Abstract
Chronic lymphocytic leukemia (CLL) is the most common adult leukemia in Western countries and is characterized by the clonal expansion of mature CD5+ B cells. There have been substantial advances in the field of CLL research in the last decade, including the identification of recurrent mutations, and clarification of clonal architectures, signaling molecules, and the multistep leukemogenic process, providing a comprehensive understanding of CLL pathogenesis. Furthermore, the development of therapeutic approaches, especially that of molecular target therapies against CLL, has markedly improved the standard of care for CLL. This review focuses on the recent insights made in CLL leukemogenesis and the development of novel therapeutic strategies.
Keywords: chronic lymphocytic leukemia, multistep leukemogenesis, BCR signaling, novel drugs
INTRODUCTION
Chronic lymphocytic leukemia (CLL) is the most common adult leukemia. It is characterized by the clonal expansion of CD5+ mature B cells in the blood, bone marrow, and lymphoid tissues. 1 CLL typically affects elderly people. 2 Recent advances in next-generation sequencing (NGS) technologies revealed recurrent somatic mutations and identified the molecular pathways involved in CLL pathogenesis. Furthermore, the analysis of clonal architecture clarified that CLL genomes exhibit heterogeneity between patients with CLL and within cells of individual patients. 3 Such advances in genetic lesion analysis significantly improved our understanding of the leukemogenic process of CLL. Moreover, CLL leukemogenesis has been described as a multistep process originating from immature hematopoietic stem cells (HSCs). 4 , 5 Thus, our understanding of CLL biology has significantly improved in the last decade. The clinical efficacy of novel drugs against CLL, such as the tyrosine kinase inhibitor ibrutinib and the B cell lymphoma 2 (BCL2) antagonist venetoclax, has markedly improved the standard of care for specific subsets of patients with CLL. This review focuses on recent insights into CLL leukemogenesis, emphasizing the immunological aspects of B cell receptors (BCRs), genetic lesions, and the process of multistep CLL leukemogenesis. It also describes the advances in the field of basic CLL research and the development of novel therapeutic strategies against CLL.
BIOLOGICAL AND IMMUNOLOGICAL FEATURES OF CLL
CLL is a B cell malignancy characterized by accumulating mature clonal CD5-expressing B cells. 6 - 8 The CLL prevalence markedly increases with age. CLL cells express functional BCRs on their surfaces. 6 , 9 , 10 CLL is classified into two subgroups based on the presence of somatic hypermutations within the variable regions of the immunoglobulin heavy chain gene (IGHV). Patients with CLL with mutated IGHV (IGHV-M CLL) have a more favorable prognosis than those with unmutated IGHV (IGHV-UM CLL). 11 The somatic hypermutations occur in the process of normal B cell development in the germinal centers during the naïve-to-memory B cell transition. Initial studies proposed distinct origins of the two types of CLL, with IGHV-UM CLL originating from naïve B cells and IGHV-M CLL originating from antigen-experienced B cells, including memory B cells. However, immunological analysis of CLL-BCRs revealed that both types recognized self-antigens, at least in vitro, suggesting that CLL originates from self-reactive B cell precursors, irrespective of the IGHV mutation status. 12 , 13 The self-reactivity of BCRs from CLL is one of the most extensively investigated biological features of human CLL. BCR signaling is constitutively activated and is an important biological feature of CLL cells, 14 providing rationale for BCR signaling-targeted therapies for CLL. Of note, the BCRs of ~1% of CLL cases express a nearly identical amino acid sequence of heavy chain complementarity-determining region 3 (HCDR3), and the BCRs of CLL cells can be classified as specific stereotyped BCRs in ~30% of CLL cases. 15 - 17 These observations led to the hypothesis that some common self-antigens recognized by CLL-BCRs drive clonal expansion and play a significant role in CLL pathogenesis. Consistent with this hypothesis, recent studies successfully identified common auto- or exoantigens, including myosin heavy chain IIA, 18 β-(1,6)-glucan, 19 and rheumatoid factors (RFs). 20 , 21 Such antigens are considered drivers of the expansion of CLL cells via BCR signaling.
In addition to the antigen-dependent mechanisms of CLL-BCR activation, two recent studies clarified the antigen-independent BCR activation mechanism in CLL. Duhren-von Minden et al. revealed that HCDR3 of CLL-BCRs recognizes the specific amino acids of the second framework region of immunoglobulins (Igs), inducing Ca2+ signaling independent of specific antigens. 22 Consistent with this theory, Binder et al. identified the alternative epitope recognized by CLL-BCRs in the third framework region of Igs. 23 Thus, CLL-BCRs have unique HCDR3 rearrangements that ensure basal BCR signaling activity via the self-recognition of CLL-BCRs. This characteristic may be specific to CLL-BCRs because BCRs from other B cell malignancies lack the self-recognition of Igs. 22 Furthermore, it is consistent with CLL-BCRs being able to bind the Igs component, acting like RFs. 20 , 21 , 24 , 25 Collectively, these studies suggest the presence of antigen-dependent and -independent mechanisms for the constitutive activation of BCR signaling in CLL.
CLL INTERACTION WITH THE MICROENVIRONMENT
In addition to cell-intrinsic molecular mechanisms that regulate the survival and proliferation of CLL cells, the interaction between CLL cells and their microenvironment also plays an essential role in CLL progression. 26 , 27 CLL cells recirculate between peripheral blood and secondary lymphoid tissues, where CLL cells proliferate at a daily birth rate of 0.1%–2.0% of all CLL clones in humans. 28 The lymphoid tissues of CLL exhibit unique histopathological features termed proliferation centers or pseudofollicles. 29 The homing process to such lymphoid tissues is essential for CLL propagation and is tightly regulated by cytokines, chemokines, chemokine receptors, and adhesion molecules. CLL cells secrete several cytokines that attract accessory cells, such as T cells and monocytes/macrophages, thereby altering their anti-leukemic activity. 30 - 33 T cells from CLL patients exhibit features of exhaustion but retain their capacity for cytokine production, 34 and such T cells demonstrate impaired immunological synapse formation. 35 CLL cells alter T cell gene expression via direct cell–cell contact. 36 Of note, alterations in T cell subsets, such as helper T cells and Th17 cells, have been reported in CLL patients with autoimmune cytopenia. 37
Nurse-like cells (NLCs) represent an important component of the CLL microenvironment. They are derived from monocytic cells and are found in the lymphoid organs of CLL patients. 38 , 39 NLCs exhibit gene expression patterns similar to M2 macrophages and tumor-associated macrophages, 40 , 41 which support the propagation of solid tumors. Of note, CLL cells produce extracellular nicotinamide phosphoribosyltransferase, which is involved in the induction of monocyte polarization to M2 macrophages secreting tumor cytokines and inhibiting T cell response. 42 Gene expression analysis revealed that the interaction between CLL cells and NLCs activated BCR and NF-κB signaling pathways in CLL. 43 , 44 Consistent with the alteration of gene expression signatures, CLL-BCR recognizes some antigens, such as vimentin and calreticulin, that are highly expressed on NLCs. 45 Thus, the interaction between CLL cells and NLCs is one of the CLL-specific mechanisms driving BCR signaling in CLL.
In addition to hematopoietic cells, mesenchymal stromal cells are also involved in CLL pathogenesis as one of the essential components of the CLL-specific microenvironment. Bone marrow stromal cells (BMSCs) support CLL cell survival through direct cellular interaction, 46 and protect CLL cells from spontaneous and drug-induced apoptosis. 47 Similar to NLCs, direct cellular interactions between CLL cells and BMSCs induce BMSC activation via the protein kinase C (AKT)-βII/NF-κB signaling pathway. 48 Furthermore, CLL cells release microvesicles carrying signaling molecules that activate the AKT pathway in BMSCs. 49 , 50 Thus, both direct and indirect interactions result in bidirectional crosstalk between CLL cells and BMSCs. These studies highlight the significance of the microenvironment in CLL pathogenesis. Thus, further studies targeting the CLL-specific microenvironment will aid in the development of novel therapeutic strategies.
GENETIC AND EPIGENETIC CHARACTERISTICS OF CLL
Although the majority of CLL cases develop sporadically, an inherited predisposition to CLL has been reported. 51 Relatives of CLL patients have an increased risk of CLL and non-Hodgkin’s lymphoma. 52 Furthermore, the incidence of CLL is highest in Europe and individuals of European descent worldwide, whereas the lowest incidence is in East Asia, including Japan. Furthermore, a study of migrants revealed a low incidence of CLL in Asian individuals born in the United States. 53 These observations suggest the presence of a genetically inherited predisposition to CLL. Several genome-wide association studies (GWASs) identified multiple risk loci for CLL; 54 - 58 however, the statistical power of individual GWASs was limited due to the modest effect size of each genetic variant.
Chromosomal abnormalities in CLL have been extensively investigated and utilized for risk assessments of CLL, in addition to the genetic predisposition of CLL revealed by GWAS. 59 CLL cells harbor immunoglobulin heavy chain (IgH)-related translocations at markedly low frequencies compared with other types of mature B cell malignancies. This genetic characteristic distinguishes CLL from other mature B cell malignancies.
The most frequently observed genetic lesions in CLL are 13q14 deletions (del13q14), found in 50%–60% of cases. 59 , 60 These are mostly monoallelic and more frequently observed in IGHV-M CLL cases than in IGHV-UM CLL cases. Del13q14 is generally associated with a favorable prognosis, but the clinical course of CLL is accelerated in patients with large 13q14 deletions involving the retinoblastoma gene (RB1). 61 The long non-coding RNAs DLEU2 and DLEU1 and the microRNA clusters miR15A–miR16-1 are found in the minimal deleted region of del13q14. 62 - 66 Model mice harboring deletions of the corresponding murine locus developed clonal B cell lymphoproliferative disorders, suggesting the significant role of microRNA in CLL pathogenesis. 67 Furthermore, the deletion of microRNAs miR-15A–miR-16-1 resulted in BCL2 overexpression, 68 providing rationale for CLL therapeutic strategies targeting BCL2.
The second common chromosomal abnormality of CLL is deletions in the 11q22–q23 (del11q) chromosomal region, detected in ~15% of cases. 59 , 60 , 69 Del11q leads to the loss of tumor suppressor gene ataxia telangiectasia mutated (ATM), which encodes the DNA damage response (DDR) kinase ATM. 70 Of note, ~25% of CLL patients with del11q deletions harbor mutations in the remaining ATM allele, and the combination of del11q and ATM mutation is associated with a poor prognosis. 71
The third frequently observed chromosomal abnormality is trisomy 12, found in ~15% of patients with CLL. 59 , 60 Trisomy 12 is a genetic lesion yielding intermediate risk. However, the coexistence of NOTCH1 mutations and trisomy 12 is associated with poor survival. 72 Moreover, CLL patients with trisomy 12 have a higher risk of transformation into Richter syndrome (RS). 73 - 75 As RS has a poor clinical prognosis, further studies are recommended to identify the molecular mechanisms by which trisomy 12 increases the risk of RS transformation.
Deletions in the 17p13 chromosomal locus (del17p) are observed in ~10% of patients with CLL 59 , 60 , 69 and are more frequently observed in IGHV-UM CLL cases than in IGHV-M CLL cases. 59 Del17p deletions usually involve the entire short arm of chromosome 17, leading to the loss of the tumor suppressor gene TP53. 76 Missense mutations in the remaining TP53 allele are found in ~80% of patients with CLL and del17p. 77 , 78 Consistent with the inactivation of TP53, patients with del17p exhibit high genomic complexity and a poorer overall prognosis than those with wild-type TP53. 59 , 76 - 80 Clinically, the assessment of del17p and TP53 mutation status is essential to select the appropriate therapeutic strategies against CLL.
In addition to the extensive chromosomal abnormalities described above, advances in NGS technologies revealed recurrent driver mutations in CLL such as SF3B1, ATM, TP53, NOTCH1, POT1, CHD2, XPO1, BIRC3, BRAF, MYD88, EGR2, MED12, FBXW7, ASXL1, KRAS, NRAS, MAP2K1, NFKBIE, TRAF3, RPS15, and DDX3X. 5 , 60 , 81 - 84
The most frequently mutated gene in CLL is SF3B1 (10%–15% of cases) and the SF3B1 K700E mutation is the most common. 81 - 83 SF3B1 mutations cause alternative splicing in CLL cells and induce RNA changes affecting multiple CLL-associated pathways, including DDR, telomere maintenance, and NOTCH signaling. 85 In addition to SF3B1, several genes involved in RNA splicing, processing, and transport, such as DDX3X and XPO1, are mutated in CLL at lower frequencies, suggesting that deregulated RNA processing is one of the major pathogenic pathways in CLL development.
NOTCH1 is the second most commonly mutated gene in CLL (~10% of cases) 60 , 84 , 86 and NOTCH1 mutations are preferentially observed in IGHV-UM CLL. Of note, ~40% of patients with NOTCH1-mutated CLL harbor trisomy 12, implying a relationship of these two genetic aberrations with CLL development. 72 , 87 The majority of NOTCH1 mutations in CLL increase the nuclear NOTCH intracellular domain by abrogating the PEST domain, which is necessary for F-box/WD repeat-containing protein 7-mediated proteasomal degradation of NOTCH1. 3 , 84 , 88 FBXW7-inactivating mutations have also been observed in ~3% of patients with CLL without NOTCH1 mutations, demonstrating an analogous outcome of increased NOTCH1 signaling. Moreover, NOTCH1 activation independent of NOTCH1 mutations has been reported in CLL cells. 89 , 90 Thus, multiple mechanisms activate the NOTCH1 pathway in CLL pathogenesis. 91
Somatic mutations affecting inflammation-related genes, such as MYD88, NFKBIE, BIRC3, and TRAF3, have been identified as recurrent mutations in CLL. 5 , 83 , 92 Mutations in the TLR/MYD88 pathway can be used to identify a subset of young CLL patients with a favorable outcome. 93 Thus, pathological pathways involved in CLL leukemogenesis include inflammatory pathways and their downstream signaling. POT1 mutations are found in 3%–7% of CLL patients and are frequently observed in IGHV-UM CLL. 60 , 81 , 82 , 84 , 94 POT1 plays an essential role in telomere protection 95 and is required to maintain the self-renewal capacity of HSCs in normal hematopoiesis. 96 POT1 mutations in CLL alter the telomeric DNA-binding domain, leading to structural aberrations and chromosomal instability. 94 Thus, the disruption of genes involved in DDR, such as TP53, ATM, and POT1, plays a significant role in CLL development.
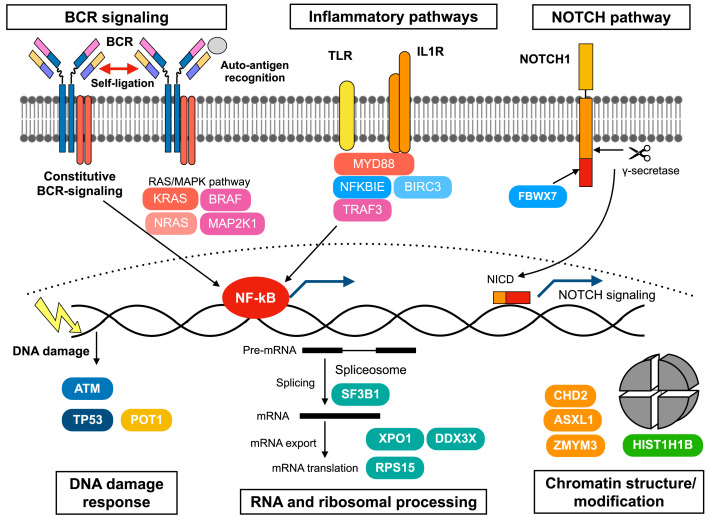
In addition to the identification of recurrent mutations in CLL cells, a recent global epigenomic status analysis revealed the regulatory chromatin landscape of CLL, and clarified that IGHV-UM CLL cells harbor more active and open chromatin than IGHV-M CLL cells. Furthermore, de novo active regions in CLL cells are enriched for NFAT, FOX, and TCF/LEF transcription factors. 97 Of note, most genetic alterations are not associated with specific epigenetic profiles, and CLL with MYD88 mutations and trisomy 12 exhibit distinct chromatin configurations. 97 In summary, genetic lesions in CLL can be categorized according to several biological pathways such as BCR signaling, inflammatory, NOTCH1 signaling, DDR, RNA and ribosomal processing, genome/chromatin structure, NF-κB signaling, cell cycle, and apoptosis. 60 , 84 These deregulated biological pathways coordinately drive CLL leukemogenesis in humans (Figure 1).
Fig. 1.
Summary of the pathways and molecules involved in CLL pathogenesis. These deregulated biological pathways are affected by genetic and non-genetic mechanisms, and coordinately drive CLL leukemogenesis.
MULTISTEP CLL LEUKEMOGENESIS IS INITIATED BY SELF-RENEWING HUMAN HSCs
After describing the important molecular pathways identified in NGS studies that are involved in CLL pathogenesis, we next focused on how such oncogenic events initiate and accumulate during leukemogenesis. The pathogenesis of other types of human leukemia, including acute myeloid leukemia, acute lymphoblastic leukemia, and chronic myeloid leukemia, is directly related to HSCs and immature progenitor cells. However, CLL is the exception because it is thought to directly originate from mature B cells. When tracing the origins of human CLL, it must be noted that it is not always monoclonal. 98 , 99 Moreover, a large cohort study demonstrated that nearly all patients with CLL had prior monoclonal B cell lymphocytosis (MBL), 100 a preleukemic state of CLL with the asymptomatic proliferation of clonal B cells and circulating numbers <5,000/μl. 101 MBL prevalence increases with age, 100 , 102 ranging from <1% 103 , 104 to 18%. 105 Of note, human MBL sometimes comprises oligoclonal B cell clones. 106 - 110
Although the progression from MBL to CLL is a stepwise process, the stage at which the first oncogenic event occurs remains unknown. The existence of oligoclonal B cell clones in both patients with CLL and MBL suggests that the first oncogenic events occur as far back as progenitor cells or HSCs. These observations led us to evaluate the primitive HSC fraction in patients with CLL and we found that the propensity to generate clonal mature B cells was present in HSCs. Although CLL cells were never directly engrafted in xenograft models, HSCs from patients with CLL caused abnormal monoclonal or oligoclonal mature B cells in vivo. 111 Moreover, NGS studies confirmed that CD34+CD19− hematopoietic stem/progenitor cells (HSPCs) and myeloid cells from patients with CLL shared somatic mutations identical to those detected in CLL cells. Such recurrent mutations included NOTCH1, SF3B1, BRAF, TP53, XPO1, MED12, NFKBIE, and EGR2. 5 , 112 , 113 Whole-genome sequencing also confirmed shared mutations between MBL/CLL cells and their respective polymorphonuclear cells, suggesting that the acquisition of some somatic mutations occurs before disease onset, likely at the HSC stage. 114 Moreover, the activation of NOTCH1 pathways is deeply involved in CLL leukemogenesis. 91 The NOTCH1 signaling pathway is aberrantly activated in HSPCs from patients with CLL regardless of NOTCH1 mutation status (compared with the HSPC levels in healthy donors), suggesting that NOTCH1 activation is an early event in CLL leukemogenesis that may lead to the development of aberrant HSPCs in patients with CLL. 113 Consistent with this, advances in the analysis of IgH genes using NGS technology confirmed the presence of independent oligoclonal B cell clones (even in immunophenotypically monoclonal CLL patients). 115 Thus, the initial oncogenic events target self-renewing HSCs in CLL in humans.
Recent studies clarified that the initial oncogenic events target HSPCs in several types of human mature lymphoid malignancies in addition to CLL. 116 - 119 Several studies using a mouse model of mature lymphoid malignancies revealed that the lymphoma-specific oncogenes expressed in HSPCs can initiate lymphomagenesis more effectively than those expressed in mature B cells, 116 , 118 , 120 - 122 supporting the model of multistep leukemogenesis or lymphomagenesis initiation from HSPCs. These studies provided novel models of leukemogenesis/lymphomagenesis. Thus, the cellular stages of tumor initiation and final transformation are distinct, and the stage-specific oncogenic events coordinately propagate the tumor in humans. Further studies are necessary to elucidate the detailed molecular mechanisms underlying the multistep leukemogenesis/lymphomagenesis of mature lymphoid malignancies in humans.
NOVEL DRUGS FOR CLL TREATMENT
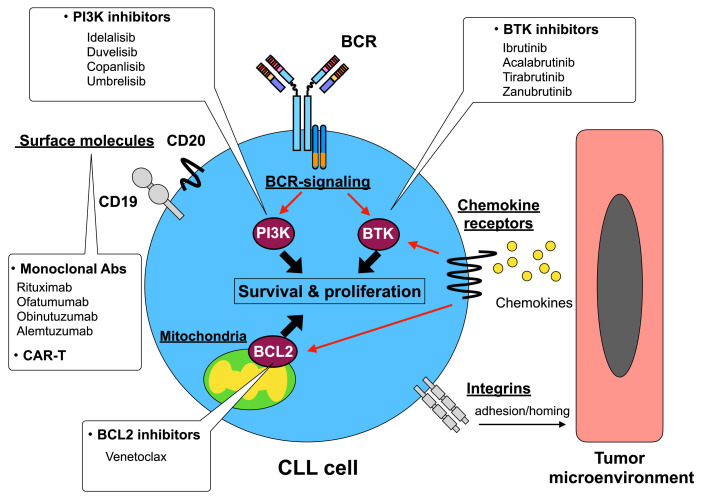
Chemoimmunotherapy using anti-CD20 monoclonal antibodies, such as the fludarabine, cyclophosphamide, and rituximab regimen, 123 remains the standard reference treatment for CLL patients aged <65 years with good health and low-risk prognostic factors. Recent advances in the understanding of CLL pathogenesis markedly improved the range of therapeutic applications for CLL treatment. Therapeutic strategies targeting BCR signaling have been developed due to the essential role of BCR signaling in CLL pathogenesis. Furthermore, venetoclax, a BCL2 inhibitor, markedly altered the therapeutic strategy for CLL treatment. The novel drugs, and their target molecules and pathways in CLL are summarized in Figure 2.
Fig. 2.
Summary of novel drugs and their target molecules and pathways.
Regarding surface molecules, CD19, CD20, and CD52 have been extensively investigated as therapeutic target molecules for CLL. CD19, a member of the Ig superfamily, is a B cell lineage specific surface molecule involved in BCR signal transduction. 124 The expression of CD19 is restricted to the B cell lineage and HSCs and the majority of hematopoietic cells lack its expression; therefore, CD19 represents a specific therapeutic target for B cell malignancies. T cells bearing a chimeric antigen receptor (CAR T cells) have been developed as a new cellular therapy. 125 The most advanced CAR T cells developed to date target human CD19; tisagenlecleucel and axicabtagene ciloleucel. The Food and Drug Administration (FDA) approved tisagenlecleucel for the treatment of refractory B-ALL and refractory diffuse large B cell lymphoma (DLBCL), and axicabtagene ciloleucel for refractory DLBCL. The efficacy of CAR T cells against CLL was first reported in 2011 126 and the number of CLL patients treated using CAR T cells increased in a series of clinical trials. 127 - 131 CLL was one of the first diseases for which CAR T cells were used; however, the therapeutic experience of CAR T cells is less extensive for CLL due to the relatively lower efficacy against CLL than against B-ALL or DLBCL. 132 CAR T cell therapy is highly effective, but it can induce substantial toxicities such as cytokine release syndrome and neurotoxicity. To overcome these problems, a recent study employed CAR natural killer cells (CAR NK cells), and demonstrated the rapid and persistent efficacy of CAR NK cells against B cell malignancies, including CLL, without the development of major toxicities. 133
CD20 is a surface glycoprotein expressed on mature B cells and its expression is restricted to the B cell lineage. HSCs and the majority of hematopoietic cells lack its expression; therefore, anti-CD20 monoclonal antibodies, such as rituximab, ofatumumab, and obinutuzumab, have been developed and utilized in the treatment of mature B cell malignancies. Anti-CD20 antibodies are classified into two groups, type I and type II, based on the differences in the epitope and binding mode. 134 Rituximab and ofatumumab belong to the type I group. Type I antibodies can stabilize CD20 molecules on lipid rafts, leading to increased C1q binding and the induction of strong complement-dependent cytotoxicity (CDC). In contrast, type II antibodies, such as obinutuzumab, cannot stabilize CD20 on lipid rafts, resulting in reduced binding potential to C1q and lower levels of CDC; however, they may directly induce cell death. 134 , 135 A recent study revealed the differential binding mechanisms of therapeutic anti-CD20 antibodies, including rituximab, ofatumumab, and obinutuzumab. 136
Rituximab, an anti-CD20 chimeric monoclonal antibody, has revolutionized the therapeutic strategies for mature B cell malignancies, including CLL. Rituximab was demonstrated as effective and tolerable as monotherapy for non-Hodgkin’s lymphoma (NHL); 137 - 140 however, rituximab monotherapy was less effective against CLL. 141 In contrast to rituximab monotherapy, chemoimmunotherapy using rituximab, such as the fludarabine, cyclophosphamide, and rituximab regimen, is significantly effective against CLL. 123 , 142
Ofatumumab is a human monoclonal anti-CD20 antibody that targets a small-loop, membrane-proximal epitope of the CD20 molecule. 143 Ofatumumab monotherapy is an effective, well-tolerated treatment for fludarabine-refractory CLL patients. 144 The safety and efficacy of combination therapies, 145 , 146 and maintenance therapy 147 using ofatumumab were investigated in several studies.
Obinutuzumab is a humanized, afucosylated, type II anti-CD20 antibody. A phase 1/2 clinical study revealed that obinutuzumab monotherapy is effective for patients with heavily pretreated relapse/refractory CLL. 148 A randomized phase 3 trial demonstrated that the addition of obinutuzumab to chlorambucil significantly prolonged overall survival compared with chlorambucil monotherapy in untreated CLL patients not eligible for intensive chemotherapy. 149
Alemtuzumab is an anti-human CD52 humanized IgG1 monoclonal antibody. A variety of human lymphoid malignancies and normal lymphocytes express CD52 antigens. The efficacy of alemtuzumab was investigated in previously-treated 150 , 151 and untreated CLL patients, 152 , 153 and the FDA approved it for the treatment of fludarabine-refractory CLL in 2001. The major toxicities of alemtuzumab treatment include infusion reactions, myelosuppression, and immunosuppression. In particular, infectious events were observed in several clinical trials 151 , 154 due to immunosuppression by CLL and T cell depletion by alemtuzumab.
In addition to surface molecules, therapeutic strategies targeting BCR signaling have been developed for CLL treatment. Ibrutinib, a Bruton tyrosine kinase (BTK) inhibitor, is an orally bioavailable small molecule that covalently bonds to the cysteine-481 residue of BTK. Ibrutinib exhibited potent activity against previously treated CLL or CLL with TP53 aberrations, 155 - 158 and is increasingly used as monotherapy or tested in combination with other regimens. Of note, BTK inhibitors exert anti-CLL activity by inhibiting BCR signaling, and the interaction between CLL cells and their microenvironment. 159 Ibrutinib treatment releases CLL cells from their microenvironment, where they are required for CLL proliferation, into peripheral blood, leading to apoptosis via the downregulation of several adhesion molecules. 160 - 162 Ibrutinib also alters the immunosuppressive CLL microenvironment by inhibiting signal transducer and activator of transcription 3 pathways. 163 Although it has high clinical efficacy, disease progression during ibrutinib treatment has been reported. Ibrutinib resistance is due to CLL clones harboring mutations in BTK and PLCG2, a downstream molecule of BTK in the BCR signaling pathway, which drive the clonal expansion of CLL during ibrutinib treatment. 164 - 167 New BTK inhibitors, such as acalabrutinib, tirabrutinib, and zanubrutinib, have been developed, and their efficacies and safety profiles were clarified in clinical trials. 168 - 171 Recent studies investigating acalabrutinib, a second generation BTK inhibitor, confirmed the efficacy of combination therapy consisting of acalabrutinib and obinutuzumab in patients with treatment naïve and relapse/refractory CLL. 168 , 172 Further studies will aid in the development of safe and effective therapeutic strategies using BTK inhibitors.
Phosphatidylinositol 3-kinases (PI3Ks) integrate and transduce signals from BCR, chemokine receptors, and adhesion molecules. 173 - 175 They are subdivided into classes I, II, and III. Class I PI3Ks comprise four isoforms, PI3K α, β, γ, and δ. PI3Kδ expression is primarily restricted to hematopoietic cells, and it plays an essential and nonredundant role in BCR signaling. 176 - 178 Idelalisib is a potent and selective PI3Kδ inhibitor 179 , 180 that exerts anti-CLL effects by suppressing BCR signaling, and the interaction between CLL cells and their microenvironment. 180 Oral idelalisib therapy exhibited a favorable safety profile and rapidly induced stable disease control in most heavily pretreated CLL patients. 181 The combination therapy of idelalisib and rituximab resulted in a higher overall response rate than rituximab monotherapy in relapsed CLL patients. 182 , 183 Another combination therapy of idelalisib, bendamustine, and rituximab improved progression-free survival compared with bendamustine plus rituximab alone in patients with relapsed or refractory CLL, but an increased risk of infection was reported in the idelalisib group. 184 Next-generation PI3K inhibitors, such as duvelisib, copanlisib, and umbralisib, were previously developed. 185 - 188 Duvelisib, a dual inhibitor of PI3Kδ and PI3Kγ, was approved by the FDA for relapsed or refractory CLL/small lymphocytic lymphoma in 2018 based on the results of the phase 3 DUO trial. 189
In addition to novel drugs targeting BCR signaling pathways, such as BTK and PI3K inhibitors, the BCL2 inhibitor venetoclax has also markedly altered CLL treatment. This BH3 domain mimic prevents the interaction between BCL2 and BH3, and inhibits the anti-apoptotic effects of BCL2. As constitutively activated BCR signaling and the most frequently observed chromosomal abnormality del13q14 cause high levels of BLC2 expression, BCL2 represents a reasonable therapeutic target in CLL. The efficacy and safety of daily oral venetoclax for relapsed or refractory CLL was reported in a phase 1 dose-escalation study. 190 A phase 2 study of venetoclax monotherapy in patients with relapsed or refractory CLL with del17p reported an overall response rate of 79.4% at a median follow-up of 12.1 months. 191 The phase 3 MURANO trial in patients with relapsed or refractory CLL compared the combination of venetoclax/rituximab with that of bendamustine/rituximab therapy, resulting in a 2-year progression-free survival rate of 84.9% and 36.3%, respectively. 192 This study evaluated the minimal/measurable residual disease (MRD) status using multicolor flow cytometry and polymerase chain reaction assays, and the frequencies of the patients who achieved a negative MRD status were significantly higher in the venetoclax/rituximab treatment group than in the bendamustine/rituximab treatment group. 192 Similarly, the recent phase 2 CLARITY trial investigating the combination of ibrutinib and venetoclax for relapsed or refractory CLL reported a high rate of MRD eradication. 193 Based on these studies, an MRD-guided treatment strategy may be the standard of care for CLL in the near future. Further studies will be helpful to establish therapeutic strategies using such novel drugs and improve the clinical outcome of CLL.
CONCLUSIONS AND PERSPECTIVES
Our understanding of the pathogenesis of CLL has markedly improved in the last decade regarding recurrent mutations, immunological aspects of CLL-BCR, and the initiation of multistep leukemogenesis by HSCs. Furthermore, the development of novel drugs targeting molecules essential for CLL has significantly improved the clinical outcome of CLL patients. Based on both basic and clinical studies, we plan to further investigate CLL biology to overcome this disease.
Footnotes
CONFLICT OF INTEREST
The author has no conflicts of interest related to this article.
REFERENCES
- 1. Ghia P, Ferreri AJM, Caligaris-Cappio F. Chronic lymphocytic leukemia. Crit Rev Oncol Hematol. 2007; 64: 234-246. [DOI] [PubMed] [Google Scholar]
- 2. Dighiero G, Hamblin TJ. Chronic lymphocytic leukaemia. Lancet. 2008; 371: 1017-1029. [DOI] [PubMed] [Google Scholar]
- 3. Fabbri G, Dalla-Favera R. The molecular pathogenesis of chronic lymphocytic leukaemia. Nat Rev Cancer. 2016; 16: 145-162. [DOI] [PubMed] [Google Scholar]
- 4. Kikushige Y, Miyamoto T. Pre-malignant lymphoid cells arise from hematopoietic stem/progenitor cells in chronic lymphocytic leukemia. Int J Hematol. 2015; 102: 528-535. [DOI] [PubMed] [Google Scholar]
- 5. Damm F, Mylonas E, Cosson A, et al. Acquired initiating mutations in early hematopoietic cells of CLL patients. Cancer Discov. 2014; 4: 1088-1101. [DOI] [PubMed] [Google Scholar]
- 6. Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005; 352: 804-815. [DOI] [PubMed] [Google Scholar]
- 7. Zenz T, Mertens D, Küppers R, Döhner H, Stilgenbauer S. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev Cancer. 2010; 10: 37-50. [DOI] [PubMed] [Google Scholar]
- 8. Hallek M, Shanafelt TD, Eichhorst B. Chronic lymphocytic leukaemia. Lancet. 2018; 391: 1524-1537. [DOI] [PubMed] [Google Scholar]
- 9. Stevenson FK, Caligaris-Cappio F. Chronic lymphocytic leukemia: revelations from the B-cell receptor. Blood. 2004; 103: 4389-4395. [DOI] [PubMed] [Google Scholar]
- 10. Caligaris-Cappio F, Ghia P. Novel insights in chronic lymphocytic leukemia: are we getting closer to understanding the pathogenesis of the disease? J Clin Oncol. 2008; 26: 4497-4503. [DOI] [PubMed] [Google Scholar]
- 11. Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999; 94: 1848-1854. [PubMed] [Google Scholar]
- 12. Hervé M, Xu K, Ng YS, et al. Unmutated and mutated chronic lymphocytic leukemias derive from self-reactive B cell precursors despite expressing different antibody reactivity. J Clin Invest. 2005; 115: 1636-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rosenwald A, Alizadeh AA, Widhopf G, et al. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J Exp Med. 2001; 194: 1639-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gobessi S, Laurenti L, Longo PG, et al. Inhibition of constitutive and BCR-induced Syk activation downregulates Mcl-1 and induces apoptosis in chronic lymphocytic leukemia B cells. Leukemia. 2009; 23: 686-697. [DOI] [PubMed] [Google Scholar]
- 15. Widhopf GF, II, Rassenti LZ, Toy TL, et al. Chronic lymphocytic leukemia B cells of more than 1% of patients express virtually identical immunoglobulins. Blood. 2004; 104: 2499-2504. [DOI] [PubMed] [Google Scholar]
- 16. Stamatopoulos K, Belessi C, Moreno C, et al. Over 20% of patients with chronic lymphocytic leukemia carry stereotyped receptors: pathogenetic implications and clinical correlations. Blood. 2007; 109: 259-270. [DOI] [PubMed] [Google Scholar]
- 17. Messmer BT, Raphael BJ, Aerni SJ, et al. Computational identification of CDR3 sequence archetypes among immunoglobulin sequences in chronic lymphocytic leukemia. Leuk Res. 2009; 33: 368-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chu CC, Catera R, Hatzi K, et al. Chronic lymphocytic leukemia antibodies with a common stereotypic rearrangement recognize nonmuscle myosin heavy chain IIA. Blood. 2008; 112: 5122-5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoogeboom R, van Kessel KPM, Hochstenbach F, et al. A mutated B cell chronic lymphocytic leukemia subset that recognizes and responds to fungi. J Exp Med. 2013; 210: 59-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hoogeboom R, Wormhoudt TA, Schipperus MR, et al. A novel chronic lymphocytic leukemia subset expressing mutated IGHV3-7-encoded rheumatoid factor B-cell receptors that are functionally proficient. Leukemia. 2013; 27: 738-740. [DOI] [PubMed] [Google Scholar]
- 21. Kostareli E, Gounari M, Janus A, et al. Antigen receptor stereotypy across B-cell lymphoproliferations: the case of IGHV4-59/IGKV3-20 receptors with rheumatoid factor activity. Leukemia. 2012; 26: 1127-1131. [DOI] [PubMed] [Google Scholar]
- 22. Duhren-von Minden M, Übelhart R, Schneider D, et al. Chronic lymphocytic leukaemia is driven by antigen-independent cell-autonomous signalling. Nature. 2012; 489: 309-312. [DOI] [PubMed] [Google Scholar]
- 23. Binder M, Müller F, Frick M, et al. CLL B-cell receptors can recognize themselves: alternative epitopes and structural clues for autostimulatory mechanisms in CLL. Blood. 2013; 121: 239-241. [DOI] [PubMed] [Google Scholar]
- 24. Sthoeger ZM, Wakai M, Tse DB, et al. Production of autoantibodies by CD5-expressing B lymphocytes from patients with chronic lymphocytic leukemia. J Exp Med. 1989; 169: 255-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bröker BM, Klajman A, Youinou P, et al. Chronic lymphocytic leukemic (CLL) cells secrete multispecific autoantibodies. J Autoimmun. 1988; 1: 469-481. [DOI] [PubMed] [Google Scholar]
- 26. Caligaris-Cappio F, Bertilaccio MTS, Scielzo C. How the microenvironment wires the natural history of chronic lymphocytic leukemia. Semin Cancer Biol. 2014; 24: 43-48. [DOI] [PubMed] [Google Scholar]
- 27. ten Hacken E, Burger JA. Microenvironment interactions and B-cell receptor signaling in Chronic Lymphocytic Leukemia: implications for disease pathogenesis and treatment. Biochim Biophys Acta. 2016; 1863: 401-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Messmer BT, Messmer D, Allen SL, et al. In vivo measurements document the dynamic cellular kinetics of chronic lymphocytic leukemia B cells. J Clin Invest. 2005; 115: 755-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Caligaris-Cappio F. Role of the microenvironment in chronic lymphocytic leukaemia. Br J Haematol. 2003; 123: 380-388. [DOI] [PubMed] [Google Scholar]
- 30. Dal Bo M, Tissino E, Benedetti D, et al. Microenvironmental interactions in chronic lymphocytic leukemia: the master role of CD49d. Semin Hematol. 2014; 51: 168-176. [DOI] [PubMed] [Google Scholar]
- 31. Ghia P, Strola G, Granziero L, et al. Chronic lymphocytic leukemia B cells are endowed with the capacity to attract CD4+, CD40L+ T cells by producing CCL22. Eur J Immunol. 2002; 32: 1403-1413. [DOI] [PubMed] [Google Scholar]
- 32. Maffei R, Bulgarelli J, Fiorcari S, et al. The monocytic population in chronic lymphocytic leukemia shows altered composition and deregulation of genes involved in phagocytosis and inflammation. Haematologica. 2013; 98: 1115-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zucchetto A, Tripodo C, Benedetti D, et al. Monocytes/macrophages but not T lymphocytes are the major targets of the CCL3/CCL4 chemokines produced by CD38 + CD49d + chronic lymphocytic leukaemia cells. Br J Haematol. 2010; 150: 111-113. [DOI] [PubMed] [Google Scholar]
- 34. Riches JC, Davies JK, McClanahan F, et al. T cells from CLL patients exhibit features of T-cell exhaustion but retain capacity for cytokine production. Blood. 2013; 121: 1612-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ramsay AG, Johnson AJ, Lee AM, et al. Chronic lymphocytic leukemia T cells show impaired immunological synapse formation that can be reversed with an immunomodulating drug. J Clin Invest. 2008; 118: 2427-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Görgün G, Holderried TAW, Zahrieh D, Neuberg D, Gribben JG. Chronic lymphocytic leukemia cells induce changes in gene expression of CD4 and CD8 T cells. J Clin Invest. 2005; 115: 1797-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lad DP, Varma S, Varma N, et al. Regulatory T-cell and T-helper 17 balance in chronic lymphocytic leukemia progression and autoimmune cytopenias. Leuk Lymphoma. 2015; 56: 2424-2428. [DOI] [PubMed] [Google Scholar]
- 38. Burger JA, Tsukada N, Burger M, et al. Blood-derived nurse-like cells protect chronic lymphocytic leukemia B cells from spontaneous apoptosis through stromal cell–derived factor-1. Blood. 2000; 96: 2655-2663. [PubMed] [Google Scholar]
- 39. Tsukada N, Burger JA, Zvaifler NJ, Kipps TJ. Distinctive features of “nurselike” cells that differentiate in the context of chronic lymphocytic leukemia. Blood. 2002; 99: 1030-1037. [DOI] [PubMed] [Google Scholar]
- 40. Filip AA, Ciseł B, Koczkodaj D, et al. Circulating microenvironment of CLL: are nurse-like cells related to tumor-associated macrophages? Blood Cells Mol Dis. 2013; 50: 263-270. [DOI] [PubMed] [Google Scholar]
- 41. Ysebaert L, Fournié JJ. Genomic and phenotypic characterization of nurse-like cells that promote drug resistance in chronic lymphocytic leukemia. Leuk Lymphoma. 2011; 52: 1404-1406. [DOI] [PubMed] [Google Scholar]
- 42. Audrito V, Serra S, Brusa D, et al. Extracellular nicotinamide phosphoribosyltransferase (NAMPT) promotes M2 macrophage polarization in chronic lymphocytic leukemia. Blood. 2015; 125: 111-123. [DOI] [PubMed] [Google Scholar]
- 43. Burger JA, Quiroga MP, Hartmann E, et al. High-level expression of the T-cell chemokines CCL3 and CCL4 by chronic lymphocytic leukemia B cells in nurselike cell cocultures and after BCR stimulation. Blood. 2009; 113: 3050-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Herishanu Y, Pérez-Galán P, Liu D, et al. The lymph node microenvironment promotes B-cell receptor signaling, NF-κB activation, and tumor proliferation in chronic lymphocytic leukemia. Blood. 2011; 117: 563-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Binder M, Léchenne B, Ummanni R, et al. Stereotypical chronic lymphocytic leukemia B-cell receptors recognize survival promoting antigens on stromal cells. PLoS One. 2010; 5: e15992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lagneaux L, Delforge A, Bron D, De Bruyn C, Stryckmans P. Chronic lymphocytic leukemic B cells but not normal B cells are rescued from apoptosis by contact with normal bone marrow stromal cells. Blood. 1998; 91: 2387-2396. [PubMed] [Google Scholar]
- 47. Kurtova AV, Balakrishnan K, Chen R, et al. Diverse marrow stromal cells protect CLL cells from spontaneous and drug-induced apoptosis: development of a reliable and reproducible system to assess stromal cell adhesion-mediated drug resistance. Blood. 2009; 114: 4441-4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lutzny G, Kocher T, Schmidt-Supprian M, et al. Protein kinase c-β-dependent activation of NF-κB in stromal cells is indispensable for the survival of chronic lymphocytic leukemia B cells in vivo. Cancer Cell. 2013; 23: 77-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ghosh AK, Secreto CR, Knox TR, et al. Circulating microvesicles in B-cell chronic lymphocytic leukemia can stimulate marrow stromal cells: implications for disease progression. Blood. 2010; 115: 1755-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ghosh AK, Secreto C, Boysen J, et al. The novel receptor tyrosine kinase Axl is constitutively active in B-cell chronic lymphocytic leukemia and acts as a docking site of nonreceptor kinases: implications for therapy. Blood. 2011; 117: 1928-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Speedy HE, Sava G, Houlston RS. Inherited susceptibility to CLL. Adv Exp Med Biol. 2013; 792: 293-308. [DOI] [PubMed] [Google Scholar]
- 52. Goldin LR, Björkholm M, Kristinsson SY, Turesson I, Landgren O. Elevated risk of chronic lymphocytic leukemia and other indolent non-Hodgkin’s lymphomas among relatives of patients with chronic lymphocytic leukemia. Haematologica. 2009; 94: 647-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gale RP, Cozen W, Goodman MT, Wang FF, Bernstein L. Decreased chronic lymphocytic leukemia incidence in Asians in Los Angeles County. Leuk Res. 2000; 24: 665-669. [DOI] [PubMed] [Google Scholar]
- 54. Di Bernardo MC, Crowther-Swanepoel D, Broderick P, et al. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat Genet. 2008; 40: 1204-1210. [DOI] [PubMed] [Google Scholar]
- 55. Crowther-Swanepoel D, Broderick P, Di Bernardo MC, et al. Common variants at 2q37.3, 8q24.21, 15q21.3 and 16q24.1 influence chronic lymphocytic leukemia risk. Nat Genet. 2010; 42: 132-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Speedy HE, Di Bernardo MC, Sava GP, et al. A genome-wide association study identifies multiple susceptibility loci for chronic lymphocytic leukemia. Nat Genet. 2014; 46: 56-60. [DOI] [PubMed] [Google Scholar]
- 57. Berndt SI, Skibola CF, Joseph V, et al. Genome-wide association study identifies multiple risk loci for chronic lymphocytic leukemia. Nat Genet. 2013; 45: 868-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Slager SL, Skibola CF, Di Bernardo MC, et al. Common variation at 6p21.31 (BAK1) influences the risk of chronic lymphocytic leukemia. Blood. 2012; 120: 843-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Döhner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000; 343: 1910-1916. [DOI] [PubMed] [Google Scholar]
- 60. Landau DA, Tausch E, Taylor-Weiner AN, et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015; 526: 525-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ouillette P, Collins R, Shakhan S, et al. The prognostic significance of various 13q14 deletions in chronic lymphocytic leukemia. Clin Cancer Res. 2011; 17: 6778-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Palamarchuk A, Efanov A, Nazaryan N, et al. 13q14 deletions in CLL involve cooperating tumor suppressors. Blood. 2010; 115: 3916-3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kalachikov S, Migliazza A, Cayanis E, et al. Cloning and gene mapping of the chromosome 13q14 region deleted in chronic lymphocytic leukemia. Genomics. 1997; 42: 369-377. [DOI] [PubMed] [Google Scholar]
- 64. Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002; 99: 15524-15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Migliazza A, Bosch F, Komatsu H, et al. Nucleotide sequence, transcription map, and mutation analysis of the 13q14 chromosomal region deleted in B-cell chronic lymphocytic leukemia. Blood. 2001; 97: 2098-2104. [DOI] [PubMed] [Google Scholar]
- 66. Hammarsund M, Corcoran MM, Wilson W, et al. Characterization of a novel B-CLL candidate gene--DLEU7--located in the 13q14 tumor suppressor locus. FEBS Lett. 2004; 556: 75-80. [DOI] [PubMed] [Google Scholar]
- 67. Klein U, Lia M, Crespo M, et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010; 17: 28-40. [DOI] [PubMed] [Google Scholar]
- 68. Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005; 102: 13944-13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wierda WG, O’Brien S, Wang X, et al. Multivariable model for time to first treatment in patients with chronic lymphocytic leukemia. J Clin Oncol. 2011; 29: 4088-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013; 14: 197-210. [PubMed] [Google Scholar]
- 71. Skowronska A, Parker A, Ahmed G, et al. Biallelic ATM inactivation significantly reduces survival in patients treated on the United Kingdom Leukemia Research Fund Chronic Lymphocytic Leukemia 4 trial. J Clin Oncol. 2012; 30: 4524-4532. [DOI] [PubMed] [Google Scholar]
- 72. Del Giudice I, Rossi D, Chiaretti S, et al. NOTCH1 mutations in +12 chronic lymphocytic leukemia (CLL) confer an unfavorable prognosis, induce a distinctive transcriptional profiling and refine the intermediate prognosis of +12 CLL. Haematologica. 2012; 97: 437-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fabbri G, Khiabanian H, Holmes AB, et al. Genetic lesions associated with chronic lymphocytic leukemia transformation to Richter syndrome. J Exp Med. 2013; 210: 2273-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chigrinova E, Rinaldi A, Kwee I, et al. Two main genetic pathways lead to the transformation of chronic lymphocytic leukemia to Richter syndrome. Blood. 2013; 122: 2673-2682. [DOI] [PubMed] [Google Scholar]
- 75. Strati P, Abruzzo LV, Wierda WG, et al. Second cancers and Richter transformation are the leading causes of death in patients with trisomy 12 chronic lymphocytic leukemia. Clin Lymphoma Myeloma Leuk. 2015; 15: 420-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Döhner H, Fischer K, Bentz M, et al. p53 gene deletion predicts for poor survival and non-response to therapy with purine analogs in chronic B-cell leukemias. Blood. 1995; 85: 1580-1589. [PubMed] [Google Scholar]
- 77. Zenz T, Kröber A, Scherer K, et al. Monoallelic TP53 inactivation is associated with poor prognosis in chronic lymphocytic leukemia: results from a detailed genetic characterization with long-term follow-up. Blood. 2008; 112: 3322-3329. [DOI] [PubMed] [Google Scholar]
- 78. Gonzalez D, Martinez P, Wade R, et al. Mutational status of the TP53 gene as a predictor of response and survival in patients with chronic lymphocytic leukemia: results from the LRF CLL4 trial. J Clin Oncol. 2011; 29: 2223-2229. [DOI] [PubMed] [Google Scholar]
- 79. Yu L, Kim HT, Kasar SN, et al. Survival of Del17p CLL depends on genomic complexity and somatic mutation. Clin Cancer Res. 2017; 23: 735-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ouillette P, Fossum S, Parkin B, et al. Aggressive chronic lymphocytic leukemia with elevated genomic complexity is associated with multiple gene defects in the response to DNA double-strand breaks. Clin Cancer Res. 2010; 16: 835-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wang L, Lawrence MS, Wan Y, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011; 365: 2497-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Quesada V, Conde L, Villamor N, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2012; 44: 47-52. [DOI] [PubMed] [Google Scholar]
- 83. Puente XS, Pinyol M, Quesada V, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011; 475: 101-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Puente XS, Beà S, Valdés-Mas R, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015; 526: 519-524. [DOI] [PubMed] [Google Scholar]
- 85. Wang L, Brooks AN, Fan J, et al. Transcriptomic characterization of SF3B1 mutation reveals its pleiotropic effects in chronic lymphocytic leukemia. Cancer Cell. 2016; 30: 750-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Fabbri G, Rasi S, Rossi D, et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J Exp Med. 2011; 208: 1389-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Riches JC, O’Donovan CJ, Kingdon SJ, et al. Trisomy 12 chronic lymphocytic leukemia cells exhibit upregulation of integrin signaling that is modulated by NOTCH1 mutations. Blood. 2014; 123: 4101-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kopan R, Ilagan MXG. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009; 137: 216-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Fabbri G, Holmes AB, Viganotti M, et al. Common nonmutational NOTCH1 activation in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2017; 114: E2911-E2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Arruga F, Gizdic B, Serra S, et al. Functional impact of NOTCH1 mutations in chronic lymphocytic leukemia. Leukemia. 2014; 28: 1060-1070. [DOI] [PubMed] [Google Scholar]
- 91. Rosati E, Baldoni S, De Falco F, et al. NOTCH1 aberrations in chronic lymphocytic leukemia. Front Oncol. 2018; 8: 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Rossi D, Fangazio M, Rasi S, et al. Disruption of BIRC3 associates with fludarabine chemorefractoriness in TP53 wild-type chronic lymphocytic leukemia. Blood. 2012; 119: 2854-2862. [DOI] [PubMed] [Google Scholar]
- 93. Martínez-Trillos A, Pinyol M, Navarro A, et al. Mutations in TLR/MYD88 pathway identify a subset of young chronic lymphocytic leukemia patients with favorable outcome. Blood. 2014; 123: 3790-3796. [DOI] [PubMed] [Google Scholar]
- 94. Ramsay AJ, Quesada V, Foronda M, et al. POT1 mutations cause telomere dysfunction in chronic lymphocytic leukemia. Nat Genet. 2013; 45: 526-530. [DOI] [PubMed] [Google Scholar]
- 95. Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004; 11: 1223-1229. [DOI] [PubMed] [Google Scholar]
- 96. Hosokawa K, MacArthur BD, Ikushima YM, et al. The telomere binding protein Pot1 maintains haematopoietic stem cell activity with age. Nat Commun. 2017; 8: 804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Beekman R, Chapaprieta V, Russiñol N, et al. The reference epigenome and regulatory chromatin landscape of chronic lymphocytic leukemia. Nat Med. 2018; 24: 868-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Sanchez ML, Almeida J, Gonzalez D, et al. Incidence and clinicobiologic characteristics of leukemic B-cell chronic lymphoproliferative disorders with more than one B-cell clone. Blood. 2003; 102: 2994-3002. [DOI] [PubMed] [Google Scholar]
- 99. Kikushige Y, Miyamoto T. Hematopoietic stem cell aging and chronic lymphocytic leukemia pathogenesis. Int J Hematol. 2014; 100: 335-340. [DOI] [PubMed] [Google Scholar]
- 100. Landgren O, Albitar M, Ma W, et al. B-cell clones as early markers for chronic lymphocytic leukemia. N Engl J Med. 2009; 360: 659-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Marti GE, Rawstron AC, Ghia P, et al. Diagnostic criteria for monoclonal B-cell lymphocytosis. Br J Haematol. 2005; 130: 325-332. [DOI] [PubMed] [Google Scholar]
- 102. Rawstron AC, Bennett FL, O’Connor SJM, et al. Monoclonal B-cell lymphocytosis and chronic lymphocytic leukemia. N Engl J Med. 2008; 359: 575-583. [DOI] [PubMed] [Google Scholar]
- 103. Shim YK, Vogt RF, Middleton D, et al. Prevalence and natural history of monoclonal and polyclonal B-cell lymphocytosis in a residential adult population. Cytometry B Clin Cytom. 2007; 72B: 344-353. [DOI] [PubMed] [Google Scholar]
- 104. Rachel JM, Zucker ML, Fox CM, et al. Monoclonal B-cell lymphocytosis in blood donors. Br J Haematol. 2007; 139: 832-836. [DOI] [PubMed] [Google Scholar]
- 105. Marti GE, Carter P, Abbasi F, et al. B-cell monoclonal lymphocytosis and B-cell abnormalities in the setting of familial B-cell chronic lymphocytic leukemia. Cytometry B Clin Cytom. 2003; 52B: 1-12. [DOI] [PubMed] [Google Scholar]
- 106. Nieto WG, Almeida J, Romero A, et al. Increased frequency (12%) of circulating chronic lymphocytic leukemia–like B-cell clones in healthy subjects using a highly sensitive multicolor flow cytometry approach. Blood. 2009; 114: 33-37. [DOI] [PubMed] [Google Scholar]
- 107. Dagklis A, Fazi C, Sala C, et al. The immunoglobulin gene repertoire of low-count chronic lymphocytic leukemia (CLL)–like monoclonal B lymphocytosis is different from CLL: diagnostic implications for clinical monitoring. Blood. 2009; 114: 26-32. [DOI] [PubMed] [Google Scholar]
- 108. Lanasa MC, Allgood SD, Volkheimer AD, et al. Single-cell analysis reveals oligoclonality among ‘low-count’ monoclonal B-cell lymphocytosis. Leukemia. 2010; 24: 133-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Shim YK, Rachel JM, Ghia P, et al. Monoclonal B-cell lymphocytosis in healthy blood donors: an unexpectedly common finding. Blood. 2014; 123: 1319-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Klinger M, Zheng J, Elenitoba-Johnson KSJ, et al. Next-generation IgVH sequencing CLL-like monoclonal B-cell lymphocytosis reveals frequent oligoclonality and ongoing hypermutation. Leukemia. 2016; 30: 1055-1061. [DOI] [PubMed] [Google Scholar]
- 111. Kikushige Y, Ishikawa F, Miyamoto T, et al. Self-renewing hematopoietic stem cell is the primary target in pathogenesis of human chronic lymphocytic leukemia. Cancer Cell. 2011; 20: 246-259. [DOI] [PubMed] [Google Scholar]
- 112. Quijada-Álamo M, Hernández-Sánchez M, Robledo C, et al. Next-generation sequencing and FISH studies reveal the appearance of gene mutations and chromosomal abnormalities in hematopoietic progenitors in chronic lymphocytic leukemia. J Hematol Oncol. 2017; 10: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Di Ianni M, Baldoni S, Del Papa B, et al. NOTCH1 is aberrantly activated in chronic lymphocytic leukemia hematopoietic stem cells. Front Oncol. 2018; 8: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Agathangelidis A, Ljungström V, Scarfò L, et al. Highly similar genomic landscapes in monoclonal B-cell lymphocytosis and ultra-stable chronic lymphocytic leukemia with low frequency of driver mutations. Haematologica. 2018; 103: 865-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Brazdilova K, Plevova K, Skuhrova Francova H, et al. Multiple productive IGH rearrangements denote oligoclonality even in immunophenotypically monoclonal CLL. Leukemia. 2018; 32: 234-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Quivoron C, Couronné L, Della Valle V, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011; 20: 25-38. [DOI] [PubMed] [Google Scholar]
- 117. Chung SS, Kim E, Park JH, et al. Hematopoietic stem cell origin of BRAFV600E mutations in hairy cell leukemia. Sci Transl Med. 2014; 6: 238ra71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Horton SJ, Giotopoulos G, Yun H, et al. Early loss of Crebbp confers malignant stem cell properties on lymphoid progenitors. Nat Cell Biol. 2017; 19: 1093-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Sakata-Yanagimoto M, Enami T, Yoshida K, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet. 2014; 46: 171-175. [DOI] [PubMed] [Google Scholar]
- 120. Green MR, Vicente-Dueñas C, Romero-Camarero I, et al. Transient expression of Bcl6 is sufficient for oncogenic function and induction of mature B-cell lymphoma. Nat Commun. 2014; 5: 3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Vicente-Dueñas C, Fontán L, Gonzalez-Herrero I, et al. Expression of MALT1 oncogene in hematopoietic stem/progenitor cells recapitulates the pathogenesis of human lymphoma in mice. Proc Natl Acad Sci USA. 2012; 109: 10534-10539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Vicente-Dueñas C, Romero-Camarero I, González-Herrero I, et al. A novel molecular mechanism involved in multiple myeloma development revealed by targeting MafB to haematopoietic progenitors. EMBO J. 2012; 31: 3704-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Hallek M, Fischer K, Fingerle-Rowson G, et al. Addition of rituximab to fludarabine and cyclophosphamide in patients with chronic lymphocytic leukaemia: a randomised, open-label, phase 3 trial. Lancet. 2010; 376: 1164-1174. [DOI] [PubMed] [Google Scholar]
- 124. Stamenkovic I, Seed B. CD19, the earliest differentiation antigen of the B cell lineage, bears three extracellular immunoglobulin-like domains and an Epstein-Barr virus-related cytoplasmic tail. J Exp Med. 1988; 168: 1205-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. June CH, O’Connor RS, Kawalekar OU, Ghassemi S, Milone MC. CAR T cell immunotherapy for human cancer. Science. 2018; 359: 1361-1365. [DOI] [PubMed] [Google Scholar]
- 126. Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N Engl J Med. 2011; 365: 725-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Brentjens RJ, Rivière I, Park JH, et al. Safety and persistence of adoptively transferred autologous CD19-targeted T cells in patients with relapsed or chemotherapy refractory B-cell leukemias. Blood. 2011; 118: 4817-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Kalos M, Levine BL, Porter DL, et al. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci Transl Med. 2011; 3: 95ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Porter DL, Hwang WT, Frey NV, et al. Chimeric antigen receptor T cells persist and induce sustained remissions in relapsed refractory chronic lymphocytic leukemia. Sci Transl Med. 2015; 7: 303ra139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Fraietta JA, Beckwith KA, Patel PR, et al. Ibrutinib enhances chimeric antigen receptor T-cell engraftment and efficacy in leukemia. Blood. 2016; 127: 1117-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Brudno JN, Somerville RPT, Shi V, et al. Allogeneic T cells that express an anti-CD19 chimeric antigen receptor induce remissions of B-cell malignancies that progress after allogeneic hematopoietic stem-cell transplantation without causing graft-versus-host disease. J Clin Oncol. 2016; 34: 1112-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Lemal R, Tournilhac O. State-of-the-art for CAR T-cell therapy for chronic lymphocytic leukemia in 2019. J Immunother Cancer. 2019; 7: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Liu E, Marin D, Banerjee P, et al. Use of CAR-transduced natural killer cells in CD19-positive lymphoid tumors. N Engl J Med. 2020; 382: 545-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Glennie MJ, French RR, Cragg MS, Taylor RP. Mechanisms of killing by anti-CD20 monoclonal antibodies. Mol Immunol. 2007; 44: 3823-3837. [DOI] [PubMed] [Google Scholar]
- 135. Mössner E, Brünker P, Moser S, et al. Increasing the efficacy of CD20 antibody therapy through the engineering of a new type II anti-CD20 antibody with enhanced direct and immune effector cell–mediated B-cell cytotoxicity. Blood. 2010; 115: 4393-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Kumar A, Planchais C, Fronzes R, Mouquet H, Reyes N. Binding mechanisms of therapeutic antibodies to human CD20. Science. 2020; 369: 793-799. [DOI] [PubMed] [Google Scholar]
- 137. Coiffier B, Haioun C, Ketterer N, et al. Rituximab (anti-CD20 monoclonal antibody) for the treatment of patients with relapsing or refractory aggressive lymphoma: a multicenter phase II study. Blood. 1998; 92: 1927-1932. [PubMed] [Google Scholar]
- 138. Davis TA, White CA, Grillo-López AJ, et al. Single-agent monoclonal antibody efficacy in bulky non-Hodgkin’s lymphoma: results of a phase II trial of rituximab. J Clin Oncol. 1999; 17: 1851-1857. [DOI] [PubMed] [Google Scholar]
- 139. Hainsworth JD, Litchy S, Burris HA, III, et al. Rituximab as first-line and maintenance therapy for patients with indolent non-hodgkin’s lymphoma. J Clin Oncol. 2002; 20: 4261-4267. [DOI] [PubMed] [Google Scholar]
- 140. Witzig TE, Vukov AM, Habermann TM, et al. Rituximab therapy for patients with newly diagnosed, advanced-stage, follicular grade I non-Hodgkin’s lymphoma: a phase II trial in the North Central Cancer Treatment Group. J Clin Oncol. 2005; 23: 1103-1108. [DOI] [PubMed] [Google Scholar]
- 141. Huhn D, von Schilling C, Wilhelm M, et al. Rituximab therapy of patients with B-cell chronic lymphocytic leukemia. Blood. 2001; 98: 1326-1331. [DOI] [PubMed] [Google Scholar]
- 142. Byrd JC, Rai K, Peterson BL, et al. Addition of rituximab to fludarabine may prolong progression-free survival and overall survival in patients with previously untreated chronic lymphocytic leukemia: an updated retrospective comparative analysis of CALGB 9712 and CALGB 9011. Blood. 2005; 105: 49-53. [DOI] [PubMed] [Google Scholar]
- 143. O’Brien S, Österborg A. Ofatumumab: a new CD20 monoclonal antibody therapy for B-cell chronic lymphocytic leukemia. Clin Lymphoma Myeloma Leuk. 2010; 10: 361-368. [DOI] [PubMed] [Google Scholar]
- 144. Wierda WG, Kipps TJ, Mayer J, et al. Ofatumumab as single-agent CD20 immunotherapy in fludarabine-refractory chronic lymphocytic leukemia. J Clin Oncol. 2010; 28: 1749-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Robak T, Warzocha K, Govind Babu K, et al. Ofatumumab plus fludarabine and cyclophosphamide in relapsed chronic lymphocytic leukemia: results from the COMPLEMENT 2 trial. Leuk Lymphoma. 2017; 58: 1084-1093. [DOI] [PubMed] [Google Scholar]
- 146. Hillmen P, Robak T, Janssens A, et al. Chlorambucil plus ofatumumab versus chlorambucil alone in previously untreated patients with chronic lymphocytic leukaemia (COMPLEMENT 1): a randomised, multicentre, open-label phase 3 trial. Lancet. 2015; 385: 1873-1883. [DOI] [PubMed] [Google Scholar]
- 147. van Oers MHJ, Kuliczkowski K, Smolej L, et al. Ofatumumab maintenance versus observation in relapsed chronic lymphocytic leukaemia (PROLONG): an open-label, multicentre, randomised phase 3 study. Lancet Oncol. 2015; 16: 1370-1379. [DOI] [PubMed] [Google Scholar]
- 148. Cartron G, de Guibert S, Dilhuydy MS, et al. Obinutuzumab (GA101) in relapsed/refractory chronic lymphocytic leukemia: final data from the phase 1/2 GAUGUIN study. Blood. 2014; 124: 2196-2202. [DOI] [PubMed] [Google Scholar]
- 149. Goede V, Fischer K, Busch R, et al. Obinutuzumab plus chlorambucil in patients with CLL and coexisting conditions. N Engl J Med. 2014; 370: 1101-1110. [DOI] [PubMed] [Google Scholar]
- 150. Osterborg A, Dyer MJ, Bunjes D, et al. Phase II multicenter study of human CD52 antibody in previously treated chronic lymphocytic leukemia. European Study Group of CAMPATH-1H Treatment in Chronic Lymphocytic Leukemia. J Clin Oncol. 1997; 15: 1567-1574. [DOI] [PubMed] [Google Scholar]
- 151. Keating MJ, Flinn I, Jain V, et al. Therapeutic role of alemtuzumab (Campath-1H) in patients who have failed fludarabine: results of a large international study. Blood. 2002; 99: 3554-3561. [DOI] [PubMed] [Google Scholar]
- 152. Österborg A, Fassas AS, Anagnostopoulos A, et al. Humanized CD52 monoclonal antibody campath-1H as first-line treatment in chronic lymphocytic leukaemia. Br J Haematol. 1996; 93: 151-153. [DOI] [PubMed] [Google Scholar]
- 153. Lundin J, Kimby E, Björkholm M, et al. Phase II trial of subcutaneous anti-CD52 monoclonal antibody alemtuzumab (Campath-1H) as first-line treatment for patients with B-cell chronic lymphocytic leukemia (B-CLL). Blood. 2002; 100: 768-773. [DOI] [PubMed] [Google Scholar]
- 154. Rai KR, Freter CE, Mercier RJ, et al. Alemtuzumab in previously treated chronic lymphocytic leukemia patients who also had received fludarabine. J Clin Oncol. 2002; 20: 3891-3897. [DOI] [PubMed] [Google Scholar]
- 155. Byrd JC, Furman RR, Coutre SE, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2013; 369: 32-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Byrd JC, Brown JR, O’Brien S, et al. Ibrutinib versus ofatumumab in previously treated chronic lymphoid leukemia. N Engl J Med. 2014; 371: 213-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Farooqui MZH, Valdez J, Martyr S, et al. Ibrutinib for previously untreated and relapsed or refractory chronic lymphocytic leukaemia with TP53 aberrations: a phase 2, single-arm trial. Lancet Oncol. 2015; 16: 169-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Advani RH, Buggy JJ, Sharman JP, et al. Bruton tyrosine kinase inhibitor ibrutinib (PCI-32765) has significant activity in patients with relapsed/refractory B-cell malignancies. J Clin Oncol. 2013; 31: 88-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Maffei R, Fiorcari S, Martinelli S, et al. Targeting neoplastic B cells and harnessing microenvironment: the “double face” of ibrutinib and idelalisib. J Hematol Oncol. 2015; 8: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. de Rooij MFM, Kuil A, Geest CR, et al. The clinically active BTK inhibitor PCI-32765 targets B-cell receptor– and chemokine-controlled adhesion and migration in chronic lymphocytic leukemia. Blood. 2012; 119: 2590-2594. [DOI] [PubMed] [Google Scholar]
- 161. Ponader S, Chen SS, Buggy JJ, et al. The Bruton tyrosine kinase inhibitor PCI-32765 thwarts chronic lymphocytic leukemia cell survival and tissue homing in vitro and in vivo. Blood. 2012; 119: 1182-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Niemann CU, Herman SEM, Maric I, et al. Disruption of in vivo chronic lymphocytic leukemia tumor-microenvironment interactions by ibrutinib–Findings from an investigator-initiated phase II study. Clin Cancer Res. 2016; 22: 1572-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Kondo K, Shaim H, Thompson PA, et al. Ibrutinib modulates the immunosuppressive CLL microenvironment through STAT3-mediated suppression of regulatory B-cell function and inhibition of the PD-1/PD-L1 pathway. Leukemia. 2018; 32: 960-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Woyach JA, Furman RR, Liu TM, et al. Resistance mechanisms for the Bruton’s tyrosine kinase inhibitor ibrutinib. N Engl J Med. 2014; 370: 2286-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Furman RR, Cheng S, Lu P, et al. Ibrutinib resistance in chronic lymphocytic leukemia. N Engl J Med. 2014; 370: 2352-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Burger JA, Landau DA, Taylor-Weiner A, et al. Clonal evolution in patients with chronic lymphocytic leukaemia developing resistance to BTK inhibition. Nat Commun. 2016; 7: 11589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Ahn IE, Underbayev C, Albitar A, et al. Clonal evolution leading to ibrutinib resistance in chronic lymphocytic leukemia. Blood. 2017; 129: 1469-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Sharman JP, Egyed M, Jurczak W, et al. Acalabrutinib with or without obinutuzumab versus chlorambucil and obinutuzumab for treatment-naive chronic lymphocytic leukaemia (ELEVATE-TN): a randomised, controlled, phase 3 trial. Lancet. 2020; 395: 1278-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Rule SA, Cartron G, Fegan C, et al. Long-term follow-up of patients with mantle cell lymphoma (MCL) treated with the selective Bruton’s tyrosine kinase inhibitor tirabrutinib (GS/ONO-4059). Leukemia. 2020; 34: 1458-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Danilov AV, Herbaux C, Walter HS, et al. Phase Ib study of Tirabrutinib in combination with Idelalisib or Entospletinib in previously treated chronic lymphocytic leukemia. Clin Cancer Res. 2020; 26: 2810-2818. [DOI] [PubMed] [Google Scholar]
- 171. Tam CS, Trotman J, Opat S, et al. Phase 1 study of the selective BTK inhibitor zanubrutinib in B-cell malignancies and safety and efficacy evaluation in CLL. Blood. 2019; 134: 851-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Woyach JA, Blachly JS, Rogers KA, et al. Acalabrutinib plus Obinutuzumab in treatment-naïve and relapsed/refractory chronic lymphocytic leukemia. Cancer Discov. 2020; 10: 394-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Niiro H, Clark EA. Regulation of B-cell fate by antigen-receptor signals. Nat Rev Immunol. 2002; 2: 945-956. [DOI] [PubMed] [Google Scholar]
- 174. Srinivasan L, Sasaki Y, Calado DP, et al. PI3 kinase signals BCR-dependent mature B cell survival. Cell. 2009; 139: 573-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Okkenhaug K, Vanhaesebroeck B. PI3K in lymphocyte development, differentiation and activation. Nat Rev Immunol. 2003; 3: 317-330. [DOI] [PubMed] [Google Scholar]
- 176. Jou ST, Carpino N, Takahashi Y, et al. Essential, nonredundant role for the phosphoinositide 3-kinase p110δ in signaling by the B-cell receptor complex. Mol Cell Biol. 2002; 22: 8580-8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Durand CA, Hartvigsen K, Fogelstrand L, et al. Phosphoinositide 3-kinase p110 δ regulates natural antibody production, marginal zone and B-1 B cell function, and autoantibody responses. J Immunol. 2009; 183: 5673-5684. [DOI] [PubMed] [Google Scholar]
- 178. Okkenhaug K, Bilancio A, Farjot G, et al. Impaired B and T cell antigen receptor signaling in p110δ PI 3-kinase mutant mice. Science. 2002; 297: 1031-1034. [DOI] [PubMed] [Google Scholar]
- 179. Lannutti BJ, Meadows SA, Herman SEM, et al. CAL-101, a p110δ selective phosphatidylinositol-3-kinase inhibitor for the treatment of B-cell malignancies, inhibits PI3K signaling and cellular viability. Blood. 2011; 117: 591-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Hoellenriegel J, Meadows SA, Sivina M, et al. The phosphoinositide 3′-kinase delta inhibitor, CAL-101, inhibits B-cell receptor signaling and chemokine networks in chronic lymphocytic leukemia. Blood. 2011; 118: 3603-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Brown JR, Byrd JC, Coutre SE, et al. Idelalisib, an inhibitor of phosphatidylinositol 3-kinase p110δ, for relapsed/refractory chronic lymphocytic leukemia. Blood. 2014; 123: 3390-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Furman RR, Sharman JP, Coutre SE, et al. Idelalisib and rituximab in relapsed chronic lymphocytic leukemia. N Engl J Med. 2014; 370: 997-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Sharman JP, Coutre SE, Furman RR, et al. Final results of a randomized, phase III study of rituximab with or without idelalisib followed by open-label idelalisib in patients with relapsed chronic lymphocytic leukemia. J Clin Oncol. 2019; 37: 1391-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Zelenetz AD, Barrientos JC, Brown JR, et al. Idelalisib or placebo in combination with bendamustine and rituximab in patients with relapsed or refractory chronic lymphocytic leukaemia: interim results from a phase 3, randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2017; 18: 297-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Balakrishnan K, Peluso M, Fu M, et al. The phosphoinositide-3-kinase (PI3K)-delta and gamma inhibitor, IPI-145 (Duvelisib), overcomes signals from the PI3K/AKT/S6 pathway and promotes apoptosis in CLL. Leukemia. 2015; 29: 1811-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Paul J, Soujon M, Wengner AM, et al. Simultaneous inhibition of PI3Kδ and PI3Kα induces ABC-DLBCL regression by blocking BCR-dependent and -independent activation of NF-κB and AKT. Cancer Cell. 2017; 31: 64-78. [DOI] [PubMed] [Google Scholar]
- 187. Dong S, Guinn D, Dubovsky JA, et al. IPI-145 antagonizes intrinsic and extrinsic survival signals in chronic lymphocytic leukemia cells. Blood. 2014; 124: 3583-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Burris HA, III, Flinn IW, Patel MR, et al. Umbralisib, a novel PI3Kδ and casein kinase-1ε inhibitor, in relapsed or refractory chronic lymphocytic leukaemia and lymphoma: an open-label, phase 1, dose-escalation, first-in-human study. Lancet Oncol. 2018; 19: 486-496. [DOI] [PubMed] [Google Scholar]
- 189. Flinn IW, Hillmen P, Montillo M, et al. The phase 3 DUO trial: duvelisib vs ofatumumab in relapsed and refractory CLL/SLL. Blood. 2018; 132: 2446-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Roberts AW, Davids MS, Pagel JM, et al. Targeting BCL2 with venetoclax in relapsed chronic lymphocytic leukemia. N Engl J Med. 2016; 374: 311-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Stilgenbauer S, Eichhorst B, Schetelig J, et al. Venetoclax in relapsed or refractory chronic lymphocytic leukaemia with 17p deletion: a multicentre, open-label, phase 2 study. Lancet Oncol. 2016; 17: 768-778. [DOI] [PubMed] [Google Scholar]
- 192. Seymour JF, Kipps TJ, Eichhorst B, et al. Venetoclax-rituximab in relapsed or refractory chronic lymphocytic leukemia. N Engl J Med. 2018; 378: 1107-1120. [DOI] [PubMed] [Google Scholar]
- 193. Hillmen P, Rawstron AC, Brock K, et al. Ibrutinib plus venetoclax in relapsed/refractory chronic lymphocytic leukemia: the CLARITY Study. J Clin Oncol. 2019; 37: 2722-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]