Abstract
Genetic factors undoubtedly affect the development of congenital heart disease (CHD) but still remain ill defined. We sought to identify genetic risk factors associated with CHD and to accomplish a functional analysis of SNP-carrying genes. We performed a genome-wide association study (GWAS) of 4034 White patients with CHD and 8486 healthy controls. One SNP on chromosome 5q22.2 reached genome-wide significance across all CHD phenotypes and was also indicative for septal defects. One region on chromosome 20p12.1 pointing to the MACROD2 locus identified 4 highly significant SNPs in patients with transposition of the great arteries (TGA). Three highly significant risk variants on chromosome 17q21.32 within the GOSR2 locus were detected in patients with anomalies of thoracic arteries and veins (ATAV). Genetic variants associated with ATAV are suggested to influence the expression of WNT3, and the variant rs870142 related to septal defects is proposed to influence the expression of MSX1. We analyzed the expression of all 4 genes during cardiac differentiation of human and murine induced pluripotent stem cells in vitro and by single-cell RNA-Seq analyses of developing murine and human hearts. Our data show that MACROD2, GOSR2, WNT3, and MSX1 play an essential functional role in heart development at the embryonic and newborn stages.
Keywords: Cardiology, Genetics
Keywords: Cardiovascular disease, Molecular genetics, iPS cells
Introduction
Congenital heart disease (CHD) accounts for approximately 28% of all congenital anomalies worldwide (1), with a CHD frequency of 9.1 per 1000 live births (2). Currently, CHD represents a major global health challenge, causing more than 200,000 deaths worldwide per year (3).
Although major progress has been made in the field of genetics during the past few decades, the exact etiologic origins of CHD still remain only partially understood. Causal genes have been identified in uncommon syndromic forms, such as TBX5 for Holt-Oram syndrome (4). CHD may also be associated with major chromosomal syndromes (5), de novo mutations (6), aneuploidy, and copy number variants (7–9). Each of these genetic abnormalities is associated with roughly 10% of CHDs, while the majority of cases seem to represent a complex multifactorial disease with unknown etiology (9). Studies have implicated an increasing number of candidate genes in causing CHD (10–12), and genetic variations suggest obvious heterogeneity (13–15). Furthermore, these studies strongly support the idea that certain variants are inherited and may cause a pronounced pathology.
Several genome-wide association studies (GWAS) have previously been conducted to determine potential genetic risk factors for CHD (14, 16–19). For atrial septal defects (ASDs), 4p16 was identified as a risk locus (19, 20). For tetralogy of Fallot (TOF), regions of interest have been reported on chromosomes 1, 12, and 13 (21, 22). Agopian and colleagues have shown an association of a single intragenetic SNP with left ventricular obstructive defects (16). For other major clinical subcategories, no risk loci have been identified to date.
We sought to identify genetic risk loci in CHD and clinical subpopulations thereof by GWAS, given the proven success of this approach (23). We conducted GWAS in more than 4000 unrelated White patients diagnosed with CHD who were classified according to the standards and categories defined by the Society of Thoracic Surgeons (STS) (24, 25). We identified 1 risk variant for CHD in general and detected an association of single or clustered SNPs in 5 major subpopulations. We determined risk loci in patients with transposition of the great arteries (TGA) and anomalies of the thoracic arteries and veins (ATAV). In addition, we demonstrate differential expression of candidate genes during differentiation of murine and human pluripotent stem cells and determined their expression in pediatric and adult aortic and atrial tissue. Finally, we document the functional role of candidate genes by single-cell RNA-Seq (scRNA-Seq) analyses in the developing murine and human heart in vivo.
Results
Association analysis in the overall population of patients with CHD and subgroups defined by STS classification.
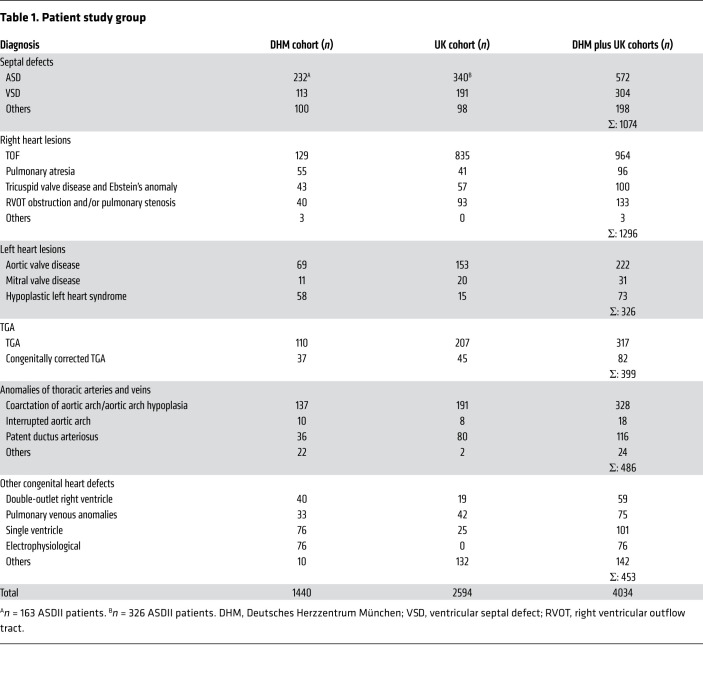
We performed a GWAS in 4034 patients with CHD (n = 2089 males, n = 1945 females) and 8486 controls (n = 4224 males, n = 4262 females) to detect possible candidate SNPs. The first group consisted of 1440 patients treated at the German Heart Center Munich. Data on 2 additional groups of 2594 patients have previously been published (19, 21). To obtain clearly defined clinical subgroups of patients, we classified all patients with CHD according to the STS Congenital Heart Surgery Database (CHSD) recommendations. This classification was established under the leadership of the International Society for Nomenclature of Pediatric and Congenital Heart Disease as a clinical data registry but also reflects common developmental etiologies and is therefore a well accepted tool for research on CHD (22, 23). The distribution of the subgroups is shown in Table 1.
Table 1. Patient study group.
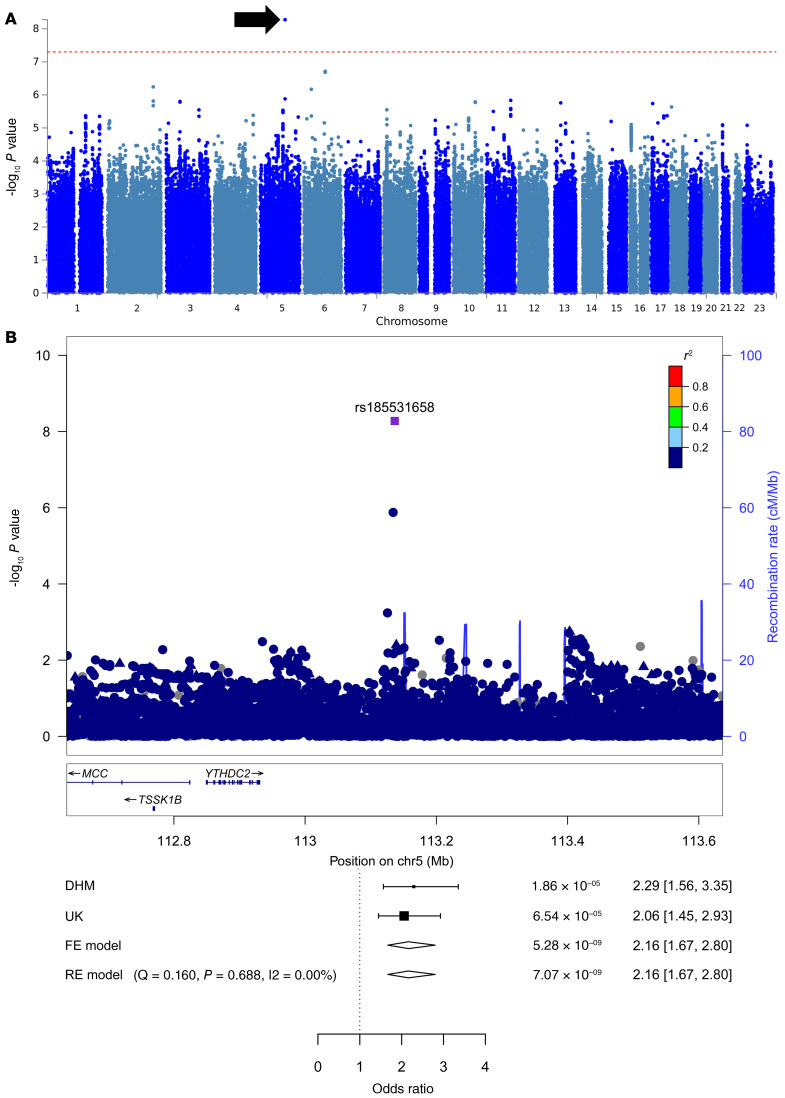
We first performed an analysis across all 4034 patients with CHD and identified 1 SNP on chromosome 5 with genome-wide significance (rs185531658; Figure 1). To exclude a false-positive signal due to genotyping errors, we validated this variation on all SNP-carrying patients by Sanger sequencing and confirmed it in more than 95% of the samples. Two representative chromatograms of patients carrying the identified SNP and chromatograms of 2 WT patients are shown in Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI141837DS1 In terms of P values, this signal was mostly driven by the septal defects, however, we cannot assume this locus to be a septal defect–specific locus based on our data.
Figure 1. Identification of SNPs with genome-wide significance across the entire CHD study group.
(A) Manhattan plot of genome-wide P values for association with the entire CHD study group. (B) LocusZoom plot of the genomic region of rs185531658 on chromosome 5 (chr5). The index SNPs are indicated by a purple diamonds. The forest plot shows the significance of the SNP and the ORs of both collectives separately and together. Circles represent imputed SNPs and triangles represent genotyped SNPs. FE, fixed effects; RE, random effects. Patients with CHD, n = 2594; control participants, n = 8486. –log10 P values were determined by association statistics from the GWAS (logistic regression).
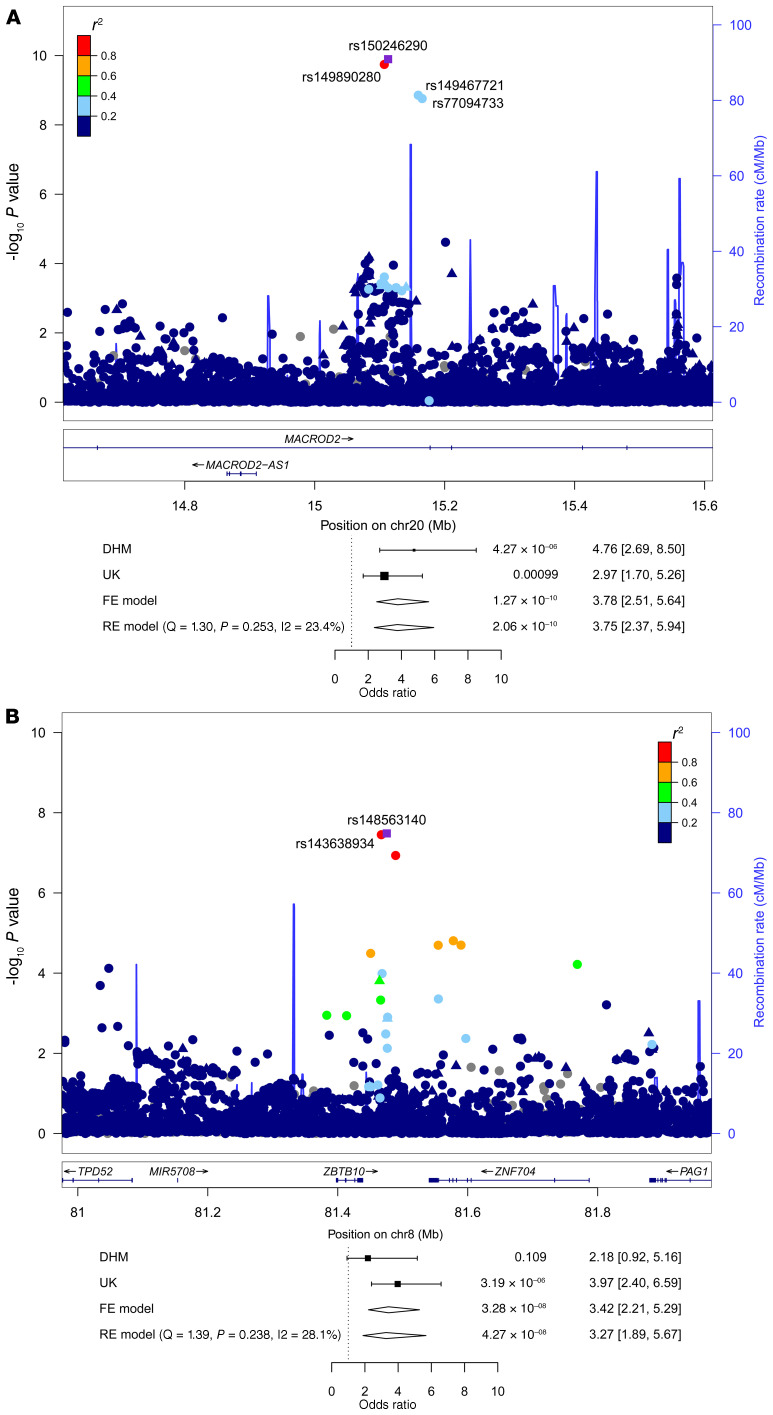
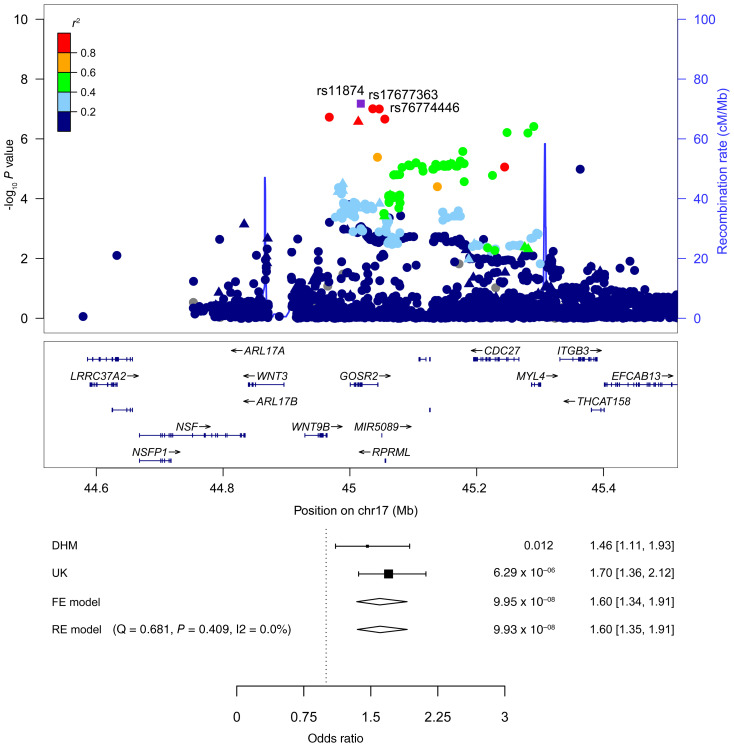
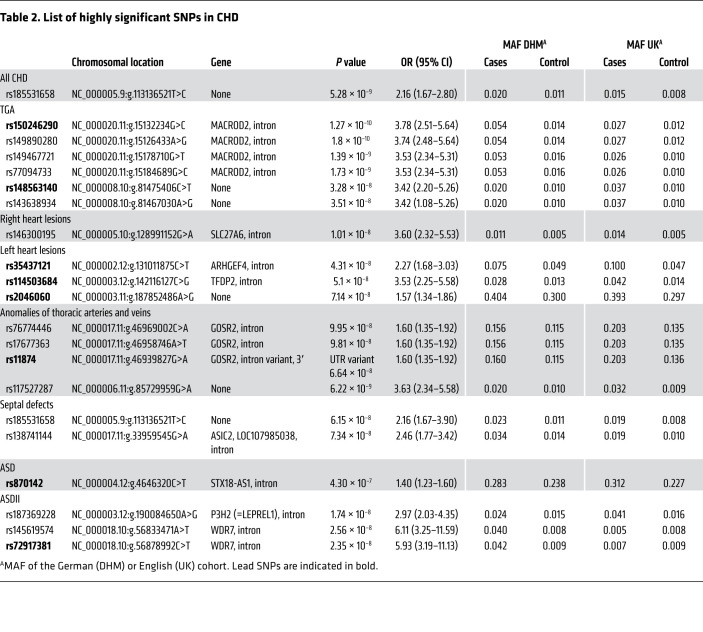
Subsequently, we examined 5 diagnostic subgroups in our cohort: TGA (n = 399), right heart lesions (n = 1296), left heart lesions (n = 326), septal defects (n = 1074), and ATAV (n = 486). In the TGA subgroup, we identified SNPs on chromosomes 20 and 8. The lead SNP (rs150246290) and 3 variants on chromosome 20, all with genome-wide significance, mapped to the MACROD2 gene (Figure 2A) implicated in chromosomal instability (26) and transcriptional regulation (27). Two SNPs (rs149890280 and rs150246290) are suggested to be possible causal variants (Supplemental Table 1). The identified risk locus on chromosome 8 close to ZBTB10 included 2 SNPs (rs148563140, rs143638934), both with genome-wide significance (Figure 2B). Given the high levels of linkage disequilibrium (LD) between these SNPs, they are indicative of the same association signal in both loci. Unexpectedly, we found that 2 risk variants at 12q24 and 13q32, previously shown to be associated with TOF (21), could not be substantiated in the German cohort (Supplemental Figure 2, A and B, and Supplemental Table 2). A single SNP (rs146300195) on chromosome 5 at the SLC27A6 locus with genome-wide significance was evident in this subgroup (Supplemental Figure 2C). In left heart lesions, 3 variants (rs3547121, on chromosome 2 and rs114503684 and rs2046060, on chromosome 3 reached genome-wide significance (Supplemental Figure 3). The same SNP on chromosome 5 (rs185531658), indicative for the whole CHD population, also appeared in the subpopulation of septal defects with near-genome-wide significance (Supplemental Figure 4A). A second SNP (rs138741144) was evident on chromosome 17 within the ASIC2 locus (Supplemental Figure 4B). Restricting the analysis to ASDs, we confirmed the previously reported significance of the lead SNP (rs870142) and multiple variants on chromosome 4p16 (ref. 19 and Supplemental Figure 5). Limiting ASD patients to those diagnosed with ASD type II (ASDII) (n = 489), we identified 2 SNPs (rs145619574 and rs72917381) on chromosome 18, in the vicinity of WDR7, and another variant (rs187369228) on chromosome 3, located close to LEPREL1 (also known as P3H2) (Supplemental Figure 6, A and B). In patients with ATAV, we found that 3 SNPs were apparent on chromosome 17 with subgenome-wide significance (rs17677363, rs11874, and rs76774446), all located within the GOSR2 locus (Figure 3). All 3 variants are predicted to be possibly causal (Supplemental Table 1). In addition, GeneHancer analyses suggested that rs11874 may affect the expression of GOSR2 and that WNT3 may be a topologically associated region (Supplemental Table 3). One additional SNP mapped to chromosome 6 (rs117527287) without a nearby gene (the closest was TBX18, approximately 0.3 Mb away) (Supplemental Figure 7). This SNP was also validated independently by Sanger sequencing (Supplemental Figure 1B). Table 2 summarizes all detected SNPs and their significance. Genes located within the LD region of each locus are listed in Supplemental Table 4.
Figure 2. SNPs associated with TGA.
(A) LocusZoom plot of the MACROD2 region on chromosome 20. (B) LocusZoom plot of the ZBTB10 region on chromosome 8. The index SNPs are indicated by purple diamonds, and the other SNPs are color coded depending on their degree of correlation (r2). Circles represent imputed SNPs and triangles genotyped SNPs. Patients with TGA, n = 399. –log10 P values were determined by association statistics from the GWAS (logistic regression).
Figure 3. SNPs associated with anomalies of thoracic arteries and veins.
LocusZoom plot of the GOSR2 region on chromosome 17. The index SNPs are indicated by purple diamonds, and the other SNPs are color coded depending on their degree of correlation (r2). Circles represent imputed SNPs and triangles genotyped SNPs. Patients with ATAV, n = 486. –log10 P values were determined by association statistics from the GWAS (logistic regression).
Table 2. List of highly significant SNPs in CHD.
Genes with genome-wide significant SNPs (listed in Table 2) and further significantly enriched variants with P values below 0.0005 (listed in Supplemental Table 5) that fell into the gene region underwent gene set enrichment analysis (GSEA). Terms related to cell-cell signaling, embryonic development, and morphogenesis showed the highest significance (Supplemental Table 6), and the well-known cardiac transcription factors GATA3, GATA4, and WNT9B were involved in all signaling cascades (Supplemental Figure 8).
Expression of SNP-carrying candidate genes during cardiac differentiation of murine embryonic stem cells.
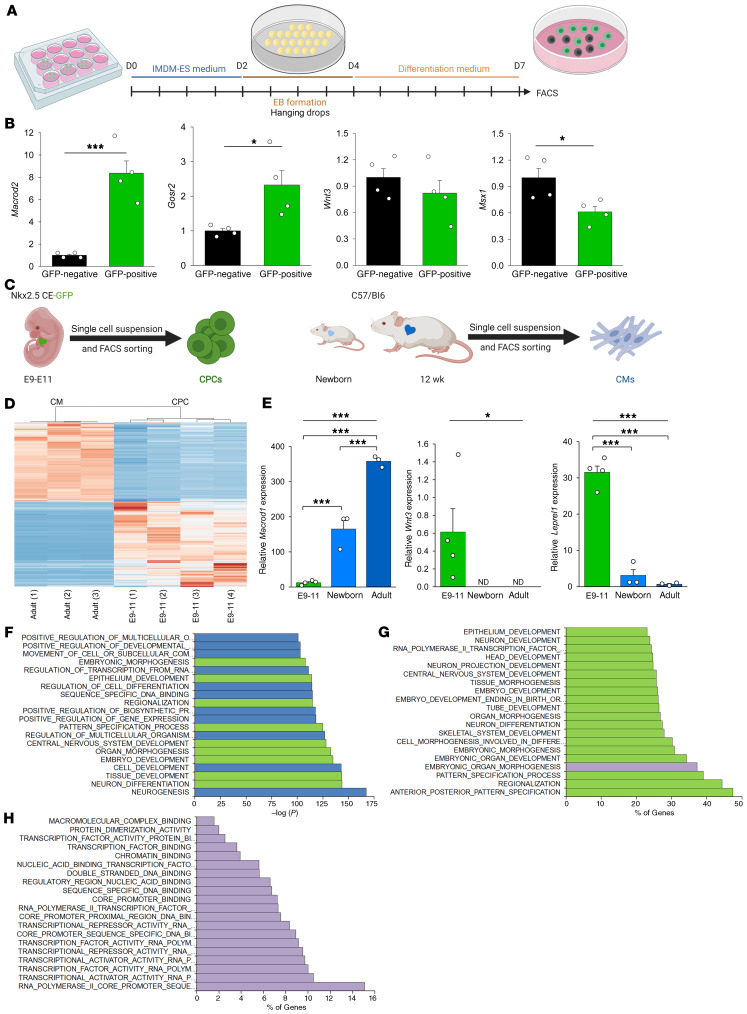
We addressed the question of whether SNP-carrying genes might be expressed by multipotent GFP-positive cardiac progenitor cells (CPCs) during differentiation of embryonic stem cells (ESCs) (Figure 4A) derived from the Nkx2.5 cardiac enhancer (CE) EGFP transgenic mouse line (28). Interestingly, we found that Macrod2 and Gosr2 were significantly enriched in beating GFP-positive CPCs compared with their GFP-negative stage-matched counterparts, in contrast to Wnt3 and Msx1 (Figure 4B).
Figure 4. Role of SNP-carrying candidate genes in murine cardiac development.
(A) Schedule of differentiation of murine ESCs. (B) Relative gene expression of Macrod2, Gosr2, Wnt3, and Msx1 in GFP-negative cells and GFP-positive CPCs (n = 4 each). Data represent the mean ± SEM. (C) Schematic representation of the enrichment of murine CPCs and postnatal CMs. (D) Heatmap of genes differentially expressed in embryonic CPCs and adult CMs. (E) Expression of Macrod1, Wnt3, and Leprel1 in E9–E11 CPCs (n = 4), newborn CMs (n = 3), and adult CMs (n = 3). Data represent the mean ± SEM. ND, no expression detected. (F–H) Results of GSEA of 1649 genes overlapping between CHD-associated SNP-carrying genes and genes upregulated in CPCs according to (F) significance of GO terms, (G) coverage of GO terms, and (H) second-level GO terms showing molecular functions. *P < 0.05 and ***P < 0.001, by unpaired, 2-tailed Student’s t test or Mann-Whitney rank-sum test (B) and 1-way ANOVA or the Kruskal-Wallis test (E), correcting for multiple testing using the Holm-Sidak method.
Role of SNP-carrying genes in murine prenatal cardiac progenitors and cardiomyocytes in vivo.
We then analyzed our existing RNA-Seq data from purified murine CPCs and postnatal cardiomyocytes (CMs) (29) (Figure 4C), clearly separated by their global expression patterns (Figure 4D), to search for SNP-carrying candidate genes that were significantly upregulated in either cell population. Both newborn and adult CMs expressed Macrod1, a paralog of Macrod2, at a much higher level than did embryonic CPCs (Figure 4E). Furthermore, Wnt3 and Leprel1 were both abundantly expressed in CPCs but barely expressed or undetectable in CMs of newborn or adult mice (Figure 4E).
The global RNA-Seq analysis (Figure 4D and Supplemental Data File 1) identified 1915 and 1155 significantly upregulated genes (>2-fold, P < 0.05) specific for CPCs and CMs, respectively. We speculated that the gene loci of the SNPs identified in our CHD cohort might be associated with either of these 2 gene pools. Therefore, we compared the genes of the entire SNP-carrying CHD cohort with the lists of genes upregulated in CPCs or CMs. We applied MAGMA, a tool that allows the simultaneous analysis of multiple gene sets (30). We performed a gene-set level association test, which showed that the GWAS signals were significantly enriched in genes upregulated in CPCs (n = 1649, P = 0.0078), but not in genes upregulated in CMs (P = 0.471) (Supplemental Data File 1). After GSEA of these 1649 genes, gene ontology (GO) terms related to neural development showed the highest significance, followed by pathways regulating tissue, cell, embryo, and organ morphogenesis (Figure 4F). Investigation of the deposited GO gene set revealed high coverage for embryonic and neural development (Figure 4G). Since “embryonic” gene sets contain many genes in common, we selected embryonic organ morphogenesis to allow a closer look at the molecular function in a second-level GO analysis. The top 20 categories all referred to DNA binding or transcription factor activity (Figure 4H). Network-based functional enrichment analysis highlighted several pathways directly involved in cardiac development, such as ventricular septum development and aortic valve, right ventricle, and atrium morphogenesis (Supplemental Figure 9).
Expression of SNP-carrying candidate genes in mouse embryonic cardiogenic tissue.
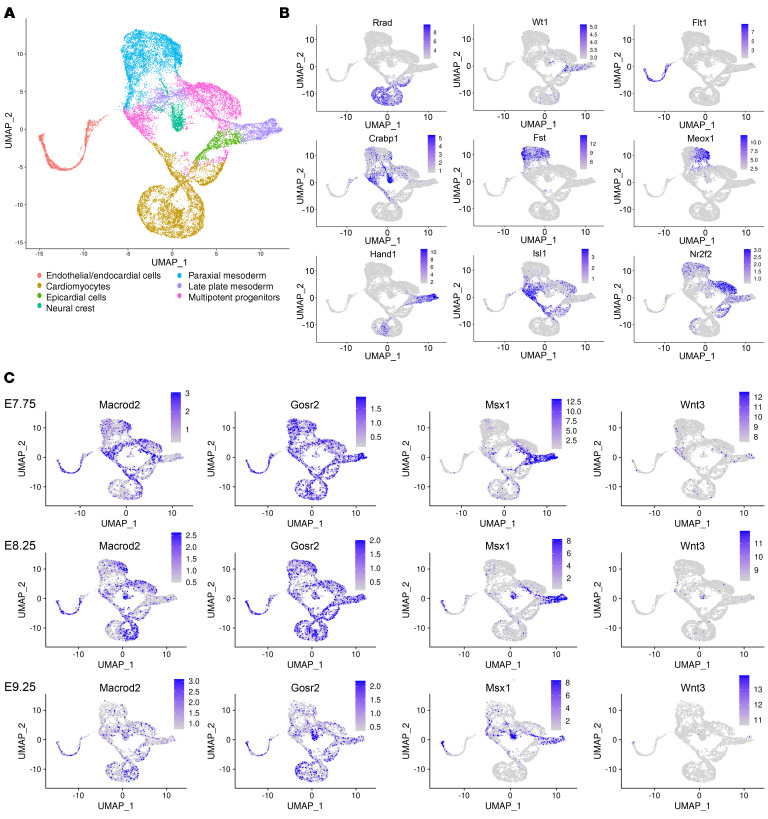
To track the expression of our candidate genes, we reanalyzed a data set of more than 56,000 cells from the cardiogenic region of mouse embryos collected at E7.75, E8.25, and E9.25 previously published by de Soysa et al. (31). Recapitulating their approach, we strictly excluded all endodermal and ectodermal cells identified by their expression of appropriate marker genes (Supplemental Figure 10, A–C). After reclustering (Supplemental Figure 10D), the remaining mesodermal cells (n = 21,745) were superimposable, comparing WT and Hand2-null embryos (Supplemental Figure 10E). The 7 distinct mesodermal cell populations (Figure 5A) were distinguished by appropriate marker genes (Figure 5B), and each showed a characteristic gene expression pattern (Supplemental Figure 10F). Macrod2 was predominantly expressed in the multipotent progenitors at E7.75 and started to concentrate in the CMs at later time points (Figure 5C). We detected Gosr2 expression in all clusters at E7.75 and E8.25. At E9.25, we observed that expression was predominantly restricted to the neural crest and CMs (Figure 5C). Msx1 showed strong expression in the late plate mesoderm at E7.75, gradually decreasing until E9.25, whereas the pattern in the neural crest was reversed (Figure 5C). Wnt3 showed a scattered expression pattern at E7.75 and was only rarely detectable in individual cells at E9.25 (Figure 5C).
Figure 5. Expression of SNP-candidate genes during murine embryonic cardiogenesis.
(A) UMAP plot of all mesodermal and neural crest cells of the cardiogenic region (n = 21,745). (B) Expression of marker genes in individual clusters. (C) Expression of Macrod2, Gosr2, Msx1, and Wnt3 in cardiogenic tissue at E7.75, E8.25, and E9.25.
Expression of SNP-carrying candidate genes during cardiac differentiation of human induced pluripotent stem cells.
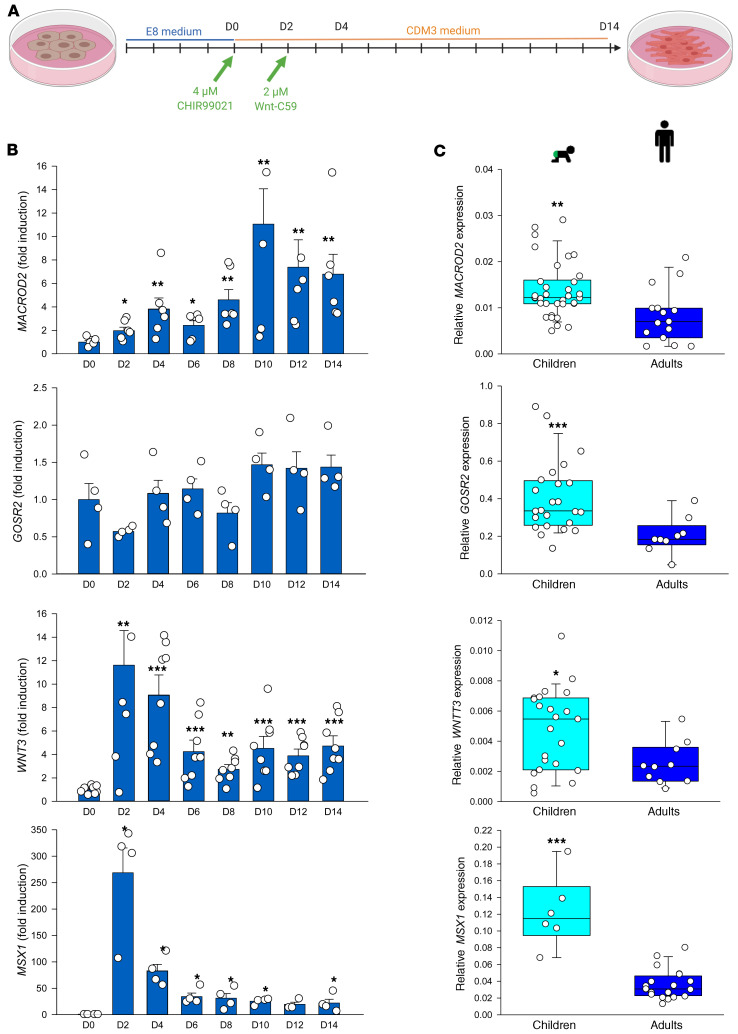
We then investigated the role of all candidate genes during cardiac differentiation of human induced pluripotent stem cells (iPSCs) (Figure 6A). Expression of MACROD2 gradually increased and peaked around day 10, whereas the expression of GOSR2 did not substantially change at any time point (Figure 6B). ATAC-Seq (assay for transposase-accessible chromatin with high-throughput sequencing) analyses suggested a potential interaction of GOSR2 variants with WNT3 and STX18-AS1 variants with MSX1, respectively, early during cardiac differentiation of human ESCs (32). In line with these results, both genes were most strongly upregulated on day 2 during differentiation of human iPSCs (Figure 6B). STX18 and LEPREL1 also peaked early, while expression of all other candidate genes was not substantially changed (Supplemental Figure 11).
Figure 6. Expression of SNP-carrying candidate genes during differentiation of human iPSCs and in pediatric and adult aortic tissue.
(A) Schedule of directed cardiac differentiation of human iPSCs. (B) Expression of MACROD2, GOSR2, WNT3, and MSX1 during directed cardiac differentiation of human iPSCs. Data represent the mean ± SEM of at least 2 independent experiments, each run in duplicate. *P < 0.05, **P < 0.01, and ***P < 0.001 versus day 0 (D0), by unpaired, 2-tailed Student’s t test. (C) Expression of MACROD2, GOSR2, WNT3, and MSX1 in aortic tissues of pediatric patients (n = 35, 24, 23, and 6, respectively) and adult surgical patients (n = 15, 9, 10, and 20, respectively). Data represent the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001, by Mann-Whitney rank-sum test.
Expression of SNP-carrying candidate genes in CHD patient tissue.
We first analyzed whether the presence of the risk variant might influence expression of the affected gene. However, the genotype did not alter expression of MACROD2, GOSR2, or WNT3 (Supplemental Figure 12). Therefore, we compared the expression of all candidate genes in aortic and atrial tissue of patients with CHD (Supplemental Table 7) with the expression in tissue of adult surgical patients (Supplemental Table 8). We found that MACROD2, GOSR2, WNT3, and MSX1 were clearly expressed at higher levels in the tissues of patients with CHD (Figure 6C). In addition, ARHGEF4, STX18-AS1, STX18, and WDR7 also showed significantly higher expression levels in pediatric aortic tissue (Supplemental Figure 13). In atrial tissue, expression of SLC27A6, MSX1, LEPREL1, and WDR7 was significantly higher in CHD samples (Supplemental Figure 14). Though not a direct proof, it is however tempting to speculate that the majority of our candidate genes may also have a role in early cardiac development.
Expression of SNP-carrying candidate genes in human fetal and adult heart tissue.
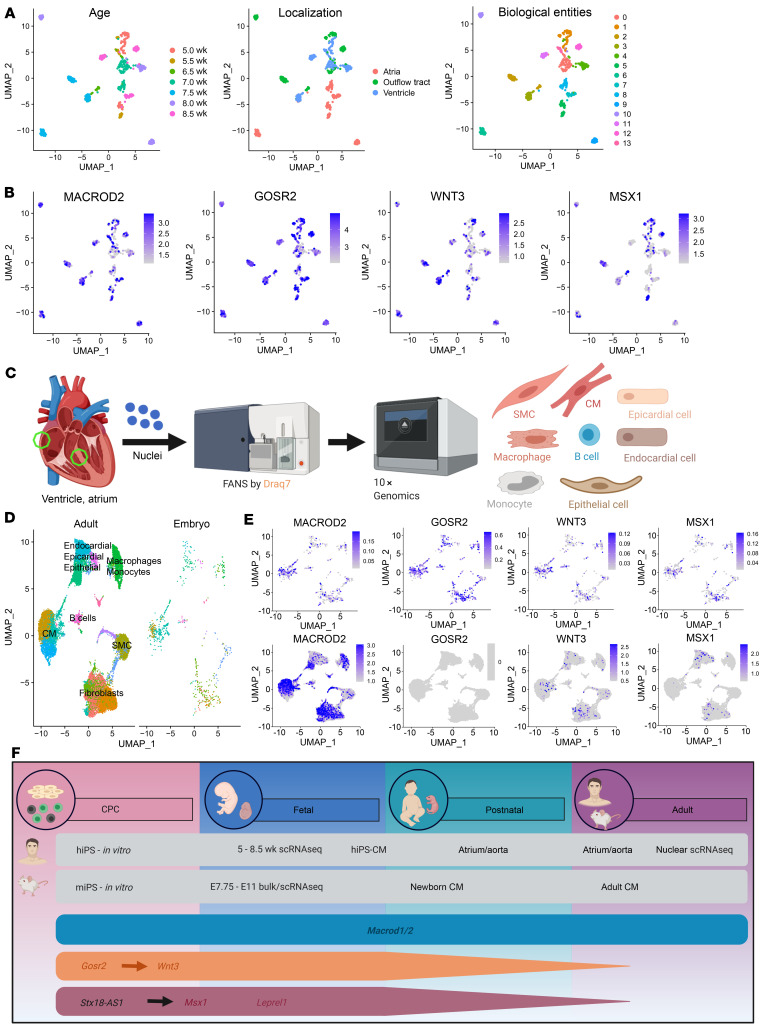
We extended our analysis and revisited a published scRNA-Seq data set for 669 human embryonic cardiac cells (33). Using principal component analysis (PCA) and unsupervised clustering, we could classify cells into distinct biological entities, defined by their gestational age and anatomical region (Figure 7A). High expression among all 14 clusters was detected for MACROD2, and especially for GOSR2, with higher relative gene expression (Figure 7B). Expression of WNT3 and MSX1 appeared broad throughout all developmental stages, (Figure 7B), albeit more concentrated on fibroblasts and myocytes (Figure 7E).
Figure 7. Role of SNP-carrying candidate genes in human cardiac development.
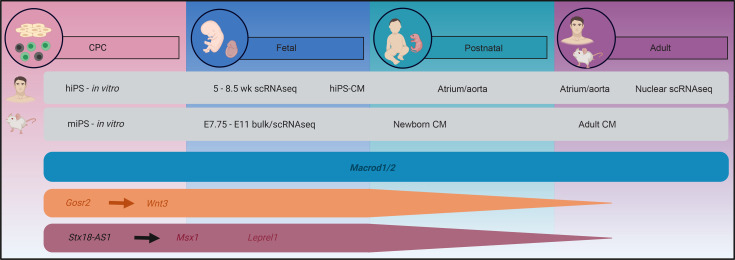
(A) Unbiased clustering of embryonic cells (n = 669) into biological entities. Cells are labeled on the basis of age as well as anatomical localization for purposes of visualization. (B) Relative expression of MACROD2, GOSR2, WNT3, and MSX1 in embryonic heart cells. (C) Schedule of scRNA-Seq analysis of cells from atria and ventricles (n = 17,782). (D) Clustering of embryonic and adult cells and identification of cell types. (E) Expression of candidate genes in the integrated data set split by embryonic cells (upper panel) and adult cells (lower panel). (F) Expression of candidate genes associated with TGA (turquoise), ATAV (orange), or septal defects (red) in vitro and in vivo during different stages of the developing murine and human heart. hiPS, human iPSCs; miPS, murine iPSCs; SMC, smooth muscle cells.
To pursue age-dependent differences in the expression of our candidate genes, we conducted additional scRNA-Seq experiments with 17,782 cells from samples of adult human atria and ventricles (Figure 7C). Integrating the data from adult and embryonic hearts, we could identify different cell types on the basis of their expression of defined marker genes (Supplemental Figure 15). Of note, cells from both adult and embryonic hearts yielded perfectly superimposable clusters (Figure 7D). MACROD2 shows robust expression in all adult cardiac cell types. By stark contrast, GOSR2, widely expressed throughout the embryonic heart, could not be detected in any adult cell (Figure 7E). WNT3 and especially MSX1 are expressed in cells of the adult heart, although at a much lower level compared with embryonic cells, given the much higher number of adult cells analyzed. Although WNT3 and MSX1 showed similar expression patterns in fetal and adult cell types, the expression of MSX1 appeared virtually absent in adult myocytes (Figure 7E). Thus, the 4 candidate genes analyzed may play a role in the developing human heart, while MACROD2 may still be important at a later point. Figure 7F summarizes the expression of candidate genes in vitro and at different stages of the developing murine and human heart in vivo.
Discussion
We performed a GWAS on more than 4000 White patients with CHD, which represents the largest genetic study of European individuals to date. Across 5 major clinical subgroups, we detected approximately 20 SNPs associated with genome-wide significance (P < 5 × 10–8).
A careful evaluation of the genes related to the identified SNPs showed no cardiac phenotype in monogenic knockout mouse models (Supplemental Table 9), which is probably due to the multigenic etiology of almost all congenital heart malformations. Nevertheless, our downstream analyses of these SNPs within the subgroups of TGA, ATAV, and ASD showed a clear functional association of the closely related genes during murine and human heart development using different in vitro and in vivo experimental strategies.
Humans and mice share similarities in the basal sequence of cardiac development (34), especially for the most key developmental checkpoints. Single-cell transcriptome analysis revealed species-shared genes in the 4 different cardiac cell types, with CMs being the most similar cell type. However, the best overlap for each cell type appeared at different time points during cardiac development because of the asynchronous cardiac development in these 2 species (35). The shown functional relevance of the identified SNPs in both species underlines the general impact of these genes during cardiac development rather than a species-specific relevance.
TGA and MACROD2.
In the TGA subgroup, 4 SNPs with genome-wide significance mapped to MACROD2, which has been linked to adipogenesis and hypertension (26, 36). Microdeletions in this gene have been implicated as a cause of chromosomal instability in cancers (37), and de novo deletion of exon 5 causes Kabuki syndrome (38). Chromosomal imbalance is also frequently seen in patients with CHD with different morphologies (39–42) including TGA (43), but so far the MACROD2 locus has not been associated with CHD.
Expression of Macrod2 was significantly enhanced in early murine CPCs derived from murine pluripotent stem cells (Figure 4B). Macrod1 was abundantly expressed in newborn and adult CMs, but negligibly so in embryonic CPCs at E9–E11 (Figure 4E). This is in line with the murine single-cell data (Figure 5) showing an enriched early expression of Macrod2 in multipotent progenitor cells, which clearly shifted over time to a predominate expression in CMs. Macrod1 and Macrod2 are paralogs with substantial structural similarity (44) and common biological activities (45), potentially suggesting similar functions during cardiac development. Regardless of the genotype of the patient, we observed no major difference in expression of MACROD2 (or of GOSR2 or WNT3). This might be because our tissue samples were unfortunately limited to patients with a heterozygous genotype. scRNA-Seq data indicated MARCOD2 expression during human embryonic development within ventricular and outflow tract cells (Figure 7B). We also detected MACROD2 expression in CMs, which is in line with its later expression during directed cardiac differentiation of human iPSCs. Even more important for structural developmental defects was a high expression level of MACROD2 during embryonic development in fibroblasts and endothelial cells (Figure 7, D and E, upper panel). The expression of MACROD2 was not limited to the embryonic stage but was high in different adult cardiac cell types (Figure 7, D and E, lower panel).
Genetic variants of MACROD2 are associated with different diseases (27), although the exact mechanisms remain unclear. We can only speculate how this locus might be linked to the development of TGA. Our data show prevalent expression of MACROD2 in human embryonic cardiac cells (Figure 7B), where it could act as a transcriptional regulator (27). In addition, the long noncoding RNA RPS10P2-AS1 was transcribed from an intronic region of the MACROD2 locus, and its expression was consistently higher than that of MACROD2 throughout adult and embryonic human tissues, including fetal heart (46). RPS10P2-AS1 has been shown to modulate the expression of multiple genes in neuronal progenitor cells (46). Importantly, a recent report suggests that one-third of patients with CHD develop neurodevelopmental disorders (14). Thus, it is conceivable that the expression of an array of different genes may be similarly affected in embryonic cardiac progenitor cells, thereby contributing, at least in part, to the development of TGA.
ATAV and GOSR2.
One risk region comprises 3 highly significant SNPs mapping to GOSR2, which is involved in directed movement of macromolecules between Golgi compartments (47). Genetic variants of GOSR2 have been implicated in coronary artery disease (48) and myocardial infarction, with contradictory results (49, 50). The ATAV subgroup included patients diagnosed with coarctation of the aorta, an interrupted/hypoplastic aortic arch, or patent ductus arteriosus. These CHD malformations all share a common origin within the aortic sac and the stepwise emerging aortic arches during embryonic development (51). The proximal aorta and portions of the outflow tract derive from the bulbus cordis.
Applying ATAC-Seq analysis, Zhang et al. described a potential interaction between GOSR2 and WNT3 during cardiac differentiation of human ESCs (32). Our expression analysis showed significantly enhanced Gosr2 expression in isolated murine CPCs, while Wnt3 showed similar expression levels in CPCs and developmentally stage-matched cells (Figure 4B), suggesting a specific role of Gosr2 during embryonic cardiac development. Nevertheless, Wnt3 was clearly detectable in embryonic CPCs but absent in newborn or adult CMs, indicating a more distinct role for Wnt3 during embryonic development. Furthermore, we could clearly detect expression of GOSR2 in human embryonic cells of the outflow tract (Figure 7B) by scRNA-Seq analysis, suggesting a potential association of this gene with the development of ATAV. In contrast, we could not detect GOSR2 expression in the adult human heart, supporting our hypothesis that GOSR2 exerts its biological role during embryonic cardiac development. The specific developmental role of Gosr2 and Wnt3 during cardiogenesis was further substantiated by the analysis of murine embryonic single-cell data (Figure 5C). Both Gosr2 and Wnt3 were mainly expressed at E7.75 and diminished over time.
ASD and STX18/MSX1.
We identified the SNP rs185531658 in patients with septal defects with high significance (P = 6.15 × 10–8). The same SNP was also strongly associated with CHD risk in general, with YTHDC2, an RNA helicase involved in meiosis, as the closest gene (52). The second SNP for septal defects is related to ASIC2, whose loss leads to hypertension in null mice (53). Restricting the patient cohort to ASDs, we confirmed the SNP rs870142, which we had previously identified (19). As this SNP appeared with a much lower significance in the German cohort (Supplemental Figure 5), its significance was lower compared with the original study (P = 4.3 × 10–7 vs. 2.6 × 10–10). Narrowing the cohort to patients with ASDII, we identified 2 risk loci. The genes in the affected loci, WDR7 and LEPREL1, are associated with growth regulation and tumor suppression in breast cancer (54, 55), but without cardiovascular importance. Lin and colleagues reported on several risk loci for septal defects in a Chinese cohort (17). We could validate 1 variant, rs490514, in our CHD population (Supplemental Table 10), supporting the validity of our GWAS results.
Zhang et al. also described a functional association between STX18 (SNP rs870142) and MSX1 (32). This interaction is also supported by our findings of significantly higher expression levels of STX18 and MSX1 during cardiac differentiation of human iPSCs at early stages. Furthermore, Msx1 was expressed at comparable levels in CPCs and developmentally stage-matched cells, suggesting a role for Msx1 during embryonic development. The similar expression levels in GFP-positive CPCs and GFP-negative developmentally stage-matched cells could either be explained by an expression not exclusively restricted to embryonic cardiac development or a predominant expression of Msx1 in second heart field (SHF) progenitors and cells of the outflow tract (56), which were not necessarily captured by our Nkx2.5 CE transgenic mouse model (28).
Even more important, extensive scRNA-Seq analyses in cells from the murine cardiogenic region showed a predominant expression of Msx1 in late plate mesodermal cells that decreased over time. Furthermore, scRNA-Seq analyses showed overlapping expression of MSX1 in cells of the outflow tract during embryonic human heart development, with CMs and fibroblasts being the main cell types at this stage. The role of MSX1 in CMs seemed to be restricted to embryonic development, whereas we could still detect MSX1 expression in fibroblasts end endothelial cells of the adult heart. This finding is in line with our comparative analysis of MACROD2, GOSR2, WNT3, and MSX1 expression in pediatric and adult aortic tissues (Figure 6C).
A second SNP, closely related to LEPREL1, was associated with the ASDII subgroup. Leprel1 was clearly detectable in embryonic CPCs but barely evident in newborn or adult CMs. Furthermore, we observed substantially elevated expression of LEPREL1 early during cardiac differentiation of human iPSCs, suggesting a role during early cardiac development. Comparing the expression of LEPREL1 in adult and pediatric atrial tissue, we could show significantly (P = 0.005) enhanced expression in pediatric samples, again suggesting a potential role during early cardiac development.
Strength and limitations of the study.
A major strength of our study is the large, homogenous cohort with a representative profile of more than 4000 European patients with CHD that yielded results with high confidence and power. At the same time, this strength turned into a limitation: an appropriate ethnically matched control cohort is presently not available, and our results may not be generally translated to cohorts of different ethnic origins. The newly discovered risk loci for TGA and ATAV, both rarely occurring pathologies, are thus still based on relatively small numbers that need to be substantiated in a larger number of patients. Finally, the genotyping of the German and United Kingdom (UK) cohort was run on different platforms that used slightly different quality parameters.
In summary, our GWAS identified multiple risk loci for all major clinical CHD subgroups. We detected genetic variants in the MACROD2 and GOSR2 loci that were strongly associated with the phenotype of TGA and ATAV, respectively. The use of murine and human pluripotent stem cells and the ex vivo results from tissues of patients with CHD underline the functional role of several candidate genes during cardiac differentiation. Finally, scRNA-Seq analyses provided strong in vivo evidence that MACROD2, GOSR2, WNT3, and MSX1 play important roles during embryonic development of the human heart.
Methods
Patients and controls.
The complete cohort of patients with CHD comprised 4034 participants. The first cohort of 1440 patients (n = 769 males, n = 671 females, mean age 17 years) were enrolled at the German Heart Center Munich between March 2009 and June 2016. The German ethnicity of the participants was confirmed by analysis of the genotype data using multidimensional scaling. In addition, 2 previously analyzed patient cohorts with a mixed CHD history (mean age, 20 years) (17) and TOF (mean age, 15 years) (19), comprising 2594 patients (n = 1320 males, n = 1274 females), were included. Patients in whom neurodevelopmental or genetic abnormalities were apparent were excluded, but since some probands were recruited as babies or young children, this would not have been evident in all cases. Genotypes were compared with 3554 (n = 1726 males, n = 1828 females) and 4932 (n = 2498 males, n = 2434 females) controls for the German and British cohorts, respectively. The German control participants were recruited from the well-established KORA (Cooperative Health Research in the Region of Augsburg) F4 and S3 cohorts used in numerous studies as a control group (57). Genotyping was performed at the Helmholtz Zentrum (Munich, Germany) and the Centre National de Genotypage (Evry, France) using the Affymetrix Axiom Genome-Wide Human array or the Illumina 660wQUAD array, respectively. The German samples were genotyped on the Affymetrix Axiom CEU array according to the Axiom GT best practices protocol and the manufacturer’s recommendation. The KORA controls were genotyped by Affymetrix on the same chip type.
Genotype calling.
Genotype calling was done following the Axiom Genotyping Solution Data Analysis Guide (http://tools.thermofisher.com/content/sfs/manuals/axiom_genotyping_solution_analysis_guide.pdf). It provides a standard workflow to perform quality control analysis for samples and plates, SNP filtering prior to downstream analysis, and advanced genotyping methods. The workflow utilizes 3 software systems, including Axiom, Analysis Suite, Power Tools (APT), and SNPolisher R package. Initially, we had 20 plates and 1921 individual samples in total. Of those, 1803 arrays passed all quality control steps (sample DishQC [DQC] >82%, sample call rate >97%). In order to obtain high-quality genotype calling, only “PolyHighRes” and “MonoHighRes” samples were kept for the next steps.
Quality control, imputation, and association analysis.
All statistical analyses and quality control procedures for the 2 British cohorts are described in detail in the 2 respective publications (19, 21). For the German cohort, a standardized 8-step GWAS quality control procedure was developed and applied to the genetic data (Supplemental Figures 16 and 17). Prior to imputation, samples were excluded from further analysis for the following reasons: the call rate was less than 98%, the sex call was incorrect or ambiguous, or the sample was potentially contaminated. In addition, the thresholds for relatedness and population outliers were set at a pihat of 0.09 or greater in an identical-by-descent (IBD) analysis, and a 2 or higher SD was applied in the multidimensional scaling (MDS) analysis. SNPs were excluded if their missing rate was higher than 3%, if the minor allele content (MAC) was less than 5, if the P value for the Hardy-Weinberg equilibrium was 1 × 10–5 or less in controls, or if the SNPs failed the cluster quality check. The population structures were evaluated using a set of pruned autosomal variants with a minor allele frequency (MAF) > 0.05, P < 1 × 10–5, and r2 ≤ 0.2 between pairs of variants (--indep-pairwise 50 5 0.2). For the principal component analysis (PCA) in PLINK (version 1.90b3.36) (58), a total of 119,381 independent SNPs were pruned (Supplemental Figure 17B and C) except for the quality cluster check, for which Affymetrix SNPolisher (version 1.5.2) (59) was used.
Genome-wide imputation was conducted on the basis of the Haplotype Reference Consortium using the Sanger Imputation Service. All individual samples were imputed on the Sanger imputation server (https://imputation.sanger.ac.uk/) with the Haplotype Reference Consortium panel and Eagle, version 2.4.1 (https://data.broadinstitute.org/alkesgroup/Eagle/) and positional Burrows-Wheeler transform (PBWT) pipelines. Imputed variants with an AF of less than 0.005 and/or an information score of less than 0.7 were excluded from the statistical analysis. The application of these filters resulted in a total of 20,441,516 high-quality SNPs available for the meta-analysis of up to 1495 patients and 3554 control samples. Because of the sex mismatch and inappropriate diagnoses, the number of samples for the final analysis had to be reduced to 1440. For the British cohort, 11,356,134 high-quality SNPs were available. The shared set used for the meta-analysis included 9,216,527 SNPs. The information on the imputation score of all lead SNPs is shown in Supplemental Table 11. The analysis of single SNP genetic association was performed with SNPTEST, version 2.5.2 (https://mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.html) via logistic regression using probabilistic imputed allele dosages with adjustment for age, sex, and the first 10 ancestry principal components. We have estimated the effective number of independent markers (Meff) by calculating the reciprocal of the variance of the off-diagonal elements of the genetic relatedness matrix (60, 61). The genome-wide significance cutoffs were 9.5 × 10–8 and 1.9 × 10–7, with a q value of 0.05 and 0.1, respectively. In accordance with the majority of published GWAS analyses, we used 5 × 10–8 and 1 × 10–5 as genome-wide and suggestive significance cutoffs. The value of the inflation factor λ for all CHD cases and subgroups is indicated in Supplemental Table 12. The GWAS.PC package (version 1.0) in R was used to confirm that data from each subgroup could be obtained with sufficient power (Supplemental Table 13).
Meta-analysis.
The quality of summary statistics of each GWAS data set was controlled with the EasyQC pipeline, version 8.5 (http://www.genepi-regensburg.de/easyqc). For the meta-analysis, we used the fixed-effect, inverse variance method with METAL, release 2011-03-25 (http://csg.sph.umich.edu/abecasis/metal/). Genomic control was done separately in each study prior to meta-analysis by calculating the inflation factor λ and adjusting for it. Lead SNPs of independent genome-wide significant signals in the meta-analysis results were defined by LD-based independent “clumps” in PLINK (version 1.90b3.36), with P < 1 × 10–5, r2 > 0.05, and a clumping distance of less than 500 kb. The heterogeneity of lead SNPs was estimated with random-effects meta-analysis using METASOFT, version 2.0.1 (http://genetics.cs.ucla.edu/meta/).
Identification of potentially causal variants by CAVIARBF.
To prioritize the possible causal variants identified by our GWAS, the fine-mapping tool CAVIARBF (https://bitbucket.org/Wenan/caviarbf/src/default/) was applied. This tool uses an approximate Bayesian method that allows for multiple causal variants (62). We used the 74 baseline annotations in a stratified LD score regression (63). SNPs within a 50 kb radius of a lead SNP and with a MAF of greater than 0.01 were considered. 1000 Genomes was used as the reference panel, and 0.2 was added to the main diagonal of the LD as a suggested correction. The exact Bayes factor was averaged over prior variances of 0.01, 0.1, and 0.5. The elastic net parameters were selected via 10-fold cross-validation.
GeneHancer annotation.
To detect the putative regulatory implication of the association signals, we annotated the significant SNPs to the GeneHancer database (64). The records of regulatory elements and linked genes were downloaded from UCSC’s table browser. A SNP is linked to a regulatory element by the colocalization for both the SNP and its proxy SNPs, which is defined with an R2 of greater than 0.6 in the 1000 Genomes EUR reference panel.
GSEA.
For the analysis of genome-wide and highly significant SNPs, the Broad Institute’s GO was used (http://software.broadinstitute.org/gsea/msigdb/annotate.jsp). The functional analysis was performed by ClueGO (https://cytoscape.org/), a network-based functional enrichment method that can generate new functional groups by measuring the similarity between different pathways and terms. The method will produce both term- and group-based enrichment scores for better visualization and interpretation. Gene-level enrichment was performed using ClueGO (version 2.5.4) in Cytoscape 3.7.1 (with GO [Biological Processes, version from April 24, 2019], GO term levels 3–8; GO terms with 2 genes and 2% total genes associated; GO terms were grouped by κ score with default settings). A Bonferroni-corrected P value of less than 0.1 was considered the cutoff for significant enrichment. For the GSEA analysis in Supplemental Table 5, a cutoff of P < 0.0005 was chosen to control the FDR at 0.05 for the gene selection by Benjamini-Hochberg correction. There, the lowest P value was assigned to the gene for P value adjustment, which was equal to snp-wise=top, 1 in MAGMA (30).
Genotyping of patients for gene expression in cardiac tissue and validation of SNPs.
To measure gene expression in cardiac tissue, we analyzed a number of patients who had not been genotyped by GWAS. In these cases, genomic DNA from peripheral blood was amplified by PCR using the following conditions: 95°C for 2 minutes, 40 cycles of 95°C for 30 seconds, 60°C for 30 seconds, and 72°C for 90 seconds using FastStart High Fidelity Enzyme Blend (Roche Diagnostics) and a final primer concentration of 0.4 μM. Identical cycling conditions were used for the validation of SNPs rs185531658 and rs117527287. PCR products were purified using the High Pure PCR Purification kit (Roche Diagnostics), and sequences were verified by conventional Sanger sequencing. The exact sequences of all primers are listed in Supplemental Table 14.
qRT-PCR analysis of gene expression in cardiac tissue.
Tissue samples were obtained during the operation, immediately snap-frozen in liquid nitrogen, and kept at –196°C until further use. RNA was extracted using the Rneasy Plus Universal kit (QIAGEN) according to the manufacturer’s recommendation. cDNA was synthesized from 100 ng total RNA using M-MLV reverse transcriptase (100 U), 250 ng random hexamer primers, 10 mM DTT, deoxynucleotide triphosphates (dNTPs) (0.5 mM each), 15 mM MgCl2, 375 mM KCl, and 250 mM Tris-HCl, pH 8.3, in a final volume of 30 μL. Quantitative real-time PCR (qRT-PCR) analyses were performed on a QuantStudio 3 (Thermo Fisher Scientific) under the following conditions: 95°C for 10 minutes, 40 cycles of 95°C for 15 seconds, and 60°C for 1 minute using 0.3 μM of each primer. The expression of ACTB (β-actin) was used to normalize expression levels in the individual samples. The exact sequences of all primers are indicated in Supplemental Table 14.
Spontaneous differentiation of murine embryonic stem cells.
Murine ESCs were differentiated according to a standard “hanging drop” protocol (65). Cells were grown for 2 days on gelatin-coated 6-well plates in IMDM-ES medium (Biochrom) supplemented with 20% FCS (Thermo Fisher Scientific), 0.1 mM 1-thioglycerol (MilliporeSigma), and 103 U/mL leukemia inhibitory factor (LIF) (MilliporeSigma). Hanging drops (1000 cells per droplet) were prepared on 15 cm cell culture dishes in differentiation medium (IMDM supplemented with 20% FCS, 0.1 mM 1-thioglycerol, 0.05 mg/mL l-ascorbic acid [MilliporeSigma] and antibiotics). Culture dishes were cultured upside-down for 2 days to allow embryoid body (EB) formation. Then, EBs were flooded with differentiation medium and cultured with a medium change every other day. On day 7, GFP-positive cardiac progenitors and their GFP-negative counterparts were sorted by FACS. RNA purification and cDNA production were performed as described above.
Directed cardiac differentiation of human iPSCs.
The human iPSC line S was established in our laboratory from PBMCs of a healthy 34-year-old male proband using Sendai virus according to the manufacturer’s protocol (Invitrogen, Thermo Fisher Scientific) and met all criteria of fully reprogrammed iPSCs. Differentiation into human CMs was performed according to a previously published protocol (66). Human iPSCs were seeded into 24-well plates and grown to confluence in normal mTeSR E8 medium (STEMCELL Technologies). On day 0, the medium was switched to RPMI 1640 supplemented with Oryza sativa–derived recombinant human albumin (500 μg/mL, MilliporeSigma) and l-ascorbic acid 2-phosphate (213 μg/mL, MilliporeSigma), referred to here as CDM3. From days 0 to 2, CDM3 was supplemented with 4 μM CHIR99021 (LC Laboratories), and from days 2 to 4, the cells received CDM3 and 2 μM WNT-C59 (Selleckchem). Thereafter, CDM3 was replaced every other day. Every second day, cells in duplicate wells were lysed with RNA lysis buffer (PEQLAB) and purified, and cDNA was produced as described above.
RNA-Seq analysis in murine CPCs and CMs.
We screened our previously published RNA-Seq data (29) to identify SNP-carrying candidate genes that were significantly upregulated in either CPCs or CMs. Original sequencing data were deposited in the NCBI’s Sequence Read Archive (SRA) (PRJNA229481). For this study, CPCs and CMs were isolated. CPCs were obtained from E9–E11 embryonic hearts from the Nkx2.5 CE-EGFP transgenic mouse line (28). Embryos were cut into small pieces and digested in a collagenase II (10,000 U/mL, Worthington Biochemical) and DNase I (10,000 U/μL, Roche, Molecular Systems) solution for 1 hour at 37°C to obtain a single-cell suspension. Cells were washed and resuspended in PBS with 0.5% BSA and 2 mM EDTA for flow cytometric analysis. GFP-positive CPCs were isolated with a FACSAria III Flow Cytometer (BD Biosciences). Dead cells were excluded by propidium iodide staining (2 μg/mL, MilliporeSigma). Forward scatter (FSC) pulse width was used to exclude doublets from the sorting. For RNA-Seq, cells were sorted into RLTplus Buffer (QIAGEN) containing β-mercaptoethanol (10 μL/mL) to extract DNA and total RNA.
CMs were obtained from C57/Bl6 mice at 12 weeks of age. Hearts were retrogradely perfused with digestion buffer for 12 minutes. The enzymatic digest was stopped by addition of 5% FCS and gentle dissociation. Cells were passed through a 100 μm filter. CMs were identified by a high FSC signal, and viable cells were discriminated by Draq5 (Cell Signaling Technology). Polyadenylated RNA was isolated from total RNA using magnetic beads [NEBNext Poly(A) mRNA Magnetic Isolation Module, New England Biolabs]. Libraries were constructed using the NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs) according to the manufacturer′s instructions. A heatmap of differentially regulated genes was generated with ClustVis software (https://biit.cs.ut.ee/clustvis_large/).
scRNA-Seq analysis of the mouse embryonic cardiogenic region.
We reanalyzed a previously published single-cell RNA-Seq data set obtained after dissection of the whole cardiogenic region at E7.75, E8.25, and E9.25. Technical details on the dissection, library preparation, sequencing, and transcript assignment were previously described (31). The raw data have been deposited in the NCBI’s Gene Expression Omnibus (GEO) database (GEO GSE126128; https://www.ncbi.nlm.nih.gov/geo/). Raw sequencing reads were processed through the 10X Genomics CellRanger pipeline generating gene expression matrices. After PCA and unsupervised clustering, we excluded all endodermal and ectodermal cells, which were identified by their expression of appropriate marker genes. The remaining cells were reclustered, and 7 major cell populations (endothelial/endocardial cells, CMs, and epicardial, neural crest, paraxial mesoderm, late plate mesoderm, multipotent progenitors) were identified using the appropriate marker genes. The Seurat object was split into the 3 developmental stages (E7.75, E8.25, and E9.25) for gene expression analysis of Macrod2, Gosr2, Wnt3, and Msx1.
scRNA-Seq analysis of human embryonic cells and cells from adult atria and ventricles.
Samples from right atrium and interventricular septum were collected from 2 patients with no history of coronary artery disease at the German Heart Center Munich and directly snap-frozen in liquid nitrogen in the operating room. Tissue samples were minced and nuclei extracted in lysis buffer containing 5 mM CaCl2, 3 mM magnesium acetate, 2 mM EDTA, 0.5 mM EGTA, 10 mM Tris, 0.2% Triton X-100, protease inhibitors, and DTT. Nuclei were centrifuged in 1 M sucrose and resuspended in PBS. After staining with Draq7, the samples were purified by fluorescence-activated nuclei sorting (FANS). Nuclei were counted under the microscope and diluted for subsequent addition to 10× Genomics Chromium Next GEM Single Cell 3′ Solution v3. Barcoding, cDNA amplification, and gene expression library construction were done according to the manufacturer’s recommendations. Library sequencing was conducted at the EMBL Heidelberg Genomics Core Facility. The sequencing parameters were 28 bp for read1, 8 bp for the index, and 56 bp for informative read2.
Single-cell RNA-Seq data from human embryonic cardiac cells have previously been published by Sahara et al. (33). Raw data were deposited in the NCBI’s SRA (accession no. PRJNA510181; https://www.ncbi.nlm.nih.gov/sra/). Single-Cell RNA-Seq data from 676 individual cells were uploaded to the Galaxy web platform (67), and we used the public Galaxy Europe server (usegalaxy.eu) for data preprocessing and alignment. Data sets were trimmed using TrimGalore (68) and aligned with RNA STAR (69) against Genome Reference Consortium Human Build 38 (hg38). Aligned reads were processed with MarkDuplicates (70), and count matrices were generated with FeatureCounts (71). Samples from adult patients were subjected to the Cellranger pipeline from 10× Genomics with default settings using a pre-mRNA reference, as detailed by the manufacturer.
Seurat (72) objects for Count matrices for all samples were created for downstream analyses. After quality filtering, the data were normalized and scaled, and variable features were detected using SCTransform (73). Data from embryonic and adult cardiac tissue were integrated as described by Stuart et al. (74). PCA and uniform manifold approximation and projection (UMAP) for dimension reduction were used to cluster cells into distinct biological identities. Cell types were identified on the basis of the expression of known markers. For expression analysis of MACROD2, GOSR2, WNT3, and MSX1, the Seurat object was split into adult and embryonic cardiac cell populations, retaining the clustering information of the integrated data set. The Seurat command FeaturePlot was used for visualization of gene expression with min.cutoff = ‘q10’ and max.cutoff = ‘q90’ settings.
Data availability.
The RNA-Seq data for single cells obtained from adult human atria and ventricles have been deposited in the NCBI’s GEO database (GEO GSE161016; https://www.ncbi.nlm.nih.gov/geo/).
Statistics.
The expression levels during directed cardiac differentiation of human iPSCs, in human tissue samples, murine ESCs, CPCs, and CMs were determined with SigmaPlot,version 13.0, applying an unpaired, 2-tailed Student’s t test or the Mann-Whitney rank-sum test if the equal variance or normality test failed. For comparisons of 3 groups, 1-way ANOVA (Macrod1 and Leprel1) or the Kruskal-Wallis test (Wnt3) was applied. A correction for multiple testing was performed between these results across genes using the Holm-Sidak method. Significance within genes for the pairwise comparisons was also determined using a Holm-Sidak approach. In all instances, P values of less than 0.05 were considered statistically significant. Statistical analyses for the GWAS are described in detail in the relevant sections above.
Study approval.
Ethics approval for the German cohort was obtained from the local ethics review board of the Medical Faculty of the Technical University of Munich (projects 5943/13 and 375/14). For the British cohort, approval was obtained from the local IRBs of all participating centers (19 and 21). Written informed consent was obtained from the participants or their parents or legal guardians.
Author contributions
HL, MJ, MD, NB, CAA, IN, ED, SAD, HJC, and BDK acquired data and materials. HL, MD, FW, NB, OB, IN, ZZ, SAD, PL, and GE conducted molecular and cellular experiments. NP, JC, MB, KCK, JZ, EM, TM, JH, PE, JRP, HJC, BDK, and MK acquired and analyzed clinical and bioinformatics data. MJ, FW, RG, LH, JRP, and BMM performed bioinformatics analyses. MJ, MD, SAD, RG, LH, JH, PE, JRP, RL, TM, HJC, and BDK reviewed and edited the manuscript. HL, MJ, BMM, and MK wrote the manuscript. All authors commented on, edited, and approved the manuscript. BMM and MK supervised the study. The order of the shared co–first authorship was determined in a discussion and a mutual agreement of all first co–first authors and the senior scientists.
Supplementary Material
Acknowledgments
We gratefully acknowledge the support of Stefan Eichhorn for his help with biobank issues and Elisabeth Zierler for her support with the genotyping of samples. The authors acknowledge the support of the Freiburg Galaxy Team: Mehmet Tekman and Rolf Backofen (Bioinformatics, University of Freiburg, Freiburg, Germany), funded by the Collaborative Research Centre 992 Medical Epigenetics (DFG grant no. SFB 992/1 2012) and the German Federal Ministry of Education and Research (BMBF grant no. 031 A538A de.NBI-RBC). Parts of Figures 3 and 4 were created with BioRender.com and exported under a paid subscription. BMM and MK had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. MK is supported by the Deutsche Stiftung für Herzforschung (grant no. F/37/11), the Deutsches Zentrum für Herz Kreislauf Forschung (grant no. DZHK_B 19 SE), and the Deutsche Forschungsgemeinschaft (grant no. KR3770/11-1 and KR3770/14-1). BMM is supported by the European Union’s Horizon 2020 Research and Innovation Programme (Marie Skłodowska-Curie grant, agreement no. 813533). BDK is supported by a British Heart Foundation personal chair (grant no. CH/13/2/30154).
Version 1. 11/17/2020
In-Press Preview
Version 2. 01/19/2021
Electronic publication
Funding Statement
to M.K.
to M.K.
to M.K.
to B. M.-M.
to BD. K.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(2):e141837.https://doi.org/10.1172/JCI141837.
See the related Commentary at Unraveling the genomic basis of congenital heart disease.
Contributor Information
Harald Lahm, Email: lahm@dhm.mhn.de.
Meiwen Jia, Email: jmwcathy@gmail.com.
Martina Dreßen, Email: dressen@dhm.mhn.de.
Nazan Puluca, Email: puluca@dhm.mhn.de.
Ralf Gilsbach, Email: gilsbach@vrc.uni-frankfurt.de.
Julie Cleuziou, Email: cleuziou@dhm.mhn.de.
Nicole Beck, Email: beck@dhm.mhn.de.
Olga Bondareva, Email: olga.bondareva@pharmakol.uni-freiburg.de.
Elda Dzilic, Email: dzilic@dhm.mhn.de.
Melchior Burri, Email: burri@dhm.mhn.de.
Karl C. König, Email: koenigc@dhm.mhn.de.
Johannes A. Ziegelmüller, Email: ziegelmueller@dhm.mhn.de.
Claudia Abou-Ajram, Email: abou@dhm.mhn.de.
Irina Neb, Email: neb@dhm.mhn.de.
Zhong Zhang, Email: dor_zhangzhong@163.com.
Stefanie A. Doppler, Email: doppler@dhm.mhn.de.
Elisa Mastantuono, Email: mastantuonoelisa@gmail.com.
Peter Lichtner, Email: lichtner@helmholtz-muenchen.de.
Gertrud Eckstein, Email: eckstein@helmholtz-muenchen.de.
Jürgen Hörer, Email: hoerer@dhm.mhn.de.
Peter Ewert, Email: ewert@dhm.mhn.de.
James R. Priest, Email: jpriest@stanford.edu.
Lutz Hein, Email: lutz.hein@pharmakol.uni-freiburg.de.
Rüdiger Lange, Email: lange@dhm.mhn.de.
Thomas Meitinger, Email: meitinger@helmholtz-muenchen.de.
Heather J. Cordell, Email: heather.cordell@newcastle.ac.uk.
Bertram Müller-Myhsok, Email: bmm@psych.mpg.de.
References
- 1.Dolk H, et al. Congenital heart defects in Europe: prevalence and perinatal mortality, 2000 to 2005. Circulation. 2011;123(8):841–849. doi: 10.1161/CIRCULATIONAHA.110.958405. [DOI] [PubMed] [Google Scholar]
- 2.van der Linde D, et al. Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J Am Coll Cardiol. 2011;58(21):2241–2247. doi: 10.1016/j.jacc.2011.08.025. [DOI] [PubMed] [Google Scholar]
- 3.Lozano R, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the global burden of disease study. Lancet. 2012;380(9859):2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basson CT, et al. Mutations in human TBX5 cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet. 1997;15(1):30–35. doi: 10.1038/ng0197-30. [DOI] [PubMed] [Google Scholar]
- 5.Weismann CG, Gelb BD. The genetics of congenital heart disease: a review of recent developments. Curr Opin Cardiol. 2007;22(3):200–206. doi: 10.1097/HCO.0b013e3280f629c7. [DOI] [PubMed] [Google Scholar]
- 6.Zaidi S, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498(7453):220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soemedi R, et al. Contribution of global rare copy-number variants to the risk of sporadic congenital heart disease. Am J Hum Genet. 2012;91(3):489–501. doi: 10.1016/j.ajhg.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glessner JT, et al. Increased frequency of de novo copy number variants in congenital heart disease by integrative analysis of single nucleotide polymorphism array and exome sequence data. Circ Res. 2014;115(10):884–896. doi: 10.1161/CIRCRESAHA.115.304458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaidi S, Brueckner M. Genetics and genomics of congenital heart disease. Circ Res. 2017;120(6):923–940. doi: 10.1161/CIRCRESAHA.116.309140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bayrak CS, et al. De novo variants in exomes of congenital heart disease patients identify risk genes and pathways. Genome Med. 2019;12(1):9. doi: 10.1186/s13073-019-0709-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Page DJ, et al. Whole exome sequencing reveals the major contributors to nonsyndromic tetralogy of Fallot. Circulation. 2019;124(4):553–563. doi: 10.1161/CIRCRESAHA.118.313250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li AH, et al. Whole exome sequencing in 342 congenital cardiac left sided lesion cases reveals extensive genetic heterogeneity and complex inheritance patterns. Genome Med. 2017;9(1):95. doi: 10.1186/s13073-017-0482-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cristo F, et al. Functional study of DAND5 variant in patients with congenital heart disease and laterality defects. BMC Med Genet. 2017;18(1):77. doi: 10.1186/s12881-017-0444-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin SC, et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet. 2017;49(11):1593–1601. doi: 10.1038/ng.3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arrington CB, et al. Family-based studies to identify genetic variants that cause congenital heart defects. Future Cardiol. 2013;9(4):507–518. doi: 10.2217/fca.13.40. [DOI] [PubMed] [Google Scholar]
- 16.Agopian AJ, et al. Genome-wide association studies and meta-analyses for congenital heart defects. Circ Cardiovasc Genet. 2017;10(3):e001449. doi: 10.1161/CIRCGENETICS.116.001449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin Y, et al. Association analysis identifies new risk loci for congenital heart disease in Chinese populations. Nat. Commun. 2015;6:8082. doi: 10.1038/ncomms9082. [DOI] [PubMed] [Google Scholar]
- 18.Hu Z, et al. A genome-wide association study identifies two risk loci for congenital heart malformations in Han Chinese populations. Nat Genet. 2013;45(7):818–821. doi: 10.1038/ng.2636. [DOI] [PubMed] [Google Scholar]
- 19.Cordell HJ, et al. Genome-wide association study of multiple congenital heart disease phenotypes identifies a susceptibility locus for atrial septal defect at chromosome 4p16. Nat Genet. 2013;45(7):822–824. doi: 10.1038/ng.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao L, et al. Association between the European GWAS-identified susceptibility locus at chromosome 4p16 and the risk of atrial septal defect: a case-control study in Southwest China and a meta-analysis. PLoS One. 2015;10(4):e0123959. doi: 10.1371/journal.pone.0123959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cordell HJ, et al. Genome-wide association study identifies loci on 12q24 and 13q32 associated with Tetralogy of Fallot. Hum Mol Genet. 2013;22(7):1473–1481. doi: 10.1093/hmg/dds552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soemedi R, et al. Phenotype-specific effect of chromosome 1q21.1 rearrangements and GJA5 duplications in 2436 congenital heart disease patients and 6760 controls. Hum Mol Genet. 2012;21(7):1513–1520. doi: 10.1093/hmg/ddr589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Visscher PM, et al. Five years of GWAS discovery. Am J Hum Genet. 2012;90(1):7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobs ML, et al. The society of thoracic surgeons congenital heart surgery database: 2019 update on research. Ann Thorac Surg. 2019;108(3):671–679. doi: 10.1016/j.athoracsur.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. The Society of Thoracic Surgeons Congenital Heart Database. Data collection form version 3.3. https://www.sts.org/sites/default/files/documents/CongenitalDCF_v3_3_Annotated_Updated20160119.pdf Updated January 19, 2016. Accessed August 18, 2020.
- 26.Jin N, Burkard ME. MACROD2, an original cause of CID? Cancer Discov. 2018;8(8):921–923. doi: 10.1158/2159-8290.CD-18-0674. [DOI] [PubMed] [Google Scholar]
- 27.Chang YC, et al. Genome-wide scan for circulating vascular adhesion protein-1 levels: MACROD2 as a potential transcriptional regulator of adipogenesis. J Diabetes Invest. 2018;9(5):1067–1074. doi: 10.1111/jdi.12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu SM, et al. Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell. 2006;127(6):1137–1150. doi: 10.1016/j.cell.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 29.Nothjunge S, et al. DNA methylation signatures follow preformed chromatin compartments in cardiac myocytes. Nat Commun. 2017;8(1):1667. doi: 10.1038/s41467-017-01724-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Leeuw CA, et al. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11(4):e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Soysa TY, et al. Single-cell analysis of cardiogenesis reveals basis for organ-level developmental defects. Nature. 2019;572(7767):120–124. doi: 10.1038/s41586-019-1414-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, et al. 3D chromatin architecture remodeling during human cardiomyocyte differentiation reveals a novel role of HERV-H in demarcating chromatin domains. Nat Genet. 2019;51(9):1380–1388. doi: 10.1038/s41588-019-0479-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sahara M, et al. Population and single-cell analysis of human cardiogenesis reveals unique LGR5 ventricular progenitors in embryonic outflow tract. Dev Cell. 2019;48(4):475–490.e7. doi: 10.1016/j.devcel.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 34.Krishnan A, et al. A detailed comparison of mouse and human cardiac development. Pediatr Res. 2014;76(6):500–507. doi: 10.1038/pr.2014.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cui Y, et al. Single-cell transcriptome analysis maps the developmental track of the human heart. Cell Rep. 2019;26(7):1934–1950.e5. doi: 10.1016/j.celrep.2019.01.079. [DOI] [PubMed] [Google Scholar]
- 36.Slavin TP, et al. Two-marker association tests yield new disease associations for coronary artery disease and hypertension. Hum Genet. 2011;130(6):725–733. doi: 10.1007/s00439-011-1009-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakthianandeswaren A, et al. MACROD2 haploinsufficiency impairs catalytic activity of PARP1 and promotes chromosome instability and growth of intestinal tumors. Cancer Discov. 2018;8(8):988–1005. doi: 10.1158/2159-8290.CD-17-0909. [DOI] [PubMed] [Google Scholar]
- 38.Maas NM, et al. The C20orf133 gene is disrupted in a patient with Kabuki syndrome. J Med Genet. 2007;44(9):562–569. doi: 10.1136/jmg.2007.049510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao W, et al. High-resolution analysis of copy number variants in adults with simple-to-moderate congenital heart disease. Am J Med Genet A. 2013;161A(12):3087–3094. doi: 10.1002/ajmg.a.36177. [DOI] [PubMed] [Google Scholar]
- 40.Hitz MP, et al. Rare copy number variants contribute to congenital left-sided heart disease. PLoS Genet. 2012;8(9):e1002903. doi: 10.1371/journal.pgen.1002903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fakhro KA, et al. Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proc Natl Acad Sci U S A. 2015;108(7):2915–2920. doi: 10.1073/pnas.1019645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Costain G, et al. Genome-wide rare copy number variations contribute to genetic risk for transposition of the great arteries. Int J Cardiol. 2016;204:115–121. doi: 10.1016/j.ijcard.2015.11.127. [DOI] [PubMed] [Google Scholar]
- 44.Li N, Chen J. ADP-ribosylation: activation, recognition, and removal. Mol Cells. 2014;37(1):9–16. doi: 10.14348/molcells.2014.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mohseni M, et al. MACROD2 overexpression mediates estrogen independent growth and tamoxifen resistance in breast cancers. Proc Natl Acad Sci U S A. 2014;111(49):17606–17611. doi: 10.1073/pnas.1408650111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bilinovich SM, et al. The long noncoding RNA RPS10P2-AS1 is implicated in autism spectrum disorder risk and modulates gene expression in human neuronal progenitor cells. Front Genet. 2019;10:970. doi: 10.3389/fgene.2019.00970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hay JC, et al. Protein interactions regulating vesicle transport between the endoplasmic reticulum and Golgi apparatus in mammalian cells. Cell. 1997;89(1):149–158. doi: 10.1016/S0092-8674(00)80191-9. [DOI] [PubMed] [Google Scholar]
- 48.Pan S, et al. G-T haplotype established by rs3785889-rs16941382 in GOSR2 gene is associated with coronary artery disease in Chinese Han population. Oncotarget. 2017;8(47):82165–82173. doi: 10.18632/oncotarget.19280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meyer TE, et al. GOSR2 Lys67Arg is associated with hypertension in whites. Am J Hypertens. 2009;22(2):163–168. doi: 10.1038/ajh.2008.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pan S, et al. A haplotype of the GOSR2 gene is associated with myocardial infarction in Japanese men. Genet Test Mol Biomarkers. 2013;17(6):481–488. doi: 10.1089/gtmb.2012.0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kau T, et al. Aortic development and anomalies. Semin Intervent Radiol. 2007;24(2):141–152. doi: 10.1055/s-2007-980040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jain D, et al. ketu mutant mice uncover an essential meiotic function for the ancient RNA helicase YTHDC2. Elife. 2018;7:e30919. doi: 10.7554/eLife.30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu Y, et al. The ion channel ASIC2 is required for baroreceptor and autonomic control of the circulation. Neuron. 2009;64(6):885–897. doi: 10.1016/j.neuron.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tian J, et al. Calycosin inhibits the in vitro and in vivo growth of breast cancer cells through WDR7-7-GPR30 signaling. J Exp Clin Cancer Res. 2017;36(1):153. doi: 10.1186/s13046-017-0625-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shah R, et al. The prolyl 3-hydroxylases P3H2 and P3H3 are novel targets for epigenetic silencing in breast cancer. Br J Cancer. 2009;100(10):1687–1696. doi: 10.1038/sj.bjc.6605042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen YH, et al. Msx1 and Msx2 regulate survival of secondary heart field precursors and post-migratory proliferation of cardiac neural crest in the outflow tract. Dev Biol. 2007;308(2):421–437. doi: 10.1016/j.ydbio.2007.05.037. [DOI] [PubMed] [Google Scholar]
- 57.Holle R, et al. KORA—a research platform for population based health research. Gesundheitswesen. 2005;67(Suppl1):S19–S25. doi: 10.1055/s-2005-858235. [DOI] [PubMed] [Google Scholar]
- 58.Nicolazzi EL, et al. AffyPipe: an open-source pipeline for Affymetrix Axiom genotyping workflow. Bioinfomatics. 2014;30(21):3118–3119. doi: 10.1093/bioinformatics/btu486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goddard ME, et al. Using the genomic relationship matrix to predict the accuracy of genomic selection. J Anim Breed Genet. 2011;128(6):409–421. doi: 10.1111/j.1439-0388.2011.00964.x. [DOI] [PubMed] [Google Scholar]
- 61.Lee JJ, et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet. 2018;50(8):1112–1121. doi: 10.1038/s41588-018-0147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen W, et al. Fine mapping causal variants with an approximate bayesian method using marginal test statistics. Genetics. 2015;200(3):719–736. doi: 10.1534/genetics.115.176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pickrell JK. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am J Hum Genet. 2014;94(4):559–573. doi: 10.1016/j.ajhg.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fishilevich S. et al. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford) 2017:bax028. doi: 10.1093/database/bax028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang X, Wu SM. Isolation and functional characterization of pluripotent stem cell-derived cardiac progenitor cells. Curr Protoc Stem Cell Biol. 2010;Chapter 1:Unit 1F.10. doi: 10.1002/9780470151808.sc01f10s14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Burridge PW, et al. Chemically defined generation of human cardiomyocytes. Nat Methods. 2014;11(8):855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Afgan E, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46(W1):W537–W544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17(1):10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 69.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. BroadInstitue. Picard tools: MarkDuplicates. https://broadinstitute.github.io/picard/ Accessed August 15,2020.
- 71.Liao Y, et al. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 72.Butler A, et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20(1):296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stuart T, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-Seq data for single cells obtained from adult human atria and ventricles have been deposited in the NCBI’s GEO database (GEO GSE161016; https://www.ncbi.nlm.nih.gov/geo/).