Abstract
Expanded human skin fibroblast cells from four different aged healthy individuals, 11-year-old female, 1-year-old male, 2-month-old male, and 8-year-old male, were used to generate integration-free induced pluripotent stem cell (iPSC) lines TRNDi021-C, TRNDi023-D, TRNDi024-D, and TRNDi025-A, respectively, by exogenous expression of four reprogramming factors, human SXO2, OCT3/4, C-MYC, KLF4. The authenticity of established iPSC lines was confirmed by the expressions of stem cell markers, karyotype analysis, and teratoma formation. These iPSC lines could serve as young healthy controls for the studies involving patient-specific iPSCs.
1. Resource Table:
| Unique stem cell lines identifier | TRNDi021-C |
| TRNDi023-D | |
| TRNDi024-D | |
| TRNDi025-A | |
| Alternative names of stem cell lines | HT725C (TRNDi021-C)HT727D |
| (TRNDi023-D)HT728D | |
| (TRNDi024-D)HT729A | |
| (TRNDi025-A) | |
| Institution | National Center for Advancing Translational Sciences |
| Contact information of distributor | wei.zheng@nih.gov |
| Type of cell lines | iPSC |
| Origin | Human |
| Cell Source | Skin fibroblasts |
| Clonality | Clonal |
| Method of reprogramming | Integration-free Cytotune 2.0 SeV kit |
| Multiline rationale | Control iPSC lines from young healthy donors |
| Gene modification | NO |
| Type of modification | N/A |
| Associated disease | N/A |
| Gene/locus | N/A |
| Method of modification | N/A |
| Name of transgene or resistance | N/A |
| Inducible/constitutive system | N/A |
| Date archived/stock date | 4/25/2019 |
| Cell line repository/bank | N/A |
| Ethical approval | NIGMS Informed Consent Form was obtained from four healthy individuals at time of sample submission. |
| Confidentiality Certificate: CC-GM-15–004 |
2. Resource utility
These human iPSC lines could be a useful cell resource as young healthy controls for the studies involving patient-specific iPSCs and for research related to evaluations of compound toxicities.
3. Resource details
Accumulating evidences suggest that almost all types of diseased iPSCs can be generated, even in certain cases where reprogramming was previously thought to be very difficult or impossible. Most publications and cell resources are focused on the collection of patient-specific or disease-specific iPSC lines. However, there is still a shortage of control iPSC lines from of healthy, different-aged individuals. Here we report the generation of four human iPSC lines from skin fibroblasts of different-aged young healthy individuals. We have used a non-integrating viral vector containing human OCT3/4, KLF4, SOX2 and C-MYC transcription factors to transduce the fibroblasts according to the method described previously (Beers et al., 2012, 2015). All these four healthy iPSC lines demonstrated a classic embryonic stem cell morphology (Fig. 1A) and normal karyotypes (46, XY or 46, XX), as confirmed by G-banding karyotyping at passage 9 (Supplemental Fig. S1A), and exhibited positive expression of NANOG, TRA-1-60 and SSEA4, as measured by flow cytometry, and of OCT4A, NANOG, SOX2 and SSEA-4, as determined by immunofluorescence staining (Fig. 1B). Moreover, the pluripotency of these iPSCs lines was confirmed by a teratoma formation assay that showed their ability to differentiate into tissues of all three germ layers, including neural epithelium (N.E.) and pigment epithelium (P.E.) from ectoderm, cartilage from mesoderm, and gut from endoderm, in vivo (Fig. 1C). Short tandem repeat (STR) DNA profiling analysis showed the genotypes of these four iPSC lines matched those of the parental fibroblasts (submitted in archive with journal). Mycoplasma tests in these iPSC lines showed the negative results (Supplemental Fig. S1B) (Table 2).
Fig. 1.
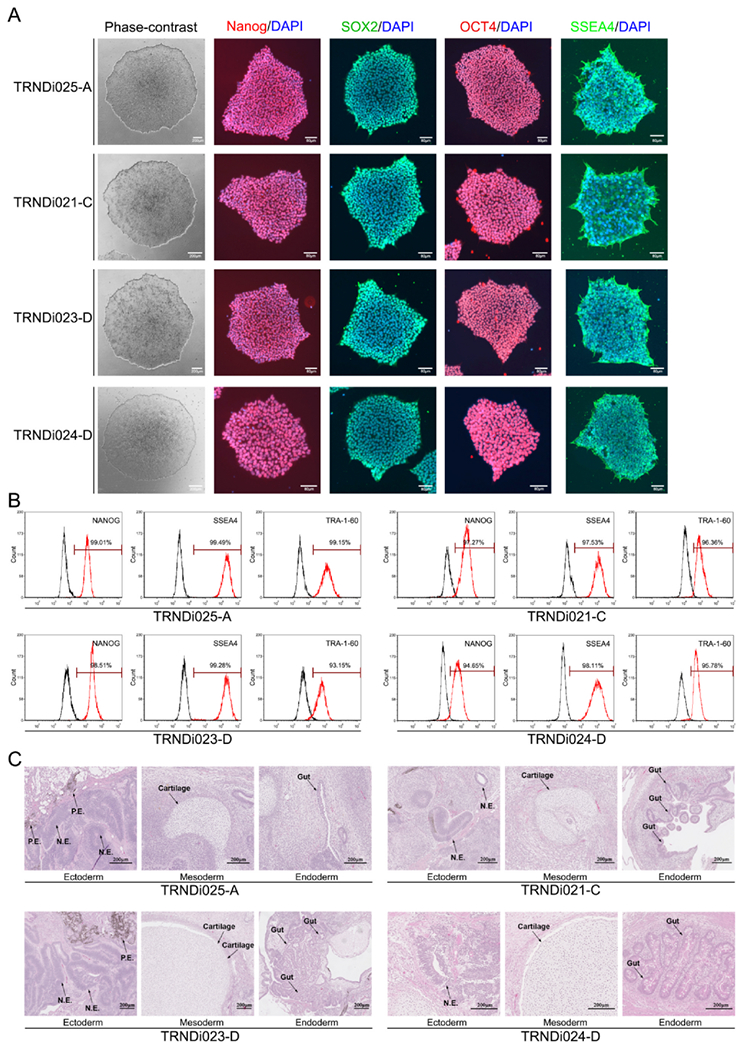
(A) Images of phase contrast microscopy and immunofluorescence staining of pluripotency markers of 4 iPSC lines. (B) Flow cytometry analysis of pluripotency markers of 4 iPSC lines. (C) Teratoma formation assay shows all 4 iPSC liness can generate three germ layers, including neural epithelium (N.E.) and pigment epithelium (P.E.) from ectoderm, cartilage from mesoderm, and gut from endoderm, in vivo.
Table 2.
Characterization and validation.
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Photography | Normal | Fig. 1A |
| Phenotype | Qualitative analysis: Immunocytochemistry Qantitative analysis: Flow Cytometry |
SOX2, OCT3/4, NANOG, SSEA-4 TRNDi021-C (TRA-1–60: 96.36% SSEA-4: 97.53% NANOG: 97.27%) TRNDi023-D (TRA-1–60: 93.15% SSEA-4: 99.28% NANOG: 98.51%) TRNDi024-D (TRA-1–60: 95.78% SSEA-4: 98.11% NANOG: 94.65%), TRNDi025-A (TRA-1–60: 99.15% SSEA-4: 99.49% NANOG: 99.01%) |
Fig. 1A Fig. 1B |
| Genotype | Karyotype (G-banding) and resolution | 46,XY or 46, XX Resolution: 425–475 | Supplemental Fig. S1A |
| Identity | Microsatellite PCR (mPCR) OR STR analysis |
not performed | N/A |
| STR analysis | 16 or 18 sites tested, all sites matched | Submitted in archive with journal | |
| Mutation analysis (IF APPLICABLE) | Sequencing Southern Blot OR WGS |
N/A N/A |
N/A N/A |
| Microbiology and virology | Mycoplasma | Mycoplasma testing by luminescence. Negative | Supplementary Fig. S1B |
| Differentiation potential | e.g. Embryoid body formation OR Teratoma formation OR Scorecard OR Directed differentiation | Teratoma formed with tissues of all three germ layers (Ectoderm, Mesoderm, Endoderm) in vivo | Fig. 1C |
| Donor screening (OPTIONAL) | HIV 1 + 2 Hepatitis B, Hepatitis C | N/A | N/A |
| Genotype additional info (OPTIONAL) | Blood group genotyping HLA tissue typing |
N/A N/A |
N/A N/A |
4. Materials and methods
4.1. Cell culture
Skin fibroblasts derived from healthy donors (GM02036 from 11-year female, GM05659 from 1-year male, GM05756 from 2-month male, and GM08398 from 8-year male) were obtained from Coriell Institute for Medical Research (Camden, NJ). Fibroblasts were cultured in DMEM (Life Technologies 11195–073) supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin and 100 μg/ml streptomycin (Life technologies 10378–016) in a humidified incubator with 5% CO2 and 20% O2 at 37 °C. Cells were routinely trypsinized (0.05% trypsin/EDTA) and passaged until reprogramming. Human iPSCs were maintained on Matrigel (Corning, #354230) in the chemically-defined Essential 8 medium (Life Technologies, #A1517001) and passaged using 0.5 mM EDTA/DPBS, as described previously (Beers et al., 2012) (Table 1).
Table 1.
Summary of lines.
| iPSC line names | Abbreviation in figures | Gender | Age | Ethnicity | Genotype of locus | Disease |
|---|---|---|---|---|---|---|
| TRNDi021-C | TRNDi021-C | Female | 11-year-old | Caucasian | Wild-type | N/A |
| TRNDi023-D | TRNDi023-D | Male | 1-year-old | Caucasian | Wild-type | N/A |
| TRNDi024-D | TRNDi024-D | Male | 2-month-old | Caucasian | Wild-type | N/A |
| TRNDi025-A | TRNDi025-A | Male | 8-year-old | Caucasian | Wild-type | N/A |
4.2. iPSC generation
Human iPSCs were established from skin fibroblasts using non-integrating Sendai virus technology as previously described (Takahashi and Yamanaka, 2006; Beers et al., 2012, 2015). Briefly, fibroblasts were infected according to the kit manual (#A16517, ThermoFisher). Infected cells were cultured in DMEM + 10%FBS + Pen/Strep medium for 4 days, before being re-plated onto Matrigel coated plates containing reprogramming medium and cultured until 25 days into reprogramming. Reprogramming medium is made by adding 64 mg/L L-ascorbic acid, 13.6 μg/L sodium selenite, 10 μg/mL Holo-transferrin, 100 ng/mL bFGF, 20 μg/mL insulin, 100 μM sodium butyrate to DMEM/F12 (Beers et al., 2015). Cells were switched to Essential 8 medium at day 20 after Sendai virus infection. Finally, iPSC colonies were picked based on human embryonic stem cell-like morphology, expanded, and cryopreserved by passage 3.
4.3. Immunocytochemistry
Immunofluorescence staining of iPSC colonies were performed as previously described (Lin et al., 2017). iPSCs were fixed in 2% paraformaldehyde (PFA) for 10 min at room temperature, washed with DPBS, and permeabilized with 0.2% Triton X-100/DPBS for 3 min. After blocking using 10 mg/ml bovine serum albumin (BSA) for 30 min, samples were incubated with primary antibody for SOX2, OCT4, NANOG, or SSEA4 for 1 h at room temperature. iPSCs were then washed with DPBS and incubated with Alexa 488 or 594-conjugated secondary antibodies for 1 h. Then nuclei staining was performed with 49,6-diamidino-2-phenylindole (DAPI). Cell images were captured using an EVOS FL cell imaging station (Thermo Fisher). The primary and secondary antibodies are listed in Table 3.
Table 3.
Reagents details.
| Antibodies used for immunoeytoehemistry/flow-eytometry | |||
|---|---|---|---|
| Antibody | Dilution | Company Cat # and RRID | |
| Pluripotency Markers | Mouse anti-SOX2 | 1:50 | R & D systems, Cat# MAB2018, RRID: AB_358009 |
| Pluripotency Markers | Rabbit anti-NANOG | 1:400 | Cell signaling, Cat# 4903, RRID: AB_10559205 |
| Pluripotency Markers | Rabbit anti-OCT4 | 1:400 | Thermo Fisher, Cat# 701756, RRID: AB_2633031 |
| Pluripotency Markers | Mouse anti-SSEA4 | 1:1000 | Cell signaling, Cat# 4755, RRID: AB_1264259 |
| Secondary Antibodies | Donkey anti-Mouse IgG (Alexa Fluor 488) | 1:400 | Thermo Fisher, Cat# A21202, RRID: AB_141607 |
| Secondary Antibodies | Donkey anti-Rabbit IgG (Alexa Fluor 594) | 1:400 | Thermo Fisher, Cat# A21207, RRID: AB_141637 |
| Flow Cytometry Antibodies | Anti-Tra-1–60-DyLight 488 | 1:50 | Thermo Fisher, Cat# MA1-023-D488X, RRID: AB_2536700 |
| Flow Cytometry Antibodies | Anti-Nanog-Alexa Fluor 488 | 1:50 | Millipore, Cat# FCABS352A4, RRID: AB_10807973 |
| Flow Cytometry Antibodies | anti-SSEA-4-Alexa Fluor 488 | 1:50 | Thermo Fisher, Cat# 53–8843-41, RRID: AB_10597752 |
| Flow Cytometry Antibodies | Mouse-IgM-DyLight 488 | 1:50 | Thermo Fisher, Cat# MA1-194-D488, RRID: AB_2536969 |
| Flow Cytometry Antibodies | Rabbit IgG-Alexa Fluor 488 | 1:50 | Cell Signaling, Cat# 4340S, RRID: AB_10694568 |
| Flow Cytometry Antibodies | Mouse IgG3-FITC | 1:50 | Thermo Fisher, Cat# 11–4742-42, RRID: AB_2043894 |
4.4. Flow Cytometry
iPSCs were suspend in permeabilization buffer (2% FBS and 0.2% Tween-20 in DPBS) for 10 min at room temperature and then incubated with the fluorophore-conjugated antibodies for 1 h at 4 °C. Nonspecific rabbit or mouse IgG served as the negative control (Antibodies used are listed in Table 3). After washing, the cells were detected, and the fluorescence signals were determined using a FACScan flow cytometer (BD Biosciences) at 488 nm. At least 10,000 events were collected from the cell gate. The data were then analyzed on a BD Accuri™ C6 Flow-Cytometry system (BD Biosciences).
4.5. G-banding karyotyping
G-banding karyotyping was performed by WiCell Cytogenetics lab (Madison, WI) using twenty randomly selected metaphases.
4.6. Short tandem repeat (STR) analysis
STR analysis for TRNDi021-C, TRNDi023-D, and GM5659 fibroblast was performed by Johns Hopkins University GRCF DNA Services using PowerPlex 18D System (Promega). STR analysis for GM2036 fibroblast, GM5756 fibroblast, GM8398 fibroblast, TRNDi024-D, and TRNDi025-A was performed by WiCell Cytogenetics lab using Powerplex 16 System (Promega). Genomic DNA used in STR analysis was extracted from the fibroblasts and iPSCs using DNeasy Blood and Tissue Kit (Qiagen).
4.7. Mycoplasma detection
Mycoplasma testing were performed following MycoAlert kit instructions (Lonza). By measuring the level of ATP in an iPSC line both before (read A) and after the addition of the MycoAlert substrate (read B), a ratio can be obtained which is indicative of the presence or absence of mycoplasma. Ratio B/A > 1.2 indicates mycoplasma positive and Ratio B/A < 0.9 indicates mycoplasma negative.
4.8. Teratoma formation assay
Human iPSCs were collected after 0.5 mM EDTA/DPBS treatment and subcutaneously injected into NSG mice (JAX #005557) with cold Matrigel (Corning #354277). Visible tumors typically appeared 6–8 weeks post injection. Palpable tumors were removed and fixed in 10% Neutral Buffer Formalin. They were then embedded in paraffin and stained with H&E according to standard procedures to check their differentiation capacity into all three germ layers in vivo.
Supplementary Material
Acknowledgements
This work was supported by the Intramural Research Programs of National Heart, Lung, and Blood Institute to Dr. Jizhong Zou (Funding Number: ZIC HL006145) and National Center for Advancing Translational Sciences to Dr. Wei Zheng (Funding Number: ZIA TR000018-05) at NIH.
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2020.102011.
References
- Beers J, Gulbranson DR, George N, Siniscalchi LI, Jones J, Thomson JA, Chen G, 2012. Passaging and colony expansion of human pluripotent stem cells by enzymefree dissociation in chemically defined culture conditions. Nat. Protoc 7, 2029–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beers J, Linask KL, Chen JA, Siniscalchi LI, Lin Y, Zheng W, Rao M, Chen G, 2015. A cost- effective and efficient reprogramming platform for large-scale production of integration-free human induced pluripotent stem cells in chemically defined culture. Sci. Rep 5, 11319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Linask KL, Mallon B, Johnson K, Klein M, Beers J, Xie W, Du Y, Liu C, Lai Y, Zou J, Haigney M, Yang H, Rao M, Chen G, 2017. Heparin promotes cardiac differentiation of human pluripotent stem cells in chemically defined albumin-free medium, enabling consistent manufacture of cardiomyocytes. Stem Cells Transl. Med 6 (2), 527–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S, 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126 (4), 663–706. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


