Abstract
Objective
To develop a process for translating semi-structured clinical decision support (CDS) into shareable, computer-readable CDS.
Materials and Methods
We developed a systematic and transparent process using publicly available tools (eGLIA, GEM Cutter, VSAC, and the CDS Authoring Tool) to translate an evidence-based clinical pathway (CP) into a Clinical Quality Language (CQL)-encoded CDS artifact.
Results
We produced a 4-phase process for translating a CP into a CQL-based CDS artifact. CP content was extracted using GEM into discrete clinical concepts, encoded using standard terminologies into value sets on VSAC, evaluated against workflows using a wireframe, and finally structured as a computer readable CDS artifact using CQL. This process included a quality control step and intermediate products to support transparency and reuse by other CDS developers.
Discussion
Translating a CP into a shareable, computer-readable CDS artifact was accomplished through a systematic process. Our process identified areas of ambiguity and gaps in the CP, which generated improvements in the CP. Collaboration with clinical subject experts and the CP development team was essential for translation. Publicly available tools were sufficient to support most translation steps, but expression of certain complex concepts required manual encoding.
Conclusion
Standardized development of CDS from a CP is feasible using a systematic 4-phase process. CPs represent a potential reservoir for developers of evidence-based CDS. Aspects of CP development simplified portions of the CDS translation process. Publicly available tools can facilitate CDS development; however, enhanced tool features are needed to model complex CDS statements.
Keywords: clinical decision support; clinical quality language; pathways, dissemination, Clostridoides difficile
INTRODUCTION
Integrating evidence into clinical practice remains an ongoing challenge. Clinical decision support (CDS) within the electronic health record (EHR) can help bridge the evidence–practice gap.1–8 However, EHR-based CDS is expensive to develop and maintain,9,10 presenting a significant barrier for adoption by many health settings.11 One alternative is for organizations to create shareable, publicly available CDS artifacts. Although numerous barriers exist to using these CDS artifacts, including differences in patient populations, local EHR settings, and unique organizational workflow,3,12–14 recent advances in standards for data exchange and interoperability15,16 and public investment in infrastructure17 have created new opportunities for creation and use of these artifacts.11,18
CDS artifacts can be described by the degree to which they are executable and interoperable across health settings. Boxwala et al developed a 4-tiered schema to describe levels of CDS knowledge abstraction and translation.19 Briefly, Level 1 (L1) CDS artifacts contain mainly narrative information (eg, clinical practice guideline). Level 2 (L2) artifacts contain semi-structured information and are human readable (eg, algorithm, flow diagram). Level 3 (L3) artifacts have defined data elements, structure, and clinical concepts mapped to standardized clinical vocabularies. They are typically developed by knowledge engineers, are human readable, and can be computer readable/interpretable. Level 4 (L4) artifacts coded programs are executable by a local CDS computer system.19
Ideally, shareable CDS artifacts intended for EHR implementation should exist as L3 artifacts20,21 because they offer several advantages over L1 or L2 artifacts for settings that are considering implementation. L3 artifacts decrease divergent interpretations during implementation by requiring less knowledge abstraction. Furthermore, they reduce the need for specialized knowledge engineers within the implementation setting, allowing locations to focus primarily on the time intensive tasks of workflow analysis, integrating, testing, end user training, and implementation.19,22–25 However, L3 artifacts have limitations, because they do not natively function with all EHRs and require mapping of local codes.19
In 2019, the Agency for Healthcare Quality and Research (AHRQ) Evidence-based Practice Center (EPC) Program26 released a call for methods for improving implementation of EPC reports into clinical practice. ECRI and the Penn Medicine Center for Evidence-based Practice, which together are an AHRQ EPC member institution, had recently developed a clinical pathway (CP) for Clostridioides (formerly Clostridium) difficile infection (CDI) treatment.27 We sought to build upon ECRI’s expertise in developing shareable CDS from evidence-based recommendations12,28–30 to develop a systematic and transparent process to translate this CP into structured CDS.
MATERIALS AND METHODS
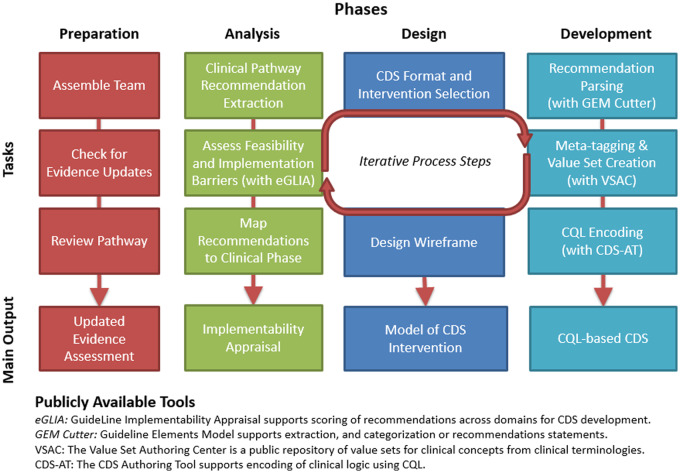
We developed an iterative process to translate clinical content from a CP for CDI treatment (L2) into structured CDS (L3). This process was divided into 4 phases, each with clearly identifiable tasks (Table 1). Completion of tasks led to interim deliverables for use in later phases (Figure 1). Development of the underpinning CP by the Penn Medicine Center for Evidence-based Practice has been previously described.27 Briefly, the CP provides guidance for clinicians on management of CDI for the spectrum of clinical presentations (eg, severity, recurrence status). It provides an algorithmic approach to determining the appropriate combination of medications, imaging studies, consultations, and procedures.31,32 The full CP and related materials have been made freely available at https://cds.ahrq.gov/cdsconnect/artifact/clostridoides-difficile-c-diff-infection-cdi-treatment-pathway.
Table 1.
CDS design and development tasks
| Phase | Description |
|---|---|
| Preparation | Assemble team and assign roles |
| Preparation | Confirm evidence base is current |
| Preparation | Review most recent version of the CP and supporting materials |
| Analysis | Extract recommendation statements from CP and assess recommendation statements for inclusion eligibility. |
| Analysis | Assess feasibility and barriers for conversion to L3 CDS artifact |
| Analysis | Assign clinical phase and target interventions to each statement |
| Design | Select CDS channel/intervention format for recommendation statements |
| Design | Create workflow based wireframes |
| Development | Parse recommendation statements and restructure content for encoding |
| Development | Meta-tagging and creation of standardized value sets |
| Development | Encode recommendation statements using CQL |
Abbreviations: CDS, clinical decision support; CP, clinical pathway; CQL, clinical quality language.
Figure 1.
CP (L2) to CQL (L3) translation process overview.
Preparation
We assembled a team with expertise across clinical and technical domains, including a physician informaticist, a guideline methodologist, physician subject matter experts (including infectious disease and hospital medicine), and a nurse–scientist with expertise in evidence translation and implementation science. To ensure CDS artifacts were being developed from the most recent evidence base, we conducted a targeted literature search (January 20, 2017 through January 11, 2019) to update a previously completed systematic review. Finally, we assessed the source CP readiness for content translation using an approach based on the GuideLines into Decision Support project.33
Analysis
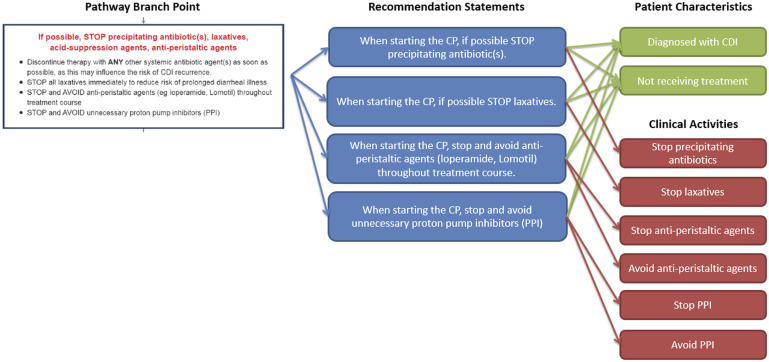
We extracted and converted CP branch points (ie, areas of the CP where a clinician would evaluate patient characteristics in order to choose between clinical activities) into 1 or more recommendation statements and evaluated barriers to EHR implementation. For example, the branch point describing initial medications to stop after a CDI diagnosis was converted into 4 separate recommendation statements, 1 for each medication to be stopped but all based on the same patient characteristics (Figure 2).
Figure 2.
Example conversion of CP branch point into recommedation statements. A single CP branch point is broken out into 4 recommendation statements. Each recommendation statement has 2 patient characteristics and 1–2 associated clinical activities.
We used the Guideline Elements Model (GEM) and GEM Cutter to facilitate content extraction.30,34 GEM Cutter promotes the categorization of clinical concepts as decision variables (triggers) and actions (clinical activities) from narrative text. Additionally, it maintains the link to the narrative source which supports process transparency. Using GEM, it is possible to separate complex recommendation statements into component parts which can facilitate future CDS development.35 We next classified actions according to action-type (eg, test, refer, monitor).36 We reviewed recommendation statements to identify and exclude those that would not be translated into executable CDS (eg, low clinical priority, non-actionable, and/or requiring data not available in an EHR).
Recommendation statements were next analyzed using the electronic Guideline Implementability Appraisal (eGLIA) instrument, a web-based tool for assessment of barriers and facilitators to implementation of guideline recommendations.37 Team members individually evaluated each recommendation statement across key domains for CDS development, including executability, decidability, measurability, and computability. One team member, who was a trained eGLIA facilitator, moderated discussions to address all discordant responses and generated a report of barriers and facilitators to implementation of recommendation statements.
Results from the eGLIA process were reviewed by the CP developers. Areas noted to have ambiguity, vagueness, or barriers to implementation (eg, consideration of a clinical action, decisions reliant on data that would not be located in an EHR) were addressed with revisions to the CP. As soon as changes were made to the CP, this updated content was provided to the CDS development team for incorporation into the CDS.
Design
To focus CQL encoding, we analyzed how recommendation statements would be applied in clinical practice and considered various EMR interface designs to support provider workflow.38 We used the “CDS 5 Rights Framework” to evaluate each recommendation statement and identified a preferred CDS format for implementation (eg, alerts or reminders for “avoid” recommendation statements and order sets for prescribing CP recommendation statements). The team reviewed the CDS format options, considered local culture, workflow, technical team availability for implementation, and build efficiency.
Development
We expressed each clinical concept using value sets (ie, lists of codes from standard clinical terminologies). We first queried the Value Set Authority Center (VSAC) to identify whether publicly maintained value sets already existed for clinical concepts in our recommendation statements.39 Three existing value sets were found, reviewed for completeness, and associated with the relevant clinical concepts. We then accessed a locally maintained database containing codes for all major publicly available clinical terminology standards (eg, ICD-10, Snomed, RxNorm), which had been populated according to instructions on the Unified Medical Language System (UMLS) website.40 We generated lists of codes for representing each remaining clinical concept. Once all lists were developed, they were reviewed by a subject matter expert. Final lists were published on the VSAC.
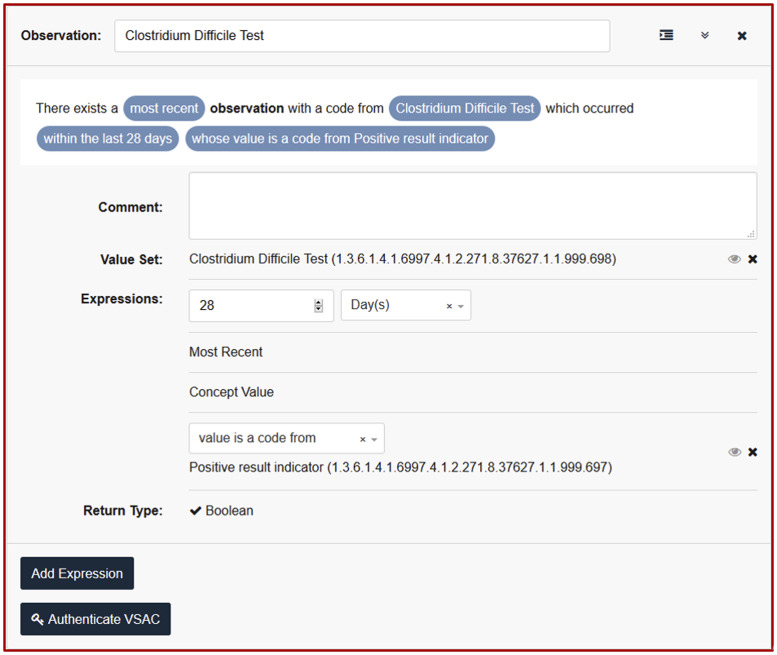
We next used the CDS Authoring Tool (CDS-AT), available through CDS Connect, to translate each recommendation statement into 1 or more CQL statements (Figure 3). CDS Connect is a publicly accessible AHRQ supported repository for CDS artifacts, many of which are contributed by CDS developers and have been made freely available. At the time of this work, CDS Connect was just beginning to support CQL authoring by external users. After an initial pass, we identified notable limitations to expressing recommendation statements within CQL using the CDS-AT. We resolved these limitations through manual edits of the CQL guided by a CQL specialist. For many of the issues identified we suggested alternatives and enhancements for the CDS-AT. The complete artifact developed during this project is available at https://cds.ahrq.gov/cdsconnect/artifact/clostridoides-difficile-c-diff-infection-cdi-treatment-pathway.
Figure 3.
CDS-AT being used to encode content as CQL.
RESULTS
Preparation & analysis phases
Our updated literature search identified 29 citations. These were reviewed for relevance by a clinician analyst. No studies were identified that necessitated updates to the CP.
We extracted 24 recommendation statements from the CP41 and classified each by clinical phase, clinical activity, and GEM action type (Table 2). Use of multiple categorizing schemes allowed for consideration of recommendation statements from multiple perspectives. Clinical phase addressed at what point in the workflow a clinician would carry out a recommendation (eg, treatment initiation, treatment selection etc), while clinical activity described the activity in further detail (eg, stop antibiotics or stop laxatives). The GEM action type is a classifier which aligned recommendation statements with a CDS function (eg, order, diagnose).30,34
Table 2.
Recommendation statements extracted from the CP
| CP recommendation statement | Clinical phase | Clinical Activity | GEM Action Type |
|---|---|---|---|
| Begin CP if inpatient, positive C. diff test, AND clinical signs/symptoms consistent with CDI | Pre Pathway Initiationa | Identify Patient | Diagnose |
| Begin CP if inpatient and high clinical suspicion (eg, fever, high white blood cell count, ≥ 3 documented liquid stools in 24 hours) | Pathway Initiationa | Identify Patient | Diagnose |
| When starting the CP, if possible STOP precipitating antibiotic(s). Discontinue therapy with inciting antibiotic agent(s) as soon as possible, as this may influence the risk of CDI recurrence. | Treatment Initiation | Stop Antibiotics | Prescribe |
| When starting the CP, if possible, STOP laxatives. | Treatment Initiation | Stop Laxatives | Prescribe |
| When starting the CP, stop and avoid anti-peristaltic agents (loperamide, Lomotil) throughout treatment course. | Treatment Initiation | Stop anti-peristaltic agents; Avoid anti-peristaltic agents | Prescribe |
| When starting the CP, stop and avoid unnecessary PPI. | Treatment Initiation | Stop PPI; Avoid PPI | Prescribe |
| Diagnose as recurrent CDI if positive C. diff test with recurrent symptoms attributable to CDI within 8 weeks of successfully completing treatment for previous CDI that was associated with interval improvement. | Treatment Selection | Diagnose as recurrent CDI | Prescribe |
| Diagnose as refractory CDI if lack of symptomatic improvement to appropriate prescribed treatment for CDI. | Treatment Selection | Diagnose as refractory CDI | Diagnose |
| If refractory CDI is suspected, consider alternative causes for infection. | Treatment Selection | Management of Refractory CDI | Consider |
| If refractory CDI is suspected, consult infectious disease. | Treatment Selection | Management of Refractory CDI | Refer/ Consult |
| If first (ie, nonrecurrent) CDI, obtain OR ensure has obtained within the last 24 hours CBC and BMP. | Treatment Selection | Evaluation, first episode | Test |
| Diagnose patient presenting with non-recurrent CDI as “CDI, non-severe” if WBC < 15 000 cells/mL AND Cr <1.5 mg/dl. | Treatment Selectionb | Diagnose as CDI Non-Severe | Diagnose |
| Diagnose patient presenting with non-recurrent CDI as “CDI, severe” if WBC>=15 000 cells/mL or Cr >=1.5 mg/dl. | Treatment Selectionb | Diagnose as CDI Severe | Diagnose |
| Diagnose patient presenting with non-recurrent CDI as “CDI, fulminant” if sepsis with acute organ dysfunction OR septic shock OR abdominal signs/symptoms (vomiting, distention) concerning for ileus, toxic megacolon. | Treatment Selection | Diagnose as CDI Fulminant | Diagnose |
| If CDI and not on antibiotics, treat with vancomycin, 125 mg q6h for 10 days. | Treatment Selection | Manage CDI Non-Fulminant, off antibiotics | Prescribe |
| If CDI and on antibiotics, treat with vancomycin, 125 mg q6h for 10 days minimum but consider extending the treatment course for 7 days beyond the current course of treatment. | Treatment Selection | Manage CDI Non-Fulminant, on antibiotics | Prescribe |
| If diagnosed with fulminant CDI, order a C. diff test to confirm. | Treatment Selection | Manage CDI Fulminant | Prescribe |
| If diagnosed with fulminant CDI, abdominal x-ray or CT is recommended if abdominal signs/symptoms (vomiting, distention) concerning for ileus, toxic megacolon. | Treatment Selection | Manage CDI Fulminant | Prescribe |
| If diagnosed with fulminant CDI, surgical and infectious disease consults are recommended. | Treatment Selection | Manage CDI Fulminant | Refer/ Consult |
| If diagnosed with fulminant CDI, and no significant abdominal findings treat with vancomycin 500 mg PO/NG Q6h x14 days and metronidazole 500 mg IV Q8H x 14 days. | Treatment Selection | Mange CDI Fulminant | Prescribe |
| If diagnosed with fulminant CDI, and significant abdominal findings treat with vancomycin 500 mg PO/NG Q6h x14 days vancomycin retention enema 500 mg in 100 mL sterile water q6h x14 days and metronidazole 500 mg IV Q8H x 14 days. | Treatment Selection | Mange CDI Fulminant | Prescribe |
| If recurrent CDI and first recurrence then treat with vancomycin 125 mg PO Q6H for 10 days (especially if previously treated with metronidazole) or vancomycin tapered regimen | Treatment Selection | Mange first recurrence | Prescribe |
| If recurrent CDI and not first recurrence then treat with vancomycin taper, consider infectious disease consult, and consider fecal microbiota transplantation. | Treatment Selection | Manage multiple recurrences | Prescribe |
| If following CP and no improvement within 5 days consider alternative diagnosis and consult infectious disease | Treatment Monitoringc | Monitor | Monitor |
Abbreviations: BMP, basic metabolic panel; CBC, complete blood count; CDI, Clostridioides difficile infection; CDS, clinical decision support; PPI, proton pump inhibitors; WBC, white blood cell.
Removed during CP revisions but retained in the CDS.
Removed during CP revisions and removed from CDS.
Excluded from CDS translation as out of scope for project.
Evaluating recommendation statements with eGLIA identified areas where CP guidance was imprecise, redundant, and/or lacked relevant details for implementation. This led to 10 changes to the CP (See Supplementary Appendix A). Alterations addressed imprecision (n = 6), incomplete guidance such as treatment for patients with vancomycin allergy (n = 3), removed redundancy (n = 2), and supported usability (n = 1). Two alterations had more than 1 clearly identified reason for initiating the CP change. The updated CP was published on CDS Connect.32
Based on input from clinical subject experts, we identified 23 of 24 recommendation statements as high priority. A single recommendation statement related to treatment monitoring was deemed out of scope because it did not address decision making related to initial treatment for CDI. Two recommendation statements related to CP initiation were removed from the revised CP, but retained in the CDS because they were necessary for identifying patients. Two additional recommendation statements were included in the initial extraction, but later removed due to CP revisions and not translated into CDS. In total, 21 of 24 recommendation statements were retained for CDS development.
Design
CDS format and intervention selection
The majority of recommendation statements pertained to medication selection. Because of this we chose an order set intervention format as the most appropriate to support clinicians and limit selection of non-preferred treatments (eg, fidaxomicin for vancomycin tolerant patients). For recommendation statements preceding treatment initiation we considered using alerts or guidance text. Ultimately, recommendation statements preceding treatment initiation were included as guidance text within an order set, eliminating the need for an implementing organization to deploy a series of alerts for this CDS.
Wireframe design
The wireframe for an EHR order set interface was developed by our team’s physician informaticist. All 21 retained recommendation statements were incorporated into the order set wireframe as orders or informational statements (Supplementary Appendix B). To support handling of related orders, orders were grouped by disease state (ie, first episode, fulminant episode, first recurrence, and multiple recurrence). At the top of the order set, we embedded CDI classification guidance, recommendation statements related to treatment initiation, and a hyperlink to the CP. The wireframe was iteratively refined by the project team. The final product was reviewed by infectious disease clinicians for additional input. This served as a model for the CQL encoding.
Development
Recommendation statement parsing
Using GEM, we identified 40 unique clinical concepts within recommendation statements extracted from the CP necessary for expressing logic within a L3 CDS artifact. This included 21 trigger conditions (eg, vancomycin allergy, abdominal pain) and 19 clinical activities (eg, prescribe fidaxomicin). The GEM report also allowed for identification of all relationships between clinical concepts that would need to be encoded in the CDS. Numerous (n = 42) logic relationships (eg, “and” and “or”), 6 evaluations (eg, occurrence count of loose stool greater than or equal to 3), and 3 temporal relationships (eg, starts less than 4 weeks before the start of) were identified as relevant for encoding.
Meta-tagging & value set creation
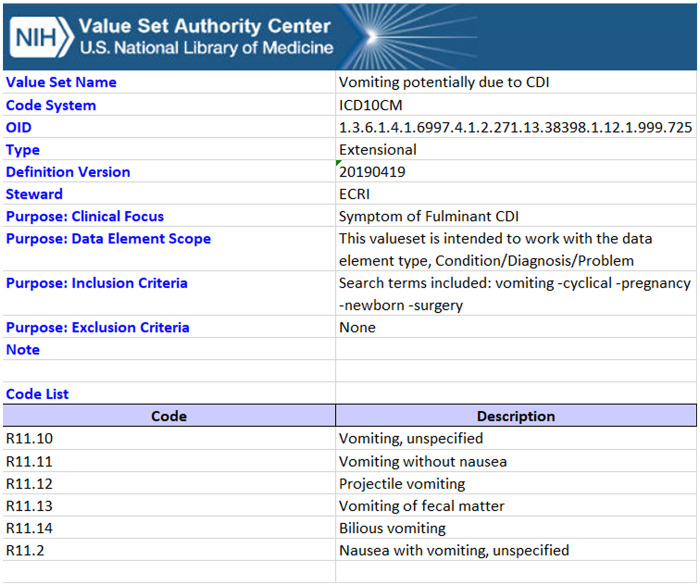
As previously noted, querying VSAC identified 3 existing relevant value sets maintained by reputable sources which sufficiently encapsulated clinical concepts. We developed an additional 61 value sets, of which 50 were published through VSAC. Eleven value sets we created were not published due to changes in the underlying CP that rendered them unnecessary. Including the 3 previously existing value sets, 53 value sets were utilized for our list of clinical concepts. All published value sets for this project are listed in Appendix C, and an example value set for Vomiting potentially due to CDI is provided in Figure 4.
Figure 4.
Example value set for “Vomiting potentially due to CDI.” This value set includes codes from ICD-10 and is a subset of the codes representing the clinical focus of “symptoms of fulminant CDI.” The inclusion criteria indicate the search string used to generate this value set, allowing for complete transparency of the value set development.
Some concepts used multiple value sets (a value set is limited to codes from a single terminology) and certain concepts were expressed simultaneously in multiple terminologies [eg, ICD-10 and Snomed-CT]. Each concept expressed with more than 1 value set required an additional ‘grouper’ value set within VSAC. Our clinical concept encoding included 8 grouper value sets comprising 22 internal value sets.
During the quality control (QC) review task, the team identified 4 distinct clinical concept codes from 3 different value sets that were clinically inappropriate (eg, related to pediatric indications or diseases when the CP clearly noted adult context). The team also identified 1 clear omission in the value set for “computed tomography (CT) abdomen.” Upon review, we discovered a source code description was misspelled within the Snomed-CT terminology and consequently not captured in the lookup. After incorporating QC findings, value sets were published on VSAC.
CQL encoding
We expressed the clinical concept codes and value sets using CQL statements using the CDS-AT (Figure 3). The CDS-AT guides users through defining several different types of CDS statements to help authors organize and develop relationships between content. We selected 4 different types of CDS statements for this project: Inclusion Criteria, Subpopulations, Base Elements, and Recommendations. Inclusion Criteria define populations for whom the CQL is relevant. Subpopulations define groups within the population for which a specific Recommendation is applicable. Base Elements allow for creation of reusable content, which simplifies development and project maintenance (eg, from a vancomycin base element, we could develop other CQL constructs such as allergy to vancomycin, vancomycin taper, and vancomycin enema). Recommendations link a subpopulation, or groups of subpopulations with clinical advice. In total we used 1 Inclusion Criterion, 15 Subpopulations, 20 Base Elements, and 16 Recommendations. Each Inclusion Criteria, Subpopulation, and Base Element statement referenced 1 or more value sets we developed. Value sets were used both to identify clinical concepts and express allowable values (eg, value sets for the test concept “Test for C. difficile” and the result concept “Positive result”).
The clinical concept of CDI was encoded using an Inclusion Criterion since it fully defined the relevant patient population. Other trigger conditions were encoded using Subpopulations. Fifteen Subpopulations (eg, “receiving laxatives,” “recent CDI”) were needed to express the clinical concepts. Of note, 3 Subpopulations were simply the inverse of other Subpopulations. At the time, the CDS-AT did not yet support negation of Subpopulations. For example, it was necessary to develop separate Subpopulations for “receiving laxatives” and “not receiving laxatives” to express the laxative recommendation statement.
We used Base Elements for 2 purposes: 1) reusable portions of compound or complex trigger conditions and 2) a placeholder for executable representation of clinical activities. Compound statements included those with temporal relationships (eg, CDI started less than 8 weeks after end of previous CDI episode) or other complex relationships. Compound CQL statements contained between 2 and 5 Base Elements. Clinical activities were expressed using Base Elements because the CDS-AT did not natively support linking actions in Recommendations. Clinical activities were expressed with a similar level of complexity as other CQL statements (between 1 and 5 clinical concepts per statement).
Logic specifying when each action should occur was encoded using the Recommendation form. A single subpopulation, or in 6 instances 2 subpopulations, were linked to clinical activity Base Elements via the development of Recommendation CQL statements.
DISCUSSION
Our work demonstrates that transparent development of shareable CDS from a CP using publicly available software is feasible and can be conducted using a systematic process. We followed a 4-phase process to translate content that included preparation, analysis, design, and development. Output from each phase informed the next task. Not surprisingly, organizations have communicated the desire to independently review evidence prior to implementing CDS. Our process provides products at each intermediate step which can be evaluated and support rapid assessment by potential users from other organizations.
We are unaware of prior studies documenting a systematic approach to translation of a CP (an L2 CDS artifact) into a more structured CDS (L3). Because our methods are adapted from processes for quality measure development,29 we believe these methods for translation can be similarly adapted for other targets or from other intermediate CDS artifacts. We noted 4 key lessons from this work: 1) benefits of utilizing publicly available software and tools, 2) benefits of starting from an existing CP, 3) collaboration and iteration to improve the CDS artifact and the CP, and 4) a systematic process that supports transparency and QC.
Publicly available software can facilitate development of CDS
Our work used many publicly available tools including Yale’s GEM Cutter and eGLIA software, the National Library of Medicine’s VSAC, and AHRQ’s CDS-AT. These applications facilitate transparent CDS development and support long-term CDS maintenance by creating intermediate end products.35,37,39,42 Use of existing value sets from VSAC decreased development effort and allowed alignment with other related projects. Additionally, by not creating redundant value sets, we avoided contributing to proliferation of overlapping value sets (eg, multiple “sepsis” value sets each maintained by different organizations), reducing confusion and divergence in concept definitions. Similarly, publishing through VSAC allows others to utilize our end products and build upon our work.
For implementation, we utilized the CDS-AT to express content using CQL statements.43 The CDS-AT is in early stages of development and all CQL encoding permutations are not yet supported.42 In particular, branch points containing temporal relationships were difficult to express within the CDS-AT. For example, 1 portion of the recurrent CDI definition included the concept “CDI that starts after the start of vancomycin treatment.” This statement contained 2 clinical concepts and a temporal relationship between them. As another example, the CDS-AT triggers only a single and first-matched Recommendation from a list of Recommendations, and therefore the order in which Recommendations are listed controls which Recommendation is triggered. However, the CP supported multiple clinical activities for each set of trigger criteria. Consultation with a CQL specialist (MITRE Corporation, McLean, VA) was needed to ensure the CQL appropriately represented the CP. We manually edited the CQL to allow for multiple recommendation statements to be concurrently triggered to modify Base Elements once they had been used and to allow Boolean data types from empty sets that resulted as True. Further use and testing with pragmatic projects will be helpful to identify other areas for enhancement to the CDS-AT.
Starting with a CP offered notable benefits
We noted clear benefits from starting with a CP as the primary content source for translation into an L3 CDS artifact instead of a clinical practice guideline (CPG) or other narrative source. While ideally CPGs would be developed alongside CDS, we note this rarely occurs, and translation of CPGs into CDS is an arduous process.44,45 Assumptions made during CPG to CDS adaptation can have far-reaching consequences and may lead to biases in care delivery.46 During CP development, the development team had already considered clinical workflows and incorporated several design principles to optimize iterations.47 For instance, all likely patient scenario combinations (ie, “recurrent” and “fulminant” CDI) and relevant care decisions had been considered. Because this is a requisite step for developing executable CDS, this directly reduced the work required by the CDS development team in identifying relevant patient populations and inclusion criteria. While some CPs are developed concurrently with CDS or with a quality measure, many are not directly executable within an EHR (eg, paper-based) or are not expressed using a structured format.47 Our work suggests that high-quality evidence-based CPs may represent a largely untapped reservoir for development of executable CDS.
Close collaboration and iteration improved end products and the source CP
A close working relationship across team members with expertise in CDS development, clinical care, and knowledge of CP development was key for efficiency and resulted in improvements to the source CP. For instance, the CP development team provided input during a first-pass evaluation of recommendation statements and identified priority recommendations allowing the CDS development team to avoid using resources to extract and encode low-priority guidance.
Unlike CPGs which are developed to meet the needs of stakeholders across various settings, CPs can be rapidly updated to address context-specific clinical workflows. Our translation process revealed several areas within the CP which could be improved. Specifically, eGLIA evaluation and GEM Cutting identified areas of ambiguity, gaps, and redundancies within the CP. Resulting changes made by the CP development team improved clarity and supported development of a revised, more actionable version of the CP.
A systematic process supports transparency and quality control activities
QC is key for all CDS development work. Our QC processes support identification of errors of omission and commission, both of which can compromise clinical care. For example, during value set development, we identified incorrect codes within our preliminary value sets (eg, pediatric-specific codes for megacolon). Our systematic and retained searches allowed for easy revision of criteria and elimination of erroneous codes from the final product. We also noted a misspelling in 1 code within a source terminology, which resulted in a code being temporarily missed. We reported this discrepancy to the source for correction. These QC processes are important for the trustworthiness of value sets and support shareable CDS by improving quality of publicly available value sets.
We primarily used a locally maintained database derived from UMLS, rather than accessing the UMLS website directly to support these QC activities. This allowed our team better control over search strategy retention and supported our QC process. Value set authoring is natively supported in VSAC, and instructions on developing a clinical terminology standards database are available on the UMLS website. Other groups looking to replicate our methods would have the ability to use this publicly available infrastructure.
Limitations
Our methodology has several important limitations. Because we used CQL, which is not yet supported at Penn Medicine for EHR integration, we could not perform production testing to clinically evaluate the match between CP and CDS guidance. As a proxy, we used a list of system-generated patients with prespecified characteristics to evaluate the final CDS. However, some CDS anomalies may not emerge until testing against real-world data can occur. Thus, despite our QC process, it is possible that value sets may not match the data stored within EHRs, and proxies for missing data may be needed to support local implementation. CQL is an L3 specification, and therefore sites interested in implementing would also need to adapt this work to local EHR implementations.
Our work describes adaption of a single CP, which was based on a systematic review of the evidence. Using CPs developed under less rigorous conditions could impact the translation process, leading to increased barriers to adaptation. For instance, less rigorously developed content sources may have deficiencies that impact implementation or include more areas of ambiguity and vagueness requiring resolution. The process outlined in this manuscript would benefit from validation with additional CPs to demonstrate reproducibility. We sought to make this process as transparent and feasible as possible with publicly available tools in order to support other CDS developers and CQL authors in completing similar translation work.
Future directions
Tools to facilitate the use of standards for interoperability and public investment in the development of shareable L3 CDS artifacts could create opportunities for widespread CDS use and adaptation. Specifically, key enhancements could be made to the CDS-AT to unleash the full potential of CQL. Features necessary to fully express logic from a CP would include handling of relationships between clinical concepts, selectable instances of clinical concepts, and the ability to define a lookback period from a relative date rather than from the current date.
CONCLUSION
Using evidence-based CPs as a source for structured CDS development represents another method for bringing evidence closer to the point of care. This work exemplifies translation of a CP into CDS using a systematic, transparent process. Additionally, because CPs are more easily mutable than CPGs, translation of a CP into CDS helped to improve the source CP as a byproduct of the translation work. Publicly available software can support analysis, encoding, and CQL development, however, key enhancements are needed to improve widespread adoption and support shareable CDS.
FUNDING
This work was supported by the Agency for Healthcare Research and Quality contract No. 290-2015-00005-I.
AUTHOR CONTRIBUTIONS
All authors participated in the design, development, and conceptualization of the project. JM organized and contributed to manuscript authorship, performed the CDS development and encoding, and functioned as facilitator for barriers to implementation analysis. EF and NM led the design and pathway updates, participated in implementation barrier analysis, and participated in manuscript authorship. LD provided subject matter expertise, manuscript review, and participated in implementation barrier analysis. AT provided project leadership in addition to manuscript authorship and review of CDS content development.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
The authors gratefully acknowledge the following individuals for their contributions to this project: ECRI: Jane Jue, MD, MSc, Amber Moran, MA, Kariann Hudson, MEd, Jennifer Maslin, and Nicholas Lepkowski; MITRE (CDS Connect): Sharon Sebastian, David Winters, PhD, and Chris Moesel, and AHRQ: Kim Wittenberg, MA.
CONFLICT OF INTEREST STATEMENT
None declared.
REFERENCES
- 1. Van de Velde S, Kunnamo I, Roshanov P, et al. The GUIDES checklist: development of a tool to improve the successful use of guideline-based computerised clinical decision support. Implement Sci 2018; 13 (1): 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boaz A, Baeza J, Fraser A, the European Implementation Score Collaborative Group (EIS). Effective implementation of research into practice: an overview of systematic reviews of the health literature. BMC Res Notes 2011; 4 (1): 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sittig DF, Wright A, Osheroff JA, et al. Grand challenges in clinical decision support. J Biomed Inform 2008; 41 (2): 387–92. Epub 2007/11/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eric GP, Blumenthal D, Jaggi T, Honour MM, Bates DW, Kaushal R.. Overcoming the barriers to the implementing computerized physician order entry systems in US hospitals: perspectives from senior management. AMIA Annu Symp Proc 2003; 2003: 975. [PMC free article] [PubMed] [Google Scholar]
- 5. Forrest CB, Fiks AG, Bailey LC, et al. Improving adherence to otitis media guidelines with clinical decision support and physician feedback. Pediatrics 2013; 131 (4): e1071–e1081. [DOI] [PubMed] [Google Scholar]
- 6. Ash JS, Sittig DF, Dykstra R, et al. Identifying best practices for clinical decision support and knowledge management in the field. Stud Health Technol Inform 2010; 160(Pt 2): 806–10. [PMC free article] [PubMed] [Google Scholar]
- 7. Bell LM, Grundmeier R, Localio R, et al. Electronic health record-based decision support to improve asthma care: a cluster-randomized trial. Pediatrics 2010; 125 (4): e770. [DOI] [PubMed] [Google Scholar]
- 8. Heselmans A, Van de Velde S, Donceel P, et al. Effectiveness of electronic guideline-based implementation systems in ambulatory care settings: a systematic review. Implement Sci 2009; 4: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Middleton B, Sittig D, Wright A.. Clinical decision support: a 25 year retrospective and a 25 year vision. Yearb Med Inform 2016; 25 (S 01): S103–S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Field TS, Rochon P, Lee M, et al. Costs associated with developing and implementing a computerized clinical decision support system for medication dosing for patients with renal insufficiency in the long-term care setting. J Am Med Inform Assoc 2008; 15 (4): 466–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wright A, Sittig DF, Ash JS, et al. Clinical decision support capabilities of commercially-available clinical information systems. J Am Med Inform Assoc 2009; 16 (5): 637–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Orenstein EW, Yun K, Warden C, et al. Development and dissemination of clinical decision support across institutions: standardization and sharing of refugee health screening modules. J Am Med Inform Assoc 2019; 26 (12): 1515–24. doi:10.1093/jamia/ocz124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Paterno MD, Maviglia SM, Ramelson HZ, et al. Creating Shareable Decision Support Services: An Interdisciplinary Challenge. AMIA Symp Proc 2010; 2010: 5. [PMC free article] [PubMed] [Google Scholar]
- 14. Hongsermeier T, Maviglia S, Tsurikova L, et al. A legal framework to enable sharing of Clinical Decision Support knowledge and services across institutional boundaries. AMIA Annu Symp Proc 2011; 2011: 925–33. [PMC free article] [PubMed] [Google Scholar]
- 15. Mandel JC, Kreda DA, Mandl KD, et al. SMART on FHIR: a standards-based, interoperable apps platform for electronic health records. J Am Med Inform Assoc 2016; 23 (5): 899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.HL7 International. FHIR Release 4. HL7; 2019. https://www.hl7.org/implement/standards/product_brief.cfm?product_id=491 Accessed July 20, 2020.
- 17.Agency for Healthcare Research and Quality (AHRQ). CDS Connect. U.S. Department of Health & Human Services; 2017. https://cds.ahrq.gov/ Accessed July 20, 2020.
- 18.Agency for Healthcare Research and Quality (AHRQ). Clinical Decision Support. U.S. Department of Health & Human Services; 2017. https://healthit.ahrq.gov/ahrq-funded-projects/current-health-it-priorities/clinical-decision-support-cds
- 19. Boxwala AA, Rocha BH, Maviglia S, et al. A multi-layered framework for disseminating knowledge for computer-based decision support. J Am Med Inform Assoc 2011; 18 (Supplement 1): i132–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shankar P, Anderson N.. Advances in sharing multi-sourced health data on decision support science 2016–2017. Yearb Med Inform 2018; 27 (01): 016–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Marco-Ruiz L, Bellika JG.. Semantic interoperability in clinical decision support systems: a systematic review. Stud Health Technol Inform 2015; 216: 958.Epub 2015/08/12. [PubMed] [Google Scholar]
- 22. Lomotan EA, Meadows G, Michaels M, et al. To share is human! Advancing evidence into practice through a national repository of interoperable clinical decision support. Appl Clin Inform 2020; 11 (01): 112–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thayer JG, Miller JM, Fiks AG, et al. Assessing the safety of custom web-based clinical decision support systems in electronic health records: a case study. Appl Clin Inform 2019; 10 (02): 237–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li AC, Kannry JL, Kushniruk A, et al. Integrating usability testing and think-aloud protocol analysis with “near-live” clinical simulations in evaluating clinical decision support. Int J Med Inform 2012; 81 (11): 761–72. [DOI] [PubMed] [Google Scholar]
- 25. Press A, McCullagh L, Khan S, et al. Usability testing of a complex clinical decision support tool in the emergency department: lessons learned. JMIR Hum Factors 2015; 2 (2): e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agency for Healthcare Research and Quality. Evidence-based Practice Center (EPC) Program Overview Rockville, MD Content last reviewed. 2020. https://www.ahrq.gov/research/findings/evidence-based-reports/overview/index.html Accessed July 20, 2020.
- 27. Flores EJ, Jue JJ, Giradi G, et al. AHRQ EPC series on improving translation of evidence: use of a clinical pathway for C. Difficile treatment to facilitate the translation of research findings into practice. Jt Comm J Qual Patient Saf 2019; 45 (12): 822–8. [DOI] [PubMed] [Google Scholar]
- 28. Sledge GW Jr., Miller RS, Hauser R.. CancerLinQ and the future of cancer care. Am Soc Clin Oncol Educ Book 2013; (33): 430–4. [DOI] [PubMed] [Google Scholar]
- 29.AAO-HNSF. Reg-ent MIPS 2018 Reporting - Quality Measures. AAO-HNSF; 2018. Accessed July 20, 2020.
- 30. Shiffman RN, Karras BT, Agrawal A, et al. GEM: A proposal for a more comprehensive guideline document model using XML. J Am Med Inform Assoc 2000; 7: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Michel J, Flores E, Mull N, et al. Translation of a Clinical Pathway for C. Difficile Treatment Into a Machine-Readable Clinical Decision Support Artifact Prototyped for Electronic Health Record Integration (Publication No. 20-EHC001-EF). Rockville, MD: AHRQ; 2019. [PubMed] [Google Scholar]
- 32. Flores E. Clostridoides Difficile (C. diff) Infection (CDI) Treatment Pathway. University of Pennsylvania Health System: CDS Conenct; 2018. https://cds.ahrq.gov/cdsconnect/artifact/clostridoides-difficile-c-diff-infection-cdi-treatment-pathway Accessed July 20, 2020.
- 33. Dixon MM, Shiffman RN. GuideLines Into DEcision Support (GLIDES). Yale University; 2013. https://medicine.yale.edu/cmi/glides/ Accessed July 20, 2020.
- 34. Michel G. GEM Cutter II. Yale Center for Medical Informatics; 2007.
- 35. Hajizadeh N, Kashyap N, Michel G, et al. GEM at 10: A decade’s experience with the Guideline Elements Model. AMIA Annu Symp Proc 2011; 2011: 520–9. [PMC free article] [PubMed] [Google Scholar]
- 36. Essaihi A, Michel G, Shiffman RN.. Comprehensive categorization of guideline recommendations: Creating an action palette for implementers. AMIA Annu Symp Proc 2003; 2003: 220–4. [PMC free article] [PubMed] [Google Scholar]
- 37. Shiffman RN, Dixon J, Brandt C, et al. The GuideLine Implementability Appraisal (GLIA): development of an instrument to identify obstacles to guideline implementation. BMC Med Inform Decis Mak 2005; 5 (1): 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sirajuddin AM, Osheroff JA, Sittig DF, et al. Implementation pearls from a new guidebook on improving medication use and outcomes with clinical decision support. Effective CDS is essential for addressing healthcare performance improvement imperatives. J Healthc Inf Manag 2009; 23 (4): 38–45. Epub 2009/11/10. [PMC free article] [PubMed] [Google Scholar]
- 39.National Library of Medicine. Value Set Authority Center. National Library of Medicine; 2012. [updated 7/16/2016]. https://vsac.nlm.nih.gov/ Accessed July 20, 2020.
- 40. Humphreys BL, Lindberg DA, Schoolman HM, et al. The Unified Medical Language System: an informatics research collaboration. J Am Med Inform Assoc 1998; 5 (1): 1–11. Epub 1998/02/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Flores E, Jue JJ, Giradi G, et al. Use of a clinical pathway to facilitate the translation and utilization of AHRQ EPC Report Findings. Rockville (MD): Agency for Healthcare Research and Quality (US); 2018. [PubMed]
- 42.Agency for Healthcare Research and Quality (AHRQ). CDS Authoring Tool. U.S. Department of Health & Human Services; 2017. https://cds.ahrq.gov/cdsconnect/authoring Accessed July 20, 2020.
- 43.HL7 Standard: Clinical Quality Language Specification, Release 1 STU3 (CQL 1.3). Health Level Seven International; 2018. https://cql.hl7.org/STU3/
- 44. Tso GJ, Tu SW, Oshiro C, et al. Automating Guidelines for Clinical Decision Support: Knowledge Engineering and Implementation. AMIA Annu Symp Proc 2016; 2016: 1189–98. [PMC free article] [PubMed] [Google Scholar]
- 45.Institute of Medicine. Clinical Practice Guidelines We Can Trust. Washington, DC: The National Academies Press; 2011.
- 46. Gagliardi AR, Brouwers MC, Palda VA, et al. An exploration of how guideline developer capacity and guideline implementability influence implementation and adoption: study protocol. Implement Sci 2009; 4: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kinsman L, Rotter T, James E, et al. What is a clinical pathway? Development of a definition to inform the debate. BMC Med 2010; 8 (1): 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.