Abstract
Background
The allele frequency of epidermal growth factor receptor (EGFR) mutations could be a potential molecular biomarker for the outcome of osimertinib therapy.
Objective
The purpose of our study was to assess the clinical relevance of the allele frequency of EGFR mutations in plasma-based circulating tumor DNA (ctDNA) before starting osimertinib therapy in patients with advanced EGFR-mutated non-small cell lung cancer (NSCLC) who had progressed under treatment with EGFR tyrosine kinase inhibitors (TKIs).
Patients and Methods
We enrolled 141 patients with advanced EGFR T790M-positive NSCLC who underwent second-line osimertinib treatment. Plasma ctDNA was tested for EGFR-activating mutations (EGFR deletions in exon 19, L858R, L861Q, S768I) and T790M by means of droplet digital polymerase chain reaction (ddPCR).
Results
The allele frequency of EGFR-activating mutations in plasma ctDNA before osimertinib initiation ranged from 0 to 81,543 copies/ml and was independently associated with progression-free survival (PFS) and overall survival (OS) after adjusting for known clinicopathological risk factors (PFS: adjusted hazard ratio [HR] 1.26, 95% confidence interval [CI] 1.15–1.39, P < 0.0001; OS: adjusted HR 1.32, 95% CI 1.18–1.47, P < 0.0001). The allele frequency of T790M in plasma ctDNA before starting osimertinib therapy ranged from 0 to 38,092 copies/ml. Multivariate analyses showed that a higher T790M allele frequency was associated with a trend towards a shorter PFS (adjusted HR 1.19, 95% CI 0.99–1.42, P = 0.05) and a significantly shorter OS (adjusted HR 1.25, 95% CI 1.02–1.53, P = 0.03) of the patients.
Conclusion
A higher allele frequency of EGFR mutations, particularly EGFR-activating mutations, in plasma ctDNA is a poor prognostic marker. Further studies on the clinical utility of liquid biopsy are needed.
Key Points
| Our findings suggest that plasma analyses prior to initiation of second-line osimertinib therapy may be clinically relevant. |
| This study, by reporting a large series of patients, provides real-life data, indicating that a higher allele frequency of epidermal growth factor receptor (EGFR) mutations, particularly EGFR-activating mutations, in plasma circulating tumor DNA before start of osimertinib treatment is associated with poor prognosis. |
| Further studies on the clinical utility of liquid biopsy are needed. |
Introduction
Current guidelines recommend epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs), including osimertinib, as standard treatment for patients with advanced EGFR-mutated lung adenocarcinoma. Although first-line osimertinib may now be considered as the preferred option for non-small cell lung cancer (NSCLC) patients with EGFR-activating mutations, sequential first- or second-generation EGFR TKI followed by osimertinib is still a valid chemotherapy-free regimen that could be offered to patients [1, 2]. Therefore, fine-tuning of second-line treatment using molecular biomarkers for patients who have progressed under treatment with EGFR TKIs may further improve the clinical outcome in these patients.
The allele frequency of EGFR mutations could be a potential molecular biomarker for response to osimertinib. Previously, it has been shown that the allele frequency of the T790M mutation in tumor biopsies may be an important predictor of rociletinib response. Patients with the highest proportion of T790M-positive cells in their pre-treatment biopsies had the most significant tumor shrinkage [3]. Similar results were obtained in the plasma of the patients. Contradictory results have been published in patients who received osimertinib. While quantification of the allele frequency of EGFR mutations was not significantly associated with response in an exploratory analysis of the AURA phase I trial [4], other small studies showed that a lower allele frequency of EGFR-activating mutations or the T790M mutation was associated with a greater decrease in tumor volume under osimertinib therapy [5, 6].
Based on this interesting observation, the allele frequency of EGFR-activating and/or EGFR resistance mutations could be an important biomarker for response to osimertinib. In the present study, we assessed the clinical relevance of the quantification of the allele frequency of EGFR mutations in plasma circulating tumor DNA (ctDNA) before initiation of osimertinib therapy in patients who had been pre-treated with EGFR TKIs.
Patients and Methods
Patients
Patients with advanced T790M-positive NSCLC who received second-line osimertinib were recruited at the Department of Respiratory and Critical Care Medicine, and Ludwig Boltzmann Institute of COPD and Respiratory Epidemiology, Otto Wagner Hospital, Vienna, between May 2015 and January 2019. All patients had histologically confirmed lung adenocarcinoma with EGFR mutations in their initial biopsy at diagnosis. Blood samples for plasma genotyping were collected under EGFR TKI therapy within the scope of diagnostic routine procedures in all patients. Plasma genotyping using droplet digital polymerase chain reaction (ddPCR) was performed at the Institute of Cancer Research, Department of Medicine I, Medical University of Vienna. Sample collection and analysis was performed in accordance with the local ethics committee (EK No. 1132/2016). All patients gave their written informed consent for providing blood specimens for plasma genotyping. These patients have been included in previous plasma ctDNA studies [7–10].
Plasma Genotyping
Blood processing for plasma preparation and storage was performed as previously described [7]. Briefly, blood samples were collected in Cell-Free DNA BCT blood collection tubes (Streck, La Vista, NE) or Cell-Free DNA Collection Tubes (Roche, Pleasanton, CA). One blood sample (8 ml) was collected from all patients at each time point.
To isolate plasma, whole blood was centrifuged at 200 g for 10 min and subsequently at 1600 g for 10 min. Finally, the supernatant was collected and centrifuged at 1900 g for 10 min.
For ddPCR, ctDNA was extracted from 2 ml of plasma using the QIAamp circulating nucleic acid kit (Qiagen, Venlo, The Netherlands) according to the manufacturer’s instructions.
EGFR deletions in exon 19, L858R, L861Q, S768I, T790M, and C797S mutations were assessed by using the QX-200™ ddPCR system (Bio-Rad, Hercules, CA) according to the manufacturer’s instructions.
Primers and probes for ddPCR assays were from Bio-Rad or custom-made by Life Technologies (Carlsbad, CA) and were used as previously described [7]. Analysis of ddPCR data was performed with QuantaSoft analysis software (Bio-Rad). All ddPCR assays were performed blinded to the study endpoint and analyzed in triplicate; results were reported as copies of mutant allele per milliliter of plasma. The threshold for positivity was > 1 copy/ml for all mutations. The median copy number of EGFR-activating mutations, 10 copies/ml for T790M, and a value of 0.22 for the EGFR T790M/activating mutation ratio were pre-specified cut-offs to classify patients [6, 7].
Statistical Analyses
The primary endpoint was progression-free survival (PFS) as determined by investigator assessment. PFS was defined as the time from first osimertinib dose to disease progression or death from any cause, whichever came first. Overall survival (OS) and response rate (RR) were secondary endpoints. OS was defined as the time from first osimertinib dose to death from any cause. RR was defined as the percentage of patients with response (complete or partial) at restaging after osimertinib initiation. Tumor response was assessed by a contrast-enhanced computed tomography (CT) scan of the chest and abdomen by the local radiologist, according to institutional practice. In addition, response was confirmed post hoc using Response Evaluation Criteria in Solid Tumors (RECIST) 1.1. The scan intervals were usually between 6 and 8 weeks at the treating physician’s discretion.
Patient and tumor characteristics included age, gender, presence or absence of extra-pulmonary metastases, tissue genotype at diagnosis, and first-line EGFR TKI therapy. Associations of plasma genotyping results with clinical parameters, including treatment response, were assessed by the chi-square test or Mann–Whitney U test. Survival probabilities were calculated with the product limit method according to Kaplan–Meier. Differences between survival curves were analyzed by means of the log-rank test. Hazard ratios (HRs) and their confidence intervals (CIs) were estimated using Cox proportional hazards regression models. For the multivariate analyses, we used full models that included age (as a continuous variable), gender (male, female), presence or absence of extra-pulmonary metastases (pulmonary, extra-pulmonary), tissue genotype at diagnosis (EGFR deletions in exon 19, L858R, other EGFR mutations), first-line EGFR TKI therapy (afatinib, erlotinib, gefitinib, more than one EGFR TKI), and allele frequency of EGFR mutations or EGFR T790M/activating mutation ratio (as a continuous variable). All reported P values are two sided. All analyses were performed using IBM SPSS Statistics software, version 25 (SPSS, IBM Corporation, Armonk, NY, USA).
Results
Analyses of Plasma Samples
Plasma samples of 141 patients were centrally analyzed by ddPCR for EGFR exon 19 deletions, L858R, L861Q, S768I, and the T790M mutation. Before starting second-line osimertinib treatment, all 141 patients were T790M positive in plasma ctDNA and/or tumor tissue. In addition to the 141 plasma samples, tissue re-biopsies were also available in 53 patients. Of these 53 samples, 35 (66%) were T790M positive in plasma and tissue, seven (13%) were positive only in tissue, and 11 (21%) were only positive in plasma. A total of 121 of 141 patients (86%) were also positive for the corresponding EGFR-activating mutations. Of the 53 patients for whom tissue re-biopsies were also available, 29 (55%) were positive for the EGFR-activating mutations in plasma and tissue, 12 (23%) were only positive in tissue, 11 (21%) were only positive in plasma, and in one patient the EGFR-activating mutation was not detectable in tissue and plasma. The 20 patients who were T790M positive but in whom the EGFR-activating mutation was not detectable were also treated with osimertinib. The allele frequency of EGFR-activating mutations and the T790M mutation in plasma ctDNA ranged from 0 to 81,543 copies/ml and from 0 to 38,092 copies/ml, respectively. The median copy number of EGFR-activating mutations was 40 copies/ml, and this cut-off was used to classify patients with low and high allele frequencies.
Allele Frequency of EGFR Mutations and Patient Characteristics
Patient characteristics are summarized in Table 1. All patients had lung adenocarcinoma histology, stage IV disease, had been pre-treated with at least one EGFR TKI, and received osimertinib as second-line treatment. The tissue genotype at diagnosis included EGFR exon 19 deletions in 90 patients (64%), L858R in 40 (28%), and other EGFR mutations in 11 patients (8%). We observed no correlations between clinicopathological variables and the allele frequency of EGFR-activating mutations (Table 1) or the T790M mutation.
Table 1.
Patient characteristics
| Characteristics | No. of patients (N = 141) |
EGFR-activating mutation < 40 copies/ml (N = 71) |
EGFR-activating mutation ≥ 40 copies/ml (N = 70) |
P value |
|---|---|---|---|---|
| Age | ||||
| Median (range), years | 67 (38–88) | 69 (38–84) | 67 (43–88) | 0.42 |
| < 65years | 53 (38%) | 25 (35%) | 28 (40%) | 0.56 |
| ≥ 65 years | 88 (62%) | 46 (65%) | 42 (60%) | |
| Gender | ||||
| Female | 106 (75%) | 54 (76%) | 52 (74%) | 0.81 |
| Male | 35 (25%) | 17 (24%) | 18 (26%) | |
| Metastases | ||||
| Lung | 43 (31%) | 26 (37%) | 17 (24%) | 0.11 |
| Bone | 11 (8%) | 2 (3%) | 9 (13%) | |
| Brain | 8 (6%) | 4 (6%) | 4 (6%) | |
| Other | 79 (56%) | 39 (55%) | 40 (57%) | |
| EGFR tissue genotype | ||||
| Exon 19 deletion | 90 (64%) | 46 (65%) | 44 (63%) | 0.36 |
| L858R | 40 (28%) | 20 (28%) | 20 (29%) | |
| L861Q | 6 (4%) | 1 (1%) | 5 (7%) | |
| L858R/exon 18 mutation | 1 (1%) | 0 (0%) | 1 (1%) | |
| Exon 18 | 1 (1%) | 1 (1%) | 0 | |
| G719X, S768I | 1 (1%) | 1 (1%) | 0 | |
| L858R, S768I | 1 (1%) | 1 (1%) | 0 | |
| L858R, L861Q | 1 (1%) | 1 (1%) | 0 | |
| First-line EGFR TKI therapy | ||||
| Afatinib | 60 (43%) | 34 (48%) | 26 (37%) | 0.41 |
| Erlotinib | 12 (9%) | 7 (10%) | 5 (7%) | |
| Gefitinib | 41 (29%) | 19 (27%) | 22 (31%) | |
| > 1 EGFR TKI | 28 (20%) | 11 (16%) | 17 (24%) |
Percentages may not total 100, because of rounding
EGFR epidermal growth factor receptor, TKI tyrosine kinase inhibitor
Response to Second-Line Osimertinib
Response to osimertinib therapy was evaluable in 139 of 141 patients, and 72% of the patients responded to osimertinib. The RR was not significantly different between patients with ≥ 40 copies and those with < 40 copies of the EGFR-activating mutations per milliliter of plasma (68% vs 79%, P = 0.13). In addition, a Mann–Whitney U test was performed to determine if there were any differences in the distribution of the allele frequency between responders and non-responders. There was a statistically significant difference in allele frequency between both groups—patients who responded to osimertinib had a significantly lower allele frequency of the EGFR-activating mutations than patients who did not respond (P = 0.03). In the case of the T790M mutation, the RR was similar in patients with ≥ 10 copies/ml and in patients with < 10 copies/ml (79% vs 69%, P = 0.20) and was not associated with the T790M allele frequency (P = 0.37).
Association with PFS and OS
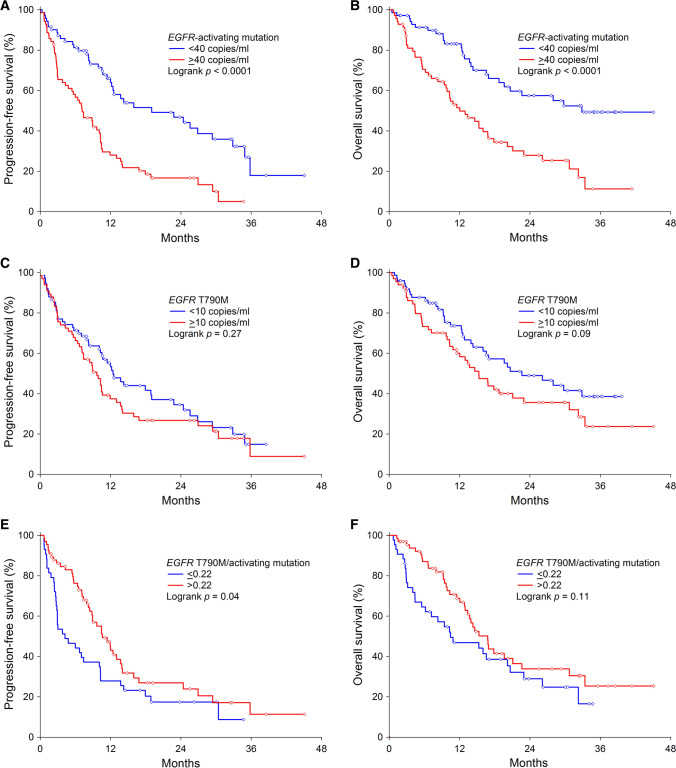
At a median follow-up of 29.8 months (95% CI 24.7–34.9), 97 of 141 patients (69%) had progressed and 76 of 108 (54%) had died. Median PFS was 10.5 months (95% CI 8.8–12.3), and median OS was 17.8 months (95% CI 13.1–22.6). In univariate analyses, age, gender, and first-line EGFR TKI therapy had no impact on PFS and OS. Absence of extra-pulmonary metastases (P = 0.005) and EGFR deletions in exon 19 (P = 0.048) were significantly associated with longer PFS. Absence of extra-pulmonary metastases also correlated with longer OS (P = 0.02) (Table 2). Patients with ≥ 40 copies/ml of the EGFR-activating mutations had a shorter PFS (median 7.2 months vs 19.0 months, P < 0.0001) (Fig. 1a) and OS (median 12.0 months vs 32.9 months, P < 0.0001) (Fig. 1b) compared to patients with < 40 copies/ml. A higher allele frequency of EGFR-activating mutations was associated with shorter PFS (HR 1.24, 95% CI 1.12–1.36, P < 0.0001) and OS (HR 1.29, 95% CI 1.15–1.44, P < 0.0001) (Table 2). The allele frequency of EGFR-activating mutations remained independently associated with PFS (HR 1.26, 95% CI 1.15–1.39, P < 0.0001) and OS after adjusting for known clinicopathological risk factors (HR 1.32, 95% CI 1.18–1.47, P < 0.0001) (Table 2).
Table 2.
Univariate and multivariate Cox proportional hazards models
| Progression-free survival, HR (95% CI); P value | Overall survival, HR (95% CI); P value | |||
|---|---|---|---|---|
| Univariate | Multivariate | Univariate | Multivariate | |
| EGFR-activating mutation | ||||
| Age | 1.00 (0.98–1.02); 0.73 | 1.01 (0.98–1.03); 0.60 | 1.00 (0.98–1.02); 0.99 | 1.02 (0.99–1.04); 0.23 |
| Gender | 1.00 (0.63–1.58); 0.99 | 1.35 (0.84–2.19); 0.22 | 0.91 (0.55–1.50); 0.70 | 1.30 (0.77–2.22); 0.33 |
| Metastases | 2.06 (1.24–3.41); 0.005 | 2.07 (1.23–3.47); 0.006 | 1.96 (1.11–3.45); 0.02 | 2.03 (1.13–3.65); 0.02 |
| EGFR tissue genotype | 1.39 (1.00–1.93); 0.048 | 1.64 (1.16–2.33); 0.005 | 1.36 (0.94–1.95); 0.10 | 1.57 (1.07–2.30); 0.02 |
| First-line EGFR TKI therapy | 1.06 (0.89–1.26); 0.52 | 1.13 (0.95–1.36); 0.17 | 1.16 (0.96–1.42); 0.13 | 1.27 (1.04–1.56); 0.02 |
| Activating mutation lg10 | 1.24 (1.12–1.36); < 0.0001 | 1.26 (1.15–1.39); < 0.0001 | 1.29 (1.15–1.44); < 0.0001 | 1.32 (1.18–1.47); < 0.0001 |
| EGFR T790M | ||||
| Age | 1.00 (0.98–1.02); 0.73 | 1.00 (0.98–1.02); 0.92 | 1.00 (0.98–1.02); 0.99 | 1.01 (0.99–1.03); 0.46 |
| Gender | 1.00 (0.63–1.58); 0.99 | 1.13 (0.70–1.82); 0.63 | 0.91 (0.55–1.50); 0.70 | 1.03 (0.61–1.74); 0.92 |
| Metastases | 2.06 (1.24–3.41); 0.005 | 2.12 (1.27–3.56); 0.004 | 1.96 (1.11–3.45); 0.02 | 2.03 (1.14–3.63); 0.02 |
| EGFR tissue genotype | 1.39 (1.00–1.93); 0.048 | 1.56 (1.10–2.20); 0.01 | 1.36 (0.94–1.95); 0.10 | 1.54 (1.05–2.26); 0.03 |
| First-line EGFR TKI therapy | 1.06 (0.89–1.26); 0.52 | 1.06 (0.88–1.28); 0.54 | 1.16 (0.96–1.42); 0.13 | 1.20 (0.97–1.48); 0.09 |
| T790M lg10 | 1.19 (0.99–1.42); 0.06 | 1.19 (0.99–1.42); 0.05 | 1.25 (1.02–1.53); 0.03 | 1.25 (1.02–1.53); 0.03 |
| EGFR T790M/activating mutation | ||||
| Age | 1.00 (0.98–1.02); 0.73 | 1.01 (0.98–1.03); 0.62 | 1.00 (0.98–1.02); 0.99 | 1.02 (0.99–1.05); 0.24 |
| Gender | 1.00 (0.63–1.58); 0.99 | 1.21 (0.73–2.02); 0.46 | 0.91 (0.55–1.50); 0.70 | 1.23 (0.71–2.15); 0.47 |
| Metastases | 2.06 (1.24–3.41); 0.005 | 2.03 (1.32–3.64); 0.02 | 1.96 (1.11–3.45); 0.02 | 2.00 (1.04–3.83); 0.04 |
| EGFR tissue genotype | 1.39 (1.00–1.93); 0.048 | 1.93 (1.31–2.84); 0.001 | 1.36 (0.94–1.95); 0.10 | 1.87 (1.22–2.86); 0.004 |
| First-line EGFR TKI therapy | 1.06 (0.89–1.26); 0.52 | 1.24 (1.02–1.51); 0.03 | 1.16 (0.96–1.42); 0.13 | 1.33 (1.07–1.65); 0.01 |
| EGFR T790M/activating mutation | 0.73 (0.52–1.04); 0.08 | 0.72 (0.51–1.02); 0.07 | 0.83 (0.60–1.15); 0.26 | 0.83 (0.60–1.15); 0.25 |
Variables were coded as described in “Patients and Methods”
CI confidence interval, EGFR epidermal growth factor receptor, HR hazard ratio, TKI tyrosine kinase inhibitor
Fig. 1.
Kaplan–Meier curves for progression-free survival (a, c) and overall survival (b, d) according to the allele frequency of EGFR-activating mutations or the T790M mutation, and progression-free survival (e) and overall survival (f) according to the EGFR T790M/activating mutation ratio in plasma circulating tumor DNA before starting second-line osimertinib therapy. EGFR epidermal growth factor receptor
Patients with ≥ 10 copies/ml of the T790M mutation had a similar median PFS (9.7 months vs 12.4 months, P = 0.27) (Fig. 1c) and showed only a trend towards a shorter median OS (15.3 months vs 22.6 months, P = 0.09) (Fig. 1d) compared to patients with < 10 copies/ml. The allele frequency of T790M in plasma ctDNA before osimertinib initiation was associated with poor PFS (HR 1.19, 95% CI 0.99–1.42, P = 0.06) and OS (HR 1.25, 95% CI 1.02–1.53, P = 0.03). Multivariate analysis showed that a higher T790M allele frequency was associated with a trend towards a shorter PFS (adjusted HR 1.19, 95% CI 0.99–1.42, P = 0.05) and a significantly shorter OS (adjusted HR 1.25, 95% CI 1.02–1.53, P = 0.03) of the patients (Table 2).
Patients with an EGFR T790M/activating mutation ratio of ≤ 0.22 had a shorter PFS (median 4.2 months vs 10.6 months, P = 0.04) (Fig. 1e) and showed a trend towards a shorter OS (median 10.4 months vs 16.6 months, P = 0.11) (Fig. 1f) than patients with a value of > 0.22. In univariate Cox regression analyses, EGFR T790M/activating mutation ratio as a continuous variable did not correlate with PFS (HR 0.73, 95% CI 0.52–1.04, P = 0.08) or OS (HR 0.83, 95% CI 0.60–1.15, P = 0.26) (Table 2). In the multivariate analyses, the EGFR T790M/activating mutation ratio did not provide any independent prognostic information in addition to the recognized prognostic factors (PFS: adjusted HR 0.72, 95% CI 0.51–1.02, P = 0.07; OS: adjusted HR 0.83, 95% CI 0.60–1.15, P = 0.25) (Table 2).
Discussion
The results of our present study suggest that quantifying the allele frequency of EGFR mutations in plasma ctDNA using ddPCR prior to second-line therapy with osimertinib in patients with advanced EGFR-mutated NSCLC is clinically relevant. A higher allele frequency of EGFR-activating mutations and the T790M mutation in plasma ctDNA is a poor prognostic factor.
Similar findings have been reported by other groups [5, 6]. Remon et al. investigated the correlation between RECIST radiological response to osimertinib and the allele frequency of EGFR-activating mutations, T790M, and the ratio of the allele frequency of EGFR-activating mutation and T790M [5]. Although the level of statistical significance was not reached in this study, a trend towards a larger decrease of target-lesion size was observed for lower allele frequencies of EGFR-activating mutation and T790M. The lack of statistical significance in this study may be due to the low number of patients. However, we were able to confirm this result in our study on a much larger patient population. We showed that patients who responded to osimertinib had a significantly lower allele frequency of the EGFR-activating mutations than patients who did not respond. In the case of the T790M mutation, no correlation with response to osimertinib was observed. Del Re et al. also reported that the allele frequency of EGFR-activating mutations and the ratio of the allele frequency of EGFR-activating mutation and T790M were associated with response to osimertinib and PFS of patients [6]. Consistent with our results, a lower allele frequency of the EGFR-activating mutations was seen in patients who responded to osimertinib, while the T790M allele frequency was not associated with the response. Moreover, we confirmed their observation that a higher allele frequency of EGFR-activating mutations in plasma ctDNA is associated with shorter PFS.
In addition to the allele frequency of EGFR mutations, other EGFR-dependent and EGFR-independent mechanisms may influence the response to osimertinib. For example, somatic copy-number alterations (SCNAs) can also be relevant to the effectiveness of the therapy [11, 12]. Therefore, the factors that affect the response to osimertinib are complex and a comprehensive plasma analysis of patients who progressed under EGFR TKI therapy could provide insights into the various mechanisms of intrinsic and acquired drug resistance and identify patients who benefit most from osimertinib. Due to the fact that a liquid biopsy is less burdensome than a tissue biopsy and that in many advanced NSCLC patients tissue sampling is clinically not feasible, plasma may be the preferred source for molecular profiling in the future. Blood samples are easily obtainable and can be taken repeatedly even in short time intervals. In addition, genetic heterogeneity of the progressing tumor may lead to an incomplete picture of the tumor genome if only single tissue biopsies are obtained. Furthermore, blood-based analytic approaches may allow real-time monitoring of the total tumor burden and the detection of upcoming mutations that arise during clinical treatment through serial blood sampling and analysis.
Patients with a higher allele frequency of EGFR-activating mutations in plasma ctDNA prior to osimertinib therapy may require a change in treatment because of their poor outcome. They may benefit from the addition of chemotherapy to osimertinib or from the addition of chemo-immunotherapy. The combination of chemotherapy with an EGFR TKI was investigated in phase III trials in the first-line setting, in which the combination of gefitinib with chemotherapy resulted in longer PFS and OS compared to gefitinib alone [13, 14]. The treatment strategy of adding chemo-immunotherapy to osimertinib is supported by an exploratory analysis of the IMpower150 trial, which indicated longer OS for the addition of atezolizumab to chemotherapy plus bevacizumab compared to chemotherapy plus bevacizumab in patients with advanced non-squamous EGFR-mutated NSCLC [15]. Therefore, these treatment strategies warrant further investigation within clinical trials among patients with a higher allele frequency of EGFR-activating mutations in plasma ctDNA prior to osimertinib therapy.
In summary, our findings suggest that plasma analyses before the start of second-line osimertinib therapy are clinically relevant. Further studies on the clinical utility of liquid biopsy are needed.
Declarations
Funding
Open access funding provided by Medical University of Vienna. The study was funded by AstraZeneca Austria.
Conflict of Interest
Anna Buder has received honoraria from AstraZeneca. Maximilian J. Hochmair has received honoraria from AstraZeneca, Bristol-Myers Squibb, Boehringer Ingelheim, Merck Sharp & Dohme, Pfizer, and Roche and had consulting or advisory roles with Boehringer Ingelheim, Merck Sharp & Dohme, Novartis, and Roche. Martin Filipits has received honoraria for advisory boards from Astra Zeneca, Bayer, Biomedica, Biorad, Boehringer Ingelheim, Myriad Genetics Inc., Pfizer, and Roche.
Ethics Approval
Sample collection and analysis was performed in accordance with the local ethics committee (EK No. 1132/2016). All patients gave their written informed consent for providing blood specimens for plasma genotyping.
Consent to Participate
All authors agreed to participate.
Consent for Publication
All authors have approved the final version submitted.
Availability of Data and Material
Not applicable.
Code availability
Not applicable.
Authors’ contributions
All authors made substantial contributions to the conception and design of the study, acquisition of data, analysis and interpretation of data, drafting the article, and revising it critically for important intellectual content.
References
- 1.Planchard D, Popat S, Kerr K, Novello S, Smit EF, Faivre-Finn C et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2018; 29: iv192–iv237. [DOI] [PubMed]
- 2.Hochmair MJ, Morabito A, Hao D, Yang CT, Soo RA, Yang JC, et al. Sequential afatinib and osimertinib in patients with EGFR mutation-positive non-small-cell lung cancer: updated analysis of the observational GioTag study. Fut Oncol. 2019;15:2905–2914. doi: 10.2217/fon-2019-0346. [DOI] [PubMed] [Google Scholar]
- 3.Piotrowska Z, Niederst MJ, Karlovich CA, Wakelee HA, Neal JW, Mino-Kenudson M, et al. Heterogeneity underlies the emergence of EGFRT790 wild-type clones following treatment of T790M-positive cancers with a third-generation EGFR inhibitor. Cancer Discov. 2015;5:713–722. doi: 10.1158/2159-8290.CD-15-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oxnard GR, Thress KS, Alden RS, Lawrance R, Paweletz CP, Cantarini M, et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J Clin Oncol. 2016;34:3375–3382. doi: 10.1200/JCO.2016.66.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Remon J, Caramella C, Jovelet C, Lacroix L, Lawson A, Smalley S, et al. Osimertinib benefit in EGFR-mutant NSCLC patients with T790M-mutation detected by circulating tumour DNA. Ann Oncol. 2017;28:784–790. doi: 10.1093/annonc/mdx017. [DOI] [PubMed] [Google Scholar]
- 6.Del Re M, Bordi P, Rofi E, Restante G, Valleggi S, Minari R, et al. The amount of activating EGFR mutations in circulating cell-free DNA is a marker to monitor osimertinib response. Br J Cancer. 2018;119:1252–1258. doi: 10.1038/s41416-018-0238-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buder A, Hochmair MJ, Schwab S, Bundalo T, Schenk P, Errhalt P, et al. Cell-free plasma DNA-guided treatment with osimertinib in patients with advanced EGFR-mutated NSCLC. J Thorac Oncol. 2018;13:821–830. doi: 10.1016/j.jtho.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 8.Buder A, Setinek U, Hochmair MJ, Schwab S, Kirchbacher K, Keck A, et al. EGFR mutations in cell-free plasma DNA from patients with advanced lung adenocarcinoma: improved detection by droplet digital PCR. Target Oncol. 2019;14:197–203. doi: 10.1007/s11523-019-00623-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hochmair MJ, Buder A, Schwab S, Burghuber OC, Prosch H, Hilbe W, et al. Liquid-biopsy-based identification of EGFR T790M mutation-mediated resistance to afatinib treatment in patients with advanced EGFR mutation-positive NSCLC, and subsequent response to osimertinib. Target Oncol. 2019;14:75–83. doi: 10.1007/s11523-018-0612-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buder A, Hochmair MJ, Setinek U, Pirker R, Filipits M. EGFR mutation tracking predicts survival in advanced EGFR-mutated non-small cell lung cancer patients treated with osimertinib. Transl Lung Cancer Res. 2020;9:239–245. doi: 10.21037/tlcr.2020.03.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knebel FH, Bettoni F, Shimada AK, Cruz M, Alessi JV, Negrao MV, et al. Sequential liquid biopsies reveal dynamic alterations of EGFR driver mutations and indicate EGFR amplification as a new mechanism of resistance to osimertinib in NSCLC. Lung Cancer. 2017;108:238–241. doi: 10.1016/j.lungcan.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Le X, Puri S, Negrao MV, Nilsson MB, Robichaux J, Boyle T, et al. Landscape of EGFR-dependent and -independent resistance mechanisms to osimertinib and continuation therapy beyond progression in EGFR-mutant NSCLC. Clin Cancer Res. 2018;24:6195–6203. doi: 10.1158/1078-0432.CCR-18-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosomi Y, Morita S, Sugawara S, Kato T, Fukuhara T, Gemma A, et al. Gefitinib alone versus gefitinib plus chemotherapy for non-small-cell lung cancer with mutated epidermal growth factor receptor: NEJ009 Study. J Clin Oncol. 2020;38:115–123. doi: 10.1200/JCO.19.01488. [DOI] [PubMed] [Google Scholar]
- 14.Noronha V, Patil VM, Joshi A, Menon N, Chougule A, Mahajan A, et al. Gefitinib versus gefitinib plus pemetrexed and carboplatin chemotherapy in EGFR-mutated lung cancer. J Clin Oncol. 2020;38:124–136. doi: 10.1200/JCO.19.01154. [DOI] [PubMed] [Google Scholar]
- 15.Reck M, Mok TSK, Nishio M, Jotte RM, Cappuzzo F, Orlandi F, et al. Atezolizumab plus bevacizumab and chemotherapy in non-small-cell lung cancer (IMpower150): key subgroup analyses of patients with EGFR mutations or baseline liver metastases in a randomised, open-label phase 3 trial. Lancet Respir Med. 2019;7:387–401. doi: 10.1016/S2213-2600(19)30084-0. [DOI] [PubMed] [Google Scholar]