Abstract
Objective
Clinical trials ensure that pharmaceutical treatments are safe, efficacious, and effective for public consumption, but are extremely complex, taking up to 10 years and $2.6 billion to complete. One main source of complexity arises from the collaboration between actors, and network science methodologies can be leveraged to explore that complexity. We aim to characterize collaborations between actors in the clinical trials context and investigate trends of successful actors.
Materials and Methods
We constructed a temporal network of clinical trial collaborations between large and small-size pharmaceutical companies, academic institutions, nonprofit organizations, hospital systems, and government agencies from public and proprietary data and introduced metrics to quantify actors’ collaboration network structure, organizational behavior, and partnership characteristics. A multivariable regression analysis was conducted to determine the metrics’ relationship with success.
Results
We found a positive correlation between the number of successful approved trials and interdisciplinary collaborations measured by a collaboration diversity metric (P < .01). Our results also showed a negative effect of the local clustering coefficient (P < .01) on the success of clinical trials. Large pharmaceutical companies have the lowest local clustering coefficient and more diversity in partnerships across biomedical specializations.
Conclusions
Large pharmaceutical companies are more likely to collaborate with a wider range of actors from other specialties, especially smaller industry actors who are newcomers in clinical research, resulting in exclusive access to smaller actors. Future investigations are needed to show how concentrations of influence and resources might result in diminished gains in treatment development.
Keywords: complex system, science of science, clinical research, collaboration network, network science
INTRODUCTION
Background and Significance
Drug research and development (R&D) is a complex, expensive undertaking that is prone to failure. Given that on average it may take over 10 years and cost up to $2.6 billion to develop a single approved molecule,1 drug R&D has become a collaborative effort. During a drug’s lifespan, it is common for a spectrum of actors including government, academic, nonprofit organizations, pharmaceutical, and biotechnology companies to conduct phases of the basic, preclinical, and clinical research, with each contributing toward the development of a drug that is eventually approved by a regulatory agency. Furthermore, these actors collaborate to increase their research capabilities through access to key technologies or specialized knowledge developed or possessed by other actors.2–4 Collaboration can include vertical alliance networks in which each actor performs a relatively distinct set of activities along the value chain.5 Success or achievement in drug development is dependent upon these collaboration networks.6 Examining collaboration networks and the actors involved can yield valuable insight into the drug R&D process by identifying the behavior and patterns of successful actors and capturing the emergence of the collective structure.
Network analysis is useful for studying the network externalities of collaboration and transfer of knowledge and information between actors in the network. Some studies have examined collaborative networks based on contractual alliances within the pharmaceutical industry,7,8 while others have studied knowledge networks by mapping the dissemination of knowledge via patent citations.9–12 However, few studies have examined the collaboration network of actors in a clinical trial context.
Previous research has vaguely defined network “cohesion” as a basic property used to characterize the connectivity level of an entire network (global) or around an actor (local). In some studies, they found that cohesion impacts the speed and reach of knowledge transfer among actors that facilitate research breakthroughs. It is known that a weakly connected network with low cohesion usually has a larger path length between each actor which is evident in many network flow problems. Thus, on the one hand, in a weakly connected network, knowledge transfer would be slow because the information has to pass through more intermediate actors to reach another actor. On the other hand, a highly cohesive network may reach a point where excess connections lead to frequent transfers of redundant knowledge, which eventually impedes research efficiency.13–15 This theory is referred to as the echo chamber effect and is supported by previous studies such as policy positions regarding climate.16 Cohesion generally can be measured by the clustering coefficient.11,17,18 Guler and Nerkar19 used this metric to investigate the relationship between innovation and network cohesion and found that local cohesion was beneficial to innovation. This theory implies that actors who have similar ideological positions make connections to a similar set of actors resulting in a reinforcement of ideals. We investigate this effect in the clinical research environment and measure whether cohesion (social embeddedness) of an actor impedes research efficiency.
In addition to cohesion, organizational characteristics of the actors and their collaborators play an important role in knowledge acquisition and creation that contributes to higher clinical research output (ie, the number of drugs approved and research efficiency). These organizational characteristics include the type of organization (eg, nonprofit, academic) and their research portfolio which reflects their experience. Research indicates there are advantages to partnering with a diverse network of collaborators.20–23 One reason is that collaboration with a partner that has uniquely different knowledge bases and research portfolios allows an actor to explore domains that were previously outside their own expertise which are potentially difficult or impossible to access without a knowledgeable partner.3 Another reason that a diverse network may be useful is that a wider knowledge base might help actors maintain alliance ties.24 Additionally, research indicates actors may seek to obtain or exploit innovations developed by partners,4 but to truly innovate, an actor must be able to combine preexisting knowledge with new knowledge that was obtained through collaboration.10 Therefore, diverse collaborations through networks comprised of heterogeneous actors may serve to expand an actor’s knowledge base or portfolio, which may be beneficial for drug research and development.
OBJECTIVE
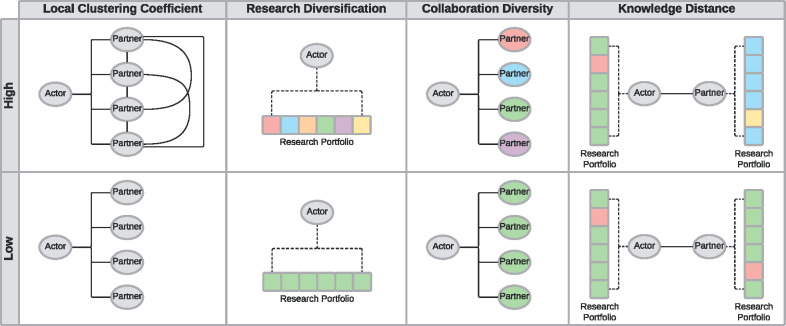
In this study, we characterize actors' roles within collaborations using network and diversity metrics to examine the extent to which their collaborative behavior fosters the development of new drugs. Figure 1 shows a selection of measures that we used to quantify each actor (see Materials and Methods and Supplementary Appendix for more details). Rather than consider the networks formed through patents or contractual alliances, this is the first study to examine the networks formed through collaborations on running clinical trials. We focus our investigation on the local network structure of each actor, research portfolio diversity, and collaborators' characteristics relative to the actor in question.
Figure 1.
Diagram of selected metrics that are used to characterize each actor in our analysis: local clustering coefficient (cohesion), research diversification, collaboration diversity, and knowledge distance. To illustrate the differences in the metric values, the top row represents high values of each metric, while the bottom row represents the low values. The ovals represent the observed actor and their partners. Research portfolios are represented as stacked squares, with each square symbolizing a trial in their portfolio. The fill color in each square symbolizes the therapeutic area of a clinical trial. Therefore, the more colors there are in an actor's research portfolio, the more diversity there is in an actor's research portfolio. Likewise, the fill color in an oval designates the therapeutic area in which an actor has expertise.
MATERIALS AND METHODS
Data sources and collaboration network construction
We focused our investigation on empirical data from 4494 organizations and 18 040 trials extracted from the Aggregate Analysis of ClinicalTrials.gov25 and BioMedTracker Pharma Intelligence databases (see Supplementary Appendix S1 for data collection and processing methods).26 Using these data, we were able to construct a 2-mode affiliation network that included the actors (sponsors and partners) and clinical trials (distinguished by the national clinical trial [NCT] identifier number). We then transformed the 2-mode affiliation network using a bipartite projection into a one-mode collaboration network that ranged from January 2006 to January 2016. The 1-mode network defines nodes as a single actor, and a link represents at least 1 instance of collaboration on a clinical trial between a pair of actors. We conducted an egocentric analysis on a dynamic, 1-mode collaboration network in which the focus is on the organizations or actors over a period of time.
We accounted for temporal differences by generating multiple time-dependent networks to capture the evolution of the network structure over the considered time period. The network was generated for every month between January 2006 to January 2016, which resulted in 121 monthly snapshots. A node or link is active at a given snapshot if the respective actor and collaborator are involved in at least 1 trial during the observed month. Based on this network, we developed several metrics to measure organizational, collaboration, and structural characteristics of each actor in the clinical trials collaboration network for each month.
Network measures and actors' attributes
In order to quantify the effects of the actor's network and research characteristics on research performance, we developed 2 metrics: cumulative trial successes, (research output); and trial success rate (research efficiency). Cumulative trial successes is defined as the cumulative number of clinical trials that an actor has been involved in as a sponsor or collaborator that eventually led to an Food and Drug Administration–approved drug. The trial success rate for each actor is the cumulative trial successes divided by the overall number of trials, which is the number of times that an actor has been classified as either a sponsor or collaborator.
We classified the actors into 6 organization-types: academic, government, nonprofit, industry, hospital system, and large pharmaceutical companies. determined the classifications based on additional data gathering efforts using publicly available sources and other methods (see Supplementary Appendix S3.1 and Supplementary Table S2 for organization type classification definition). We differentiated between large pharmaceutical companies and industry actors by selecting the companies that were ranked as either the top 25 with the highest market capitalization in 2016, top 15 revenue in 2016, or top 15 R&D budgets in 2016. The complete listing for large pharmaceutical companies is included in Supplementary Table S3. This allowed us to stratify and add fixed effects to control for the organization type in our regression.
For each actor on the network, we computed several node-specific metrics that quantified expertise, structural, organizational, and collaboration characteristics. Expertise is determined by designating each actor as having specialization in one particular therapeutic area (see Supplementary Appendix S3B). Structural characteristics, such as betweenness centrality and local clustering coefficient, are local network characteristics of the actor (see Supplementary Appendix S3C). Organizational characteristics are based on the clinical research experience of the actor (see Supplementary Appendix S3D). Collaboration characteristics are based on the relative comparison of all collaborators vs the observed actor (see S3E in Supplement).
Structural characteristics
Betweenness centrality is defined as
| (1) |
where the denominator σjk represents the shortest path between nodes j and k in node set, V, which includes all possible pairwise combination of nodes in the network, and the numerator σjk(i) represents the number of shortest paths from node j to node k that goes through node i.27 This metric is useful for measuring the extent to which a node acts as a “bridge” between 2 communities.
The local clustering coefficient, CCit, measures the extent to which an actor's neighbors are connected to each other for actor i at time t. If an actor's local neighborhood, which includes itself and its neighbor, is fully connected as a clique (fully connected subgraph), then the clustering coefficient would be 1, while a completely unconnected local network would be 0. Formally, the local clustering coefficient for an undirected graph is defined as
| (2) |
where Lit represents the number of links between the neighbors of actor i at time t, and δit represents the number of degrees of actor i at time t.
Organizational characteristics
Research diversification, , gives us an impression of the level of interdisciplinary experience in an actor's clinical trials portfolio. We quantified using an entropic measure that measures the heterogeneity of actor i's knowledge mix vector .
| (3) |
We assumed that a company that has completed 0 trials will have an entropy of 0.
Using RDit defined in equation 3, we can determine the mean neighbor research diversification of all partnering organizations j that collaborate with actor at any given time period . The mean research diversification 〈RD〉it is simply
| (4) |
where RDjt is the research diversification of actor i's partner, actor j.
Knowledge is developed through an actor's experience which can be quantified as the number of trials conducted in each therapeutic area (eg, neurology). By stratifying knowledge by therapeutic area, we can determine the relative competencies of each actor in the network and measure the extent to which their knowledge is concentrated and distributed.
Collaboration characteristics
Vaccario et al28 defined the knowledge distance as the Euclidean distance between organizations i and j at time t. In other economic literature, this is known as the technological distance29 and is formally defined as
| (5) |
where xidt represents an element of the knowledge mix vector xit (see equation 1 in Supplementary Appendix S3D.1) with element xidt representing the fraction of clinical trials conducted in therapeutic area d at time t.
We adopted KDijt as a metric to measure the research differences between a pair of organizations' portfolios (ie, the distribution of trial experience in each therapeutic area). The knowledge distance is at a maximum (KDijt = √2) when actors are concentrated in 2 exclusively, different therapeutic areas. When 2 firms are concentrated in the same therapeutic area, the knowledge distance equals to 0 because they are identical in expertise. Therefore, a higher KD corresponds with a larger difference between the 2 research portfolios.
In our analysis, we calculate the mean knowledge distance, 〈KD〉it, based on equation 3, for all incident links to actor i at time t and used it as a variable in our regression. We can define this as
| (6) |
where is the number of degrees for actor at time .
We also classified each actor as an “expert” in one therapeutic area, which corresponds to the therapeutic area that has the highest number of trials. Once we designated each actor as an expert in a particular therapeutic area, we measured collaboration diversity by using an entropic measure of diversity,
| (7) |
The variable zidt is the number experts in therapeutic area d ∈ D that actor is actively collaborating with at timeT. The set D includes all the therapeutic areas that were defined in the BioMedTracker Pharma Intelligence database. This entropic measure is commonly used to quantify diversity in many fields that range from biology to production portfolios.30,31
MULTIVARIATE REGRESSION ANALYSIS
We conducted a regression analysis on 2 response variables that relate to research output and efficiency: cumulative trial successes and trial success rate. We ran separate regressions on each response variable with 1-, 2-, and 5-year lag to capture the delay of knowledge adoption and implementation. Robustness of our regression analysis was verified with 3 separate sets of regression: (1) regression with all measures, (2) regression with selected measures based on statistical significance and reduced collinearity, and (3) regression with only control variables. This resulted in a total of 9 models (see Supplementary Tables S5-S7 and S9-S11 for all regression results). The variables Trialsi(t-k), PrevSucci(t-k), and PrevExpi(t-k)and the fixed effects γt and κi are considered to be the control variables.
The lagged regression that examined cumulative trial successes (CTSit) with respect to each actor i at time period t utilizes a negative binomial generalized linear model. We chose a negative binomial generalized linear model because the distribution of CTSit was overdispersed (see Supplementary Appendix S4 for details). The regression with selected variables is defined as
| (8) |
We add a control variable, PrevSucci(t-k), which takes on a value of 1 if the actor has achieved at least 1 success before time t - k. Additionally, time and actor-type are also controlled with fixed effects, γt and κi. The response variable is lagged k years, therefore all the covariates corresponding with each actor are at an earlier time t - k.
Because the trial success rate, ranges from 0 to 1, we used a lagged beta regression. However, beta regressions are only used to predict values in the (0,1) domain which excludes 0 and 1. We conducted a mathematical transformation on (see section Supplementary S4B in Supplement) to convert the 0 and 1 values to be within the prescribed range. The trial success rate is defined as the cumulative number of trial successes normalized against the cumulative number of trials. The regression can be shown as
| (9) |
The dummy variable PrevExpi(t-k) takes on binary values and represents whether the actor has conducted at least 6 trials (average number of trials) before time t - k.
Robust checks include conducting regressions on control and all measured variables for all 3 lag times. The results of these other models are located in the Supplementary Tables S5-S7 and S9-S11.
RESULTS
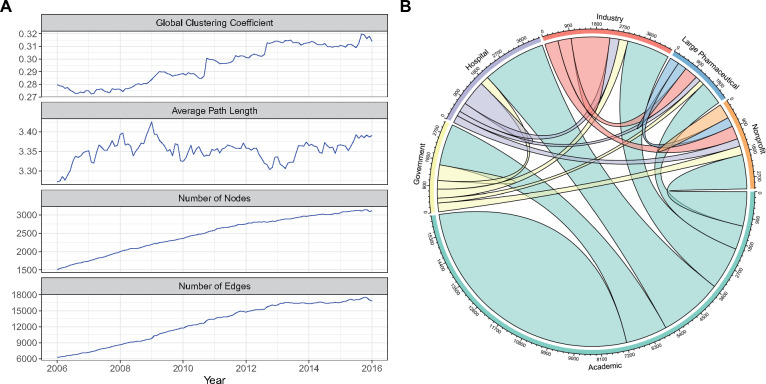
The clinical trials collaboration network consisted of 4494 unique nodes (actors). From January 2016 to January 2016, the number of edges increased from 6287 to 16 821, and nodes increased from 1509 to 3108 (Figure 2A). In Figure 2A, we observed that the network is becoming more cohesive over time by the global clustering coefficient and average path length measures. The average degree of each node is 27.7 ± 39.9. Consistent through all time periods, academic actors have the largest number of edges in the network compared with other organization-types (Figure 2B). The descriptive statistics for all network measures are summarized in Supplementary Table S4.
Figure 2.
Global network characteristics. (A) Overall (global) characteristics of the network for each month from January 2006 to January 2016 for 4 measures: average path length, global clustering coefficient, number of edges, and number of nodes. (B) Chord diagram illustrates the number of collaboration links between and within the 6 organization types in January 2015. The width of the links is scaled based on the volume of collaborations. Academic actors tend to have more collaborative links across all organization types. This is not surprising since academic centers offer resources and infrastructure for clinical trials that are not available to other actor types. As a result, academic collaborations tend to dominate this collaboration network in terms of connections. Many principal investigators on clinical trials also have an academic appointment, even if the trial is sponsored by industry, which explains the higher count of academic actors.
The regression analysis shown in Table 1 suggests that more successful actors have highly diversified research portfolios and collaborate with a diverse set of experts from a variety of specialties. In the short term (1- and 2-year windows), actors that collaborate with partners that have similar research portfolios have higher research output. More research-efficient actors tend to have lower cohesion in their local networks, higher research diversification, and collaborators that also have highly diversified research portfolios. Not surprisingly, we found that previous success and cumulative trials conducted had the largest impact on cumulative trial successes which represents research output, while the previous experience dummy variable was the strongest predictor for trial success rate, a measure of research efficiency.
Table 1.
Standardized coefficient estimates for cumulative trial successes and trial success rate
| Cumulative Trial Successes (Research Output) |
Trial Success Rate (Research Efficiency) |
|||||
|---|---|---|---|---|---|---|
| Variable | 1-y Lag | 2-y Lag | 5-y Lag | 1-y Lag | 2-y Lag | 5-y Lag |
| Previous success | 2.572c | 2.029c | 1.127c | |||
| (0.038) | (0.035) | (0.051) | ||||
| Previous experience | 0.238c | 0.196c | 0.055 | |||
| (0.053) | (0.061) | (0.107) | ||||
| Cumulative trials conducted | 0.290c | 0.251c | 0.092c | |||
| (0.009) | (0.010) | (0.021) | ||||
| Collaboration diversity | 0.182c | 0.213c | 0.186c | −0.020 | −0.029 | −0.040a |
| (0.017) | (0.017) | (0.022) | (0.017) | (0.018) | (0.021) | |
| Local clustering coef. | −0.064c | −0.074c | −0.128c | −0.041c | −0.044c | −0.041b |
| (0.014) | (0.014) | (0.019) | (0.015) | (0.015) | (0.018) | |
| Mean knowledge distance | −0.248c | −0.194c | 0.092c | 0.028b | 0.038c | 0.038b |
| (0.018) | (0.018) | (0.021) | (0.014) | (0.014) | (0.018) | |
| Mean neighbor research diversification | 0.041c | 0.056c | 0.068c | 0.102c | 0.118c | 0.113c |
| (0.013) | (0.014) | (0.018) | (0.014) | (0.014) | (0.018) | |
| Betweenness centrality | 0.015 | 0.026b | 0.125c | 0.0003 | 0.006 | 0.033 |
| (0.011) | (0.013) | (0.022) | (0.018) | (0.019) | (0.024) | |
| Research diversification | 0.135c | 0.128c | 0.159c | 0.097c | 0.109c | 0.100c |
| (0.015) | (0.015) | (0.022) | (0.020) | (0.021) | (0.027) | |
| Constant | −2.460c | −1.723c | −0.743c | −0.422c | −0.658c | −0.740c |
| (0.130) | (0.112) | (0.092) | (0.147) | (0.118) | (0.089) | |
Standardized coefficient estimates with standard errors are shown for response variables, cumulative trial successes and trial success rate. The coefficients for cumulative trial successes are estimated by the negative binomial regression, while the coefficient estimates of trial success rate are the result of a beta regression. We show the coefficients of each response variable for 3 lag lengths (1 year, 2 years, and 5 years) to account for the dynamic effects. Refer to Supplementary Tables S6-S8 and S10-S12 for fit statistics.
P < .1.
P < .05.
P < .01.
Mean knowledge distance has a negative impact on cumulative trial success for 1- and 2-year lags while having a slightly positive impact 5-year lag, which suggests that collaborations with actors collaborating with dissimilar actors in a longer time window have some beneficial effects on research output. Mean knowledge distance has a positive and significant impact on research efficiency for the 2-year lag.
Although the local clustering coefficient is statistically significant—except for the trial success rate with a 5-year lag—we did not observe a large effect on research output and efficiency. Nevertheless, the local clustering coefficient does have a negative trend between research efficiency and output which suggests there is some sort of relationship. Also, the local clustering coefficient has a larger and more significant effect when compared with the other structural characteristic, betweenness centrality.
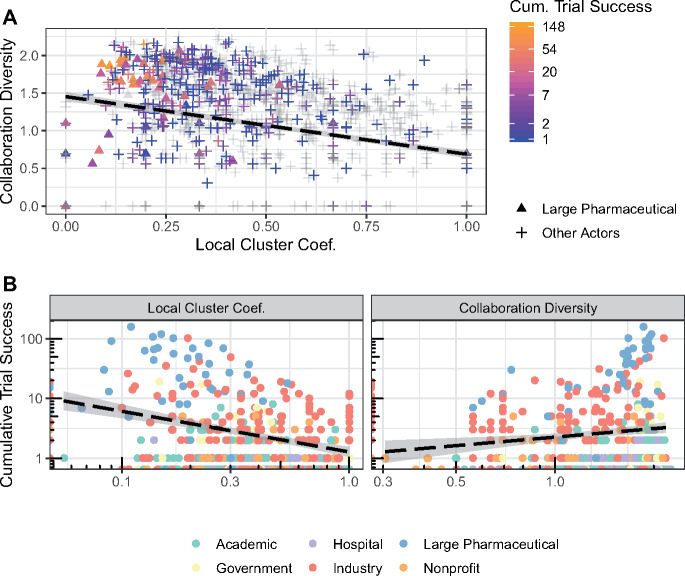
Based on our research question and regression analysis, we dove deeper into the relationship between the local clustering coefficient (network cohesion) and collaboration diversity. Figure 3A illustrates a slight inverse relationship between collaboration diversity and local clustering coefficient, which is indicated by the negative-sloped linear trend line. This indicates that actors that collaborate more diversely are less embedded in the network. In Figure 3B, we observe that most actors with lower local clustering coefficient and higher collaboration diversity tend to be large pharmaceutical actors that have achieved more research output.
Figure 3.
Relating cumulative trial success with local clustering coefficient and collaboration diversity. (A) Scatterplot of collaboration diversity vs local clustering coefficient in January 2015. This distribution is similar for 2006 to 2016 (see Supplementary Figure S11). For all time periods, large pharmaceutical companies that have more success are distinguished as triangles. The black dashed trend line shows a linear negative correlation between the clustering coefficient and collaboration diversity. The color gradient represents cumulative trial successes. This plot highlights the research performance relative to collaboration diversity and the local clustering coefficient. The gray points represent actors that are active during January 2015 but have not subsequently participated in a successful clinical trial. (B) Scatterplot showing the relationships of cumulative trial successes with respect to local clustering coefficient and collaboration for January 2015. Each organization type is distinguished by color.
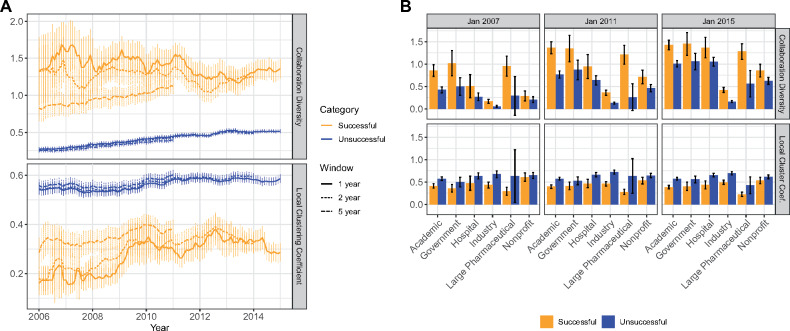
Figure 4A shows the differences in collaboration diversity and clustering coefficients between successful and unsuccessful actors. Successful actors tend to have a lower clustering coefficient than their unsuccessful counterparts while having more collaboration diversity. If we focus on organization types in Figure 4B, we will notice that nonprofit organizations tend to have a higher clustering coefficient. However, even the successful nonprofit vs unsuccessful nonprofit organizations have observable differences in their level of cohesion.
Figure 4.
Comparing successful vs unsuccessful actors. (A) The plot shows the average collaboration diversity (top) and average local clustering coefficient (bottom) for dynamically defined successful and unsuccessful actors in the network for each month. Successful actors are dynamically defined at each time period t as organizations that will achieve at least 1 successful trial within the forward time window ranging from t to t + k, where k is the lag length. Otherwise, the organizations are characterized as unsuccessful. The standard deviations are shown as error bars. (B) We divide into 2 static sets: successful actors (n = 888) and unsuccessful actors (n = 3606). The average collaboration diversity (top) and average local clustering (bottom) are stratified by actor type and success for 3 time periods. The error bars show the 95% confidence interval.
Figure 4B also shows that government and academic actors are the ones with the highest collaboration diversity, with industry actors being the least diverse in collaboration. This is expected because academic and government institutions are usually responsible for leading and sponsoring many clinical trials across therapeutic disciplines. Furthermore, industry actors tend to have lower collaboration diversity because they collaborate with fewer partners, in general (see Supplementary Figure S5). We also notice that successful large pharmaceutical companies are more likely to collaborate with a diverse set of actors.
DISCUSSION
We found that collaboration diversity is related to higher research output and efficiency. The collaboration network shows signs of preferential attachment that occurs when actors are attracted to other actors that have a demonstrated record of successes, resulting in a feedback loop known as the Matthew effect.2 Large pharmaceutical companies that are historically successful benefit from preferential attachment when they attract a wide range of actors with varying therapeutic expertise and experience, which would explain the correlation between success and collaboration diversity. Figure 3 shows this effect, in which actors classified as large pharmaceutical companies tend to have lower clustering coefficients with more collaboration diversity. Others have found that highly connected nodes bridge the gap between disciplines in science.32 As the network evolves, the preferential attachment to these large pharmaceutical companies will create a more evident hub formation that decreases the local clustering coefficient and increase their collaboration diversity.
Our evidence suggests that there is potentially an echo chamber effect in which actors that have a cohesive local collaboration network with nondiverse partners perform poorly. In contrast, actors do benefit from exposure to diverse ideas and knowledge through clinical trial collaboration due to the knowledge exchanged with partners on the peripheries of the network that have few collaborators. Our work suggests that local network cohesion captured by the local clustering coefficient is negatively correlated with collaboration diversity, research output, and efficiency. This network metric demonstrates the association of network position in relation to innovation.
Academic institutions, government agencies, and large pharmaceutical companies are the ones with the lowest clustering coefficient and highest collaboration diversity. Government and academic actors provide resources and personnel to other actors, which explains their network structure and higher diversity in collaboration. We observed in our network that large pharmaceutical companies collaborate more often with several smaller industrial partners than other actor types resulting in their higher collaboration diversity. These private-sector actors, oftentimes, recognize the benefit of knowledge diversity and are strategically motivated to collaborate with diverse actors that complement their core expertise. Large pharmaceutical companies collaborate with many smaller, new market entrants that are not as embedded in the collaboration network, like biotechnology startups.33 Furthermore, large pharmaceutical companies are proficient at absorbing distinct knowledgebases of more specialized actors that are newcomers which results in a lower clustering coefficient. This is reflected in increased instances of public-private partnerships.34 Given that large pharmaceutical companies have resources to exploit comparative advantages of alliances, they are more likely to partner with actors that are relatively new players that complement their ability and are not yet embedded in the system.35 Eventually, large pharmaceutical companies may even acquire these companies because they would also acquire the intellectual property that helps produce better therapeutic products. Existing evidence of biotechnology companies searching for novel knowledge from various scientific communities has been supported by previous studies.36
Knowledge distance between actors and their partners tend to be larger for more efficient actors, which indicates that actors that cooperate with other actors that have a contrasting research portfolio may result in a higher rate of success. Because smaller actors had less output in the short run (1- and 2-year lags), these actors had larger knowledge distance. However, as small actors become more successful in the long run (5-year lag), we see that knowledge distance begins to benefit them because the larger knowledge distances are associated with more collaborations with smaller actors that have only 1 specialization. In our case, knowledge distance supports the notion of comparative advantage in partnerships. Research indicates that collaborating with experts in different fields increases the actor’s knowledge base and ability to innovate by combining varying knowledge; this phenomenon is known as knowledge absorption, which has been studied extensively within the pharmaceutical industry.37,38 However, our study contributes to the body of knowledge by observing this phenomenon within the clinical R&D context. Links in our network embodied the knowledge, resource, and data exchange via clinical trial collaboration between partners.
It should be noted that an approved drug may not necessarily be indicative of novel knowledge creation, since it may be a marginal improvement on an existing drug; many drugs are considered “me-too drugs”' which are not breakthroughs in therapeutic effectiveness.39 Given the availability of data, we found it difficult to find a quantitative indication of true innovation. Although some studies use patents as indicators for innovation, the pharmaceutical industry files multiple patents for any compound that may have higher marketing potential to protect them from generic drug competition.40 Nevertheless, we argue that a company is developing knowledge by accumulating clinical research data, personnel, and patient base.
The findings of our study suggest that the system is moving toward more influence of larger actors over pharmaceutical R&D, such as large pharmaceutical companies. As these select groups of actors become loci of innovation, future research needs to investigate how disproportional concentration of institutionalized knowledge, resource, and expertise might result in diminished efficiency and productivity for the pharmaceutical industry overall. Furthermore, more investigations into the role of diversity within these biomedical specialties will shed light on how subdiscipline diversity within these biomedical specialties impact the productivity and efficiency of treatment development.
Limitations
One issue with our study included the existence of actors that have no successes, which may introduce zero inflation in the regression analysis. Although the 0-1 inflation distribution may have been a better fit to our data, we decided that it was more appropriate to employ the Smithson and Verkuilen41 transformed beta distribution for our research question because we were not dealing with actors that were perpetually unsuccessful or successful. Furthermore, we attempted to rectify this issue by introducing fixed effect variables for previous successes and previous experience to predict cumulative trial successes and trial success rate.
Because many actors exist in multiple snapshots, serial correlation is present within each actor between time periods, which may magnify certain actor’s behaviors in our regression analysis, especially if they are active in for many years. However, we have many actors who were active throughout the entire duration of our analysis, thus minimizing the unbalance in our panel dataset.
CONCLUSION
Our study showed that large pharmaceutical companies and other successful organizations tend to have lower cohesion in their local network. Large pharmaceutical companies are productive at absorbing novel knowledge by searching for diverse partners. The role of outsiders and new actors such as startup biotechnology or life science companies will be crucial because the system is moving toward more cohesion, thus saturating the existing distribution of knowledge.
FUNDING
The project was generously funded through gifts from Bloomberg Philanthropies, Blakeley Investment Corporation, the Argosy Foundation, and the Kelly Family Foundation.
AUTHOR CONTRIBUTIONS
GL conceived this study. GL, TI, and TA designed the research; GL performed research; GL collected and prepared the data; GL and TI analyzed the data; and GL, SS, JB, DAM, LG, TA, and TI wrote and edited the manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Felipe Feijoo, Michele Palopoli, Viva Dadwal, Francisco Del Canto, Smile Indias, Patricia Natalie, Eleanor Carlough, Justin Cook, and Patty Lauria for their contributions. The ideas in this article were developed as a result of the broader Clinical Trials System Project, a multiyear effort led by Massachusetts Institute of Technology Collaborative Initiatives in partnership with Johns Hopkins University, Rhode Island School of Design, and Maryland Institute College of Art.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- 1. DiMasi JA, Grabowski HG, Hansen RW.. Innovation in the pharmaceutical industry: new estimates of R&D costs. J.Health Econ 2016; 47: 20–33. [DOI] [PubMed] [Google Scholar]
- 2. Gay B, Dousset B.. Innovation and network structural dynamics: study of the alliance network of a major sector of the biotechnology industry. Res Policy 2005; 34 (10): 1457–75. [Google Scholar]
- 3. Bercovitz J, Feldman M.. Entrepreneurial universities and technology transfer: a conceptual framework for understanding knowledge-based economic development. J Technol Transfer 2006; 31 (1): 175–88. [Google Scholar]
- 4. Fabrizio KR. Absorptive capacity and the search for innovation. Res Policy 2009; 38 (2): 255–67. [Google Scholar]
- 5. Stuart TE, Ozdemir SZ, Ding WW.. Vertical alliance networks: The case of university–biotechnology–pharmaceutical alliance chains. Res Policy 2007; 36 (4): 477–98. [Google Scholar]
- 6. Chiaroni D, Chiesa V, Frattini F.. Patterns of collaboration along the bio-pharmaceutical innovation process. J Bus Chem 2008; 5 (1): 7–22. [Google Scholar]
- 7. Powell WW, Koput KW, Smith-Doerr L.. Interorganizational collaboration and the locus of innovation: networks of learning in biotechnology. Adm Sci Q 1996; 41 (1): 116–45. [Google Scholar]
- 8. Zhang J, Baden-Fuller C, Mangematin V.. Technological knowledge base, R&D organization structure and alliance formation: evidence from the biopharmaceutical industry. Res Policy 2007; 36 (4): 515–28. [Google Scholar]
- 9. Phelps C, Heidl R, Wadhwa A.. Knowledge, networks, and knowledge networks: a review and research agenda. J Manage 2012; 38 (4): 1115–66. [Google Scholar]
- 10. Singh H, Kryscynski D, Li X, Gopal R.. Pipes, pools, and filters: how collaboration networks affect innovative performance. Strat Manage J 2016; 37: 1649–66. [Google Scholar]
- 11. Schilling MA, Phelps CC.. Interfirm collaboration networks: the impact of large-scale network structure on firm innovation. Manage Sci 2007; 53 (7): 1113–26. [Google Scholar]
- 12. Guan J, Zhao Q.. The impact of university–industry collaboration networks on innovation in nanobiopharmaceuticals. Technol Forecasting Soc Change 2013; 80 (7): 1271–86. [Google Scholar]
- 13. Burt RS. Structural holes and good ideas. Am J Sociol 2004; 110 (2): 349–99. [Google Scholar]
- 14. Filieri R, Alguezaui S.. Structural social capital and innovation. Is knowledge transfer the missing link? J Knowledge Manage 2014; 18 (4): 728–57. [Google Scholar]
- 15. Granovetter MS. The Strength of Weak Ties. Am J Sociol 1973; 78 (6): 1360–80. [Google Scholar]
- 16. Jasny L, Waggle J, Fisher DR.. An empirical examination of echo chambers in US climate policy networks. Nature Clim Change 2015; 5 (8): 782–6. [Google Scholar]
- 17. Watts DJ, Strogatz SH.. Collective dynamics of ‘small-world’networks. Nature 1998; 393 (6684): 440–2. [DOI] [PubMed] [Google Scholar]
- 18. Fritsch M, Kauffeld-Monz M.. The impact of network structure on knowledge transfer: an application of social network analysis in the context of regional innovation networks. Ann Reg Sci 2010; 44 (1): 21–38. [Google Scholar]
- 19. Guler I, Nerkar A.. The impact of global and local cohesion on innovation in the pharmaceutical industry. Strat Manage J 2012; 33 (5): 535–49. [Google Scholar]
- 20. Lee D, Kirkpatrick-Husk K, Madhavan R.. Diversity in alliance portfolios and performance outcomes: A meta-analysis. J Manage 2017; 43 (5): 1472–97. [Google Scholar]
- 21. Cohen SK, Caner T.. Converting inventions into breakthrough innovations: the role of exploitation and alliance network knowledge heterogeneity. J Eng Technol Manage 2016; 40: 29–44. [Google Scholar]
- 22. Rodan S, Galunic C.. More than network structure: how knowledge heterogeneity influences managerial performance and innovativeness. Strat Manage J 2004; 25 (6): 541–62. [Google Scholar]
- 23. Sampson RC. R&D alliances and firm performance: the impact of technological diversity and alliance organization on innovation. Acad Manage J 2007; 50 (2): 364–86. [Google Scholar]
- 24. Parkhe A. Interfirm diversity, organizational learning, and longevity in global strategic alliances. J Int Bus Stud 1991; 22 (4): 579–601. [Google Scholar]
- 25. Tasneem A, Aberle L, Ananth H, et al. The database for aggregate analysis of ClinicalTrials.gov (AACT) and subsequent regrouping by clinical specialty. PloS One 2012; 7 (3): e33677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thomas DW, Burns J, Audette J, Carroll A, Dow-Hygelund C, Hay M.. Clinical development success rates 2006–2015. BIO Ind Anal 2016; 1: 16. [Google Scholar]
- 27. Freeman L. The gatekeeper, pair-dependency and structural centrality. Qual Quantity 1980; 14: 585–92. [Google Scholar]
- 28. Vaccario G, Tomasello MV, Tessone CJ, Schweitzer F.. Quantifying knowledge exchange in R&D networks: a data-driven model. J Evol Econ 2018; 28 (3): 461–93. [Google Scholar]
- 29. Bar T, Leiponen A.. A measure of technological distance. Econ Lett 2012; 116 (3): 457–9. [Google Scholar]
- 30. Jost L. Entropy and diversity. Oikos 2006; 113 (2): 363–75. [Google Scholar]
- 31. Miller DJ. Technological diversity, related diversification, and firm performance. Strat Manage J 2006; 27 (7): 601–19. [Google Scholar]
- 32. Varga A. Shorter distances between papers over time are due to more cross-field references and increased citation rate to higher-impact papers. Proc Natl Acad Sci U S A 2019; 116 (44): 22094–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang L, Plump A, Ringel M.. Racing to define pharmaceutical R&D external innovation models. Drug Discov Today 2015; 20: 361–70. [DOI] [PubMed] [Google Scholar]
- 34. Widdus R. Public-private partnerships for health: their main targets, their diversity, and their future directions. Bull World Health Organ 2001; 79 (8): 713–20. [PMC free article] [PubMed] [Google Scholar]
- 35. Makri M, Hitt MA, Lane PJ.. Complementary technologies, knowledge relatedness, and invention outcomes in high technology mergers and acquisitions. Strat Manage J 2010; 31: 602–28. [Google Scholar]
- 36. Powell WW, White DR, Koput KW, Owen-Smith J.. Network dynamics and field evolution: The growth of interorganizational collaboration in the life sciences. Am J Sociol 2005; 110 (4): 1132–205. [Google Scholar]
- 37. Nieto M, Quevedo P.. Absorptive capacity, technological opportunity, knowledge spillovers, and innovative effort. Technovation 2005; 25 (10): 1141–57. [Google Scholar]
- 38. Mangematin V, Nesta L.. What kind of knowledge can a firm absorb? Int J Technol Manage 1999; 18 (3/4): 149–72. [Google Scholar]
- 39. Garattini S. Are me-too drugs justified? J Nephrol 1997; 10 (6): 283–94. [PubMed] [Google Scholar]
- 40. Ouellette LL. How many patents does it take to make a drug-follow-on pharmaceutical patents and university licensing. MTTLR 2010; 17: 299. [Google Scholar]
- 41. Smithson M, Verkuilen J.. A better lemon squeezer? Maximum-likelihood regression with beta-distributed dependent variables. Psychol Methods 2006; 11 (1): 54–71. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.