Abstract
TH17 cell subpopulations have been defined that contribute to inflammation and homeostasis, yet the characteristics of TH17 cells that contribute to host defense against infection are not clear. To elucidate the antimicrobial machinery of the TH17 subset, we studied the response to Cutibacterium acnes, a skin commensal that is resistant to IL-26, the only known TH17-secreted protein with direct antimicrobial activity. We generated C. acnes–specific antimicrobial TH17 clones (AMTH17) with varying antimicrobial activity against C. acnes, which we correlated by RNA sequencing to the expression of transcripts encoding proteins that contribute to antimicrobial activity. Additionally, we validated that AMTH17-mediated killing of C. acnes and bacterial pathogens was dependent on the secretion of granulysin, granzyme B, perforin, and histone H2B. We found that AMTH17 cells can release fibrous structures composed of DNA decorated with histone H2B that entangle C. acnes that we call T cell extracellular traps (TETs). Within acne lesions, H2B and IL-17 colocalized in CD4+ T cells, in proximity to TETs in the extracellular space composed of DNA decorated with H2B. This study identifies a functionally distinct subpopulation of TH17 cells with an ability to form TETs containing secreted antimicrobial proteins that capture and kill bacteria.
Keywords: Immunology
Keywords: T cells
Introduction
T cell responses represent an important component of the adaptive immune response and contribute to host defense against microbial pathogens by secreting cytokines that activate antimicrobial effector pathways and proteins that directly lyse infected targets (1). Classically, CD4+ T cell subsets with diverse immunological functions have been distinguished based on unique cytokine secretion patterns and transcription factor profiles (2–7). TH17 cells express the transcriptional factor RORγt and secrete IL-17, IL-22, and IL-26, among others. Cytokines such as TGF-β, IL-1β, and IL-6 are involved in TH17 cell differentiation (8–10), and signals from these cytokines result in the activation of the transcription factor STAT3, which directly regulates downstream genes involved in TH17 differentiation (11, 12). Defective TH17 cell responses in STAT3-deficient patients have been associated with increased susceptibility to bacterial infections, indicating that the TH17 subset has a major role in host defense (13–16).
The ability of T cells to lyse infected target cells can be accompanied by the release of antimicrobial effector molecules that kill both intracellular and extracellular bacteria. Two major mechanisms are responsible for T cell–mediated cytolytic activity. The first involves the secretion of lytic granules containing perforin and granzymes by T cells upon contact with a target, and the second involves the interaction of membrane-bound Fas ligand on T cells with the Fas protein on the target cell (17, 18). In addition to CD8+ T cells, several in vivo studies have demonstrated that cytolytic CD4+ T cells can play a protective role in viral clearance, antimicrobial activity against intracellular bacteria (19), and elimination of tumors (20–25).
The identification of CD4+ T cell–mediated killing of target cells has been described within the entire heterogeneous CD4+ T cell population and little is known about the extent to which CD4+ T subsets are involved in CD4+ T cell–mediated antimicrobial activity. In the case of the TH17 cells, most of the work has focused on defining their role in pathologic inflammation and disease (26, 27). It is unclear what distinguishes inflammatory TH17 cells elicited by pathogens from tissue-resident TH17 cells induced by commensals. Two functionally distinct populations of TH17 cells can simultaneously reside within the gut during pathogen-induced inflammation; notably, TH17 cells induced by segmented filamentous bacteria were shown to induce noninflammatory homeostatic TH17 cells, whereas Citrobacter rodentium–induced TH17 cells exhibited a high inflammatory cytokine profile reflecting an inflammatory effector potential (28). Similarly, we and others have demonstrated that Cutibacterium acnes is a potent inducer of IL-17 and IFN-γ in CD4+ T cells, and that IL-17+ cells are present in perifollicular infiltrates of acne lesions, indicating that TH17 cells contribute to the pathogenesis of the disease (8, 29). Moreover, acne-associated (CA) and healthy-associated (CH) strains of C. acnes differentially modulate the CD4+ T cell responses to induce IL-17/IFN-γ– or IL-17/IL-10–secreting TH17 cells, respectively. However, little is known about how TH17 cells contribute to the killing of C. acnes, as TH17-mediated release of the antimicrobial protein IL-26 did not reduce bacterial viability (30).
Here, we used RNA sequencing (RNA-seq) to determine the mechanism(s) involved in antimicrobial TH17 cell–mediated killing of bacteria, initially studying the immune response to C. acnes. We generated C. acnes–specific antimicrobial TH17 clones (hereafter termed AMTH17) with varying antimicrobial activity against C. acnes. We show that C. acnes–induced AMTH17 cells represent a subset of CD4+ TEM and TEMRA cells. RNA-seq analysis indicated that cytotoxic gene expression in AMTH17 clones correlates with both protein secretion and antimicrobial activity against C. acnes and is dominated by a number of known antimicrobial proteins. We found that AMTH17 cells release histone-rich T cell extracellular traps (TETs) in conjunction with antimicrobial proteins that can entangle and kill bacteria. Our results suggest that AMTH17-mediated killing of bacteria may be a general mechanism that contributes to homeostatic regulation of bacterial colonization.
Results
C. acnes–specific AMTH17 are highly enriched in cytotoxic genes.
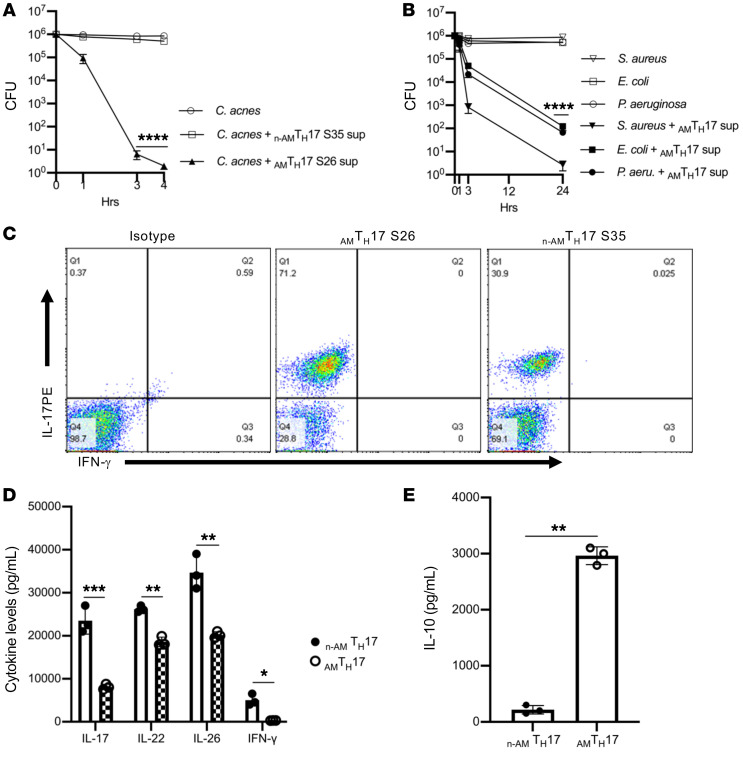
Besides CD8+ cytotoxic T lymphocytes (CTLs), human CD4+ T cells with cytolytic functions have been reported in response to viral infections (31–34). CD4+ T cells are able to function as CTLs ex vivo and can be detected following vaccinations, including against poliovirus, small pox, and in response to vaccines against HIV infection (35, 36), and are likely to play a role in host defense (37). During the propagation of long-term CD4+ T cell lines in the absence of cloning, cells expressing cytotoxic and antimicrobial activity are lost after several weeks of culture (17); therefore, we developed a cloning strategy that involves the use of whole C. acnes bacteria to stimulate immune cells, and used sterile cell sorting to select for C. acnes–specific TH17 cells (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI141594DS1). We generated and maintained short-term cultures of stable C. acnes strain–specific TH17 clones, which enabled us to recapitulate the spectrum of the biology present in ex vivo TH17 cells (28). The quick expansion also permitted the analysis of transcripts associated with TH17 cells. We first compared the antimicrobial activity of supernatants derived from these clones against C. acnes and several bacterial strains. We identified AMTH17 cells that had antimicrobial activity against C. acnes and other Gram-positive and Gram-negative bacteria (Figure 1, A and B). We also identified non-antimicrobial TH17 clones, hereafter termed n-AMTH17, that lacked antimicrobial potency (Figure 1A). Both the AMTH17 and n-AMTH17 clones were able to secrete IL-17 upon stimulation with α-CD3/α-CD28 antibodies (Figure 1C). In further comparisons of the cytokine secretion patterns of the AMTH17 and n-AMTH17 clones, we observed that the secretion of IL-17, IL-22, IL-26, and IFN-γ (P ≤ 0.001) was higher in the n-AMTH17 than in the AMTH17 clones (Figure 1D). On the other hand, IL-10 levels were elevated within the AMTH17 compared with the n-AMTH17 clones (Figure 1E), suggesting that the AMTH17 subset likely produces IL-10 in addition to other cytokines as an important regulatory molecule to dampen excessive inflammation.
Figure 1. AMTH17 cells secrete TH17-associated cytokines and are antimicrobial against Cutibacterium acnes and other bacterial strains.
(A) Observed CFU activity against C. acnes strain HL005PA1 after 4-hour incubation with AMTH17 clone S26 and n-AMTH17 clone S35 supernatants. (B) Observed CFU activity against several bacterial strains after 24-hour incubation with AMTH17 clone S26 and n-AMTH17 clone S35 supernatants. Data are shown as mean ± SEM (n > 3). ****P < 0.0001 by repeated-measures 1-way ANOVA for treatment groups compared with n-AMTH17 supernatants in panel A and C. acnes in panel B. (C) AMTH17 and n-AMTH17 clones were stimulated with α-CD3/α-CD28 for 5 hours and IL-17 and IFN-γ expression determined by flow cytometry (n > 3). (D and E) Cytokine levels in AMTH17 clones (S26, S27, and S28) and n-AMTH17 clones (S35, S38, and S44), as determined by ELISA. Data are shown as mean ± SEM (n > 3). *P < 0.05, **P < 0.01, ***P < 0.001 by 2-tailed Student’s t test.
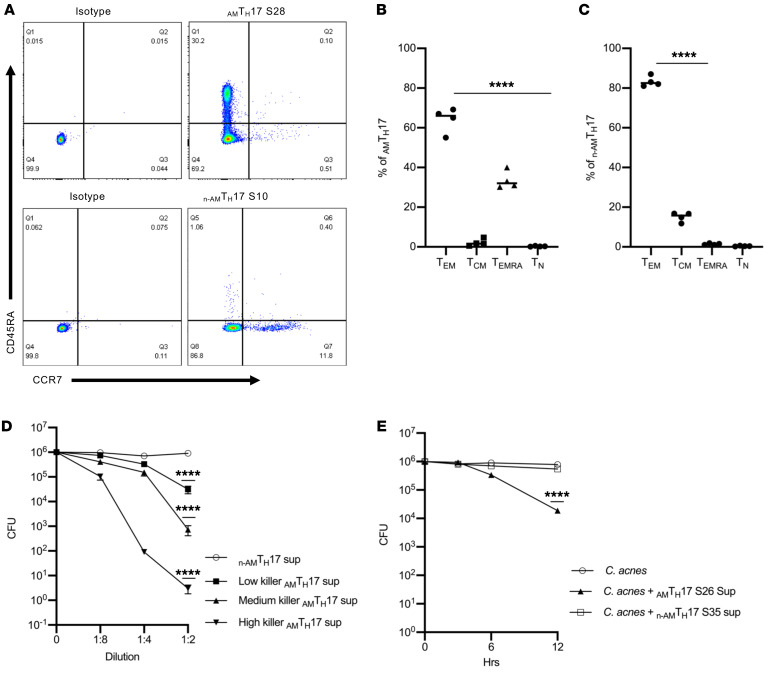
We next investigated the phenotype of the TH17 clones via flow cytometry. We analyzed 15 AMTH17 clones and discovered that a mean of 64% of the AMTH17 were enriched in the CD4+ TEM (CD4+CD45RA–CCR7–) and 34% within the TEMRA (CD4+CD45RA+CCR7–) subsets (Figure 2, A and B). On the other hand, of the 5 n-AMTH17 clones that we analyzed, 82% were highly enriched within the CD4+ TEM and 15% within the CD4+ TCM (CD4+CD45RA–CCR7+) subsets (Figure 2, A and C). In addition, these clones expressed transcripts associated with tissue-resident memory T cells such as CXCR6, ITGAE (CD103), KFL2, and S1PR1 (Supplemental Figure 2 and refs. 38, 39). These data suggest that the AMTH17 cells are at an advanced stage of differentiation, and may have the ability to exert antimicrobial activity as they home to peripheral nonlymphoid tissues such as the skin.
Figure 2. AMTH17 are CD4+ TEM and TEMRA cells and demonstrate antimicrobial activity as early as 6 hours.
(A) AMTH17 and n-AMTH17 clones were stimulated with α-CD3/α-CD28 and stained with antibodies against CD4, CD45RA, and CCR7. The AMTH17 clones consisted of primarily CD4+CD45RA+CCR7–RA (TEM) and CD4+CD45RA–CCR7– (TEMRA), whereas the n-AMTH17 clones consisted mainly of TEM and CD4+CD45RA–CCR7+ (TCM). Data are representative of 4 independent experiments using clones derived from 4 different donors. (B and C) Analysis of memory markers in AMTH17 clones (S5, S16, S26, and S28) and n-AMTH17 clones (S10, S13, S35, and S38) by flow cytometry (n = 4). ****P < 0.0001 by repeated-measures 1-way ANOVA for TEM compared with TCM, TEMRA, and naive T cells (TN). (D) Several AMTH17 and n-AMTH17 clones were stimulated with α-CD3/α-CD28 and supernatants used for CFU assays against C. acnes strain HL096PA1. The AMTH17 clones were subsequently stratified into High, Medium, and Low based on the results of the CFU assays. ****P < 0.001 by repeated-measures 1-way ANOVA, Low, Medium, and High killer AMTH17 compared with n-AMTH17. (E) Observed antimicrobial kinetics of supernatants derived from activated AMTH17 clones against several C. acnes strains (HL110PA1, HLA110PA3, HL043PA1, HL096PA1 HL005PA2, and ATCC6919) in CFU assays. Data are shown as mean ± SEM (n > 3). ****P < 0.0001 by repeated-measures 1-way ANOVA for treatment groups compared with C. acnes control.
AMTH17 cells exhibit antimicrobial activity as early as 6 hours.
T cells are generally thought to contribute to antimicrobial activity either by releasing cytokines, which recruit and activate other cells, or by major histocompatibility complex–restricted (MHC-restricted) lysis of infected host cells (40). The fact that only supernatants derived from activated AMTH17 clones had the ability to kill C. acnes in in vitro CFU assays suggested that these T cells were producing soluble bactericidal product(s). To further understand the mechanism(s) of TH17 cell–mediated killing, we used RNA-seq to determine differential antimicrobial gene expression in AMTH17 and n-AMTH17 clones. To this end, we took advantage of the finding that AMTH17 clones had varying levels of antimicrobial activity, which we termed Low, Medium, and High based on the results of C. acnes CFU assays. Against C. acnes strain HL005PA2 (Figure 2D), greater than 5-log, 3-log, and 1-log reductions in CFU were observed using undiluted supernatants derived from activated High, Medium, and Low AMTH17 clones, respectively. In contrast, supernatants from activated n-AMTH17 clones did not exhibit antimicrobial activity against the 3 C. acnes strains that we tested (HL005PA2, HL096PA1, and HL110PA1). We next determined the killing kinetics of AMTH17 supernatants against C. acnes. As shown in Figure 2E, we established that antimicrobial activity was detectable after 6 hours, reaching a 2-log reduction after 12 hours of incubation. In contrast, supernatants derived from activated n-AMTH17 clones lacked antimicrobial activity against C. acnes even after 24 hours of incubation. Thus, in subsequent bulk RNA-seq experiments, 15 AMTH17 clones with varying antimicrobial activity were stimulated with α-CD3/α-CD28 for 6 hours and 12 hours, and as a control, we used 5 n-AMTH17 clones (Figure 2D).
Identification of antimicrobial proteins of AMTH17 by RNA-seq.
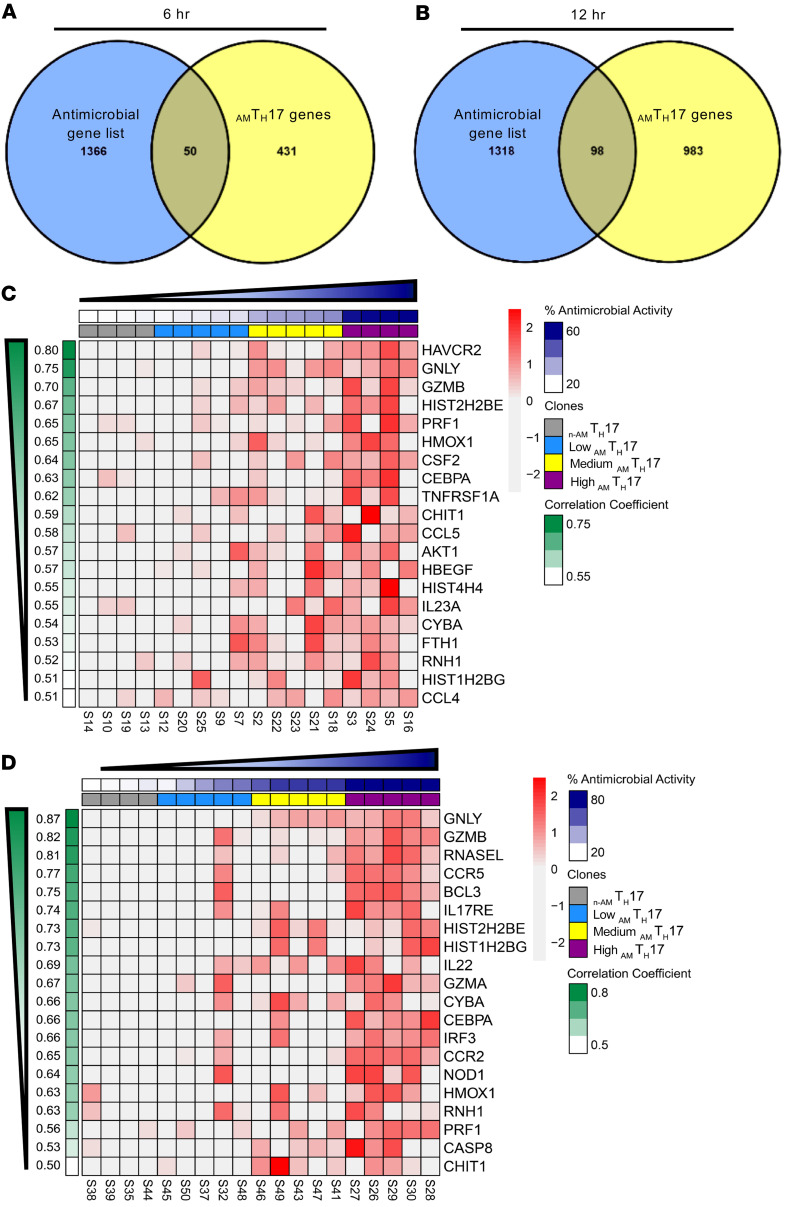
Using the transcriptome sequencing data, we next correlated the genes that had a greater than 2-fold expression within AMTH17 over n-AMTH17 with antimicrobial activity, as determined by in vitro C. acnes CFU activity. We identified 431 and 983 genes in the AMTH17-specific signatures for the 6- and 12-hour time points, respectively (Figure 3, A and B). Subsequently, overlap of these genes with an antimicrobial gene list from the GeneCards database revealed 50 and 98 common genes with significantly higher expression in AMTH17 compared with the n-AMTH17 clones at the 6- and 12-hour time points, respectively (Figure 3, A and B). These common genes included cytotoxic granule and antimicrobial protein genes encoding granulysin (GNLY), granzyme B (GZMB), granzyme A (GZMA), perforin (PRF1), and histones H2B and H4, and were also highly enriched in the High-killer AMTH17 as compared with the n-AMTH17 clones (Figure 3, C and D, and Supplemental Tables 1 and 2). Transcripts encoding transcription factors and receptors related to TH17 cells such as RORc and IL17RE were also expressed at high levels in the AMTH17 clones (Supplemental Figure 3). The GNLY transcript had the highest mean expression in AMTH17 compared with the n-AMTH17 clones at both the 6- and 12-hour time points (Supplemental Figure 3, A and B). GNLY is linked to the cytotoxic function of natural killer and CD8+ T cells, and has a wide range of antimicrobial activity against bacteria and fungi (18, 19).
Figure 3. Antimicrobial transcripts are highly expressed in AMTH17.
(A and B) AMTH17 genes with a log2(fold change) greater than 2 and positively correlated with percentage antimicrobial activity (r > 0.5) were overlapped with an antimicrobial gene list from the GeneCards database. (C and D) Heatmaps of the top 20 highest correlated genes with percentage antimicrobial activity found in the AMTH17 clones with Low (sky blue), Medium (yellow), and High (purple) antimicrobial activity against C. acnes at 6 hours (C) and 12 hours (D). Annotation for percentage antimicrobial activity and correlation coefficient values for each sample and gene are displayed on top (dark blue) and on the left (green). Gene expression values are displayed as z scores of log10-normalized counts.
Cytotoxic gene expression in AMTH17 clones is highly correlated with protein secretion and antimicrobial activity.
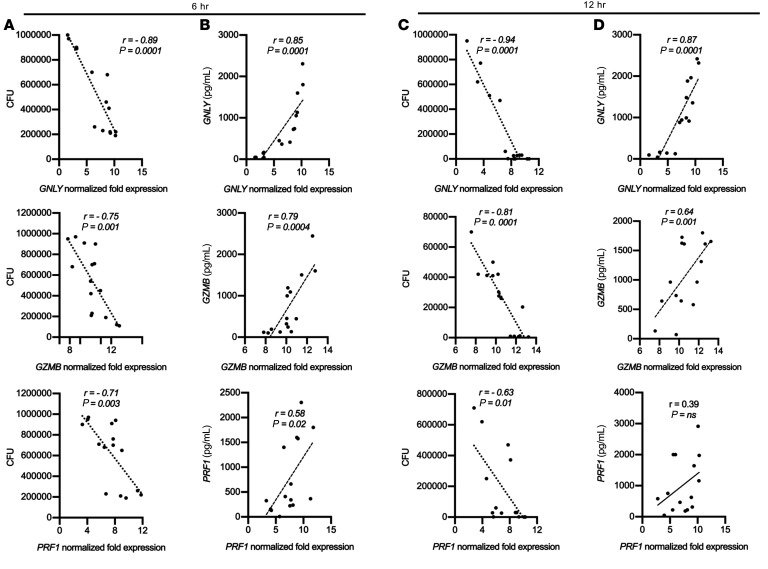
To assess the functional capacity of AMTH17, we confirmed the expression of some of the cytotoxicity-related transcripts (GNLY, GZMB, and PRF1) at the protein level following 6- and 12-hour in vitro stimulation with α-CD3/α-CD28 antibodies. We found that gene expression of GNLY, GZMB, and PRF1, as determined by RNA-seq, had a high positive correlation with the protein secretion data (r = 0.85, 0.79, and 0.58, respectively, at 6 hours and r = 0.87, 0.64, and 0.39, respectively, at 12 hours) (Figure 4, B and D). We then performed CFU experiments using the same supernatants that were measured using ELISA. We observed a negative correlation in GNLY, GZMB, and PRF1 gene expression and antimicrobial activity in AMTH17 (r = –0.89, –0.75, and –0.71, respectively, at 6 hours and r = –0.94, –0.81, and –0.63, respectively, at 12 hours) (Figure 4, A and C), suggesting that the products of these genes may play an important role in AMTH17-mediated antimicrobial activity against a wide variety of pathogens. The correlation between PRF1 gene expression and perforin protein secretion decreased from 6 to 12 hours. In looking at a dynamic process in which the transcripts and proteins are induced and degraded with different kinetics, this correlation may vary with time (41). We further validated that granulysin, granzyme B, and perforin are highly enriched within the AMTH17 and not the n-AMTH17 clones (Supplemental Figure 4, A and B). Therefore, our combined transcriptomics, protein analysis, and antimicrobial CFU data suggest that AMTH17-mediated killing is a general mechanism, and just like CD8+ CTLs, AMTH17 can secrete granulysin, granzymes, perforin, and other molecules as part of their antimicrobial arsenal, and these molecules can act synergistically to target C. acnes and a multitude of other cutaneous pathogens.
Figure 4. Antimicrobial gene expression in AMTH17 clones highly correlate with both protein secretion and antimicrobial CFU activity.
(A–D) Correlation plots of GNLY, PRF1, and GZMB expression in stimulated AMTH17, as determined by RNA-seq. Specific AMTH17 gene signatures with 2-fold or greater expression in comparison with the n-AMTH17 clones and that highly correlated with C. acnes CFU activity (A and C) and ELISA protein secretion (B and D) are shown for the 6-hour and 12-hour time points. P values by Student’s t test (n = 15).
Histone H2B contributes to AMTH17-mediated antimicrobial activity.
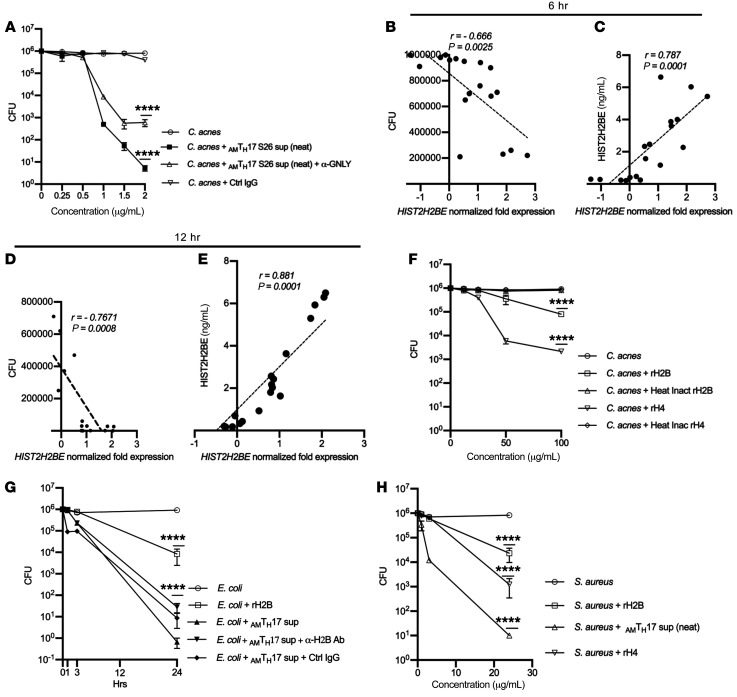
RNA-seq data revealed that GNLY was the top gene expressed in activated AMTH17 compared with n-AMTH17. The high values of GNLY expression are consistent with the role of granulysin as a protein with broad-spectrum antimicrobial activity against microbial pathogens (1). Neutralizing the effect of granulysin using a monoclonal antibody led to a 2-log reduction but not a complete abrogation in bacterial CFU (Figure 5A). We therefore reasoned that the AMTH17-mediated killing can involve a complex of other molecules and further mined the RNA-seq data to gain a global view of additional genes highly expressed in AMTH17 in comparison with n-AMTH17 clones. We discovered that histones (HIST2H2BE, HIST4H4, and HIST1H2BG) were among the top genes that were highly expressed after stimulation of AMTH17. Specifically, we observed a negative correlation in HIST2H2BE gene expression and antimicrobial activity in AMTH17 (r = –0.67 at 6 hours and r = –0.77 at 12 hours) (Figure 5, B and D). We also found that expression of HIST1H2BE, as determined by RNA-seq, had a high positive correlation with the protein secretion data (r = 0.79 at 6 hours and r = 0.88 at 12 hours) (Figure 5, C and E). These data therefore suggested that histone H2B contributes to the AMTH17-mediated antimicrobial activity.
Figure 5. Histone H2B is a component of AMTH17 antimicrobial activity.
(A) Supernatants derived from activated AMTH17 clone S26 were incubated with α-granulysin neutralizing antibody or control IgG for 1 hour prior and used in CFU assay against C. acnes strain HL005PA1. Data are shown as mean ± SEM (n > 3). ****P < 0.0001 by repeated-measures 1-way ANOVA for treatment groups compared with C. acnes control. (B–E) Correlation plots of HIST2H2BE gene expression in AMTH17, as determined in RNA-seq against CFU assays and ELISA protein secretion after 6 hours (B and C) and 12 hours (D and E). P values by Student’s t test (n = 20). (F) Observed CFU activity against C. acnes strain HLA110PA3 after 4-hour incubation with recombinant histones H2B and H4 and heat-inactivated controls. Data are representative of 4 independent experiments. ****P < 0.0001 by repeated-measures 1-way ANOVA for treatment groups compared with C. acnes control. (G) Supernatants derived from activated AMTH17 clone S26 were incubated with α-H2B neutralizing antibody or control IgG for 1 hour prior and used in CFU assay against E. coli. Data are representative of 3 independent experiments. ****P < 0.0001 by repeated-measures 1-way ANOVA for treatment groups compared with E. coli control. (H) S. aureus after 24-hour incubation with recombinant histones H2B and H4. Data show average CFU from 3 independent experiments. ****P < 0.0001 by repeated-measures 1-way ANOVA for treatment groups compared with S. aureus control.
Histone proteins share essential traits of cationic antimicrobial peptides (CAMPS), and are a major antimicrobial component of neutrophil extracellular traps (NETs) (42). To confirm that histone H2B can negatively affect C. acnes growth, we used recombinant histone H2B and H4 and performed CFU assays. Indeed, a 1- and 2.5-log reduction in bacterial CFU was observed when C. acnes was incubated with recombinant H2B and H4, respectively (Figure 5F). Treatment of AMTH17 supernatants with neutralizing antibodies against histone H2B and H4 led to a 1- to 2-log reduction in bacterial CFU (Supplemental Figure 5A). A pronounced decrease in CFU was observed against E. coli, and against Staphylococcus aureus (Figure 5, G and H) treated with recombinant histone H4. Histones have been reported in mitochondria, cytosolic granules, and on the cell surface (43), and based on this we reasoned that the AMTH17 clones may have the ability to secrete histones upon exposure to bacteria and that these extranuclear histones can play an important role in host defense. We therefore stained the AMTH17 clones with α–histone H2B antibodies and DAPI and found that histone H2B could localize to the cell surface of AMTH17 clones (Supplemental Figure 5, B–D). To address whether AMTH17 secrete histone H2B, we stimulated AMTH17 and n-AMTH17 clones, harvested the supernatants and lysates, and performed ELISA and Western blots. Indeed, the AMTH17 and not the n-AMTH17 clones were able to secrete histones (Supplemental Figure 6). Together, these data support our notion that histones can be secreted by AMTH17 and that they are antimicrobial against both Gram-positive and Gram-negative bacteria.
Previous studies detected DNA in supernatants of peripheral blood mononuclear cells (PBMCs) stimulated with phytohemagglutinin (44, 45). Although both human and mouse CD4+ T cells could release DNA and histones, it was not determined which T cell subset was involved (46). We therefore next examined the ability of TH1 and TH2 cells to secrete histones. We demonstrated that TH1 and TH2 cell lines release the signature cytokines IFN-γ and IL-4, respectively, upon stimulation with phorbol 12-myristate 13-acetate (PMA) (Supplemental Figure 6, D and E). However, both cell lines lacked the ability to secrete histones and subsequently kill C. acnes in vitro (Supplemental Figure 6F). In addition, histone-DNA complex formation by these cells was undetectable by confocal microcopy (Supplemental Figure 7, A and B). These data indicate that the ability of AMTH17 cells to secrete histone-coated extracellular traps as part of an antimicrobial response is specific to this T cell subpopulation.
AMTH17 cells release TETs that entangle C. acnes.
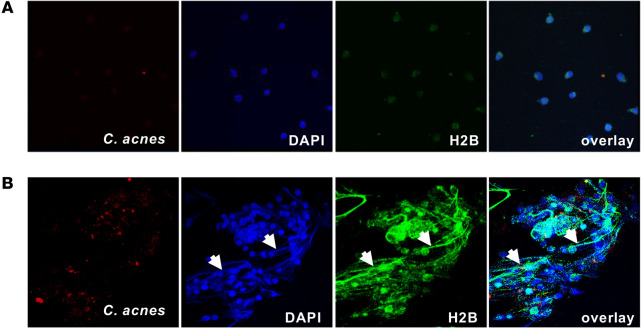
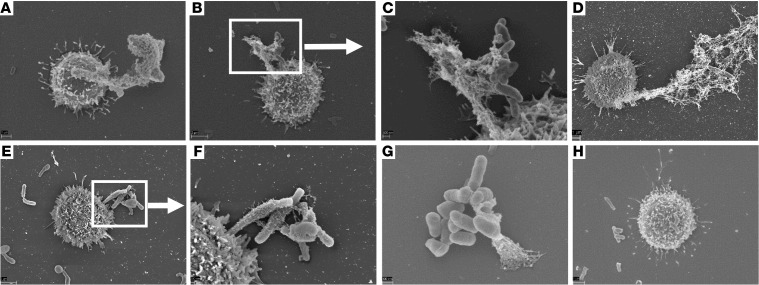
Based on the fact that the AMTH17 were viable after histone secretion, we hypothesized that the mechanism of histone secretion involves an early nonlytic extracellular trap formation that can be induced by the recognition of bacterial stimuli or products. As shown previously, the formation of extracellular traps by immune cells is an important mechanism in the innate immune response (42, 47). Extracellular traps are composed of chromatin coated with histones, proteases, and cytosolic proteins that not only ensnare bacteria, fungi, and protozoans, but also provide a high concentration of antimicrobial molecules that help trap and kill bacteria and fungi (42, 48–51). To study the mechanism of TH17 extracellular trap formation, we stimulated AMTH17 and n-AMTH17 clones with PMA, α-CD3/α-CD28 antibodies, or C. acnes, either in the presence or in the absence of deoxyribonuclease (DNase). Confocal staining showed that histone H2B accumulated in the cytoplasm and on the cell surface of AMTH17, suggesting that traps can mediate T cell antimicrobial activity (Figure 6 and Supplemental Figure 5, C and D). Furthermore, confocal microscopy demonstrated that activated AMTH17 form TETs that are fibrous structures composed of DNA prominently decorated with histone H2B (Figure 6 and Supplemental Figures 7 and 8). To closely visualize the TETs, we used scanning electron microscopy (SEM) and found that AMTH17 are able to externalize a meshwork of extracellular traps into the extracellular space that entangle C. acnes (Figure 7 and Supplemental Figure 9). We next tested the TET-forming characteristics of AMTH17 and n-AMTH17 clones activated only by contact with C. acnes. We observed that C. acnes were able to induce TETs in AMTH17 and not the n-AMTH17 (Figure 7, E and F), and that these structures could trap bacteria (Figure 7F). Because extracellular traps are degraded by treatment with DNase (42), this enzyme was added to PMA-activated AMTH17 followed by addition of C. acnes. Treatment of AMTH17 with DNase led to a reduction in TET formation (Supplemental Figure 9, G and H).
Figure 6. AMTH17 extracellular structures are prominently coated with histone H2B.
(A and B) n-AMTH17 clone S13 (A) and AMTH17 clone S16 (B) were stimulated with PMA for 2 hours as previously described (42) and incubated with PKH26-labeled C. acnes (red) (1:1). Cells were fixed and then stained with DAPI (blue) and α–histone H2B (green). Confocal staining images are shown. White arrows indicate T cell extracellular traps and ensnared C. acnes. Original magnification, ×63.
Figure 7. Antimicrobial TH17 release extracellular traps that entangle C. acnes.
Scanning electron microscopy of the interaction of AMTH17 and C. acnes at different time points. (A–H) AMTH17 clones were stimulated with PMA for 30 minutes (A), PMA and C. acnes for 30 minutes (B and C), α-CD3/α-CD28 for 30 minutes (D), C. acnes for 30 minutes (E and F), PMA and C. acnes for 40 minutes (G), and PMA, C. acnes, and DNase for 40 minutes (42) (H). Extended and released TETs can be seen attached to bacteria.
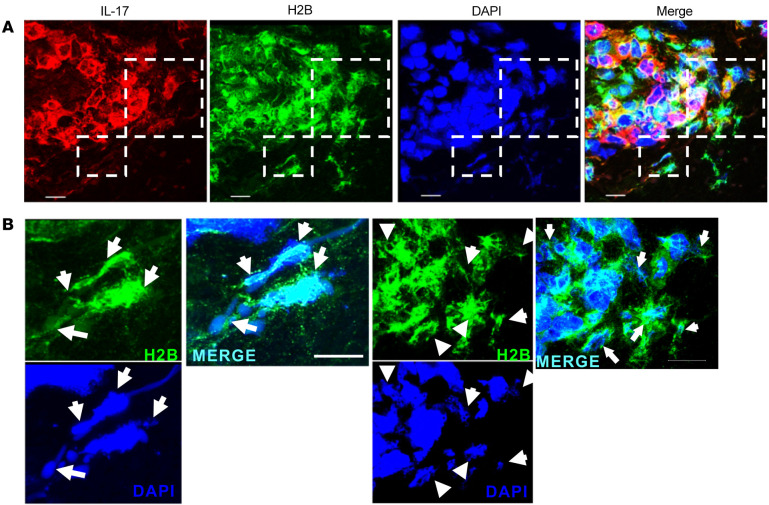
To explore the disease relevance of TH17 TET formation in vitro, we investigated whether such extracellular structures could be detected in vivo in biopsy specimens from patients with acne. We detected H2B and IL-17 in the inflammatory infiltrate in acne lesions but not in normal skin (Supplemental Figure 10). We further investigated the presence of extracellular traps in acne lesions using confocal microscopy after labeling CD4, IL-17, and H2B as well as staining with DAPI. We identified CD4+ T cells expressing IL-17 in acne lesions (Supplemental Figure 11). The area containing CD4+IL-17+ cells was selected and H2B visualized (Figure 8). IL-17 and H2B colocalized with DNA in fibrous structures in the extracellular space proximal to the CD4+ T cells, indicative of extracellular trap formation. Identical structures were detected in a second acne biopsy sample (Supplemental Figure 12). The isotype controls for both samples were negative (Supplemental Figure 13). In summary, our data demonstrate that, as in several innate immune cells (42, 47, 52–54), AMTH17 cells can release traps composed of DNA decorated with lysine-rich histones such as H2B, providing a mechanism by which the adaptive T cell response can monitor and regulate commensals such as C. acnes and invading pathogens including S. aureus.
Figure 8. Expression of T cell extracellular traps in acne lesions.
(A) Confocal images of IL-17 (red), histone H2B (green), and nuclei (DAPI, blue) in acne lesions. Dashed-line boxes identify the area further studied at higher power. Original magnification, ×63 with ×2 zoom from lower magnification in Supplemental Figure 9. (B) Higher-power magnification (×4 zoom of images in A) of the delineated regions marked in A showing H2B (green) and DAPI (red) only. White arrows indicate TETs in proximity to CD4+IL-17+H2B+ triple-positive cells within acne lesions. TETs are visualized as fibrous structures containing DNA (DAPI, blue) decorated with histone H2B (green) in the extracellular space. The images are projections of confocal Z-stacks generated from sections of 10-μm thickness. Data are from 3 individual samples. Scale bars: 10 μm (enlarged insets).
Discussion
Most of our understanding about mechanisms of host defense against infectious disease has come from exploration of the response to pathogenic microbes. However, the vast majority of microbial encounters are those resulting from commensal and/or symbiotic relationship with the microbiota. In the case of acne vulgaris, while most humans harbor C. acnes on their skin, the loss of the skin microbial diversity together with the action of the innate immune response, in particular, is thought to drive the chronic inflammatory condition (55). The ability of C. acnes strains to induce differential activation of both the innate and adaptive arms of the immune response are most likely due to differences in lineage-specific genetic elements among the strains (56). In this study, we identify AMTH17 cells as a population within the CD4+ TH17 subset, discovered through RNA-seq and functional analysis that utilizes a combination of antimicrobial molecules to kill C. acnes and other microbial pathogens. Importantly, AMTH17 cells have the ability to form TETs in vitro, which were also detected in vivo in acne lesions.
Previous studies have reported the presence of extracellular traps in neutrophils, mast cells, macrophages, and basophils (42, 48, 57–59). The extracellular traps entrap not only Gram-positive and Gram-negative bacteria, such as S. aureus, Salmonella typhimurium, Streptococcus pneumoniae, and group A streptococci, but also pathogenic fungi, such as Candida albicans (42, 49, 60–62). However, it is not known whether T cells form extracellular traps and trap bacteria such as C. acnes. Notably, we observed that C. acnes can activate AMTH17, leading to the formation of TETs, fibrous structures composed of DNA that are prominently decorated with histone H2B, and that upon release the TETs form a meshwork in the extracellular space that have the capacity to entangle C. acnes. After entrapment, we determined through SEM that most of the C. acnes were killed. However, some bacteria have developed strategies to reduce trapping and killing by repelling CAMPs in extracellular traps (63) or by degrading the DNA backbone with DNase (60, 62). Treatment of TETs with commercial DNase rendered them ineffective, suggesting that DNA is required for TET structure and function. As part of the pathogenesis of acne, disruption of the pilosebaceous unit results in the entry of C. acnes into the dermis, which contributes to the induction of an inflammatory response. We have previously determined that IL-17+ cells are present in the perifollicular infiltrate of inflamed acne lesions (64). Herein, we visualized TETs in vivo in biopsy specimens from acne lesions, observing the colocalization of fibrous structures composed of DNA and H2B in proximity to CD4+ T cells expressing IL-17. We demonstrate that these TETs can contribute to an antimicrobial response against C. acnes, but may also contribute to inflammation.
We further characterized the full repertoire of antimicrobial molecules expressed by AMTH17 in our RNA-seq data set, and identified histones as a component of AMTH17-mediated immunity. Histones have been reported to coat extracellular traps of neutrophils and other innate immune cells (42, 47). Four core histones (H2A, H2B, H3, and H4) form an octamer, around which DNA is wrapped in nucleosomes. These histones can display biological activities different from nucleosome structures and form an important part of skin defense (65). Histones are hydrophobic, cationic, and can form amphipathic α-helical structures and therefore share essential traits of CAMPS. Lysine-rich histones H2A and H2B are present on the epithelial surface of the placenta, providing the placenta and fetus protection against microbial infection (43). In addition, histones H2A and H2B both possess the capacity to neutralize endotoxin (66) and in our study, we demonstrate antimicrobial activity against E. coli, S. aureus, and C. acnes. Therefore, the observation that histone H2B gene expression highly correlated with granulysin activity in CFU assays is consistent with its antimicrobial action (67), but how this increased antimicrobial response is activated in vivo is unknown. We also observed high expression of the arginine-rich histone H4 in AMTH17. Histone H4 is known to mediate antimicrobial activity through the destruction of the cell membrane, and human sebocytes can release H4, which displays bactericidal activity against S. aureus and C. acnes. The antibacterial activity of H4 is enhanced by the presence of fatty acids on the skin (67).
TH17 cells are well known for their host-protective role against fungal infections in barrier tissues, in particular, those caused by C. albicans and in protection against extracellular bacteria (68–71). Our findings highlight the relevance of TH17 immunity to the skin commensal C. acnes and other bacterial strains. Direct comparison of AMTH17 and n-AMTH17 clones confirmed that AMTH17 displayed antimicrobial activity against both Gram-positive and Gram-negative bacteria. We show that the antimicrobial activity of AMTH17 is associated with a rapid expression and induction of antimicrobial transcripts, the impact of which is underscored by the finding that the top most abundantly secreted antimicrobial molecules of AMTH17 (granulysin, histone H2B) alone accounted for nearly 50% of the antimicrobial killing. We further identified multiple antimicrobial transcripts/molecules that are functionally important to immune defense. These results suggest that the antimicrobial molecules including granulysin, granzyme B, and perforin can act synergistically as part of the antimicrobial arsenal of AMTH17. It therefore seems more likely that AMTH17 cells are a functionally distinct population that serve a protective role during infection. We suggest a model where, in the case of extracellular bacteria, AMTH17 cells can secrete granulysin that is then attracted to the bacterial cell wall by ionic interactions mediated by positively charged arginine residues. These residues interact with the negatively charged phospholipids on the surface of the pathogen, and granulysin can then alter membrane permeability, leading to osmotic lysis by itself. We also envisage a scenario where granulysin can colocalize with the pore-forming molecule perforin and act synergistically with granzyme B, histone H2B, and histone H4, leading to osmotic lysis. Both mechanisms can allow granulysin to access the intracellular compartments in which the pathogens reside, leading to bacterial killing. Additionally, the impact of other antimicrobial cytokines such as IL-26 cannot be definitively excluded in the bacterial killing even though we did not see significant differences in IL-26 expression between the AMTH17 and n-AMTH17 clones.
Extracellular traps have been observed in diseases such as human appendicitis (42), and in sinusoids of the liver and lungs during sepsis (72). The evidence presented in the current study shows that those TH17 cells that express histones make TETs and secrete a combination of molecules that have antimicrobial activity against extracellular bacteria. TETs were detected in acne lesions, linking them to the site of disease where they could contribute to the antimicrobial response in the extracellular environment, such as in the extracellular matrix, for example. Human genetic studies indicate that alterations in TH17 cell differentiation due to STAT3 mutations, or deletion of IL-17 receptors predisposes to multiple infections, such that these mechanisms are necessary for host defense (13, 69). Patients with hyperimmunoglobulin E syndrome (HIES), caused by mutations in STAT3, have few detectable TH17 cells in peripheral blood (69), and a failure of CD4+ TH17 cell differentiation in vitro (13, 15, 73–75). Therefore, it is not possible to study the role of TETs in STAT3-deficient TH17 cells in humans. In addition, murine and human genetic approaches point strongly to a model in which IL-17RA/RC signaling in nonmyeloid cells is a necessary in vivo effector mechanism of TH17 cells (76, 77). However, these studies do not indicate whether TH17 production of IL-17 is sufficient for host defense. It will be difficult to assess whether TETs are also necessary for host defense, as inherited mutations in HIST2H2BE have not been reported. Nevertheless, it is possible that both IL-17 and TETs contribute to host defense against extracellular bacteria. It is likely that there is redundancy in the immune response such that several antimicrobial mechanisms work additively or in synergy in vivo to destroy extracellular bacteria.
Although our data indicate that TETs are involved in antimicrobial responses, as are other extracellular traps, we cannot exclude the possibility that TETs contribute to pathology. In psoriasis, NETs may contribute to TH17 induction as part of the disease pathogenesis (78). In addition, a correlation between the presence of NET-associated DNA and pathology has also been implicated in other diseases (42, 70), and whether this is true for C. acnes–induced TETs remains to be explored. A charge-mediated mechanism whereby cationic antimicrobial molecules and histones such as H2B and H4 in TETs trap negatively charged commensals such as C. acnes seems plausible. The fact that TH17 cells can release traps implies that these cells can act as an important link between the innate and adaptive responses targeting efficient clearance of invading pathogens. Taken together, our data identify a functionally distinct subpopulation of TH17 cells with the ability to secrete antimicrobial proteins and TETs to capture and kill extracellular bacteria.
Methods
Bacterial strains.
C. acnes strains used in this study were obtained from Biodefense and Emerging Infections Research Resources Repository (BEI Resources) and cultured as previously described (30). S. aureus SA113, Pseudomonas aeruginosa PAO1, and E. coli DH5α were grown in Luria broth (LB) overnight at 37°C with agitation. Overnight bacterial cultures were subcultured and incubated until mid-log phase was reached, which was determined to be OD600 = 0.4. Cultures were washed in sterile PBS and renormalized to OD600 = 0.4 in culture media.
PBMC isolation, stimulation, and cytokine ELISAs.
PBMCs were obtained from healthy donors with written informed patient consent. PBMCs were then isolated using Ficoll-Paque gradients (GE Healthcare) as previously described (30). Briefly, cells were cultured in T cell media (RPMI 1640, 10% heat-inactivated human serum [Gemini], 2 mM L-glutamine, 10 U/mL penicillin, and 100 μg/mL streptomycin) and stimulated with different strains of C. acnes at 1 multiplicity of infection (1 MOI). Levels of cytokines accumulated in culture supernatants were measured by ELISA. As a positive control for NET formation, neutrophil isolation was done using the Neutrophil Isolation Kit (Miltenyi Biotec) following the manufacturer’s protocol and assessed for spontaneous NET formation (incubated with RPMI medium with 2% fetal calf serum for 130 minutes) and for NET formation after stimulation with 20 nM PMA for 80, 100, and 130 minutes, as previously described (42).
Sterile cell sorting, TH17 cloning, and neutrophil isolation.
We developed a cloning system that uses C. acnes microbes and autologous monocytes as antigen-presenting cells. This cloning approach provides a large number of antigens and a variety of stimuli to innate receptors to elicit polarizing cytokines for TH17 differentiation. Briefly, PBMCs were stimulated for 16 hours with C. acnes strains, and cytokine secretion determined using IL-17 cytokine secretion capture assay following the manufacturer’s protocol (Miltenyi Biotec). After IL-17 staining, the cells were further stained with α-CD4 antibodies (BD, clone RPA-T4) and the CD4+IL-17+ cells sorted under sterile conditions using a BD FACSVantage. Dead cells were excluded by DAPI staining. Sorted cells were cloned in Terasaki plates (Nunc Microwell, Sigma-Aldrich) as previously described (30) and maintained in T cell media supplemented with 100 U/mL IL-2 and 2 ng/mL IL-23. To avoid the effect of long-term culture, TH17 cell clones were expanded for a maximum of 13 days, aliquoted, and frozen, and/or used immediately for RNA-seq and subsequent functional experiments. Samples were acquired on a BD Biosciences FACScan, and analyzed using FlowJo software (v7.6). In additional experiments, human TH1 and TH2 cells were isolated using CD4+ T cell isolation kits (Miltenyi Biotec) and cultured in the presence of IL-2 and AB serum, as previously described (79). Levels of IFN-γ and IL-4 were determined by ELISA (R&D Systems).
Bacterial CFU assay.
C. acnes strains were grown under anaerobic conditions in Reinforced Clostridial Medium (Oxoid) for 2 days and collected in mid-log phase. The bacteria were washed 3 times with the assay buffer (10 mM Tris pH 7.4, supplemented with 0.03% v/v trypticase soy broth [Tris-TSB]), and enumerated by applying a conversion factor of 7.5 × 107 bacteria per mL = 1 OD600. TH17 culture supernatants were diluted in Tris-TSB and the CFU assays performed as previously described (80, 81). For the S. aureus, E. coli, and P. aeruginosa CFU assays, bacteria were grown as described above and resuspended in RPMI 1640. Depletion of granulysin was performed by incubating supernatants with 10 μg/mL neutralizing α-granulysin monoclonal antibody (BioLegend, clone DH10) or an isotype monoclonal antibody for 12 hours at 4°C. Reactions (100 μL of bacteria plus TH17 supernatants or rhIL-26 or α-granulysin or α-H2B, or α-H4; Abcam) were added to 1.5-mL tubes and incubated at 37°C with shaking for 1, 3, or 24 hours. After the specified incubation periods, 10-fold serial dilutions were plated on LB plates to quantify surviving CFU.
Bulk RNA-seq library and sequencing.
Fifteen AMTH17 and 5 n-AMTH17 clones (control) generated from 6 healthy donors were stimulated with α-CD3/α-CD28 (BD) in T cell media. Total RNA was isolated at 2 time points (6 and 12 hours) after treatment using RLT buffer supplemented with 1% β-mercaptoethanol (QIAGEN). RNA extraction was performed on a total of 40 samples according to the manufacturer’s instructions using an RNeasy Micro Kit (QIAGEN), including the on-column DNase treatment step. Extracted RNA was quantified with a Quant-iT RiboGreen RNA Assay Kit (Invitrogen) and RNA quality was assessed using the Agilent 2200 TapeStation (RNA assay). mRNA libraries were prepared using the Illumina TruSeq mRNA Library Prep kit following the manufacturer’s protocol. Briefly, total RNA was subjected to poly-A selection to purify mRNA, and then fragmented and converted into double-stranded cDNA. Double-stranded cDNA was then end repaired, ligated to adapters, and amplified. Final libraries were quantified using PicoGreen (Invitrogen) and the quality was assessed using the Agilent 2200 TapeStation (D1000 assay). Libraries were pooled (4 per lane) at equimolar quantities (10 μM each library) and sequenced on a HiSeq 2000 sequencer (Illumina) with a 50-bp single-end protocol. The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO GSE144852) (82).
Bioinformatics methods.
The alignment of the samples was performed using STAR 2.5.3 (83) using the human genome (GRCh38.90). We explored the data to check for outliers and one sample (S31) was removed from downstream analyses. For each experiment, the 19 samples were divided into groups (Low, Medium, High, and n-AMTH17) based on in vitro C. acnes CFU killing assays. We filtered reads for low counts and the remaining were normalized using TMM (trimmed mean of M values) in the edgeR package (84) in R. Reads were then processed by voomwithqualityweight in Limma to convert into log2 counts per million (logCPM) with associated precision weights (84, 85), followed by contrast comparisons. A total of 11,995 genes and 12,040 genes were kept for contrast comparisons in the 6-hour and 12-hour stimulation experiments, respectively. For CFU correlation analysis, AMTH17 and n-AMTH17 (controls) clones were stimulated with α-CD3/α-CD28 (BD), and total RNA was isolated (6 and 12 hours) and processed for RNA-seq. Specific AMTH17 gene signatures with a 2-fold or greater expression in comparison with the n-AMTH17 clones were used in a correlation analysis with percentage antimicrobial activity determined by in vitro C. acnes activity. Genes with a coefficient of correlation (r) greater than 0.5 were overlapped with a list of antimicrobial-related molecules obtained from the GeneCards database (https://www.genecards.org/).
SEM.
TH17 clones were adhered on silicon wafers (Ted Pella Inc.) treated with 0.01% poly-L-lysine (Sigma-Aldrich). C. acnes added at a 1:1 ratio were incubated for 20, 40, 60, and 90 minutes at room temperature. Samples were rinsed with warm fixative (2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4) and then incubated with fresh fixative for 1 hour on ice. Next, samples were rinsed 5 times (2 minutes each) with 0.1 M sodium cacodylate and then postfixed with 2% osmium tetroxide in 0.1 M sodium cacodylate for 30 minutes on ice. Following the incubation with osmium, the samples were rinsed 5 times (2 minutes each) with deionized H2O and then dehydrated by incubating with an ascending series of ethanol concentrations (30%, 50%, 70%, 85%, and 95%; 2 minutes each). Dehydration was completed by washing the samples in 3 changes (2 minutes each) of 100% anhydrous ethanol. Next, samples were loaded into a Tousimis Autosamdri-810 critical point dryer and dried at the critical point of CO2 before mounting the silicon wafers onto aluminum SEM stubs with double-sided carbon tape and transferring them to an ion-beam sputter coater and coating with approximately 5 nm of iridium. Finally, secondary electron images were acquired with a Zeiss Supra 40VP scanning electron microscope set to 3.5 kV accelerating voltage. All reagents were purchased from Electron Microscopy Sciences.
Cell culture, immunoperoxidase and immunofluorescence labeling.
C. acnes were labeled with PKH26 (Sigma-Aldrich) following the manufacturer’s protocol. TH17 clones were then treated with PMA, PKH26-labeled C. acnes, or left untreated in T cell medium for 3 hours. Following stimulation, both TH17 clones and PKH26-labeled C. acnes were adhered to poly-L-lysine–coated transwells for 1 hour. Cells were then washed and fixed for 30 minutes with BD Cytofix/Cytoperm (BD Biosciences) before being washed again. Next, cells were blocked with normal goat serum for 20 minutes, and immunolabeled with primary antibodies against histone H2B (Abcam, ab1790) for 1 hour. Following washing, cells were stained with secondary antibodies for 1 hour, washed, and mounted with DAPI. Immunofluorescence of cell cultures was examined using a Leica TCS-SP8 MP inverted single confocal laser-scanning microscope at the Advanced Microscopy/Spectroscopy Laboratory Macro-Scale Imaging Laboratory (California NanoSystems Institute, UCLA). For immunoperoxidase labeling, deidentified normal skin and acne lesion specimens were obtained from the UCLA Translational Pathology Core Laboratory after obtaining written informed consent. Staining for histone H2B and IL-17 (Abcam, ab189377) was performed using the standard streptavidin-biotin technique, using the commercial kit HRP-AEC system following the manufacturer’s recommendations (R&D Systems). For confocal imaging of acne tissues, immunofluorescence labeling was performed by serially incubating cryostat tissue sections with anti-human monoclonal antibodies for 2 hours and washed 3 times with 1× PBS, followed by incubation with specific, fluorochrome-labeled (A488, A568, A647) goat anti–mouse immunoglobulin antibodies (Molecular Probes) for 90 minutes. Controls included staining with isotype-matched antibodies. Nuclei were stained with DAPI (Thermo Fisher Scientific). Immunofluorescence of skin sections was examined using the Leica TCS-SP8 MP as described above.
Histone H2B Western blot analysis.
Western blot assays were performed using supernatants and whole-cell lysates from TH17 clones. Protein concentrations were estimated by the Bradford method (Thermo Fisher Scientific). Briefly, lysates prepared from cells in a lysis buffer containing protease inhibitor cocktail (Roche) were resolved by SDS-PAGE, transferred to PVDF membranes, and immunoblotted. Immunoblots were performed with lysates and supernatants using antibodies against H2B (1:1000, Abcam) and β-actin as an internal control (1:5000, Abcam, ab8227) overnight.
Statistics.
Data obtained from at least 3 independent experiments were analyzed using GraphPad Prism software version 8. If data sets were not normally distributed, a nonparametric test was used to determine significance. If more than 2 data sets were compared, 1-way analysis of variance (ANOVA) was used to compare variances within groups. A post hoc 2-tailed Student’s t test was used for comparison between 2 groups. For comparisons among 3 or more groups, we used repeated-measures 1-way ANOVA with the Greenhouse-Geisser correction, along with Tukey’s multiple-comparison test, with individual variances computed for each comparison. Significant differences were considered for those probabilities < 5% (P < 0.05).
Study approval.
This study was conducted according to the principles expressed in the Declaration of Helsinki. The study was approved by the UCLA IRB (no. 118-00193). All donors and patients with acne provided written informed consent for the collection of peripheral blood and subsequent analysis.
Author contributions
GWA conceived and designed the experiments, analyzed the data, wrote the manuscript, and performed most of the experiments. RMBT and TW participated in confocal and SEM experiments. AM, PRA, and MM helped with RNA-seq and bioinformatics analyses. RLM and MP supervised the study, provided critical suggestions and discussions throughout the study, and revised the manuscript.
Supplementary Material
Acknowledgments
We kindly thank the UCLA Neuroscience Genomics Core for their assistance in RNA-seq library preparation and valuable advice, and Haile Salem for assistance with the flow cytometry and cell sorting and Nairy Ceja-Garcia for help with bacterial culture and CFU assays. AM and MM were supported by QCB Collaboratory Postdoctoral Fellowships (UCLA). We used computational and storage services associated with the Hoffman2 Shared Cluster provided by UCLA Institute for Digital Research and Education’s Research Technology Group. Confocal laser-scanning microscopy was performed at the Advanced Light Microscopy/Spectroscopy Laboratory and the Leica Microsystems Center of Excellence at the California NanoSystems Institute at UCLA with funding support from NIH Shared Instrumentation grant S10OD025017 and NSF Major Research Instrumentation grant CHE-0722519. Flow cytometry was performed in the UCLA Jonsson Comprehensive Cancer Center (JCCC) and Center for AIDS Research Flow Cytometry Core Facility. This work was supported by NIH grant K01AR071479 and Burroughs Wellcome Collaborative Research Travel Grant 1019954 (to GWA).
Version 1. 11/19/2020
In-Press Preview
Version 2. 01/19/2021
Print issue publication
Footnotes
Conflict of interest: the authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(2):e141594.https://doi.org/10.1172/JCI141594.
See the related Commentary at Keeping the peace: commensal Cutibacterium acnes trains CD4+ TH17 cells to trap and kill.
Contributor Information
George W. Agak, Email: Gagak@mednet.ucla.edu.
Alice Mouton, Email: alicemoutonresearch@gmail.com.
Marco Morselli, Email: marco.murslegn@gmail.com.
Priscila R. Andrade, Email: prandrade86@gmail.com.
Matteo Pellegrini, Email: matteop@mcdb.ucla.edu.
Robert L. Modlin, Email: rmodlin@mednet.ucla.edu.
References
- 1.Ernst WA, et al. Granulysin, a T cell product, kills bacteria by altering membrane permeability. J Immunol. 2000;165(12):7102–7108. doi: 10.4049/jimmunol.165.12.7102. [DOI] [PubMed] [Google Scholar]
- 2.Hirahara K, et al. Mechanisms underlying helper T-cell plasticity: implications for immune-mediated disease. J Allergy Clin Immunol. 2013;131(5):1276–1287. doi: 10.1016/j.jaci.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakayamada S, et al. Helper T cell diversity and plasticity. Curr Opin Immunol. 2012;24(3):297–302. doi: 10.1016/j.coi.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41(4):529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crotty S. A brief history of T cell help to B cells. Nat Rev Immunol. 2015;15(3):185–189. doi: 10.1038/nri3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korn T, et al. IL-17 and Th17 cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 7.Vignali DA,et al. How regulatory T cells work. Nat Rev Immunol. 2008;8(7):523–532. doi: 10.1038/nri2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agak GW, et al. Propionibacterium acnes induces an IL-17 response in acne vulgaris that is regulated by vitamin A and vitamin D. J Invest Dermatol. 2014;134(2):366–373. doi: 10.1038/jid.2013.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrington LE, et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6(11):1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 10.Zhou L, et al. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007;8(9):967–974. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- 11.Ivanov, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126(6):1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 12.Durant L, et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 2010;32(5):605–615. doi: 10.1016/j.immuni.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma CS, et al. Deficiency of Th17 cells in hyper IgE syndrome due to mutations in STAT3. J Exp Med. 2008;205(7):1551–1557. doi: 10.1084/jem.20080218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meller S, et al. T(H)17 cells promote microbial killing and innate immune sensing of DNA via interleukin 26. Nat Immunol. 2015;16(9):970–979. doi: 10.1038/ni.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Beaucoudrey L, et al. Mutations in STAT3 and IL12RB1 impair the development of human IL-17-producing T cells. J Exp Med. 2008;205(7):1543–1550. doi: 10.1084/jem.20080321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basu R, et al. The Th17 family: flexibility follows function. Immunol Rev. 2013;252(1):89–103. doi: 10.1111/imr.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams NS, Engelhard VH. Identification of a population of CD4+ CTL that utilizes a perforin- rather than a Fas ligand-dependent cytotoxic mechanism. J Immunol. 1996;156(1):153–159. [PubMed] [Google Scholar]
- 18.Stenger S, et al. An antimicrobial activity of cytolytic T cells mediated by granulysin. Science. 1998;282(5386):121–125. doi: 10.1126/science.282.5386.121. [DOI] [PubMed] [Google Scholar]
- 19.Ochoa MT, et al. T-cell release of granulysin contributes to host defense in leprosy. Nat Med. 2001;7(2):174–179. doi: 10.1038/84620. [DOI] [PubMed] [Google Scholar]
- 20.Wilkinson TM, et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med. 2012;18(2):274–280. doi: 10.1038/nm.2612. [DOI] [PubMed] [Google Scholar]
- 21.Yasukawa M, et al. Differential in vitro activation of CD4+CD8- and CD8+CD4- herpes simplex virus-specific human cytotoxic T cells. J Immunol. 1989;143(6):2051–2057. [PubMed] [Google Scholar]
- 22.Eichelberger M, et al. Clearance of influenza virus respiratory infection in mice lacking class I major histocompatibility complex-restricted CD8+ T cells. J Exp Med. 1991;174(4):875–880. doi: 10.1084/jem.174.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muller D, et al. LCMV-specific, class II-restricted cytotoxic T cells in beta 2-microglobulin-deficient mice. Science. 1992;255(5051):1576–1578. doi: 10.1126/science.1347959. [DOI] [PubMed] [Google Scholar]
- 24.Frey AB. Rat mammary adenocarcinoma 13762 expressing IFN-gamma elicits antitumor CD4+ MHC class II-restricted T cells that are cytolytic in vitro and tumoricidal in vivo. J Immunol. 1995;154(9):4613–4622. [PubMed] [Google Scholar]
- 25.Verma S, et al. Cytomegalovirus-specific CD4 T cells are cytolytic and mediate vaccine protection. J Virol. 2016;90(2):650–658. doi: 10.1128/JVI.02123-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bunte K, Beikler T. Th17 cells and the IL-23/IL-17 axis in the pathogenesis of periodontitis and immune-mediated inflammatory diseases. Int J Mol Sci. 2019;20(14):3394. doi: 10.3390/ijms20143394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kryczek I, et al. Human TH17 cells are long-lived effector memory cells. Sci Transl Med. 2011;3(104):104ra100. doi: 10.1126/scitranslmed.3002949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Omenetti S, et al. The intestine harbors functionally distinct homeostatic tissue-resident and inflammatory Th17 cells. Immunity. 2019;51(1):77–89.e6. doi: 10.1016/j.immuni.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kistowska M, et al. Propionibacterium acnes promotes Th17 and Th17/Th1 responses in acne patients. J Invest Dermatol. 2015;135(1):110–118. doi: 10.1038/jid.2014.290. [DOI] [PubMed] [Google Scholar]
- 30.Agak GW, et al. Phenotype and antimicrobial activity of Th17 cells induced by propionibacterium acnes strains associated with healthy and acne skin. J Invest Dermatol. 2018;138(2):316–324. doi: 10.1016/j.jid.2017.07.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Juno JA, et al. Cytotoxic CD4 T cells-friend or foe during viral infection? Front Immunol. 2017;8:19. doi: 10.3389/fimmu.2017.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Appay V, et al. Characterization of CD4(+) CTLs ex vivo. J Immunol. 2002;168(11):5954–5958. doi: 10.4049/jimmunol.168.11.5954. [DOI] [PubMed] [Google Scholar]
- 33.Zaunders JJ, et al. Identification of circulating antigen-specific CD4+ T lymphocytes with a CCR5+, cytotoxic phenotype in an HIV-1 long-term nonprogressor and in CMV infection. Blood. 2004;103(6):2238–2247. doi: 10.1182/blood-2003-08-2765. [DOI] [PubMed] [Google Scholar]
- 34.Norris PJ, et al. Beyond help: direct effector functions of human immunodeficiency virus type 1-specific CD4(+) T cells. J Virol. 2004;78(16):8844–8851. doi: 10.1128/JVI.78.16.8844-8851.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Leeuwen EM, et al. Emergence of a CD4+CD28- granzyme B+, cytomegalovirus-specific T cell subset after recovery of primary cytomegalovirus infection. J Immunol. 2004;173(3):1834–1841. doi: 10.4049/jimmunol.173.3.1834. [DOI] [PubMed] [Google Scholar]
- 36.Brown DM. Cytolytic CD4 cells: direct mediators in infectious disease and malignancy. Cell Immunol. 2010;262(2):89–95. doi: 10.1016/j.cellimm.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patil VS, et al. Precursors of human CD4+ cytotoxic T lymphocytes identified by single-cell transcriptome analysis. Sci Immunol. 2018;3(19):eaan8664. doi: 10.1126/sciimmunol.aan8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clark RA. Resident memory T cells in human health and disease. Sci Transl Med. 2015;7(269):269rv1. doi: 10.1126/scitranslmed.3010641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y, et al. Tissue-specific control of tissue-resident memory t cells. Crit Rev Immunol. 2018;38(2):79–103. doi: 10.1615/CritRevImmunol.2018025653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levitz SM, et al. Direct antimicrobial activity of T cells. Immunol Today. 1995;16(8):387–391. doi: 10.1016/0167-5699(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 41.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13(4):227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brinkmann V, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303(5663):1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 43.Kim HS, et al. Endotoxin-neutralizing antimicrobial proteins of the human placenta. J Immunol. 2002;168(5):2356–2364. doi: 10.4049/jimmunol.168.5.2356. [DOI] [PubMed] [Google Scholar]
- 44.Rogers JC, et al. Excretion of deoxyribonucleic acid by lymphocytes stimulated with phytohemagglutinin or antigen. Proc Natl Acad Sci U S A. 1972;69(7):1685–1690. doi: 10.1073/pnas.69.7.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rogers JC. Identification of an intracellular precursor to DNA excreted by human lymphocytes. Proc Natl Acad Sci U S A. 1976;73(9):3211–3215. doi: 10.1073/pnas.73.9.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Costanza M, et al. DNA threads released by activated CD4+ T lymphocytes provide autocrine costimulation. Proc Natl Acad Sci U S A. 2019;116(18):8985–8994. doi: 10.1073/pnas.1822013116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Daniel C, et al. Extracellular DNA traps in inflammation, injury healing. Nat Rev Nephrol. 2019;15(9):559–575. doi: 10.1038/s41581-019-0163-2. [DOI] [PubMed] [Google Scholar]
- 48.Guimarães-Costa AB, et al. Leishmania amazonensis promastigotes induce and are killed by neutrophil extracellular traps. Proc Natl Acad Sci U S A. 2009;106(16):6748–6753. doi: 10.1073/pnas.0900226106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Urban CF, et al. Neutrophil extracellular traps capture and kill Candida albicans yeast and hyphal forms. Cell Microbiol. 2006;8(4):668–676. doi: 10.1111/j.1462-5822.2005.00659.x. [DOI] [PubMed] [Google Scholar]
- 50.Urban C, Zychlinsky A. Netting bacteria in sepsis. Nat Med. 2007;13(4):403–404. doi: 10.1038/nm0407-403. [DOI] [PubMed] [Google Scholar]
- 51.Fuchs TA, et al. Novel cell death program leads to neutrophil extracellular traps. J Cell Biol. 2007;176(2):231–241. doi: 10.1083/jcb.200606027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.von Köckritz-Blickwede M, et al. Phagocytosis-independent antimicrobial activity of mast cells by means of extracellular trap formation. Blood. 2008;111(6):3070–3080. doi: 10.1182/blood-2007-07-104018. [DOI] [PubMed] [Google Scholar]
- 53.Yousefi S, et al. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat Med. 2008;14(9):949–953. doi: 10.1038/nm.1855. [DOI] [PubMed] [Google Scholar]
- 54.Ueki S, et al. Eosinophil extracellular trap cell death-derived DNA traps: their presence in secretions and functional attributes. J Allergy Clin Immunol. 2016;137(1):258–267. doi: 10.1016/j.jaci.2015.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dréno B, et al. Cutibacterium acnes (Propionibacterium acnes) and acne vulgaris: a brief look at the latest updates. J Eur Acad Dermatol Venereol. 2018;32 suppl 2:5–14. doi: 10.1111/jdv.15043. [DOI] [PubMed] [Google Scholar]
- 56.Tomida S, et al. Pan-genome and comparative genome analyses of propionibacterium acnes reveal its genomic diversity in the healthy and diseased human skin microbiome. mBio. 2013;4(3):e00003–e00013. doi: 10.1128/mBio.00003-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wong KW, Jacobs WR. Mycobacterium tuberculosis exploits human interferon to stimulate macrophage extracellular trap formation and necrosis. J Infect Dis. 2013;208(1):109–119. doi: 10.1093/infdis/jit097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Möllerherm H, et al. Antimicrobial activity of mast cells: role and relevance of extracellular DNA traps. Front Immunol. 2016;7:265. doi: 10.3389/fimmu.2016.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yousefi S, et al. Basophils exhibit antibacterial activity through extracellular trap formation. Allergy. 2015;70(9):1184–1188. doi: 10.1111/all.12662. [DOI] [PubMed] [Google Scholar]
- 60.Beiter K, et al. An endonuclease allows Streptococcus pneumoniae to escape from neutrophil extracellular traps. Curr Biol. 2006;16(4):401–407. doi: 10.1016/j.cub.2006.01.056. [DOI] [PubMed] [Google Scholar]
- 61.Wartha F, Henriques-Normark B. ETosis: a novel cell death pathway. Sci Signal. 2008;1(21):pe25. doi: 10.1126/stke.121pe25. [DOI] [PubMed] [Google Scholar]
- 62.Buchanan JT, et al. DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr Biol. 2006;16(4):396–400. doi: 10.1016/j.cub.2005.12.039. [DOI] [PubMed] [Google Scholar]
- 63.Wartha F, et al. Capsule and D-alanylated lipoteichoic acids protect Streptococcus pneumoniae against neutrophil extracellular traps. Cell Microbiol. 2007;9(5):1162–1171. doi: 10.1111/j.1462-5822.2006.00857.x. [DOI] [PubMed] [Google Scholar]
- 64.Kawasaki H, et al. A protein with antimicrobial activity in the skin of Schlegel’s green tree frog Rhacophorus schlegelii (Rhacophoridae) identified as histone H2B. Biochem Biophys Res Commun. 2003;312(4):1082–1086. doi: 10.1016/j.bbrc.2003.11.052. [DOI] [PubMed] [Google Scholar]
- 65.Hoeksema M, et al. Histones as mediators of host defense, inflammation and thrombosis. Future Microbiol. 2016;11(3):441–453. doi: 10.2217/fmb.15.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee DY, et al. Histone H4 is a major component of the antimicrobial action of human sebocytes. J Invest Dermatol. 2009;129(10):2489–2496. doi: 10.1038/jid.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Acosta-Rodriguez EV, et al. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat Immunol. 2007;8(6):639–646. doi: 10.1038/ni1467. [DOI] [PubMed] [Google Scholar]
- 68.Milner JD, et al. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature. 2008;452(7188):773–776. doi: 10.1038/nature06764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Happel KI, et al. Divergent roles of IL-23 and IL-12 in host defense against Klebsiella pneumoniae. J Exp Med. 2005;202(6):761–769. doi: 10.1084/jem.20050193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cho JS, et al. IL-17 is essential for host defense against cutaneous Staphylococcus aureus infection in mice. J Clin Invest. 2010;120(5):1762–1773. doi: 10.1172/JCI40891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Clark SR, et al. Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nat Med. 2007;13(4):463–469. doi: 10.1038/nm1565. [DOI] [PubMed] [Google Scholar]
- 72.Renner ED, et al. Novel signal transducer and activator of transcription 3 (STAT3) mutations, reduced T(H)17 cell numbers, and variably defective STAT3 phosphorylation in hyper-IgE syndrome. J Allergy Clin Immunol. 2008;122(1):181–187. doi: 10.1016/j.jaci.2008.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jiao H, et al. Novel and recurrent STAT3 mutations in hyper-IgE syndrome patients from different ethnic groups. Mol Immunol. 2008;46(1):202–206. doi: 10.1016/j.molimm.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 74.Freeman AF, Holland SM. The hyper-IgE syndromes. Immunol Allergy Clin North Am. 2008;28(2):277–280. doi: 10.1016/j.iac.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen K, et al. IL-17 Receptor signaling in the lung epithelium is required for mucosal chemokine gradients and pulmonary host defense against K. pneumoniae. Cell Host Microbe. 2016;20(5):596–605. doi: 10.1016/j.chom.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Conti HR, et al. Signaling through IL-17C/IL-17RE is dispensable for immunity to systemic, oral and cutaneous candidiasis. PLoS One. 2015;10(4):e0122807. doi: 10.1371/journal.pone.0122807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lambert S, et al. Neutrophil extracellular traps induce human Th17 cells: effect of psoriasis-associated TRAF3IP2 genotype. J Invest Dermatol. 2019;139(6):1245–1253. doi: 10.1016/j.jid.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Uyemura K, et al. Limited T-cell receptor beta-chain diversity of a T-helper cell type 1-like response to Mycobacterium leprae. Infect Immun. 1992;60(11):4542–4548. doi: 10.1128/IAI.60.11.4542-4548.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schmidt NW, et al. Pentobra: a potent antibiotic with multiple layers of selective antimicrobial mechanisms against Propionibacterium acnes. J Invest Dermatol. 2015;135(6):1581–1589. doi: 10.1038/jid.2015.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McInturff JE, et al. Granulysin-derived peptides demonstrate antimicrobial and anti-inflammatory effects against Propionibacterium acnes. J Invest Dermatol. 2005;125(2):256–263. doi: 10.1111/j.0022-202X.2005.23805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Edgar R, et al. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Law CW, et al. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu R, et al. Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res. 2015;43(15):e97. doi: 10.1093/nar/gkv412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.