Abstract
Urine can accumulate systemic changes with no mechanism to be stable, which may reflect early changes associated with physiological or pathophysiological processes. To explore the potential value of the urine proteome, two rat models were established by intrahepatic injection of two different hepatoma cell lines, CBRH-7919 and RH-35. Urine samples were collected and analyzed. Compared with controls, the two models exhibited different numbers and types of differentially expressed urinary proteins despite having similar histological results. The results were compared with the urine proteome of a Walker 256 (W-256) liver tumor model. The differentially expressed urinary protein patterns in the three models were different. These findings demonstrate that changes in the urine proteomes of the two models can be detected at early stages and that the patterns of differentially expressed urinary proteins can differ even when the histological results are similar. Urinary proteins have potential utility for distinguishing among different tumor cells grown in the same organ.
1. Introduction
Urine, an ideal biomarker resource, can accumulate systemic changes and sensitively reflect changes associated with physiological or pathophysiological processes at early stages [1]. Due to the lack of control by a homeostatic mechanism, urine can tolerate more and higher-magnitude changes than other body fluids, including blood and cerebrospinal fluid [1]. It can also be obtained easily and noninvasively in large quantities [2]. However, urine is affected by various factors, including sex, age, diet, exercise, and lifestyle [3]. It is difficult to sort out disease factors due to the changes in clinical urine samples caused by other factors. The use of animal models can help avoid the effects of such factors in related research [4]. Specifically, animal models can minimize the influences of confounding factors and can be used to monitor whole processes from disease onset, which will be helpful for discovery of early biomarker candidates and future clinical validation. Animal models such as myocarditis [5], Alzheimer's [6], liver fibrosis [7], glioma [8], pulmonary fibrosis [9], intracerebral W-256 [10], and chronic pancreatitis models [11] have been widely used to research different diseases. The related studies have indicated that animal models are effective tools for identification of early urinary protein biomarkers, which can help elucidate the starting point of a disease and the dynamic changes in the urine proteome that occur throughout disease development.
Recent studies have shown that the urine proteome has great potential not only for discovering early biomarker candidates but also for distinguishing some subtle differences, such as the differences between the same types of tumor cells grown in different organs [12]. Different animal models have been established by injection of W-256 carcinosarcoma cells into different organs, including the W-256 subcutaneous model [2], the W-256 intracerebral tumor model [10], the W-256 liver tumor model [12], and the W-256 lung metastasis model [13]. For these four models, changes in the urinary proteome can be identified at early stages, even before histological examination or magnetic resonance imaging (MRI) reveals changes. Previous research has indicated that urinary proteins can be used to differentiate among tumor cells of the same type grown in different organs [12]. In addition, the urine proteome has been reported to reflect pathophysiological changes with extremely high sensitivity [14]. In one study, changes in the urine proteome could be sensitively detected even when only approximately ten tumor cells were injected subcutaneously in rats, and changes in biological pathways have been reported to be associated with tumors. These findings led us to investigate the sensitivity of urinary proteins. We hypothesized that urinary proteins have the potential value for distinguishing among different types of tumor cells grown in the same organ.
In this study, we established two rat models by intrahepatic injection of two different hepatoma cell lines, CBRH-7919 and RH-35, at the same dose. Urine samples were collected and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) on days 0, 5, 7, 14, and 28. Dynamic changes in the urinary proteome were analyzed and compared with those of the W-256 liver tumor model [12]. This study aimed to discover dynamic changes in urinary proteins during the growth of two hepatoma cell lines in the liver and to investigate the usefulness of the urine proteome for differentiating among different tumor cell types grown in the same organ.
2. Materials and Methods
2.1. Animal Models
All experiments in this study were approved by the Institutional Animal Care, Use and Welfare Committee of the Institute of Basic Medical Sciences, Peking Union Medical College (Animal Welfare Assurance Number: ACUC-A02-2014-007), and all methods were performed in accordance with the relevant guidelines and regulations. Two different tumor-bearing animal models were established in this study. Fifty male Wistar rats (180 ± 20 g) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. The animal license was SCXK (Beijing) 2016-0006. All Wistar rats were randomly divided into different groups: the control group (n = 10), the CBRH-7919 experimental group (n = 20), and the RH-35 experimental group (n = 20). Food and water were withheld from all rats for 12 h before urine samples were collected for the modeling experiments.
The two rat hepatoma cell lines, CBRH-7919 and RH-35 (China Infrastructure of Cell Line Resource), were all cultured in DMEM (Gibco) supplemented with 10% fetal bovine serum (Gibco), 100 IU/mL penicillin G, and 100 μg/mL streptomycin. The cells were maintained in a humidified atmosphere with 5% CO2 and 95% air. Before each implantation procedure, the viability of cells was tested via trypan blue staining (confirming >90% cell viability for each tumor implantation procedure). The cell suspension concentration was 1 × 108 cells/mL. In the CBRH-7919 model, all rats were anesthetized with sodium pentobarbital first; then, the rat abdominal cavity was opened, and the left medial lobe of the liver was exposed. CBRH-7919 hepatoma cell suspension (0.1 mL) was visually injected under the hepatic capsule into this lobe. An equal volume of PBS was also injected into the same location in each of the control rats. After the injection, the wound was sutured. In the RH-35 model, all procedures were the same as those described above.
2.2. Histological Analysis
The livers of twenty-four experimental rats and two control rats were randomly harvested at 5, 7, 14, 21, 28, and 35 days after injection, and the rest of the rats were sacrificed at day 42. The livers were used for histopathology. All the samples were fixed in formalin, embedded in paraffin, sectioned, and evaluated with hematoxylin and eosin (H&E) staining.
2.3. Urine Sample Preparation
Urine samples were collected from all rats before sacrifice, including the experimental rats who were randomly selected for histopathology at different time points. During urine collection, food and water were withheld from all rats. The rats were placed in metabolic cages alone overnight to collect urine. The urine samples were temporarily stored at −80°C for later use. Before LC-MS/MS analysis, the urine samples were thawed and centrifuged at 12,000 ×g for 30 min to remove impurities. The supernatants were transferred to new centrifuge tubes and mixed with three volumes of precooled ethanol. The mixtures were precipitated at −20°C for 12 h. Then, the samples were centrifuged for 30 min, the supernatant was discarded, the precipitates were dried, and lysis buffer (8 mol/L urea, 2 mol/L thiourea, 50 mmol/L Tris, and 25 mmol/L dithiothreitol (DTT)) was added until the precipitates dissolved. The samples were again centrifuged at 12,000 ×g for 30 min, and the supernatant was retained. The protein concentration was measured by Bradford assay. All proteins were digested by the FASP method [15]. One hundred micrograms of each sample were added to a 10 kDa filter device (Pall, Port Washington, NY, USA), and prepared urea buffer (UA; 8 mol/L urea and 0.1 mol/L Tris-HCl, pH 8.5) and 25 mmol/L NH4HCO3 were used to wash the protein several times in sequence. Then, 20 mmol/L DTT (Sigma) was added to the protein samples, and the samples were incubated at 37°C for 1 h. Next, 50 mmol/L iodoacetamide (IAA, Sigma) was added to the samples, which were incubated for 40 min in the dark. After centrifuging the samples at 14,000 ×g for 40 min, UA and NH4HCO3 were added again to wash the protein several times, trypsin (enzyme-to-protein ratio of 1 : 50) was added to the protein, and the samples were digested at 37°C overnight. The collected peptide mixtures were desalted with Oasis HLB cartridges (Waters, Milford, MA) and dried by vacuum evaporation. Then, all peptides were diluted to 0.5 μg/μL with 0.1% formic acid (FA). The concentrations were determined by BCA assay.
A total of 76 samples were chosen for LC-MS/MS analysis: 8 urine samples from control rats; 32 samples from CBRH-7919 experimental rats on days 5, 7, 14, and 28; and 36 samples from RH-35 experimental rats on days 5, 7, 14, and 28. In the CBRH-7919 model, a pooled sample of equal amounts of polypeptides from the 40 individuals (8 control samples and 32 CBRH-7919 model samples) was used for library generation, whereas the 40 individual samples were used to analyze the urinary proteins. The 44 samples for the RH-35 model were also treated according to the method mentioned above. The two pooled samples of polypeptides were separated with a Pierce High pH Reversed-Phase Peptide Separation Kit (catalog number: 84868, Thermo Fisher Scientific, USA) according to the manufacturer's instructions. Ten effluents of each model mixture were obtained after separation and evaporation to dryness in vacuo, labeled, and stored at −80°C.
2.4. LC-MS/MS Analysis
Both models were treated before LC-MS/MS. A calibration kit (iRT kit, Ki3002, Biognosys AG, Switzerland), which contained a mixture of nonnaturally occurring synthetic peptides, was used in this study. iRT was added to each sample before measurement at a ratio of 1 : 10. For data-dependent acquisition (DDA), ten isolated samples from each model obtained by the above method were added to 25 μL of 0.1% FA and centrifuged at 14,000 ×g and 4°C for 30 min. Twenty microliters of each sample was mixed with 2 μL of the iRT polypeptide. Then, 2 μL of each peptide sample was loaded on the trap column and separated on a reverse-phase C18 analytical column with an EASY-nLC 1200 HPLC system (Thermo Fisher Scientific, USA) at 0.25 μL/min (the column flow rate) for 90 min. A Thermo Orbitrap Fusion Lumos Tribrid Mass Spectrometer (Thermo Fisher Scientific, USA) was used for the analysis. The MS data were acquired using the following parameters: a spray voltage of 2.4 kV, an ion transfer tube temperature of 320°C, a first-level full scan range of 350–1,550 m/z with a resolution of 60,000, a secondary scan range of 200–2,000 m/z with a resolution of 30,000, a cycle time of 3 s, and 30% HCD collision energy.
For data-independent acquisition (DIA), to prepare a mixed sample for quality control for each model, 3 μL of each peptide sample was taken, and the iRT polypeptide was added at a ratio of 1 : 10 (20 μL of each sample was mixed with 2 μL of the iRT polypeptide). The 40 samples from the CBRH-7919 model and the 44 samples from the RH-35 model were processed by DIA individually to assess proteome differences. The variable window parameter was set to 36 isolation windows, and the maximum injection time was 50 ms for the full scan and DIA scans. The MS1 parameter settings were as follows: a resolution of 60,000, a range of 350–1400 m/z, and an automatic gain control (AGC) of 1e6. The MS2 parameters included a resolution of 30,000.
2.5. Data Analysis
For each model, 10 raw files from DDA were searched using Proteome Discoverer software (PD, version 2.1, Matrix Science, UK) with the SwissProt_2017_02 database (taxonomy: Rattus; containing 7992 sequences). The search conditions included trypsin digestion, two missed cleavage sites, cysteine as the fixed modification, methionine oxidation as the variable modification, 10 ppm as the peptide mass tolerance, and 0.02 Da as the fragment mass tolerance. The applied false discovery rate (FDR) cutoff was 0.01 at the protein level. Then, the PD results and 10 DDA raw files were imported into Spectronaut X software (Biognosys, Switzerland) to generate the spectral library used for DIA data analysis. All the single DIA raw files were also analyzed using Spectronaut X software. The k-nearest neighbor (K-NN) method was used to fill the missing values of protein abundance.
2.6. Statistical Analysis
Comparisons between two groups were conducted using two-sided unpaired t-tests. The selection criteria for differentially expressed urinary proteins were a fold change ≥2 or ≤0.5 and a P value <0.05. All results are expressed as the mean ± standard deviation. The results for the W-256 liver tumor model were used in this study for comparisons [12]. All the differentially expressed urinary proteins in these three models and their biological processes were compared.
2.7. Bioinformatics Analysis
For functional annotation, all differentially expressed proteins were analyzed with the Database for Annotation, Visualization and Integrated Discovery (DAVID) 6.8 (https://david.ncifcrf.gov/). The proteins were described according to three categories: biological process, cellular component, and molecular function. For ingenuity pathway analysis (IPA), the UniProt accession numbers of differentially expressed proteins were uploaded to IPA software (Qiagen, USA). The proteins were mapped to available canonical pathways and ranked by P values.
3. Results
3.1. Bodyweight (BW) and Histological Features over Time
All rats in the two models except for one rat from the CBRH-7919 model that died after anesthesia lived until they were sacrificed on day 42. The BW and daily behavior changes in the two models were observed and recorded after modeling. Compared with controls, the two models exhibited no significant differences in daily behaviors. On day 42, 7 rats in the CBRH-7919 experimental group, 8 rats in the RH-35 experimental group, and 8 rats in the control group were sacrificed. Statistical analysis was performed on the BW data of all the rats. On day 0, the mean BWs of the CBRH-7919, RH-35, and control rats were 199.4 g (±5.8 g), 206.3 g (±16.0 g), and 205.6 g (±8.3 g), respectively; on day 7, the BWs of the RH-35, CBRH-7919, and control rats were 255.3 g (±10.2 g), 243.1 g (±16.9 g), and 266.8 g (±18.4 g), respectively; and on day 42, the BWs of the CBRH-7919, RH-35, and control rats were 415.6 g (±16.6 g), 400.1 g (±20.4 g), and 447.1 g (±41.8 g), respectively. In the CBRH-7919 model, there were no significant differences in BW during the 42 days (Figure 1(a)). In the RH-35 model, there was a significant difference in BW between the tumor-bearing and control rats on day 7 (Figure 1(b)).
Figure 1.
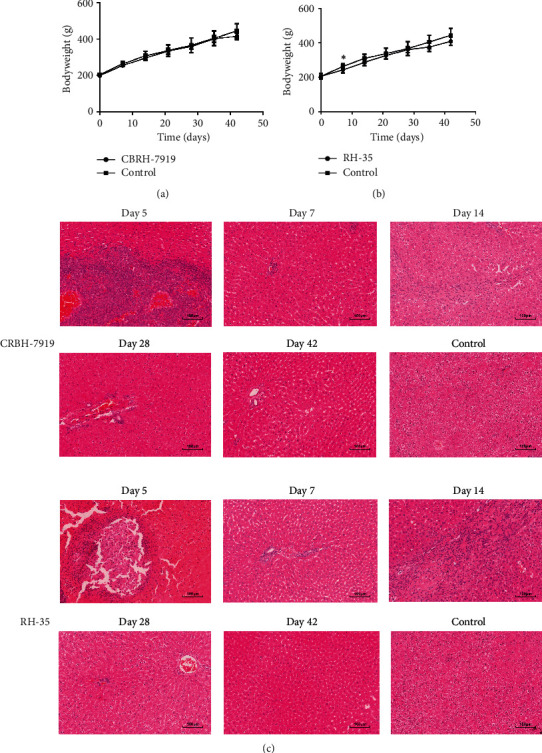
BW and histopathological characterization of the two models. (a) Change in BW in the CBRH-7919 model (CBRH-7919 group: n = 7; control group: n = 8). (b) Change in BW in the RH-35 model (RH-35 group: n = 8; control group: n = 8). ∗P value < 0 : 05. (c) H&E staining of a control rat and two model rats.
Two rats from each experimental group (CBRH-7919 and RH-35) were randomly selected for histological examinations on days 5, 7, 14, and 28. Two control rats were also randomly selected for histological examination on day 5. The remaining rats in the two models were sacrificed on day 42, and their livers were used for histological examination. H&E staining showed that the histological results of the two models were similar when the two different hepatoma cell lines grew in the liver over the whole experimental period (Figure 1(c)). On day 5, white lesions were clearly seen on the liver. H&E staining revealed the presence of numerous hepatoma cells and necrosis of some liver cells in the two models. On days 7, 14, and 28, the presence of some inflammatory cells was observed in the two models. Some necrotic and apoptotic hepatocytes were also stained partially with hematoxylin. Moreover, no obvious hepatoma cells were found. On day 42, no hepatoma cells appeared in the livers from the two models.
3.2. Changes in the Urine Proteomes of the Two Groups
Seventy-six urine samples from eight CBRH-7919 rats, nine RH-35 rats, and eight control rats at four time points (on days 5, 7, 14, and 28) were selected for analysis. Urine samples from before day 28 were selected for investigation of the early changes in the urine proteome caused by injection of the two different types of hepatoma cells because the histological features of the liver did not change substantially thereafter.
In the CBRH-7919 model, a total of 973 urinary proteins were identified (Supplementary Table 1). The screening criteria were a fold change ≥2 or ≤0.5 and a P value < 0.05. Differentially expressed urinary proteins were selected that satisfied the following criteria: the peak area for the rats (n ≥ 5) in the upregulation group was greater than that for the rats in the downregulation group, or the peak area for the rats (n ≥ 5) in the downregulation group was less than that for the rats in the upregulation group. With changes in the numbers of rats (e.g., n ≥ 6, 7, and 8), the numbers of differentially expressed urinary proteins varied (Table 1). A total of 133 differentially expressed proteins had human orthologs (Table 2). Moreover, 97, 28, 16, and 29 differentially expressed proteins with human orthologs were altered on days 5, 7, 14, and 28, respectively. On day 5, 64 upregulated differentially expressed proteins and 33 downregulated differentially expressed proteins were identified; on day 7, 7 upregulated and 21 downregulated differentially expressed proteins were identified; on day 14, 3 upregulated and 13 downregulated differentially expressed proteins were identified; and on day 28, 5 upregulated and 24 downregulated differentially expressed proteins were identified. The Venn diagram in Figure 2(a) shows the overlapping differentially expressed proteins at different time points.
Table 1.
The number of differentially proteins under different screening criteria in the two models.
| Model | Time | Fold change (FC ≥ 2.0 or ≤0.5) | N ≥ 5 | N ≥ 6 | N ≥ 7 | N ≥ 8 | N = 9 |
|---|---|---|---|---|---|---|---|
| CBRH-7919 | Day 5 | 132 | 115 | 86 | 57 | 19 | — |
| Day 7 | 52 | 41 | 27 | 21 | 8 | — | |
| Day 14 | 23 | 18 | 11 | 8 | 5 | — | |
| Day 28 | 36 | 30 | 24 | 18 | 10 | — | |
|
| |||||||
| RH-35 | Day 5 | 64 | 35 | 30 | 22 | 18 | 4 |
| Day 7 | 29 | 17 | 12 | 10 | 9 | 6 | |
| Day 14 | 36 | 24 | 20 | 16 | 8 | 4 | |
| Day 28 | 69 | 53 | 47 | 42 | 27 | 10 | |
Note. N indicates the number of rats; — indicates no data reach the criteria.
Table 2.
Differential urinary proteins in the CBRH-7919 model.
| Accession | Protein names | Trend | Fold changes | Reported to hepatocellular carcinoma | Reported to other cancers | |||
|---|---|---|---|---|---|---|---|---|
| D5 | D7 | D14 | D28 | |||||
| Q9Y694 | Solute carrier family 22 member 7 | ↑ | 3.41 | — | — | — | Colorectal cancer | |
| P53004 | Biliverdin reductase A | ↑ | 3.34 | 2.29 | — | — | Breast cancer | |
| O76082 | Solute carrier family 22 member 5 | ↑ | 3.33 | 3.17 | 2.17 | — | Breast cancer | |
| P48507 | Glutamate–cysteine ligase regulatory subunit | ↑ | 3.13 | 2.09 | — | — | ||
| O00338 | Sulfotransferase 1C2 | ↑ | 3.08 | — | — | — | ||
| P11234 | Ras-related protein Ral-B | ↑ | 2.83 | 2.32 | — | — | Nonsmall cell lung cancer | |
| Q01523 | Neutrophil antibiotic peptide NP-4 | ↑ | 2.80 | — | — | — | ||
| B1AK53 | Espin | ↑ | 2.77 | — | — | — | ||
| Q96KP4 | Cytosolic nonspecific dipeptidase | ↑ | 2.76 | — | — | — | ||
| P0C0L4 | Complement C4 | ↑ | 2.69 | — | — | — | Serum | |
| P32929 | Cystathionine gamma-lyase | ↑ | 2.64 | 2.04 | — | — | Melanoma | |
| P14174 | Macrophage migration inhibitory factor | ↑ | 2.62 | — | — | — | Serum | Gastrointestinal tract (GIT) malignancy |
| P21399 | Cytoplasmic aconitate hydratase | ↑ | 2.57 | — | — | — | ||
| Q86YJ6 | Threonine synthase-like 2 | ↑ | 2.50 | — | — | — | ||
| P02511 | Alpha-crystallin B chain | ↑ | 2.49 | — | — | — | Tissue | Colorectal cancer |
| O94760 | Dimethylargininase-1 | ↑ | 2.48 | — | — | — | Tissue | Prostate cancer |
| Q96S37 | Solute carrier family 22 member 12 | ↑ | 2.48 | — | — | — | ||
| Q8N5Z0 | 2-Aminoadipate transaminase | ↑ | 2.47 | — | — | — | ||
| Q13113 | PDZK1-interacting protein 1 | ↑ | 2.46 | — | — | — | ||
| O95154 | Aflatoxin B1 aldehyde reductase member 3 | ↑ | 2.45 | — | — | — | ||
| O00299 | Chloride intracellular channel protein 1 | ↑ | 2.44 | — | — | — | Tissue | Gastric, colon, lung, and glioblastoma cancers |
| P13866 | Sodium/glucose cotransporter 1 | ↑ | 2.42 | — | — | — | Pancreatic cancer | |
| P36543 | V-type proton ATPase subunit E 1 | ↑ | 2.40 | — | — | — | Pancreatic cancer | |
| Q9Y696 | Chloride intracellular channel protein 4 | ↑ | 2.39 | — | — | — | ||
| Q96FL8 | Multidrug and toxin extrusion protein 1 | ↑ | 2.38 | 2.10 | — | — | ||
| O43708 | Maleylacetoacetate isomerase | ↑ | 2.36 | — | — | — | ||
| P35579 | Myosin-9 | ↑ | 2.35 | — | — | — | Colorectal cancer | |
| P26038 | Moesin | ↑ | 2.34 | — | — | — | Breast cancer | |
| P12955 | Xaa-Pro dipeptidase | ↑ | 2.33 | — | — | — | ||
| Q15599 | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 | ↑ | 2.27 | — | — | — | ||
| P08133 | Annexin A6 | ↑ | 2.27 | — | — | — | Tissue | Melanoma, cervical cancer, epithelial carcinoma, breast cancer, and gastric cancer |
| P80723 | Brain acid soluble protein 1 | ↑ | 2.26 | — | — | — | Tissue | Pancreatic cancer and cervical cancer |
| P48506 | Gamma-glutamylcysteine synthetase | ↑ | 2.26 | — | — | — | ||
| Q9UHI7 | Solute carrier family 23 member 1 | ↑ | 2.25 | — | — | — | ||
| Q03154 | Aminoacylase-1A | ↑ | 2.25 | — | — | — | ||
| Q96KN2 | Beta-Ala-His dipeptidase | ↑ | 2.24 | — | — | — | Cancer cachexia | |
| O15400 | Syntaxin-7 | ↑ | 2.23 | — | — | — | ||
| Q99685 | Monoglyceride lipase | ↑ | 2.23 | — | — | — | Tissue | Gastrointestinal stromal tumor |
| Q14894 | Ketimine reductase mu-crystallin | ↑ | 2.23 | — | — | — | ||
| Q6ZQN7 | Solute carrier family 21 member 20 | ↑ | 2.23 | — | — | — | ||
| O00161 | Synaptosomal-associated protein 23 | ↑ | 2.22 | — | — | — | ||
| P49189 | 4-Trimethylaminobutyraldehyde dehydrogenase | ↑ | 2.21 | — | — | — | ||
| Q9Y6I3 | Epsin-1 | ↑ | 2.20 | — | — | 2.18 | Prostate cancer | |
| Q9UBR1 | Beta-ureidopropionase | ↑ | 2.17 | — | — | — | Serum | |
| Q14847 | LIM and SH3 domain protein1 | ↑ | 2.16 | — | — | — | Tissue | Gastric cancer |
| O43704 | Sulfotransferase family cytosolic 1B member 1 | ↑ | 2.15 | — | — | — | ||
| Q9UNQ0 | ATP-binding cassette subfamily G member 2 | ↑ | 2.15 | — | — | — | Tissue | Right-sided colon cancer |
| P30041 | Peroxiredoxin-6 | ↑ | 2.14 | — | — | — | ||
| O14745 | Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | ↑ | 2.13 | — | — | — | ||
| P68032 | Actin, alpha cardiac muscle 1 | ↑ | 2.12 | — | — | — | Glioblastoma | |
| P27449 | V-type proton ATPase 16 kDa proteolipid subunit | ↑ | 2.11 | — | — | — | Prostate cancer | |
| Q5T2W1 | Na(+)/H(+) exchange regulatory cofactor NHE-RF3 | ↑ | 2.09 | — | — | — | ||
| Q16348 | Solute carrier family 15 member 2 | ↑ | 2.09 | — | — | — | ||
| Q9H0W9 | Ester hydrolase C11orf54 homolog | ↑ | 2.08 | — | — | — | Renal cell carcinoma | |
| P60709 | Actin, cytoplasmic 1 | ↑ | 2.08 | — | — | — | Tissue | |
| P09467 | Fructose-1,6-bisphosphatase 1 | ↑ | 2.08 | — | — | — | Tissue | |
| P46721 | Solute carrier organic anion transporter family member 1A1 | ↑ | 2.07 | — | — | — | ||
| P05062 | Fructose-bisphosphate aldolase B | ↑ | 2.07 | — | — | — | ||
| P29972 | Aquaporin-1 | ↑ | 2.06 | — | — | — | Tissue | Bladder, brain, breast, cervix, colon, lung, nasopharynx, and prostate cancers |
| Q08257 | Quinone oxidoreductase | ↑ | 2.03 | — | — | — | ||
| O75083 | WD repeat-containing protein 1 | ↑ | 2.03 | — | — | — | Ovarian cancer | |
| Q9H8S9 | MOB kinase activator 1A | ↑ | 2.02 | — | — | — | ||
| Q14019 | Coactosin-like protein | ↑ | 2.02 | — | — | — | Small cell lung cancer | |
| Q8TF66 | Leucine-rich repeat-containing protein 15 | ↑ | 2.02 | — | — | — | Solid tumors (e.g., breast, head and neck, lung, and pancreatic) | |
| P05090 | Apolipoprotein D | ↑ | — | 3.38 | — | 2.43 | Tissue | Breast cancer and prostate cancer |
| Q12792 | Twinfilin-1 | ↑ | — | — | 2.48 | — | Lung adenocarcinoma | |
| P55259 | Glycoprotein 80 | ↑ | — | — | 2.47 | 4.00 | ||
| P06870 | Prostatic glandular kallikrein-6 | ↑ | — | — | — | 3.37 | ||
| P02741 | C-reactive protein | ↑ | — | — | — | 2.16 | Serum | Gastric cancer |
| O00115 | Deoxyribonuclease-2-alpha | ↓ | 0.50 | — | — | — | ||
| P37173 | TGF-beta receptor type-2 | ↓ | 0.49 | — | — | — | ||
| Q9BRK5 | 45 kDa calcium-binding protein | ↓ | 0.49 | — | — | — | ||
| O94985 | Calsyntenin-1 | ↓ | 0.49 | — | — | — | Lung adenocarcinoma | |
| P10909 | Clusterin | ↓ | 0.48 | — | — | — | Serum | Lung adenocarcinoma |
| P07339 | Cathepsin D | ↓ | 0.47 | — | — | — | Tissue | |
| Q6P4A8 | Phospholipase B-like 1 | ↓ | 0.46 | — | — | — | ||
| P34096 | Ribonuclease 4 | ↓ | 0.44 | — | — | — | ||
| P04233 | MHC class II-associated invariant chain | ↓ | 0.42 | — | — | — | ||
| Q8NFL0 | Beta-1,3-N-acetylglucosa-minyltransferase 7 | ↓ | 0.42 | — | — | — | Breast cancer | |
| P30740 | Leukocyte elastase inhibitor A | ↓ | 0.41 | — | — | — | ||
| Q8TD33 | Secretoglobin family 1C member 1 | ↓ | 0.40 | 0.31 | — | 0.42 | ||
| Q16674 | Melanoma-derived growth regulatory protein | ↓ | 0.40 | — | — | — | ||
| P15309 | Prostatic acid phosphatase | ↓ | 0.40 | — | — | — | ||
| P01834 | Ig kappa chain C region, A allele | ↓ | 0.40 | 0.46 | 0.49 | — | ||
| P01859 | Ig gamma-2A chain C region | ↓ | 0.38 | — | — | — | ||
| Q14315 | Filamin-C | ↓ | 0.38 | 0.49 | — | — | Tissue | Prostate cancer |
| P16444 | Dipeptidase 1 | ↓ | 0.38 | — | — | — | Colorectal cancer | |
| P09237 | Matrilysin | ↓ | 0.38 | — | — | — | Tissue | |
| P27797 | Calreticulin | ↓ | 0.37 | 0.42 | — | — | ||
| Q13217 | DnaJ homolog subfamily C member 3 | ↓ | 0.36 | — | — | — | ||
| Q9UGM3 | Deleted in malignant brain tumors 1 protein | ↓ | 0.36 | — | — | — | Tissue | |
| Q9NZU0 | Leucine-rich repeat transmembrane protein FLRT3 | ↓ | 0.35 | 0.47 | — | — | ||
| Q08188 | Protein-glutamine gamma-glutamyltransferase E | ↓ | 0.34 | — | — | — | ||
| Q3LXA3 | Triokinase/FMN cyclase | ↓ | 0.33 | 0.18 | 0.13 | — | ||
| Q9BY76 | Angiopoietin-related protein 4 | ↓ | 0.29 | — | — | — | Serum | Colorectal cancer |
| P08118 | Beta-microseminoprotein | ↓ | 0.23 | 0.16 | 0.23 | — | Prostate cancer | |
| P15086 | Carboxypeptidase B | ↓ | 0.21 | — | — | — | ||
| Q99895 | Chymotrypsin-C | ↓ | 0.19 | — | — | — | ||
| P34913 | Bifunctional epoxide hydrolase 2 | ↓ | 0.18 | 0.32 | — | — | ||
| P05109 | Protein S100-A8 | ↓ | 0.16 | 0.29 | — | — | Breast cancer | |
| P06702 | Protein S100-A9 | ↓ | 0.14 | 0.23 | — | — | Urine | Esophageal adenocarcinoma |
| Q9UPX8 | SH3 and multiple ankyrin repeat domains protein 2 | ↓ | 0.09 | — | — | — | ||
| P36896 | Activin receptor type-1B | ↓ | — | 0.49 | — | 0.42 | ||
| P08253 | 72 kDa type IV collagenase | ↓ | — | 0.48 | — | 0.50 | Rectal cancer, laryngeal cancer | |
| P02760 | Protein AMBP | ↓ | — | 0.48 | 0.47 | — | ||
| Q6UY14 | ADAMTS-like protein 4 | ↓ | — | 0.47 | — | — | ||
| Q8N8N7 | Prostaglandin reductase 2 | ↓ | — | 0.45 | — | — | ||
| Q9Y678 | Coatomer subunit gamma-1 | ↓ | — | 0.45 | — | 0.30 | ||
| Q13145 | BMP and activin membrane-bound inhibitor homolog | ↓ | — | 0.43 | 0.50 | 0.29 | Tissue | Melanoma, colorectal cancer |
| Q66K79 | Carboxypeptidase Z | ↓ | — | 0.43 | — | 0.22 | Tissue | |
| P48745 | CCN family member 3 | ↓ | — | 0.42 | 0.41 | 0.44 | Tissue | Breast cancer, gastric cancer |
| P09382 | Galectin-1 | ↓ | — | 0.40 | — | — | Tissue | Gastric cancer, colorectal cancer, and pancreatic ductal adenocarcinoma |
| Q03403 | Trefoil factor 2 | ↓ | — | 0.33 | 0.39 | 0.34 | Tissue | Colorectal cancer |
| P0CE71 | Oncomodulin | ↓ | — | — | 0.47 | — | ||
| Q9UBC9 | Cornifin-A | ↓ | — | — | 0.45 | — | ||
| O95388 | WNT1-inducible-signaling pathway protein 1 | ↓ | — | — | 0.43 | — | Oral squamous cell carcinoma | |
| P49221 | Protein-glutamine gamma-glutamyltransferase 4 | ↓ | — | — | 0.35 | — | Prostate cancer | |
| P02452 | Collagen alpha-1 | ↓ | — | — | 0.22 | 0.16 | Colorectal cancer | |
| P57740 | Nuclear pore complex protein Nup107 | ↓ | — | — | 0.14 | 0.35 | ||
| P35443 | Thrombospondin-4 | ↓ | — | — | — | 0.49 | Tissue | Gastric cancer |
| P49747 | Cartilage oligomeric matrix protein | ↓ | — | — | — | 0.48 | Tissue | Breast cancer and prostate cancer |
| O15197 | Ephrin type-B receptor 6 | ↓ | — | — | — | 0.48 | Colorectal cancer | |
| P35613 | Basigin | ↓ | — | — | — | 0.47 | ||
| P16112 | Aggrecan core protein | ↓ | — | — | — | 0.47 | ||
| Q04721 | Neurogenic locus notch homolog protein 2 | ↓ | — | — | — | 0.46 | Laryngeal squamous cell carcinoma | |
| Q96CG8 | Collagen triple helix repeat-containing protein 1 | ↓ | — | — | — | 0.43 | ||
| Q14112 | Nidogen-2 | ↓ | — | — | — | 0.43 | Serum | Ovarian cancer |
| Q12805 | EGF-containing fibulin-like extracellular matrix protein 1 | ↓ | — | — | — | 0.42 | Meningioma | |
| Q96IU4 | Protein ABHD14B | ↓ | — | — | — | 0.42 | ||
| Q9NP85 | Podocin | ↓ | — | — | — | 0.41 | ||
| O60462 | Neuropilin-2 | ↓ | — | — | — | 0.39 | Tissue | Nonsmall cell lung cancer, squamous cell carcinoma, and melanoma |
| O95336 | 6-Phosphogluconolactonase | ↓ | — | — | — | 0.39 | Breast cancer, nonsmall cell lung cancer, and ovarian cancer | |
| P11684 | Uteroglobin | ↓ | — | — | — | 0.21 | Prostate cancer | |
Note. — indicates no data reach the criteria compared with control. The results of two parts (reported to be related to hepatocellular carcinoma and other diseases) are annotated from previous studies.
Figure 2.
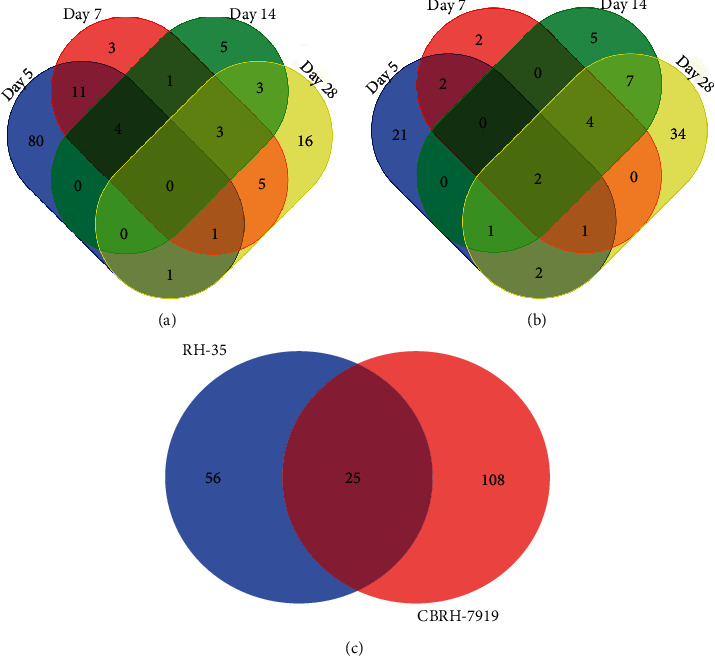
Venn diagram of differentially expressed proteins at different time points in the two models. (a) Differentially expressed proteins in the CBRH-7919 model rats at four different time points (days 5, 7, 14, and 28). (b) Differentially expressed proteins in the RH-35 model rats at four different time points (days 5, 7, 14, and 28). (c) Comparison of differentially expressed proteins between the two models.
In the RH-35 model, the screening conditions were similar to those in the CBRH-7919 model. The numbers of differentially expressed urinary proteins were different for different numbers of rats (e.g., n ≥ 5, 6, 7, 8, and 9) (Table 1). A total of 903 urinary proteins were identified (Supplementary Table 2). Eighty-one differentially expressed proteins with human orthologs were identified (Table 3). In addition, 29, 11, 19, and 51 differentially expressed proteins with human orthologs were altered on days 5, 7, 14, and 28, respectively. On day 5, 6 upregulated and 23 downregulated differentially expressed proteins were identified; on day 7, 1 upregulated and 10 downregulated differentially expressed proteins were identified; on day 14, 2 upregulated and 17 downregulated differentially expressed proteins were identified; and on day 28, 11 upregulated and 40 downregulated differentially expressed proteins were identified. The overlapping differentially expressed proteins are shown in a Venn diagram in Figure 2(b).
Table 3.
Differential urinary proteins in the RH-35 model.
| Accession | Protein names | Trend | Fold change | Reported to hepatocellular carcinoma | Reported to other cancers | |||
|---|---|---|---|---|---|---|---|---|
| D5 | D7 | D14 | D28 | |||||
| Q03154 | Aminoacylase-1A | ↑ | 2.38 | — | — | — | ||
| Q66K79 | Carboxypeptidase Z | ↑ | 2.33 | — | — | 0.49 | Tissue | |
| P02763 | Alpha-1-acid glycoprotein | ↑ | 2.18 | — | — | — | Serum | Bladder cancer and lung cancer |
| P05090 | Apolipoprotein D | ↑ | 2.11 | — | — | — | Tissue | Prostate cancer and breast cancer |
| Q9UP38 | Frizzled-1 | ↑ | 2.03 | — | — | — | Follicular thyroid carcinoma | |
| P29972 | Aquaporin-1 | ↑ | 2.03 | — | — | — | Tissue | Bladder cancer, brain cancer, and breast cancer |
| P09488 | Glutathione S-transferase Mu 2 | ↑ | — | 2.02 | 2.17 | 3.58 | Nonsmall cell lung cancer | |
| Q9H3Z4 | DnaJ homolog subfamily C member 5 | ↑ | — | — | 2.05 | 3.24 | ||
| P06870 | Prostatic glandular kallikrein-6 | ↑ | — | — | — | 5.44 | ||
| P31639 | Sodium/glucose cotransporter 2 | ↑ | — | — | — | 3.10 | ||
| P08754 | Guanine nucleotide-binding protein G | ↑ | — | — | — | 2.87 | ||
| Q15833 | Syntaxin-binding protein 2 | ↑ | — | — | — | 2.74 | ||
| O94832 | Unconventional myosin-Id | ↑ | — | — | — | 2.35 | ||
| O95968 | Prostatic steroid-binding protein C2 | ↑ | — | — | — | 2.23 | ||
| Q9H2G2 | STE20-like serine/threonine-protein kinase | ↑ | — | — | — | 2.02 | Tissue | Glioma |
| O14773 | Tripeptidyl peptidase 1 | ↑ | — | — | — | 2.02 | Tissue | |
| P02144 | Myoglobin | ↑ | — | — | — | 2.01 | Lung adenocarcinoma and breast cancer | |
| Q9H6B4 | CXADR-like membrane protein | ↓ | 0.49 | — | — | — | ||
| Q9Y2T3 | Guanine deaminase | ↓ | 0.45 | — | — | — | Serum | Gastric cancer |
| Q92932 | Receptor-type tyrosine-protein phosphatase N2 | ↓ | 0.45 | 0.39 | — | 0.48 | GRN-associated frontotemporal dementia | |
| P15309 | Prostatic acid phosphatase | ↓ | 0.44 | — | — | — | ||
| P34096 | Ribonuclease 4 | ↓ | 0.44 | — | — | — | ||
| Q99757 | Thioredoxin | ↓ | 0.43 | — | — | — | ||
| P61204 | ADP-ribosylation factor 3 | ↓ | 0.43 | — | — | — | ||
| Q9UGM5 | Fetuin-B | ↓ | 0.42 | — | — | — | ||
| P07384 | Calpain-1 catalytic subunit | ↓ | 0.42 | — | — | — | ||
| P05937 | Calbindin | ↓ | 0.41 | — | 0.40 | 0.37 | Lung cancer | |
| P01859 | Ig gamma-2A chain C region | ↓ | 0.39 | — | — | — | ||
| Q71U36 | Tubulin alpha-1A chain | ↓ | 0.39 | — | — | — | Renal cell carcinoma | |
| P37837 | Transaldolase | ↓ | 0.39 | — | — | — | Serum | Ovarian cancer |
| P08962 | CD63 antigen | ↓ | 0.39 | — | — | — | Melanoma | |
| P10909 | Clusterin | ↓ | 0.39 | 0.48 | — | — | Tissue | Lung adenocarcinoma |
| P11021 | Endoplasmic reticulum chaperone BiP | ↓ | 0.37 | — | — | — | Gastric cancer | |
| Q9BS40 | Latexin | ↓ | 0.35 | — | — | — | Tissue | Gastric cancer and prostate cancer |
| Q14315 | Filamin-C | ↓ | 0.34 | — | — | — | Tissue | Prostate cancer |
| P57740 | Nuclear pore complex protein Nup107 | ↓ | 0.27 | 0.09 | 0.21 | 0.17 | ||
| P0CE71 | Oncomodulin | ↓ | 0.25 | 0.29 | 0.33 | 0.36 | ||
| P30048 | Thioredoxin-dependent peroxide reductase | ↓ | 0.23 | — | — | — | Serum | Endometrial cancer and prostate cancer |
| P27797 | Calreticulin | ↓ | 0.22 | 0.31 | — | — | ||
| P08118 | Beta-microseminoprotein | ↓ | 0.15 | — | — | 0.21 | Prostate cancer | |
| Q6P587 | Acylpyruvase FAHD1 | ↓ | — | 0.49 | — | — | ||
| P31150 | Rab GDP dissociation inhibitor alpha | ↓ | — | 0.48 | — | — | Nonsmall cell lung cancer | |
| Q5KU26 | Collectin-12 | ↓ | — | 0.46 | 0.45 | 0.47 | ||
| Q6ZVN8 | Hemojuvelin | ↓ | — | 0.46 | 0.49 | 0.49 | Tissue | |
| Q96DG6 | Carboxymethylenebutenolidase homolog | ↓ | — | 0.45 | 0.38 | 0.39 | ||
| Q9HCB6 | Spondin-1 | ↓ | — | — | 0.50 | 0.48 | Ovarian cancer | |
| Q12805 | Fibulin-3 | ↓ | — | — | 0.49 | 0.46 | Meningioma | |
| P47755 | F-actin-capping protein subunit alpha-2 | ↓ | — | — | 0.49 | 0.43 | ||
| P20333 | Tumor necrosis factor receptor 2 | ↓ | — | — | 0.48 | 0.50 | Tissue | Cholangiocarcinoma, colorectal cancer, and myeloma |
| Q9NWV4 | CXXC motif containing zinc-binding protein | ↓ | — | — | 0.48 | — | ||
| Q14332 | Frizzled-2 | ↓ | — | — | 0.43 | — | Tissue | Prostate cancer |
| O95716 | GTP-binding protein Rab-3D | ↓ | — | — | 0.41 | — | ||
| P04003 | C4b-binding protein alpha chain | ↓ | — | — | 0.39 | 0.48 | Serum | Pancreatic cancer and epithelial ovarian carcinoma |
| Q03403 | Trefoil factor 2 | ↓ | — | — | 0.36 | 0.27 | Colorectal cancer and cholangiocarcinoma | |
| Q6P1J6 | Phospholipase B1 | ↓ | — | — | 0.19 | — | ||
| P02452 | Collagen alpha-1 | ↓ | — | — | 0.19 | — | Colorectal cancer | |
| P07108 | Acyl-CoA-binding protein | ↓ | — | — | — | 0.50 | Lung cancer | |
| Q96RD6 | Pannexin-2 | ↓ | — | — | — | 0.50 | ||
| Q9Y5Q5 | Atrial natriuretic peptide-converting enzyme | ↓ | — | — | — | 0.49 | Small cell lung cancer | |
| P13591 | Neural cell adhesion molecule 1 | ↓ | — | — | — | 0.49 | Tissue | Lung cancer |
| P07602 | Prosaposin | ↓ | — | — | — | 0.48 | Gallbladder cancer and breast cancer | |
| P51884 | Lumican | ↓ | — | — | — | 0.48 | ||
| Q04771 | Activin receptor type-1 | ↓ | — | — | — | 0.48 | ||
| P13667 | Protein disulfide isomerase A4 | ↓ | — | — | — | 0.48 | Serum | |
| Q9H461 | Frizzled-8 | ↓ | — | — | — | 0.48 | Prostate cancer and renal cell carcinoma | |
| P09382 | Galectin-1 | ↓ | — | — | — | 0.47 | Tissue | Colorectal cancer and gastric cancer |
| P51687 | Sulfite oxidase | ↓ | — | — | — | 0.46 | Serum | Prostate cancer |
| P35443 | Thrombospondin-4 | ↓ | — | — | — | 0.44 | Tissue | Gastric cancer |
| P36896 | Activin receptor type-1B | ↓ | — | — | — | 0.43 | ||
| Q96CG8 | Collagen triple helix repeat-containing protein 1 | ↓ | — | — | — | 0.43 | Tissue | Osteosarcoma and ovarian cancer |
| Q8TER0 | Sushi, nidogen, and EGF-like domain-containing protein 1 | ↓ | — | — | — | 0.43 | Papillary thyroid carcinoma | |
| Q9Y678 | Coatomer subunit gamma-1 | ↓ | — | — | — | 0.42 | ||
| O60667 | Fas apoptotic inhibitory molecule 3 | ↓ | — | — | — | 0.41 | Chronic lymphocyte leukemia | |
| P16112 | Aggrecan core protein | ↓ | — | — | — | 0.40 | ||
| Q6UY11 | Protein delta homolog 2 | ↓ | — | — | — | 0.39 | Melanoma | |
| P04155 | Trefoil factor 1 | ↓ | — | — | — | 0.36 | Prostate cancer | |
| O95336 | 6-Phosphogluconolactonase | ↓ | — | — | — | 0.32 | Breast cancer, nonsmall cell lung cancer, and ovarian cancer | |
| P36957 | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial | ↓ | — | — | — | 0.30 | ||
| Q9BQ08 | Resistin-like gamma | ↓ | — | — | — | 0.29 | ||
| P58062 | Serine protease inhibitor Kazal-type 7 | ↓ | — | — | — | 0.21 | Esophageal cancer | |
| Q9UBC9 | Cornifin-A | ↓ | — | — | — | 0.18 | ||
Note. — indicates no data reach the criteria compared with the control. The results of two parts (reported to be related to hepatocellular carcinoma and other diseases) are annotated from previous studies.
When the differentially expressed proteins of the two models were compared, 25 common proteins were identified. In addition, 108 and 56 unique proteins were identified, respectively (Figure 2(c)). Although the histological results for the two models were similar, the numbers and categories of the identified urinary proteins were mostly different. The results indicate that the patterns of differentially expressed urinary proteins are different when different tumor cells grow in the liver.
3.3. Random Allocation Statistical Analysis
To further confirm that the differentially expressed proteins were influenced by hepatoma cells, we randomly allocated the data for 16 samples (8 experimental and 8 control samples) at each time point in the CBRH-7919 model and the data for 17 samples (9 experimental and 8 control samples) at each time point in the RH-35 model. The criteria used to screen differentially expressed urinary proteins were the same as mentioned above: a fold change ≥2 or ≤0.5 and a P value <0.05. At each time point, 16 samples of the CBRH-7919 model were randomly divided into 2 groups (n = 8 in each group). A total of 6,435 random allocations at each time point were performed, and the details are shown in Supplementary Tables 3 and 4. At each time point, 17 samples of the RH-35 model were randomly divided into 2 groups (n = 8 in group 1, n = 9 in group 2). A total of 12,155 random allocations at each time point were performed, and the details are shown in Supplementary Tables 5 and 6. In each iteration, the data for group 1 were set as the control data, and the data for group 2 were used as the experimental data. Table 4 shows the results of the random allocation statistical analysis. It should be noted that the random allocation results included the actual grouping results. The results show that there were false-positives in the actual grouping. The highest false-positive rate was 0.177. However, when tumor cells grew actively in the liver, the number of differentially expressed urinary proteins was large, and the false-positive rate was low. For example, on day 5, the false-positive rates of the two models were 0.049 and 0.087, respectively. Therefore, most of the changes in differentially expressed urinary proteins were caused not by random allocation but by the tumor cells.
Table 4.
Random allocation in two models.
| Model | The numbers of differential urinary proteins | Day 5 | Day 7 | Day 14 | Day 28 |
|---|---|---|---|---|---|
| CBRH-7919 | Actual grouping | 132 | 52 | 23 | 36 |
| The average of random grouping | 6.45 | 5.07 | 4.06 | 4.69 | |
| The false-positive rate (the average of random grouping/actual grouping) | 0.049 | 0.098 | 0.177 | 0.130 | |
|
| |||||
| RH-35 | Actual grouping | 64 | 29 | 36 | 69 |
| The average of random grouping | 5.59 | 4.44 | 4.22 | 5.10 | |
| The ratio (the average of random grouping/actual grouping) | 0.087 | 0.153 | 0.117 | 0.074 | |
3.4. Functional Analysis of Differentially Expressed Proteins
By using the DAVID and the IPA database, functional analysis was performed for the identified differentially expressed proteins from the CBRH-7919 and RH-35 models. The proteins were categorized on the basis of their related biological processes, cell components, molecular functions, and canonical pathways.
In the CBRH-7919 model, a variety of biological processes were associated with the differentially expressed proteins on day 5. Specifically, the glutathione metabolic process, transport, aging, positive regulation of the intrinsic apoptotic signaling pathway, innate immune response, negative regulation of the apoptotic process, phagocytosis and engulfment, complement activation and classical pathway, defense response to bacterium, response to drug, complement activation, classical pathway, positive regulation of B cell activation, and neutrophil aggregation terms were associated with significantly changed proteins. On day 7, the differentially expressed proteins were associated mainly with the following biological process terms: intermediate filament organization, neutrophil aggregation, response to axon injury, apoptotic process, positive regulation of peptide secretion, peptidyl-cysteine S-nitrosylation, negative regulation of inflammatory response, response to activity, peripheral nervous system axon regeneration, chronic inflammatory response, and leukocyte migration involved in the inflammatory response. On day 14, the positive regulation of epithelial-to-mesenchymal transition and peptide cross-linking terms were associated with the responses to the tumor cells. On day 28, the biological process terms included the wound healing, response to radiation, endothelial cell-cell adhesion, positive regulation of protein binding, cell adhesion, intramembranous ossification, response to mechanical stimulus, and bone trabecula formation terms (Figure 3(a)). In the cellular component category, most of the differentially expressed proteins were associated with the extracellular exosome, blood microparticle, apical plasma membrane, extracellular space, brush border membrane, focal adhesion, cytosol, cytoplasm, lysosome, extracellular region, and extracellular matrix terms, which represent the main sources of urinary protein (Supplementary Figure 1(a)). In the molecular function category, the differentially expressed proteins were associated mainly with metallodipeptidase activity, antigen binding, protein homodimerization activity, transporter activity, dipeptidase activity, glutathione transferase activity, protein binding, carboxypeptidase activity, immunoglobulin receptor binding, cytoskeletal protein binding, structural molecule activity, carbohydrate binding, and Toll-like receptor 4 binding (Supplementary Figure 1(b)).
Figure 3.
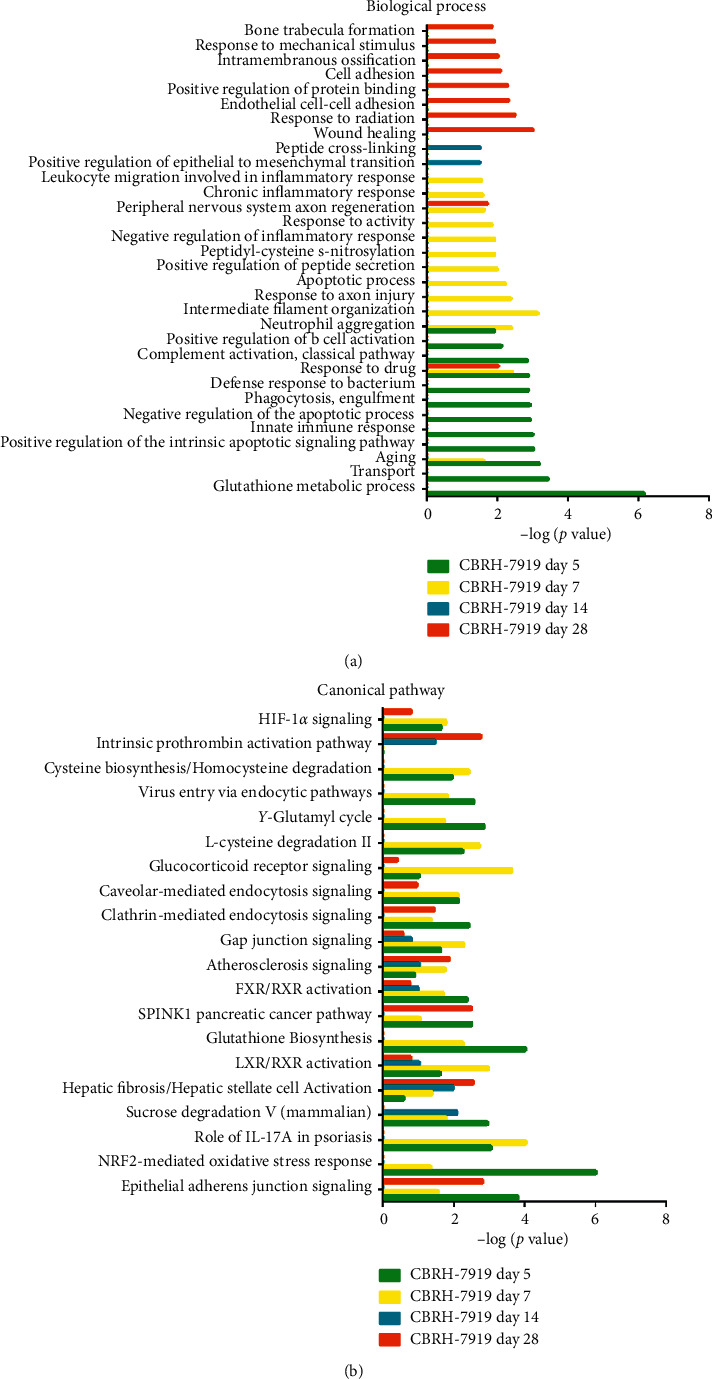
Functional analysis of differentially expressed proteins at days 5, 7, 14, and 28 in the CBRH-7919 model. (a) Biological processes for the CBRH-7919 model. (b) Canonical pathways for the CBRH-7919 model.
IPA software was used to analyze the canonical pathways associated with the differentially expressed proteins. Liver fibrosis/hepatic stellate cell activation, LXR/RXR activation, FXR/RXR activation, atherosclerosis signaling, NRF2-mediated oxidation stress response, HIF1α signaling, and gap junction signaling were enriched (Figure 3(b)). Several signaling pathways have been reported to play important roles in tumor development. First, tumor cell apoptosis and proliferation are promoted in hypoxic environments, and hypoxia-inducible factor 1α (HIF1α) is a significant hypoxia-inducing factor that is related to the movement and adhesion of liver cancer tumor cells in such environments [16]. Second, NF-E2-related factor-2 (NRF2) is an important transcription factor that can regulate oxidative stress and the expression of a series of detoxification genes and antioxidant defense genes in the liver [17]. Third, liver fibrosis/hepatic stellate cell activation is a prominent pathological feature of liver fibrosis [18]. Finally, the liver X receptor/retinoid AX receptor (LXR/RXR) activation pathway is related to the regulation of cholesterol transport, glucose metabolism, and the inflammatory response, and increasing evidence has shown the involvement of LXRs in various malignancies [19].
In the RH-35 model, the differentially expressed proteins were associated mainly with the transport, response to drug, negative regulation of endopeptidase activity, lipid metabolic process, protein transport, vesicle-mediated transport, defense response to bacterium, response to oxidative stress, and positive regulation of B cell activation terms in the biological process category on day 5. On day 7, chaperone-mediated protein folding was independently enriched. On day 14, the immune response and regulation of cytosolic calcium ion concentration terms were associated with the differentially expressed proteins. On day 28, the activin receptor signaling pathway, signal transduction by protein phosphorylation, learning or memory, cellular detoxification of nitrogen compound, nitrobenzene metabolic process, central nervous system development, xenobiotic catabolic process, cell adhesion, response to acidic pH, and multicellular organismal response to stress terms were associated with the responses to the tumor cells (Figure 4(a)). For the cellular component category, the differentially expressed proteins were derived mainly from exosomes, the extracellular region, and the extracellular space (Supplementary Figure 1(c)). For the molecular function category, on day 5, protein binding, antigen binding, metalloendopeptidase inhibitor activity, misfolded protein binding, transporter activity, immunoglobulin receptor binding, and GTP binding were overrepresented. On days 7 and 14, the differentially expressed proteins were associated with glycoprotein binding, metal ion binding, and transmembrane receptor protein serine/threonine kinase activity. On day 28, the growth factor activity, receptor signaling protein serine/threonine kinase activity, activin receptor activity, type I calcium ion binding, glycoprotein binding, activin binding, protein homodimerization activity, and transmembrane receptor protein serine/threonine kinase activity terms were enriched (Supplementary Figure 1(d)).
Figure 4.
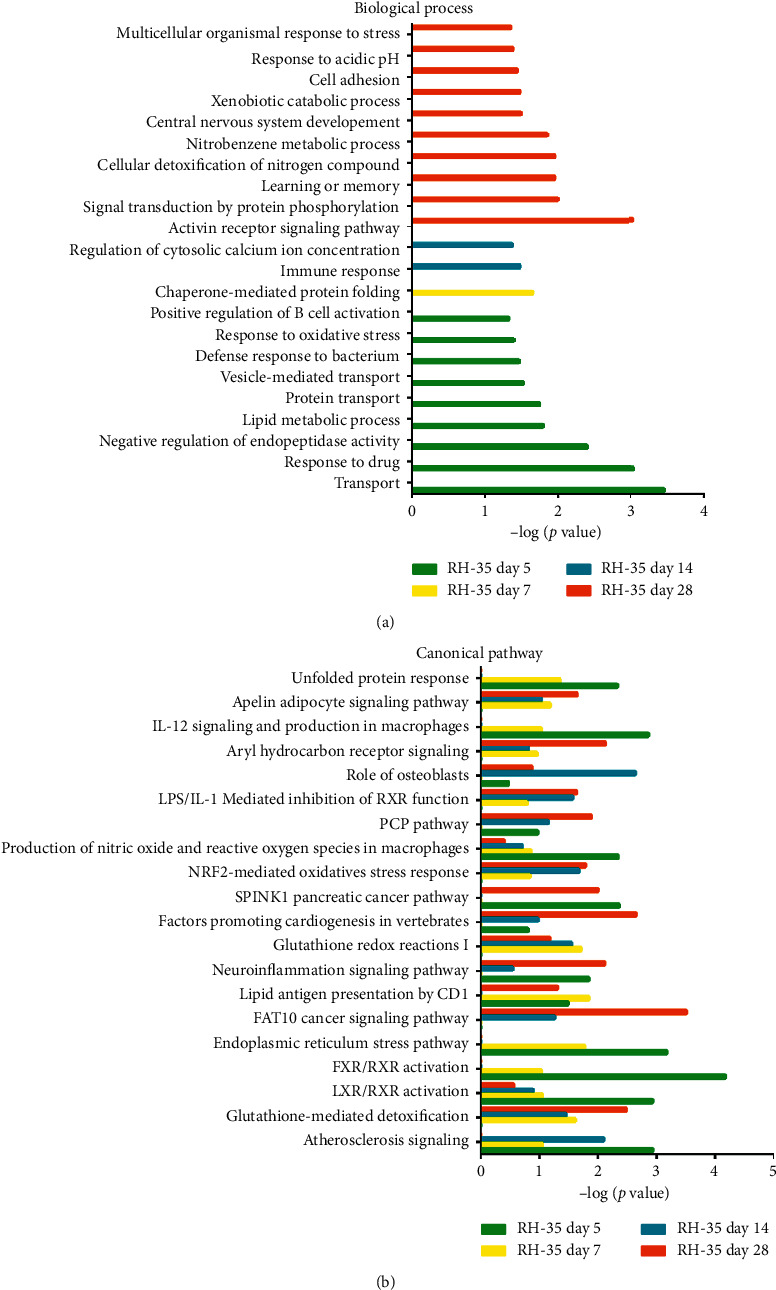
Functional analysis of differentially expressed proteins at days 5, 7, 14, and 28 in the RH-35 model. (a) Biological processes for the RH-35 model. (b) Canonical pathways for the RH-35 model.
IPA software was also employed to analyze the canonical pathways in which the differentially expressed proteins of the RH-35 model were involved. The atherosclerosis signaling, glutathione-mediated detoxification, LXR/RXR activation, FXR/RXR activation, FAT10 cancer signaling pathway, NRF2-mediated oxidative stress response, production of nitric oxide and reactive oxygen species in macrophages, and IL-12 signaling and production in macrophages pathway terms were enriched (Figure 4(b)). In addition to the pathways mentioned for the CBRH-7919 model, some other signaling pathways have also been reported to play important roles in tumor development. For example, high expression of FAT10 has been found to be positively correlated with proliferation and poor prognosis in liver cancer [19]. In addition, M2-polarized macrophages have the ability to generate nitric oxide (NO), which has different effects on different types of tumors because it can promote either the growth or death of tumor cells (depending on the cell source) [20]. Furthermore, IL-12 can induce the production of other cytokines, thereby exerting its biological functions and regulating the occurrence and development of inflammation and tumors [21]. Clinical studies have shown that IL-12 can kill tumor cells by activating and expanding natural killer (NK) cells in peripheral blood [22].
The above results indicate that when two different types of hepatoma cells are injected into the liver, there are differences in the biological processes and the main signaling pathways of the responses.
3.5. Comparison of the Urinary Proteomes of the Three Models
To investigate the differences in urinary proteins exhibited with the growth of different tumor cells in the same location of the liver, the differentially expressed urinary proteins of the CBRH-7919, RH-35, and W-256 models were compared [12]. Briefly, the differentially expressed proteins at a common time point (on days 5 and 7) were selected from each model for comparison, and the criteria used to screen the differentially expressed urinary proteins were a fold change value ≥ 2 or ≤0.5 and a P value <0.05. The comparison results for the differentially expressed proteins are shown in Figure 5. On day 5, four common urinary proteins were identified, including aminoacylase-1A (ACY1A), vomeromodulin (VOME), probasin (PBAS), and complement C4 (CO4). The numbers of unique proteins in the CBRH-7919, RH-35, and W-256 models were 25, 90, and 31, respectively (Figure 5(a)). On day 7, there were no common proteins among the three models, and 42, 43, and 22 unique proteins were identified in the CBRH-7919, RH-35, and W-256 models, respectively (Figure 5(b)). After injection of tumor cells, the histological results for the W-256 model were different from those for the CBRH-7919 and RH-35 models. The tumors grew more aggressively in the W-256 model than in the other models, which may be the main reason why its differentially expressed urinary protein pattern was different from the patterns of the other models.
Figure 5.
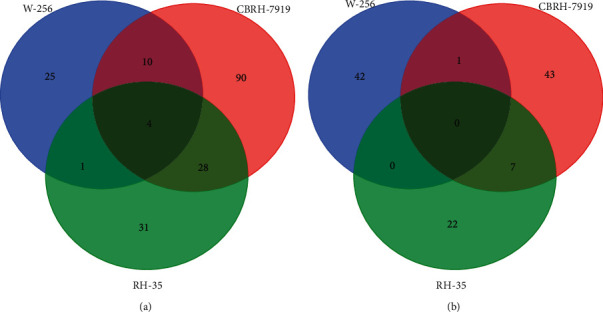
Comparison of differentially expressed proteins in the three different models. (a) Overlapping differentially expressed proteins on day 5. (b) Overlapping differentially expressed proteins on day 7.
The DAVID was applied to screen the differentially expressed proteins by using the above criteria. On day 5, the common biological processes of the three models were the negative regulation of endopeptidase activity, complement activation classical pathway, and transport processes (Figure 6(a)). The unique biological process terms for the W-256 model included the glycolytic, gluconeogenesis, and carbohydrate metabolism terms. The unique biological process terms for the CBRH-7919 model were the innate immune responses and positive regulation of B cell activation terms. The lipid metabolism process was enriched only for the RH-35 model. On day 7, there were no common biological processes among the three models (Figure 6(b)). The unique biological process terms for the W-256 model included the responses to oxidative stress, glutathione metabolic process, cellular redox homeostasis, cellular oxidant detoxification, and response to reactive oxygen species terms. The unique biological process terms for the CBRH-7919 model were the apoptotic process, phagocytosis engulfment, complement activation classical pathway, innate immune response, neutrophil aggregation, and B cell receptor signaling pathway terms. The unique biological process of the RH-35 model was chaperone-mediated protein folding. According to the above results, the W-256 model tumors grew aggressively over time. Tumor cells rapidly generate ATP and growth substrates by using aerobic glycolysis, thereby consuming large amounts of glucose [23]. The unique biological process reflects mainly tumor growth [2]. Based on the histological results, it can be inferred that the hepatoma cells of the CBRH-7919 and RH-35 models may have been recognized by the immune system and gradually cleared after the injection. Therefore, the immune response term was enriched in the biological process category for the CBRH-7919 model. The RH-35 model exhibited fewer enriched biological processes than the other models because of (1) its small number of differentially expressed proteins and (2) the diverse responses to the different hepatoma cells after the injection.
Figure 6.
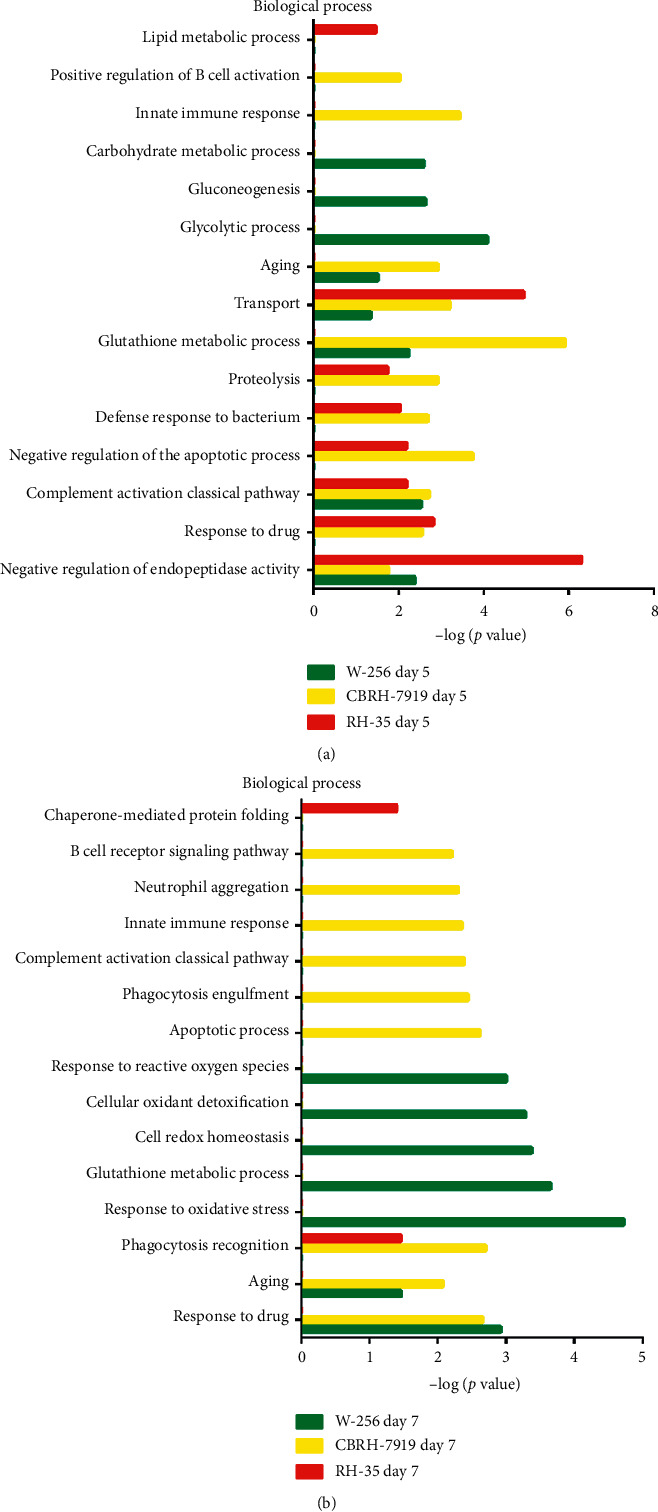
Functional analysis of differentially expressed proteins in the three different models. (a) Biological processes on day 5. (b) Biological processes on day 7.
In summary, it can be concluded that the early changes in urinary proteins are different when different tumor cells are injected into the same location of the liver.
3.6. Differentially Expressed Protein Analysis
In both models (the CBRH-7919 model and the RH-35 model), a total of 25 proteins were identified, 8 of which have been reported to be associated with liver cancer; these proteins are here described. (1) Carboxypeptidase Z (CBPZ). The specific constitutive expression of CBPZ in the liver protects against acute liver injury by combining the mechanisms that inhibit apoptosis in hepatocytes and promote cell cycle progression and proliferation [24]. (2) Apolipoprotein D (APOD). APOD may be a new tumor suppressor gene of hepatocellular carcinoma (HCC), and its expression status may be an available biomarker for predicting patient outcomes [25]. (3) Aquaporin-1 (AQP1). AQP1 may be a highly selective marker of differentiated bile duct cells that can help to diagnose liver tumors [26]. (4) Clusterin (CLUS). Abnormal expression of CLUS in the tumor tissues or serum of patients with primary liver cancer is considered a useful biomarker for diagnosis and monitoring [27]. (5) Filamin-C (FLNC). FLNC is a potential marker of the progression of HCC that can be assessed by using quantitative proteomics analysis [28]. (6) Galectin-1 (LEG1). LEG1 plays a role in the migration and invasion of HCC cells. It was initially identified for its role in the pathogenesis of HCC and has since become a potential molecular therapeutic target [29]. (7) Thrombospondin-4 (THBS4). THBS4 may play a role in the development of HCC and may become an independent prognostic biomarker and/or therapeutic target for patients with HCC [30]. (8) Collagen triple helix repeat-containing protein 1 (CTHRC1). Overexpression of CTHRC1 in solid tumors leads to enhanced tumor cell migration and invasion and is associated with decreased overall survival and disease-free survival. CTHRC1 expression is enriched in patients with hepatitis B virus infection and is highly correlated with the progression of HCC [31]. In the current study, in addition to changes in urinary proteins that have been reported as biomarkers of HCC, there were some changes in common urinary proteins that may have been related to the growth of animals during this period (28 days).
Twenty-four of the unique proteins in the CBRH-7919 model have been reported to be associated with liver cancer (Table 2), of which 17 could be clearly identified on day 5, including CO4, macrophage migration inhibitory factor (MIF), alpha-crystallin B chain (CRYAB), dimethylargininase-1 (DDAH1), chloride intracellular channel protein 1 (CLIC1), annexin A6 (ANXA6), brain acid soluble protein 1 (BASP1), monoglyceride lipase (MGLL), beta-ureidopropionase (BUP1), LIM and SH3 domain protein 1 (LASP1), ATP-binding cassette subfamily G member 2 (ABCG2), fructose-1,6-bisphosphatase 1 (F16P1), cathepsin D (CATD), matrilysin (MMP7), deleted in malignant brain tumors 1 protein (DMBT1), angiopoietin-related protein 4 (ANGL4), and protein S100-A9 (S100A9). Among them, CO4, CRYAB, CLIC1, BASP1, BUP1, LASP1, ABCG2, F16P1, and ANGL4 have been reported as biomarkers for the diagnosis or prognosis of liver cancer [32–40]. In addition, the combination of S100A9 and other biomarkers in urine has been reported to be useful for the early diagnosis of HCC [41]. The expression of MIF is significantly positively correlated with HCC; therefore, MIF plays an important role in the progression of HCC [42]. DDAH1 is important for the regulation of angiogenesis in human HCC, and its expression is increased in patients with liver cancer [43]. It has been reported that the specificity of ANXA6 is decreased in the context of human HCC, indicating its function in the development of liver cancer [44]. MGLL has been identified as a unique tumor suppressor for HCC [45]. CATD has proteolytic activity, and its expression is a prerequisite for cancer invasion. Its expression is also valuable for predicting the prognosis of HCC [46]. It has been reported that suppression of MMP-7 expression can inhibit the invasion and metastasis of HCC [47]. DMBT1 may play an important role in the proliferation of hepatic progenitor cells (HPCs) in HPV-related liver disease, and decreases in DMBT1 may increase the risk of malignant transformation of HPCs [48].
Fourteen of the unique biomarkers of the RH-35 model have been reported to be related to liver cancer (Table 3), of which 5 were clearly identified on day 5 and are here described. (1) Alpha-1-acid glycoprotein (A1AG). Notably, glycosylation of A1AG gradually changes with the progression of cirrhosis to cancer [49]. (2) Guanine deaminase (GUAD). Due to its favorable specificity, combining assessment of GUAD with assessment of other enzymes (such as AST and ALT) in liver function tests may effectively predict liver disease with a lower false-positive rate than other methods [50]. (3) Transaldolase (TALDO). TALDO can be used as a novel serum biomarker for HCC metastasis [51]. (4) Latexin (LXN). LXN has the potential to suppress tumor cells in HCC [52]. (5) Thioredoxin-dependent peroxide reductase (PRDX3). PRDX3 can be used as an early and sensitive biomarker for the early detection of HCC [53].
On day 5 after injection of hepatoma cells, large numbers of hepatoma cells were observed in the livers, and a variety of urinary proteins related to HCC were identified at the corresponding time point. These findings indicate that urinary proteins can sensitively respond to changes in hepatoma cells at an early stage.
4. Discussion
Urine is an early and sensitive source of biomarkers. It has been reported to be useful for early diagnosis in a variety of animal models. Urinary proteins have the potential to be used to distinguish among the same types of cells growing in different organs [12, 54]. In this study, two different hepatoma cell lines were injected into the livers of animal models. To reduce the impact of individual differences, during the screening process, proteins were only considered differentially expressed if they were upregulated or downregulated in more than half of the total number of animals. Although the changes in histological features were similar after the injection, the changes in urinary proteins were quite different. These findings indicate that urinary protein patterns can still be used to sensitively differentiate among tumor cells even when histological findings are similar.
In the random allocation statistical analysis, it was found that under the same screening criteria, random grouping could also generate some differentially expressed urinary proteins. As the growth of tumor cells decreased, the number of differentially expressed urinary proteins decreased and the false-positive rate increased. However, the maximum false-positive rate was 0.177, indicating that most of the identified proteins were related to the physiological response to tumor cells rather than the result of random allocation. The existence of false-positive proteins is undeniable. The number of differentially expressed urinary proteins was increased, and the false-positive rate was decreased, when tumor cells grew actively in the liver. The false-positive rate can be reduced by increasing the stringency of the screening criteria, but the false-negative rate will increase at the same time. How to select appropriate screening criteria may still be a challenge for future studies.
Compared with the previously reported W-256 model, the two models had different urinary protein patterns and associated biological processes. Even when they are grown in the same organ, tumor cells can lead to different changes in the body, which indicates that urinary proteins can be used to recognize such changes at the early stage regardless of how the tumor cells grow. Urinary protein changes may also predict the trends of subsequent tumor-related changes in the body. Different types of tumor cells may be able to be classified by their different urinary protein patterns. This study demonstrates that urinary proteins have the potential to be used to distinguish among different types of hepatoma cells. This possibility needs to be further validated in clinical samples in future studies.
Interestingly, the urinary proteins identified at each time point were different even after the same numbers of CBRH-7919 and RH-35 cells were injected into the liver. The largest number of urinary proteins was identified in the CBRH-7919 model on day 5, while the numbers of proteins decreased at the following three time points. The numbers of urinary proteins identified for the RH-35 model were decreased at the first three time points and increased on day 28. At the same time point, the types and quantities of identified proteins in the two cell lines were quite different, which may have been related to the different categories of hepatoma cells. On day 5, the unique biological processes in the CBRH-7919 model included the innate immune response, complement activation, classical pathways, and neutrophil aggregation, which indicate that the immune system quickly responded to CBRH-7919 cells with numerous protein changes after the injection. Positive regulation of B cell activation was a common biological process in the CBRH-7919 and RH-35 models on day 5. However, no other immune-related biological processes were identified in the RH-35 model, indicating that RH-35 hepatoma cells did not immediately cause a strong immune response and that relatively few changes in urinary proteins occurred. On day 7, the unique biological process terms of the CBRH-7919 model associated with the inflammatory response included the chronic inflammatory response and leukocyte migration terms. At the following time points, no biological processes were related to these immune and inflammatory reactions, and relatively few differentially expressed proteins were identified. The immune response biological process term was enriched in the RH-35 model on day 14. More proteins were identified on day 28 than at the other three time points, which may have been due to the late appearance of the immune response caused by RH-35 hepatoma cells.
5. Conclusion
In this study, changes in urinary proteins caused by the injection of two different hepatoma cell types were identified at an early stage. Even when similar histological results were obtained, the patterns of the urinary proteins could be used to sensitively distinguish the tumor cells. Some of the differentially expressed proteins have been reported to be associated with HCC. Urinary proteins have the potential to differentiate among different tumor cells grown in the same organ, which may provide clues for clinical diagnoses in the future.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2018YFC0910202 and 2016 YFC 1306300), the Fundamental Research Funds for the Central Universities (2020KJZX002), Beijing Natural Science Foundation (7172076), Beijing Cooperative Construction Project (110651103), Beijing Normal University (11100704), and Peking Union Medical College Hospital (2016-2.27).
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Supplementary Materials
Supplementary Figure 1. Functional analysis of differentially expressed proteins at days 5, 7, 14, and 28 in two models. (a) Cell component for the CBRH-7919 model. (b) Molecular function for the CBRH-7919 model. (c) Cell component for the RH-35 model. (d) Molecular function for the RH-35 model. Supplementary Table 1. All urinary proteins identified in the CBRH-7919 model. Supplementary Table 2. All urinary proteins identified in the RH-35 model. Supplementary Table 3. The details of 6435 random allocations in the CBRH-7919 model. Supplementary Table 4. The occurrence of the protein in 6435 random allocations in the CBRH-7919 model. Supplementary Table 5. The details of 12155 random allocations in the RH-35 model. Supplementary Table 6. The occurrence of the protein in 12155 random allocations in the RH-35 model.
References
- 1.YouHe G. Urine—an untapped goldmine for biomarker discovery? Science China (Life Sciences) 2013;10 doi: 10.1007/s11427-013-4574-1. [DOI] [PubMed] [Google Scholar]
- 2.Wu J., Guo Z., Gao Y. Dynamic changes of urine proteome in a walker 256 tumor-bearing rat model. Cancer Medicine. 2017;6(11):2713–2722. doi: 10.1002/cam4.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu J., Gao Y. Physiological conditions can be reflected in human urine proteome and metabolome. Expert Review of Proteomics. 2015;12(6):623–636. doi: 10.1586/14789450.2015.1094380. [DOI] [PubMed] [Google Scholar]
- 4.Gao Y. Roadmap to the urine biomarker era. MOJ Proteomics Bioinform. 2014;1:p. 5. doi: 10.15406/mojpb.2014.01.00005. [DOI] [Google Scholar]
- 5.Zhao M., Wu J., Li X., Gao Y. Urinary candidate biomarkers in an experimental autoimmune myocarditis rat model. Journal of Proteomics. 2018;179:71–79. doi: 10.1016/j.jprot.2018.02.032. [DOI] [PubMed] [Google Scholar]
- 6.Zhang F., Wei J., Li X., Ma C., Gao Y. Early candidate urine biomarkers for detecting alzheimer’s disease before amyloid-β plaque deposition in an APP (swe)/PSEN1dE9 transgenic mouse model. Journal of Alzheimer’s Disease. 2018;66(2):613–637. doi: 10.3233/jad-180412. [DOI] [PubMed] [Google Scholar]
- 7.Zhang F., Ni Y., Yuan Y., Yin W., Gao Y. Early urinary candidate biomarker discovery in a rat thioacetamide-induced liver fibrosis model. Science China Life Sciences. 2018;61(11):1369–1381. doi: 10.1007/s11427-017-9268-y. [DOI] [PubMed] [Google Scholar]
- 8.Ni Y., Zhang F., An M., Yin W., Gao Y. Early candidate biomarkers found from urine of glioblastoma multiforme rat before changes in MRI. Science China Life Sciences. 2018;61(8):982–987. doi: 10.1007/s11427-017-9201-0. [DOI] [PubMed] [Google Scholar]
- 9.Wu J., Li X., Gao Y. Early detection in urinary proteome for the effective early treatment of bleomycin-induced pulmonary fibrosis in a rat model. Proteomics Clinical Applications. 2017;11:11–12. doi: 10.1002/prca.201700103. [DOI] [PubMed] [Google Scholar]
- 10.Zhang L., Li Y., Meng W., Ni Y., Gao Y. Dynamic urinary proteomic analysis in a walker 256 intracerebral tumor model. Cancer Medicine. 2019;8(7):3553–3565. doi: 10.1002/cam4.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L., Li Y., Gao Y. Early changes in the urine proteome in a diethyldithiocarbamate-induced chronic pancreatitis rat model. Journal of Proteomics. 2018;186:8–14. doi: 10.1016/j.jprot.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 12.Yameng Z., Yufei G., Youhe G. Early changes in the urine proteome in a rat liver tumor model. bioRxiv. 2019;2019 doi: 10.1101/568246v1.568246 [DOI] [Google Scholar]
- 13.Wei J., Ni N., Meng W., Gao Y. Early urine proteome changes in the walker-256 tail-vein injection rat model. Science Reports. 2019;9:p. 13804. doi: 10.1038/s41598-019-50301-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei J., Meng W., Gao Y. Urine proteome changes in rats subcutaneously inoculated with approximately ten tumor cells. PeerJ. 2019;7 doi: 10.7717/peerj.7717.e7717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wisniewski J. R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nature Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 16.Martelli A. M., Evangelisti C., Follo M. Y, et al. Targeting the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin signaling network in cancer stem cells. Current Medicinal Chemistry. 2011;18:2715–2726. doi: 10.2174/092986711796011201. [DOI] [PubMed] [Google Scholar]
- 17.Shin S. M., Yang J. H., Ki S. H. Role of the Nrf2-ARE pathway in liver diseases. Oxidative Medicine and Cellular Longevity. 2013;2013 doi: 10.1155/2013/763257.763257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aydin M. M., Akcali K. C. Liver fibrosis. The Turkish Journal of Gastroenterology. 2018;29(1):14–21. doi: 10.5152/tjg.2018.17330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davalieva K., Kiprijanovska S., Maleva Kostovska I, et al. Comparative proteomics analysis of urine reveals down-regulation of acute phase response signaling and LXR/RXR activation pathways in prostate cancer. Proteomes. 2017;6 doi: 10.3390/proteomes6010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perrotta C., Cervia D., Di Renzo I, et al. Nitric oxide generated by tumor-associated macrophages is responsible for cancer resistance to cisplatin and correlated with syntaxin 4 and acid sphingomyelinase inhibition. Frontiers in Immunology. 2018;9:p. 1186. doi: 10.3389/fimmu.2018.01186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerkar S. P., Restifo N. P. The power and pitfalls of IL-12. Blood. 2012;119(18):4096–4097. doi: 10.1182/blood-2012-03-415018. [DOI] [PubMed] [Google Scholar]
- 22.El-Shemi A. G., Ashshi A. M., Na Y, et al. Combined therapy with oncolytic adenoviruses encoding TRAIL and IL-12 genes markedly suppressed human hepatocellular carcinoma both in vitro and in an orthotopic transplanted mouse model. Journal of Experimental and Clinical Cancer Research. 2016;35:p. 74. doi: 10.1186/s13046-016-0365-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lei S., Yang J., Chen C, et al. FLIP(L) is critical for aerobic glycolysis in hepatocellular carcinoma. Journal of Experimental and Clinical Cancer Research. 2016;35:p. 79. doi: 10.1186/s13046-016-0358-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martín-Sanz P., Casado M., Boscá L. Cyclooxygenase 2 in liver dysfunction and carcinogenesis: facts and perspectives. World Journal of Gastroenterology. 2017;23(20):3572–3580. doi: 10.3748/wjg.v23.i20.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Utsunomiya T., Ogawa K., Yoshinaga K., et al. Clinicopathologic and prognostic values of apolipoprotein D alterations in hepatocellular carcinoma. International Journal of Cancer. 2005;116(1):105–109. doi: 10.1002/ijc.20986. [DOI] [PubMed] [Google Scholar]
- 26.Mazal P. R., Susani M., Wrba F., Haitel A. Diagnostic significance of aquaporin-1 in liver tumors. Human Pathology. 2005;36(11):1226–1231. doi: 10.1016/j.humpath.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 27.Yao M., Fang M., Zheng W., Dong Z., Yao D. Role of secretory clusterin in hepatocarcinogenesis. Translational Gastroenterology and Hepatology. 2018;3:p. 48. doi: 10.21037/tgh.2018.07.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi Y., Xu F., Chen L., et al. Quantitative proteomics reveals FLNC as a potential progression marker for the development of hepatocellular carcinoma. Oncotarget. 2016;7(42):68242–68252. doi: 10.18632/oncotarget.11921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spano D., Russo R., Di Maso V., et al. Galectin-1 and its involvement in hepatocellular carcinoma aggressiveness. Molecular Medicine. 2010;16(3-4):102–115. doi: 10.2119/molmed.2009.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu H., Zhang G., Li Z., et al. Thrombospondin-4 expression as a prognostic marker in hepatocellular carcinoma. Gene. 2019;696:219–224. doi: 10.1016/j.gene.2019.02.049. [DOI] [PubMed] [Google Scholar]
- 31.Wu Q., Yang Q., Sun H. Role of collagen triple helix repeat containing-1 in tumor and inflammatory diseases. Journal of Cancer Research and Therapeutics. 2017;13(4):621–624. doi: 10.4103/jcrt.JCRT_410_17. [DOI] [PubMed] [Google Scholar]
- 32.Kim H., Kim K., Yu S. J, et al. Development of biomarkers for screening hepatocellular carcinoma using global data mining and multiple reaction monitoring. PLoS One. 2013;8 doi: 10.1371/journal.pone.0063468.e63468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang Q., Liu Y.-F., Zhu X.-J., et al. Expression and prognostic significance of the αB-crystallin gene in human hepatocellular carcinoma. Human Pathology. 2009;40(3):300–305. doi: 10.1016/j.humpath.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 34.Zhang S., Wang X.-M., Yin Z.-Y., et al. Chloride intracellular channel 1 is overexpression in hepatic tumor and correlates with a poor prognosis. APMIS. 2013;121(11):1047–1053. doi: 10.1111/apm.12093. [DOI] [PubMed] [Google Scholar]
- 35.Moribe T., Iizuka N., Miura T, et al. Identification of novel aberrant methylation of BASP1 and SRD5A2 for early diagnosis of hepatocellular carcinoma by genome-wide search. International Journal of Oncology. 2008;33(5):949–958. [PubMed] [Google Scholar]
- 36.Awan F. M., Naz A., Obaid A, et al. Identification of circulating biomarker candidates for hepatocellular carcinoma (HCC): an integrated prioritization approach. PLoS One. 2015;10 doi: 10.1371/journal.pone.0138913.e0138913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H., Li W., Jin X., Cui S., Zhao L. LIM and SH3 protein 1, a promoter of cell proliferation and migration, is a novel independent prognostic indicator in hepatocellular carcinoma. European Journal of Cancer. 2013;49(4):974–983. doi: 10.1016/j.ejca.2012.09.032. [DOI] [PubMed] [Google Scholar]
- 38.Zhang G., Wang Z., Luo W., Jiao H., Wu J., Jiang C. Expression of potential cancer stem cell marker ABCG2 is associated with malignant behaviors of hepatocellular carcinoma. Gastroenterology Research and Practice. 2013;2013 doi: 10.1155/2013/782581.782581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen S. S., Yu K. K., Ling Q. X, et al. The combination of three molecular markers can be a valuable predictive tool for the prognosis of hepatocellular carcinoma patients. Science Reports. 2016;6:p. 24582. doi: 10.1038/srep24582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ng K. T.-P., Xu A., Cheng Q., et al. Clinical relevance and therapeutic potential of angiopoietin-like protein 4 in hepatocellular carcinoma. Molecular Cancer. 2014;13(1):p. 196. doi: 10.1186/1476-4598-13-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang C.-H., Kuo C.-J., Liang S.-S., et al. Onco-proteogenomics identifies urinary S100A9 and GRN as potential combinatorial biomarkers for early diagnosis of hepatocellular carcinoma. BBA Clinical. 2015;3:205–213. doi: 10.1016/j.bbacli.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang D., Luo L., Chen W., et al. Significance of the vascular endothelial growth factor and the macrophage migration inhibitory factor in the progression of hepatocellular carcinoma. Oncology Reports. 2014;31(3):1199–1204. doi: 10.3892/or.2013.2946. [DOI] [PubMed] [Google Scholar]
- 43.Buijs N., Oosterink J. E., Jessup M., et al. A new key player in VEGF-dependent angiogenesis in human hepatocellular carcinoma: dimethylarginine dimethylaminohydrolase 1. Angiogenesis. 2017;20(4):557–565. doi: 10.1007/s10456-017-9567-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meier E. M., Rein-Fischboeck L., Pohl R., et al. Annexin A6 protein is downregulated in human hepatocellular carcinoma. Molecular and Cellular Biochemistry. 2016;418(1-2):81–90. doi: 10.1007/s11010-016-2735-9. [DOI] [PubMed] [Google Scholar]
- 45.Rajasekaran D., Jariwala N., Mendoza R. G., et al. Staphylococcal nuclease and tudor domain containing 1 (SND1 Protein) promotes hepatocarcinogenesis by inhibiting monoglyceride lipase (MGLL) Journal of Biological Chemistry. 2016;291(20):10736–10746. doi: 10.1074/jbc.m116.715359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen R. X., Song H. Y., Dong Y. Y, et al. Dynamic expression patterns of differentially expressed proteins during early invasion of hepatocellular carcinoma. PLoS One. 2014;9 doi: 10.1371/journal.pone.0088543.e88543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tu K., Dou C., Zheng X, et al. Fibulin-5 inhibits hepatocellular carcinoma cell migration and invasion by down-regulating matrix metalloproteinase-7 expression. BMC Cancer. 2014;14:p. 938. doi: 10.1186/1471-2407-14-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deng H., Gao Y.-B., Wang H.-F., Jin X.-L., Xiao J.-C. Expression of deleted in malignant brain tumours 1 (DMBT1) relates to the proliferation and malignant transformation of hepatic progenitor cells in hepatitis B virus-related liver diseases. Histopathology. 2012;60(2):249–260. doi: 10.1111/j.1365-2559.2011.04082.x. [DOI] [PubMed] [Google Scholar]
- 49.Comunale M. A., Rodemich-Betesh L., Hafner J, et al. Linkage specific fucosylation of alpha-1-antitrypsin in liver cirrhosis and cancer patients: implications for a biomarker of hepatocellular carcinoma. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012419.e12419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nishikawa Y., Fukumoto K., Watanabe F. Clinical evaluation of serum guanase activity in liver diseases. Clinical Biochemistry. 1984;17(5):327–330. doi: 10.1016/s0009-9120(84)90661-1. [DOI] [PubMed] [Google Scholar]
- 51.Wang C., Guo K., Gao D., et al. Identification of transaldolase as a novel serum biomarker for hepatocellular carcinoma metastasis using xenografted mouse model and clinic samples. Cancer Letters. 2011;313(2):154–166. doi: 10.1016/j.canlet.2011.08.031. [DOI] [PubMed] [Google Scholar]
- 52.Ni Q.-F., Tian Y., Kong L.-L., Lu Y.-T., Ding W.-Z., Kong L.-B. Latexin exhibits tumor suppressor potential in hepatocellular carcinoma. Oncology Reports. 2014;31(3):1364–1372. doi: 10.3892/or.2014.2966. [DOI] [PubMed] [Google Scholar]
- 53.Ismail S., Mayah W., Battia H. E., et al. Plasma nuclear factor kappa B and serum peroxiredoxin 3 in early diagnosis of hepatocellular carcinoma. Asian Pacific Journal of Cancer Prevention. 2015;16(4):1657–1663. doi: 10.7314/apjcp.2015.16.4.1657. [DOI] [PubMed] [Google Scholar]
- 54.Beretov J., Wasinger V. C., Millar E. K., Schwartz P., Graham P. H., Li Y. Proteomic analysis of urine to identify breast cancer biomarker candidates using a label-free LC-MS/MS approach. PLoS One. 2015;10 doi: 10.1371/journal.pone.0141876.e0141876 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Functional analysis of differentially expressed proteins at days 5, 7, 14, and 28 in two models. (a) Cell component for the CBRH-7919 model. (b) Molecular function for the CBRH-7919 model. (c) Cell component for the RH-35 model. (d) Molecular function for the RH-35 model. Supplementary Table 1. All urinary proteins identified in the CBRH-7919 model. Supplementary Table 2. All urinary proteins identified in the RH-35 model. Supplementary Table 3. The details of 6435 random allocations in the CBRH-7919 model. Supplementary Table 4. The occurrence of the protein in 6435 random allocations in the CBRH-7919 model. Supplementary Table 5. The details of 12155 random allocations in the RH-35 model. Supplementary Table 6. The occurrence of the protein in 12155 random allocations in the RH-35 model.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.


