19 were not induced by NAA in GR-axr2–1 plants (Fig. 2C; Supplementary Fig. S2C). However, both auxin-responsive genes were induced by NAA treatment in the tir1 afb1 afb4 afb5 mutant, although the level of induction was lower than observed in WT. Thus, auxin signaling in the tir1 afb1 afb4 afb5 mutant is only partially impaired.
The tir1 afb1 afb4 afb5 mutant accumulates elevated levels of IAA.
It is not immediately obvious why the tir1 afb1 afb4 afb5 mutant, which has decreased auxin-responsive gene expression, exhibits enhanced susceptibility to PtoDC3000. We hypothesized that the tir1 afb1 afb4 afb5 plants might accumulate elevated levels of IAA, due to disruption of the feedback mechanism that normally maintains IAA homeostasis in WT plants (Takato et al. 2017) and that this elevated IAA promotes susceptibility. To investigate whether the enhanced pathogen growth in tir1 afb1 afb4 afb5 plants is correlated with elevated IAA in host tissue, we quantified free IAA levels in WT Col-0 and tir1 afb1 afb4 afb5, in both PtoDC3000-infected and mock-treated plants at 24 h postinoculation. We found that tir1 afb1 afb4 afb5 mutant plants accumulated significantly higher levels of IAA than WT Col-0 plants, regardless of treatment (Fig. 2D). Similarly, elevated IAA levels have been observed in several other auxin signaling mutants (Takato et al. 2017). Thus, the elevated levels of IAA in the tir1 afb1 afb4 afb5 mutant may promote pathogen growth despite a partial impairment in auxin responsiveness.
Elevated IAA in tir1 afb1 afb4 afb5 plants is associated with reduced SA-mediated defenses.
Observations from several previous experiments suggest that stimulation of host auxin signaling promotes virulence by suppressing SA-mediated defenses (McClerklin et al. 2018; Wang et al. 2007). Since tir1 afb1 afb4 afb5 plants are still capable of responding to NAA, we hypothesized that the elevated levels of IAA in tir1 afb1 afb4 afb5 plants may promote pathogen growth by suppressing SA-mediated defenses through host auxin signaling via AFB2 and AFB3. Thus, we predicted that induction of SA-mediated defenses would be reduced in these plants compared with in WT plants. We tested this by monitoring both the accumulation of SA and the expression of the SA-responsive defense gene PR1 in infected plants. Although we observed an increase in SA levels in both WT Col-0 and tir1 afb1 afb4 afb5 plants after inoculation with PtoDC3000, the level of SA that accumulated in the mutant was significantly lower than in WT Col-0 (Fig. 3A). Furthermore, PR1 expression was strongly and reproducibly induced by 24 h postinoculation in WT Col-0 plants but was not significantly induced over levels observed in mock-treated tir1 afb1 afb4 afb5 plants (Fig. 3B). Thus, the reduction in SA levels in infected tir1 afb1 afb4 afb5 mutant plants is correlated with the lower expression of PR1 observed in these plants. These results are consistent with the hypothesis that elevated IAA levels suppress SA-mediated defenses during PtoDC3000 infection.
Fig. 3.
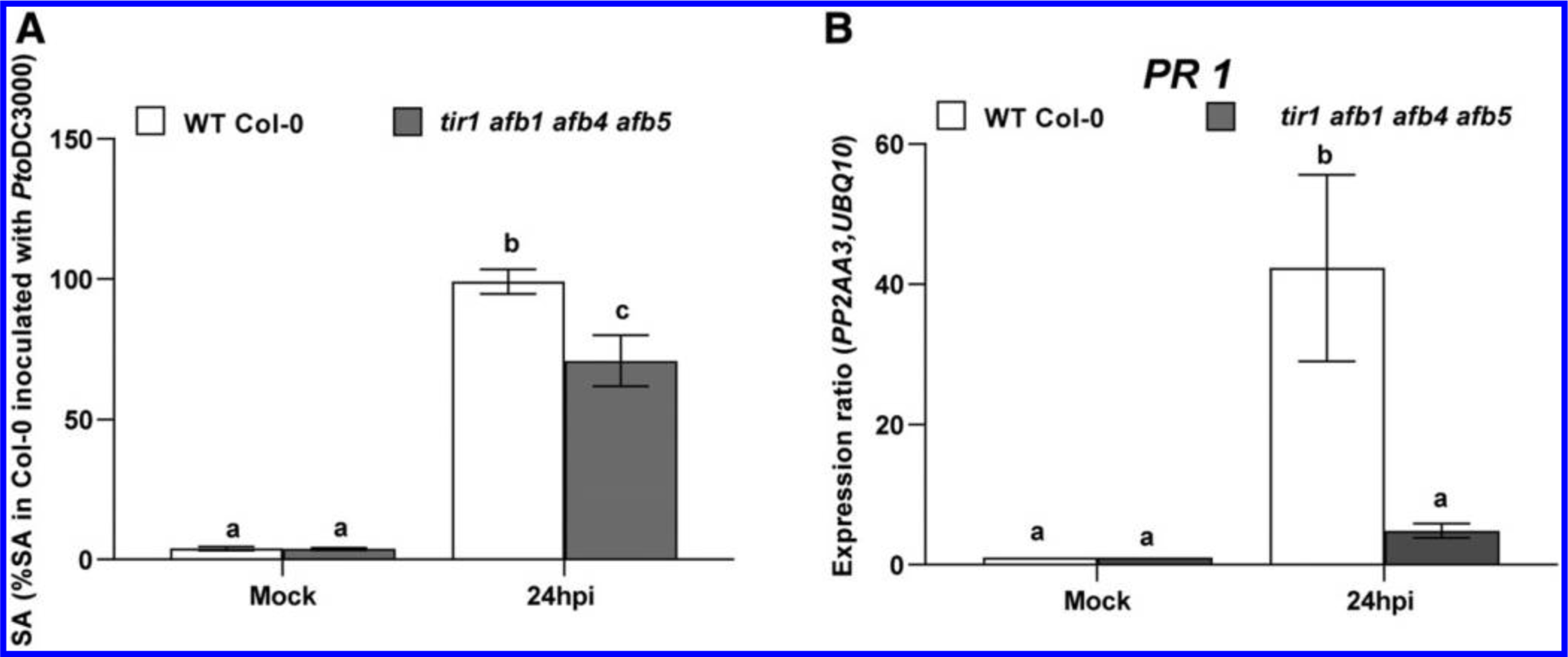
Salicylic acid (SA)-mediated defenses in wild type (WT) Col-0 and the tir1 afb1 afb4 afb5 auxin receptor mutant. A, SA levels in WT Col-0 and tir1 afb1 afb4 afb5 mutant plants, 24 h after mock treatment (10 mM MgCl2) or inoculation with PtoDC3000. Values are expressed as a percent SA accumulation in WT Col-0 24 h after inoculation with PtoDC3000, and represent an average ± standard error of the mean (SEM) for data compiled from two independent experiments, with six biological replicates per plant genotype per treatment. Results were analyzed using analysis of variation, followed by a Tukey’s post hoc test, and different letters indicate significant difference between samples with P < 0.05. B, Expression of PATHOGENESIS RELATED 1 (PR1) 24 h after mock treatment (10 mM MgCl2) or inoculation with PtoDC3000. Expression data were normalized using PP2AA3 and UBQ10 to give the relative gene expression. Values are an average ± SEM for data compiled from two independent experiments with six biological replicates in total.
However, suppression of SA-mediated defenses may not be the only mechanism by which IAA enhances disease susceptibility. Previous findings suggest that IAA can promote PtoDC3000 pathogenesis through one or more mechanisms that function independently of SA-mediated defenses (Mutka et al. 2013). Furthermore, our data show that auxin-responsive gene expression is not significantly induced in tir1 afb1 afb4 afb5 plants during infection (Fig. 2B), suggesting that this mutant does not exhibit significantly increased auxin responses despite elevated IAA levels. Therefore, we reasoned that the increased bacterial growth in the tir1 afb1 afb4 afb5 mutant plants may not be mediated solely through host auxin responses and considered the possibility that the elevated IAA may promote pathogen growth via an additional mechanism.
IAA regulates PtoDC3000 virulence gene expression in culture.
One possible mechanism by which IAA can promote pathogen growth independently of host auxin signaling is by directly impacting the pathogen, for example by modulating virulence gene expression, as has been previously shown in Agrobacterium tumefaciens (Liu and Nester 2006; Yuan et al. 2008), Dickeya didantii (formerly known as Erwinia chrysanthemi) (Yang et al. 2007), and Pseudomonas savastanoi pv. savastanoi (Aragón et al. 2014). Accordingly, we sought to determine if IAA also directly impacts PtoDC3000 virulence gene expression in culture. First, to assess if IAA has an effect on bacterial growth, we cultured PtoDC3000 in rich media (NYG [nutrient yeast glycerol medium]) for several hours before transferring cells to hrp/hrc de-repressing media (HDM), a minimal media believed to mimic growth conditions in the apoplastic space of the leaf (Anderson et al. 2014; Huynh et al. 1989), containing a variety of concentrations of IAA (Supplementary Fig. S3A). We observed that transfer of cells to HDM containing a concentration of 100 μM IAA or greater impaired the growth of the cells, as compared with growth in HDM lacking IAA (Supplementary Fig. S3A) (Mutka 2013). Thus, IAA does impact PtoDC3000 biology. We decided to analyze gene expression in cells treated with 100 μM IAA, as this concentration only minimally inhibited the growth of the cells in culture. At this concentration, in HDM at pH 5.7, the concentration of the undissociated, lipophilic form of IAA, which can presumably easily diffuse into the cell (Raven 1975), is approximately 10 μM.
Central to the pathogenesis of PtoDC3000 is the deployment of a T3SS (Alfano and Collmer 1996; Büttner and He 2009). Expression of genes involved in T3S (e.g., avrPto and hrpL, which encode a T3S effector and the RNA polymerase sigma factor responsible for transcribing T3S-related genes, respectively) can be induced in culture by growing cells in HDM (Anderson et al. 2014; Huynh et al. 1989). To test the effect of IAA on expression of virulence genes, we transferred PtoDC3000 from NYG to HDM containing 0 or 100 μM IAA, grew the cells for 90 min (Supplementary Fig. S3B), and monitored the expression of both known and putative virulence genes. The housekeeping genes gyrB and rpoD were used as an internal reference, and gene expression levels were normalized to the basal expression levels in NYG. As expected, we observed low levels of expression of the T3S genes hrpL and avrPto in NYG, and strong induction upon transfer to HDM (Fig. 4A and B). However, when the cultures were treated with IAA, the expression of hrpL and avrPto was significantly reduced (Fig. 4A and B). In contrast, the presence of IAA did not alter the expression of cmaA, a gene required for synthesis of the virulence factor coronatine (Fig. 4C), and stimulated expression of the virulence-associated gene tvrR (Preiter et al. 2005) (Fig. 4D) as well as genes predicted to be involved in type VI secretion (T6S), hcp1 and PSPTO_5415, which likely encodes a Vgr protein (Fig. 4E, F). We chose to examine these last two genes, as T6S systems have been proposed to play a role during bacteria-host interactions or in bacteria-bacteria interactions in the microbial community. Further, expression of T6S-related genes has been previously shown to be regulated by IAA in P. savastanoi (Aragón et al. 2014). Our results indicate that exposure to IAA causes transcriptional changes in PtoDC3000, leading to both up- and downregulation of distinct subsets of genes.
Fig. 4.
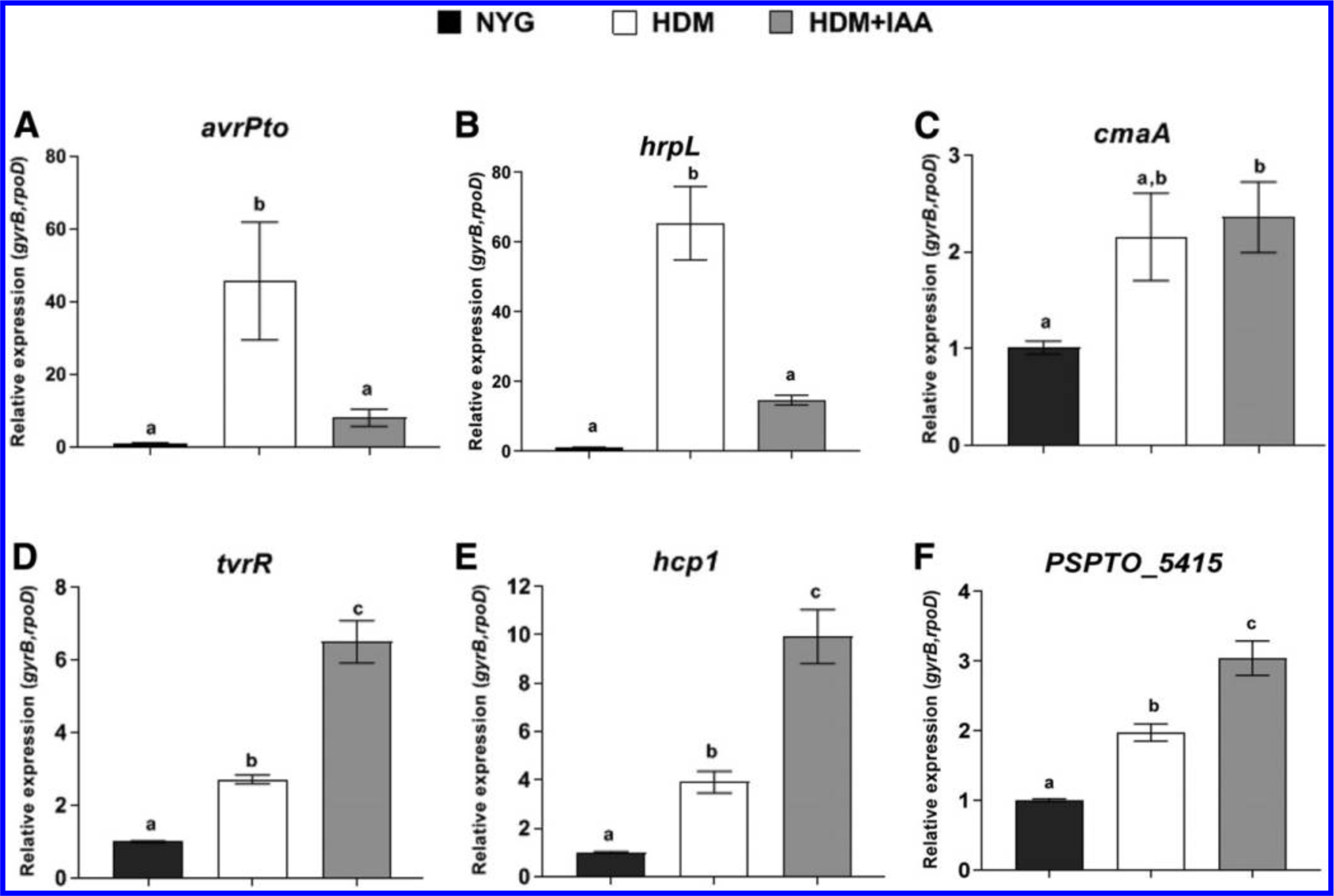
The effect of indole-3-acetic acid (IAA) treatment on PtoDC3000 virulence-related gene expression in culture. A, Expression of virulence-related genes avrPto, B, hrpL, C, cmaA, D, tvrR, E, hcp1, and F, PSPTO_5415 1.5 h after being transferred from nutrient yeast gycerol (NYG) to hrp/hrc de-repressing media (HDM) or HDM containing 100 μM IAA (HDM+IAA). Expression levels in cells growing in NYG was used as the calibrator of relative expression. The relative expression was calculated using the reference genes rpoD and gyrB. Data from two independent experiments, carried out on different days, were combined, with each datapoint representing the average of six biological replicates and error bars representing the standard error of the mean between biological replicates. Results were analyzed using analysis of variation, followed by a Tukey’s post hoc test. Different letters indicate significant difference between samples with P < 0.05. This experiment was repeated a third time with similar results.
Auxin modulates bacterial virulence-related gene expression in planta.
Our results indicate that IAA induces transcriptional changes in PtoDC3000 in culture. However, studies performed on bacteria grown in culture may not fully represent bacteria growing in host tissue during infection. Therefore, we next sought to investigate if IAA also impacts PtoDC3000 gene expression when growing in plant tissue. To accomplish this, we took advantage of the tir1 afb1 afb4 afb5 mutant described above, as it accumulates elevated IAA (Fig. 2D). WT Col-0 and tir1 afb1 afb4 afb5 mutant plants were inoculated with PtoDC3000 and leaves were collected at 24 and 48 h postinoculation (Supplementary Fig. S2B). Total RNA, including both PtoDC3000 and plant RNA, was purified from these leaves and was used to monitor expression of the bacterial virulence-associated genes assayed in culture. RNA from the inoculum used to infect these plants was used as a calibrator of relative expression.
We assessed the stability of expression of several housekeeping genes in order to choose those suitable for use as reference genes for normalization of gene expression (Smith et al. 2018) and found that recA and rpoD were expressed at very similar levels at 24 and 48 h after inoculation. Similar to what has been previously observed, the expression of avrPto and hrpL was strongly induced within 24 h after inoculation, compared with the expression level in the inoculum (Fig. 5A and B) (McAtee et al. 2018; Nobori et al. 2018; Ortiz-Martín et al. 2010), and the expression of both genes declined by 48 hpi. However, the expression of avrPto and hrpL was significantly lower in tir1 afb1 afb4 afb5 mutant plants compared with WT Col-0 plants, a finding that is consistent with our observation that IAA suppresses the induction of these genes in culture (Fig. 4A and B). Thus, elevated IAA also appears to suppress expression of T3S genes during growth in plant tissue by 24 h after inoculation. We observed that expression of cmaA also was induced in planta, but, consistent with our findings in liquid media, its expression was not significantly different in WT and tir1 afb1 afb4 afb5 plants (Fig. 5C). Furthermore, the expression of tvrR was induced in planta and was expressed at higher levels in tir1 afb1 afb4 afb5 mutant plants, with elevated expression most pronounced at 48 h after inoculation (Fig. 5D). Interestingly, although we observed a low level of expression of hcp1 and PSPTO_5415 in planta, we did not observe further upregulation of these genes in the tir1 afb1 afb4 afb5 plants (Fig. 5E and F), suggesting that, for some genes, the effects of IAA on expression in culture does not reflect what happens in planta. Overall, our results demonstrate that IAA regulates the expression of virulence-related genes in PtoDC3000, both in culture and during growth in planta.
Fig. 5.
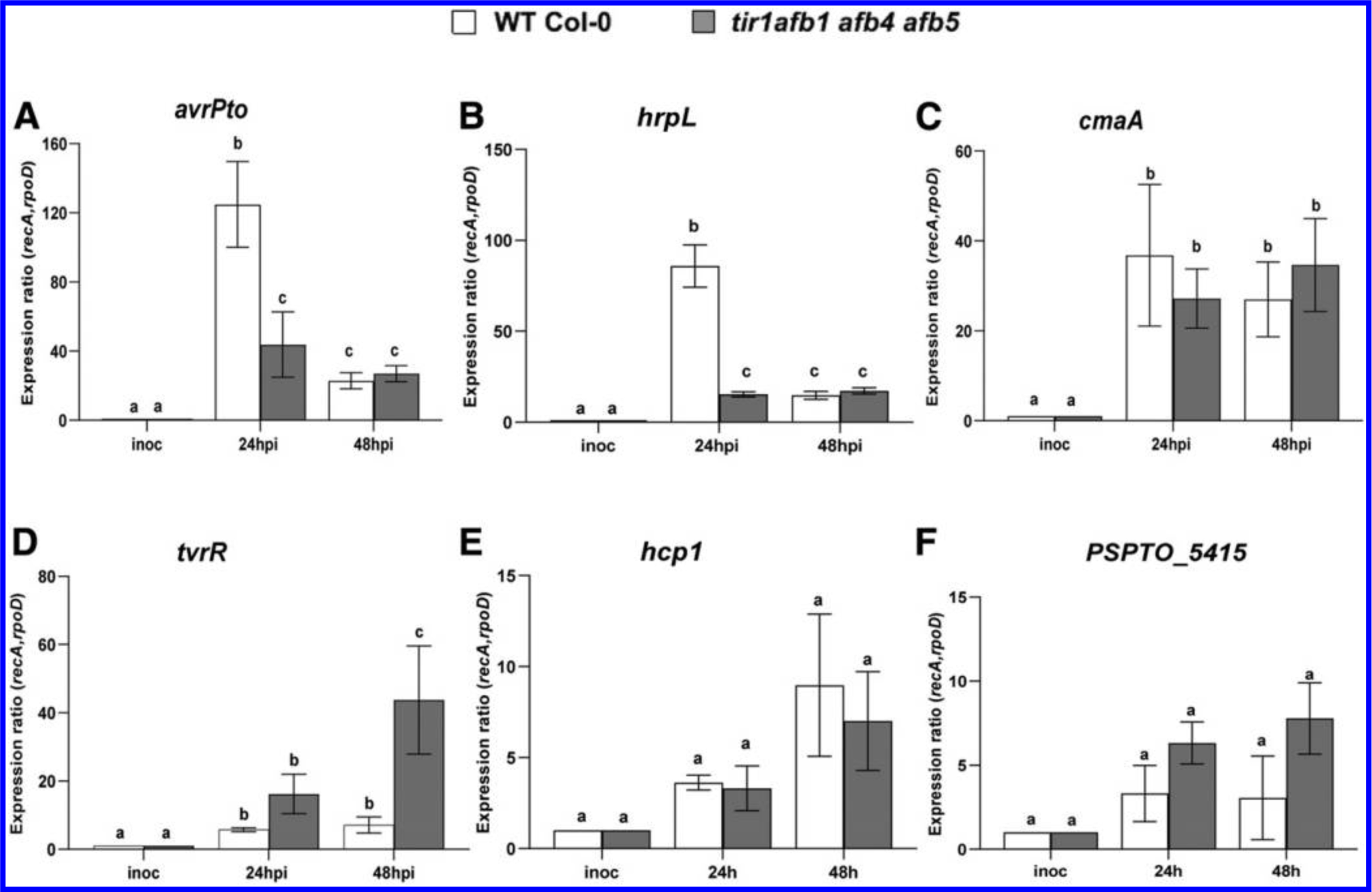
The effect of elevated indole-3-acetic acid levels on PtoDC3000 virulence-related gene expression in planta. A, Expression of virulence-related genes avrPto, B, hrpL, C, cmaA, D, tvrR, E, hcp1, and F, PSPTO_5415 in PtoDC3000 growing in wild type (WT) Col-0 and tir1 afb1 afb4 afb5 Arabidopsis thaliana plants. Infected leaves were harvested 24 and 48 h after inoculation, and total RNA was isolated and used for quantitative reverse transcription-PCR, to quantify bacterial gene expression. RNA prepared from the PtoDC3000 cell suspension used for the inoculum was used as the calibrator for relative expression. The data shown were compiled from two independent experiments carried out on different days and one experiment for PSPTO_5415 (similar results were observed in two independent experiments). The relative expression was calculated using reference genes recA and rpoD. Each datapoint is the average of six biological replicates, and error bars represent the standard error of the mean between biological replicates. Results were analyzed using analysis of variation, followed by a Tukey’s post hoc test. Different letters indicate significant difference between samples with P < 0.05.
DISCUSSION
Our investigation of the roles of auxin during pathogenesis of A. thaliana by P. syringae reveals that auxin promotes virulence of PtoDC3000 through two different mechanisms: i) activating host auxin signaling to suppress SA-mediated plant defenses and ii) directly impacting the pathogen by modulating virulence gene expression.
Host auxin signaling is required for normal susceptibility to PtoDC3000.
There is growing evidence that auxin promotes disease development in A. thaliana, and it is hypothesized this is mediated primarily through regulatory crosstalk between auxin and SA-mediated signaling pathways in the host (Denancé et al. 2013; Kazan and Manners 2009; McClerklin et al. 2018; Robert-Seilaniantz et al. 2011). Our finding that an A. thaliana transgenic line carrying the dominant axr2–1 mutation (GR-axr2–1) that impairs auxin responses exhibits reduced susceptibility to PtoDC3000 is consistent with earlier observations by Wang et al. (2007) and confirms that host auxin signaling is required for normal susceptibility to PtoDC3000. To further test the hypothesis that auxin signaling promotes disease by suppressing SA-mediated defenses, we introduced the sid2–2 mutation and quantified susceptibility to PtoDC3000 in these plants. Our finding that WT levels of disease susceptibility were restored in GR-axr2–1 sid2–2 plants (Fig. 1D) indicates that reduced susceptibility in plants with impaired auxin-signaling is due, at least in part, to elevated SA-mediated defenses. This supports the hypothesis that host auxin signaling suppresses SA-mediated defenses in P. syringae–Arabidopsis interactions and demonstrates that this is mediated through the canonical TIR1/AUX/IAA host auxin signaling pathway (Fig. 6).
Fig. 6.
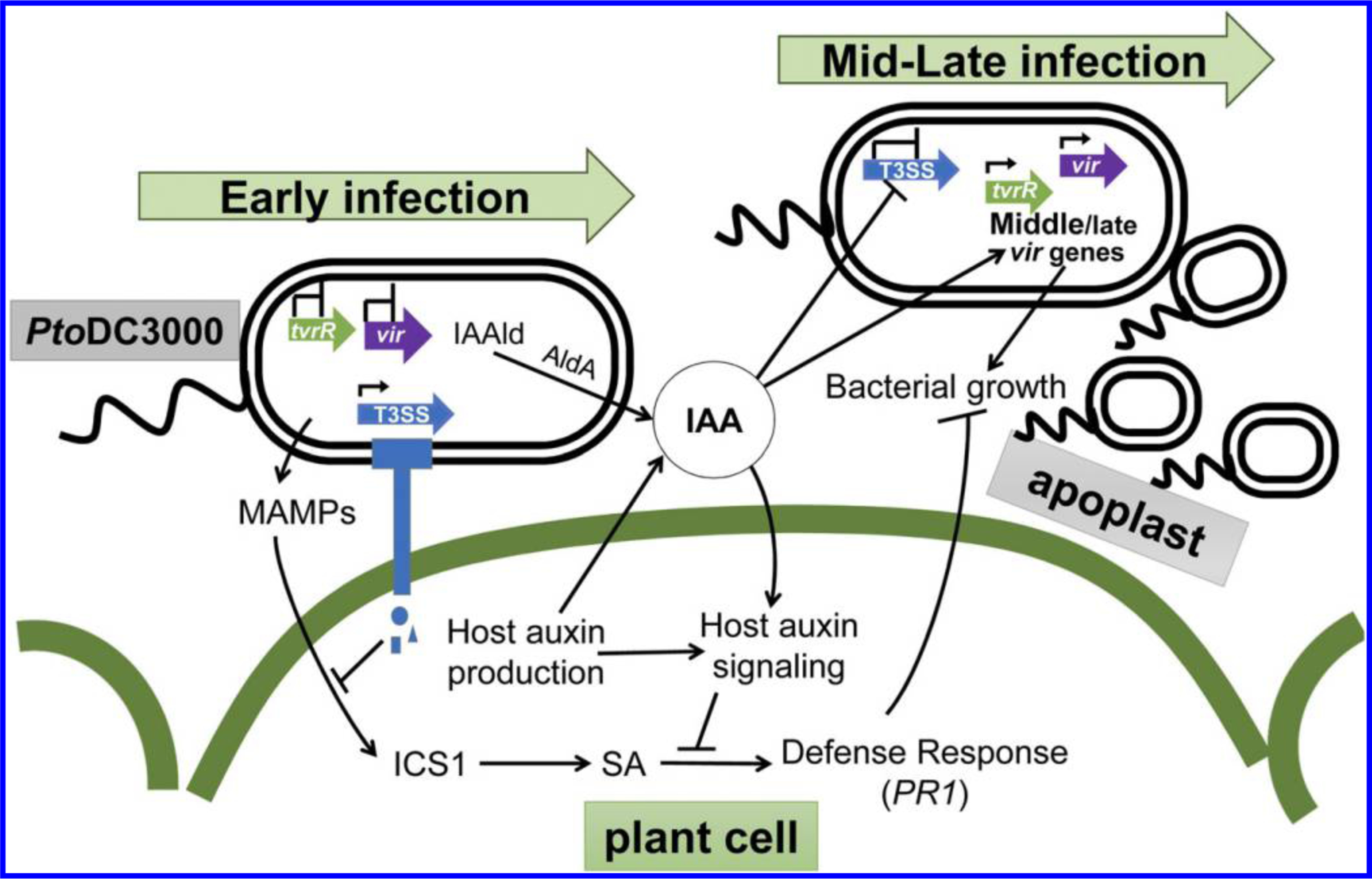
Indole-3-acetic acid (IAA) plays multiple roles during PtoDC3000 pathogenesis. A working model illustrates how IAA promotes PtoDC3000 pathogenesis via multiple mechanisms. Upon PtoDC3000 infection, detection of microbe associated molecular patterns (MAMPs) induces expression of basal host defense responses mediated by salicylic acid (SA). Early during infection expression of the T3S system (T3SS) allows delivery of effector proteins (blue shapes) into the host cell to suppress MAMP-induced defenses. PtoDC3000 infection results in elevated auxin (e.g., IAA) levels in infected tissue, possibly due to auxin synthesis by both the host and PtoDC3000 (via activity of the Indoleacetaldehyde dehydrogenase AldA (McClerklin et al. 2018). Disruption of host auxin signaling, for example by the dominant axr2–1 mutation, prevents suppression of SA defenses and results in reduced disease susceptibility. IAA also promotes the growth of PtoDC3000 independently of suppression of SA-mediated defenses, by regulating expression of pathogen virulence genes (green and purple block arrows). We hypothesize that IAA downregulates T3SS genes after they are no longer needed (e.g., 24 h postinfection), and activates virulence genes, such as tvrR, that are required at intermediate or late stages of infection. Genes that are transcribed are indicated by the small black arrowhead above the block arrows.
The tir1 afb1 afb4 afb5 auxin coreceptor mutant exhibits elevated IAA levels and reduced SA-mediated defenses.
The finding that host auxin signaling contributes to disease susceptibility raises the question of whether any specific auxin coreceptors are required for normal PtoDC3000 infection. We examined this by testing two higher order (quadruple) tir1/afb mutants for altered susceptibility and observed that both mutants supported at least WT levels of pathogen growth (Fig. 2; Supplementary Fig S2). Although we have not tested all possible combinations of auxin coreceptor quadruple mutants, our observations suggest that the combined activity of two coreceptors (AFB2 and AFB3 in tir1 afb1 afb4 afb5 or AFB4 and AFB5 in tir1 afb1 afb2 afb3) provides sufficient auxin signaling to support normal levels of pathogen growth. In keeping with this, we demonstrated that the tir1 afb1 afb4 afb5 mutant is only partially impaired in its ability to respond to the synthetic auxin NAA, likely due to the presence of the intact AFB2 and AFB3 auxin coreceptors (Fig. 2C; Supplementary Fig S2C).
Given that host auxin signaling is required for normal pathogenesis, we were surprised to find that the tir1 afb1 afb4 afb5 mutant exhibited enhanced susceptibility to PtoDC3000 (Fig. 2A; Supplementary Fig S2B). The fact that these plants accumulate elevated levels of IAA compared with WT Col-0 (Fig. 2D) provides a reasonable explanation for this result, as we have previously shown that elevated levels of endogenous auxin promotes growth of PtoDC3000 (Mutka 2013). Further, our observation that tir1 afb1 afb4 afb5 plants exhibit elevated IAA levels is consistent with recent findings that homeostasis of endogenous IAA is maintained by feedback regulation through the canonical auxin signaling pathway (Takato et al. 2017). Consistent with the role for IAA in suppressing SA signaling, we demonstrated that the tir1 afb1 afb4 afb5 mutant plants exhibited significantly reduced expression of PR-1 and reduced SA levels compared with WT plants at 24 h postinoculation (Fig. 3). These results suggest that during PtoDC3000 infection the elevated levels of IAA in the tir1 afb1 afb4 afb5 plants leads to suppression of SA-mediated defenses. However, it is not clear whether this is mediated primarily at the level of SA synthesis, accumulation, or downstream signaling responsiveness.
IAA has a direct effect on PtoDC3000 by modulating bacterial virulence-related gene expression.
Our observation that elevated IAA and increased disease susceptibility in the tir1 afb1 afb4 afb5 auxin coreceptor mutant is correlated with suppression of SA-mediated defenses conflicts with results from an earlier study, in which we demonstrated that elevated endogenous IAA levels in YUCCA1-overexpressing plants promoted growth of PtoDC3000 via a mechanism independent of SA suppression (Mutka et al. 2013). This discrepancy raised the possibility that auxin also promotes pathogenesis by having a direct effect on the pathogen, for example, by modulating virulence-related gene expression. We investigated this by testing the effect of IAA on PtoDC3000 gene expression in culture. We observed a significant reduction of hrpL and avrPto transcript levels when the cultures were treated with IAA (Fig. 4). Initially, we found this result surprising, as we expected that, if IAA impacted virulence gene expression, it would be to promote expression of these genes, which encode virulence factors that are important during early stages of tissue colonization (Büttner 2016; Ferreira et al. 2006). However, a similar negative effect of IAA on expression of T3SS-related genes has recently been reported in the gall-forming pathogen P. savastanoi (Aragón et al. 2014). We also observed that IAA did not alter the expression of cmaA (Fig. 4C), a coronatine biosynthetic gene that contributes to PtoDC3000 virulence (Brooks et al. 2004), and enhanced expression of several other virulence-associated genes, including tvrR, hcp1, and PSPTO_5415 (Fig. 4D, E, and F). Thus, IAA does not seem to cause a global shift in transcription but, rather, differentially impacts specific classes of genes.
IAA has been previously shown to influence gene expression in a variety of plant-associated microbes (Duca et al. 2014; Kunkel and Harper 2018; Liao et al. 2017; Spaepen and Vanderleyden 2011), but this had been only demonstrated in culture, and thus, the biological relevance of these findings was unclear. To investigate whether IAA regulates PtoDC3000 gene expression in planta, we monitored transcript levels of several virulence-related genes in PtoDC3000 growing in plants with normal (WT Col-0) or elevated levels of IAA (tir1 afb1 afb4 afb5 mutants) at 24 and 48 h postinoculation. As expected, expression of the virulence-related genes examined was induced in both plant genotypes by 24 h postinoculation (Fig. 5) (McAtee et al. 2018; Nobori et al. 2018; Ortiz-Martín et al. 2010). However, consistent with what we observed in the presence of IAA in culture, induction of avrPto and hrpL was significantly lower in the tir1 afb1 afb4 afb5 mutant compared with that in WT Col-0 plants at 24 h (Fig. 5A and B). Transcript levels of cmaA, hcp1, and PSPTO_5415 were not significantly different between the two genotypes at either timepoint, and expression of tvrR was elevated in tir1 afb1 afb4 afb5, but only at 48 h postinoculation. Thus, the pattern of IAA-mediated repression or induction of these genes observed in culture reflects, for the most part, the effect of elevated IAA on bacterial virulence genes in planta.
Our discovery that IAA appears to directly impact PtoDC3000 virulence-related gene expression during growth in planta is significant, as it allows us to begin to place the differential regulation of these genes into the context of pathogenesis. Further, it prompts us to hypothesize that IAA acts as a signaling molecule that coordinates expression of virulence genes required during different phases of pathogenesis (Fig. 6). Early during pathogenesis, bacteria colonizing the apoplast assemble the T3SS and secrete type III effector proteins into host cells in order to suppress basal host defenses (Büttner 2016; Xin et al. 2018). Once that is accomplished, the bacteria can obtain water and nutrients and multiply to high levels in the apoplast. At this point, most of the bacterial cells are not in direct contact with plant cells (Misas-Villamil et al. 2011) and, presumably, do not need to express the T3SS-related genes. By this stage in pathogenesis, we also speculate that local IAA levels in the infected tissue have increased to a concentration high enough to downregulate the T3S-related genes and induce expression of virulence genes involved in subsequent stages of infection (middle or late virulence genes) (Fig. 6). The observation that IAA remains high in plant tissue for several days after infection is consistent with this hypothesis (O’Donnell et al. 2003; Schmelz et al. 2004). An example of a gene induced by IAA is tvrR, which encodes a transcription factor previously shown to be required for PtoDC3000 virulence on A. thaliana (Preiter et al. 2005) that we hypothesize regulates transcription of genes required at intermediate stages of infection. Recent experiments examining the expression of P. syringae pv. actinidiae during infection of kiwi plantlets reveal three distinct phases of gene expression over the course of infection (McAtee et al. 2018). Their observation that genes encoding the T3SS or effectors are strongly induced during the first 24 h after infection and their expression levels then decline by 48 hpi is consistent with what we observed in our studies and suggests that there is a regulatory mechanism coupling virulence gene expression to the stage of pathogenesis. It is important to note that the elevated level of IAA present in the tir1 afb1 afb4 afb5 plants is not likely to inhibit expression of T3S-related genes at early timepoints during infection, as PtoDC3000 grows to high levels in these plants. Thus, we hypothesize that some physiological condition in the leaf or appearance of another signal in addition to elevated IAA levels is required for the downregulation of early virulence genes. Future analysis of PtoDC3000 virulence gene expression during the various phases of colonization and growth in susceptible host tissue will provide more insight into this fascinating but as-yet-poorly understood process.
It is now well-established that auxin promotes disease development in many plant-pathogen interactions, and in several cases this has been shown to involve suppression of host defenses (Denancé et al. 2013; Fousia et al. 2018; McClerklin et al. 2018; Robert-Seilaniantz et al. 2011). There is also a growing number of reports that auxin, specifically IAA, has a direct impact on gene expression in plant-associated bacteria (Donoso et al. 2017; Greenhut et al. 2018; Kunkel and Harper 2018), and it appears that this regulation may play a variety of different roles. In some interactions, IAA may serve as a signal to the microbe that it is in the presence of a potential plant host and that virulence genes should be induced (Spaepen and Vanderleyden 2011; Yang et al. 2007). In others, IAA may serve as a signaling molecule to coordinate gene expression in the microbe, either to induce catabolic genes required for breakdown of IAA (Donoso et al. 2017; Greenhut et al. 2018; Lin et al. 2012) or as a signal that a specific stage of infection or a minimal cell density has been achieved and, thus, that it is time to activate a new set of genes (Levy et al. 2018). It is also possible that, in the context of the larger microbial community of one or both the phyllosphere and rhizosphere, IAA acts as a microbial signal to regulate interactions between different members of the community (Hassani et al. 2018; Levy et al. 2018) and may provide an explanation for the induction of the T6SS by IAA (Aragón et al. 2014; Van Puyvelde et al. 2011). Future studies to investigate the mechanisms by which microbes recognize and respond to IAA, as well as the generation of mutants that do not properly respond to IAA, will allow us and others to more fully examine the multiple roles that auxin and auxin signaling play during plant-microbe interactions.
MATERIALS AND METHODS
Plant material and growth conditions.
All Arabidopsis thaliana WT, mutant, and transgenic lines used in this study were in the Columbia (Col-0) background. The sid2–2 mutant was obtained from M. Wildermuth (Wildermuth et al. 2001). The tir1 and afb1, afb2, afb3, afb4 and afb5 auxin receptor mutants used in this study, including various higher order mutants, have been previously described (Parry et al. 2009; Prigge et al. 2016, 2020).
As the tir1 afb1 afb2 afb3 quadruple mutant displays severe developmental abnormalities, including a high percentage of rootless seedlings or seeds that do not germinate (Parry et al. 2009), we germinated the mutant on agar plates, transplanted seedlings that produced roots to soil, and allowed them to grow to maturity for pathogen inoculation. Plants were grown on soil in a growth chamber with a short-day photoperiod (8 h of light and 16 h of dark) at 21°C and 75% relative humidity, with a light intensity of approximately 130 μEinsteins s−1 m−2.
The GR-axr2–1 lines were generated as follows. The DNA fragment encoding the fusion protein GR-axr2–1 was made by using fusion PCR, an 834-bp cDNA fragment encoding a rat glucocorticoid receptor was amplified with primers Top-oGW_GR_F1 and GR_AXR2_R1, using the pINDEX3 vectors (Aoyama and Chua 1997) as a template, and axr2–1 cDNA was amplified with primers GR_AXR2_F1 and AXR2_V1_R1 from cDNA synthesized using RNA extracted from axr2–1 mutant plants as a template. Purified DNA fragments from these two PCRs were mixed at equal molar ratio and were used as templates and primers for a third PCR to amplify the fusion cDNA GR-axr2–1. The fusion cDNA was introduced first into the Invitrogen pENTR/D-Topo vector and was then subcloned into the expression vector pEarleyGate 100 (Earley et al. 2006) downstream of a cauliflower mosaic virus 35S promoter. The resulting construct (pEarleyGate 100-GR-axr2–1) was transferred into Agrobacterium sp. strain GV3101 (pMP90) using electroporation. Transgenic Arabidopsis plants were generated using a floral-dip approach (Clough and Bent 1998).
The sid2–2 GR-axr2–1 line was generated by crossing the sid2–2 mutant to the GR-axr2–1 transgenic line. F1 progeny from the cross were allowed to self-pollinate, and F2 plants homozygous for the sid2–2 and the GR-axr2–1 transgene were identified by PCR genotyping, using primers described in Supplementary Table S1. Plants homozygous for the sid2–2 allele only yielded an amplification product of 581 bp, indicative of the presence of the sid2–2 allele (Mutka et al. 2013). Homozygous sid2–2 plants that scored positively for the presence of the GR-axr2–1 construct were then allowed to self-pollinate and were assayed for segregation of the GR-axr2–1 construct in the F3 generation. F3 families that segregated 100% for the presence of GR-axr2–1 were selected for further analysis.
Bacterial strains and culture conditions.
Pseudomonas syringae strain PtoDC3000 WT was used in this study, and was grown on NYG (Daniels et al. 1988) or in a modified HDM containing 50 mM fructose and 20 μM citrate (Anderson et al. 2014) at 28°C with 100 μg of rifampicin per milliliter, plus other antibiotics as needed (kanamycin at 25 μg ml−1, tetracycline at 16 μg ml−1). When grown in liquid medium, cultures were shaken at 200 rpm.
Auxin-responsive gene expression in Arabidopsis plants.
To monitor auxin-responsiveness in mature GR-axr2–1 transgenic and WT Col-0 plants, plants were sprayed with 0.1% ethanol (mock) or 10 μM Dex (Sigma-Aldrich, St. Louis) in 0.1% ethanol 24 h prior to auxin treatment. Fully expanded leaves were infiltrated with a needle-less syringe containing 1 μM of the synthetic auxin NAA(Sigma-Aldrich) dissolved in 0.01% DMSO or 0.01% DMSO for mock treatment (0 μM). Plants were allowed to stand at room temperature and the leaves were harvested after 3 h, were flash-frozen in liquid nitrogen, and were stored at −80°C. The frozen samples were ground using a bead beater machine (Retsch, Newtown, PA, U.S.A.), followed by total RNA extraction using the RNeasy plant mini kit (Qiagen, Germantown, MD, U.S.A.) according to manufacturer instructions. Residual genomic DNA was digested during RNA purification using an on-column DNAse I treatment (Qiagen). The purified RNA was reverse-transcribed to synthesize first-strand cDNA using SuperScript III RT (Thermo Scientific, Waltham, MA, U.S.A.) or the Revertaid premium first-strand cDNA synthesis kit (Thermo Scientific). Negative control reactions lacking RT were run in parallel to verify that there was no contamination from genomic DNA. Real time qPCR was used to monitor the expression of the auxin-responsive genes GH3.3 (AT2G23170) and IAA19 (AT3G15540) (Mutka 2013; Paponov et al. 2008), using SYBR Green Jump-Start Taq ReadyMix (Sigma-Aldrich) and ig SYBR Green real time PCR 2× master mix (Intact Genomic, St. Louis) on a CFX Connect real-time PCR detection system (Bio-Rad, Hercules, CA, U.S.A.). In each experiment, gene expression analysis was performed on three biological replicates with three technical replicates for each biological replicate.
The cycling conditions were as follows: initial denaturation for 15 min at 95°C, followed by 40 cycles of 95°C for 5 s and 58°C for 30 s, with camera capture at the end of each cycle, then 72°C extension for 30 s. To confirm the specificity of all amplifications, a melt curve was generated after 40 cycles, using the following parameters: 65°C for 5 s, 95°C for 5 min, then a slow ramp (0.5°C for 5 s), with camera capture. The relative expression was determined using Pfaffl’s relative quantification method (González-Lamothe et al. 2012; Pfaffl 2001). RT-qPCR data were normalized using Protein Phosphatase 2A subunit A3 (PP2AA3, AT1G13320) (Czechowski et al. 2005) and polyubiquitin 10 (UBQ10, AT4G05320) (Czechowski et al. 2005) as reference genes. The mock-treated sample was used as a calibrator of relative expression. Auxin-responsive gene expression was carried out in three independent experiments for WT Col-0 (treated with Dex) and in two independent experiments for GR-axr2–1 plants (treated with Dex or mock-treated) and GR-axr2–1 sid2 plants treated with Dex. All primers used in this study are described in Supplementary Table S1.
Pathogen inoculation and in planta bacterial growth.
A. thaliana Col-0 (WT), mutant, and transgenic plants were inoculated at approximately 4 to 5 weeks of age. Bacterial solutions containing approximately 106 PtoDC3000 cells per milliliter in 10 mM MgCl2, prepared from freshly growing bacterial cultures were injected into leaves using a 1-ml needle-less syringe.
To quantify bacterial growth in the plant, whole leaves were sampled 2 to 3 h after inoculation (day 0) and 3 or 4 days after inoculation, were weighed to determine leaf mass, were ground in 10 mM MgCl2, and were then plated in serial dilutions on NYG with rifampicin. Four to eight leaves were sampled per treatment, depending on the experiment and timepoint. Following incubation at 28°C for 48 h, colonies were counted to determine the number of bacteria in the leaves. For experiments involving Dex treatment, plants were sprayed with 0.1% ethanol (mock) or 10 μM DEX suspended in 0.1% ethanol 24 h prior to inoculation.
Hormone quantification in plant tissue.
To quantify the levels of free IAA and SA, WT Col-0 and tir1 afb1 afb4 afb5 (4×) mutant plants were inoculated with PtoDC3000 (106 CFU ml−1) or treated with 10 mM MgCl2 (mock). At 24 h after inoculation, approximately 100 mg of leaves were sampled for each of three biological replicates per treatment, were flash-frozen in liquid nitrogen, and were stored at −80°C. Subsequently, free IAA and SA were quantified by liquid chromatography-tandem mass spectrometry (LC-MS/MS), as described in supplemental information.
Monitoring bacterial gene expression and the effect of IAA in culture.
To monitor the effect of IAA on bacterial gene expression in culture, triplicate cultures of 10 ml of NYG broth were inoculated with PtoDC3000 and were incubated for several hours at 28°C until they reached an optical density at 600 nm of around 0.07 to 0.10, as measured by a BioTek PowerWave XS2 96-well plate reader. As a control, a 1-ml sample of bacterial cells was removed from each culture (NYG), was treated with 2 ml of RNAprotect bacteria reagent (Qiagen) following manufacturer instructions, and the samples were flash-frozen in liquid nitrogen and were stored at −80°C until further use. The remaining cultures were then transferred to a modified HDM media supplemented with 20 μM citrate (Anderson et al. 2014), to induce expression of T3SS-related genes, containing either IAA or a buffer control. This was accomplished by collecting the cells from the initial NYG cultures by centrifugation at room temperature at 5,000 × g for 5 min and resuspending each cell pellet in 10 ml of fresh media (HDM), which was then split into two 5-ml aliquots that were immediately treated with 100 μM IAA (in 0.1% DMSO) or 0.1% DMSO (no IAA control). At 1.5 h after treatment, 1 ml of each culture was removed and was treated with 2 ml of RNAprotect bacteria reagent, was flash-frozen in liquid nitrogen, and was stored at −80°C. Bacterial growth was monitored in the cultures prior to and for about 12 h after the treatment.
For each PtoDC3000 sample, RNA was extracted using the RNeasy RNA isolation kit (Qiagen). Samples stored at −80°C were thawed, the cells were lysed enzymatically by treatment with 0.1 ml lysozyme (1 mg/ml in TE buffer), and RNA was extracted, following manufacturer instructions, using the RNase-free DNase I set for on-column DNase treatment (Qiagen).
For each sample, approximately 1 μg of purified RNA was used for cDNA synthesis, using SuperScript III (Thermo Scientific) and random hexamers as primers (Integrated DNA Technologies, Coralville, IA, U.S.A.). Control reactions lacking RT were included to check the samples for genomic DNA contamination. The products from the cDNA synthesis reactions were diluted into 30 μl of 10 mM Tris buffer (pH 8) and stored at −4°C. In order to verify the quality of the cDNA and to make sure that there was no genomic DNA contamination, PCR reactions were performed on all samples, using primers for 16S rRNA (Supplementary Table S1) and the following cycling conditions: 5 min at 95°C, followed by 25 cycles of 95°C for 30 s, 58°C for 30 s, and 68°C for 3 min. The amplification product of this reaction was visualized as an approximately 1.5-kb band on an agarose gel. Only cDNA samples that exhibited no DNA contamination in the control reactions lacking RT were used for RT-qPCR.
Expression of the following virulence-related genes was monitored: avrPto (PSPTO_4001), hrpL (PSPTO_1404), cmaA (PSPTO_4709), tvrR (PSPTO_3576), hcp1 (PSPTO_2539), and PSPTO_5415. To monitor bacterial gene expression, real time PCR was performed on cDNA samples using PowerUp SYBR Green master mix (Thermo Scientific). Reactions were set up in a 20-μl final volume, and real time PCR was performed on a CFX Connect Real-Time PCR detection system (Bio-Rad). The cycling conditions were as follows: 2 min at 94°C, followed by 49 cycles of 95°C for 15 s and 58°C for 30 s, with camera capture at the end of each cycle. The specificity of all amplifications was confirmed by generating a melt curve after 40 cycles, using these parameters: 65°C for 5 s, 95°C for 5 min, then a slow ramp (0.5°C for 5 s), with camera capture. Bacterial gene expression was calculated using the comparative cycle threshold method for relative quantification (Wong and Medrano 2005). Each biological replicate was tested in technical triplicates, and the quantification cycle for each biological replicate (average of three technical replicates) was normalized to the geometric mean of two internal reference genes, namely, gyrB (PSPTO_0004) and rpoD (PSPTO_0537). Bacterial gene expression from the NYG samples collected prior to transfer of the culture to HDM was used as a calibrator of relative expression. All primers used in this study are described in Supplementary Table S1.
Monitoring bacterial gene expression in planta.
Arabidopsis WT Col-0 and tir1 afb1 afb4 afb5 mutant plants were inoculated at approximately 4 to 5 weeks of age. Whole leaves were syringe-infiltrated with PtoDC3000 (106 CFU ml−1) in 10 mM MgCl2 prepared from freshly growing bacterial cultures or treated with 10 mM MgCl2 (mock treatment for inplanta gene expression). Approximately 100 mg of leaves were collected for RNA isolation at 24 and 48 h after inoculation, were frozen immediately in liquid nitrogen, and were stored at −80°C. A combination of protocols from the RNAprotect bacteria reagent kit (Qiagen) and RNeasy plant mini kit (Qiagen) was used to isolate and enrich for bacterial RNAs from the samples. The frozen leaves were ground into a fine powder using a bead beater machine (Retsch), followed by the addition of 1.0 ml of RNAprotect bacteria reagent to each sample. The standard protocol for enzymatic lysis of bacteria from the RNAprotect bacteria reagent handbook (Qiagen protocol 1 up to step 9) was performed. Following the addition of the RLT lysis buffer, the protocol from the RNeasy plant mini kit was followed. The total RNA obtained was a mix of bacterial and plant RNAs. We used the concentration of PtoDC3000-specific 16S rRNA (PSPTO_r01) as an in-sample proxy for bacterial concentration within the mixed RNA samples, as described recently by Smith et al. (2018). In this approach, the concentrations of bacterial RNA in the total RNA isolated from infected plant tissue were standardized using PtoDC3000-specific 16S ribosomal RNA primers. For comparison, 1.0 ml of the initial inoculum was collected by centrifugation and total RNA was extracted following the protocol described above for bacteria grown in culture. The extracted RNA was used for cDNA synthesis using the Revertaid premium first strand cDNA synthesis kit (Thermo Scientific) and random hexamers with the subsequent steps exactly as mentioned above for the bacterial cultures. Real time PCR was then used to monitor the expression of avrPto, hrpL, cmaA, tvrR, hcp1, and PSPTO_5415 using ig SYBR Green real time PCR 2× master mix (Intact Genomic) on a CFX Connect real-time PCR detection system (Bio-Rad). The cycling conditions were 15 min at 95°C, followed by 40 cycles of 95°C for 5 s and 58°C for 30 s, with camera capture at the end of each cycle. A melt curve was generated after 40 cycles, using the followinge parameters: 65°C for 5 s, 95°C for 5 min, then a slow ramp (0.5°C for 5 s), with camera capture. In each experiment, gene expression analysis was performed on three biological replicates with three technical replicates for each. The relative expression was determined using Pfaffl’s relative quantitation method, as described previously (Pfaffl 2001; González-Lamothe et al. 2012). RT-qPCR data were normalized to recA (PSPTO_4033) and rpoD (PSPTO_0537), used as reference genes. The stability of recA and rpoD expression in planta at 24 and 48 h after inoculation was confirmed as described by Smith et al. (2018), using a synthetic double-stranded DNA gBlock (Integrated DNA Technologies) and PtoDC3000-specific 16S rRNA primers. The bacterial gene expression in the inoculum sample was used as a calibrator of relative expression. We also monitored the expression of PR1 and IAA-19 in these samples, in leaves harvested 24 h after inoculation, following the above protocol for auxin responsive genes.
Statistical analysis.
Datasets were statistically compared with the statistical analysis software GraphPad Prism 8.0 (GraphPad software, San Diego, CA, U.S.A.), using one-way analysis of variation, followed by the Tukey’s post hoc test. The confidence level of all analyses was set at 95%, and values with P < 0.05 were considered significant.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Z. (Alex) Li, C. Holland, A. Zimmerman, S. DeCou, and S. Sanghani for technical help with experiments. We thank C. Perrot-Rechenmann and L. Strader for helpful discussion and R. Bart for comments on the manuscript. We also thank B. Evans, J. Mattingly, J. Li, and S. Alvarez at the Proteomics and Mass Spectrometry Facility of the Donald Danforth Plant Science Center (St. Louis, MO, U.S.A.) for hormone analysis. Their analytical methods are based upon work supported by the National Science Foundation.
Funding: This work was funded by the National Science Foundation (IOS-1645908) and National Institutes of Health (GM43644). The analytical methods were based upon work supported by the National Science Foundation under grant number DBI-1427621 for acquisition of the QTRAP LC-MS/MS.
Footnotes
The e-Xtra logo stands for “electronic extra” and indicates there are three supplementary figures and supplementary materials published online.
The author(s) declare no conflict of interest.
LITERATURE CITED
- Agrios GN 1997. Plant Pathology. Academic Press, San Diego. [Google Scholar]
- Alfano JR, and Collmer A. 1996. Bacterial pathogens in plants: Life up against the wall. Plant Cell 8:1683–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JC, Wan Y, Kim YM, Pasa-Tolic L, Metz TO, and Peck SC 2014. Decreased abundance of T3S system-inducing signals in Arabidopsis mkp1 enhances resistance against Pseudomonas syringae. Proc. Natl. Acad. Sci. U.S.A. 111:6846–6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T, and Chua NH 1997. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11:605–612. [DOI] [PubMed] [Google Scholar]
- Aragón IM, Pérez-Martínez I, Moreno-Pérez A, Cerezo M, and Ramos C. 2014. New insights into the role of indole-3-acetic acid in the virulence of Pseudomonas savastanoi pv. savastanoi. FEMS Microbiol. Lett. 356:184–192. [DOI] [PubMed] [Google Scholar]
- Brooks DM, Hernández-Guzmán G, Kloek AP, Alarcón-Chaidez F, Sreedharan A, Rangaswamy V, Peñaloza-Vázquez A, Bender CL, and Kunkel BN 2004. Identification and characterization of a well-defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato DC3000. Mol. Plant-Microbe Interact. 17:162–174. [DOI] [PubMed] [Google Scholar]
- Büttner D. 2016. Behind the lines-actions of bacterial type III effector proteins in plant cells. FEMS Microbiol. Rev. 40:894–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner D, and He SY 2009. Type III protein secretion in plant pathogenic bacteria. Plant Physiol. 150:1656–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Agnew JL, Cohen JD, He P, Shan L, Sheen J, and Kunkel BN 2007. Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc. Natl. Acad. Sci. U.S.A. 104:20131–20136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, and Bent AF 1998. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16:735–743. [DOI] [PubMed] [Google Scholar]
- Cui F, Wu S, Sun W, Coaker G, Kunkel B, He P, and Shan L. 2013. The Pseudomonas syringae type III effector AvrRpt2 promotes pathogen virulence via stimulating Arabidopsis auxin/indole acetic acid protein turnover. Plant Physiol. 162:1018–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, and Scheible WR 2005. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels MJ, Dow JM, and Osbourn AE 1988. Molecular genetics of pathogenicity in phytopathogenic bacteria. Phytopathology 26:285–312. [Google Scholar]
- Denancé N, SánchezVallet A, Goffner D, and Molina A. 2013. Disease resistance or growth: The role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 4:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, and Estelle M. 2005. The F-box protein TIR1 is an auxin receptor. Nature 435:441–445. [DOI] [PubMed] [Google Scholar]
- Donati AJ, Lee HI, Leveau JH, and Chang WS 2013. Effects of indole-3-acetic acid on the transcriptional activities and stress tolerance of Bradyrhizobium japonicum. PLoS One 8:e76559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoso R, Leiva-Novoa P, Zúñiga A, Timmermann T, Recabarren-Gajardo G, and Gonzalez B. 2017. Biochemical and genetic bases of indole-3-acetic acid (Auxin Phytohormone) degradation by the plant-growth-promoting Rhizobacterium paraburkholderia phytofirmans PsJN. Appl. Environ. Microbiol. 83:e01991–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duca D, Lorv J, Patten CL, Rose D, and Glick BR 2014. Indole-3-acetic acid in plant-microbe interactions. Antonie van Leeuwenhoek 106:85–125. [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, and Pikaard CS 2006. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45:616–629. [DOI] [PubMed] [Google Scholar]
- Ferreira AO, Myers CR, Gordon JS, Martin GB, Vencato M, Collmer A, Wehling MD, Alfano JR, Moreno-Hagelsieb G, Lamboy WF, DeClerck G, Schneider DJ, and Cartinhour SW 2006. Whole-genome expression profiling defines the HrpL regulon of Pseudomonas syringae pv. tomato DC3000, allows de novo reconstruction of the Hrp cis clement, and identifies novel coregulated genes. Mol. Plant-Microbe Interact. 19:1167–1179. [DOI] [PubMed] [Google Scholar]
- Fousia S, Tsafouros A, Roussos PA, and Tjamos SE 2018. Increased resistance to Verticillium dahliae in Arabidopsis plants defective in auxin signalling. Plant Pathol. 67:1749–1757. [Google Scholar]
- González-Lamothe R, El Oirdi M, Brisson N, and Bouarab K. 2012. The conjugated auxin indole-3-acetic acid-aspartic acid promotes plant disease development. Plant Cell 24:762–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhut IV, Slezak BL, and Leveau JHJ 2018. iac gene expression in the indole-3-acetic acid-degrading soil bacterium Enterobacter soli LF7. Appl. Environ. Microbiol. 84:e01057–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassani MA, Durán P, and Hacquard S. 2018. Microbial interactions within the plant holobiont. Microbiome 6:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh TV, Dahlbeck D, and Staskawicz BJ 1989. Bacterial blight of soybean: Regulation of a pathogen gene determining host cultivar specificity. Science 245:1374–1377. [DOI] [PubMed] [Google Scholar]
- Kazan K, and Manners JM 2009. Linking development to defense: Auxin in plant-pathogen interactions. Trends Plant Sci. 14:373–382. [DOI] [PubMed] [Google Scholar]
- Kunkel BN, and Harper CP 2018. The roles of auxin during interactions between bacterial plant pathogens and their hosts. J. Exp. Bot. 69:245–254. [DOI] [PubMed] [Google Scholar]
- Levy A, Conway JM, Dangl JL, and Woyke T. 2018. Elucidating bacterial gene functions in the plant microbiome. Cell Host Microbe 24: 475–485. [DOI] [PubMed] [Google Scholar]
- Liao X, Lovett B, Fang W, and St Leger RJ 2017. Metarhizium robertsii produces indole-3-acetic acid, which promotes root growth in Arabidopsis and enhances virulence to insects. Microbiology 163: 980–991. [DOI] [PubMed] [Google Scholar]
- Lin GH, Chen HP, Huang JH, Liu TT, Lin TK, Wang SJ, Tseng CH, and Shu HY 2012. Identification and characterization of an indigo-producing oxygenase involved in indole 3-acetic acid utilization by Acinetobacter baumannii. Antonie van Leeuwenhoek 101: 881–890. [DOI] [PubMed] [Google Scholar]
- Liu P, and Nester EW 2006. Indoleacetic acid, a product of transferred DNA, inhibits vir gene expression and growth of Agrobacterium tumefaciens C58. Proc. Natl. Acad. Sci. U.S.A. 103:4658–4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAtee PA, Brian L, Curran B, van der Linden O, Nieuwenhuizen NJ, Chen X, Henry-Kirk RA, Stroud EA, Nardozza S, Jayaraman J, Rikkerink EHA, Print CG, Allan AC, and Templeton MD 2018. Re-programming of Pseudomonas syringae pv. actinidiae gene expression during early stages of infection of kiwifruit. BMC Genomics 19:822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClerklin SA, Lee SG, Harper CP, Nwumeh R, Jez JM, and Kunkel BN 2018. Indole-3-acetaldehyde dehydrogenase-dependent auxin synthesis contributes to virulence of Pseudomonas syringae strain DC3000. PLoS Pathog. 14:e1006811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misas-Villamil JC, Kolodziejek I, and van der Hoorn RA 2011. Pseudomonas syringae colonizes distant tissues in Nicotiana benthamiana through xylem vessels. Plant J. 67:774–782. [DOI] [PubMed] [Google Scholar]
- Mockaitis K, and Estelle M. 2008. Auxin receptors and plant development: A new signaling paradigm. Annu. Rev. Cell Dev. Biol. 24:55–80. [DOI] [PubMed] [Google Scholar]
- Mutka AM 2013. The roles of auxin in Pseudomonas syringae pathogenesis. PhD thesis. Washington University in St. Louis, St. Louis, MO, U.S.A. https://search.proquest.com/docview/1459240147. [Google Scholar]
- Mutka AM, Fawley S, Tsao T, and Kunkel BN 2013. Auxin promotes susceptibility to Pseudomonas syringae via a mechanism independent of suppression of salicylic acid-mediated defenses. Plant J. 74:746–754. [DOI] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, and Jones JD 2006. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312: 436–439. [DOI] [PubMed] [Google Scholar]
- Nobori T, Velasquez AC, Wu J, Kvitko BH, Kremer JM, Wang Y, He SY, and Tsuda K. 2018. Transcriptome landscape of a bacterial pathogen under plant immunity. Proc. Natl. Acad. Sci. U.S.A. 115: E3055–E3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell PJ, Schmelz EA, Moussatche P, Lund ST, Jones JB, and Klee HJ 2003. Susceptible to intolerance—A range of hormonal actions in a susceptible Arabidopsis pathogen response. Plant J. 33: 245–257. [DOI] [PubMed] [Google Scholar]
- Ortiz-Martín I, Thwaites R, Macho AP, Mansfield JW, and Beuzón CR 2010. Positive regulation of the Hrp T3S system in Pseudomonas syringae pv. phaseolicola. Mol. Plant-Microbe Interact. 23:665–681. [DOI] [PubMed] [Google Scholar]
- Paponov IA, Paponov M, Teale W, Menges M, Chakrabortee S, Murray JA, and Palme K. 2008. Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol. Plant 1:321–337. [DOI] [PubMed] [Google Scholar]
- Parry G, Calderon-Villalobos LI, Prigge M, Peret B, Dharmasiri S, Itoh H, Lechner E, Gray WM, Bennett M, and Estelle M. 2009. Complex regulation of the TIR1/AFB family of auxin receptors. Proc. Natl. Acad. Sci. U.S.A. 106:22540–22545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiter K, Brooks DM, Penaloza-Vazquez A, Sreedharan A, Bender CL, and Kunkel BN 2005. Novel virulence gene of Pseudomonas syringae pv. tomato strain DC3000. J. Bacteriol. 187:7805–7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge MJ, Greenham K, Zhang Y, Santner A, Castillejo C, Mutka AM, O’Malley RC, Ecker JR, Kunkel BN, and Estelle M. 2016. The Arabidopsis auxin receptor F-box proteins AFB4 and AFB5 are required for response to the synthetic auxin picloram. G3 (Bethesda) 6: 1383–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge MJ, Platre M, Kadakia N, Zhang Y, Greenham K, Szutu W, Pandey BK, Bhosale RA, Bennett MJ, Busch W, and Estelle M. 2020. Genetic analysis of the Arabidopsis TIR1/AFB auxin receptors reveals both overlapping and specialized functions. eLife 9:e54740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JA 1975. Transport of indoleacetic acid in plant cells in relation to pH and electrical potential gradients, and its significance for polar IAA transport. New Phytol. 74:163–172. [Google Scholar]
- Robert-Seilaniantz A, Grant M, and Jones JD 2011. Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 49:317–343. [DOI] [PubMed] [Google Scholar]
- Schmelz EA, Engelberth J, Tumlinson JH, Block A, and Alborn HT 2004. The use of vapor phase extraction in metabolic profiling of phytohormones and other metabolites. Plant J. 39:790–808. [DOI] [PubMed] [Google Scholar]
- Smith A, Lovelace AH, and Kvitko BH 2018. Validation of RT-qPCR approaches to monitor Pseudomonas syringae gene expression during infection and exposure to pattern-triggered immunity. Mol. Plant-Microbe Interact. 31:410–419. [DOI] [PubMed] [Google Scholar]
- Spaepen S, and Vanderleyden J. 2011. Auxin and plant-microbe interactions. Cold Spring Harb. Perspect. Biol. 3:a001438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takato S, Kakei Y, Mitsui M, Ishida Y, Suzuki M, Yamazaki C, Hayashi KI, Ishii T, Nakamura A, Soeno K, and Shimada Y. 2017. Auxin signaling through SCFTIR1/AFBs mediates feedback regulation of IAA biosynthesis. Biosci. Biotechnol. Biochem. 81:1320–1326. [DOI] [PubMed] [Google Scholar]
- Timpte C, Wilson AK, and Estelle M. 1994. The axr2–1 mutation of Arabidopsis thaliana is a gain-of-function mutation that disrupts an early step in auxin response. Genetics 138:1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Puyvelde S, Cloots L, Engelen K, Das F, Marchal K, Vanderleyden J, and Spaepen S. 2011. Transcriptome analysis of the rhizosphere bacterium Azospirillum brasilense reveals an extensive auxin response. Microb. Ecol. 61:723–728. [DOI] [PubMed] [Google Scholar]
- Wang D, Pajerowska-Mukhtar K, Culler AH, and Dong X. 2007. Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol 17:1784–1790. [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, and Ausubel FM 2001. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414:562–565. [DOI] [PubMed] [Google Scholar]
- Wong ML, and Medrano JF 2005. Real-time PCR for mRNA quantitation. Biotechniques 39:75–85. [DOI] [PubMed] [Google Scholar]
- Woodward AW, and Bartel B. 2005. A receptor for auxin. Plant Cell 17: 2425–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin XF, and He SY 2013. Pseudomonas syringae pv. tomato DC3000: A model pathogen for probing disease susceptibility and hormone signaling in plants. Annu. Rev. Phytopathol. 51:473–498. [DOI] [PubMed] [Google Scholar]
- Xin XF, Kvitko B, and He SY 2018. Pseudomonas syringae: What it takes to be a pathogen. Nat. Rev. Microbiol. 16:316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Zhang Q, Guo J, Charkowski AO, Glick BR, Ibekwe AM, Cooksey DA, and Yang C-H 2007. Global effect of indole-3-acetic acid biosynthesis on multiple virulence factors of Erwinia chrysanthemi 3937. Appl. Environ. Microbiol. 73:1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z-C, Haudecoeur E, Faure D, Kerr KF, and Nester EW 2008. Comparative transcriptome analysis of Agrobacterium tumefaciens in response to plant signal salicylic acid, indole-3-acetic acid and g-amino butyric acid reveals signalling cross-talk and Agrobacterium-plant co-evolution. Cell. Microbiol. 10:2339–2354. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


