Abstract
The COVID-19 corona virus has become a world pandemic which started in December 2019 in Wuhan, China with no confirmed biological source. Various countries reported the genomic sequence of different isolates obtained from infected patients. This allowed us to obtain a number of 38 isolates of full genomic sequences. Alignment of nucleotide (nt) sequence was carried out using Clustal Omega multiple alignment service at the EBI website. Alignment of nt sequence and phylogenetic relationship revealed that the COVID-19 is a new viral strain and its biological source has not been yet detected. The expected orf pattern was different among isolates obtained from the same country or different countries as well as from SARS-CoV isolates or bats CoV suggesting different virus human interaction possibilities during infection and severity. All isolates had the main five orfs (1ab, S, M, N, E), whereas they differed in the expected accessory orfs. Being with the biological source of COVID-19 undetected, the role of human endogenous retrovirus (HERVs) in the regulation of the host cell gene expression or the encoding for products that could modulate COVID-19 infection and the spectrum of its symptoms is discussed.
Keywords: COVID-19, genome, nucleotide sequence alignment, Human endogenous retroviruses (HERVs)
INTRODUCTION
Coronavirus belong to coronaviridae family, genus betacoronavirus, and subgenus sarbecovirus. Coronaviridae includes numerous birds and mammalian coronaviruses [1, 2]. Human to human coronaviruses was detected after its outbreak in Southern China in 2003 [3–5]. It was associated with severe acute respiratory symptoms (SARS), therefore it was named SARS-Coronavirus (SARS-CoV) [1, 6]. Its worldwide spread in 2003 outbreak caused above 8000 infections and more than 774 confirmed dead [1]. It was detected in the civets at the Himalayan palm [7]. Genome comparison confirmed that the civet viral isolate had 29 missing nucleotide of the open reading frame 10 (orf10) in most of characterized human isolates in the 2003 outbreak [7]. This led to the suggestion that the missing nucleotides caused the transmission of the virus from civets to human [1]. Another version of the virus was isolated from horseshoe bats [8] with 29 nucleotide insertion in orf8 (Bat-SARS-CoV) compared to most characterized human isolates. This genomic relationship suggested a common ancestor for civets, bats, and human SARS-CoV genomes [8]. After SARS outbreak in 2003, bats were considered the reservoir for future human CoV pandemics [9]. In 2012, the Middle East Respiratory coronavirus (MERS-CoV) was detected in Saudi Arabia [10, 11]. It is believed that it was transmitted from dromedary camels to human [12] but its origin was linked also to bats [13]. It caused 2521 infections and the death of 919 (35%) [14].
In 2019, a novel coronavirus (COVID-19) appeared in China (Wuhan City, Hubei Province). It is believed that COVID-19 originated from fresh seafood [15, 16]. This version of coronavirus was able to transmit from human to human [17, 18]. It has been spread in 193countries with above 10 Million confirmed infection and more than 500 000 confirmed deaths [19].
Analysis of COVID-19 full genome showed that it is similar to betacoronavirus, yet it is different from the previous SARS-CoV or MERS-CoV [15]. The COVID-19 diverged with the Bat_SARS-CoV in a separate group of sarbecovirus [15]. Genome study of COVID-19 and the Bat SARS-CoV (isolate BatCoV RaTG13) revealed that the genetic similarity between COVID-19 and RaTG13 indicated that COVID-19 is not the exact variant that led to the outbreak in China. However, the COVID-19 could have originated form the bats. Also, this study confirmed that COVID-19 did not result of recombination and not a mosaic [14]. Bioinformatics analysis using nucleotide sequence of COVID-19 genome isolated from patients revealed that the COVID-19 has 89% nt identity with Bat coronavirus (Bat SARS-like-CoVZXC21) and 82% to the SARS-CoV. Using amino acid sequence of the expected orfs of COVID-19 showed that it was diverged with bat, civet, and human SARS-CoV. Yet, unlike other coronaviruses, its orf3b produce a shorter protein and its orf8 encode a secreted protein making the source of the cOVID-19 version is undetectable [20].
Interaction between the COVID-19 spike protein (S) receptor and its host receptor angiotensin-converting enzyme 2 (ACE2) was investigated based on similar information obtained from SARS-CoV. The amino acid (aa) sequence of COVID-19 S protein including the receptor-binding domain (RBD) which interact with ACE2 is similar to that of SARS-CoV. This supports that the COVID-19 use ACE2 as its receptor and it has more affinity to human ACE2 and other animals, explaining its capability of human cell infection and human-human transmission [21].
The question now is where the COVID-19 came from and how similar are the isolates from different patients and different countries? Also, the wide spectrum of symptoms of the virus starting from no symptoms to death is a second key question. These are fundamental questions need to be answered for better understanding of the virus origin, transmission, and severity. In this study, we investigated the similarity of nucleotide sequence of 38 COVID-19 isolates from 6 countries to evaluate differences among them. Similarity among COVID-19 at the nt sequence or the predicted orfs were investigated. The role of human endogenous retroviruses (HERVs) in the COVID-19 wide range of symptoms is also discussed.
MATERIALS AND METHODS
Nucleotide and Protein Sequences
All nucleotide sequences of COVID-19 or SARS-CoV complete genome nt sequence of isolates were obtained from NCBI nucleotide database (https://www. ncbi.nlm.nih.gov/nuccore). Isolates included 17 from China, 10 from USA, 5 from Japan, 2 from Hong Kong, 2 from Taiwan, 1 from South Korea, 1 from Australia (Table 1).
Table 1. .
Nucleotide sequence identity to the first reported case from China isolateHZ-1 (Accession no. MT039873.1)
| Accession | Isolate | Country | Total Score | Query Cover, % |
Ident, % | |
|---|---|---|---|---|---|---|
| 1 | MT019532.1 | IPBCAMS-WH-04 | China | 55 092 | 100 | 100 |
| 2 | MN996528.1 | WIV04 | China | 55 092 | 100 | 100 |
| 3 | MN988668.1 | V WHU01 | China | 55 092 | 100 | 100 |
| 4 | NC_045512.2 | Wuhan-Hu-1 | China | 55 092 | 100 | 100 |
| 5 | MT019533.1 | IPBCAMS-WH-05 | China | 55 086 | 100 | 100 |
| 6 | MT019531.1 | IPBCAMS-WH-03 | China | 55 086 | 100 | 100 |
| 7 | MT066176.1 | NTU02 | Taiwan | 55 081 | 100 | 99.99 |
| 8 | MT066175.1 | NTU01 | Taiwan | 55 081 | 100 | 99.99 |
| 9 | MT027064.1 | USA-CA5 | USA | 55 081 | 100 | 99.99 |
| 10 | MN994468.1 | USA-CA2 | USA | 55 081 | 100 | 99.99 |
| 11 | MT027062.1 | USA-CA3 | USA | 55 075 | 100 | 99.99 |
| 12 | MT019529.1 | IPBCAMS-WH-01 | China | 55 075 | 100 | 99.99 |
| 13 | MN985325.1 | USA-WA1 | USA | 55 075 | 100 | 99.99 |
| 14 | MN996530.1 | WIV06 | China | 55 071 | 99 | 100 |
| 15 | LC522974.1 | TY/WK-501 | Japan | 55 070 | 100 | 99.99 |
| 16 | LC522972.1 | KY/V-029 | Japan | 55 070 | 100 | 99.99 |
| 17 | MN997409.1 | USA-AZ1 | USA | 55 070 | 100 | 99.99 |
| 18 | MT039888.1 | USA-MA1 | USA | 55 066 | 100 | 99.98 |
| 19 | MT039887.1 | USA-WI1 | USA | 55 066 | 100 | 99.99 |
| 20 | MT049951.1 | Yunnan-01 | China | 55 064 | 100 | 99.98 |
| 21 | LC522975.1 | TY/WK-521 | Japan | 55 064 | 100 | 99.98 |
| 22 | LC522973.1 | TY/WK-012 | Japan | 55 064 | 100 | 99.98 |
| 23 | MN996529.1 | WIV05 | China | 55 064 | 99 | 99.99 |
| 24 | MN975262.1 | HKU-SZ-005b | H Kong | 55 064 | 100 | 99.98 |
| 25 | MN996531.1 | WIV07 | China | 55 062 | 99 | 99.99 |
| 26 | MN988713.1 | USA-IL1 | USA | 55 062 | 100 | 99.97 |
| 27 | LR757996.1 | Wuhan, genome assembly | China | 55 060 | 99 | 100 |
| 28 | MT019530.1 | IPBCAMS-WH-02 | China | 55 058 | 100 | 99.98 |
| 29 | LR757995.1 | Wuhan, genome assembly | China | 55 057 | 99 | 99.99 |
| 30 | MT044257.1 | USA-IL2 | USA | 55 053 | 100 | 99.98 |
| 31 | MN994467.1 | USA-CA1 | USA | 55 053 | 100 | 99.98 |
| 32 | MT039890.1 | SNU01 | Korea | 55 042 | 100 | 99.97 |
| 33 | LR757998.1 | Wuhan, genome assembly | China | 55 040 | 99 | 99.99 |
| 34 | MN996527.1 | WIV02 | China | 55 025 | 99 | 99.99 |
| 35 | MN938384.1 | HKU-SZ-002a | H Kong | 55 022 | 99 | 99.99 |
| 36 | MT007544.1 | VIC01 | Australia | 55 010 | 100 | 99.96 |
| 37 | MT044258.1 | USA-CA6 | USA | 54 937 | 100 | 99.92 |
| 38 | LC521925.1 | Japan/AI/I-004 | Japan | 54 926 | 100 | 99.91 |
| 39 | MN996532.1 | BatCoV-RaTG13 | China | 48 630 | 99 | 96.11 |
| 40 | AY395003.1 | SARS coronavirus ZS-C | China (2004) | 15 213 | 88 | 82.34 |
Blast and Multiple Alignment Analysis of COVID-19 Isolates
The sequence of the first reported COVID-19 isolate from China (HZ-1, MT039873.1) was used in a BLAST search to determine the identity of its sequence with other sequences reported from China or other countries in the nucleotide database. The nt sequence of isolates were aligned using Clustal Omega (ClustalO) multiple alignment service (https://www.ebi.ac.uk/Tools/msa/ clustalo/). Phylogenetic tree of isolate sequence was constructed using the same ClustalO. Nucleotide SNPs were detected manually in the aligned sequences.
Expected ORFs of Different COVID-19 Isolates
The expected orfs of each COVID-19 isolate were obtained from the NCBI graphics view of the nucleotide accession at the NCBI nucleotide database website (https://www.ncbi.nlm.nih.gov/nuccore).
RESULTS
Nucleotide Sequence Identity of COVID-19 and Other Corona Viruses
The first Chinese reported sequence (MT039873.1) of COVID-19 was used in a BLAST search. This search revealed high identity to the other 38 COVID-19 isolates (Table 1). These included 16 other reported sequences from China, 11 form USA, 5 from Japan, 2 Hong Kong, 2 from Taiwan, and 1 from Australia. High identity of these isolates was observed to the Chinese isolate ranging from 100 to 99.91% (Table 1) with query coverage range from 99–100%. Interestingly, the Chinese first reported case showed 96.11% identity and 99% coverage with the Chinese BatCoV-RaTG13 (MN996532.1) isolate; closest identity in this study. More important, its identity to the closest isolate of SARS-CoV (AY395003.1) was 82.34% identity and 88% query coverage (Table 1).
Phylogenetic Relationship among COVID-19 Isolates
Phylogenetic relationship among the 38 COVID-19 isolates reported from different countries showed random clustering without any noticeable phylogenetic relationship on various clades of the phylogenetic tree of isolates from China or any other country (Fig. 1). Clade A has 1 Chinese isolate. Clade B has 2 Chinese isolates. Clade C has 14 isolates, 1 from Australia, 3 USA, 6 from China, 1 from Taiwan, 2 from Japan, 1 from Korea. Clade D has 3 isolates, 2 from China, 1 from USA. Clade E has 18 isolates, 7 from USA, 6 from China, 2 from Hong Kong, 3 from Japan (Fig. 1). This random distribution of isolates from the same country, specifically Chinese isolates, indicated they belong to the same strain.
Fig. 1.

Phylogenetic relationship among COVID-19 isolates from different countries.
Nucleotide Sequence Alignment of COVID-19 Isolates
Using blast search, COVID-19 first reported Chinese isolate had 3.89% difference from the closest SARS-CoV and 17.66% difference from the closest bat coronavirus isolate (Table 1), Similarly, alignment of COVID-19 and SARS-CoV isolates as one group resulted in tremendous differences in the nt sequence spread overall the genome, therefore we investigated the nucleotide SNPs among COVID-19 and SARS-CoV isolates. The 38 COVID-19 isolates and the 3 SARS-CoV isolates were compared as separate groups.
Among the 38 COVID-19 isolates, 108 nucleotide changes (103 SNPs and 5 deletions) were detected (Table 2). Seven Chinese isolates did not have any SNPs, whereas other isolates had different number of SNPs ranging from 1–9 (Table 2). The Korean isolate SNU01 came on the top with 9 SNPs, followed by USA isolate USA-IL1, USA isolate USA-IL1, and the Chinese isolate IPBCAMS-WH-02 with 8, 7, 6 SNPs consecutively (Table 2). All Japanese isolates had SNPs ranged from 3–5. Nucleotide SNPs were distributed among transition (66) and transversion (37). The number of detected SNPS indicated that the base substitution rate (SNPs) rate for all studied COVID-19 isolate was 103/1 135 284 = 9.07 × 10–5. Similar alignment among three SARS-CoV isolates (DQ182595.1; China, AY323977.2, Italy; AY310120.1, Germany) revealed that the Chinese isolate (DQ182595.1) nucleotide sequence had 99.97 and 99.95% identity with the Italian (AY323977.2) and German (AY310120.1) isolates consecutively. Nucleotide sequence alignment resulted in 12 SNPs and 1 deletion among the three SARS-CoV isolates (Table 2) indicating base substitution rate of 12/89197 = 12.22 × 10–5 among SARS-CoV isolates. This seems to be higher that the SNPs rate in COVID-19 isolates because of low number of isolates used.
Table 2. .
Summary of detected nucleotide SNPs among COVID-19 isolates
| NO | AC # | Country | Isolate | Length, nt |
Nt SNPs | SNPs |
|---|---|---|---|---|---|---|
| 1 | LR757995.1 | China | Whole genome | 29 872 | T28129C, C8767T | 2 |
| 2 | LR757996.1 | China | Whole genome | 29 868 | ||
| 3 | LR757998.1 | China | Whole genome | 29 866 | C6943A, T11739A | 2 |
| 4 | MN988668.1 | China | WHU01 | 29 881 | ||
| 5 | MN996527.1 | China | WIV02 | 29 825 | G21292A, A24292G | 2 |
| 6 | MN996528.1 | China | WIV04 | 29 891 | ||
| 7 | MN996529.1 | China | WIV05 | 29 852 | G7004A, A21125G | 2 |
| 8 | MN996530.1 | China | WIV06 | 29 854 | ||
| 9 | MN996531.1 | China | WIV07 | 29 857 | A7988C, C9521T, | 2 |
| 10 | MT019529.1 | China | IPBCAMS-WH-01 | 29 899 | A3778G, A8388G, T8987A | 3 |
| 11 | MT019530.1 | China | IPBCAMS-WH-02 | 29 889 | T104A, T111C, T112G, C119G, T120C, G124A | 6 |
| 12 | MT019531.1 | China | IPBCAMS-WH-03 | 29 899 | T6996C | 1 |
| 13 | MT019532.1 | China | IPBCAMS-WH-04 | 29 890 | ||
| 14 | MT019533.1 | China | IPBCAMS-WH-05 | 29 883 | G7866T | 1 |
| 15 | MT039873.1 | China | HZ-1, 1st case | 29 833 | ||
| 16 | NC_045512.2 | China | Wuhan-Hu-1 | 29 903 | ||
| 17 | MT049951.1 | China | Yunnan-01 | 29 903 | C75A, C8782T,G11083C, T21644A, T28144C | 5 |
| 18 | MN985325.1 | USA | USA-WA1 | 29 882 | C8782T, T28144C | 2 |
| 19 | MN994468.1 | USA | USA-CA2 | 29 883 | C17000T, G26144T | 2 |
| 20 | MT027062.1 | USA | USA-CA3 | 29 882 | G614A, A5084G, C28854T | 3 |
| 21 | MT027064.1 | USA | USA-CA5 | 29 882 | C2091T, C21707T | 2 |
| 22 | MT044258.1 | USA | USA-CA6 | 29 858 | Del 508-523, del 671-679, | 2 |
| 23 | MT039888.1 | USA | USA-MA1 | 29 882 | G3518T, C8782T,A17423G, C24034T C28854T | 5 |
| 24 | MT039887.1 | USA | USA-WI1 | 29 879 | C17373T, del 20298-20300, | 2 |
| 25 | MN988713.1 | USA | USA-IL1 | 29 882 | T490W,C3177Y, C8782Y, C24034Y, T26729Y, G28077S, T28144Y, C28854Y | 8 |
| 26 | MT044257.1 | USA | USA-IL2 | 29 882 | T490A, C3177T, C8782T, C24034T, T26729C, G28077C, T28144C | 7 |
| 27 | MN997409.1 | USA | USA-AZ1 | 29 882 | C8782T,G11083T, T28144C, C29095T | 4 |
| 28 | LC521925.1 | Japan | AI-I-004 | 29 848 | Del 351-374, C18485T, C18485T | 3 |
| 29 | LC522972.1 | Japan | KY-V-029 | 29 878 | G11554T, C15321T, C25807G, C29300T | 4 |
| 30 | LC522973.1 | Japan | TY-WK-012 | 29 878 | C2659T, C8779T C3789T, C29092T, T28141C | 5 |
| 31 | LC522974.1 | Japan | TY-WK-501 | 29 878 | C2659T, C8779T, C29092T, T28141C | 4 |
| 32 | LC522975.1 | Japan | TY-WK-521 | 29 878 | C2659T, C8779T, C29092T, G29702T, T28141C | 5 |
| 33 | MN938384.1 | H Kong | HKU-SZ-002a | 29 838 | C8750T, C29063T | 2 |
| 34 | MN975262.1 | H Kong | HKU-SZ-005b | 29 891 | C8782T,C9561T, T15607C, C29095T, T28144C | 5 |
| 35 | MT066175.1 | Taiwan | NTU01 | 29 870 | C8782T, T28144C | 2 |
| 36 | MT066176.1 | Taiwan | NTU02 | 29 870 | A9034G, C9491T | 2 |
| 37 | MT007544.1 | Ausuralia | Australia-VIC01 | 29 893 | T19065C, T22303G, G26144T,del 2974029950 | 4 |
| 38 | MT039890.1 | Korea | SNU01 | 29 903 | G2969T, C6031T, C12115T, T15597C, C20936G, C22224G, G25775T, G26144T, T26354A | 9 |
| Total | 1 135 284 | 108 | ||||
| SARS-CoV-1 | ||||||
| 39 | AY310120.1 | Germany | SARS-CoV-1-FRA | 29 740 | T18965A, C19084T, C24933T, C26660T, C28268T | 5 |
| 40 | AY323977.2 | Italy | SARS-CoV-1-HSR1 | 29 751 | G27254R | 1 |
| 41 | DQ182595.1 | China | SARS-CoV-1ZJ0301 | 29 706 | Del1-16, A12965C,T14022A, A14976T, C17478G, T17518A,C22573T | 7 |
| Total | 89 197 | 13 |
COVID-19 Open Reading Frames (orfs)
Five main orfs are usually produced by all corona virus isolates including orflab polyprotein, orfS, orfN, orfM, and orfE. Another seven orfs have been reported by various isolates including orf1a polyprotein, orf3a, orf6, orf7a, orf7b, orf8, and orf10 (Table 3). Usually, polyprotein 1ab and orf1a are processed into smaller accessory orfs (Table 4). The accessory orfs are not produced in all corona virus isolates.
Table 3. .
Common coronavirus orfs
| Accession # | orf | Genomic location | Length, aa | Function | |
|---|---|---|---|---|---|
| start nt | end nt | ||||
| YP_009724389.1 | orf1ab | 266 | 21 555 | 7.096 | Polyprotein |
| YP_009725295.1 | orf1a | 266 | 13 483 | 4.405 | Polyprotein |
| YP_009724390.1 | orfS | 21 563 | 25 384 | 1,273 | Surface glycoprotein |
| YP_009724392.1 | orfE | 2624 | 26 472 | 75 | Envelope protein |
| YP_009724397.2 | orfN (orf9) | 28 274 | 29 533 | 419 | Nucleocapsid phosphoprotein |
| YP_009724393.1 | orfM (orf5) | 26 523 | 27 191 | 222 | Membrane glycoprotein |
| YP_009724391.1 | orf3a | 25 393 | 26 220 | 275 | ORF3a protein |
| YP_009724394.1 | orf6 | 27 202 | 27 387 | 61 | ORF6 protein |
| YP_009724395.1 | orf7a | 27 394 | 27 759 | 121 | ORF7a protein |
| YP_009725296.1 | orf7b | 27 756 | 27 887 | 43 | ORF7b |
| YP_009724396.1 | orf8 | 27 894 | 8259 | 121 | ORF8 protein |
| YP_009725255.1 | orf10 | 29 558 | 29 674 | 38 | ORF10 protein |
Table 4. .
Accessory orfs produced from polyprotein orf1ab and orf1a
| Accession# | Protein name | Length (aa) | Source orf (1ab or 1a) | Function |
|---|---|---|---|---|
| YP_009725297.1 | nsp1 | 180 | ofr1ab, orf1a | Leader protein |
| YP_009725298.1 | nsp2 | 638 | ofr1ab, orf1a | – |
| YP_009725299.1 | nsp3 | 1,945 | ofr1ab, orf1a | – |
| YP_009725300.1 | nsp4 | 500 | ofr1ab, orf1a | – |
| YP_009725301.1 | nsp5 | 306 | ofr1ab, orf1a | 3C-like proteinase |
| YP_009725302.1 | nsp6 | 290 | ofr1ab, orf1a | – |
| YP_009725303.1 | nsp7 | 83 | ofr1ab, orf1a | – |
| YP_009725304.1 | nsp8 | 198 | ofr1ab, orf1a | – |
| YP_009725305.1 | nsp9 | 113 | ofr1ab, orf1a | – |
| YP_009725306.1 | nsp10 | 139 | ofr1ab, orf1a | – |
| YP_009725312.1 | nsp11 | 13 | orf1a | – |
| YP_009725307.1 | nsp12 | 932 | orf1a | RNA-dependent RNA polymerase |
| YP_009725308.1 | nsp13 | 601 | orf1a | Helicase |
| YP_009725309.1 | nsp14 | 527 | orf1a | 3'-to-5' exonuclease |
| YP_009725310.1 | nsp15 | 346 | orf1a | EndoRNAse |
| YP_009725311.1 | nsp16 | 298 | orf1a | 2'-O-ribose methyltransferase |
Expected orfs from COVID-19 Isolates
We investigated the expected orfs of different isolates from the same country or from different countries to check if different corona virus isolate differ in their expected orf pattern, although they have similar genome size and high identity in their genome nucleotide sequence (Tables 1, 2). Interestingly, orf pattern produced by isolates form the same country or from different countries differed greatly (Table 5, Fig. 2). All COVID-19, SARS-CoV, and the BatCoV-RaTG13 isolates have the five main orfs (1ab, S, E, M, N). Also, all of these isolates have orf3a except the Chinese isolate WHU01 (MN988668.1). This isolate is expected to produce only the five main orfs being the minimum orfs detected in this study. Only two Chinese isolates (Wuhan-Hu-1 and Yunnan-01) of COVID-19 38 isolates had the orf1a which is expected in three SARS-CoV isolates and the BatCoV-RaTG13 isolate (Table 5). Orf6 and orf7a are expected in all isolates except the Chinese isolate Wuhan-Hu-1. Orf7b is expected only in 7 Chinese isolates, the three SARS-CoV isolates, and the BatCoV-RaTG13 isolate, whereas orf8 is not expected in the three SARS-CoV isolates and the Chinese isolate Wuhan-Hu-1 (Table 5). Orf10 is not expected in 6 COVID-19 Chinese isolates, the three SARS-CoV isolates, and the BatCoV-RaTG13 isolate. Four extra accessory orfs (3b, 8a, 8b, 9b) are only expected in the three SARS-CoV isolates and the BatCoV-RaTG13 isolate (Table 5). Among isolates from the same country, USA isolates and Japanese Isolates did not show differences among their groups in the expected orf pattern. On the other hand, Chinese isolates showed differences in orfs 1a, 3a, 6, 7a, 7b, 8, 10 with Chinese isolate WHU01 (MN988668.1) is expected to produce only the five main orfs (Table 5). The orf pattern of selected 4 Chinese COVID-19 isolates, one SARS-CoV isolates, and the BatCoV-RaTG13 isolate is shown in Fig. 2. The first reported Chinese isolate (HZ-1, MT039873.1) has10 expected orfs of its genome including 1ab, N, S, E, M, 3a, 6, 7a, 8, 10. Orf1a is not expected from the genome of this isolate (Fig. 2). On the other hand, another Chinese isolate (Yunnan-01, MT049951.1) is expected to produce orf1a and orf7b beside the 10 orfs expected in isolate HZ-1 (Fig. 2). In addition, the Chinese isolate WIV02 (MN996527.1) expected orfs is similar to expected orf pattern of isolate Yunnan-01 except the absence of orf1a. Interestingly, bat isolate BatCoV-RaTG13 (MN996532.1) has exact similar expected orfs pattern as Chinese isolate WIV02. The Chinese isolate WHU01 (MN988668.1) only has 5 expected orfs (1ab, S, M, N, E). The Chinese SARS-CoV isolate SARS-CoV-1-ZJ0301 has expected 32 orfs including the main 5 orfs and 27 accessory orfs (Fig. 2).
Table 5. .
Summary of predicted ORFs in reported nCoV-2 isolates (+ indicates the presence of orf, – indicates the absence of orf)
| No | AC # | Country | Isolate | bp | orf | Extra orfs | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1ab | 1a | S | 3a | E | M | 6 | 7a | 7b | 8 | N | 10 | 3b | 8a | 8b | 9b | |||||
| 1 | LR757995.1* | China | Whole genome | 29 872 | ||||||||||||||||
| 2 | LR757996.1* | China | Whole genome | 29 868 | ||||||||||||||||
| 3 | LR757998.1* | China | Whole genome | 29 866 | ||||||||||||||||
| 4 | MN988668.1 | China | WHU01 | 29 881 | + | – | + | – | + | + | – | – | – | – | + | – | – | – | – | – |
| 5 | MN996527.1 | China | WIV02 | 29 825 | + | – | + | + | + | + | + | + | + | + | + | – | – | – | – | – |
| 6 | MN996528.1 | China | WIV04 | 29 891 | + | – | + | + | + | + | + | + | + | + | + | – | – | – | – | – |
| 7 | MN996529.1 | China | WIV05 | 29 852 | + | – | + | + | + | + | + | + | + | + | + | – | – | – | – | – |
| 8 | MN996530.1 | China | WIV06 | 29 854 | + | – | + | + | + | + | + | + | + | + | + | – | – | – | – | – |
| 9 | MN996531.1 | China | WIV07 | 29 857 | + | – | + | + | + | + | + | + | + | + | + | – | – | – | – | – |
| 10 | MT019529.1 | China | IPBCAMS-WH-01 | 29 899 | + | – | + | + | + | + | + | + | – | + | + | + | – | – | – | – |
| 11 | MT019530.1 | China | IPBCAMS-WH-02 | 29 889 | + | – | + | + | + | + | + | + | – | + | + | + | – | – | – | – |
| 12 | MT019531.1 | China | IPBCAMS-WH-03 | 29 899 | + | – | + | + | + | + | + | + | – | + | + | + | – | – | – | – |
| 13 | MT019532.1 | China | IPBCAMS-WH-04 | 29 890 | + | – | + | + | + | + | + | + | – | + | + | + | – | – | – | – |
| 14 | MT019533.1 | China | IPBCAMS-WH-05 | 29 883 | + | – | + | + | + | + | + | + | – | + | + | + | – | – | – | – |
| 15 | MT039873.1 | China | HZ-1, 1st case | 29 833 | + | – | + | + | + | + | + | + | – | + | + | + | – | – | – | – |
| 16 | MT039890.1 | S. Korea | SNU01 | 29 903 | + | – | + | + | + | + | + | + | – | + | + | + | – | – | – | – |
| 17 | NC_045512.2 | China | Wuhan-Hu-1 | 29 903 | + | + | + | + | + | + | + | + | + | + | + | + | – | – | – | – |
| 18 | MT049951.1 | China | Yunnan-01 | 29 903 | + | + | + | + | + | + | + | + | + | + | + | + | – | – | – | – |
| 19 | MN985325.1 | USA | USA-WA1 | 29 882 | + | – | + | + | + | + | + | + | – | + | + | + | – | – | – | – |
| 20 | MN994468.1 | USA | USA-CA2 | 29 883 | + | – | + | + | + | + | + | + | – | + | + | + | – | – | – | – |
| 21 | MT027062.1 | USA | USA-CA3 | 29 882 | + | – | + | + | + | + | + | + | – | + | + | + | – | – | – | – |
| 22 | MT027064.1 | USA | USA-CA5 | 29 882 | + | – | + | + | + | + | + | + | – | + | + | + | – | – | – | – |
| 23 | MT044258.1 | USA | USA-CA6 | 29 858 | + | + | + | + | + | + | + | + | + | + | – | – | – | – | ||
| 24 | MT039888.1 | USA | USA-MA1 | 29 882 | + | – | + | + | + | + | + | + | – | + | + | + | – | – | – | – |
| 25 | MT039887.1 | USA | USA-WI1 | 29 879 | + | – | + | + | + | + | + | + | – | + | + | + | – | – | – | – |
| 26 | MN988713.1 | USA | USA-IL1 | 29 882 | + | – | + | + | + | + | + | + | – | + | + | + | – | – | – | – |
| 27 | MT044257.1 | USA | USA-IL2 | 29 882 | + | – | + | + | + | + | + | + | + | + | + | – | – | – | – | |
| 28 | MN997409.1 | USA | USA-AZ1 | 29 882 | + | – | + | + | + | + | + | + | – | + | + | + | – | – | – | – |
| 29 | LC521925.1 | Japan | AI-I-004 | 29 848 | + | – | + | + | + | + | + | + | – | + | + | + | – | – | – | – |
| 30 | LC522972.1 | Japan | KY-V-029 | 29 878 | + | – | + | + | + | + | + | + | – | + | + | + | – | – | – | – |
| 31 | LC522973.1 | Japan | TY-WK-012 | 29 878 | + | – | + | + | + | + | + | + | – | + | + | + | – | – | – | – |
| 32 | LC522974.1 | Japan | TY-WK-501 | 29 878 | + | – | + | + | + | + | + | + | – | + | + | + | – | – | – | – |
| 33 | LC522975.1 | Japan | TY-WK-521 | 29 878 | + | – | + | + | + | + | + | + | – | + | + | + | – | – | – | – |
| 34 | MN938384.1 | H Kong | HKU-SZ-002a | 29 838 | + | – | + | + | + | + | + | + | – | + | + | – | – | – | – | – |
| 35 | MN975262.1 | H Kong | HKU-SZ-005b | 29 891 | + | – | + | + | + | + | + | + | – | + | + | + | – | – | – | – |
| 36 | MT066175.1 | Taiwan | NTU01 | 29 870 | + | – | + | + | + | + | + | + | – | + | + | + | – | – | – | – |
| 37 | MT066176.1 | Taiwan | NTU02 | 29 870 | + | – | + | + | + | + | + | + | + | + | + | – | – | – | – | |
| 38 | MT007544.1 | Ausuralia | Australia-VIC01 | 29 893 | + | – | + | + | + | + | + | + | – | + | + | + | – | – | – | – |
| Bat CoV-2 | ||||||||||||||||||||
| 39 | MN996532.1 | China | BatCoV-RaTG13 | 29 855 | + | – | + | + | + | + | + | + | + | + | + | – | ||||
| SARS-CoV | ||||||||||||||||||||
| 40 | AY310120.1 | Germany | SARS-CoV-1-FRA | 29 740 | + | + | + | + | + | + | + | + | + | – | + | – | + | + | + | + |
| 41 | AY323977.2 | Italy | SARS-CoV-1-HSR1 | 29 751 | + | + | + | + | + | + | + | + | + | – | + | – | + | + | + | + |
| 42 | DQ182595.1 | China | SARS-CoV-1ZJ0301 | 29 706 | + | + | + | + | + | + | + | + | + | – | + | – | + | + | + | + |
*Isolates number 1,2,3 have their nt sequence in the nucleotide database without their expected orfs annotated.
Fig. 2.
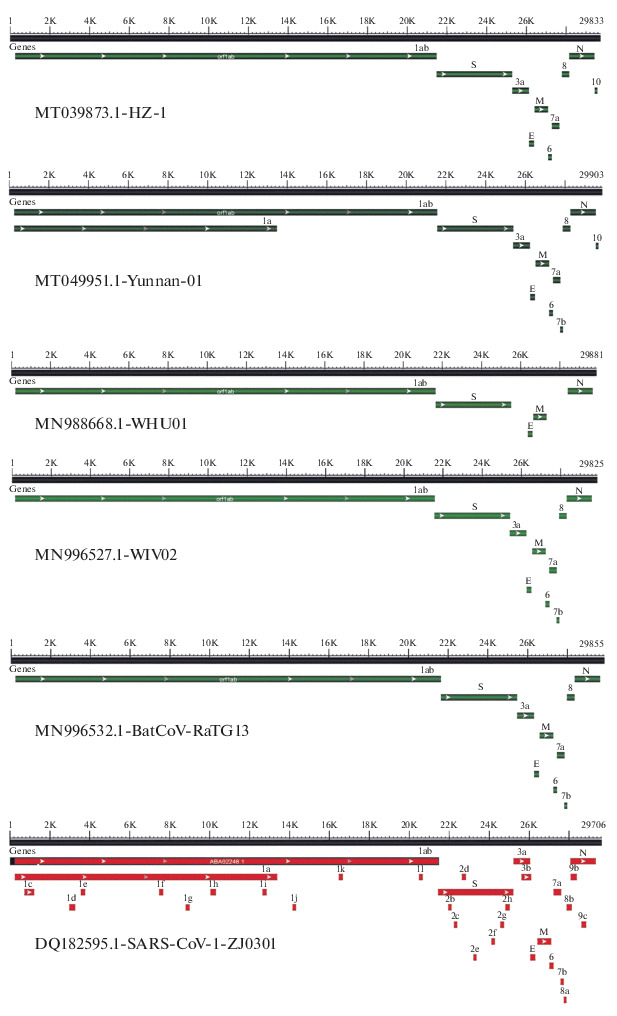
Map of expected orfs pattern of selected 4 COVID-19, 1 SARS-CoV isolate compared to the bat BatCoV-RaTG13 isolate. Accession number and isolate name are shown in each map panel.
DISCUSSION
The high identity (99.91 to 100%) in nucleotide sequence among COVID-19 isolates from various countries or the same country (Table 1) and their random clustering on the phylogenetic tree (Fig. 1) indicated that the reported COVID-19 isolates from different countries are highly similar and they belong to one COVID-19 strain. Also, the difference between COVID19 and SARS-CoV (11.66%) or COVID-19 and bat corona virus isolate BatCoV-RaTG13 (3.89%) strains distance COVID-19 as a novel viral strain that has not been identified before with different genome context. In addition, the low differences in nt sequence indicated by the nt SNPs among COVID-19 isolates and their distinction from SARS-CoV or bat corona virus support the same idea. Interestingly, collective base substitution rate for the studied isolates was 9.07 × 10–5. Base substitution rate of RNA viruses is the number of changed bases per cellular infection (generation). This is very difficult to determine because it is not known how many generations (infections) these isolates have gone before they had been sequenced, therefore this number is overestimation of SNPs rate in the studied strains because they should have gone through huge number of infections from being isolated from patients with symptoms. RNA viruses have mutation rate from 1 × 10–6 to 1 × 10–4 [22–24]. Our overestimated mutation rate of COVID-19 is still in the range of RNA viruses' mutation rate indicating that COVID-19 is a new viral strain.
COVID-19 isolates showed differences in the expected orf pattern from their highly similar genome suggesting a high level of expected complexity of the COVID-19 genome and its host cells. This is in agreeing with other previous reports. Production of extra orfs beside the main orfs by different retroviruses has been reported previously. Human endogenous retrovirus K (HERV-K) produces two variant proteins (np9, rec) of its full sequence or the 292 bp deficient gene respectively [25].
Our results are in agree with results reported from other several studies which indicated that COVID-19 is a novel corona virus and did not originate from other previous existing strains [15]. Similarly, it was reported that COVID-19 is not a mosaic virus nor did it originated from recombination events [14]. In the same line, a third study revealed that COVID-19 had 89% nt identity with Bat coronavirus (Bat SARS-like-CoVZXC21) and 82% to the SARS-CoV. Its orf3b produce a shorter protein and its orf8 encode for a secreted protein leaving the source of the COVID-19 undetectable [20].
Therefore, the most probable scenario is that this strain was transmitted from unknown organism and developed the ability to infect and transmit from human to human [16]. Based on this scenario, future studies are needed to screen wide range of animals that come in contact with human to search for the possible source of this viral strain; COVID-19. On the other hand, in the absence of its biological source, the possibility of it is being synthetic and it became public by a leakage from unknown biological facilities can not be rolled out at this time. This possibility is supported by the detection of unique isolate reported in 2004. The sequence of a new SARS-CoV strain was reported in 2004 and filled by Centre National de la Recherche Scientifique CNRS, Institut Pasteur, Universite Paris Diderot as patent to the European Patent Office (Patent no. EP1694829B1). This strain was isolated from a patient from Hanoi, Vietnam. The sequence of this strain was not deposited in the nucleotide database or anywhere else except in the patent itself. When we blasted the nt sequence of this strain against the nucleotide database it turned out the SARS-CoV Urbani isolate icSARS-MA (Acc no. MK062180.1) as the closest sequence with only 89.65% identity indicating its difference from reported SARS-CoV isolates at that time and consequently from any other reported corona virus or COVID-19 isolates.
COVID-19 Symptoms Implicate Its Unique Interaction with Human Biology
It is well known that COVID-19 has a wide range of symptoms in human ranging from no symptoms to death. The valid question here is that what makes people different in their response to COVID-19 infection? Based on the distinction of COVID-19 genome from SARS-CoV and Bat CoV, COVID-19 unique characteristics, similarity among COVID-19 isolates at the nt, some possible scenarios could be suggested for the discrepancies among humans in response to infection. In addition to age and health of the host person, some genomic scenarios are summarized in the following sections based on the current studies of human endogenous retroviruses (HERVs).
4.1.1 Human endogenous retroviruses (HERVs). HERVs are DNA sequences originated from recurrent integrations of the previous exogenous retrovirus [26, 27]. HERVs are one type of highly conserved transposable elements (TE). TE and HERVS make up 40 and 8% of our genome consecutively [28]. HERVs were first detected in the human genome in the 1970s [29]. HERVs are classified into three main groups; I (gamaretrovirus and epsilonretrovirus-like), II (betaretrovirus-like), III (spumaretrovirus-like) based on their phylogenetic relationship [30, 31]. Their integration allowed the vertical transmission of retroviral genomes along with the human genome across generation [32]. HERVs are inserted in the genome through the reverse transcription of viral RNA producing a double stranded DNA (provirus) using the viral reverse transcriptase [33] and then the integration of the provirus in the host genome by the viral integrase and other host proteins [34]. Integrated copies can be activated and become active infection. After integration, the proviral DNA produce mRNA that encodes for various viral proteins or reverse transcribed by viral reverse transcriptase into proviral DNA that has the capability of new integration cycle. HERVs have similar structure to exogenous retroviruses that is comprised of two long terminal repeats (LTRs) with internal gag (matrix protein), pro-pol (protease, reverse transcriptase, and integrase), env (envelope) viral genes [32]. Beside these main retroviral proteins, some retroviruses produce extra proteins. Accordingly, the env gene of the HERV-K encodes two different protein variants (np9, rec) using its full sequence or the 292 bp deficient variant respectively [25].
4.1.2. Impact of HERVs on human cells. HERVs have several different impacts on their host cells. Production of RNA and proteins from HERV sequence could have a role in the regulation of human genes and modulate immunity of the host [35, 36]. Although most of TEs have been silenced by accumulation of mutations or hypermethyaltion, some of them have been domesticated and still active in human biology [37]. For example, syncytins is a group of env proteins produced by different HERVs in mammals [38]. In human genome, two env genes HERV-W and HERV-FRD are involved in the production of env proteins syncytin-1 and -2, respectively [39]. They are involved in placental syncytiotrophoblast development, homeostasis [39, 40], and maternal immune tolerance to the growing fetus [41] respectively.
4.1.3. HERVs and regulation of human gene expression. At DNA level, huge number of HERV are integrated in the human genome and function as binding sites for transcription factors, alternative promoter, or splicing signals for cellular genes [37, 42–46] which indicates their role in regulation of transcription and human genome development. This could lead to upregulation, downregulation, suppression, or tissue-specific splicing of cellular genes [42, 45, 47]. Also, they represent a plethora of cis-acting regulatory elements that function as binding sites for the host trans-acting elements. The interplay between both types of elements makes up the gene regulation network in a cell [48, 49]. In the same line, the solitary LTRs, reminiscent of complete HERVs, can also regulate the host gene expression. Recurrent insertions of HERVs cause insertional mutations in the target genes and allelic homologous recombination [32]. For example, recombination between homologous HERV-I on chromosome Y cause microdeletion in the azoosperma factor and consequently male infertility [50]. In addition, HERVs can produce non-coding RNAs (ncRNAs) including microRNA and long ncRNA which furnish recognition motifs for RNA binding proteins or modulate the function of transcription factors [32]. Accordingly, HERV ncRNAs that has sequence similarity to human miRNA work as RNA sponges to bind other miRNA which are involved in the post-transcriptional regulation of gene expression [51]. This was the case in the regulation of embryonic stem cells in which an interaction of ncRNA (HPAT5) produced by HERVH to the let-7 miRNAs sequence [52]. Furthermore, in case of a HERV produces a protein which could function as regulator of the host gene expression during the virus life cycle and provide cellular functions during the cycle [36]. Interesting example is the HERV Gag and Rec proteins which are involved in the stability and translation the host cell mRNA [36]. For example, HML2 Rec was able to bind to 1 600 nt mRNAs of host embryonic cells and regulate their translation by ribosome in an early development process [53]. In the same line, Arc Gag-like protein produced by the Ty3/gypsy retrotransposon was suggested to coordinate brain neural cell communication indicating its role in the nervous system development [54, 55]. Specifically, Arc has been proposed to form capsids to carry mRNA between neuron cells via extracellular vesicles to be translated in the target neuron cell [56].
A group of HERVs spread in the human genome can form a coordinated regulatory network to regulate the expression of many host genes involved in the same pathway simultaneously [35, 47, 57]. For example, more than 30% of the human genome binding sites for the protein p53 were distributed in the genome by the HERV sequences and become the target network of p53 protein [58] leading to human genome plasticity and cellular networking. An interesting example for this plasticity is the MHC (major histocompatibility complex) locus which has been shown to have heavy integration of HERVs leading to its tremendous plasticity and hyper genetic variability [59]. Accordingly, the HERVK (HERVKC4) was integrated in the 9th intron of human complement C4A gene leading to its hyper variation [60, 61]. One vital example is the role of HERVs in the interferon (IFN) antiviral pathway in the innate immunity in the induction of adaptive immune response [62]. HERV integrations were involved in the development of INF network of INF inducible transcription enhancers in various mammalian genomes [35]. It was shown that deletion of HERV sequence near IFN gene suppressed the linked pathway [35]. Also, sequences of the HERV LTRs function as promoter or enhancer sites in response to IFN based activation [63]. The HERVK LTRs that have two IFN-stimulated response elements (ISREs) were induced by the IFN cascade in response to inflammation [64].
4.1.4. HERVs and human immune modulation. Products of ancient integrated HERV represent the border line between human self and microbial non-self molecules and can be tolerated by human immune system or induce human immunity giving rise to autoimmune diseases. The innate immune pathways induced by HERVs’ products are the ones that function in the exogenous antiviral infection [65]. In humans, Toll Like Receptors (TLRs) and cytosolic pattern recognition receptors (cytPRRs) can recognize HERV products and lead to induction of immune response. This was reported in the case of autoimmune diseases and cancer [66, 67].
Recognition of viral molecules by innate immune receptors induces inflammatory molecules including IFN, cytokines, and chemokines invoking the antiviral response. This group of molecules activates the adaptive immune response through the activation of T and B cells. Both immune responses are required to fight exogenous viral infection and finally stop this activated response after infection. In case of HERV products, their continuous presence in the host cells provokes chronic stimulation of the host immune response resembling the chronic stimulation of immune response in autoimmune and inflammatory diseases caused by exogenous retroviral molecules [67–70]. The induced antiviral response activated by HERV products cause vicious circle in which the produced inflammatory molecules and epigenetic dysregulation further upregulated HERV expression [65, 71, 72]. Also, peptides produced from HERVs were implicated in the suppression of immune response. This includes the env proteins that has immunosuppressive conserved domain (ISD) in retroviral env proteins. For example, ISD from HERVs function in the maternal immune tolerance during pregnancy [38, 41].
4.1.5. HERVs and exogenous viral infection. It is well documented that HERVs can contribute negatively or positively during exogenous viral infection [67]. Infection by some viruses including HIV, herpesviruses and influenza changed HERV expression [73–75]. In this regard exogenous infection could cooperatively upregulate the HERV expression and increase the immune response [67]. Also, HERV products could play a protective role against exogenous viral infection [36]. For example, production of HERV antisense RNA develops protection against exogenous infection by viruses with complementary RNA [65, 76]. Some studies reported that products of HERV function as pathogen-associated molecular patterns (PAMPs) which is able to induce receptors for host defense system [49, 65]. In addition, some of their products mimic antigens for stimulating specific B and T cells [77, 78]. This explains the role of HERVs in autoimmune and inflammatory diseases. On the other hand, they had a role in suppressing the immunity of host cells as they have been involved in maternal immune suppression and protection of excessive imune activation [79, 80].
Possible Role for HERVs in COVID-19 Infection and Symptoms
HERVs could modulate the infection and symptoms in the case of exogenous COVID-19 infection in different possible ways. First, HERVs or their products could compromise the immune system and facilitate the infection and penetrance of the virus to human cells. Also, individuals with high levels of the ACE2 receptor could be an easy target for the virus, especially those with high blood pressure and various types of stress. Second, different isolates of the virus can use the host cell to produce different protein sets (orf pattern) that can use the host cells and compromise the host immune system with different efficiencies. This will result in spectrum of disease severity and possibly death. In this study, different isolates from the same country (China) or from different countries are expected to produce various orf patterns. Some of the produced orfs which is the enzyme responsible for methylation of the 2' carbon of the ribose sugar of viral RNA. This modification of viral RNA makes it undetectable by the host immune system and effectively infects human cells [81]. Third, HERVs could produce protein products that complement the viral set of orfs in its entry, infection, replication, packaging, and integration in the human genome. In addition, partial proviral genomes of previous integration can produce some enzymes required for the replication of viral isolates that do not have the infection ability. For example, one animal isolate which does not have the capability to infect human could transfer to human and find in this individual’s genome some proviral genes that complement the animal strain to be infectious and able to cause the symptoms. Fourth, Corona virus genome can only produce its effective proteins for viral reproduction with -1ribosomal slippage at the translation start site. HERVs may produce proteins or miRNA that modulates the translation start for the ribosome changing the pattern of COVID-19 orfs in different human hosts. This leads to different course of symptoms and severity of the COVID-19 infection.
Long term studies are urgent to be conducted on the COVID-19 and other retroviruses that attach human to validate all of these possibilities for future safety and better management of future pandemics like COVID-19. Also, intensive studies are needed to survey human populations (expecially elders and immune compromised) for their HERV loads and link this to their predisposition for other autoimmune diseases, cancer, and their risk for exogenous viral infection.
CONCLUSIONS
Our results conclude that COVID-19 did not originate from a known biological source or other previously characterized strains. COVID-19 isolates used in this study showed high similarity at the nt sequence, yet they differed greatly in the expected orf pattern from their similar genomes. The most probable scenario is that this strain was transmitted from unknown organism and has/or has developed the ability to infect human cells as well as to transmit from human to human. On the other hand, in the absence of its biological source, the possibility of it is being synthetic and it became public from unknown biological facilities can not be rolled out at this time.
FUNDING
This work was funded by Taif University Researchers Supporting Project number (TURSP-2020/75), Taif University, Taif, Saudi Arabia.
COMPLIANCE WITH ETHICAL STANDARDS
The authors declare that they have no conflict of interest.
This article does not contain any studies involving animals or human participants performed by any of the authors.
AUTHOR CONTRIBUTIONS
Authors have contributed equally to this manuscript.
REFERENCES
- 1.Kahn J.S., McIntosh K. History and recent advances in coronavirus discovery. Pediatr. Infect. Dis. J. 2005;24:S223–S226. doi: 10.1097/01.inf.0000188166.17324.60. [DOI] [PubMed] [Google Scholar]
- 2.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Meth. Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drosten C. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 4.Ksiazek T.G. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 5.Peiris J.S. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paules C.I., Marston H.D., Fauci A.S. Coronavirus infections more than just the common cold. JAMA. 2020;323:707–708. doi: 10.1001/jama.2020.0757. [DOI] [PubMed] [Google Scholar]
- 7.Guan Y. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 8.Lau S.K. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U. S. A. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2018;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaki A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–20. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 11.Hajjar S.A., Memish Z.A., McIntosh K. Middle East respiratory syndrome coronavirus (MERS-CoV): a perpetual challenge. Ann. Saudi Med. 2013;33:427–436. doi: 10.5144/0256-4947.2013.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alagaili A.N. Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. mBio. 2014;5:e00884–14. doi: 10.1128/mBio.00884-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ithete N.L., Stoffberg S., Corman V.M. Close relative of human Middle East respiratory syndrome coronavirus in bat. South Africa. Emerg. Infect. Dis. 2013;19:1697–1699. doi: 10.3201/eid1910.130946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paraskevis D., Kostaki E.G., Magiorkinis G., Panayiotakopoulos G., Sourvinos G., Tsiodras S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect. Genet. Evol. 2020;79:104212. doi: 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu N. A novel coronavirus from patients with pneumonia in China, 2020. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perlman S. Another decade, another coronavirus. N. Engl. J. Med. 2020;382:760–762. doi: 10.1056/NEJMe2001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.World Health Organization (WHO), Geneva, Switzerland, 2020.
- 18.Hui D.S. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—the latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.World Health Organization (WHO), Novel Coronavirus (2019-nCoV) Situation Report–162, June30, 2020, Geneva, Switzerland, 2020.
- 20.Chan J.F., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microb. Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wan, Y., Shang, J., Graham, R., Baric, R.S., and Li. F., Receptor recognition y novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS, J. Virol., Pii. 2020, JVI.00127-20. 10.1128/JVI.00127-20 [DOI] [PMC free article] [PubMed]
- 22.Duffy S., Shackelton L.A., Holmes E.C. Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet. 2008;9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- 23.Sanjuan R., Nebot M.R., Chirico N., Mansky L.M., Belshaw R. Viral mutation rates. J. Virol. 2010;84:9733–9734. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Retel C., Markle H., Becks L., Feulner P.G.D. Ecological and evolutionary processes shaping viral genetic diversity. Viruses. 2019;11:220. doi: 10.3390/v11030220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Armbruester V., Sauter M., Krautkraemer E. A novel gene from the human endogenous retrovirus K expressed in transformed cells. Clin. Cancer Res. 2002;8:1800–1807. [PubMed] [Google Scholar]
- 26.Bannert N., Kurth R. The evolutionary dynamics of human endogenous retroviral families. Ann. Rev. Genom. Hum. Genet. 2006;7:149–173. doi: 10.1146/annurev.genom.7.080505.115700. [DOI] [PubMed] [Google Scholar]
- 27.Vargiu L., Rodriguez-Tomé P., Sperber G.O. Classification and characterization of human endogenous retroviruses; mosaic forms are common. Retrovirology. 2016;13:7. doi: 10.1186/s12977-015-0232-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lander E.S., Linton L.M., Birren B. Initial sequencing and analysis of the human genome. Nature. 2001;412:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 29.Bronson D.L., Fraley E.E., Fogh J., Kalter S.S. Induction of retrovirus particles in human testicular tumor (Tera-1) cell cultures: an electron microscopic study. J. Natl. Cancer Inst. 1979;63:337–339. [PubMed] [Google Scholar]
- 30.Jern P., Sperber G.O., Blomberg J. Use of endogenous retroviral sequences (ERVs) and structural markers for retroviral phylogenetic inference and taxonomy. Retrovirology. 2005;2:50. doi: 10.1186/1742-4690-2-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blomberg J., Benachenhou F., Blikstad V., Sperber G., Mayer J. Classification and nomenclature of endogenous retroviral sequences (ERVs): problems and recommendations. Gene. 2009;448:115–123. doi: 10.1016/j.gene.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 32.Grandi N., Tramontano E. Human endogenous retroviruses are ancient acquired elements still shaping innate immune responses. Front. Immunol. 2018;9:2039. doi: 10.3389/fimmu.2018.02039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esposito, F., Corona, A., and Tramontano, E., HIV-1 reverse transcriptase still remains a new drug target: structure, function, classical inhibitors, and new inhibitors with innovative mechanisms of actions, Mol. Biol. Int., 2012, p. 586401. 10.1155/2012/586401 [DOI] [PMC free article] [PubMed]
- 34.Esposito F., Tramontano E. Past and future. Current drugs targeting HIV-1 integrase and reverse transcriptase-associated ribonuclease H activity: single and dual active site inhibitors. Antivir. Chem. Chemother. 2013;23:129–144. doi: 10.3851/IMP2690. [DOI] [PubMed] [Google Scholar]
- 35.Chuong E.B., Elde N.C., Feschotte C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science. 2016;351:1083–7. doi: 10.1126/science.aad5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frank J.A., Feschotte C. Co-option of endogenous viral sequences for host cell function. Curr. Opin. Virol. 2017;25:81–89. doi: 10.1016/j.coviro.2017.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feschotte C., Gilbert C. Endogenous viruses: insights into viral evolution and impact on host biology. Nat. Rev. Genet. 2012;13:283–296. doi: 10.1038/nrg3199. [DOI] [PubMed] [Google Scholar]
- 38.Lavialle C., Cornelis G., Dupressoir A. Paleovirology of “syncytins,” retroviral env genes exapted for a role in placentation. Philos. Trans R. Soc. Lond. B. Biol. Sci. 2013;368:20120507. doi: 10.1098/rstb.2012.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mi S., Lee X., Li X., Veldman G.M. Syncytin is a captive retroviral envelope protein involved. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 40.Malassine A., Handschuh K., Tsatsaris V. Expression of HERV-WEnv glycoprotein (syncytin) in the extravillous trophoblast of first trimester human placenta. Placenta. 2005;26:556–62. doi: 10.1016/j.placenta.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 41.Mangeney M., Renard M., Schlecht-Louf G. Placental syncytins: genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins. Proc. Natl. Acad. Sci. U. S. A. 2007;104:20534–9. doi: 10.1073/pnas.0707873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van de Lagemaat L.N., Landry J.R., Mager D.L., Medstrand P. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet. 2003;19:530–536. doi: 10.1016/j.tig.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 43.Bourque G., Leong B., Vega V.B. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18:1752–1762. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sundaram V., Cheng Y., Ma Z. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 2014;24:1963–1976. doi: 10.1101/gr.168872.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trizzino M., Park Y., Holsbach-Beltrame M. Transposable elements are the primary source of novelty in primate gene regulation. Genome Res. 2017;27:1623–1633. doi: 10.1101/gr.218149.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ito J., Sugimoto R., Nakaoka H. Systematic identification and characterization of regulatory elements derived from human endogenous retroviruses. PLoS Genet. 2017;13:e1006883. doi: 10.1371/journal.pgen.1006883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feschotte, C., The contribution of transposable elements ot the evolution of regulatory networks, Nat. Rev. Genet., 2008, vol. 397–405. 10.1038/nrg2337 [DOI] [PMC free article] [PubMed]
- 48.Goke J., Ng H.H. CTRL+INSERT: retrotransposons and their contribution to regulation and innovation of the transcriptome. EMBO Rep. 2016;17:1131–1144. doi: 10.15252/embr.201642743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolff F., Leisch M., Greil R., Risch A., Pleyer L. The double-edged sword of (re)expression of genes by hypomethylating agents: from viral mimicry to exploitation as priming agents for targeted immune checkpoint modulation. Cell Commun. Signal. 2017;15:13. doi: 10.1186/s12964-017-0168-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kamp C., Hirschmann P., Voss H., Huellen K., Vogt P.H. Two long homologous retroviral sequence blocks in proximal Yq11 cause AZFa microdeletions as a result of intrachromosomal recombination events. Hum. Mol. Genet. 2000;9:2563–2572. doi: 10.1093/hmg/9.17.2563. [DOI] [PubMed] [Google Scholar]
- 51.Wang Y., Xu Z., Jiang J. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev. Cell. 2013;25:69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 52.Durruthy-Durruthy J., Sebastiano V., Wossidlo M. The primate-specific noncoding RNA HPAT5 regulates pluripotency during human preimplantation development and nuclear reprogramming. Nat. Genet. 2016;48:44–52. doi: 10.1038/ng.3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grow E.J., Flynn R. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015;522:221–5. doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mikuni T., Uesaka N., Okuno H. Arc/Arg3.1 is a postsynaptic mediator of activity-dependent synapse elimination in the developing cerebellum. Neuron. 2013;78:1024–1035. doi: 10.1016/j.neuron.2013.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang W., Wu J., Ward M.D. Structural basis of arc binding to synaptic proteins: implications for cognitive disease. Neuron. 2015;86:490–500. doi: 10.1016/j.neuron.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pastuzyn, E.D., Day, C.E., Kearns, R.B., et al. The neuronal gene arc encodes a repurposed retrotransposon gag protein that mediates intercellular RNA transfer, Cell, 2018, vol. 172, pp. 275–288. e18. [DOI] [PMC free article] [PubMed]
- 57.Chuong E.B., Elde N.C., Feschotte C. Regulatory activities of transposable elements: from conflicts to benefits. Nat. Rev. Genet. 2017;18:71–86. doi: 10.1038/nrg.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang T., Zeng J., Lowe C.B. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc. Natl. Acad. Sci. U. S. A. 2007;104:18613–8. doi: 10.1073/pnas.0703637104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Andersson G., Svensson A.C., Setterblad N., Rask L. Retroelements in the human MHC class II region. Trends Genet. 1998;14:109–114. doi: 10.1016/S0168-9525(97)01359-0. [DOI] [PubMed] [Google Scholar]
- 60.Grandi N., Cadeddu M., Pisano M.P., Esposito F., Blomberg J., Tramontano E. Identification of a novel HERV-K(HML10): comprehensive characterization and comparative analysis in non-human primates provide insights about HML10 proviruses structure and diffusion. Mob. DNA. 2017;8:15. doi: 10.1186/s13100-017-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mack M., Bender C., Schneider P.M. Detection of retroviral antisense transcripts and promoter activity of the HERV-K(C4) insertion in the MHC III region. Immunogenetics. 2004;56:321–332. doi: 10.1007/s00251-004-0705-y. [DOI] [PubMed] [Google Scholar]
- 62.Nehyba J., Hrdlicková R., Bose H.R. Dynamic evolution of immune system regulators: the history of the interferon regulatory factor family. Mol. Biol. Evol. 2009;26:2539–2550. doi: 10.1093/molbev/msp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Katoh I., Kurata S. Association of endogenous retroviruses and long terminal repeats with human disorders. Front. Oncol. 2013;3:234. doi: 10.3389/fonc.2013.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Manghera M., Ferguson-Parry J., Lin R., Douville R.N. NF-kB and IRF1 induce endogenous retrovirus K expression via interferon-stimulated response elements in its 5' long terminal repeat. J. Virol. 2016;90:9338–9349. doi: 10.1128/JVI.01503-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hurst T.P., Magiorkinis G. Activation of the innate immune response by endogenous retroviruses. J. Gen. Virol. 2015;96:1207–1218. doi: 10.1099/vir.0.000017. [DOI] [PubMed] [Google Scholar]
- 66.Grandi N., Tramontano E. HERV envelope proteins: physiological role and pathogenic potential in cancer and autoimmunity. Front. Microbiol. 2018;9:462. doi: 10.3389/fmicb.2018.00462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grandi N., Tramontano E. Type W human endogenous retrovirus (HERVW) integrations and their mobilization by L1 machinery: contribution to the human transcriptome and impact on the host physiopathology. Viruses. 2017;9:162. doi: 10.3390/v9070162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trela M., Nelson P.N., Rylance P.B. The role of molecular mimicry and other factors in the association of human endogenous retroviruses and autoimmunity. APMIS. 2016;124:88–104. doi: 10.1111/apm.12487. [DOI] [PubMed] [Google Scholar]
- 69.Nelson P., Rylance P., Roden D., Trela M., Tugnet N. Viruses as potential pathogenic agents in systemic lupus erythematosus. Lupus. 2014;23:596–605. doi: 10.1177/0961203314531637. [DOI] [PubMed] [Google Scholar]
- 70.Mameli G., Erre G.L., Caggiu E. Identification of a HERV-K env surface peptide highly recognized in Rheumatoid Arthritis (RA) patients: a cross-sectional case–control study. Clin. Exp. Immunol. 2017;189:127–131. doi: 10.1111/cei.12964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Manghera M., Douville R.N. Endogenous retrovirus-K promoter: a landing strip for inflammatory transcription factors? Retrovirology. 2013;10:16. doi: 10.1186/1742-4690-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hurst T.P., Magiorkinis G. Epigenetic control of human endogenous retrovirus expression: focus on regulation of long-terminal repeats (LTRs) Viruses. 2017;9:1–13. doi: 10.3390/v9060130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nellaker C., Yao Y., Jones-Brando L. Transactivation of elements in the human endogenous retrovirus W family by viral infection. Retrovirology. 2006;3:44. doi: 10.1186/1742-4690-3-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li F., Nellaker C., Sabunciyan S. Transcriptional derepression of the ERVWE1 locus following influenza A virus infection. J. Virol. 2014;88:4328–4337. doi: 10.1128/JVI.03628-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Young G.R., Mavrommatis B., Kassiotis G. Microarray analysis reveals global modulation of endogenous retroelement transcription by microbes. Retrovirology. 2014;11:59. doi: 10.1186/1742-4690-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gurtler C., Bowie A.G. Innate immune detection of microbial nucleic acids. Trends Microbiol. 2013;21:413–420. doi: 10.1016/j.tim.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roulois D., Loo Y.H., Singhania R. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. 2015;162:961–973. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ramasamy R., Joseph B., Whittall T. Potential molecular mimicry between the human endogenous retrovirus W family envelope proteins and myelin proteins in multiple sclerosis. Immunol. Lett. 2017;183:79–85. doi: 10.1016/j.imlet.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 79.Dupressoir A., Lavialle C., Heidmann T. From ancestral infectious retroviruses to bona fide cellular genes: role of the captured syncytins in placentation. Placenta. 2012;33:663–671. doi: 10.1016/j.placenta.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 80.Hummel J., Kammerer U., Müller N., Avota E., Schneider-Schaulies S. Human endogenous retrovirus envelope proteins target dendritic cells to suppress T-cell activation. Eur. J. Immunol. 2015;45:1748–1759. doi: 10.1002/eji.201445366. [DOI] [PubMed] [Google Scholar]
- 81.Zust R., Cervantes-Barragan L., Habjan M., Maier R., Neuman B.W., Ziebuhr J., Szretter K.J., Baker S.C., Barchet W., Diamond M.S., Siddell S.G., Ludewig B., Thiel V. Ribose 2'-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]


