Introduction
It is increasingly evident that patients with hypertension are at high risk for COVID-19 and exhibit gastrointestinal (GI) symptoms suggesting that impaired gut-lung communications could be responsible, at least in part, for the multi-organ pathologies including cardiovascular manifestations of this disease.1 Higher expression of angiotensin-converting enzyme-2 (ACE2) and transmembrane protease serine-2 (TMPRSS2), key molecules in SARS-CoV-2 infection, in gut epithelium from spontaneously hypertensive rats (SHR) supports this view2. Finally, changes in gut microbiome associated with short-chain fatty acids, particularly butyrate, is found in both hypertension and COVID-19.1 Butyrate is a histone-deacetylase (HDAC) inhibitor that maintains acetylation of histones, affecting chromatin organization and gene expression. Therefore, we sought to test the hypothesis that increased risk of COVID-19 in hypertension could, in part, be due to cumulative depletion of butyrate-producing gut bacteria leading to decreases in butyrate.1 Therefore, treatment with this short chain fatty acid would regulate ACE2 and its partners influencing antiviral genes. This could be critical in the control of gut viral infection and rebalancing of the gut-lung axis.
Results and Discussion
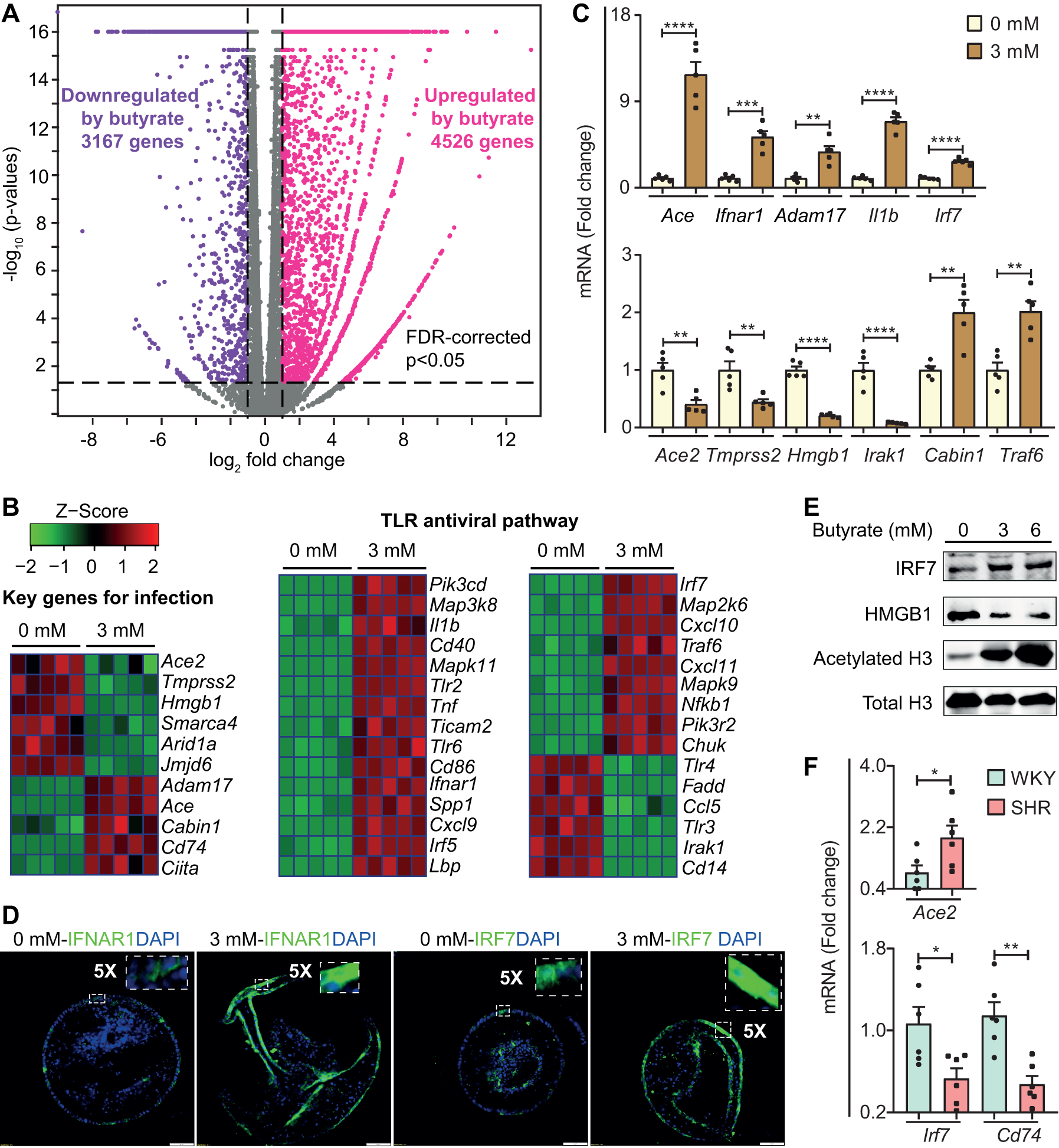
We performed high-throughput RNA-seq to determine the effects of butyrate on transcriptional program of colonic organoids. We observed 4526 upregulated genes (Pink dots) and 3167 downregulated genes (Purple dots) in butyrate-treated organoids (Figure [A]). Many of these genes were related to SARS-CoV-2 infection: reads per kilobase per million (RPKM) of Ace2, and Tmprss2 that facilitate SARS-CoV-2 entry into host cells were both significantly decreased by butyrate and qPCR confirmed this observation (Figure [B]/[C]). In contrast, Adam17, a metallopeptidase involved in shedding of ACE2, was upregulated by butyrate (Figure [B]/[C]]). These data suggest that butyrate could suppress viral infectivity by decreasing membrane ACE2, via downregulating its transcription and increasing its shedding, and decreasing activation of viral spike protein. Butyrate increased expression of angiotensin‐converting enzyme (ACE) (Figure [B]/[C]), which could have deleterious or beneficial consequences: increased angiotensin-II is vasoconstrictive while decreased bradykinin is beneficial in controlling inflammation in COVID-19 patients.1 Bradykinin-B2 receptor antagonist, icatibant, improves oxygenation in early stage of COVID-19.1 High-mobility group protein-1 (HMGB1) regulates transcription under conditions of stress, and is critical for SARS-CoV-2 replication at the post-entry phase.3 We found RPKM of Hmgb1 decreased ~5-fold by butyrate (Figure [B]/[C]). HMGB1 protein was decreased ~3-fold by butyrate (Figure [E]). These results indicate that butyrate may control SARS-CoV-2 replication through downregulation of Hmgb1 expression.
Figure. Butyrate regulates SARS-COV-2 infection and TLR antiviral pathway relevant genes in gut epithelium.
Colonic three-dimensional (3D) organoid culture: Primary colonic crypts were isolated from proximal colons of male 14-week-old Wister Kyoto rats (WKY, Charles River) with gentle cell dissociation reagent (STEMCELL Technologies) including 2 mM EDTA for 50 mins, grown and maintained as 3D-spheroid cultures in Matrigel (BD Biosciences) containing organoids growth medium (STEMCELL Technologies) with growth factors (Noggin, HGF, FGF and IGF-I; Biolegend) as described previously.2 Colonic organoids were cultured for 4 days, then treated with 3.0 mM butyrate (Sigma-Aldrich) for 24 hours in following experiments. RNA-seq: Total RNA was extracted from colonic organoids with RNeasy Plus Mini Kit (Qiagen). cDNA was generated using a SMART-Seq HT kit (Takara Bio) and RNA-Seq libraries were constructed with Nextera DNA Flex Library Prep kit and Nextera DNA Unique Dual Indexes Set A (Illumina) and sequenced on NovaSeq6000 instrument (Illumina) at the University of Florida NextGen DNA Sequencing Core Facility. RNA seq analysis was performed using CLC genomics workbench (Qiagen).
(A) Volcano plot of genes altered by butyrate in colonic organoids. (False discovery rate (FDR)-corrected p<0.05, absolute fold change≥2, purple dots show downregulated genes by butyrate, pink dots show upregulated genes by butyrate). Gray plots indicate genes that were not differentially expressed (n=5).
(B) Heat map of gene clusters of differentially expressed genes between control and 3 mM butyrate treatment in organoids (All the genes shown in the heat map, p<0.05).
(C) mRNA levels of representative genes by qPCR. (n=5, 0 mM and 3 mM). Fold change relative to control treatment. Values are means ±SEM. **p<0.01, ***p<0.001 and ****p<0.0001, unpaired T test. qPCR: total RNA was purified with RNeasy Plus Mini Kit (Qiagen) and reverse-transcribed using iScript Reverse Transcription Supermix (Bio-Rad). Finally, qPCR (ABI Prism 7600) was performed with Taqman universal PCR master mix and specific probes (Ace, Ifnar1, Adam17, II1b, Irf7, Ace2, Tmprss2, Hmgb1, Irk1, Cabin1 and Traf6). B2m (Beta-2-Microglobulin) was used to as the reference gene for normalization.
(D) Confocal images showing expression of IFNAR1 and IRF7 (20X objective, Scale bar: 50 μm). Immunofluorescence imaging: organoids were fixed, permeabilized, and blocked, followed by incubation with IFNAR1 (Novus) and IRF7 (Invitrogen) antibodies. The organoids were then stained with DAPI and Alexa Fluro 488 secondary antibody (Thermo Fisher Scientific) and imaged with a confocal microscope (Olympus IX81).
(E) Protein levels of IRF7, HMGB1, acetylated-histone H3 (Lys9) and Histone H3. Western blot: organoids were treated with 3 and 6 mM butyrate, homogenized in 2% SDS-Tris buffer (pH=7.5). Protein samples (6.7 mg/ml) were separated on 12% TGX precast gels and transferred to Polyvinylidene fluoride membranes (Bio-Rad). Membranes were incubated with HMGB1 (Invitrogen), IRF7 (Invitrogen), acetylated-histone H3 (Lys9) (Cell signaling) and Histone-H3 antibody (Cell signaling) followed by Rabbit and Mouse IRDY 680RD secondary antibody (Li-Cor Biosciences). Protein bands were detected using the Odyssey infrared imaging system.
(F) mRNA levels of Ace2, Cd74 and Irf7 by qPCR in proximal colon of SHR and WKY rats (n=6), mean systolic blood pressures 209±5 and 132±6 mmHg, respectively, measured by tail-cuff plethysmography. Fold change relative to WKY. Gapdh was used as a reference gene for normalization. Values are means ±SEM. *p<0.05 and **p<0.01, unpaired T test.
MHC class-II transactivator (CIITA) activates expression of CD74 isoform-p41, which inhibits SARS-CoV-2 and Ebola virus entry by blocking endosomal entry pathway.4 Butyrate increased expression of Ciita and Cd74 by ~5- and ~32-times, respectively (Figure [B]). Additionally, calcineurin binding protein-1 (Cabin1) is antiviral in SARS-CoV-2-infected Vero-E6 cells,3 and upregulated by butyrate (Figure [B]/[C]).
Another clinical feature of COVID-19 patients is drastically impaired antiviral immunity with decreased type-I and -III interferons.5 Our analysis disclosed that toll-like receptor (TLR) signaling pathway, an innate immune system targeting viruses, was significantly enriched by butyrate (NES=1.6329, adjusted p value= 0.013). Heat map showed elevated expression of genes in this antiviral pathway (Figure [B]), that were inhibited in the GI tract of COVID-19 patients such as Il1b (Interleukin-1-beta) and Irf7 (interferon regulatory factor-7).5 Further validation confirmed upregulation of Il1b, Irf7 and Ifnar1 (Interferon-alpha/beta receptor) by butyrate at mRNA and protein levels (Figure [C]/[D]/[E]). In addition, cluster of differentiation-14 (Cd14) and chemokine ligand-5 (Ccl5) expression were inhibited by butyrate (Figure [B]); blocking their expression is beneficial to COVID-19 patients.6
Butyrate is a HDAC inhibitor that increased acetylation of histone-H3 (H3) while it decreased total H3 in colonic organoids (Figure [E]). Additionally, other DEGs may be associated with HDAC inhibition: histone arginine-demethylase Jmjd6, chromatin-remodeling complex members Smarca4 and Arid1a, whose expression exacerbates SARS-CoV-2-induced cell death,3 were downregulated by butyrate (Figure [B]/[C]), suggesting that butyrate may have the potential to prevent cell death by downregulating these genes in gut epithelium.
Hypertension, one of the most common comorbidities with unfavorable outcomes shares many common pathophysiological features with COVID-19 including inflammation, endothelial dysfunction, gut microbial dysbiosis, decreased butyrate-producing bacteria, increased gut leakiness and altered renin-angiotensin system.1 Our data showed that mRNA for Ace2 was increased while Cd74 and Irf7 was decreased in proximal colon of SHR (Figure [F]). These data are consistent with the premise of greater infectivity of SARS-COV-2 with hypertension.4,5 Together, they suggest that dysregulation of SARS-COV-2 infection-relevant genes in the colon could alter epithelial-microbiota communication in hypertension. Therefore, GI regulation of butyrate may be key in increased comorbidity with hypertension in COVID-19.
In conclusion, we have demonstrated that butyrate downregulates genes essential for SARS-CoV-2 infection but also upregulates TLR and other antiviral pathways. Therefore, this implies that increased risk of COVID-19 in hypertension is partly due to cumulative depletion of butyrate-producing bacteria in the gut. Further studies are warranted to validate this concept using commensal-derived butyrate and effective butyrate derivatives against SARS-CoV-2 in preclinical and early stage COVID-19 settings.
Sources of Funding
This work was supported by the following grants: NIH NHLBI grants HL033610, HL110170 and HL132448
Footnotes
Disclosures
None
References
- 1.Sharma RK, Stevens BR, Obukhov AG, Grant MB, Oudit GY, Li Q, Richards EM, Pepine CJ and Raizada MK ACE2 (Angiotensin-Converting Enzyme 2) in Cardiopulmonary Diseases: Ramifications for the Control of SARS-CoV-2. Hypertension 2020; 76(3): 651–661. 10.1161/HYPERTENSIONAHA.120.15595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li J, Stevens BR, Richards EM and Raizada MK SARS-CoV-2 Receptor ACE2 (Angiotensin-Converting Enzyme 2) Is Upregulated in Colonic Organoids From Hypertensive Rats. Hypertension 2020; 76(3): e26–e28. 10.1161/HYPERTENSIONAHA.120.15725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei J, Alfajaro M, Hanna R, DeWeirdt P, Strine M, Lu-Culligan W, Zhang SM, Graziano V, Schmitz C, Chen J, et al. Genome-wide CRISPR screen reveals host genes that regulate SARS-CoV-2 infection. Biorxiv 2020; 10.1101/2020.06.16.155101 [DOI] [Google Scholar]
- 4.Bruchez A, Sha K, Johnson J, Chen L, Stefani C, McConnell H, Gaucherand L, Prins R, Matreyek KA, Hume AJ, et al. MHC class II transactivator CIITA induces cell resistance to Ebola virus and SARS-like coronaviruses. Science 2020; doi: 10.1126/science.abb3753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Møller R, Jordan TX, Oishi K, Panis M, Sachs D, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 2020;181(5):1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin TR, Wurfel MM, Zanoni I, Ulevitch R. Targeting innate immunity by blocking CD14: novel approach to control inflammation and organ dysfunction in COVID-19 illness. EBioMedicine. 2020; 57(1):102836 10.1016/j.ebiom.2020.102836. [DOI] [PMC free article] [PubMed] [Google Scholar]


