Abstract
Early appearance of neutralizing antibodies during acute hepatitis C virus (HCV) infection is associated with spontaneous viral clearance. However, the longitudinal changes in antigen-specific memory B cell (MBCs) associated with divergent HCV infection outcomes remain undefined. We characterized longitudinal changes in E2 glycoprotein-specific MBCs from subjects who either spontaneously resolved acute HCV infection or progressed to chronic infection, using single-cell RNA-seq and functional assays. HCV-specific antibodies in plasma from chronically infected subjects recognized multiple E2 genotypes, while those from spontaneous resolvers exhibited variable cross-reactivity to heterotypic E2. E2-specific MBCs from spontaneous resolvers peaked early after infection (4–6 months), following expansion of activated circulating T follicular helper cells (cTfh) expressing interleukin 21. In contrast, E2-specific MBCs from chronically infected subjects, enriched in VH1-69, expanded during persistent infection (> 1 year), in the absence of significantly activated cTfh expansion. Early E2-specific MBCs from spontaneous resolvers produced monoclonal antibodies (mAbs) with fewer somatic hypermutations and lower E2 binding but similar neutralization as mAbs from late E2-specific MBCs of chronically infected subjects. These findings indicate that early cTfh activity accelerates expansion of E2-specific MBCs during acute infection, which might contribute to spontaneous clearance of HCV.
Keywords: Immunology, Infectious disease
Keywords: Adaptive immunity, B cells, Hepatitis
Introduction
Mechanisms of immune protection are not well defined for many rapidly evolving RNA viruses, such as HIV and hepatitis C virus (HCV), which affect millions worldwide without effective vaccines (1, 2). It has been challenging to study the kinetics and protective roles of neutralizing antibodies (nAbs) during acute infection by these viruses in humans. HCV is a blood-borne virus and people who inject drugs have the highest risk for infection, making them a suitable cohort for studying the immune response to HCV (3). Moreover, HCV infection is an interesting model for comparing immune responses associated with divergent infection outcomes, as approximately 75% of acutely infected subjects develop chronic infection, while 25% resolve spontaneously (4).
Antigen-specific CD4+ and CD8+ T cells are essential for clearance of acute HCV infection. Loss of CD4+ T helper (Th) cells during acute infection and exhaustion of CD8+ T cells are associated with persistence of viremia and virus escape (5, 6). However, a recent phase 1/2 trial (NCT01436357) found that viral vectors encoding HCV nonstructural (NS) proteins, designed to induce HCV-specific T cell responses (a prime-boost strategy), did not significantly curb HCV persistence in people who inject drugs (7). This might be because the vaccine was ineffective at inducing cross-reactive, virus-specific CD4+ T cells. Furthermore, these results suggest that T cell responses alone are insufficient to clear HCV infection.
Early appearance of serum nAbs is associated with spontaneous clearance of acute HCV infection and reinfection in humans (8, 9). In contrast, subjects with delayed nAb responses often develop persistent infection (9, 10). Elucidation of the crystal structure of HCV envelope glycoprotein E2, peptide mapping, and computerized modelling of antibodies against E1E2 heterodimer has led to identification of conserved nAb binding epitopes on E1 and E2 (11–13). Despite these advances, little is known about the dynamics and function of antigen-specific B cells during acute HCV infection. For example, it is not clear which signals induce activation and expansion of antigen-specific B cells that produce nAbs and contribute to HCV clearance. More importantly, attempts to characterize the changes in phenotypes and functions of B cells during HCV infection have been restricted to analyses of bulk B cell populations due to a limited capacity for detecting antigen-specific B cells directly ex vivo (14–16). Moreover, interactions between HCV-specific B cells and T follicular helper (Tfh) cells, a critical component of germinal centers (GCs), require further characterization in the context of acute and persistent HCV infection.
Tfh cells mediate affinity maturation and differentiation of GC B cells into antibody-secreting and/or memory B cells (MBCs) in response to antigens, including chronic infection with viruses such as lymphocytic choriomeningitis, HIV, and simian immunodeficiency virus (17). Tfh cells from GCs in lymphoid tissues are difficult to sample in humans, but circulating Tfh (cTfh) cells can be collected from peripheral blood and studied (18, 19). Nevertheless, only a few studies have characterized cTfh cell function during human HCV infection. Frequency of HCV-specific activated cTfh cell correlates with increased plasma HCV NS4-specific antibody responses during acute HCV infection, and their frequency decreases as viremia persists (20). Interestingly, HCV elimination with direct-acting antiviral therapy results in a sharp increase of HCV-specific CD4+ T cells in the blood, and a shift from a Th1 to a Tfh cell profile (21). However, it has not been possible to simultaneously analyze the dynamics of cTfh and HCV-specific B cells in subjects with divergent acute infection outcomes.
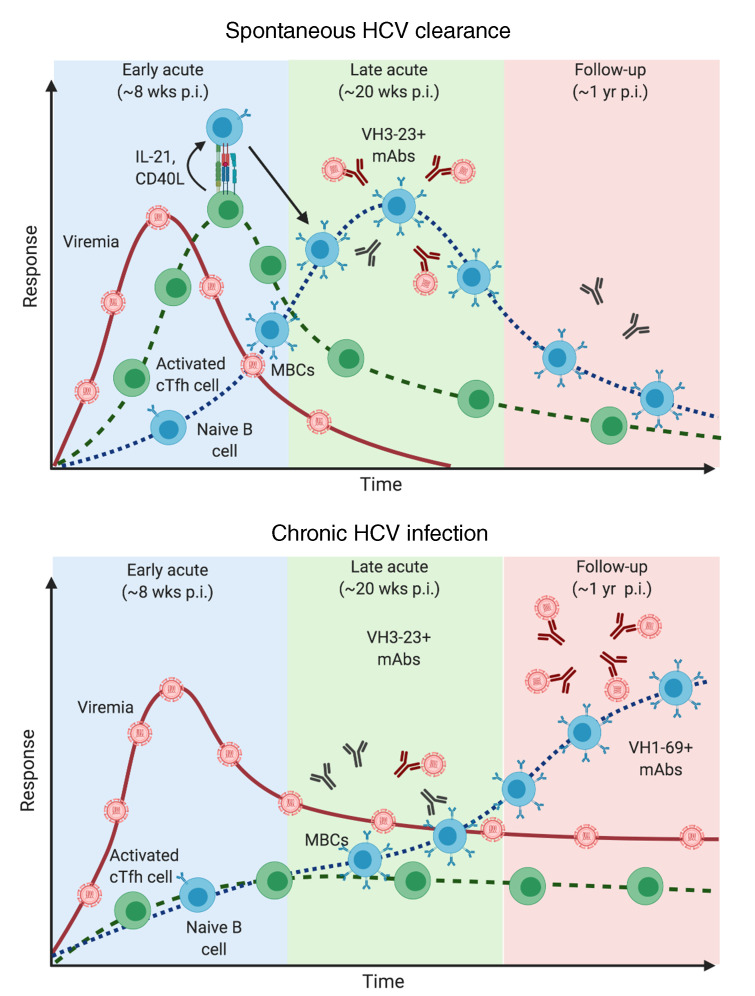
We used an HCV E2 tetrameric bait to study phenotypes, gene expression patterns, B cell receptor (BCR) repertoire, and functions of antigen-specific MBCs in people who inject drugs who spontaneously resolved acute HCV infection (resolvers) or became chronically infected. We report that resolvers experienced earlier expansion of E2-specific MBCs during late acute infection (4–6 months after estimated date of infection [EDI]), whereas E2-specific MBCs expanded during the follow-up time point in chronically infected individuals (>1 year after EDI). HCV persistence led to a shift of VH3–23 to enrichment of VH1–69 gene usage and increased somatic hypermutation (SHM) in the BCR repertoire of E2-specific MBCs. mAbs derived from chronically infected subjects at follow-up had increased binding to E2 antigen and increased SHM, but neutralized HCV pseudoparticles (HCVpp) as efficiently as mAbs with low SHM derived from resolvers during late acute infection. Importantly, cTfh cell activation and effector function increased before E2-specific B cell expansion in resolvers, while no significant changes in cTfh cells were observed in chronically infected subjects at any time point analyzed. Our findings indicate that early cTfh cell activity is associated with earlier expansion of E2-specific B cells during acute infection, which might contribute to resolution of acute HCV infection.
Results
HCV-specific antibodies in plasma of chronically infected individuals are more abundant and react to a wider breadth of HCV genotypes than antibodies from resolvers.
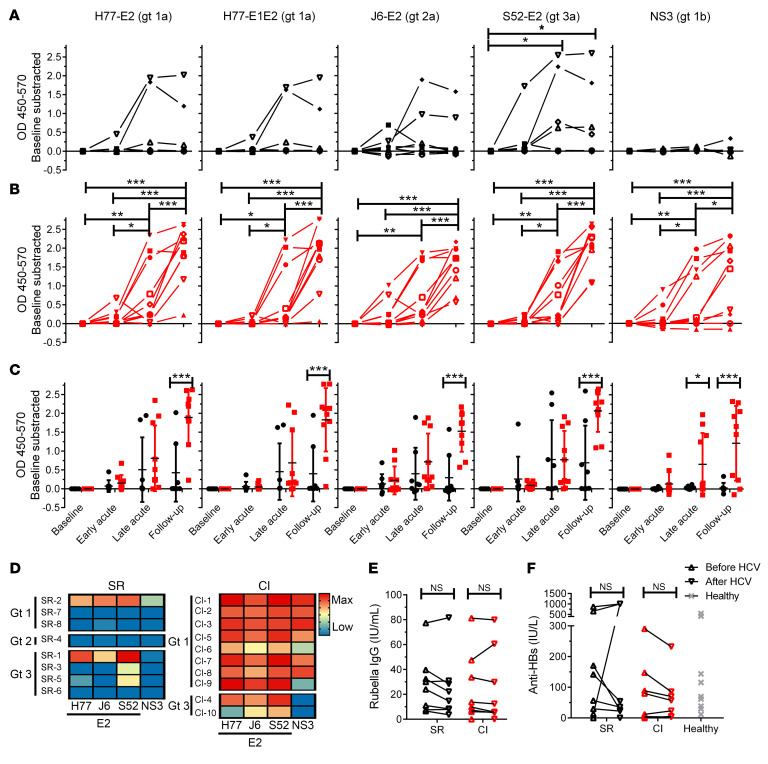
High-risk subjects with acute HCV infection were recruited among people who inject drugs who were participating in the Montreal Acute Hepatitis C Cohort Study (HEPCO) (22). Subjects’ clinical and demographic data are summarized in Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI140590DS1 EDI was defined as the median date between the last negative and the first positive HCV test (RNA or antibody). We collected data before HCV infection (baseline, ≥ 8 weeks before the first positive HCV test), at early acute infection (8 ± 2 weeks after EDI), at late acute infection (20 ± 4 weeks after EDI), and at the follow-up examination (≥ 51 weeks after EDI). We compared polyclonal HCV-specific antibody responses between resolvers (n = 8) and chronically infected individuals (n = 10) by measuring relative levels of E2- and NS3-specific antibodies (IgG) in plasma, over time, by ELISA. Importantly, the HCV genotypic diversity in resolvers was higher than that of chronically infected subjects, as 3, 1, and 4 of 8 resolvers were infected with genotypes (gts) 1, 2, and 3, respectively, compared with 8 and 2 subjects chronically infected with gts 1 and 3, respectively. We detected a positive response in 5 of 8 resolvers, with 2 of 8 subjects (one each for gts 1 and 3) displaying a strong and broad response to multiple E2 genotypes, and 3 of 8 (all gt 3) having a weaker response directed mostly to autologous E2 protein (Figure 1A). In contrast, plasma antibodies from 9 of 10 chronically infected subjects (8 for gt 1 and 1 for gt 3) reacted strongly to autologous and heterologous E2 proteins at late acute and follow-up time points (Figure 1B). Moreover, E2-specific antibody responses in plasma from chronically infected subjects were more robust at follow-up than antibody responses from resolvers (Figure 1C).
Figure 1. HCV-specific antibodies in the plasma of chronically infected subjects are more abundant and react to a wider breadth of HCV genotypes than antibodies in plasma from resolvers.
(A and B) Longitudinal anti-E2 and NS3 IgG responses in plasma measured by ELISA and represented as OD450–570 with subtraction of the baseline sample for each subject (baseline > 8 weeks before EDI; early acute, 8 ± 2 weeks after EDI; late acute, 20 ± 4 weeks after EDI; follow-up, > 51 weeks after EDI). Antigens are indicated on top of the graphs. Each symbol represents a single subject. (A) Resolvers (black, n = 8). (B) Chronically infected subjects (red, n = 10). (C) Combined data from A and B, presented as mean ± SD for each group. (D) Heatmaps showing the magnitude of the ELISA response at follow-up against different E2 and NS3 proteins. Infecting HCV genotype for each subject is indicated on the left. Key: blue, low or no response; yellow, medium response; red, maximum response. (E and F) Anti-rubella virus IgG response (E) and HBsAg (F) for resolvers (SR, n = 8) and chronically infected subjects (CI, n = 10). Values for healthy donor group (n = 10) were only available for HBsAg (gray). (C, E, and F) Data are shown as means for each group of subjects and error bars represent SD. Two-way repeated measure ANOVA with Tukey’s post hoc test. *P < 0.05; **P < 0.01; ***P < 0.001; NS, P > 0.05.
Plasma antibody reactivity to NS3 was nearly indistinguishable from baseline in 7 of 8 resolvers (Figure 1A). In contrast, plasma from chronically infected subjects had a significant increase in NS3-specific antibody responses at late acute in 4 of 10 subjects, which increased to 6 of 10 subjects (all gt 1) at follow-up (Figure 1B). Moreover, the antibody response to NS3 in plasma of chronically infected subjects was significantly higher than that of resolvers at late acute and follow-up (Figure 1C). To highlight the differences in magnitude and breadth of the antibody responses between resolvers and chronically infected subjects at follow-up, we generated a heatmap based on the OD values for each antigen (Figure 1D). The map shows high reactivity to all HCV antigens in most chronically infected subjects (shown in red) but limited and lower responses (in yellow / blue) in resolvers.
To determine whether differences in antibody responses between both groups is due to a generalized impairment in the humoral response, we quantified plasma titers of antibodies against common vaccines before and after HCV infection. We tested a common childhood vaccine (rubella, Figure 1E) and hepatitis B surface antigen (HBsAg, Figure 1F) because 19 of 20 subjects from our study reported immunization with the TWINRIX vaccine (combined hepatitis A and hepatitis B virus vaccine). Plasma antibody responses against rubella or HBsAg did not differ significantly between baseline and follow-up in resolvers or chronically infected subjects (Figure 1, E and F). There was also no difference in antibody titers against HBsAg or rubella between both groups (Figure 1, E and F). Our findings indicate that antibodies in plasma of resolvers display variable reactivity against heterotypic E2, whereas HCV-specific antibodies in chronically infected subjects react strongly to autologous and heterologous genotypes of E2. Moreover, plasma antibodies from resolvers reacted weakly to gt 1b NS3, while antibody reactivity to NS3 from chronically infected subjects was robust, consistent with previous findings (23, 24). These differences in responses are not due to overall impairments in antibody responses in these subjects.
Plasma from chronically infected subjects neutralizes H77 HCVpp more effectively than plasma from resolvers.
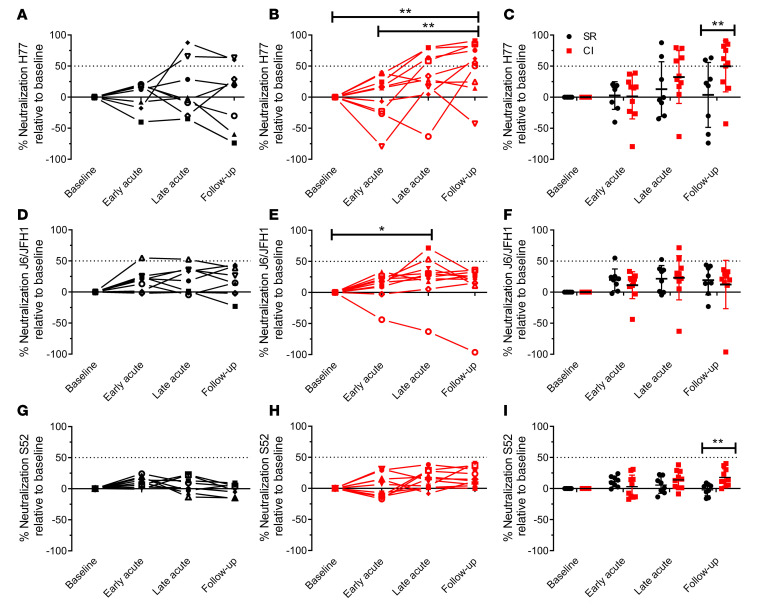
We evaluated longitudinal neutralizing activity of plasma from resolvers and chronically infected subjects against HCVpp from genotypes 1a (H77), 2a (J6/JFH1), and 3a (S52). For H77 HCVpp, neutralizing activity in plasma of resolvers and chronically infected subjects resembled kinetics observed for their respective E2 antibody plasma responses (Figure 1). Specifically, neutralizing activity of plasma from resolvers against H77 HCVpp was generally low, as plasma from only 2 of 8 resolvers (same subjects with high ELISA response to H77 E2) had greater than 50% neutralization (Figure 2A). In contrast, plasma from 4 of 10 chronically infected subjects had strong neutralizing activity against H77 HCVpp at late acute, and this increased to 6 of 10 at follow-up (Figure 2B). At this later time point, neutralizing activity against H77 HCVpp was significantly higher in chronically infected subjects compared with resolvers (Figure 2C).
Figure 2. Plasma from chronically infected subjects neutralizes H77 HCVpp more effectively than plasma from resolvers.
Longitudinal plasma neutralizing activity from resolvers (black, n = 8) and chronically infected subjects (red, n = 10) against H77 HCVpp (A and B), J6/JFH1 HCVpp (D and E), and S52 HCVpp (G and H) at 1:50 dilution, presented as percentage of neutralization relative to baseline (see Figure 1 for time point definitions). (C) Combined data from A and B. (F) Combined data from D and E. (I) Combined data from G and H. All data are presented as the mean ± SD for each group. Dotted line indicates the 50% neutralization threshold. Results are presented as the mean of 3 independent experiments and error bars represent SD. Two-way repeated measure ANOVA with Tukey’s post hoc test. *P < 0.05; **P < 0.01; ***P < 0.001; NS, P > 0.05.
Neutralization of J6/JFH1 HCVpp by plasma from resolvers and chronically infected subjects was weak and did not differ significantly between groups (Figure 2, D–F). Plasma from only 1 of 8 resolvers had greater than 50% neutralization at early acute and late acute time points, whereas plasma from 2 of 10 chronically infected subjects neutralized J6/JFH1 HCVpp above the 50% threshold at late acute (Figure 2, D and E). Surprisingly, all subjects, including those infected with gt 3 HCV, neutralized S52 (gt 3) HCVpp poorly (< 50% neutralization) for all time points (Figure 2, G–I). Furthermore, neutralization of S52 HCVpp by plasma from chronically infected subjects was significantly higher than that of resolvers at follow-up time point, although it was still well below the 50% threshold (Figure 2I). These findings indicate that nAbs to H77 HCVpp in plasma are mostly present at follow-up in chronically infected subjects, but are not readily detected in resolvers. Moreover, antibodies from chronically infected subjects reacted to E2 from HCV gts 1a, 2a, and 3a, but only neutralized gt 1a HCVpp effectively.
E2-specific MBCs undergo earlier expansion in resolvers than in chronically infected subjects.
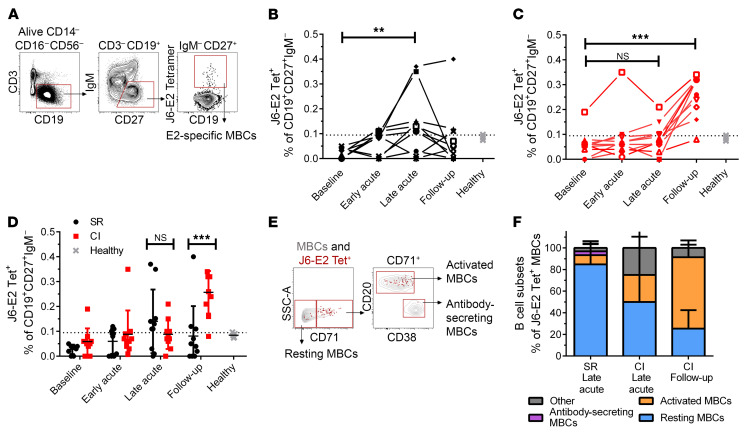
We examined longitudinal changes in E2-specific, class-switched MBCs in blood from resolvers and chronically infected subjects using fluorescent J6 E2 tetramers (Figure 3A). E2-specific MBCs (CD19+CD27+IgM-E2 tet+) frequencies peaked at late acute infection compared with baseline in 7 of 10 resolvers (Figure 3B), whereas only 4 of 10 chronically infected subjects had E2-specific MBCs above the threshold at the same time point (Figure 3C). At follow-up, E2-specific MBCs decreased in most resolvers, but their numbers peaked in 9 of 10 chronically infected subjects (Figure 3C) and were also significantly higher than in resolvers (Figure 3D). These differences in E2-specific MBC frequencies were not a by-product of fluctuations in total MBCs, because the total frequency of MBCs remained consistent in both groups throughout infection (Supplemental Figure 1A). Importantly, while our J6 E2 tetramer enables us to detect cross-reactive, E2-specific MBCs from subjects infected with different HCV genotypes, we cannot exclude the possibility that MBCs expressing BCRs with restricted genotype specificity (other than gt 2a) may not be detected.
Figure 3. E2-specific MBCs undergo earlier expansion in resolvers than in chronically infected subjects.
(A) Representative gating strategy of class-switched, E2-specific MBCs (CD3–CD14–CD16–CD56–CD19+CD27+IgM–E2tet+) in total PBMCs. (B and C) Longitudinal frequencies of E2-specific (J6-E2 Tet+) MBCs in resolvers (B, n = 10, black) or chronically infected (CI) subjects (C, n = 10, red) at indicated time points (see Figure 1). Data from healthy individuals (controls) are gray (n = 5). Threshold of detection of E2-specific B cells (dotted line) was 0.095% (mean detection from healthy individuals + 2 SD) (35). (D) Combined data from B and C, presenting the mean ± SD for each group. (E) Representative gating of resting MBCs (CD71–), activated MBCs (CD71+CD20hiCD38int-lo), antibody-secreting MBCs (CD71+CD20loCD38hi) shown for the total MBCs population (gray contour plots), and HCV E2-specific MBCs (red dots). (F) Phenotypes of E2-specific MBCs in resting (blue), activated (orange), or antibody-secreting (purple) states (cells that did not meet these categories are designated as Other and shown in gray) for resolvers (SR) at late acute (n = 5) and CI subjects (n = 5) at late acute and follow-up time points. (B–D) Data are shown as means for each group of subjects and error bars represent SD. Two-way repeated measure ANOVA with Tukey’s post hoc test, **P < 0.01; ***P < 0.001; NS, P > 0.05.
Activated, E2-specific MBCs from chronically infected subjects increase at follow-up.
We evaluated activation of E2-specific MBCs in a subset of subjects (n = 5 for each group) at late acute and follow-up. Eighty percent of E2-specific MBCs from resolvers had a resting phenotype (CD71–) (25). The phenotype of E2-specific MBCs in chronically infected subjects was heterogeneous at late acute, but 65% had an activated phenotype (CD71+CD20hiCD38int-lo) at follow-up, indicative of viral replication and antigen stimulation (Figure 3, E and F). Interestingly, bulk-activated MBCs (as frequencies of total MBCs) from resolvers did not change from baseline throughout acute infection, whereas total bulk activated MBCs increased significantly, compared with baseline, in chronically infected subjects during early and late acute infection (Supplemental Figure 1B). Frequencies of bulk-activated MBCs were also significantly higher in chronically infected subjects compared with resolvers at late acute (Supplemental Figure 1B). These data show that the proportion of activated E2-specific MBCs in resolvers is relatively low during their peak expansion at the late acute time point, whereas levels of total and activated, E2-specific MBCs continue to increase in chronically infected subjects as acute infection progresses to chronicity. This late expansion of activated E2-specific MBCs in chronically infected subjects begins during late acute infection, concomitant with peak expansion of total, activated MBCs.
E2-specific MBCs from resolvers and chronically infected subjects sorted at their respective peaks of expansion have similar patterns of gene expression.
We investigated factors that affect kinetics of E2-specific MBCs expansion and activation between resolvers and chronically infected subjects. We sorted single E2-specific MBCs from resolvers at late acute infection (n = 6 subjects) and chronically infected subjects at follow-up (n = 5 subjects) and compared gene expression profiles (Supplemental Figure 2A). To mitigate batch effects, we analyzed 3 plates that had E2-specific MBCs from 1 chronically infected subject and 1 resolver on the same plate and compared gene expression patterns between resolvers (3 subjects, 47 single cells) and chronically infected subjects (3 subjects, 95 single cells) (Supplemental Table 2). Most cells expressed mRNAs encoding the same B cell–specific surface markers (such as CD19, CD27, CD20) that define class-switched MBCs, which we sorted by flow cytometry, thereby confirming our target population (Supplemental Figure 2B). Most cells also expressed low levels of genes associated with B cell exhaustion, such as FCRL4, SOX5, CD11C, RUNX1, RUNX2, RANKL, and DLL, with the exception of FGR (Supplemental Figure 2C) (26). Few cells expressed genes associated with GC responses (such as AID, BCL6, and FAS), the cell cycle (cyclins E, D1, A2, B1, and B2, cyclin-dependent kinases), or proliferation (MKI67), with the exception of C-MYC (Supplemental Figure 2, D and E) (27). These results indicate that E2-specific MBCs from resolvers and chronically infected subjects have similar gene expression profiles and are not functionally impaired, as they do not express markers of B cell exhaustion.
Enrichment of heavy and light chain variable regions differs between E2-specific MBCs from resolvers vs.
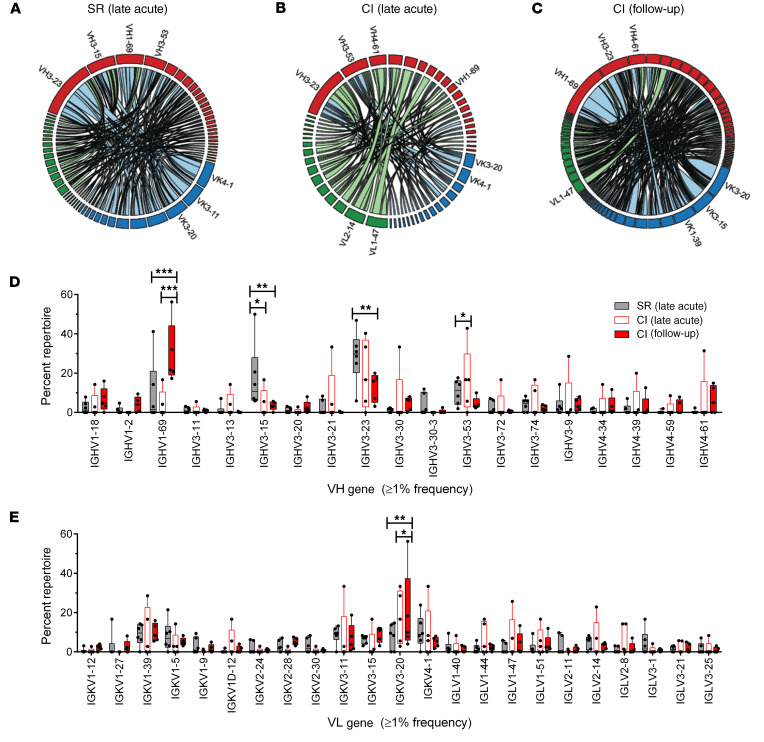
chronically infected subjects at different stages of infection. We compared differences in BCR repertoires of single, E2-specific MBCs sorted from resolvers (n = 6) at the late acute time point vs. E2-specific MBCs sorted from chronically infected subjects at the late acute and follow-up time points (n = 5). Paired Ig chains from scRNA-seq and Sanger sequences of PCR samples from resolvers and chronically infected subjects were combined. Among all groups, the most frequent IgH isotypes identified were IgG1 and IgG2, but a dominant bias of kappa light chain usage was evident (Supplemental Figure 3A). We also compared variable regions of heavy (VH) and light (VL) chains of all BCRs from both groups. We observed enrichment of VH3-23 and VH3-15 among BCRs from 4 of 6 resolvers at late acute infection (Figure 4A and Supplemental Figure 3B). In contrast, although VH3-23 was initially enriched in chronically infected subjects during late acute infection (Figure 4B and Supplemental Figure 3C), VH1-69 was significantly enriched in BCRs from all chronically infected subjects (5/5) at follow-up (Figure 4C and Supplemental Figure 3D) compared with all other VH families. This indicates selection of VH1-69 in the E2-specific MBCs population over time. We also observed significantly higher frequencies of VH3-23 and VH3-15 in pooled BCRs from resolvers at the late acute time point compared with pooled BCRs from chronically infected subjects at follow-up, but not in pooled BCRs from chronically infected subjects at late acute infection (Figure 4D). Conversely, VH1-69 was significantly higher in pooled BCRs from chronically infected subjects at follow-up compared with VH1-69 in pooled BCRs from both groups at late acute infection (Figure 4D). VL chains were not enriched in BCRs from either group at late acute infection. In contrast, VK3-20 usage was significantly increased in BCRs from chronically infected subjects at follow-up and was significantly higher than BCRs from resolvers (Figure 4E).
Figure 4. VH1-69 usage is higher in BCR repertoire of E2-specific MBCs from chronically infected subjects compared with BCR of resolvers.
(A–C) Circos diagrams showing relative frequencies of V gene pairs from heavy and light chains of pooled E2-specific MBCs from resolvers (SR, n = 6 subjects, 219 pooled cells, A) at late acute stage and chronically infected (CI) subjects (n = 5) at late acute (123 pooled cells, B) and follow-up (403 pooled cells, C) time points. The width of each ribbon indicates the frequency of the VH–VK/VL pairing. The length of the arc corresponds to V gene frequency. (D and E) Relative abundances of heavy (D) or light (E) chain V genes in BCRs of pooled E2-specific MBCs from resolvers (dark gray) at late acute and CI subjects at late acute (white, red border) or follow-up (solid red) time points, presented as mean percentages of repertoire ± SD. Two-way ANOVA with Bonferroni’s post hoc test (D and E), unpaired, Mann-Whitney U test (F). *P < 0.05; **P < 0.01; ***P < 0.001; NS, P > 0.05.
We quantified the frequencies of all combinations of paired heavy and light chains from BCRs of both groups (Figure 4A and Supplemental Figure 3B) The heavy and light chain pair with the highest frequency in BCRs from resolvers at late acute infection was VH3-23 and VK3-11. For BCRs from chronically infected subjects, the heavy and light chain pairs with the highest frequencies were VH4-39 and VL1-47 and VH1-69 and VK3-20, for late acute and follow-up, respectively (Figure 4, B and C, Supplemental Figure 3C and D). These results indicate that VH3-23 and VH1-69 are enriched in BCRs from resolvers at late acute infection and chronically infected subjects at follow-up, respectively.
Monoclonal antibodies derived from E2-specific MBCs of chronically infected subjects at follow-up bind more strongly to J6 E2 than mAbs from resolvers at late acute infection.
To evaluate the functionality of antibodies encoded by BCRs of E2-specific MBCs, we cloned paired heavy and light chains from 3 resolvers at late acute (SR-2-LA, SR-4-LA, and SR-6-LA) and 3 chronically infected subjects at follow-up (CI-3-FU, CI-1-FU, and CI-9-FU) and expressed them as human IgG1 mAbs. For each subject, we cloned, expressed, and purified 10 mAbs and calculated their binding to varying concentrations of HCV J6 E2 glycoprotein by biolayer interferometry. We tested binding to J6 E2 specifically because these mAbs were derived from E2-specific MBCs that were captured with J6 E2 tetramer, thus BCRs (and mAbs) from these cells should cross-react to J6 E2, regardless of the subject’s infecting HCV genotype. Individual binding curves for all mAbs are shown in Supplemental Figure 4, A–F. MAbs from chronic CI-1-FU had significantly higher binding to J6 E2 than mAbs from all 3 resolvers, whereas mAbs from CI-3-FU had significantly higher binding to E2 than mAbs from SR-4-LA and SR-6-LA (Supplemental Figure 4G). However, E2-specific binding of mAbs from chronic CI-9-FU did not differ significantly from binding efficiency of mAbs from any resolvers (Supplemental Figure 4G). Nevertheless, the mean binding of pooled mAbs from all chronically infected subjects was significantly higher than binding of mAbs from pooled resolvers (Figure 5A). These results indicate that mAbs produced by E2-specific MBCs of chronically infected subjects present at follow-up bind to J6 E2 more strongly than mAbs produced during late acute infection in resolvers.
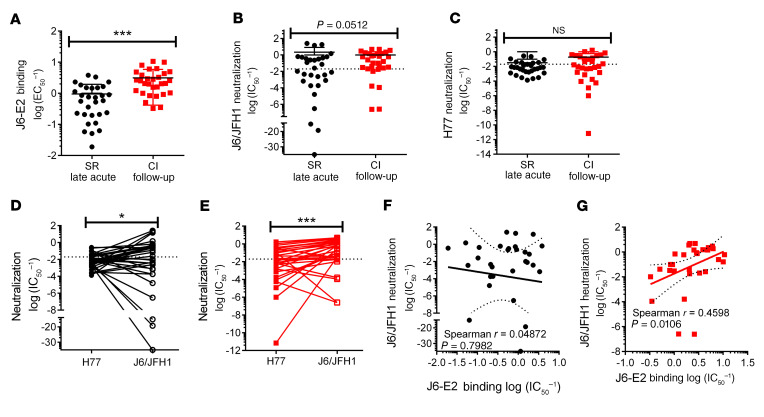
Figure 5. E2-specific mAbs from resolvers neutralize HCVpp as efficiently as mAbs from chronically infected subjects despite weaker J6 E2 binding.
(A) Relative binding strength of pooled mAbs from resolvers (SR, black circles, n = 3 subjects, 10 mAbs per subject) and CI subjects (red squares, n = 3 subjects, 10 mAbs per subject) to J6 E2 protein, quantified by biolayer interferometry and expressed as mean log EC50–1 ± SD. (B and C) Neutralization against J6/JFH1 (B) or H77 (C) HCVpp of pooled mAbs from resolvers or CI subjects. Dotted line indicates the threshold of IC50–1 = 0.02. (D and E) Comparison of neutralizing activity against H77 and J6/JFH1 by individual mAbs from resolvers (D) and CI subjects (E). (F and G) Correlation between J6/JFH1 HCVpp neutralization and binding abilities by individual mAbs of resolvers (F) or CI subjects (G). Results for binding experiments represent means of pooled mAbs and error bars represent SD. Results for neutralization experiments represent the mean of 3 independent experiments. Unpaired, Mann-Whitney U test (A–D), Spearman’s correlation (E–H). **P < 0.01; ***P < 0.001; NS, P > 0.05. In all graphs, each dot represents a single mAb.
MAbs derived from E2-specific MBCs of resolvers and chronically infected subjects neutralize HCVpp with comparable efficiencies.
We investigated whether differences in E2 binding between mAbs from resolvers and chronically infected subjects affect their ability to neutralize 2 different genotypes of HCVpp (J6/JFH1 and H77). We followed a similar rationale for testing neutralization against J6/JFH1 HCVpp, as stated for our binding assays in that mAbs from both resolvers and chronically infected subjects cross-react with J6 E2 (the genotype of our E2 tetramer). Individual J6/JFH1 neutralization curves for all mAbs are shown in Supplemental Figure 5, A–F. mAbs from SR-2-LA had significantly higher neutralizing activity than mAbs from SR-4-LA, SR-6-LA and CI-9-FU (Supplemental Figure 5G). Fifteen of 30 mAbs from resolvers and 20 of 30 mAbs from chronically infected subjects neutralized J6/JFH1 HCVpp above the threshold (IC50–1 = 0.02) (Figure 5B). However, overall neutralizing activity of pooled mAbs did not differ significantly between resolvers and chronically infected subjects (Figure 5B). For H77 HCVpp, less than 50% of pooled mAbs from resolvers (8/30) and chronically infected subjects (13/30) neutralized H77 HCVpp above the threshold (Figure 5C). Individual H77 neutralization curves for all mAbs are shown in Supplemental Figure 6, A–F. There was no significant difference in neutralization of H77 HCVpp between pooled mAbs from resolvers and chronically infected subjects (Figure 5C, Supplemental Figure 6G). We also compared each individual mAb’s neutralizing activity against H77 and J6/JFH1 HCVpp. mAbs from both resolvers and chronically infected subjects neutralized J6/JFH1 HCVpp with significantly higher efficiency than H77 HCVpp (Figure 5, D and E). This may potentially result from a preferential binding of isolated mAbs to J6 E2, since our E2 tetramer is of J6 (gt 2a) origin.
We found no correlation between J6 E2 binding and neutralizing activity against J6/JFH1 HCVpp for mAbs from resolvers (Figure 5F). In contrast, we observed a positive correlation between J6 E2 binding and neutralization of J6/JFH1 HCVpp for mAbs from chronically infected subjects (Figure 5G). These findings indicate that mAbs from resolvers neutralize HCVpp as effectively as mAbs from chronically infected subjects, even though mAbs from resolvers bind E2 less efficiently than mAbs from chronically infected subjects. Supplemental Tables 3 and 4 summarize mAb binding, neutralization, and other BCR characteristics for resolvers and chronically infected subjects.
mAbs from resolvers at late acute have fewer somatic hypermutation than mAbs from chronically infected subjects at follow-up.
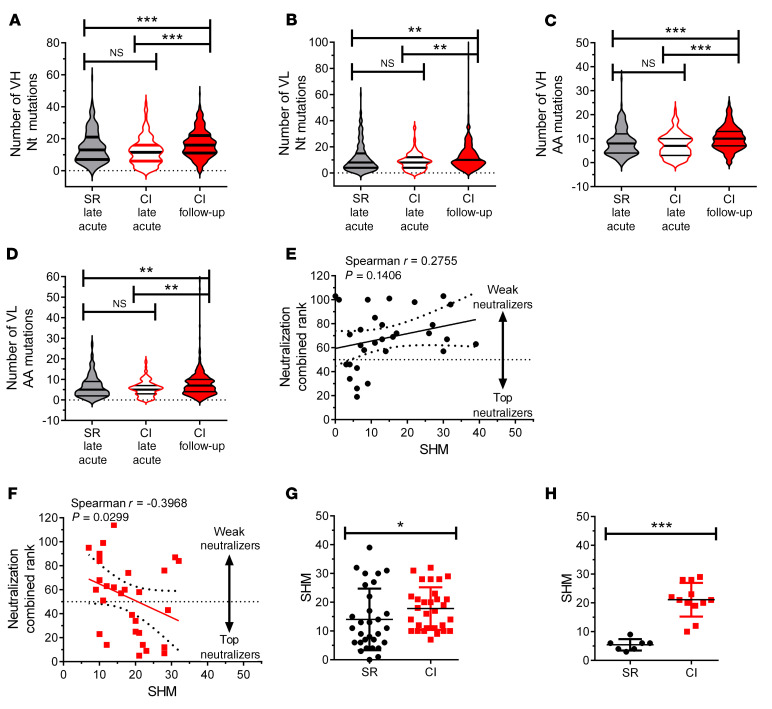
Binding strength of mAbs to their cognate antigen is typically determined by accumulation of SHMs in the BCR as antigen-specific B cells undergo multiple cycles of GC reactions (28). BCRs from E2-specific MBCs of chronically infected subjects at follow-up contained significantly higher combined nucleotide and amino acid mutations in heavy and light chains than BCRs from E2-specific MBCs of resolvers and chronically infected subjects at late acute infection (Figure 6, A–D), possibly resulting from continued antigen stimulation. However, the number of SHMs in BCRs of E2-specific MBCs from resolvers at late acute infection was similar to that of BCRs from E2-specific MBCs from chronically infected subjects at the same time point (Figure 6, A–D). These results indicate that BCRs from E2-specific MBCs from chronically infected subjects at follow-up accrue significantly more SHMs than BCRs from both groups at late acute infection.
Figure 6. mAbs from resolvers at late acute stage have fewer somatic hypermutation than mAbs from chronically infected subjects at follow-up.
(A–D) SHMs in pooled, single E2-specific MBCs (n = 219) from resolvers (n = 6 subjects, gray violins) and CI subjects (n = 123 cells, n = 5 subjects, white violins with red border) at late acute and CI subjects (n = 5) at follow-up (n = 403 cells, red violins) time points, presented as the number of nucleotide substitutions (A and B) and amino acid mutations (C and D) in heavy (A and C) and light (B and D) chains plotted as absolute number of mutations. (E and F) Correlation between mAbs neutralization combined rank (rank of 1 = best neutralizer, rank of 114 = worst neutralizer, established from H77 and J6/JFH1 neutralization, see Figure 5) and number of SHMs in CDR3H and CDR3L regions of mAbs derived from resolvers (n = 3 subjects, 30 mAbs, E) or chronically infected subjects (n = 3, 30 mAbs, F). (G and H) Number of SHMs in CDR3 regions of all cloned mAbs (G) or the top neutralizers (H, combined rank < 50). (E–H) Each dot represents the rank value for an individual mAb. Results represent means of pooled mAbs and error bars represent SD 1-way ANOVA with Tukey’s post hoc test (A–D) or paired Student’s t test (G and H). *P < 0.05; **P < 0.01; ***P < 0.001; NS, P > 0.05.
Since we observed stronger J6 E2 binding and higher SHM in nAbs from chronically infected subjects, we hypothesized that a correlation might exist between HCVpp neutralization and SHMs. Thus, we ranked combined neutralization of H77 and J6/JFH1 HCVpp for each mAb from highest neutralizer (rank close to 2) to lowest neutralizer (rank close to 120) (Supplemental Table 3 and Supplemental Table 4) and correlated ranks of individual mAbs with their respective SHMs. Interestingly, we found no correlation between SHM and neutralization rank of mAbs from resolvers (Figure 6E) but we found a negative correlation between SHMs and neutralization rank from chronically infected subject–derived mAbs, indicating that the best neutralizing mAbs have high numbers of SHMs (Figure 6F). Moreover, the mean number of SHMs of all mAbs tested for chronically infected subjects was significantly higher than the mean number of SHMs of all mAbs from resolvers (Figure 6G). This difference was even greater when we compared the top neutralizing mAbs (combined rank <50) (Figure 6H). Thus, mAbs from resolvers neutralize HCVpp as efficiently as mAbs from chronically infected subjects even though mAbs from resolvers have fewer SHMs.
Activated cTfh cells expand during early acute infection in resolvers but not in chronically infected subjects.
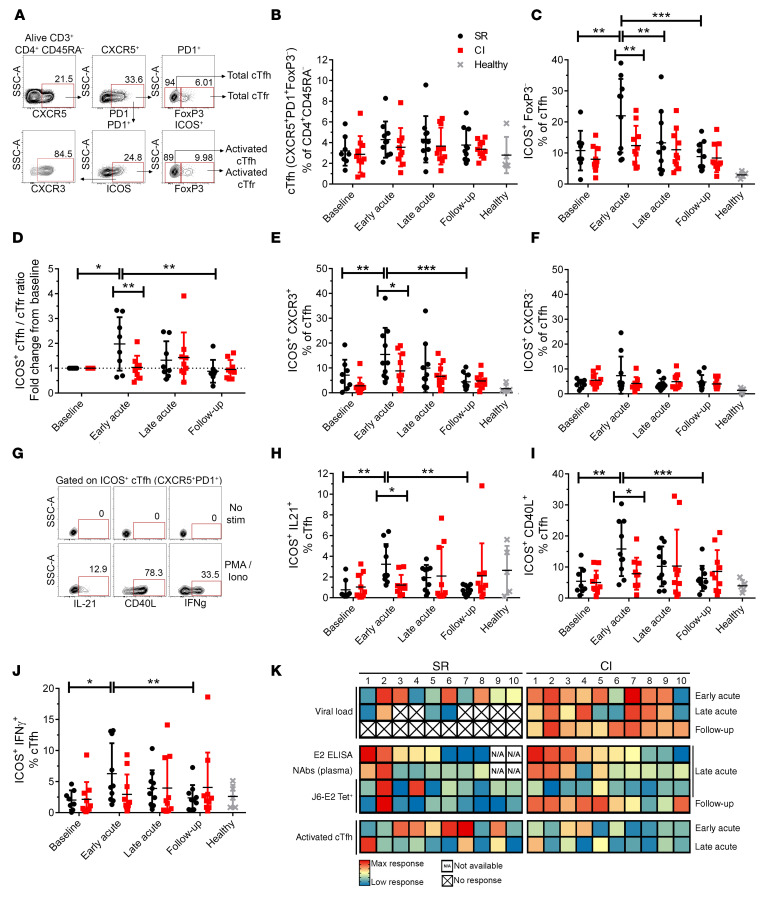
Given that the overall gene expression pattern of E2-specific MBCs did not differ significantly between both groups, we investigated whether Tfh cells promote expansion of E2-specific B cells in resolvers. We found no significant changes in total cTfh cell populations (CD4+CD45RA–CXCR5+PD1+) (Figure 7A) throughout HCV infection in either group (Figure 7B). However, significantly higher frequencies of cTfh cells from resolvers were activated (ICOS+) at early acute infection, compared with all other time points. This difference was not observed in cTfh cells from chronically infected subjects (Figure 7C, Supplemental Figure 7A and B). There was also a significantly higher frequency of activated cTfh in resolvers as compared with chronically infected subjects at the early acute time point (Figure 7C). Regulatory cTfh cells (cTfr, FoxP3+) can reduce cTfh activity, and a high cTfh/cTfr ratio indicates increased GC reaction potential (29, 30). The frequencies of total or activated (ICOS+) cTfr cells did not change over time in any group (Supplemental Figure 7C and D), but the ratio of activated cTfh/cTfr significantly increased at early acute infection only in resolvers (Figure 7D, Supplemental Figure 7E and F). cTfh can be further stratified into different profiles based on expression of additional chemokine markers. Th2-like cTfh (CXCR3–) were shown to provide better help to B cells as compared with Th1-like cTfh (CXCR3+) in healthy donor cells, stimulated with SEB (18, 19). However, in the context of influenza vaccine, Th1-like cTfh cells were activated after vaccination and correlated with antibody production (31). We investigated the profile of activated cTfh in acute HCV infection. Similar to influenza vaccination, Th1-like cTfh cells were significantly activated in early acute HCV in resolvers (Figure 7E, Supplemental Figure 7G and H) while no activation of Th2-like cTfh was observed throughout the infection period in either group (Figure 7F). These results demonstrate that more activated cTfh cells with a Th1-like phenotype are present at early acute infection in resolvers compared with chronically infected subjects and this could affect the kinetics of B cell responses.
Figure 7. Activated cTfh cells expand early during acute infection in resolvers but not in chronically infected subjects.
(A) Representative gating strategy of total cTfh cells (CD3+CD4+CD45RA–CXCR5+PD1+FoxP3–), activated cTfh cells (ICOS+), total cTfr cells (CD3+CD4+CD45RA–CXCR5+PD1+FoxP3+), and activated cTfr cells (ICOS+), from PBMCs of HCV-infected subjects. (B) Frequencies of total cTfh cells at different time points in resolvers (n = 10, black) or chronically infected subjects (n = 10, red; see Figure 1). Data from 5 healthy donors (controls) are shown in gray. (C) Frequencies of activated cTfh cells at different time points. (D) Ratio of activated cTfh over cTfr cells at different time points, shown as fold-change from baseline. (E) Frequencies of activated Th1-like (CXCR3+) cTfh cells at different time points. (F) Frequencies of activated Th2-like (CXCR3–) cTfh cells at different time points. (G) Representative gating strategy of cytokine-producing or CD40L-expressing, activated cTfh cells before (no stim, top) and after PMA and ionomycin stimulation (5 hours, bottom). (H–J) Frequencies of ICOS+IL-21+ cells (H), ICOS+CD40L+ cells (I), and ICOS+IFN-γ+ (J) cTfh cells at different time points. (K) Summary heatmaps of the main data from this study. Each component (row) indicates the intensity of response of the indicated test (far left) at the indicated time point (far right). Each square provides data for one resolver (left) or chronically infected subject (right). An X indicates no response (values of 0); N/A, subject for which the test could not be done (no baseline blood sample available). Blue, low or no response; yellow, medium response; red, maximum response. Data are shown as means for each group of subjects and error bars represent SD. Two-way repeated measure ANOVA with Tukey’s post hoc test. *P < 0.05; **P < 0.01; ***P < 0.001; NS, P > 0.05.
Activated cTfh cells from resolvers produce higher levels of IL-21, CD40L, and interferon gamma (IFN-γ) during early acute infection.
Two major components of Tfh cell help toward B cells are the expression of CD40L for costimulation, and secretion of IL-21 (17). Tfh cells also produce IL-4, IL-17, and IFN-γ, depending on their phenotype (18). We stimulated cTfh cells from resolvers and chronically infected subjects and assessed activated cells (ICOS+) for expression of IL-21, CD40L, and IFN-γ expression (Figure 7G). We observed a significant increase in the frequency of activated cTfh cells that express IL-21 (Figure 7H, Supplemental Figure 7, I and J), CD40L (Figure 7I, Supplemental Figure 7, K and L), and IFN-γ (Figure 7J, Supplemental Figure 7, M and N) at early acute infection in resolvers but not in chronically infected subjects. Moreover, frequencies of ICOS+IL-21+ and ICOS+CD40L+ cTfh cells were significantly higher in resolvers compared with chronically infected subjects at the early acute time point (Figure 7, H and I). These results indicated that activated cTfh cells expand faster in resolvers and are more functional than cTfh cells from chronically infected subjects, and this might contribute to the early activation of E2-specific B cells in subjects who resolve HCV infection.
Discussion
We performed an in-depth, longitudinal characterization of HCV-specific responses of B cells, at the single-cell level, integrating analyses of humoral responses including antigen-specific antibodies in plasma, BCR repertoire, and mAbs derived from single antigen-specific B cells. We also examined the kinetics and function of cTfh cells to determine interactions between activities of cTfh cells and HCV-specific B cell responses. We demonstrated that early expansion of HCV-specific B cells producing nAbs correlated with resolution of primary infection in 7 of 10 resolvers. This seems to require the help of other immune components, such as Tfh cells, to foster early emergence of antigen-specific B cells. Failure to do so potentially leads to persistence of infection despite the generation of antigen-specific B cells with high levels of SHM and strong binding to cognate antigen, from which the virus appears to escape.
Our results demonstrate that antibodies from plasma of resolvers react with and neutralize a limited breadth of E2 proteins. These observations differ from other studies, which found that plasma from resolvers had an increased neutralization breadth, compared with plasma from chronically infected subjects, during acute HCV infection (9, 11, 32). However, it is important to note that the majority of subjects from the cohorts in these studies were predominantly infected with gt 1 HCV, while the resolvers in our study were infected with a greater diversity of HCV genotypes (3 infected with gt 1, 2 with gt 2, and 4 with gt 3) compared with chronically infected subjects (8 infected with gt 1 and 2 with gt 3), which may explain why plasma from more chronically infected subjects neutralized H77 HCVpp.
Interestingly, plasma from both resolvers and chronically infected subjects infected with gt 3 HCV failed to effectively neutralize autologous HCVpp. Variations in neutralizing activity of antibodies from human and chimpanzee plasma against different HCV genotypes have also been reported by other studies (33, 34). Plasma from humans and chimpanzees infected with gt 1a HCV generally contains stronger nAbs against autologous gt 1 and heterologous gts 4a, 5a, and 6a HCVpp, but not against gts 2a and 3a HCVpp (33, 34). Meunier et al. also reported that neutralization of gt 3 HCVpp by plasma of HCV-infected humans and chimpanzees was the weakest for all 6 genotypes tested (33). These studies also noted a weak nAb presence in plasma from both resolvers and chronically infected subjects infected with gt 1a HCV during acute infection that later increased during persistent infection, similar to our findings. Thus, the nature of the subject’s infecting HCV genotype may significantly impact neutralization efficiency and breadth against autologous and heterologous HCVpp.
While E2-specific MBCs expanded earlier in 7 of 10 resolvers compared with those from chronically infected subjects, plasma neutralization of HCVpp was relatively weak, possibly relating to a low nAb abundance at these time points. Moreover, neutralizing activity from plasma of chronically infected subjects only improved at the follow up time point, as a correlate of increasing E2-specific antibodies in plasma. Nevertheless, E2-specific mAbs, isolated from single MBCs, from both resolvers and chronically infected subjects neutralized autologous and heterologous HCVpp with similar efficiencies at their respective peak of E2-specific MBC expansion. This is an important observation because it suggests that although the total nAb titer in plasma is relatively low in resolvers at the peak of E2-specific MBC expansion, individual mAbs from resolvers neutralize HCVpp as efficiently as mAbs from chronically infected subjects that arise much later during persistent infection. Our data are consistent with recent observations by Chen et al. (35) demonstrating that nAbs from nonhuman primates immunized with E1/E2 were not present in plasma at sufficient levels for broad neutralization of HCVpp, but mAbs isolated from sorted plasma cells and MBCs exhibited high levels of neutralization. Thus, we posit that the timing and quality of nAbs produced by rapidly expanding antigen-specific B cells constitutes a more pivotal factor in the resolution of acute HCV infection than the quantity of nAbs present in plasma.
A possible factor affecting differences in antibody kinetics between resolvers and chronically infected subjects is the skewed proportion of activated B cells during acute infection. Activated B cells (including E2-specific MBCs) steadily increase during acute infection in chronically infected subjects, concurrent with an increase in HCV-specific IgG in plasma. We surmise that many of these activated B cells differentiate into plasmablasts or plasma cells that secrete HCV-specific antibodies. We and others observed an increase in HCV-specific, antibody-secreting cells in PBMCs from chronically infected subjects that coincides with an increase in HCV-specific antibody titers in plasma (14, 36, 37). In contrast, fewer activated B cells and HCV-specific antibodies are present in PBMCs from resolvers. This reduced production of HCV-specific IgG in plasma from resolvers cannot be attributed to impaired B cell function during acute infection, because antibodies against rubella virus or HBsAg in plasma of resolvers are similar to those of chronically infected subjects, before and after HCV infection. This indicates that HCV infection does not disrupt general B cell responses in resolvers. Furthermore, E2-specific MBCs from resolvers are not functionally exhausted and have similar gene expression patterns as E2-specific MBCs from chronically infected subjects at their respective expansion peaks.
An increasing dominance of VH1-69+ antibodies in the BCR repertoire of E2-specific MBCs as acute infection transitions to persistence might account for the broad reactivity of HCV-specific antibody responses in chronically infected subjects. Previous studies have reported an increased presence of HCV-specific, VH1-69+ mAbs in chronically infected subjects as viremia persists and shown that these mAbs have broadly neutralizing activity against HCVpp and HCV cell culture isolates (38–41). Moreover, there is some evidence that certain HCV antigens, such as E2, are specifically recognized by VH1-69+ B cells, as prime boost immunization with recombinant E1E2 promotes expansion of VH1-69+ antigen-specific B cells (35). Accordingly, our data show that VH1-69+ E2-specific mAbs from chronically infected subjects have the highest neutralizing activity against 2 different genotypes of HCVpp. However, VH1-69+ E2-specific B cells do not comprise most E2-specific B cells in resolvers or chronically infected subjects during acute infection—rather, enrichment of VH3-23+ B cells is observed in both groups. Although the role of VH3-23+ mAbs in controlling HCV infection is unclear, they neutralize and protect against infections by related flaviviruses like Zika and Dengue (42).
As mAbs from both resolvers and chronically infected subjects were enriched in VH3-23 at the late acute time point, there are possibly other factors besides V gene usage that could influence the function of mAbs from HCV-specific B cells. SHMs, for example, correlate with neutralizing activity in mAbs of chronically infected subjects. However, SHM does not affect the neutralizing activity of nAbs in resolvers, because mAbs with few SHMs neutralize HCVpp as effectively as mAbs with high SHMs isolated from chronically infected subjects. Similar findings by Bailey et al. support this observation (12). Antibodies with low levels of SHMs can be generated earlier, since they require fewer rounds of mutations and selection by Tfh cells. This is associated with early expansion of circulating E2-specific MBCs in circulation in resolvers and might contribute to clearance.
cTfh cells may potentially drive E2-specific MBC expansion in resolvers, as they increase sharply during early acute infection. These cells also produce increased levels of IL-21 and CD40L, which could stimulate antigen-specific B cells to enter GC reactions and differentiate into MBCs or plasma cells. While our findings are focused on total rather than HCV-specific cTfh cells, previous studies have demonstrated that a large subset of activated cTfh cells during acute infection are HCV-specific and are associated with an increased NS4 antibody response (20). Further studies of the relationship between HCV E2-specific cTfh cells and development of nAbs in acute resolving infection will remain a high priority to guide future vaccine design. Even though we could not detect a significant increase of activated cTfh cells in blood from chronically infected subjects during acute infection, this does not mean that they are completely absent. We observed a sharp increase in frequency of activated cTfh cells in 2 of 10 chronically infected subjects at late acute infection. Moreover, SHMs in E2-specific MBCs of chronically infected subjects significantly increased at follow-up stages, indicating rounds of active replication and mutation in GCs, possibly induced by Tfh cells. Other studies have identified activated cTfh cells in livers of chronically infected individuals (20, 21, 43). Moreover, cTfh cells expand following direct-acting antiviral therapy and loss of virus replication (21). It is therefore possible that transient activation of cTfh cells in chronically infected subjects occurs during late acute infection, or periodically during persistence, in response to fluctuating levels of viremia.
Our findings support a model in which cTfh cells become activated during early acute HCV in resolvers and specifically activate B cells that react with HCV antigens, via IL-21 secretion and CD40L interactions (Figure 7K). This stimulates HCV-specific B cell entry into limited GC cycles, where many of them switch to VH3-23+ IgG1 or IgG2 isotype and accrue SHMs. A fraction of these cells differentiates into short-lived plasmablasts that secrete HCV-specific antibodies in the serum, whereas others exit as MBCs during late acute infection. It is possible that many of these antigen-specific MBCs eventually differentiate into plasmablasts or plasma cells that secrete nAbs against HCV antigen as acute infection resolves. In the case of chronically infected subjects, an early absence of activated cTfh cells leads to a slower activation of antigen-specific B cells by an unknown mechanism (Figure 7K). These “helpless” B cells possibly differentiate directly into plasmablasts that secrete antibodies against autologous and heterologous HCV antigens in sera. As infection persists, more antigen-specific B cells, many of which express VH1-69, undergo multiple GC reactions, possibly induced by delayed cTfh cell activity. This results in the accumulation of SHMs in their BCRs, which increases binding to cognate HCV antigen. Antigen-specific MBCs increase over time, seeding more plasmablasts that secrete HCV antibodies with limited neutralization breadth in sera.
It is important to note that even with early cTfh cell activation, the relative abundance and breadth of HCV-specific nAbs in resolvers is limited. nAbs are probably therefore not the main determinant of viral clearance, which is likely to also involve robust HCV-specific CD8+ and CD4+ Th-cell responses that eliminate HCV infection together. Increasing our understanding of the interactions between functions of T and B cells could contribute to development of an efficacious vaccine against HCV infection.
Methods
Human study participants.
Study subjects were recruited among people who inject drugs who were participating in HEPCO (22). All subjects were HIV negative. Acute HCV infection was identified as a positive HCV RNA or antibody test following a negative test in the past 3 months. EDI was defined as the median point between the last negative and the first positive test. Chronic HCV infection was defined as a positive result from an HCV RNA test at 6 months after EDI. Twenty subjects were selected for longitudinal analysis: 10 resolvers (HCV Ab+ and HCV RNA–) and 10 who developed a chronic infection (HCV Ab+ and HCV RNA+). Time points were baseline (negative for HCV RNA and antibodies, ≥ 8 weeks before EDI), early acute (8 ± 2 weeks after EDI), late acute (20 ± 4 weeks after EDI), and follow-up (≥ 51 weeks after EDI). PBMCs from 5 healthy donors were obtained from the leukapheresis study of the Fonds de recherche du Québec – Santé (FRQS) AIDS and Infectious Disease Network (Réseau SIDA-MI). An additional 10 HCV-naive people who inject drugs from the HEPCO cohort were added for the analysis of B cell subsets. Participants’ demographics and clinical characteristics are listed in Supplemental Table 1.
Cell lines and cell culture.
Human embryonic kidney 293T/17 (HEK293T/17, ATCC) and human hepatoma Huh-7.5 cells (generated in-house) (13) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin and maintained at 37°C and 5% CO2. Human Expi293F cells (Thermo Fisher Scientific) were cultured in Expi293 expression medium (Thermo Fisher Scientific) without FBS or antibiotics at 37°C and 8% CO2 while shaking at a constant speed of 80 rpm.
HCV E2 and NS3 ELISA.
Flat-bottom 96-well plates (Corning) were coated with 0.5 μg/mL H77 E2 and J6 E2 (both generated in-house)(13), S52 E2 and H77 E1E2 (both provided by Michael Houghton, John Law, and Michael Logan, University of Alberta, Edmonton, Canada) or NS3 protein (Abcam, recombinant NS3 (gt 1b) amino acids 1192 to 1456) overnight at 4°C. Plates were washed twice with PBS-T (PBS 1X, 0.05% Tween-20; Thermo Fisher Scientific) and blocked with 10% normal goat serum (Jackson ImmunoResearch Laboratories, catalog 005-000-121) in PBS-T overnight at 4°C. Plates were washed twice and 100-μL diluted plasma samples from the subjects was added to each well (starting dilution was 1:250 with 4-fold dilutions up to 1:16,000 in 0.1% normal goat serum in PBS-T) and incubated for 2 hours at room temperature. Plates were washed 6 times and HRP-conjugated anti-human IgG (BD Biosciences, clone G18-145, catalog 555788) was added at a dilution of 1:5000 for 90 minutes at room temperature. Plates were washed again 6 times and developed with Tetramethylbenzidine (TMB) substrate (BD Biosciences) for 15 minutes at room temperature before adding 1 volume of stop solution (2N H2SO4). Absorbances were read at 450 and 570nm.
Heatmaps.
Heatmaps were generated with GraphPad Prism, using the actual values of indicated tests and a 3-colors scheme, where red indicates the highest value, blue the lowest value, and yellow the median value of each test.
Measurement of antibodies against rubella and HBsAg.
Plasma IgG levels against rubella and HBsAg were evaluated by the clinical laboratory of the Montreal University Hospital.
Generation of HCV pseudoparticles.
HCVpp for genotype 1a (H77), genotype 2a (J6/JFH1) E1E2, and genotype 3a (S52) were generated in-house as described. Briefly, HEK293T/17 were seeded at 9.0 × 106 cells/dish on 15 cm dishes precoated with poly-l-lysine solution (MilliporeSigma). The following day, cells were transfected with pV1-Gluc reporter, HIV-gagpol, and HCV H77, J6/JFH1, or S52 Env plasmids for a total of 15 μg plasmid DNA using 92 μL Trans-IT-LT1 transfection reagent (Mirus Bio) following the manufacturer’s instructions. Media were changed 24 hours later, and supernatants were collected at 72 and 120 hours after transfection. Supernatants were centrifuged and filtered through a 0.45 μm filter. Supernatants were then centrifuged at 131,100 g for 4 hours over a 20% sucrose cushion at 4°C to concentrate HCVpp. Viral pellets were resuspended in a reduced volume of DMEM supplemented with 3% fetal calf serum (FCS), separated into aliquots and stored at –80°C until future use.
HCVpp neutralization assays.
Neutralization assays were performed as previously described (44). Huh7.5 cells were plated on wells of sterile 96-well plates (Corning) precoated with collagen solution (0.01%, MilliporeSigma) at a density of 1 × 104 cells/well. The next day, plasma aliquots were incubated at 56°C for 30 minutes to inactivate complement. Fixed plasma dilutions (1:25) were added to an equal volume of either H77, J6/JFH1, or S52 E1E2 HCVpp (2-fold concentrated). For assays involving mAbs cloned from human subjects, each individual mAb was diluted into five 5-fold serial dilutions in DMEM/3% FCS in 96-well round-bottom plates. Dilutions of human polyclonal IgG (MilliporeSigma, catalog AG711) and mouse anti-HCV E2 mAb (2C1, generated in-house; ref. 36) were included as negative and positive controls, respectively. Plasma-HCVpp or mAb-HCVpp complexes (final plasma dilution 1:50) were incubated at 37°C for 1 hour. Fifty microliters of plasma/mAb-HCVpp mixture was added to cell monolayers for 16–18 hours, after which the monolayers were washed and cultured in fresh media. After 48 hours, cells were lysed in 50 μL Renilla Luciferase Assay Lysis Buffer (Promega), and 30 μL of cell lysates was mixed with 30 μL of Renilla Luciferase Assay Substrate (1X, Promega). Luminescence was detected on a Synergy H1 Microplate Reader (Biotek Instruments). Percent neutralization was calculated and normalized to baseline using the following equation: % neutralization = [1 – (RLUinfection time point – RLUmock)/(RLUbaseline – RLUmock)] × 100%.
For assays involving subject mAbs, a neutralization threshold was set to an inverse IC50 value (IC50–1) of 0.02 (or IC50 = 50 μg/mL) as designated by previous studies (44). IC50 values were calculated from 4PL dose response nonlinear regression curves fitted to percentage of neutralization as a function of increasing mAb concentrations. mAbs from all subjects regardless of infection outcome were ranked from 1 to 60 according to neutralizing activity for H77 and J6/JFH1 HCVpp, as measured by inverse IC50 values. The highest neutralizing mAb had the highest inverse IC50 value and was ranked 1, whereas the lowest neutralizing mAb had the lowest IC50 value and was ranked 60. Ranks were assigned using the RANK function in Microsoft Excel. The combined rank was calculated as the sum of ranks for H77 and J6/JFH1 neutralization. Three independent experiments were performed for all neutralization assays.
Flow cytometry.
Cryopreserved PBMCs were thawed, washed with FACS buffer: PBS 1X, 1% FBS (MilliporeSigma), 0.01% sodium azide (Thermo Fisher Scientific) then either stained directly or stimulated for functional analysis. Cells (2 × 106) were stimulated with phorbol myristate acetate (PMA, 2.5 ng/mL) and ionomycin (1 μg/mL) in snap cap tubes (BD Biosciences) for 5 hours at 37°C. Brefeldin-A and monensin (both 10 μg/mL) were added 1 hour after stimulation.
Biotinylated E2 monomers (generated in-house, ref. 36) were used to prepare tetramers by adding phycoerythrin-labeled (PE-labeled) ExtrAvidin (MilliporeSigma) (5 aliquots of 7.5 μL, 10 minutes incubation at room temperature each). Tetramer staining was performed for 30 minutes at room temperature. Cells were then stained with surface marker (antibodies used are listed in Supplemental Table 5) and live cells were identified using LIVE/DEAD fixable aqua dead cell stain kit (Thermo Fisher Scientific) for 30 minutes at 4°C. Cells were permeabilized using either BD Cytofix/Cytoperm (BD Biosciences) for 15 minutes at 4°C or FoxP3 fixation and permeabilization buffer (Thermo Fisher Scientific) for 30 minutes at 4°C for functional or phenotypic analyses, respectively. Intracellular staining was done for 30 minutes at 4°C. Cells were fixed with 1% paraformaldehyde (PFA, MilliporeSigma) in PBS before FACS analysis. Multiparameter flow cytometry was performed at the flow cytometry core of the Centre de Recherche du Centre Hospitalier de l’Université de Montréal using a BD LSRII instrument equipped with violet (405 nm), blue (488 nm), yellow-green (561 nm), and red (633 nm) lasers and FACSDiva version 8.0.1 (BD Biosciences). FCS data files were analyzed using FlowJo version 10.0.8 for Mac (Tree Star).
Sorting and reverse-transcription PCR of single E2-specific MBCs.
Cryopreserved PBMCs were thawed and stained with fluorescently labeled J6 E2 tetramer and cell surface markers as described above. Single E2-specific MBCs were sorted on a FACS Aria Cell Sorter into individual wells of 96-well PCR plates (Bio-Rad Laboratories) containing 10 μL of one of 2 formulations of “catch” buffer: RLT buffer (QIAGEN) supplemented with 2-mercaptoethanol (MilliporeSigma) for single-cell RNA-seq; or 10 mM Tris pH 8.0 supplemented with RNase inhibitor rRNAsin (Promega) for samples that were exclusively used for BCR sequencing and cloning. Plates were centrifuged briefly, immediately frozen on dry ice, and stored at –80°C.
Reverse transcription PCR of sorted, single cells was performed using Maxima First Strand cDNA Synthesis Kit for RT-qPCR (Thermo Fisher Scientific) as previously outlined (45). Single cell lysates were thawed on ice and mixed with 5X Reaction Buffer, Maxima Enzyme Mix, and 5% IGEPAL. RNA was transcribed to cDNA under the following conditions: 25°C for 10 minutes, 50°C for 30 minutes, 85°C for 5 minutes, and 4°C hold. Two microliters cDNA were used in subsequent PCR reactions for amplifying heavy and light chain Ig genes.
Immunoglobulin bioinformatic analyses.
Ig reconstruction of paired heavy and light chains from single cells was carried out using the IG-mapped_Unmapped filtering method within the BCR Assignment of Lineage using De novo Reconstruction (BALDR) pipeline (46). The default trimmomatic v0.36 parameters were modified to only include reads with a minimum length of 50. Gnu parallel was used to run parallel BALDR reconstructions.
As part of the BALDR pipeline, the reads were mapped to the human reference genome GRCh38 (Ensembl release 86 primary assembly) using STAR v 2.5.2b (47, 48). The reads mapping to Ig loci and unmapped reads were combined using samtools v1.7 and seqtk v1.2-r94 and then assembled using Trinity v2.3.2 (49, 50). The assembled reads were annotated with IgBLAST v1.6.1 (51) using the International Immunogenetics Information System (IMGT) database (52, 53). The reads were mapped to the assembled sequences using bowtie2 v2.3.0 (54). The assembled sequences were ranked based on the number of mapped reads and the best heavy and light chain sequences were selected manually. The Circos plots were generated using the Circos v0.69-6 command-line software (55).
Cloning of monoclonal antibodies.
Paired heavy (IgH) and light chain (IgK/L) genes were amplified from cDNA of single E2-specific MBCs using a nested PCR approach using DreamTaq Green PCR Master Mix (Thermo Fisher Scientific) or Q5 High Fidelity 2X Master Mix (New England Biolabs) and V gene-specific primers (Supplemental Table 6) and (45, 56, 57). PCR products were purified and concentrated by column purification using a DNA Clean and Concentrator-100 kit (Zymo Research). Purified PCR products and human heavy (pFUSEss-CHIg-hG1; InvivoGen) or light chain (pFUSE2ss-CLIg-hK for kappa chain or pFUSE2ss-CLIg-hL2 for lambda chain; InvivoGen) plasmids were digested with EcoRI-HF and NheI-HF for IgH chain, EcoRI-HF and BsiWI-HF for IgK chain, or EcoRI-HF and AvrII for IgL chain (New England Biolabs) and ligated into respective heavy or light chain vectors in-frame with and upstream of human IgG1 Fc region. IgH and IgK/L constructs were transformed into NEB 5α Competent E. coli (New England Biolabs) and plated on LB Zeocin (InvivoGen) or LB Blasticidin (InvivoGen) plates for selection. Colonies were screened for presence of V gene insert by colony PCR using the appropriate primers (Supplemental Table 6). Positive clones were grown in 5 mL LB overnight and 37°C, and plasmid DNA was purified using Qiaprep Spin Miniprep Kit (QIAGEN). Plasmids were further screened by DraI (New England Biolabs) restriction digest and sequenced for proper insertion of heavy or light V chain gene.
Expression and purification of monoclonal antibodies.
Expi293F cells (Thermo Fisher Scientific) were transfected with Expifectamine (Thermo Fisher Scientific) and separate plasmids encoding paired heavy and light chain genes in a 1:1 ratio using the Expi293 Expression System (Thermo Fisher Scientific) according to the manufacturer’s instructions. Supernatants containing secreted mAbs were harvested 7 days after transfection and centrifuged to remove cells and cell debris. mAbs were purified as previously described (45). mAbs were precipitated by overnight incubation with Pierce Protein A Agarose beads (Thermo Fisher Scientific) in Pierce immunoprecipitation IP buffer (25 mM Tris, 150 mM NaCl, pH 7.2). mAb-Protein A complexes were transferred to Mini Bio-Spin Chromatography Columns (Bio-Rad) and washed once with 1M NaCl, followed by 2 washes with 1X PBS. mAbs were eluted with 0.1M glycine-HCl (pH 2.5) into tubes containing 1M Tris pH 9.0. mAb eluates were further concentrated in sterile 1X PBS buffer by centrifugation in Amicon Ultra 0.5 mL Centrifugal Filter (MilliporeSigma). IgG yield was measured using a NanoDrop Spectrophotometer (Thermo Fisher Scientific).
E2 binding assays.
Binding of mAbs to J6 E2 glycoprotein was measured by biolayer interferometry (BLI) using an Octet RED96 instrument (Fortebio). Individual mAbs were diluted in binding buffer (0.1% BSA, 0.02% Tween-20, 0.05% NaN3 in 1X PBS) at a fixed concentration of 10 μg/mL and immobilized on Anti-hIgG Fc Capture (AHC) biosensor (Fortebio) for 300 seconds. Baseline readings were recorded for 60 seconds. Sensors were then submerged into varying concentrations (1000 nM, 333 nM, 111 nM, 37 nM, 12.3 nM, and 0 nM) of HCV J6 E2 glycoprotein (aa 368 – 713) for 300 seconds at 30°C. Sensor-mAb-E2 complexes were then allowed to dissociate for 600 seconds while shaking at 1000 rpm. Sensors were stripped of mAbs by 3 cycles of immersion in regeneration buffer (10 mM glycine-HCl, pH 1.7) and binding buffer. Polyclonal human IgG (10 μg/mL; MilliporeSigma, catalog AG711) was included as a negative control. Binding strength, defined as the fraction of E2-mAb complexes (measured by changes in optical intensity) over total bound and unbound mAbs formed at increasing concentrations of J6 E2, were calculated using ForteBio Data Analysis 9.0 software. Binding strength was expressed as inverse EC50 values (EC50–1) calculated from 4PL dose response nonlinear regression curves fitted to response intensities as a function of increasing J6 E2 molar concentrations.
Statistics.
Statistical analyses were performed with SigmaPlot (Systat Software) or Prism versions 5.0, 7.0, or 8.0 (GraphPad). Details of tests (including number of data points (n) and P values) are provided in each figure legend. Differences between groups in longitudinal analyses were determined by 2-way repeated measure ANOVA with Tukey’s post hoc test. Analyses that did not include multiple time points were determined by Mann-Whitney test or 1-way ANOVA (Kruskal-Wallis test with Dunn’s multiple comparisons test) as indicated. For all statistical tests, P values less than 0.05 were considered significant.
Data code and availability.
Gene expression data are deposited in the NCBI Gene Expression Omnibus (GEO) repository under accession number GSE157447. The code used to obtain Ig sequences is available at https://github.com/BosingerLab/HCV_manuscript The Ig sequences are deposited in GenBank under accessions MW176150–MW177530.
Graphical illustrations.
The graphical abstract was created with BioRender.com.
Study approval.
This study was approved by the Institutional Ethics Committee of the Centre Hospitalier de l’Université de Montréal, Montréal (protocol SL05.014), Montreal, Quebec, Canada. and performed in accordance with the Declaration of Helsinki. All subjects provided written informed consent.
Author contributions
ES, MB, NHS, and AG conceived and designed the study. ES and MB performed experiments, analyzed the data, and wrote the manuscript. AAU, SEB, NHS, and AG analyzed the data and wrote the manuscript. JM, CAD, and MJE contributed essential reagents and equipment. NB and SAN performed experiments. JB contributed essential clinical samples and data.
Supplementary Material
Acknowledgments
We would like to thank the Yerkes and the Research Center of Montreal University Hospital flow cytometry cores for their assistance in cell sorting and the Yerkes Non-human Primate Genomics Core laboratory for processing samples for single-cell RNA-seq. We thank Dennis Jang and Carson “Elliot” Anderson at Emory University for their assistance in cloning IgG plasmids for expression of mAbs.
This study was funded by NIH grants R01AI136533, R01AI124680, R01AI096882, and R01AI126890 (to AG); Office of Research Infrastructure Programs/Office of the Director P51OD011132 (formerly National Center for Research Resources P51RR000165) to the Yerkes National Primate Research Center; 5U01AI131313 from NIH (to NHS); the Canadian Institutes of Health Research (CIHR) (PJT-173467; to NHS and JB); Alberta Innovates-Health Solutions (to NHS and JB); Fonds de recherche du Québec – Santé (FRQS) AIDS and Infectious Disease Network (Réseau SIDA-MI) (to NHS and JB); and the Intramural Research Programs of the National Institute of Allergy and Infectious Diseases (to JM). MB received postdoctoral fellowships from the FRQS, the American Liver Foundation, and the Canadian Network on Hepatitis C (CanHepC). CanHepC is funded by a joint initiative of the CIHR (NHC142832) and the Public Health Agency of Canada. JB is the Canada Research Chair in Addiction Medicine. This work was facilitated by the Immunology and Flow Cytometry Core of the Center for AIDS Research at Emory University (P30AI050409).
Version 1. 01/19/2021
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(2):e140590.https://doi.org/10.1172/JCI140590.
Contributor Information
Eduardo Salinas, Email: esalin2@emory.edu.
Maude Boisvert, Email: maude.boisvert.chum@ssss.gouv.qc.ca.
Amit A. Upadhyay, Email: amit.anil.upadhyay@emory.edu.
Nathalie Bédard, Email: nathalie.bedard.chum@ssss.gouv.qc.ca.
Sydney A. Nelson, Email: sydney.anne.nelson@emory.edu.
Julie Bruneau, Email: julie.bruneau@umontreal.ca.
Cynthia A. Derdeyn, Email: cderdey@emory.edu.
Joseph Marcotrigiano, Email: joseph.marcotrigiano@nih.gov.
Matthew J. Evans, Email: matthew.evans@mssm.edu.
Steven E. Bosinger, Email: steven.bosinger@emory.edu.
Naglaa H. Shoukry, Email: naglaa.shoukry@umontreal.ca.
Arash Grakoui, Email: arash.grakoui@emory.edu.
References
- 1.Tomaras GD, Plotkin SA. Complex immune correlates of protection in HIV-1 vaccine efficacy trials. Immunol Rev. 2017;275(1):245–261. doi: 10.1111/imr.12514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shoukry NH. Hepatitis C vaccines, antibodies, and T cells. Front Immunol. 2018;9:1480. doi: 10.3389/fimmu.2018.01480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suryaprasad AG, et al. Emerging epidemic of hepatitis C virus infections among young nonurban persons who inject drugs in the United States, 2006-2012. Clin Infect Dis. 2014;59(10):1411–1419. doi: 10.1093/cid/ciu643. [DOI] [PubMed] [Google Scholar]
- 4.Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. 2002;36(5 Suppl 1):S21–S29. doi: 10.1053/jhep.2002.36227. [DOI] [PubMed] [Google Scholar]
- 5.Grakoui A, et al. HCV persistence and immune evasion in the absence of memory T cell help. Science. 2003;302(5645):659–662. doi: 10.1126/science.1088774. [DOI] [PubMed] [Google Scholar]
- 6.Shoukry NH, et al. Memory CD8+ T cells are required for protection from persistent hepatitis C virus infection. J Exp Med. 2003;197(12):1645–1655. doi: 10.1084/jem.20030239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox AL, et al. LB10. A randomized, double-blind, placebo-controlled efficacy trial of a vaccine to prevent chronic hepatitis C virus infection in an at-risk population. Open Forum Infect Dis. 2019;6(Suppl 2):S997 [Google Scholar]
- 8.Kinchen VJ, et al. Plasma deconvolution identifies broadly neutralizing antibodies associated with hepatitis C virus clearance. J Clin Invest. 2019;130(11):4786–4796. doi: 10.1172/JCI130720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osburn WO, et al. Clearance of hepatitis C infection is associated with the early appearance of broad neutralizing antibody responses. Hepatology. 2014;59(6):2140–2151. doi: 10.1002/hep.27013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osburn WO, et al. Spontaneous control of primary hepatitis C virus infection and immunity against persistent reinfection. Gastroenterology. 2010;138(1):315–324. doi: 10.1053/j.gastro.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colbert MD, et al. Broadly neutralizing antibodies targeting new sites of vulnerability in hepatitis C virus E1E2. J Virol. 2019;93(14):e02070-18. doi: 10.1128/JVI.02070-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bailey JR, et al. Broadly neutralizing antibodies with few somatic mutations and hepatitis C virus clearance. JCI Insight. 2017;2(9):e92872. doi: 10.1172/jci.insight.92872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan AG, et al. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature. 2014;509(7500):381–384. doi: 10.1038/nature13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sugalski JM, Rodriguez B, Moir S, Anthony DD. Peripheral blood B cell subset skewing is associated with altered cell cycling and intrinsic resistance to apoptosis and reflects a state of immune activation in chronic hepatitis C virus infection. J Immunol. 2010;185(5):3019–3027. doi: 10.4049/jimmunol.1000879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Racanelli V, et al. Antibody V(h) repertoire differences between resolving and chronically evolving hepatitis C virus infections. PLoS One. 2011;6(9):e25606. doi: 10.1371/journal.pone.0025606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oliviero B, et al. Skewed B cells in chronic hepatitis C virus infection maintain their ability to respond to virus-induced activation. J Viral Hepat. 2015;22(4):391–398. doi: 10.1111/jvh.12336. [DOI] [PubMed] [Google Scholar]
- 17.Crotty S. T Follicular helper cell biology: a decade of discovery and diseases. Immunity. 2019;50(5):1132–1148. doi: 10.1016/j.immuni.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morita R, et al. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 2011;34(1):108–121. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Locci M, et al. Human circulating PD-1+CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity. 2013;39(4):758–769. doi: 10.1016/j.immuni.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raziorrouh B, et al. Virus-specific CD4+ T cells have functional and phenotypic characteristics of follicular T-helper cells in patients with acute and chronic HCV infections. Gastroenterology. 2016;150(3):696–706.e3. doi: 10.1053/j.gastro.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 21.Smits M, et al. Follicular T helper cells shape the HCV-specific CD4+ T cell repertoire after virus elimination. J Clin Invest. 2020;130(2):998–1009. doi: 10.1172/JCI129642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grebely J, et al. Cohort profile: the international collaboration of incident HIV and hepatitis C in injecting cohorts (InC3) study. Int J Epidemiol. 2013;42(6):1649–1659. doi: 10.1093/ije/dys167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Araujo AC, Astrakhantseva, Fields HA, Kamili S. Distinguishing acute from chronic hepatitis C virus (HCV) infection based on antibody reactivities to specific HCV structural and nonstructural proteins. J Clin Microbiol. 2011;49(1):54–57. doi: 10.1128/JCM.01064-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beld M, et al. Quantitative antibody responses to structural (Core) and nonstructural (NS3, NS4, and NS5) hepatitis C virus proteins among seroconverting injecting drug users: impact of epitope variation and relationship to detection of HCV RNA in blood. Hepatology. 1999;29(4):1288–1298. doi: 10.1002/hep.510290442. [DOI] [PubMed] [Google Scholar]
- 25.Ellebedy AH, et al. Defining antigen-specific plasmablast and memory B cell subsets in human blood after viral infection or vaccination. Nat Immunol. 2016;17(10):1226–1234. doi: 10.1038/ni.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ehrhardt GR, Hijikata A, Kitamura H, Ohara O, Wang JY, Cooper MD. Discriminating gene expression profiles of memory B cell subpopulations. J Exp Med. 2008;205(8):1807–1817. doi: 10.1084/jem.20072682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein U, et al. Transcriptional analysis of the B cell germinal center reaction. Proc Natl Acad Sci U S A. 2003;100(5):2639–2644. doi: 10.1073/pnas.0437996100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan TD, Brink R. Affinity-based selection and the germinal center response. Immunol Rev. 2012;247(1):11–23. doi: 10.1111/j.1600-065X.2012.01118.x. [DOI] [PubMed] [Google Scholar]
- 29.Sage PT, Alvarez D, Godec J, von Andrian UH, Sharpe AH. Circulating T follicular regulatory and helper cells have memory-like properties. J Clin Invest. 2014;124(12):5191–5204. doi: 10.1172/JCI76861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sage PT, Francisco LM, Carman CV, Sharpe AH. The receptor PD-1 controls follicular regulatory T cells in the lymph nodes and blood. Nat Immunol. 2013;14(2):152–161. doi: 10.1038/ni.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bentebibel SE, et al. Induction of ICOS+CXCR3+CXCR5+ TH cells correlates with antibody responses to influenza vaccination. Sci Transl Med. 2013;5(176):176ra32. doi: 10.1126/scitranslmed.3005191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kinchen VJ, et al. Broadly neutralizing antibody mediated clearance of human hepatitis C virus infection. Cell Host Microbe. 2018;24(5):717–730.e5. doi: 10.1016/j.chom.2018.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meunier JC, et al. Evidence for cross-genotype neutralization of hepatitis C virus pseudo-particles and enhancement of infectivity by apolipoprotein C1. Proc Natl Acad Sci U S A. 2005;102(12):4560–4565. doi: 10.1073/pnas.0501275102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Logvinoff C, et al. Neutralizing antibody response during acute and chronic hepatitis C virus infection. Proc Natl Acad Sci U S A. 2004;101(27):10149–10154. doi: 10.1073/pnas.0403519101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen F, et al. Antibody responses to immunization with HCV envelope glycoproteins as a baseline for B-Cell-based vaccine development. Gastroenterology. 2020;158(4):1058–1071.e6. doi: 10.1053/j.gastro.2019.11.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boisvert M, et al. Novel E2 glycoprotein tetramer detects hepatitis C virus-specific memory B cells. J Immunol. 2016;197(12):4848–4858. doi: 10.4049/jimmunol.1600763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Racanelli V, et al. Antibody production and in vitro behavior of CD27-defined B-cell subsets: persistent hepatitis C virus infection changes the rules. J Virol. 2006;80(8):3923–3934. doi: 10.1128/JVI.80.8.3923-3934.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olbrich A, et al. Repertoire and neutralizing activity of antibodies against hepatitis C virus E2 peptide in patients with spontaneous resolution of hepatitis C. J Infect Dis. 2019;220(7):1209–1218. doi: 10.1093/infdis/jiz274. [DOI] [PubMed] [Google Scholar]
- 39.Chen F, Tzarum N, Wilson IA, Law M. VH1-69 antiviral broadly neutralizing antibodies: genetics, structures, and relevance to rational vaccine design. Curr Opin Virol. 2019;34:149–159. doi: 10.1016/j.coviro.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tzarum N, et al. Genetic and structural insights into broad neutralization of hepatitis C virus by human VH1-69 antibodies. Sci Adv. 2019;5(1):eaav1882. doi: 10.1126/sciadv.aav1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flyak AI, et al. HCV broadly neutralizing antibodies use a CDRH3 disulfide motif to recognize an E2 glycoprotein site that can be targeted for vaccine design. Cell Host Microbe. 2018;24(5):703–716.e3. doi: 10.1016/j.chom.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robbiani DF, et al. Recurrent potent human neutralizing antibodies to Zika virus in Brazil and Mexico. Cell. 2017;169(4):597–609.e11. doi: 10.1016/j.cell.2017.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spaan M, et al. CD4+ CXCR5+ T cells in chronic HCV infection produce less IL-21, yet are efficient at supporting B cell responses. J Hepatol. 2015;62(2):303–310. doi: 10.1016/j.jhep.2014.09.024. [DOI] [PubMed] [Google Scholar]
- 44.Bailey JR, Urbanowicz RA, Ball JK, Law M, Foung SKH. Standardized method for the study of antibody neutralization of HCV pseudoparticles (HCVpp) Methods Mol Biol. 2019;1911:441–450. doi: 10.1007/978-1-4939-8976-8_30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ho IY, et al. Refined protocol for generating monoclonal antibodies from single human and murine B cells. J Immunol Methods. 2016;438:67–70. doi: 10.1016/j.jim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Upadhyay AA, et al. BALDR: a computational pipeline for paired heavy and light chain immunoglobulin reconstruction in single-cell RNA-seq data. Genome Med. 2018;10(1):20. doi: 10.1186/s13073-018-0528-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cunningham F, et al. Ensembl 2015. Nucleic Acids Res. 2015;43(Database issue):D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ye J, Ma N, Madden TL, Ostell JM. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41(Web Server issue):W34–W40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giudicelli V, Brochet X, Lefranc MP. IMGT/V-QUEST: IMGT standardized analysis of the immunoglobulin (IG) and T cell receptor (TR) nucleotide sequences. Cold Spring Harb Protoc. 2011;2011(6):695–715. doi: 10.1101/pdb.prot5633. [DOI] [PubMed] [Google Scholar]
- 53.Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36(Web Server issue):W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith K, et al. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat Protoc. 2009;4(3):372–384. doi: 10.1038/nprot.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods. 2008;329(1-2):112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.