Abstract
FOXP3+ Tregs rely on fatty acid β-oxidation–driven (FAO-driven) oxidative phosphorylation (OXPHOS) for differentiation and function. Recent data demonstrate a role for Tregs in the maintenance of tissue homeostasis, with tissue-resident Tregs possessing tissue-specific transcriptomes. However, specific signals that establish tissue-resident Treg programs remain largely unknown. Tregs metabolically rely on FAO, and considering the lipid-rich environments of tissues, we hypothesized that environmental lipids drive Treg homeostasis. First, using human adipose tissue to model tissue residency, we identified oleic acid as the most prevalent free fatty acid. Mechanistically, oleic acid amplified Treg FAO–driven OXPHOS metabolism, creating a positive feedback mechanism that increased the expression of FOXP3 and phosphorylation of STAT5, which enhanced Treg-suppressive function. Comparing the transcriptomic program induced by oleic acid with proinflammatory arachidonic acid, we found that Tregs sorted from peripheral blood and adipose tissue of healthy donors transcriptomically resembled the Tregs treated in vitro with oleic acid, whereas Tregs from patients with multiple sclerosis (MS) more closely resembled an arachidonic acid transcriptomic profile. Finally, we found that oleic acid concentrations were reduced in patients with MS and that exposure of MS Tregs to oleic acid restored defects in their suppressive function. These data demonstrate the importance of fatty acids in regulating tissue inflammatory signals.
Keywords: Autoimmunity, Immunology
Keywords: Adipose tissue, Fatty acid oxidation, Multiple sclerosis
Introduction
Metabolic signatures of T cells are intricately linked to their differentiation and activation status. Metabolic pathways facilitate cellular functions, therefore linking metabolic remodeling to the development, activation, differentiation, and survival of T cells. Upon activation, quiescent, naive T cells become rapidly dividing effector T cells (Teffs) and switch their metabolic program from oxidative phosphorylation (OXPHOS) to aerobic glycolysis, known as the Warburg effect, in order to meet the increase in demand for cellular energy and biomass (1, 2). However, despite having similar developmental origins, Tregs rely predominantly on a fatty acid β-oxidation–driven (FAO-driven) OXPHOS metabolic program to maintain their suppressive phenotype, which is further promoted by the expression of FOXP3 (3–5). Forced expression of FOXP3 in T cells suppresses glycolysis-related genes, while inducing lipid and oxidative metabolism–related genes that are required for maximum suppression (4). Cytokines that promote Treg differentiation, such as TGF-β (6), activate AMPK (7) and promote FAO to skew naive T cells to a Treg phenotype (3, 8). Furthermore, Treg differentiation and suppression are reduced by inhibiting FAO (3), highlighting the importance of FAO-driven OXPHOS in the initiation and maintenance of the Treg phenotype.
The suppressive function of Tregs is critical for controlling immune responses and preventing autoimmunity. We have previously identified functional Treg defects in patients with autoimmune disease (9). However, a second critical function for Tregs is the regulation of tissue homeostasis that can have a secondary impact on organismal biology. In mice and humans, Tregs infiltrate tissues not only during inflammatory conditions or injury, but also during homeostasis (10–12), and reside within tumors (13, 14). Resident Tregs adapt to perform tissue-specific functions in order to regulate inflammation and perform homeostatic functions, such as wound repair and maintenance of metabolic indices (10, 12, 15). Specifically, in visceral adipose tissue (VAT), VAT-resident Tregs possess unique epigenetic and transcriptional profiles that allow Tregs to survive in lipotoxic environments and enable the utilization of FAO as metabolic fuel (10, 16). Despite these findings, the signals that act to balance canonical Treg function and Treg adaptations of tissue-specific signals remain unknown.
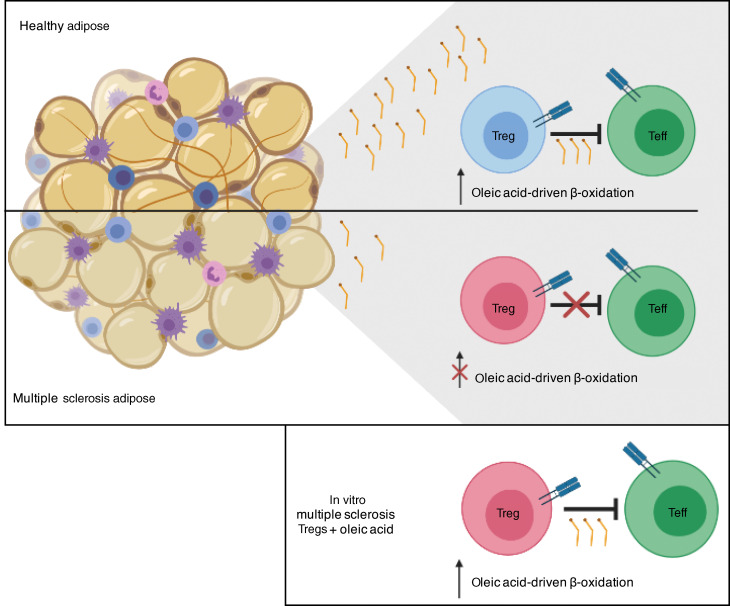
Dissecting the signals that act to either promote or inhibit Treg adaptation in tissues is essential to our understanding of tissue Treg biology. An example are the signals that prevent loss of FOXP3 or promote the generation of Th-like Tregs (9, 17–21). While both local antigens and the cytokine milieu play a role in tissue adaptation (22), we hypothesized that environmental cues, such as lipids, may be critical, considering their role in shaping the Treg metabolic/functional axis. Here, we identify oleic acid as the most prevalent long-chain free fatty acid (FFA) in human adipose tissue and dissect its involvement in the maintenance of Treg function. Oleic acid amplifies Treg FAO-driven OXPHOS metabolism, creating a positive feedback mechanism that induces the expression of FOXP3 and enhances the phosphorylation of STAT5 (p-STAT5), which act to stabilize the Treg lineage and increase suppressive function (23–27). We compared the transcriptomic program induced by oleic acid with that of proinflammatory arachidonic acid (28, 29) and derived a computational transcriptome signature to quantify the similarity of the Treg RNA profile to either state. We found that Tregs sorted from peripheral blood and adipose tissue of healthy donors transcriptomically resembled the Tregs treated in vitro with oleic acid, whereas Tregs obtained from patients with relapsing-remitting multiple sclerosis (MS) more closely resembled the arachidonic acid–treated Treg profile. A similar trend was observed when we compared patients with MS in the treated and untreated groups. Finally, we found that oleic acid concentrations were reduced in the fat tissue of patients with MS and that exposure of dysfunctional MS Tregs to oleic acid partially restored their suppressive function, highlighting the importance of fatty acids in regulating inflammatory signaling in tissues.
Results
Oleic acid upregulates fatty acid β-oxidation in Tregs.
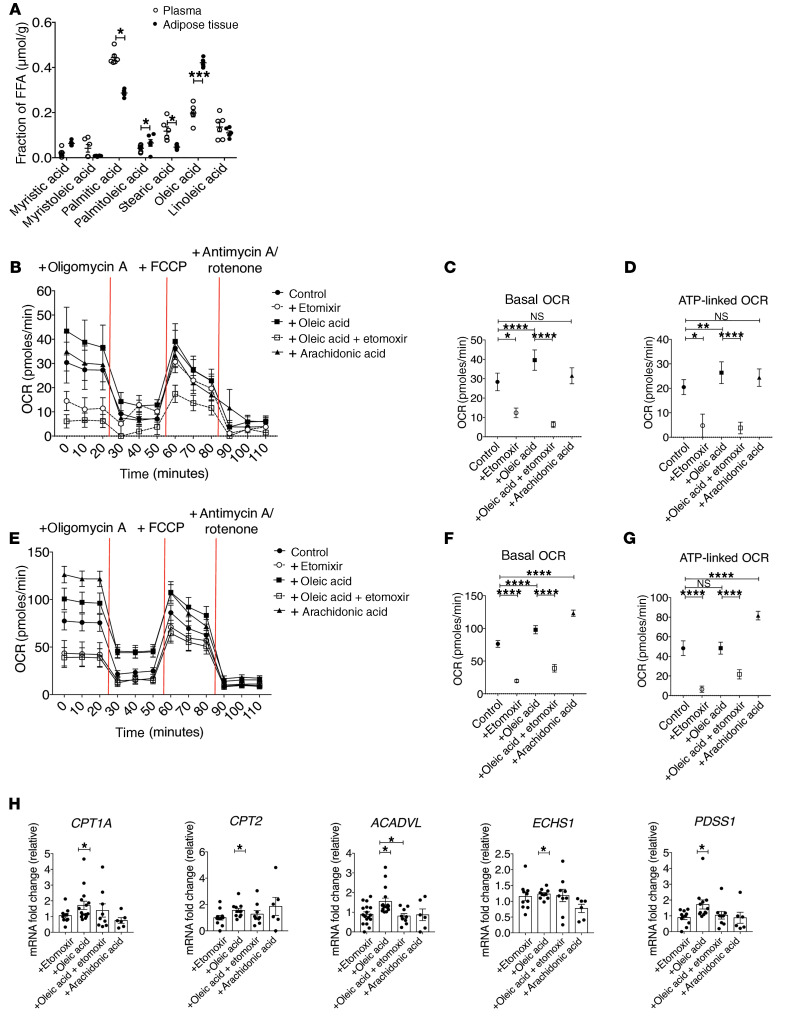
In order to determine which FFAs Tregs may be responding to in human adipose tissue, we performed mass spectrometry on the supernatant from healthy human adipose tissue and paired serum samples. In agreement with previous reports (30–33), we found oleic acid to be the most abundant FFA in adipose tissue (Figure 1A). Oleic acid is a monounsaturated, omega-9 long-chain fatty acid (LCFA) found in most animal and vegetable sources. In animal tissues, oleic acid is one of the most abundant FFAs, regardless of tissue or species (30–33), implying that tissue-resident Tregs sense oleic acid and that oleic acid may be an important signal for tissue-resident Tregs. LCFAs can alter cellular proliferation and viability in a dose-dependent manner (3, 34–40). However, at a concentration of 10 μM, oleic acid, but not arachidonic acid, increases FOXP3 expression (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/JCI138519DS1), without disrupting Treg viability (Supplemental Figure 1C) or proliferation (Supplemental Figure 1D), compared with vehicle. FOXP3 expression began to increase 48 hours after activation in the presence of oleic acid, but reached maximum expression at 72 hours (Supplemental Figure 1E). We also measured FOXP3 expression in Tregs after exposure to LCFA species of varying lengths and saturations and detected upregulation of FOXP3 only in the presence of oleic acid (Supplemental Figure 1F), further supporting our detailed investigation using this specific LCFA species. We thus compared the effects of oleic acid with those of arachidonic acid, since arachidonic acid is known to induce a proinflammatory phenotype in T cells (28, 29). Furthermore, LCFA treatment did not affect FOXP3 demethylation (Supplemental Figure 1G), suggesting that the stability of FOXP3 expression was unchanged under these conditions.
Figure 1. Oleic acid increases FAO in Tregs.
(A) Mass spectrometric analysis of LCFAs in supernatant from human adipose samples compared with blood plasma (n = 6). *P < 0.05 and ***P < 0.001, by paired Student’s t test. (B) OCR of Tregs stimulated for 72 hours in the presence or absence of etomoxir (50 μM), oleic acid (10 μM), oleic acid plus etomoxir, or arachidonic acid (10 μM) (n = 12). Summary of (C) basal OCR and (D) ATP-linked OCR of Tregs stimulated as in B (n = 12). *P < 0.05, **P < 0.01, and ****P < 0.0001, by paired Student’s t test corrected for multiple-hypothesis testing using the Holm-Sidak method. (E) OCR of Teffs stimulated as described in B (n = 12). Summary data for (F) basal OCR and (G) ATP-linked OCR of Teffs stimulated under the same conditions as in B (n = 12). ****P < 0.0001, by paired Student’s t test corrected for multiple-hypothesis testing using the Holm-Sidak method. (H) Fold change of mRNA expression relative to vehicle in Tregs stimulated for 72 hours as described in B. *P < 0.05, by paired Student’s t test corrected for multiple hypothesis testing using the Holm-Sidak method. Data represent the mean ± SEM.
To determine whether FAO drives Treg differentiation and function (3–5), we examined the metabolic phenotype of CD25+CD127lo/– Tregs in the presence of LCFAs. First, we performed Seahorse analysis to determine the oxygen consumption rate (OCR) as a measurement of OXPHOS and found that oleic acid specifically increased the OCR in Tregs (Figure 1B). Tregs cultured with oleic acid showed elevated basal respiration (Figure 1C) and an ATP-linked OCR (Figure 1D), effects that were blocked by the inhibition of CPT1A via etomoxir, suggesting that the effects of oleic acid were mediated by its entry into FAO. We also observed small but significant increases in mitochondrial mass (Supplemental Figure 2A), but not ROS, in the presence of oleic acid (Supplemental Figure 2B). Thus, the observed OCR increases could be attributed to increases in mitochondria rather than to increases in the respiratory rates of individual mitochondria. Basal respiration was greater than or equal to maximal respiration rates (Figure 1, B and C, and Supplemental Figure 3A) in most donor Tregs, which is consistent with previous reports involving human and murine Tregs (37, 41–43). We observed a decrease in the overall spare respiratory capacity in the presence of oleic acid (Supplemental Figure 3B). However, the addition of etomoxir to stimulated Tregs increased the spare respiratory capacity (Supplemental Figure 3B), indicating that oleic acid–driven FAO might act to deplete this capacity in Tregs.
We compared oleic acid and arachidonic acid and observed that both increased basal respiration in Tregs (Figure 1, E and F), whereas only arachidonic acid increased ATP-linked OCR in CD127+CD25lo/– Teffs (Figure 1G), and both fatty acids were negated by the addition of etomoxir. Unlike Tregs, the maximum OCR and spare respiratory capacity of Teffs increased with oleic acid, also in a CPT1A-dependent manner (Supplemental Figure 3, C and D), whereas we observed no differences in mitochondrial mass (Supplemental Figure 2C) or ROS (Supplemental Figure 2D). Given reports of off-target effects of etomoxir at high doses (44, 45), we also tested lower concentrations of etomoxir in our experiments and found no differences in our results (Supplemental Figure 4, A–D), suggesting that our observations were based on CPT1-dependent mechanisms. These data demonstrate that, although both Tregs and Teffs were targeting lipids toward FAO, they exhibited different abilities to respond to an excess of lipid uptake. These differences might reflect unique metabolic requirements of effector versus regulatory functions, or the inherent ability of Tregs versus Teffs to metabolically adapt to niche tissue environments.
We then measured the expression of mitochondrial metabolic genes in Tregs by quantitative PCR (qPCR) and confirmed CPT1A, CPT2, ACADVL, ECHS1, and PDSS1 were all upregulated in the presence of oleic acid (Figure 1H). In contrast, expression of ACADVL and PDSS1 was dependent on FAO, as it was downregulated by the addition of etomoxir (Figure 1H). Conversely, the same genes showed no significant effects in Teffs (Supplemental Figure 5). These data demonstrate that FFAs may have differential effects on Tregs compared with Teffs because of the reliance of Tregs on FAO-driven OXPHOS, and are supported by the uptake of extracellular lipids (3, 46). Notably, the expression of PPARG and ACACB, which are measurements of global lipid metabolism and storage and fatty acid synthesis, respectively, trended upward in Tregs (Supplemental Figure 6), whereas ACACB trended downward in Teffs (Supplemental Figure 5), providing further evidence that the 2 cell types were utilizing oleic acid differently. Together, our data show that oleic acid drives a unique gene expression profile in Tregs, characterized by the upregulation of FAO-driven OXPHOS.
Given the role of PPAR transcription factors in mediating and responding to global changes in lipid metabolism (47), we hypothesized that PPAR has a role in controlling oleic acid–mediated effects. We examined Treg suppression in the presence of the PPARα and PPARγ inhibitors GW6471 and GW9662, respectively (48–50), and found that inhibition of PPARγ decreased Treg suppression (Supplemental Figure 7, A and B) and reversed oleic acid–mediated increases in Treg suppression (Supplemental Figure 7, A and B). Similarly, inhibition of PPARα or PPARγ decreased FOXP3 protein and mRNA expression in Tregs (Supplemental Figure 7C) and downregulated expression of the FAO genes CPT1A and ACADVL (Supplemental Figure 7D). These data support a general role for PPAR transcription factors in regulating Treg function and metabolism.
Previous reports have found human Tregs to be engaged in both FAO and glycolysis to support their expansion (41, 51). To determine whether LCFAs specifically upregulate FAO as opposed to simply increasing the activation status of cells, we measured glycolysis activity via the extracellular acidification rate by Seahorse analysis. We found increases in Tregs (Supplemental Figure 8, A and B) and Teffs (Supplemental Figure 8, C and D) cultured with LCFAs, suggesting that these cells were engaging in glycolysis upon exposure to LCFAs and were activated.
Oleic acid drives Treg suppression by upregulating FAO-dependent Treg genes.
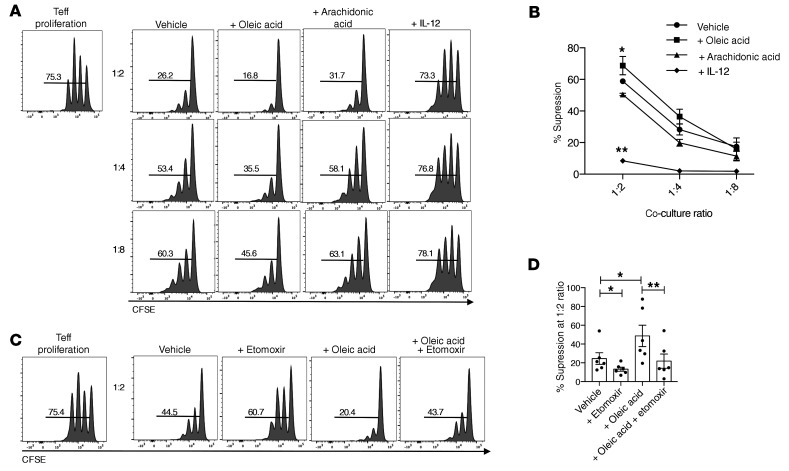
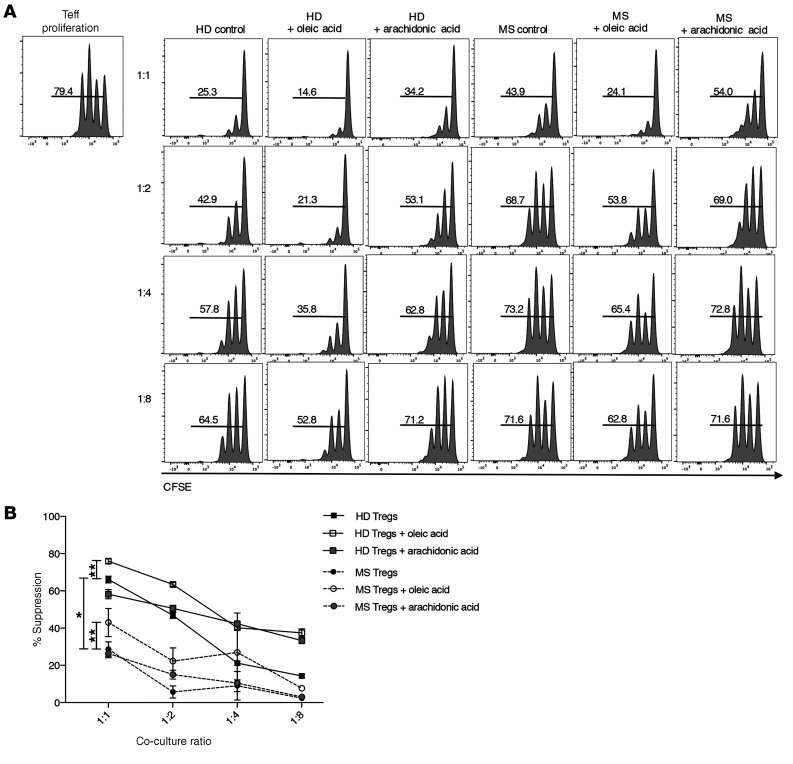
In order to determine whether oleic acid influences Treg function, we performed a suppression assay with Tregs preincubated with oleic acid, arachidonic acid, or IL-12 as a negative control (9). We found that oleic acid specifically increased Treg suppression (Figure 2, A and B), which was dependent on oleic acid–driven FAO, as etomoxir inhibited oleic acid–driven Treg suppression (Figure 2, C and D). Other omega-9 fatty acids tested, including elaidic acid, gondoic acid, or eurcic acid, did not have any effect on Treg suppression (Supplemental Figure 9, A and B), suggesting that the effects observed were oleic acid specific and not a general attribute of omega-9 fatty acids.
Figure 2. Oleic acid increases the suppressive capacity of Tregs.
(A) CFSE measurement of the proliferation of Teffs cocultured with Tregs that were preincubated with vehicle, 10 μM oleic acid, 10 μM arachidonic acid, or 25 ng/mL IL-12 for 72 hours. Proliferation was measured after 4 days of coculture with Tregs (n = 10). Histograms are from 1 representative experiment. (B) Summary of the suppression data described in A based on 5 independent experiments. *P < 0.05 and **P < 0.01, by paired Student’s t test corrected for multiple-hypothesis testing using the Holm-Sidak method. (C) CFSE measurement of the proliferation of Teffs cocultured with Tregs that were preincubated with vehicle, 50 μM etomoxir, 10 μM oleic acid, or oleic acid plus etomoxir for 72 hours. Proliferation was measured after 4 days of coculture with Tregs (n = 8). Histograms are from 1 representative experiment. (D) Summary of suppression data at the 1:2 ratio. *P < 0.05 and **P < 0.01, by paired t test corrected for multiple-hypothesis testing using the Holm-Sidak method. Data represent the mean ± SEM.
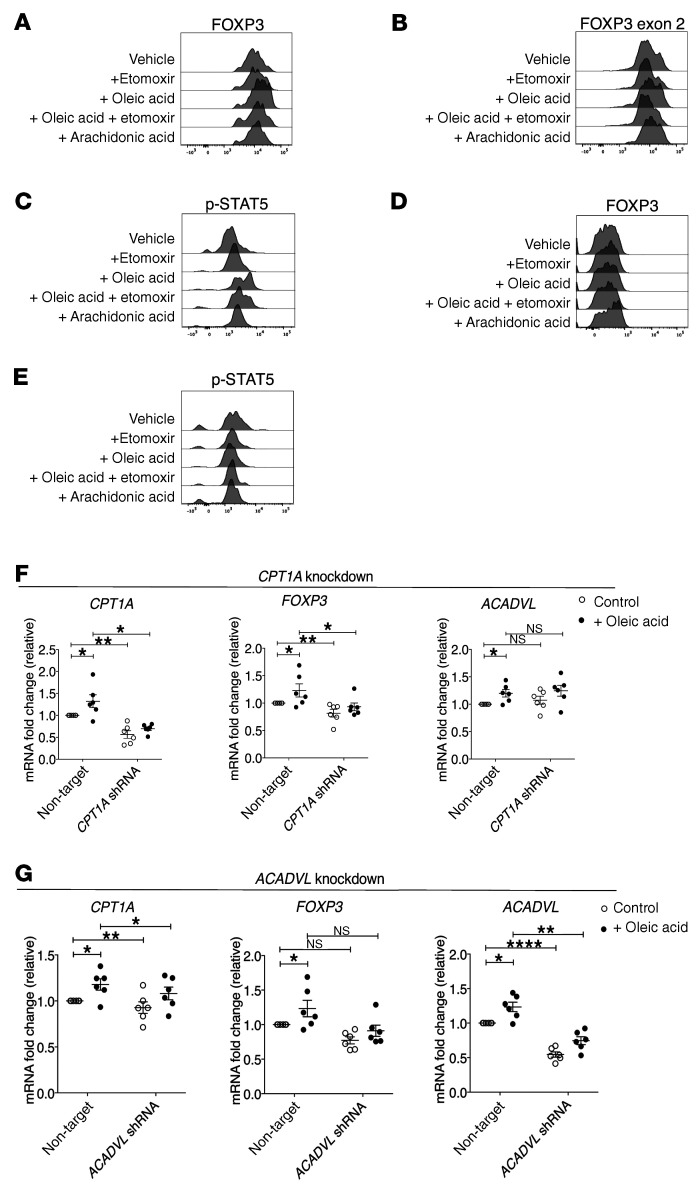
Treg differentiation requires an intrinsic metabolic switch to FAO, and expression of FOXP3 promotes lipid- and OXPHOS-related gene expression (3–5). In order to understand how oleic acid–driven FAO enhances Treg suppression, we measured protein expression of FOXP3 and the phosphorylation status of STAT5 (p-STAT5), which promotes Treg lineage stability via FOXP3 demethylation (25–27). Oleic acid increased the expression of FOXP3 and, specifically, FOXP3 exon 2 (52–54) (Figure 3, A and B, and Supplemental Figure 10, A and B), as well as p-STAT5 (Figure 3C and Supplemental Figure 10, C and D), which are dependent on oleic acid entry into FAO. Again, oleic acid did not increase the expression of FOXP3 (Figure 3D and Supplemental Figure 11A) or p-STAT5 (Figure 3E Supplemental Figure 11B) in Teffs. However, lentiviral knockdown of CPT1A (Figure 3F and Supplemental Figure 12, A and B) or ACADVL (Figure 3G) reduced the expression of FOXP3 (Figure 3, F and G and Supplemental Figure 12, A and B), further demonstrating that oleic acid–driven FAO was driving tissue-resident Treg function.
Figure 3. Oleic acid–driven FAO drives FOXP3 expression in Tregs.
Representative experiment assessing (A) FOXP3 and (B) FOXP3 exon 2 expression in Tregs treated with vehicle, etomoxir (50 μM), oleic acid (10 μM), oleic acid plus etomoxir, or arachidonic acid (10 μM) after 72 hours (n = 12). (C) Representative experiment measuring p-STAT5 in Tregs after 6 hours of stimulation in the presence of the same agents described in A (n = 10). Representative experiment assessing (D) FOXP3 protein expression (n = 12) and (E) p-STAT5 levels after 6 hours (n = 8) in Teffs treated for 72 hours with the same agents described in A. mRNA expression of the indicated genes relative to the control in Tregs cultured in the presence of absence of 10 μM oleic acid after lentiviral knockdown of (F) CPT1A or (G) ACADVL (n = 8). *P < 0.05, **P < 0.01, and ****P <0.0001, by paired t test corrected for multiple hypothesis testing using the Holm-Sidak method. Data represent the mean ± SEM.
The enhanced phosphorylation of STAT5 led us to investigate CD25 expression on Tregs. On day 3, we found that oleic acid increased the total expression of CD25 on Tregs, which could explain the observed increases in p-STAT5 (Supplemental Figure 13, A and B), as IL-2 drives STAT5 phosphorylation (25, 27). Further, inhibition of different components of the electron transport chain inhibited the oleic acid–driven increase and overall expression of CD25 (Supplemental Figure 13, A and B), providing evidence that oleic acid–driven mitochondrial respiration drives the CD25/STAT5 axis and provides an additional layer of stabilization to FOXP3 and the Treg lineage (25, 27, 55). In total, oleic acid–driven increases in mitochondrial metabolism drove Treg-suppressive functions through stabilization of the Treg lineage by increasing p-STAT5 and upregulating FOXP3 expression.
Global transcriptomic effects of oleic acid versus arachidonic acid.
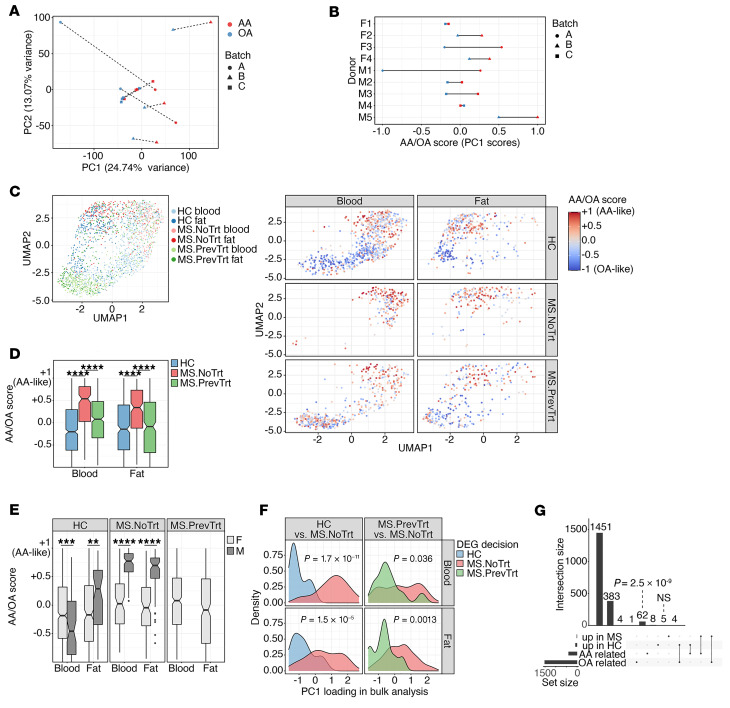
Given the observed oleic acid–induced changes in mitochondrial respiration and suppressive function in Tregs, we sought to understand the coinciding transcriptional changes, and how they might contribute to the tissue-resident Treg phenotype. We first defined the expression profile of Tregs isolated from the peripheral blood of healthy donors and treated with oleic or arachidonic acid. We then compared each profile with the transcriptional program of Tregs isolated from the blood and adipose tissue of healthy donors or patients with MS.
First, we isolated Tregs from the peripheral blood of 9 healthy donors, cultured them in vitro with oleic or arachidonic acid, and performed bulk RNA-Seq (Supplemental Figure 14 and Supplemental Table 1). We observed considerable donor-specific effects that obstructed attempts to infer differentially expressed genes (DEGs) between the control and treatment groups through inclusion of the donor as a nuisance covariate of a generalized linear model (data not shown). Therefore, we took advantage of the experimental paired design by subtracting the high-dimensional gene expression vector of the vehicle state from either treatment, producing 2 vectors for each patient that captured the differential effect of oleic or arachidonic acid treatment (see Methods). The first principal component (PC1) computed over this set of vectors captured the differential effect for each fatty acid (PC1 accounts for 24.7% of the variance) (Figure 4A). The dynamic range and magnitude of the PC1 effect were donor specific, but the overall trend captured by PC1 was robust for the entire donor group (Figure 4B). The gene loadings on PC1 were also consistent with a gene-wise linear model designed to directly compare the effects of oleic acid versus arachidonic acid (Supplemental Figure 15, Methods, and Supplemental Table 2). Overall, despite considerable donor specificity in the Treg response to fatty acids, aggregation of the signal across multiple genes successfully discerned a transcriptomic effect that was consistent across donors and a primary source of variability in the data.
Figure 4. Oleic acid transcriptomic signature characterizes healthy but not MS Tregs.
(A and B) Tregs from peripheral blood of healthy donors were stimulated in the presence or absence of 10 μM oleic acid (OA) or arachidonic acid (AA). Each donor is represented by 2 dots, which correspond to the difference between oleic acid and the control or arachidonic acid and the control. (A) PCA was performed; each patient’s symbols are connected by a dashed line. Donors were partitioned into 3 separate date groups for tissue collection, denoted as batches (batch A took place over 2 consecutive days). (B) PC1 is scaled to [–1,1]; each line represents 1 donor (donor’s sex is indicated on the left). M1, male 1; F1, female 1. (C) Single-cell transcriptomes of Tregs were sorted from peripheral blood or adipose tissue of healthy donors (HC) or patients with MS and projected to a bidimensional space with UMAP (left). The same data are shown on the right, stratified by tissue and donor group. Colors indicate a computational signature of similarity to oleic acid– or arachidonic acid–stimulated blood Tregs (see the main text and Methods). (D and E) The computational signatures of single cells were aggregated and stratified by tissue, treatment group, and donor sex. Significance was determined by 2-sided Welch’s t test. (F) Densities of genes upregulated in healthy donors versus untreated patients with MS with respect to the loadings of PC1 shown in panel A (left) and a similar comparison of treated and untreated patients with MS (right). Significance was determined by 2-sided Welch’s t test. (G) Upset plot (101) showing the overlap of DEGs upregulated in healthy or MS (aggregating treated and untreated patients) states in the vivo single-cell RNA-Seq, and genes belonging to the oleic or arachidonic acid modules in the in vitro bulk RNA. The P value in G was calculated by hypergeometric enrichment test. NoTrt, no treatment; PrevTrt, previous treatment.
On the basis of the loadings of PC1, we defined 250 genes with the lowest loading as the module of oleic acid–related genes, and the 250 genes with the highest loadings as the module of arachidonic acid–related genes. The oleic acid module was enriched for genes related to LCFA FAO, mitochondrial integrity, and promotion of Treg generation. For example, ACBD7, ACOT11, and ACSL6 bind and convert LCFAs to acyl-CoA for degradation by FAO (56–59). LPIN3 has a general role in regulating fatty acid metabolism (60), and LETM1 regulates mitochondrial dynamics and maintains normal mitochondrial morphology (61). We also observed upregulation of C5AR2, which has been shown to promote peripherally induced Tregs in human and murine models.
Conversely, the arachidonic acid module was enriched for genes related to both glycolysis and mitochondrial respiration, as well as proinflammatory Teff subsets. PGK1 and PGM1 catalyze the breakdown of glucose (62–64), and SLC2A7 is a glucose transporter (65). SIRT4 regulates FAO via inhibition of PPARα transcription (66) and FABP5, an intracellular lipid chaperone (67, 68). Enrichment of CD69 and CD9 indicates that arachidonic acid–treated Tregs are more activated (69–71). Notably, we detected genes that promote proinflammatory T cells, such as CKS2, LDHA, and LDHB, which promote Th17 (72, 73) and Th1 (74) differentiation, respectively. In contrast to genes enriched in the oleic acid module, arachidonic acid–related genes implied more activated Tregs that were potentially adopting a more proinflammatory phenotype.
Adipose tissue–derived Tregs from healthy individuals, not patients with MS, are more similar in profile to oleic acid–treated Treg profiles.
To study the relevance of the fatty acid milieu in vivo, we examined the transcriptional profile of Tregs isolated from the adipose tissue of healthy donors and patients diagnosed with an inflammatory autoimmune disease associated with altered Treg function. Our laboratory has previously shown that Tregs isolated from patients with MS express IFNG and are less suppressive in vitro (9). However, Th1-like Tregs have also been described in models of chronic infection (75–77) and in patients with type 1 diabetes (T1D) (19), and childhood obesity is a risk factor for the development of MS (78, 79). Therefore, using MS as our model, we performed single-cell RNA-Seq to profile 1334 Tregs from peripheral blood and 805 Tregs from adipose tissue of 8 healthy donors and 8 patients with MS, 2 of whom were untreated (for more than ~1.5 years at the time of the procedure), and the other patients had previously received disease-modifying treatments (DMTs) (>6 months prior to the procedure, but were currently off treatment) (Supplemental Figure 14, Supplemental Figure 16, and Supplemental Table 3). We assigned a quantitative score to each cell on the basis of the PC1 axis we inferred from the in vitro data above (Figure 4, C and D, and see Methods). This score represented the similarity between the single cell’s transcriptome and the bulk data set from either oleic acid– or arachidonic acid–treated blood Tregs. The scores ranged from –1 to +1, which represented more similarity to oleic acid or arachidonic acid, respectively.
Tregs collected from the adipose tissue of healthy donors had a significantly lower quantitative score than Tregs collected from the adipose tissue of previously treated patients with MS, which in turn had a significantly lower score than Tregs isolated from untreated patients with MS (Figure 4D; 2-sided Welch’s t test). This suggests that oleic acid might be more prevalent in the lipid milieu encountered by Tregs in healthy individuals compared with that of patients with MS. Moreover, immunotherapy may partially restore the transcriptomic Treg state that is characteristic of a healthy donor, consistent with previous observations regarding the link between transcriptome restoration in response to treatment (80). However, these observations alone do not necessarily indicate a causative role for fatty acids in the development of autoimmunity, since the patients with MS in our sample were on average older and had higher BMIs than did the healthy donors. Although the distribution of scores reflecting the similarity to oleic acid– or arachidonic acid–treated Tregs was sex specific (Figure 4E), the generalizability of these particular data is unclear, given the small number of donors in the current study.
Significant overlap of oleic acid–responsive genes and genes suppressed in MS.
We further examined the relevance of the in vitro–derived oleic acid similarity score to human autoimmune disease by computing the set of DEGs between Tregs isolated from the blood and adipose tissue of healthy donors and Tregs isolated from the same tissues of untreated patients with MS based on single-cell RNA-Seq (Supplemental Table 4, and see Methods). We found that the genes upregulated in healthy donors had significantly lower PC1 loadings with respect to the in vitro PC1 derived above (Figure 4F, left), similar to the results obtained when comparing the previously treated and untreated patients with MS (Figure 4F, right). Further, comparison of healthy donors and patients with MS (both previously treated and untreated) showed a significant overlap between genes upregulated in the healthy state and those belonging to the oleic acid gene module defined above, based on the in vitro PC1 (Figure 4G; 1.65-fold, hypergeometric P = 2.5 × 10–9). In contrast, we observed no significant overlap between genes upregulated in the MS state and those in the arachidonic acid module (hypergeometric P = 0.4). These data corroborate our findings that, compared with MS patients, the transcriptional signature observed in Tregs isolated from the blood and adipose tissue of healthy donors was significantly more similar to blood Tregs stimulated with oleic acid than to those stimulated with arachidonic acid. These data suggest that Tregs in the blood and adipose tissue of healthy donors may be exposed to a lipid milieu that contains larger amounts of oleic acid than can be found in patients with MS. Moreover, these data provide evidence that the exposure of peripherally derived Tregs to oleic acid can, to a certain degree, recapitulate clinically relevant aspects of the adipose tissue–derived phenotype. Thus, oleic acid may counteract inflammatory signals in the tissues by reinforcing the canonical Treg program and suppressive function.
Oleic acid–driven FAO partially restores defective Treg function.
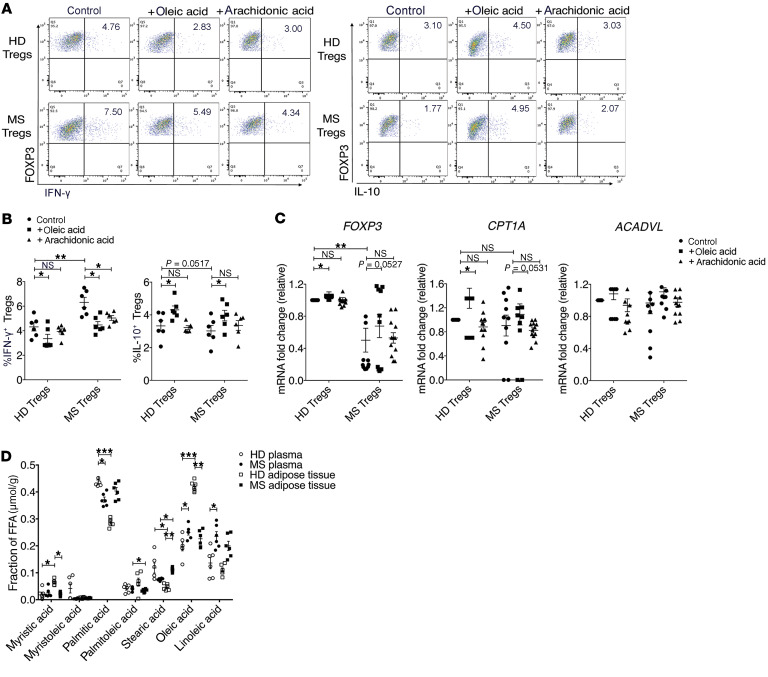
To address the physiological importance of oleic acid–driven Treg stability and function in the tissues and to address the hypothesis that lipids, as a metabolic cue, are able to buffer inflammatory signals, we cultured Tregs isolated from untreated patients with MS (no treatment for ~1.5 years at the time of the blood draw) with oleic acid. Oleic acid partially restored Treg suppression compared with healthy, aged-matched control Tregs (Figure 5, A and B), and oleic acid decreased the percentage of IFN-γ+ Tregs, while increasing the percentage of IL-10+ Tregs from patients with MS (Figure 6, A and B). Furthermore, exposure to oleic acid upregulated FOXP3 and CPT1A transcripts in MS Tregs (Figure 6C), suggesting that oleic acid–driven FAO was negating Treg dysfunction. To test the ability of oleic acid to counteract inflammatory signals, we cultured Tregs in the presence of both IL-12 and oleic acid. Our results suggest that oleic acid can partially reduce the expression of IL-12–driven IFNG (Supplemental Figure 17, A and B) and that oleic acid can partially inhibit the development of inflammatory Th1-like Tregs.
Figure 5. Oleic acid partially restores suppressive defects in MS Tregs.
(A) CFSE was used to measure the proliferation of Teffs cultured with Tregs sorted from frozen PBMCs obtained from either healthy donors (HD) or patients with MS. Tregs were preincubated in the presence or absence of 10 μM oleic or arachidonic acid for 72 hours, and proliferation was measured after 4 days in coculture (n = 8). Histograms are from 1 representative experiment. (B) Summary of suppression data from 4 independent experiments (n = 8). *P < 0.05 and **P < 0.01, by paired Student’s t test corrected for multiple-hypothesis testing using the Holm-Sidak method. Data represent the mean ± SEM.
Figure 6. Oleic acid partially negates the MS Treg proinflammatory phenotype.
(A) Representative dot plot of healthy donor or MS Tregs sorted from frozen PBMCs and stimulated in the presence or absence of 10 μM oleic acid or arachidonic acid for 72 hours. Intracellular staining for IFN-γ (top) or IL-10 (bottom) was measured after a 4-hour stimulation with PMA and ionomycin in the presence of GolgiStop. (B) Summary of IFN-γ+ and IL-10+ healthy and MS Tregs after treatment with 10 M oleic acid or arachidonic acid for 3 days (n = 8). *P < 0.05 and **P < 0.01, by paired Student’s t test corrected for multiple-hypothesis testing using the Holm-Sidak method. (C) mRNA expression of the indicated genes relative to control, measured in the healthy and MS Tregs described in A (n = 10). *P < 0.05 and **P < 0.01, by paired Student’s t test corrected for multiple-hypothesis testing using the Holm-Sidak method. (D) Mass spectrometric analysis of LCFAs in supernatant from human adipose tissue samples compared with blood plasma of healthy donors or patients with MS (n = 6) *P < 0.05, **P < 0.01, and ***P < 0.001, by paired Student’s t test corrected for multiple-hypothesis testing using the Holm-Sidak method. Data represent the mean ± SEM.
To determine whether FAO-driven OXPHOS can influence Treg IFN-γ and IL-10 expression, we measured the percentage of IFN-γ+ and IL-10+ Tregs from healthy donors in the presence of electron transport chain inhibitors. In the presence of etomoxir, the percentage of IFN-γ+ Tregs increased, suggesting that FAO normally acts to inhibit IFN-γ expression (Supplemental Figure 18, A and B). Conversely, the percentage of IL-10+ Tregs decreased when CPT1A and complex I were inhibited by etomoxir and rotenone, respectively, indicating that FAO and the electron transport chain promoted IL-10 expression (Supplemental Figure 18, C and D). Together, these data suggest that pro- versus antiinflammatory cytokine expression is regulated in an FAO-dependent manner in Tregs and support previous studies involving Th1 Teffs (81).
To directly address the question of whether adipose tissue–resident Tregs are exposed to different concentrations of oleic acid compared with those in patients with MS, we measured LCFA concentrations in the plasma and adipose supernatant of MS donors. The fraction of oleic acid present in the adipose supernatant of patients with MS was strikingly reduced compared with that of healthy donors, whereas in the plasma, oleic acid was increased in patients with MS (Figure 6D). We observed inverse trends in the fraction of oleic acid present in healthy versus MS tissue compartments. For healthy donors, we detected more oleic acid in adipose supernatant relative to plasma, but for patients with MS, we found no significant differences. This suggests either the existence of a basal state of inflammation or an inherent dysregulation of LCFA uptake and storage in patients with MS. Further, we observed an overall change in LCFA composition between healthy participants and patients with MS (Figure 6D), consistent with an intrinsic defect in the regulation of LCFAs in the autoimmune and, in this case, the MS disease state.
Discussion
Tissue-resident Treg populations possess unique epigenetic and transcriptional profiles that allow them to fine-tune their tissue-specific functions (10, 12, 82–84). Tregs in muscle expand upon injury and play a role in the maintenance of homeostasis and tissue regeneration (12, 85, 86). In the skin, Tregs are necessary for preventing skin lesions, hypersensitivity, and atopic dermatitis (87–90). In adipose tissue, VAT-resident Tregs are enriched within the VAT CD4+ T cell compartment (10) and express genes that specifically allow the adaptation of VAT Tregs to survive in lipotoxic environments through the utilization of FAO as metabolic fuel (16). However, the modes of Treg metabolic adaptation to different tissue environments, and the signals that act to balance adaptation and the canonical Treg program are unknown.
Here, we show that oleic acid is the most prevalent LCFA in human adipose tissue and a critical environmental signal that stabilizes FOXP3 and drives Treg suppression by enhancing the FAO-OXPHOS metabolic program and that oleic acid treatment can partially restore Treg suppression in patients with MS. Furthermore, the oleic acid–derived gene signature in Tregs more closely resembles the expression profile of blood- and adipose tissue–derived Tregs from healthy donors rather than from patients with MS. Interestingly, the transcriptomic analysis suggests that the balance of oleic and arachidonic acids in the extracellular environment modulates the Treg phenotype. The high degree of variability with respect to both the range and magnitude of the LCFA effect may be attributed to the highly variable nature of human donors, such as dietary or lifestyle patterns that may affect lipid uptake or metabolic adaptations of Tregs.
We provide evidence of environmental factors influencing tissue-resident immune cell function and show that LCFA composition fluctuates between healthy and autoimmune disease states. We observed differences in the gene expression profile of Tregs isolated from the adipose tissue of healthy donors compared with that of patients with MS. When we compared these signatures with those of peripheral blood Tregs exposed to oleic acid, we found that the oleic acid signature was more reflective of healthy adipose tissue–resident Tregs than were MS adipose tissue–resident Tregs. Conversely, the proinflammatory arachidonic acid signature more closely resembled Tregs isolated from MS adipose tissue and was supported by the significant overlap observed between genes upregulated in the healthy state and genes expressed following oleic acid treatment. These data demonstrate that exposure of peripherally derived Tregs to oleic acid can partially recapitulate the transcriptional profile of adipose tissue–resident Tregs and perhaps identify a new signal necessary for maintenance of the canonical Treg program in tissue-resident Tregs, especially during inflammation. We did not observe similar trends with arachidonic acid, supporting our in vitro data that LCFAs might be metabolized differently, resulting in different functional effects. Arachidonic acid is known to be metabolized into proinflammatory lipid mediators, and further experiments are needed to understand how these 2 lipids function in Tregs.
We hypothesized that tissue-specific environmental signals allow both adaptation and acquisition of unique, tissue-specific functions and stabilize and promote canonical Treg functions. Without this balance, Tregs can acquire Th-effector properties and lose their suppressive functions, as seen in environments of chronic inflammation (9, 19, 75–77). Considering the lipid-rich environment of most tissues, we posit that environmental lipids, specifically oleic acid, are an important signal in striking a functional balance in tissue-resident Tregs, as our data demonstrate that oleic acid engagement of FAO-driven OXPHOS reinforced canonical regulators of the Treg lineage and Treg-suppressive function. We observed distinct lipid composition profiles in the plasma and adipose tissue compartments between healthy participants and patients with MS, suggesting that there could be an inherent LCFA regulatory or storage defect in MS adipose tissue, or a basal state of inflammation that contributes to the contrasting lipid profiles. A reduction in oleic acid may be one mechanism by which Tregs in patients with MS are more susceptible to dysfunction in environments of chronic inflammation, as the exposure to oleic acid partially restores Treg-suppressive function. Of note, MS donors had a higher average BMI than healthy donors, so dietary and other lifestyle choices could also explain these differences. Although the link between obesity or lipid dysregulation and development of MS is not well understood, childhood obesity has been reported to be a risk factor for MS (78, 79, 91, 92), and Mangalam et al. define metabolic differences, and most significantly lipid alterations, associated with MS disease progression in a relapsing-remitting experimental autoimmune encephalomyelitis (RR-EAE) model (93). Further studies will be needed to better understand the relationship between lipid regulation, obesity, and inflammation in adipose tissue in relation to the risk of autoimmune disease.
Our data reinforce the importance of FAO and OXPHOS in promoting Treg survival and functions (3). However, recent reports revealed different metabolic requirements of Tregs during development versus those for the proper functioning of established Tregs. In human Tregs, OXPHOS and glycolytic engagement upon activation have been reported (41, 51), and it is known that glycolysis must be engaged to prevent enolase-1 suppression of FOXP3 (94). However, Tregs have a lower measured extracellular acidification rate (ECAR) relative to that of other Th-effector cell subsets (95), and CD45RO+ Tregs have greater mitochondrial mass relative to that of CD45RO+ Teffs (51). It can be considered that glycolytic restriction or FAO engagement favors Treg development, then, once the Treg program is established, glycolysis is re-engaged in order to preserve the phenotype. In this regard, enolase-1 has been shown to suppress FOXP3, and acetylation, a by-product of FAO and OXPHOS, enhances FOXP3 stability (94, 96). Nevertheless, FAO-driven OXPHOS is still the major metabolic program, as we have provided evidence that it reinforces Treg stability in existing Treg populations.
Oleic acid–driven FAO and OXPHOS in Tregs drive the expression of FOXP3, CD25, and p-STAT5, all of which act to reinforce the Treg lineage by inducing and reinforcing demethylation of the FOXP3 Treg-specific demethylated region (TSDR), even under inflammatory conditions (25, 27, 97–99). CD25 also increases sensitivity to IL-2, enhances Treg lineage stability via STAT5 occupancy at the FOXP3 enhancer region, and is a critical mechanism by which a stable Treg phenotype is established, especially in the periphery (25, 27). Importantly, the increase in CD25 expression might also serve as a suppressive mechanism, as loss of environmental IL-2 deprives Teffs of a crucial growth factor while simultaneously driving the Treg lineage (100–103).
In summary, we define what we believe to be a new mechanism by which environmental lipids drive cellular-specific metabolic programs that establish a positive feedback loop designed to enhance the stability and function of Tregs via the CD25/STAT5/FOXP3 axis. These signals act to balance inflammatory and tissue-specific cues so that the canonical Treg phenotype and function can be maintained as these cells acquire tissue-specific plasticity. We show that oleic acid partially restored defects in the suppressive function of Tregs isolated from patients with MS, which further suggests the importance of fatty acid species in counteracting inflammatory signals in the tissue. Investigating the crosstalk between Tregs and tissue-resident cell populations and the mechanisms by which Treg metabolic programs orchestrate unique tissue-resident phenotypes and functions will further our understanding of tissue-resident Tregs development and, perhaps, the treatment of autoimmune disorders associated with Treg dysfunction.
Methods
Study design
The objective of this study was to identify the role of environmental lipids in shaping the tissue-resident Treg phenotype. To do this, we used a combination of ex vivo computational analyses and in vitro experimental assays with human Tregs isolated from peripheral blood and adipose tissue. We designed and performed the experiments mainly in the fields of cellular immunology and computational biology.
Study participants
PBMCs for suppression assays were cryopreserved from 8 patients with relapsing-remitting MS (average age, 39 years; minimum age, 32 years; maximum age, 55 years) and 8 healthy individuals (average age, 29 years; minimum age, 21 years; maximum age, 35 years). The patients were diagnosed with either clinically isolated syndrome (CIS) or MS according to the 2010 McDonald Criteria and received no immunomodulatory therapy at the time of the blood draw (Supplemental Table 5). For bulk RNA-Seq, PBMCs were obtained from healthy individuals (average age, 25.4 years; minimum, 21 years; maximum, 33 years) (Supplemental Table 1). Adipose tissue aspirates were obtained from healthy participants (median age, 40.8 ± 12 years; BMI, 24.75 ± 3.16) and patients with MS who were off modulatory therapy for at least 6 months (median age, 49.1 ± 9 years; BMI, 29.5 ± 5.42) (Supplemental Table 3). These same samples were used to quantify LCFA concentrations by mass spectrometry. PBMCs form healthy participants for experiments not matched with PBMCs from patients with MS were freshly isolated.
Human T cell isolation and culturing
PBMCs from donors were isolated by Ficoll-Paque PLUS (GE Healthcare) or Lymphoprep (STEMCELL Technologies) gradient centrifugation. Total CD4+ T cells were isolated by negative magnetic selection using a CD4+ T Cell Isolation Kit (STEMCELL Technologies), and CD4+CD25hiCD127lo-neg Tregs were sorted on a FACSAria (BD Biosciences). Tregs were cultured in RPMI 1640 medium supplemented with 5% human serum, 2 nM l-glutamine, 5 mM HEPES, 100 U/mL penicillin, 100 μg/mL streptomycin, 0.5 mM sodium pyruvate, 0.05 mM nonessential amino acids, and 5% human AB serum (Gemini Bio-Products). Ninety-six-well round-bottomed plates (Corning) were precoated with anti–human CD3 (UCHT1, BD Biosciences, 1 μg/mL) and used for Treg in vitro culturing with soluble anti–human CD28 (BD Biosciences, 28.2, 1 μg/mL) and human IL-2 (50 U/mL). Human IL-2 was obtained through the NIH’s AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID). Th1 Tregs were induced with human recombinant IL-12 (R&D Systems, 20 ng/mL). FFAs (10 μM) were solubilized by DMSO and conjugated in 250 μM BSA–lipid-free RPMI (Gibco, Thermo Fisher Scientific, 27016021) containing oleic acid (MilliporeSigma, O1008), arachidonic acid (MilliporeSigma, A3611), elaidic acid (MilliporeSigma E4637), gondoic acid (MilliporeSigma E3635), and eurcic acid (MilliporeSigma, E3385). The following metabolic inhibitors were used: 50 μM etomoxir (MilliporeSigma, E1905), 250 μM 2-deoxy-d-glucose (2DG) (MilliporeSigma, D6134), 250 μM 5-aminoimidazole-4-carboxamide 1-β-d-ribofuranoside, acadesine, N1-(β-d-ribofuranosyl)-5-aminoimidazole-4-carboxamide (AICAR) (MilliporeSigma, A9978), 1 μM oligomycin A (MilliporeSigma, 75351), 5 μg/mL 5-(tetradecyloxy)-2-furoic acid (TOFA) (MilliporeSigma, T6575), 1 M rotenone (MilliporeSigma, R8875), 1 μM antimycin A (MilliporeSigma, A8674), and 10 mM dimethyl malonate (MilliporeSigma, 136441). The following PPAR inhibitors were used: 5 μM GW9662 (MilliporeSigma, M6191) and 5 μM GW6471 (MilliporeSigma, G5045).
Suppression assay
CD4+CD25+ Tregs were sorted from peripheral blood on a FACSAria Cell Sorter (BD Biosciences) and stimulated with anti-CD3 and anti-CD28 in the presence or absence of IL-12, oleic acid, arachidonic acid, elaidic acid, gondoic acid, eurcic acid, etomoxir, GW9662, or GW6471 for 3 days. The cells were then washed and cocultured with 104 CFSE-labeled responder CD4+CD25lo/negCD127+ T cells at different Treg/Teff ratios. Treg Inspector Beads (Miltenyi Biotec) were used as a stimulus at a 1:2 cell/bead ratio. On day 4, the cocultures were stained to assess viability with LIVE/DEAD Far Red Fixable Viability Dye (Thermo Fisher Scientific) and fixed using FOXP3 staining buffer (eBioscience), and viable responder T cell proliferation was analyzed on a Fortessa Flow Cytometer (BD Biosciences).
qPCR
Total RNA was extracted using the RNeasy Micro Kit (QIAGEN). RNA was treated with DNase and reverse transcribed using TaqMan Reverse Transcription Reagents (Applied Biosystems). cDNAs were amplified with TaqMan probes (TaqMan Gene Expression Arrays) and TaqMan Fast Advanced Master Mix on a StepOne Real-Time PCR System (Applied Biosystems) according to the manufacturer’s instructions. mRNA expression was measured relative to B2M expression. Values are presented as the difference in Ct values normalized to β2-microglobulin for each sample according to the following formula: relative RNA expression = (2−ΔCt) × 1000. The following primers and probes were used for the TaqMan assays: ACACB (Hs00153715), ACADVL (Hs00825606), ACOX1 (Hs01074241), B2M (Hs00187842), CPT1A (Hs00912671), CPT2 (Hs0418816), ECHS1 (Hs00187943), FOXP3 (Hs01085834), HADHA (Hs00426191), PDSS1 (Hs00372008), POLRMT (Hs04187596), PPARG (Hs01115513), and PPRC1 (Hs01563925).
Flow cytometric analysis
PBMCs were prepared and stained with fixable viability dye for 20 minutes at room temperature, followed by staining with surface antibodies for 30 minutes, also at room temperature. For intracellular staining, cells were fixed and permeabilized with the FOXP3 Fix/Perm Buffer Set (eBioscience) for 20 minutes at room temperature, followed by staining with intracellular antibodies. For cytokine staining, cells were stimulated with PMA (50 ng/mL) and ionomycin (250 ng/mL) in the presence of GolgiStop (BD Biosciences) for 4 hours at 37°C. The following antibodies and reagents were used for flow cytometric analysis: anti–CD25 phycoerythrin (PE) (BD Biosciences, M-A251), anti-CD127 APC (BD Biosciences, HIL-7R-M21), anti-FOXP3 (eBioscience, PCH101), anti–FOXP3 exon 2 (BioLegend, AD2), anti–IFN-γ (BioLegend, 4S.B3), anti–IL-10 (Thermo Fisher Scientific, JES3-9D7), anti-CPT1A (Thermo Fisher Scientific, 8F6AE9), and LIVE/DEAD Far Red Fixable Viability Dye (Thermo Fisher Scientific). Stained samples were analyzed with a BD Fortessa Flow Cytometer (BD Biosciences). The number of viable cells analyzed ranged from 3500 to 6500 per experiment. Data were analyzed using FlowJo software (Tree Star).
Phosphorylation staining
Freshly isolated Tregs and Teffs were stimulated for the indicated durations with 50 nM PMA (MilliporeSigma) and 250 nM ionomycin (MilliporeSigma) in the specified conditions. After stimulation, the cells were fixed with BD Cytofix Buffer (BD Biosciences, 554655), followed by a 10-minute incubation at 37°C. Fixed cells were permeabilized with ice-cold BD PhosphoFlow Perm Buffer III (BD Biosciences, 558050) and stained with the mAb STAT5 (Tyr694) (eBioscience, 11-9010-42). After a 45-minute incubation, the cells were washed and acquired with a BD Fortessa Flow Cytometer.
Metabolic assays
For ROS and mitochondrial mass measurements, freshly isolated Tregs and Teffs were stimulated for 3 days as described above, in specified conditions. Cells were then stained with LIVE/DEAD Far Red Fixable Viability Dye at 1:10,000× for 20 minutes at room temperature. Cells were then incubated with MitoTracker Green FM and MitoTracker Orange CM-H2TMRos at 250 nM (Life Technologies, Thermo Fisher Scientific) for 20 minutes at 37°C for 20 minutes and then analyzed by flow cytometry. For the Seahorse assay, 400,000 freshly isolated Tregs or Teffs were stimulated for 3 days as described above, in specified conditions, and the OCR and ECAR were measured on a Seahorse XF96 Analyzer (Agilent Technologies) in the presence of the mitochondrial inhibitor oligomycin (1.5 μM), the mitochondrial uncoupler carbonyl cyanide p-(tri-fluromethoxy)phenyl-hydrazone (FCCP) (1 μM), and the respiratory chain inhibitor antimycin A/rotenone (0.5 μM).
Proliferation assay
Cells were sorted on a FACSAria Cell Sorter, spun down, and resuspended at 106 cells/mL in room-temperature PBS with 0.1% BSA. CFSE was added to the cells at 1 μL/mL, inverted to mix, and immediately incubated at 37°C for 5 minutes. Cells were then incubated in ice-cold RPMI complete media for 10 minutes, after which the cells were spun, counted, and resuspended to the desired numbers.
FOXP3 demethylation
The methylation status of the FOXP3 gene was determined from DNA purified from frozen aliquots of sorted CD25hiCD127lo/neg Tregs cultured in the presence of anti–human CD3, soluble anti–human CD28, and IL-2 in the presence or absence of oleic acid and arachidonic acid as described above. Samples were sent to Epiontis (Berlin, Germany) for bisulfite modification and quantification of TSDR methylation by epigenetic human FOXP3 qPCR assay.
Lentiviral transduction for shRNA gene silencing
Lentiviral plasmids encoding shRNAs were obtained from MilliporeSigma, and all-in-one vectors carrying CTNNB1 sgRNA/Cas9 with a GFP reporter were obtained from Applied Biological Materials. Each plasmid was transformed into One Shot Stbl3 Chemically Competent cells (Invitrogen, Thermo Fisher Scientific) and purified using the ZymoPURE plasmid Maxiprep Kit (Zymo Research). Lentiviral pseudoparticles were obtained after plasmid transfection of 293FT cells using Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific). The lentivirus-containing media were harvested 48 or 72 hours after transfection and 40- to 50-fold concentrated using Lenti-X Concentrator (Takara Clontech). Sorted Tregs were stimulated with plate-bound anti-CD3 (1 μg/mL) and soluble anti-CD28 (1 μg/mL) for 24 hours and transduced with lentiviral particles by spinfection (1000g for 90 minutes at 32°C) in the presence of polybrene (5 μg/mL) on the plates coated with retronectin (50 μg/mL) (Clontech, Takara Bio) and anti-CD3 (1–2 μg/mL). Human Tregs were directly transduced with lentiviral particles by spinfection. Twenty-four hours after transduction, 10 μM oleic acid was added to culture. Five days after transduction, cells were sorted according to the expression of GFP, and gene expression was measured using the qPCR methods described above. The shRNA constructs for gene knockdown were: CPT1A (TRCN0000036282) and ACADVL (TRCN0000245178) (MilliporeSigma).
Mass spectrometric analysis of FFAs
Fatty acid concentrations.
Nonesterified fatty acid (NEFA) concentrations in plasma and WAT (2:1 chloroform/methanol extraction) were determined using the Wako HR Series NEFA-HR2 in vitro enzymatic colorimetric assay (Fufjifilm, Wako Diagnostics).
Fatty acid profiles.
Plasma and adipose tissue were extracted into chloroform/methanol (2:1), and NEFAs were isolated using a Kinesis TELOS aminopropyl-NH2 weak anion exchange column (200 mg, 3 mL) to determine fatty acid profiles in the samples. Neutral lipids were eluted with chloroform-2-propanol (2:1), and the NEFA fraction was then eluted with 2% formic acid in diethyl ether, dried under N2 gas, and derivatized to the fatty acid methyl ester with boron trifluoride methanol 14% (MilliporeSigma). Fatty acid methyl esters (C14 to C18) were determined by gas chromatography–mass spectrometry (GC-MS) analysis (chemical ionization [CI] mode) and selective ion monitoring of masses 241 to 299 with an HP-1 column (25 m, 0.2 mm ID, 0.33 μm film) at a temperature gradient from 100°C to 220°C. The concentration of each LCFA in μmol/g was determined by multiplying the LCFA concentration (μmol/mL) by the sample volume, divided by the weight of each sample (mg), multiplied by 1000. The ratio of each LCFA per sample was generated by dividing the concentration of each LCFA by the sum of LCFA concentrations measured in each sample. The sum of each value for LCFAs measured per sample equals 1.
RNA-Seq library preparation and data analysis
Preparation of cells for RNA-Seq.
For bulk RNA-Seq analysis, CD4+CD25hiCD127lo/neg Tregs from healthy donors were sorted by FACS and cultured with 10 μM oleic acid, 10 μM arachidonic acid, or control conditions in the presence of anti–human CD3 (UCHT1, 1 μg/mL), soluble anti–human CD28 (28.2, 1 μg/mL), and anti–human IL-2 (50 U/mL) for 3 days as described above. Cells were harvested and RNA isolated using the RNeasy Micro Kit (QIAGEN) followed by immediate processing for cDNA preparation. Samples were collected from 9 healthy individuals for identification of FFA-induced signatures.
For single-cell sequencing, both peripheral blood and adipose tissue were collected from healthy participants and patients with MS. PBMCs were isolated from peripheral blood, and Tregs were single sorted into 96-well plates by FACS using the CD4+CD25hiCD127lo/neg antibodies described above. For adipose tissue, we first performed collagenase digestion (104). Next, the lymphocytes were isolated from the sample by Ficoll gradient and centrifugation (as done for PBMC isolation) at a 9 mL sample/4 mL Ficoll ratio. The lymphocyte layer was removed and washed, and the Tregs were then single sorted by FACS using the CD4+CD25hiCD127lo/neg antibodies described above.
cDNA and library preparation and sequencing.
cDNA was generated from sorted and harvested cells for bulk RNA-Seq using the SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing (Clontech, Takara Bio). Barcoded libraries were generated using the Nextera XT DNA Library Preparation Kit (Illumina) and sequenced with a 2 × 100 bp paired-end protocol on the HiSeq 2000 Sequencing System (Illumina).
Bulk RNA-Seq data analysis.
Libraries were pseudoaligned with Kallisto (version 0.46.0) (105) to the Ensembl (version 96) human transcriptome. Transcripts per million (TPM) were aggregated and summed for gene-level analysis and then log2-transformed; non–protein-coding transcripts were excluded. This resulted in 3 gene expression vectors per donor for Tregs stimulated in the presence of oleic acid, arachidonic acid, or none (control). To mitigate donor-specific effects, we computed 2 vectors per donor by subtracting the control vector from either treatment (note that subtraction was done in log space, and it therefore represents the ratio between the original signals). Coordinates of these vectors were z scaled for the purpose of principal component analysis (PCA) computation. An alternative analysis was performed by subtracting a donor’s arachidonic acid vector from their oleic acid vector. The limma R package (106) was used to fit a linear regression model, with the vector as the dependent variable, an intercept, and 2 sum-to-zero nuisance covariates to regress out the division of the donors into the 3 collection batches, followed by an empirical Bayes moderation of the gene-wise sample variances with mean variance trend (the limma trend method, as described in ref. 107). A moderated t test was used to determine whether the intercept coefficient, which is supposed to capture the differential effect on that gene (comparing between oleic acid and arachidonic acid), was significantly different than zero.
Single-cell RNA-Seq data analysis.
Single-cell libraries were pseudoaligned as described above for bulk libraries and processed with the Seurat sctransform workflow, version 3 (108). We identified non-Treg cell clusters in the data, excluded them, and repeated the processing to eliminate their effect on the subsequent workflow. DEGs were called with a Wilcoxon rank-sum test as implemented in Seurat’s FindAllMarkers function with default parameters.
We defined a transcriptomic signature based on the in vitro bulk RNA data. As described above, we defined the top-250 genes with the lowest (most negative) loadings with respect to PC1 of the bulk data as the oleic acid–related module and the top-250 genes with the highest loading as the arachidonic acid module. The score for a single cell is the dot product of its normalized gene expression vector with a vector that contains –1 for the oleic acid module genes, +1 for arachidonic acid module genes, and 0 otherwise. The resulting scores were rank transformed and then scaled to a [–1,+1] range, with –1 representing the most oleic acid–like and +1 the most arachidonic acid–like cell in the data.
Sequencing data.
The bulk and single-cell sequencing data from this study were deposited in the NCBI’s Gene Expression Omnibus (GEO) database (GEO GSE152543).
Statistics
Significance was determined by a paired, 2-tailed Student’s t test and corrected for multiple hypothesis testing using the Holm-Sidak method. All statistical tests were performed with GraphPad Prism, version 8 (GraphPad Software). Data are shown as the mean ± SEM. P values of 0.05 or less were considered significant.
Study approval
All experiments were approved by the Yale Institutional Review Board, and all subjects provided written informed consent prior to participation in the study. The study conformed to the principles set out in the World Medical Association (WMA) Declaration of Helsinki and the US Department of Health and Human Services Belmont Report.
Author contributions
SLP performed in vitro experiments with the help of MDV. AW and NY analyzed the RNA-Seq data. JL and AK designed protocols, collected, and sequenced human adipose tissue samples. SLP performed data analysis and wrote the manuscript with AW under the supervision of NY, MDV, and DAH. MDV and DAH supervised the overall study.
Supplementary Material
Acknowledgments
We would like to thank Gary Cline and the Mouse Metabolic Phenotyping Center at the Yale University School of Medicine for mass spectrometric analysis of fat supernatant and serum samples, and members at the Yale Center for Genome Analysis for cDNA library preparation and sequencing of samples. We also wish to thank Erin Longbrake and Sarah Wesley for obtaining adipose aspirates from the healthy participants and patients with MS. This work was supported by grants to DAH from the NIH (U19 AI089992, R25 NS079193, P01 AI073748, U24 AI11867, R01 AI22220, UM 1HG009390, P01 AI039671, P50 CA121974, and R01 CA227473); the National Multiple Sclerosis Society (NMSS) (CA 1061- A-18, RG-1802-30153); the Nancy Taylor Foundation for Chronic Diseases; and Race to Erase MS. This study was supported by a training grant from the NIH (T32AI07019) and the Yale Interdisciplinary Immunobiology Training Program.
Version 1. 11/10/2020
In-Press Preview
Version 2. 01/19/2021
Print issue publication
Footnotes
Conflict of interest: DAH has received funding for his laboratory from Bristol Myers Squibb and Genentech. A child of DAH is an employee of Sanofi. Further information regarding funding is available at: https://openpaymentsdata.cms.gov/physician/166753/general-payments.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(2):e138519.https://doi.org/10.1172/JCI138519.
Contributor Information
Saige L. Pompura, Email: saige.pompura@yale.edu.
Allon Wagner, Email: revsor@gmail.com.
Alexandra Kitz, Email: alexandra.kitz@yale.edu.
Jacob LaPerche, Email: jacoblaperche@gmail.com.
Nir Yosef, Email: niryosef@eecs.berkeley.edu.
Margarita Dominguez-Villar, Email: m.dominguez-villar@imperial.ac.uk.
References
- 1.Warburg O, et al. The metabolism of tumors in the body. J Gen Physiol. 1927;8(6):519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heiden MGV, et al. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michalek RD, et al. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol. 2011;186(6):3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerriets VA, et al. Foxp3 and Toll-like receptor signaling balance Treg cell anabolic metabolism for suppression. Nat Immunol. 2016;17(12):1459–1466. doi: 10.1038/ni.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angelin A, et al. Foxp3 reprograms T cell metabolism to function in low-glucose, high-lactate environments. Cell Metab. 2017;25(6):1282–1293.e7. doi: 10.1016/j.cmet.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Travis MA, Sheppard D. TGF-β activation and function in immunity. Annu Rev Immunol. 2014;32:51–82. doi: 10.1146/annurev-immunol-032713-120257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie M, et al. A pivotal role for endogenous TGF-beta-activated kinase-1 in the LKB1/AMP-activated protein kinase energy-sensor pathway. Proc Natl Acad Sci U S A. 2006;103(46):17378–17383. doi: 10.1073/pnas.0604708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gualdoni GA, et al. The AMP analog AICAR modulates the Treg/Th17 axis through enhancement of fatty acid oxidation. FASEB J. 2016;30(11):3800–3809. doi: 10.1096/fj.201600522R. [DOI] [PubMed] [Google Scholar]
- 9.Dominguez-Villar M, Baecher-Allan CMD. Identification of T helper type 1-like, Foxp3+ regulatory T cells in human autoimmune disease. Nat Med. 2011;17(6):673–675. doi: 10.1038/nm.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feuerer M, et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat Med. 2009;15(8):930–939. doi: 10.1038/nm.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng WF, et al. Human CD4(+)CD25(+) cells: a naturally occurring population of regulatory T cells. Blood. 2001;98(9):2736–2744. doi: 10.1182/blood.V98.9.2736. [DOI] [PubMed] [Google Scholar]
- 12.Burzyn D, et al. A special population of regulatory T cells potentiates muscle repair. Cell. 2013;155(6):1282–1295. doi: 10.1016/j.cell.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmadzadeh M, et al. Tumor-infiltrating human CD4+ regulatory T cells display a distinct TCR repertoire and exhibit tumor and neoantigen reactivity. Sci Immunol. 2019;4(31):aao4310. doi: 10.1126/sciimmunol.aao4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Simone M, et al. Transcriptional landscape of human tissue lymphocytes unveils uniqueness of tumor-infiltrating T regulatory cells. Immunity. 2016;45(5):1135–1147. doi: 10.1016/j.immuni.2016.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deiuliis J, et al. Visceral adipose inflammation in obesity is associated with critical alterations in tregulatory cell numbers. PLoS One. 2011;6(1):e16376. doi: 10.1371/journal.pone.0016376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cipolletta D, et al. Appearance and disappearance of the mRNA signature characteristic of Treg cells in visceral adipose tissue: age, diet, and PPARγ effects. Proc Natl Acad Sci U S A. 2015;112(2):482–487. doi: 10.1073/pnas.1423486112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitz A, Dominguez-Villar M. Molecular mechanisms underlying Th1-like Treg generation and function. Cell Mol Life Sci. 2017;74(22):4059–4075. doi: 10.1007/s00018-017-2569-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitz A, et al. AKT isoforms modulate Th1-like Treg generation and function in human autoimmune disease. EMBO Reports. 2016;17(8):1169–1183. doi: 10.15252/embr.201541905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McClymont SA, et al. Plasticity of human regulatory T cells in healthy subjects and patients with type 1 diabetes. J Immunol. 2011;186(7):3918–3926. doi: 10.4049/jimmunol.1003099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butcher MJ, et al. Atherosclerosis-driven Treg plasticity results in formation of a dysfunctional subset of plastic IFNγ+ Th1/Tregs. Circ Res. 2016;119(11):1190–1203. doi: 10.1161/CIRCRESAHA.116.309764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rivas MN, et al. Regulatory T cell reprogramming toward a Th2-cell-like lineage impairs oral tolerance and promotes food allergy. Immunity. 2015;42(3):512–523. doi: 10.1016/j.immuni.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panduro M, et al. Tissue Tregs. Annu Rev Immunol. 2016;34:609–633. doi: 10.1146/annurev-immunol-032712-095948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fontenot JD, et al. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4(4):330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 24.Hori S, et al. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299(5609):1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 25.Feng Y, et al. Control of the inheritance of regulatory T cell identity by a cis element in the Foxp3 locus. Cell. 2014;158(4):749–763. doi: 10.1016/j.cell.2014.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iizuka-Koga M, et al. Induction and maintenance of regulatory T cells by transcription factors and epigenetic modifications. J Autoimmun. 2017;83:113–121. doi: 10.1016/j.jaut.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Yao Z, et al. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 2007;109(10):4368–4375. doi: 10.1182/blood-2006-11-055756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corsetto PA, et al. Effects of n-3 PUFAs on breast cancer cells through their incorporation in plasma membrane. Lipids Health Dis. 2011;10:73. doi: 10.1186/1476-511X-10-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kambe T, et al. Polyunsaturated fatty acids potentiate interleukin-1-stimulated arachidonic acid release by cells overexpressing type IIA secretory phospholipase A2. FEBS Lett. 1999;453(1-2):81–84. doi: 10.1016/S0014-5793(99)00702-4. [DOI] [PubMed] [Google Scholar]
- 30.Guest J, et al. Relationship between central and peripheral fatty acids in humans. Lipids Health Dis. 2013;12:79. doi: 10.1186/1476-511X-12-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pilitsis JG, et al. Quantification of free fatty acids in human cerebrospinal fluid. Neurochem Res. 2001;26(12):1265–1270. doi: 10.1023/A:1014227231130. [DOI] [PubMed] [Google Scholar]
- 32.Takigawa H, et al. Deficient production of hexadecenoic acid in the skin is associated in part with the vulnerability of atopic dermatitis patients to colonization by Staphylococcus aureus. Dermatology. 2005;211(3):240–248. doi: 10.1159/000087018. [DOI] [PubMed] [Google Scholar]
- 33.Body DR. The lipid composition of adipose tissue. Prog Lipid Res. 1988;27(1):39–60. doi: 10.1016/0163-7827(88)90004-5. [DOI] [PubMed] [Google Scholar]
- 34.Howie D, et al. Foxp3 drives oxidative phosphorylation and protection from lipotoxicity. JCI Insight. 2017;2(3):e89160. doi: 10.1172/jci.insight.89160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carrillo C, et al. Role of oleic acid in immune system; mechanism of action: a review. Nutr Hosp. 2012;27(4):978–990. doi: 10.3305/nh.2012.27.4.5783. [DOI] [PubMed] [Google Scholar]
- 36.Angela M, et al. Fatty acid metabolic reprogramming via mTOR-mediated inductions of PPARγ directs early activation of T cells. Nat Commun. 2016;7:13683. doi: 10.1038/ncomms13683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rueda CM, et al. High density lipoproteins selectively promote the survival of human regulatory T cells. J Lipid Res. 2017;58(8):1514–1523. doi: 10.1194/jlr.M072835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Jong AJM. Fatty acids, lipid mediators, and T-cell function. Front Immunol. 2015;5:483. doi: 10.3389/fimmu.2014.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cury-Boaventura MF, et al. Comparative toxicity of oleic and linoleic acid on human lymphocytes. Life Sci. 2006;78(13):1448–1456. doi: 10.1016/j.lfs.2005.07.038. [DOI] [PubMed] [Google Scholar]
- 40.Passos MEP, et al. Differential effects of palmitoleic acid on human lymphocyte proliferation and function. Lipids Health Dis. 2016;15(1):217. doi: 10.1186/s12944-016-0385-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Procaccini C, et al. The proteomic landscape of human ex vivo regulatory and conventional T cells reveals specific metabolic requirements. Immunity. 2016;44(2):406–421. doi: 10.1016/j.immuni.2016.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beier UH, et al. Essential role of mitochondrial energy metabolism in Foxp3(+) T-regulatory cell function and allograft survival. FASEB J. 2015;29(6):2315–2326. doi: 10.1096/fj.14-268409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ellis GI, et al. Mitochondrial and cytosolic roles of PINK1 shape induced regulatory T-cell development and function. Eur J Immunol. 2013;43(12):3355–3360. doi: 10.1002/eji.201343571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Divakaruni AS, et al. Etomoxir inhibits macrophage polarization by disrupting CoA homeostasis. Cell Metab. 2018;28(3):490–503.e7. doi: 10.1016/j.cmet.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Connor RS, et al. The CPT1a inhibitor, etomoxir induces severe oxidative stress at commonly used concentrations. Sci Rep. 2018;8(1):6289. doi: 10.1038/s41598-018-24676-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berod L, et al. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nature Medicine. 2014;20(11):1327–1333. doi: 10.1038/nm.3704. [DOI] [PubMed] [Google Scholar]
- 47.Varga T, et al. PPARs are a unique set of fatty acid regulated transcription factors controlling both lipid metabolism and inflammation. Biochim Biophys Acta. 2011;1812(8):1007–1022. doi: 10.1016/j.bbadis.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seimandi M, et al. Differential responses of PPARalpha, PPARdelta, and PPARgamma reporter cell lines to selective PPAR synthetic ligands. Anal Biochem. 2005;344(1):8–15. doi: 10.1016/j.ab.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 49.Leesnitzer LM, et al. Functional consequences of cysteine modification in the ligand binding sites of peroxisome proliferator activated receptors by GW9662. Biochemistry. 2002;41(21):6640–6650. doi: 10.1021/bi0159581. [DOI] [PubMed] [Google Scholar]
- 50.Xu HE, et al. Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha. Nature. 2002;415(6873):813–817. doi: 10.1038/415813a. [DOI] [PubMed] [Google Scholar]
- 51.Cluxton D, et al. Differential regulation of human Treg and Th17 cells by fatty acid synthesis and glycolysis. Front Immunol. 2019;10:115. doi: 10.3389/fimmu.2019.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Joly AL, et al. Foxp3 lacking exons 2 and 7 is unable to confer suppressive ability to regulatory T cells in vivo. J Autoimmun. 2015;63:23–30. doi: 10.1016/j.jaut.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 53.Wang Q, et al. Deletion of the exon 2 of mouse Foxp3 results in systemic lupus erythematosus-like disease. J Immunol. 2016;196(1 Suppl):118.24 [Google Scholar]
- 54.Du JG, et al. Exon 2 of Foxp3 contributes to the peripheral tolerance of the immune system. J Immunol. 2016;196(1 Suppl):191.23 [Google Scholar]
- 55.Yang R, et al. Hydrogen sulfide promotes Tet1- and Tet2-mediated Foxp3 demethylation to drive regulatory T cell differentiation and maintain immune homeostasis. Immunity. 2015;43(2):251–263. doi: 10.1016/j.immuni.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neess D, et al. Long-chain acyl-CoA esters in metabolism and signaling: role of acyl-CoA binding proteins. Prog Lipid Res. 2015;59:1–25. doi: 10.1016/j.plipres.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 57.Moffat C, et al. Acyl-CoA thioesterase-2 facilitates mitochondrial fatty acid oxidation in the liver. J Lipid Res. 2014;55(12):2458–2470. doi: 10.1194/jlr.M046961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lopes-Marques M, et al. Diversity and history of the long-chain acyl-CoA synthetase (Acsl) gene family in vertebrates. BMC Evol Biol. 2013;13:271. doi: 10.1186/1471-2148-13-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamashita A, et al. Acyltransferases and transacylases that determine the fatty acid composition of glycerolipids and the metabolism of bioactive lipid mediators in mammalian cells and model organisms. Prog Lipid Res. 2014;53:18–81. doi: 10.1016/j.plipres.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 60.Temprano A, et al. Redundant roles of the phosphatidate phosphatase family in triacylglycerol synthesis in human adipocytes. Diabetologia. 2016;59(9):1985–1994. doi: 10.1007/s00125-016-4018-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li YW, et al. LETM1 is required for mitochondrial homeostasis and cellular viability (Review) Mol Med Rep. 2019;19(5):3367–3375. doi: 10.3892/mmr.2019.10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yi W, et al. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science. 2012;337(6097):975–980. doi: 10.1126/science.1222278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Najjar VA, Pullman ME. The occurrence of a group transfer involving enzyme (phosphoglucomutase) and substrate. Science. 1954;119(3097):631–634. doi: 10.1126/science.119.3097.631. [DOI] [PubMed] [Google Scholar]
- 64.Pouyssegur J, et al. Induction of two transformation-sensitive membrane polypeptides in normal fibroblasts by a block in glycoprotein synthesis or glucose deprivation. Cell. 1977;11(4):941–947. doi: 10.1016/0092-8674(77)90305-1. [DOI] [PubMed] [Google Scholar]
- 65.Thorens B, Mueckler M. Glucose transporters in the 21st century. Am J Physiol Endocrinol Metab. 2010;298(2):E141–E145. doi: 10.1152/ajpendo.00712.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nasrin N, et al. SIRT4 regulates fatty acid oxidation and mitochondrial gene expression in liver and muscle cells. J Biol Chem. 2010;285(42):31995–32002. doi: 10.1074/jbc.M110.124164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Senga S, et al. Fatty acid-binding protein 5 (FABP5) promotes lipolysis of lipid droplets, de novo fatty acid (FA) synthesis and activation of nuclear factor-kappa B (NF-κB) signaling in cancer cells. Biochim Biophys Acta Mol Cell Biol Lipids. 2008;1863(9):1057–1067. doi: 10.1016/j.bbalip.2018.06.010. [DOI] [PubMed] [Google Scholar]
- 68.Field CS, et al. Mitochondrial integrity regulated by lipid metabolism is a cell-intrinsic checkpoint for Treg suppressive function. Cell Metab. 2020;31(2):422–437.e5. doi: 10.1016/j.cmet.2019.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cibrian D, Sanchez-Madrid F. CD69: from activation marker to metabolic gatekeeper. Eur J Immunol. 2017;47(6):946–953. doi: 10.1002/eji.201646837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bierer BE, et al. Synergistic T cell activation via the physiological ligands for CD2 and the T cell receptor. J Exp Med. 1988;168(3):1145–1156. doi: 10.1084/jem.168.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rocha-Perugini V, et al. Tetraspanins CD9 and CD151 at the immune synapse support T-cell integrin signaling. Eur J Immunol. 2014;44(7):1967–1975. doi: 10.1002/eji.201344235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gibson SA, et al. Protein kinase CK2 controls the fate between Th17 cell and regulatory T cell differentiation. J Immunol. 2017;198(11):4244–4254. doi: 10.4049/jimmunol.1601912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gibson SA, et al. CK2 controls Th17 and regulatory T cell differentiation through inhibition of FoxO1. J Immunol. 2018;201(2):383–392. doi: 10.4049/jimmunol.1701592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peng M, et al. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science. 2016;354(6311):481–484. doi: 10.1126/science.aaf6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.MacDonald KG, et al. Regulatory T cells produce profibrotic cytokines in the skin of patients with systemic sclerosis. J Allergy Clin Immunol. 2015;135(4):946–e9. doi: 10.1016/j.jaci.2014.12.1932. [DOI] [PubMed] [Google Scholar]
- 76.Nakagawa H, et al. Instability of helios-deficient Tregs is associated with conversion to a T-effector phenotype and enhanced antitumor immunity. Proc Natl Acad Sci U S A. 2016;113(22):6248–6253. doi: 10.1073/pnas.1604765113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bovenschen HJ, et al. Foxp3+ regulatory T cells of psoriasis patients easily differentiate into IL-17A-producing cells and are found in lesional skin. J Invest Dermatol. 2011;131(9):1853–1860. doi: 10.1038/jid.2011.139. [DOI] [PubMed] [Google Scholar]
- 78.Gianfrancesco MA, et al. Evidence for a causal relationship between low vitamin D, high BMI, and pediatric-onset MS. Neurology. 2017;88(17):1623–1629. doi: 10.1212/WNL.0000000000003849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gianfrancesco MA, et al. Causal effect of genetic variants associated with body mass index on multiple sclerosis susceptibility. Am J Epidemiol. 2017;185(3):162–171. doi: 10.1093/aje/kww120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wagner A, et al. Drugs that reverse disease transcriptomic signatures are more effective in a mouse model of dyslipidemia. Mol Syst Biol. 2015;11(3):791. doi: 10.15252/msb.20145486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bailis W, et al. Distinct modes of mitochondrial metabolism uncouple T cell differentiation and function. Nature. 2019;571(7765):403–407. doi: 10.1038/s41586-019-1311-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sefik E, et al. Mucosal immunology. Individual intestinal symbionts induce a distinct population of RORγ+ regulatory T cells. Science. 2015;49(6251):993–997. doi: 10.1126/science.aaa9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sather BD, et al. Altering the distribution of Foxp3(+) regulatory T cells results in tissue-specific inflammatory disease. J Exp Med. 2007;204(6):1335–1347. doi: 10.1084/jem.20070081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dudda JC, et al. Foxp3+ regulatory T cells maintain immune homeostasis in the skin. J Exp Med. 2008;205(7):1559–1565. doi: 10.1084/jem.20072594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Villalta SA, et al. Regulatory T cells suppress muscle inflammation and injury in muscular dystrophy. Sci Transl Med. 2014;6(258):258ra142. doi: 10.1126/scitranslmed.3009925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kuswanto W, et al. Poor repair of skeletal muscle in aging mice reflects a defect in local, interleukin-33-dependent accumulation of regulatory T cells. Immunity. 2016;44(22):355–367. doi: 10.1016/j.immuni.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Scharschmidt TC, et al. A wave of regulatory T cells into neonatal skin mediates tolerance to commensal microbes. Immunity. 2015;43(5):1011–1021. doi: 10.1016/j.immuni.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Belkaid Y, et al. CD4+CD25+ regulatory T cells control Leishmania major persistence and immunity. Nature. 2002;420(6915):502–507. doi: 10.1038/nature01152. [DOI] [PubMed] [Google Scholar]
- 89.Malhotra N, et al. RORα-expressing T regulatory cells restrain allergic skin inflammation. Sci Immunol. 2018;3(21):eaao6923. doi: 10.1126/sciimmunol.aao6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Halabi-Tawil M, et al. Cutaneous manifestations of immune dysregulation, polyendocrinopathy, enteropathy, X-linked (IPEX) syndrome. Br J Dermatol. 2009;160(3):645–651. doi: 10.1111/j.1365-2133.2008.08835.x. [DOI] [PubMed] [Google Scholar]
- 91.Gianfrancesco MA, et al. Increased body mass index is causally associated with pediatric MS onset: a Mendelian randomization study. Neurology. 2016;86(Suppl 1):P1.375 [Google Scholar]
- 92.Gianfrancesco MA, et al. Obesity and multiple sclerosis susceptibility: a review. J Neurol Neuromedicine. 2016;1(7):1–5. doi: 10.29245/2572.942X/2016/7.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Giri S, et al. Profile of circulatory metabolites in a relapsing-remitting animal model of multiple sclerosis using global metabolomics. J Clin Cell Immunol. 2014;192(1 Suppl):136.9. doi: 10.4172/2155-9899.1000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.De Rosa V, et al. Glycolysis controls the induction of human regulatory T cells by modulating the expression of FOXP3 exon 2 splicing variants. Nat Immunol. 2015;16(11):1174–1184. doi: 10.1038/ni.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gerriets VA, et al. Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J Clin Invest. 2015;125(1):194–207. doi: 10.1172/JCI76012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.van Loosdregt J, Coffer PJ. Post-translational modification networks regulating FOXP3 function. Trends Immunol. 2014;35(8):368–378. doi: 10.1016/j.it.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 97.Lio CWJ, Hsieh CS. A two-step process for thymic regulatory T cell development. Immunity. 2008;28(1):100–111. doi: 10.1016/j.immuni.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hill JA, et al. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 2007;27(5):786–800. doi: 10.1016/j.immuni.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 99.Kashiwagi I, et al. Smad2 and Smad3 inversely regulate TGF-β autoinduction in Clostridium butyricum-activated dendritic cells. Immunity. 2015;43(1):65–79. doi: 10.1016/j.immuni.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 100.Yamaguchi T, et al. Two modes of immune suppression by Foxp3(+) regulatory T cells under inflammatory or non-inflammatory conditions. Semin Immunol. 2011;23(6):424–430. doi: 10.1016/j.smim.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 101.Azizi E, et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. 2018;174(5):1293–1308.e36. doi: 10.1016/j.cell.2018.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Plitas G, et al. Regulatory T cells exhibit distinct features in human breast cancer. Immunity. 2016;45(5):1122–1134. doi: 10.1016/j.immuni.2016.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Saito T, et al. Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat Med. 2016;22(6):679–684. doi: 10.1038/nm.4086. [DOI] [PubMed] [Google Scholar]
- 104.Orr JS, et al. Isolation of adipose tissue immune cells. J Vis Exp. 2013;(75):e50707. doi: 10.3791/50707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bray NL, et al. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34(5):525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 106.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Law CW, et al. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20(1):296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.