Abstract
There has been considerable research showing populations of neurons encoding for different aspects of space in the brain. Recently, several studies using two-photon calcium imaging and virtual navigation have identified “spatially” modulated neurons in the posterior cortex. We enquire here whether the presence of such spatial representations may be a cortex-wide phenomenon and, if so, whether these representations can be organized in the absence of the hippocampus. To this end, we imaged the dorsal cortex of mice running on a treadmill populated with tactile cues. A high percentage (40–80%) of the detected neurons exhibited sparse, spatially localized activity, with activity fields uniformly localized over the track. The development of this location specificity was impaired by hippocampal damage. Thus, there is a substantial population of neurons distributed widely over the cortex that collectively form a continuous representation of the explored environment, and hippocampal outflow is necessary to organize this phenomenon.
SIGNIFICANCE STATEMENT Increasing evidence points to the role of the neocortex in encoding spatial information. Whether this feature is linked to hippocampal functions is largely unknown. Here, we systematically surveyed multiple regions in the dorsal cortex of the same animal for the presence of signals encoding for spatial position. We described populations of cortical neurons expressing sequential patterns of activity localized in space in primary, secondary, and associational areas. Furthermore, we showed that the formation of these spatial representations was impacted by hippocampal lesion. Our results indicate that hippocampal inputs are necessary to maintain a precise cortical representation of space.
Keywords: calcium imaging, neocortex, place cells, spatial navigation, two-photon
Introduction
The neural mechanisms underlying the emergence of place coding continue to elude complete understanding. In the hippocampal and parahippocampal regions reside large populations of neurons displaying specialized spatial firing patterns (Moser et al., 2015). Of particular interest are hippocampal “place cells,” which share the same anatomic substrate as episodic memory functions (O'Keefe and Dostrovsky, 1971). Hippocampal place cells fire at one or a few specific locations in an environment and collectively are believed to form the neural basis of the cognitive representation of space (O'Keefe and Conway, 1978). It is now recognized that once “allocated” to a place, mainly through self-motion-related processes (McNaughton et al., 2006), these neurons become modulated by other attributes of the current state of the brain (e.g., sensory input, motor actions, plans, affective state), a phenomenon dubbed “rate remapping” (Leutgeb et al., 2005; Sheintuch et al., 2020).
The response characteristics of place cells are indicative of complex integrations of information spanning multiple sensory modalities and levels of cortical processing. Studies have shown that place cell activity is modulated by a broad range of environmental features such as visual (O'Keefe and Conway, 1978; Muller and Kubie, 1987; O'Keefe and Speakman, 1987; Quirk et al., 1990), olfactory (Save et al., 2000), auditory (Moita et al., 2003), and tactile information (Gener et al., 2013). Hippocampal place cells can be driven by spatial orienting cues in a novel environment and still fire in the correct locations in the dark or in the absence of cues once the environment becomes familiar, supporting the notion that cortical–hippocampal interactions are involved in the formation of associations among places, movements, and cues established during the learning of an environment (O'Keefe and Speakman, 1987; McNaughton et al., 1989).
Although place cells have predominantly been described in the hippocampal formation, a number of recent studies have reported neurons with similar firing patterns as place cells in multiple neocortical regions (Nitz, 2009; Harvey et al., 2012; Fiser et al., 2016; Mao et al., 2017; Pakan et al., 2018; Saleem et al., 2018; Minderer et al., 2019). The parietal cortex presents cells with spatially modulated activity patterns important to solving navigational tasks and also presents population activity with specific choice trajectories (Nitz, 2006; Harvey et al., 2012; Wilber et al., 2014). Visual responses have been shown to be modulated by spatial location and controlled by navigational signals (Saleem et al., 2018). Additionally, retrosplenial cortex neurons have been shown to encode spatial activity sequences resembling the activity of hippocampal CA1 place cells, and the formation of these spatial sequences relies on an intact hippocampus (Mao et al., 2017, 2018; Chang et al., 2020).
Given the potential interplay between the hippocampus and neocortex at large, we surveyed and compared multiple dorsal neocortical regions (including primary, secondary, and associational areas) to investigate the extent to which other neocortical regions also express continuous spatial representations. With targeted bilateral excitotoxic lesions to the dorsal hippocampus, we examined the degree to which these representations relied on an intact hippocampus.
Materials and Methods
Experimental design
Thy1-GCaMP mice were implanted with a 5 mm cranial window above the dorsal cortex. Animals were head restrained and trained to run for reward over a 150-cm-long treadmill belt on which were mounted several tactile cues. Two-photon calcium imaging was conducted across different neocortical regions in animals with and without excitotoxic hippocampal lesions. We tracked the spatial activity across different cortical areas and evaluated how the spatial representation found in the neocortex was overall influenced by the hippocampus.
Animals
A total of eight adult transgenic Thy1-GCaMP6s mice (weight, 22–30 g; age, 2–4 months), specifically expressing the calcium indicator in excitatory neurons, were used for this study. Mice were housed in standard rodent cages, maintained at 24°C room temperature and under a 12 h light/dark cycle (lights on at 7:30 A.M.), with free access to food and water before the beginning of training. All experiments were performed between 7:30 A.M. and 7:30 P.M. Procedures were conducted in accordance with the guidelines established by the Canadian Council on Animal Care, and using protocols approved by the Animal Welfare Committee of the University of Lethbridge.
Surgery
Before surgery, mice were injected with 0.5 ml of a 5% dextrose and 0.9% saline solution mixed with atropine (3 µg/ml, s.c.), buprenorphine (0.05 mg/kg, s.c.), and dexamethasone (0.2 mg/kg, i.m.). Animals were then anesthetized with isoflurane (1–1.5%, O2; 0.5–1 L/min) and placed in a stereotactic frame with body temperature maintained at 37.0 ± 0.5°C through a heating pad with a closed-loop control system. Once the skull was exposed, a custom-made titanium headplate was fixed to the skull using adhesive cement (C&B Metabond, Parkell) and dental acrylic. Bregma was identified and marked in the cement. A 5-mm-diameter craniotomy was made following bregma-referenced coordinates: 1.5 mm anterior to −3.5 mm posterior; ±2.5 mm medial–lateral (Fig. 1A). Three layers of coverslips affixed with optical adhesive (NOA71, Norland) were implanted over the craniotomy, attached to the skull with Vetbond (3M). A rubber ring was fixed over the headplate to form a well to hold water between the imaging region and the immersion objective. Surgeries were concluded with injections of meloxicam (Metacam; 1.0 mg/kg, s.c.) and enrofloxacin (Baytril; 10 mg/kg, s.c.). The same drugs were administered once daily for 3 d. Mice were then allowed to recover for 1 week. For bilateral hippocampal excitotoxic lesions, NMDA (15 mg/ml, in 1× PBS) was injected before implanting the coverslip. Injections were made with a micropipette, with tip diameter between 20 and 30 µm, loaded on a nanoliter injector (Nanoject II, Drummond Scientific), at a speed of 9.2 nl/pulse over eight pulses (in total: 9.2 nl/pulse × 8 pulses = 73.6 nl; interpulse interval, 15 s) at each site. In total, the following four injection sites were used (two in each hemisphere): anteroposterior (AP), −2.3 mm; mediolateral (ML), 1.5 mm; dorsoventral (DV), 1.8 mm; and, AP, −3.2 mm; ML, 2.5 mm; DV, 2.0 mm. Diazepam (5 mg/kg, i.p.) and phenobarbital (30 mg/kg, s.c.) were administered right after surgery to suppress potential seizures.
Figure 1.

Cranial window implant, treadmill track, and two-photon Ca2+ imaging. A, Cranial window implant position (circle) with all the regions of neocortex imaged (black boxes; adapted from Kirkcaldie, 2012). B, Top, Basic setup for recording on treadmill track using two-photon Ca2+ imaging. Bottom, Diagram of belt lined with the tactile cues. C, Sequential activity of neocortical (RSC) spatially selective cells during running on the tactile belt. Neurons were ordered by their peak response positions. Black trace shows the position, and blue drops indicate reward. D, Cranial window of one animal (top), field of view of one imaging session (middle), and identified neurons (ROIs) from one session (bottom). E, Plot of the normalized, raw calcium ΔF/F time courses (top) and deconvolved trace smoothed using a σ = 8 s. Gaussian kernel (bottom) of 20 simultaneously imaged neurons in M2. Animal position and reward are shown below. F, Example of normalized activity of the six spatially selective cells as a function of location for multiple laps (left, raw trace; right, deconvolved). Pos., Position.
Treadmill apparatus
Once recovered, mice were water restricted throughout training and imaging sessions. During the water restriction period, mice were given ad libitum access to water for a maximum of 30 min/d in their home cages after training/imaging sessions, and their weight was monitored to be maintained to at least 85% of their baseline weight (average weight 3 d before water restriction). Animals were gradually accustomed to run on the treadmill while head fixed. The treadmill consisted of a 150 cm Velcro (Country Brook) belt with tactile cues (made of hot-glue stripes, reflective tape, and Velcro) inserted at several locations (Fig. 1B). Polyamide wheels (diameter, 10 cm) were attached to both ends of the treadmill to guide the belt, and an optical encoder (Avago Tech) attached to the wheel shaft was used to monitor belt movement. After each lap, a drop of sucrose water (10% concentration; ∼2.5 µl volume) was delivered to the animal. The reward was dispensed by a solenoid pinch valve (Bio-Chem Laboratories) whenever a photoelectric sensor (Omron) was triggered by the reflective tape attached to the opposite side of the belt. A custom-designed circuit with a microcontroller (Arduino UNO, Farnell) was used to control the reward delivery and to monitor the encoder.
Two-photon imaging
After training, imaging experiments were conducted using a Bergamo II Multiphoton Microscope (THORLABS). Samples were excited by a Ti:Sapphire pulsed laser (Coherent) tuned to a wavelength of 920 nm (∼80 mW output power measured at the sample). Focusing of the beam and light collection were achieved by a 16× water-immersion objective lens (numerical aperture, 0.8; Nikon). Scanning was conducted by Galvo-Resonant X-Y mirrors. Emitted fluorescent lights were detected by a GaAsP photomultiplier tube (Hamamatsu). Samples were acquired from a 835 × 835 µm field of view (FOV) at depths between 130 and 190 µm (layers II/III). Images were digitized at a sampling rate of 19 Hz, and at a resolution of 800 × 800 pixels. Photosensor and encoder signals from the treadmill were acquired and synchronized with imaging using Clampex software (Molecular Devices).
Imaging data from all animals were acquired from the right hemisphere. The hemisphere was divided into 16 regions partitioned by grid (Fig. 1A). Each region spanned the same dimensions as the imaging FOV (Fig. 1D). In total, 16 recording sessions were conducted per animal, one for each region, and each recording was 5–10 min in duration. The imaging experiments were performed over the course of 1 week, after at least 1 month of familiarization on the track/belt. The cranial window allowed us to image five different neocortical regions. For data analysis, we considered grids 1–3 as restrosplenial cortex (RSC); grids 4–6 as secondary motor cortex (M2); grids 10 and 11 as primary motor cortex (M1); grids 8 and 13 as posterior parietal cortex (PPC); and grids 14 and 15 as primary somatosensory cortex (SS1; Fig. 1A).
Data analysis
Image preprocessing was conducted automatically using the Suite-2P software suite (Pachitariu et al., 2017), as previously described (Mao et al., 2017, 2018). Regions of interest (ROIs) detected as candidate neurons were visually inspected. Neuropil contamination was estimated from the surround of ROIs and was subtracted (Bonin et al., 2011). To infer relative underlying firing rates for each ROI, the ΔF/F time courses were deconvolved using constrained non-negative matrix factorization (Pnevmatikakis et al., 2016). All analyses were conducted using MATLAB (R2017a, MathWorks). All subsequent analyses were conducted on the deconvolved time courses using data normalized between 0 and 1, unless stated otherwise (Fig. 1E).
Spatially selective cells were identified based on the following criteria: the length of the belt was divided into 50 equally spaced bins (3 cm each). The activity of each neuron was mapped to corresponding position bins for each lap. This position-mapped activity was normalized by the time spent in each bin (occupancy) and then filtered with a Gaussian window (4.5 cm SD). Spatial information (SI) was calculated for each cell using the following formula (Skaggs et al., 1992):
| (1) |
where N is the total number of bins, π is the occupancy probability in the ith bin, fi is the activity in the ith bin, and f is the overall activity (averaged fi across all bins). We generated a shuffled distribution of spatial information by circularly shifting the time courses 1000 times. To be classified as significantly spatially tuned, the original SI was required to be higher than the 95th percentile of the shuffled distribution, the mean activity within a place field (PF) was required to be 2.5 times higher than the mean activity outside of field, and the peak activity was required to occur inside the place field in at least a third of the laps (Fig. 1F). Note that SI is a conservative measure. For example, many hippocampal cells have several clear place fields and hence show less spatial information overall. The spatial tuning characteristics of place fields were derived by obtaining a continuous wavelet transform, W, over the spatial response curve fi of each neuron with a Mexican Hat mother wavelet, ψ, as follows:
| (2) |
where
| (3) |
The local maxima in the resulting spectrum indicate the location (given by τ) and the width (given by σ) of place fields. Peaks with values lower than three median absolute deviations from the median of the wavelet coefficients at the lowest scale (σ = 1) were rejected. Peaks that fell within the receptive field of a local maximum at a higher scale σ were rejected. The mean activity within a place field was required to be 2.5 times higher than the mean activity outside of the field. The peak activity was required to occur inside the place field in at least a third of the laps (Fig. 1F). Place fields must be wider than 5% of the length of the environment, but narrower than 80%. Sparsity, a measure of how dispersed the firing profile of a neuron is in relation to the environment, was calculated as follows:
| (4) |
where fi is the mean activity in the ith spatial bin and π is the probability of occupancy in the ith spatial bin over a total of N = 50 spatial bins (Jung et al., 1994). Sparsity ranges between 0 and 1, where smaller values indicate finer spatial tuning profiles.
Population activity was decoded using an independent Bayesian decoder (Zhang et al., 1998; Mao et al., 2018). In brief, for every time bin, we estimated the probability of the animal being at a position x given the population response of all imaged neurons as follows:
| (5) |
where fi(10) is the mean deconvolved fluorescence trace over position x and ni is the time course vector of the ith neuron within a time bin of length τ, N is the number of neurons, and C is a normalizing constant that makes the probability distribution across all positions sum to 1. Running periods of odd trials were used for training the model, and even trials were used to evaluate the error of decoding. A decoded position was defined as the position with the highest probability for any given time bin, and the absolute value of the difference between true position and decoded position was defined as the Bayesian decoding error. The direct implementation of a Bayesian decoder based on a Poisson likelihood distribution, for use with deconvolved calcium traces, poses a limitation. Indeed, we cannot assume that the underlying distribution of deconvolved firing rates is drawn from Poisson point processes. However, considering that there is a marked linear relationship between calcium-inferred firing rates and true firing rates (Pnevmatikakis et al., 2016), the estimator used in the present study is still valid as a close approximation. We expect improved and standardized methodologies to become available in future studies with the maturation of calcium imaging techniques.
The anatomic density distribution map was obtained by discretizing the entire window into 120 × 60 grids of 20 × 20 pixels (435.76 µm2) and then calculating the number of cells in each grid. The density distribution map of spatially selective cells was obtained by dividing the number of spatially tuned cells by the total number of cells detected in each grid. Data were smoothed using a σ = 2 pixel Gaussian kernel. Maps were aligned by locating bregma (marked outside the window during surgery) in the upper part of grid 4 (see Fig. 4A).
Figure 4.

Distribution of cells. A, Density distribution map of all the cells detected across the window for the four control animals (left) and the four animals with bilateral hippocampal lesions (right). B, Density distribution map of all cells detected of each group on the same map. Color bar shows the number of cells inside an area of 20 × 20 pixels (435.76 µm2). Colored boxes refer to the different neocortical regions imaged (RSC, red; M2, brown; M1, black; PPC, white; SS1, magenta). C, Box plot of all the cells detected separated per region. Line, median; box, 25th and 75th percentile; dots, values for an individual animal; whiskers, minimum and maximum values. No significant differences were found in the total number of cells detected in any neocortical region across groups. For exact p values, see Table 1.
Histology
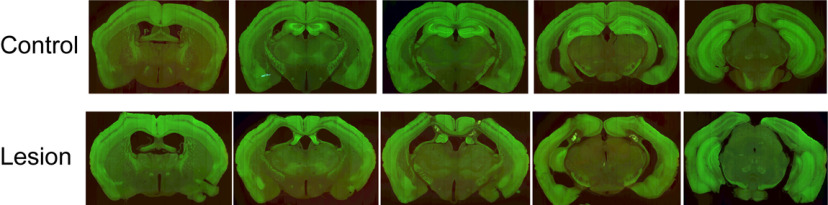
Mice were perfused with PBS, and brains were postfixed with 4% paraformaldehyde for 24 h after the end of the experiments. Brains were then cryoprotected in 30% sucrose solution (with 0.02% sodium azide) and sectioned in the coronal plane at 40 µm using a blockface imaging system composed of a sliding microtome and an Olympus MVX10 microscope. We used a NanoZoomer scanning microscope (Hamamatsu Photonics) to acquire images of sections. Images of coronal sections from −0.8 to −3.8 mm AP (Paxinos and Franklin, 2003) were used to evaluate the extent of hippocampal lesions.
Statistical analysis
All statistical tests performed in this work were conducted using MATLAB functions (catalog #R2017a, MathWorks). Further details of all statistical tests implemented in this study are provided in Table 1.
Table 1.
Summary of all statistics
| Figure | Variable | n | Test | Results |
|---|---|---|---|---|
| Figure 3C, right | Fraction of spatially selective cells per group | Value per animal; n = 4 each group | t test | All values are mean ± SEM Control, 0.60 ± 0.02; lesion, 0.26 ± 0.03; p < 0.001 |
| Figure 3C, left | Fraction of spatially selective cells per region. | Value per session recorded in each region RSC: control = 11; lesion = 9; PPC: control = 7; lesion = 5; M2: control = 10;lesion = 8; M1: control = 8; lesion = 8; SS1: control = 8; lesion = 8; |
Wilcoxon rank-sum test | All values are median (interquartile range) RSC: control, 0.52 (0.63–0.47); lesion, 0.16 (0.29–0.14); p = 0.0008 PPC: control, 0.42 (0.45–0.36); lesion, 0.23 (0.26–0.19); p = 0.048 M2: control, 0.73 (0.83–0.70); lesion, 0.25 (0.48–0.05); p = 0.0003 M1: control, 0.72 (0.76–0.61); lesion, 0.36 (0.44–0.25); p = 0.0003 SS1: control, 0.56 (0.64–0.47); lesion, 0.27 (0.37–0.18); p = 0.0002 |
| Figure 3D, left | Running speed | Value per animal | t test | All values are mean ± SEM Control, 18.4 ± 2.3 cm/s; lesion, 14.5 ± 2.1 cm/s; p = 0.249 |
| Figure 3D, right | Number of laps | Value per session recorded in all region. control = 44; lesion = 38 | Wilcoxon rank-sum test | All values are median (interquartile range) Control, 20.5 (17.0–25.0); lesion 18.0 (14.0–24.0); p = 0.2279 |
| Fig. 4C | Cells detected per region per group | Value per animal; n = 4 each group | Wilcoxon rank-sum test | All values are median (interquartile range): RSC: control, 313.0 (407.0–243.5); lesion, 403.5 (448.0–389.5); p = 0.3429 PPC: control, 125.5 (163.0–59.5); lesion, 53.0 (80.0–30.0); p = 0.2000 M2: control, 254.0 (280.5–179.5); lesion, 134.5 (252.5–94.0); p = 0.4857 M1: control, 208.5 (278.5–173.0); lesion, 321.5 (467.0–211.0); p = 0.3429 SS1: control, 154.5 (174.0–140.5); lesion, 203.5 (262.5–156.0); p = 0.3429 |
| Figure 5B, right | Place field width, spatial information, and sparsity per group | Value per animal n = 4 each group |
t test | All values are mean ± SEM: Place field width: control, 43.0 ± 2.3; lesion, 42.38 ± 2.75; p = 0.876 Spatial information: control, 1.69 ± 0.02; lesion, 1.176 ± 0.11; p = 0.004 Sparsity: control, 0.239 ± 0.005; lesion, 0.349 ± 0.025; p = 0.005 |
| Figure 5B, left | Place field width, spatial information, and sparsity of the spatially selective cells per region | Value per session recorded in each region. RSC: control = 11; lesion = 8; PPC: control = 7; lesion = 5; M2: control = 10; lesion = 7; M1: control = 8; lesion = 7; SS1: control = 8; lesion = 8; |
Wilcoxon rank-sum test | All values are median (interquartile range) Place field width: RSC: control, 48.77 (57.47–44.18); lesion, 43.69 (48.11–42.15); p = 0.07 PPC: control, 46.94 (50.40–43.58); lesion, 44.4 (63.17–40.62); p = 1.00 M2: control, 41.62 (48.43–38.67); lesion, 40.77 (53.87–34.23); p = 0.81 M1: control, 38.09 (41.73–36.33); lesion, 38.88 (44.15–34.90); p = 0.95 SS1: control, 36.64 (39.24–34.40); lesion, 40.53 (45.91–38.31); p = 0.13 Spatial information: RSC: control, 1.34 (1.53–1.25); lesion, 0.80 (1.10–0.73); p = 0.025 PPC: control, 1.49 (1.53–1.36); lesion, 0.90 (1.10–0.82); p = 0.025 M2: control, 1.79 (1.94–1.59); lesion, 1.07 (1.32–0.88); p = 0.0001 M1: control, 1.84, (1.97–1.73); lesion, 1.49 (1.66–1.24); p = 0.0093 SS1: control, 1.76 (1.93–1.71); lesion, 1.15 (1.40–1.05); p = 0.0006 Sparsity: RSC: control, 0.29 (0.31–0.25); lesion, 0.43 (0.47–0.36); p = 0.0025 PPC: control, 0.26 (0.29–0.36); lesion, 0.41 (0.43–0.34); p = 0.0101 M2: control, 0.21 (0.25–0.19); lesion, 0.36 (0.42–0.30); p = 0.0001 M1: control, 0.20 (0.22–0.19); lesion, 0.27 (0.32–0.24); p = 0.0022 SS1: control, 0.22 (0.23–0.19); lesion, 0.34 (0.35–0.29); p = 0.0002 |
| Figure 6B, bottom | Peak activity | Value per animal n = 4 each group |
t test | All values mean ± SEM Control, 3.932 ± 0.407; lesion, 2.987 ± 0.276; p = 0.011 |
| Figure 8B | Decoding error as function of position | Value per animal n = 4 each group |
Two-way ANOVA | Source of variance Group: df = 1; SS = 15,457.119; MS = 15,457.119; F = 2694.449; p < 0.001 Position: df = 49; SS = 3876.430; MS = 79.111; F = 13.790; p < 0.001 Group × position: df = 49; SS = 1725.192; MS = 35.208; F = 6.137; p < 0.001 Residual: df = 300; SS = 1720.996; F = 5.737 Total: df = 399; SS = 22,779.738; F = 57.092 |
| Figure 8C | Decoding error for each position | Value per group control = 2200; lesion = 1850 |
Kolmogorov–Smirnov test | Control vs lesion, p < 0.0001 |
| Figure 8D | Decoding error per region. | Value per animal n = 4 each group |
t test | All values mean ± SEM RSC: control, 10.01 ± 1.33; lesion, 23.1 ± 2.17; p = 0.0022 PPC: control, 16.43 ± 0.64; lesion, 22.96 ± 0.63; p = 0.0004 M2: control, 6.48 ± 1.05; lesion, 19.78 ± 3.25; p = 0.0081 M1: control, 6.44 ± 0.98; lesion, 18.38 ± 2.61; p = 0.0053 SS1: control, 7.61 ± 0.93, lesion 20.19 ± 1.40; p = 0.0003 |
Results
Using two-photon Ca2+ imaging, we systematically recorded neurons from the superficial cortical surface (layers II/III) of the right hemisphere exposed under a 5 mm craniotomy in four mice in a head-fixed, treadmill assay (Fig. 1A,B). We found neurons highly tuned to the positions of animals on the belt, as illustrated in Figure 1C. Ca2+ imaging was also conducted on a second group of four mice with dorsal hippocampal lesions (bilateral) to evaluate whether the spatial representations were dependent on an intact hippocampus. The animals in the lesion group sustained extensive neuron/tissue loss in the dorsal hippocampal formation (Fig. 2). Running speed was similar across groups (Fig. 3D, left), and no significant difference was found between the number of laps performed by each group (Fig. 3D, right).
Figure 2.
Histology. Example histology of one representative mouse from each group. Note the tissue loss in the dorsal hippocampal region in the lesion group.
Figure 3.

Distribution of spatially selective cells in the neocortex and its hippocampal dependence. A, B, Density distribution map with all the spatially modulated neurons of each animal and group, showing that cells expressing spatial coding characteristics were found in multiple areas of the neocortex and that hippocampal lesions disrupted the spatial coding in those regions. Color bar shows the fraction of cells that passed the criteria for being spatially selective cells inside an area of 20 × 20 pixels (435.76 µm2); Colored boxes refer to the different neocortical regions imaged (RSC, red; M2, brown; M1, black; PPC, white; SS1, magenta). C, Fraction of neurons with spatial selectivity for each area (left: line, median; box, 25th and 75th percentiles; dots, cell fraction for an individual session recorded in each region from all animals; whiskers, minimum and maximum values; *p < 0.05) and per group (right: mean ± SEM over animals; dots, cell fraction for individual animals; n = 4 in each group; *p < 0.05). There was a significant reduction of spatially selective cells after lesioning for all regions as well as for the regions pooled. D, Left, Speed profiles as a function of position from all trials. D, Right, Number of laps performed by each group during all sessions. Line, median; box, 25th and 75th percentiles; dots, values for an individual session recorded for all animals; whiskers, minimum and maximum values; + signs, outliers. No significant difference was found between the speed and the number of laps performed by each group. For exact n and p values, see Table 1.
Spatially selective cells are widespread in the neocortex and are dependent on an intact hippocampus
We found neurons with spatial selectivity in all neocortical areas examined, as shown in Figure 3, A and B, left. However, in the lesion group, the fraction of cells expressing high spatial information content relative to position on the belt was greatly reduced (Fig. 3A,B, right). Averaging the cell fractions over all ROIs, the control animals presented a fraction of spatially selective cells that was 2.3 times higher than in the lesion group (Fig. 3C, right). By comparing this fraction per region, the median from the control group was significantly higher than that of the lesion group in all neocortical regions (Fig. 3C, left). These results show that cells with spatial selectivity were present in all neocortical regions examined, and that the emergence of those properties was disrupted by hippocampal lesions. Cell density maps for each animal are shown in Figure 4A. Among the cells detected during linear treadmill running, there was no difference in the total number of cells across groups in any neocortical region (Fig. 4B,C).
Spatially selective neurons in the hippocampal lesion group convey less spatial information and lower sparse coding characteristics
We next evaluated whether the neurons that passed the criteria for being spatially selective exhibit specific differences between groups. For cells passing the criteria, the distribution of PF widths in both groups was very similar. However, the SI and the sparsity distribution showed that the lesion group cells exhibited lower spatial information and higher sparsity index (i.e., were less selective) when compared with the control group (Fig. 5A). On average, there were no differences in the PF width, but control animals' cells had higher spatial information, and sparser coding than the lesion group (Fig. 5B, right). These results were confirmed when the spatially selective cells were evaluated separately by region (Fig. 5B, left).
Figure 5.

Spatially selective cell characteristics with and without bilateral hippocampal lesions. Data shown were for cells that passed the spatial selectivity criteria (see Materials and Methods). A, Probability distributions of place field widths, SI, and sparsity for control group (white) and lesion group (red). B, Left, Box plot of place field widths, SI, and sparsity separated by region. Line, median; box, 25th and 75th percentiles; dots, values for an individual sessions recorded in each region from all animals; whiskers, minimum and maximum values; + signs, outliers; *p < 0.05. B, Right, Bar plots of the average place field width, SI, and sparsity for neurons per group. Error bars denote SEM over animals (control, n = 4; lesion, n = 4; *p < 0.05). The average place width did not change with the hippocampal lesion. However, lesioned animals presented cells with reduced spatial information content, along with reduced sparsity. For exact n and p values, see Table 1.
Hippocampal lesion impairs the ability of neocortex to create a uniform representation of space
Next, we checked for differences between the encoding patterns of both groups. We first evaluated the position-mapped activity for each group (Fig. 6A, top) and the aligned trial-averaged profile (Fig. 6A, bottom) for all imaged neurons regardless of spatial specificity. This analysis showed that the control group cells had more spatially compact firing patterns with sparser coding. When we averaged the unnormalized position maps and the aligned activity profiles, we observed no difference in the overall activity between groups (Fig. 6B, top), but the aligned trial-averaged activity showed that the lesion group cells had significantly reduced peak activity (Fig. 6B, bottom).
Figure 6.

Cell activity during the task with and without bilateral hippocampal lesions. A, Pooled data from all mice showing activity profiles for all imaged neurons regardless of spatial specificity. Trial-averaged activity map for all cells in both groups as a function of position (position map). Top, Activity profiles were normalized and ordered by their peak response positions. Bottom, Aligned position map of all cells (circularly shifted to be aligned in the middle) sorted by their SI values. B, Grand averages for both groups of the trial-averaged activity (non-normalized) as a function of position (top) and with their peak aligned in the middle (bottom). Shaded area represents SEM over animals (control, n = 4; lesion, n = 4; *p < 0.05). The lesion group cells had less spatially compact firing patterns, with lower peak activity of the distributions. C, Cell activity of the neurons that passed the criteria for being spatially selective cells. Left, Position map obtained from odd trials, for only the spatially selective cells of each group. Right, Same as in left but for even trials and ordered by the position of the maximum response of odd trials. D, Pearson's correlation matrices of population vectors between pairs of positional bins for both groups with only the spatially selective cells. For exact n and p values, see Table 1.
Next, we evaluated only the neurons that met our criteria for spatial selectivity (see Materials and Methods). We first cross-validated the position map by dividing the trials into two sets and by using the odd-numbered trials to find the position at which each neuron fired maximally. Then we quantified the position map of the odd and even trials sorted by the peak of the odd trials and found that the preference for position was consistent, with neurons presenting sequential firing during movement covering the entire belt for both groups. However, the lesion group presented a less uniform representation over position, with more cells having peak activity by the cues and the reward site (Fig. 6C). With the correlation map of those cells, we quantified the similarity between cells tuned to distinct locations. The control group showed cells with a high correlation between nearby locations (near the diagonal) and a steep drop-off in correlation with the distance. The lesion group cells also had a high correlation near the diagonal. Still, there appears to be a tendency in the lesion group that the correlation with the distance only drops steeply with distance (as with the control group) at the locations of cues (Fig. 6D). Analysis of the averaged position map and the trial-averaged activity aligned for each neocortical region are shown in Figure 7.
Figure 7.

Trial-averaged activity (non-normalized) as a function of position for each region. Grand averages for both groups for all detected neurons, evaluated separately by neocortical region, of the trial-averaged, not normalized, activity as a function of position (top), and with their peaks aligned in the middle (bottom). Shaded area represents SEM over animals (control, n = 4, lesion, n = 4). Generally, there is no difference in the overall activity and the peak of the aligned atrial-averaged activity is higher in the control group.
Hippocampal lesion increases decoding error of position
Finally, we applied Bayesian decoding to estimate the position with the highest probability given the activity of all imaged neurons (Fig. 8A). The decoding error as a function of position for the lesion animals was significantly higher at all positions on the belt (Fig. 8B). The probability distribution of the decoding error showed that the median error from the control group was 7.03 cm. In contrast, the median decoding error in the lesion group was 20.35 cm (Fig. 8C). By computing the error by region, we also observed a significant difference between the two groups, the average decoding error from the lesion group was at least 40% higher in all the regions evaluated (Fig. 8D). The distribution map—position decoded as a function of the actual position of the animal—also showed a substantial between-group difference in the decoding performance (Fig. 8E). The red diagonal stripe in the control group and the higher dispersion around the diagonal in the lesion group show that the representations of position corresponded more with the actual location of the animal in the control group. Similar results were obtained when the decoding error was evaluated for separate neocortical areas (Fig. 9).
Figure 8.

Decoding population activity between the control and lesion groups. A, Position (black) and position-decoded (red) traces of a session for a control and a lesioned animal. B, Decoding error as a function of position for the control group (black) and the lesion group (red) averaged across animals and regions (*p < 0.05; n = 4 per group). C, Left, Distribution of the decoding error for the control (black) and the lesion group (red); *p < 0.05. C, Right, Cumulative distributions of left. D, Mean decoding error for the five regions in the control and the lesion groups (error bars denote SEM over animals; *p < 0.05). Dots represent average decoding errors for each animal. E, Probability density map of all animals showing the distribution of position decoded from the activity of all neurons (y-axis) as a function of the location of the animal (x-axis). The red diagonal stripe indicates that the decoder from the control group gives a more accurate estimation of position. For exact n and p values, see Table 1.
Figure 9.
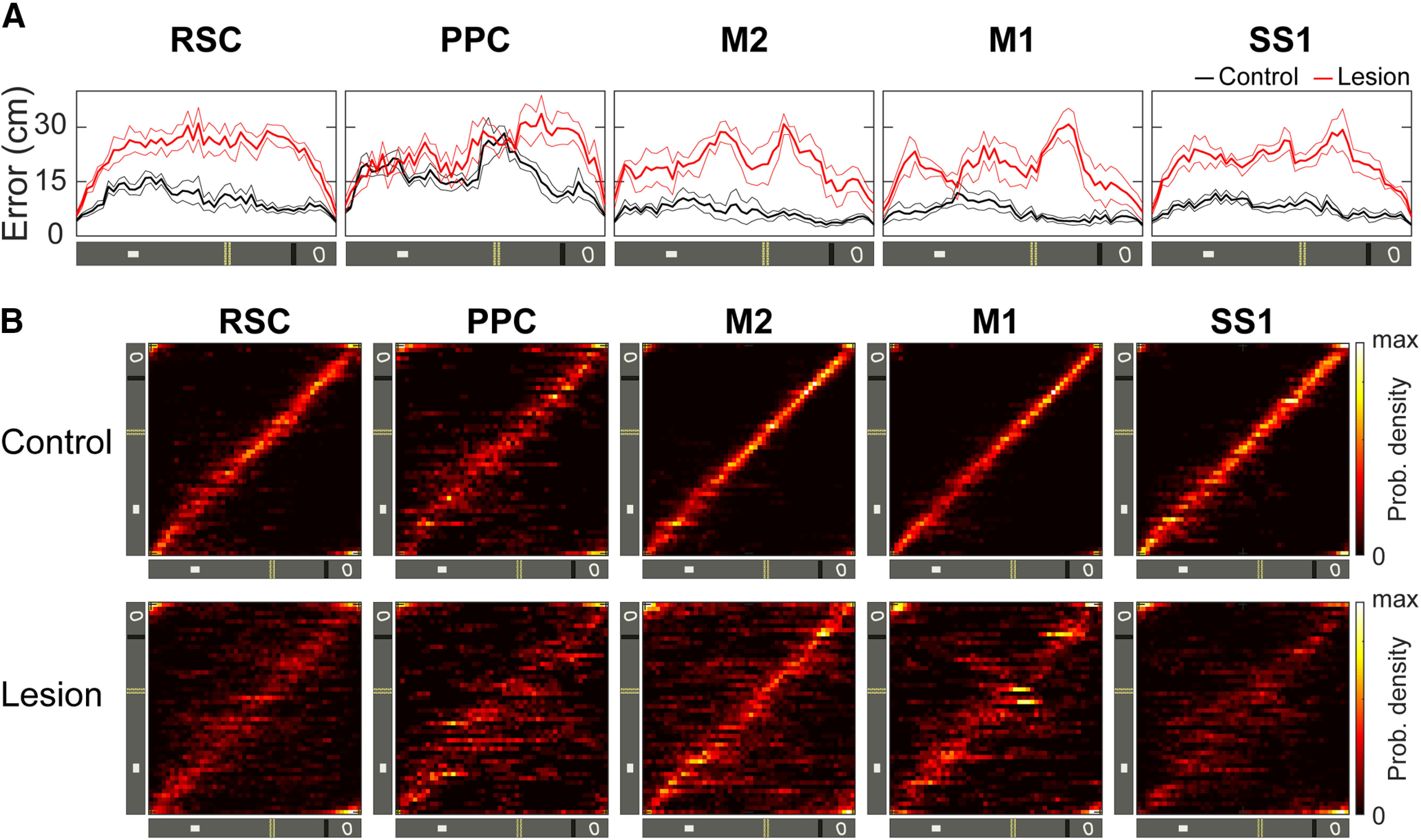
Decoding error for the five neocortical regions. A, Decoding error as a function of position for the control group (black) and the lesion group (red) averaged across animals for the five neocortical regions (n = 4/group). B, Probability density map showing the distribution of position decoded from the activity of all neurons (y-axis) as a function of the location of the animal (x-axis) for the five neocortical regions.
Discussion
By combining a 5 mm craniotomy with a treadmill assay, we found a large fraction of spatially selective neurons in the superficial layers of all neocortical regions surveyed. Bilateral lesions of the dorsal hippocampus significantly reduced the number of neurons exhibiting location-specific firing along the treadmill track. Furthermore, the place fields of the residual spatial neurons following lesion were not wider, but showed reduced sparsity and conveyed less spatial information. Together, these results suggest that, although a hippocampal lesion does not abolish spatially selective firing in neocortical areas, hippocampal inputs are necessary to maintain a precise cortical representation of space. We further supported these conclusions by showing that the positions of the animals on the track can be accurately estimated with the population activity of neocortical ensembles using a Bayesian approach, while a hippocampal lesion resulted in a significant decrease in decoding accuracy.
In our study, there was a general trend for anterior regions to express higher fractions of spatially selective neurons, with the posterior parietal cortex exhibiting the lowest numbers. Although PPC is the region less affected by the lesion, it is also the region with a lower number of cells detected (expressing spatial coding characteristics or not), and a higher decoding error in the control group. Such a discrepancy may be because of the presence of dense vascular structures over the parietal region that can impact the signal-to-noise ratio of the recordings using in vivo two‐photon Ca2+ imaging. Several studies have shown the posterior parietal cortex to be a critical component of the navigational system (Kolb et al., 1983; Parron and Save, 2004; Nitz, 2006, 2012; Whitlock et al., 2012). The PPC has been shown to integrate input from multiple sensory modalities, and to encode spatial information in both egocentric (body-centered) and allocentric (world-centered) frames of reference (McNaughton et al., 1994; Nitz, 2006; Wilber et al., 2014). The construction of spatial cognitive maps relies on the ability to translate navigational signals, which arise from a body-centered frame of reference in primary cortical areas, to a world-centered frame of reference (McNaughton et al., 2006). The PPC has been postulated as the region where the transition from an egocentric to an allocentric coding scheme occurs (Burgess et al., 1999).
Although single-unit recordings in behaving animals have revealed many forms of spatial cell types in extrahippocampal regions, the precise nature of these signals remains poorly understood (Knierim, 2006; Grieves and Jeffery, 2017). Recently, a growing body of studies that used two-photon calcium imaging and treadmill assays have described spatially tuned neurons in many neocortical areas, including the posterior parietal cortex (Nitz, 2006; Harvey et al., 2012), the retrosplenial cortex (Mao et al., 2017, 2018), and the visual cortex (Fiser et al., 2016; Pakan et al., 2018; Saleem et al., 2018; Minderer et al., 2019). In these studies, special emphasis was given to occipitoparietal areas involved in visual processing. Interest in those regions may be justified by the anatomic connectivity these areas receive from the dorsal hippocampus (Strange et al., 2014; Skelin et al., 2019). Nevertheless, in the present work, we indiscriminately surveyed multiple neocortical regions (including primary, secondary, and association areas), and demonstrated that, in general, these spatial representations do not differ in fundamental features across regions, although it may be expected that differences may emerge when cues and behavioral context are manipulated, which was not done in this survey. Furthermore, we directly investigated the link between the neocortex and the hippocampus with respect to the coding of location and characterized the extent to which cortical representations relied on hippocampal inputs.
The current results fittingly complement the previous reports by highlighting the importance of hippocampal feedback to cortical spatial coding. In fact, a good example of this has been given when a lesion to the hippocampus caused medial entorhinal grid cells to lose their spatial periodicity, with some adopting instead the response schemes of head direction cells (Bonnevie et al., 2013). However, following lesions of the medial entorhinal cortex, place cells could still be observed (Miller and Best, 1980; Brun et al., 2008; Schlesiger et al., 2015). Similarly, specific lesions of the perirhinal cortex, of the postrhinal cortex, and of the presubiculum, three other important hubs for corticohippocampal inputs, did not lead to complete disruption of the spatial specificity of place cells either (Muir and Bilkey, 2001; Calton et al., 2003; Nerad et al., 2009). These data suggest that spatial representations in the hippocampus may emerge from highly parallelized streams of information and can adapt to a substantial amount of signal loss. In contrast, spatial signals found in regions outside of the hippocampus appear to rely more heavily on hippocampal inputs.
Overall, much of what is known about the general behavior of rodents that have undergone hippocampectomy has been described by a large body of research in rats (Nadel, 1968; Mumby et al., 2002; Clark et al., 2005; Faraji et al., 2008; Ocampo et al., 2018), but not all results from rats can necessarily be generalized to mice. As expected, it has been shown that hippocampal lesions in mice also impaired spatial working memory and spatial reference memory, and disrupt some species-typical behavior (e.g., rearing, exploring, and hoarding). However, anxiety in lesioned mice was not uniformly diminished, and lesioned mice did not present an ill-groomed appearance and did not show alteration in locomotor activity (Arns et al., 1999; Deacon et al., 2002; Deacon and Rawlins, 2005). In addition, another study conducted to evaluate the effect of dorsal and ventral hippocampal lesions in mice showed that although hippocampal lesions made mice hyperactive during the habituation period in the open field, dorsal lesions were not able to produce a significant difference in the maze running time (Ammassari-Teule and Passino, 1997). Together, these works suggest that lesions of the hippocampus can produce in mice hyperactivity in a novel environment, can affect responses to external stimuli (spatial novelty), and can reduce directed exploration (rearing and head dipping), but all these effects were not because of impaired motility. At least for the task presented in this study, we could not detect any significant locomotion difference between both groups. Lesion and control groups were overall equally responsive during the entire time that they were tested, and hippocampal lesions did not detectably alter the performance of the task by the mice. The lack of difference may rather reflect the case that both groups were well habituated to the task.
One outstanding question that merits further investigation is how the experience of the animals affects the time course of the formation of spatial representations in the neocortex. Several works have shown that the hippocampal place code can emerge during the first traversal of a novel environment, that hippocampal population activity accurately represents the position of the animal after ∼10 min of experience in a new environment, and that place fields can expand with experience (Hill, 1978; Wilson and McNaughton, 1993; Mehta et al., 1997). In the neocortex, the emergence of spatial representations in the RSC has been shown to progressively improve with experience (Mao et al., 2017). But for other cortical regions, the dynamics of spatial representations still remain to be elucidated.
In summary, our results provide new insights into the importance of the hippocampus in shaping neocortical activity during the exploration of a controlled environment. Although this study offered a general perspective on the encoding of space in the neocortex, further research using more complex assays is needed to test various sensory/cognitive features to better characterize those cells and to identify region-specific attributes. We demonstrated that spatial representations are widely observed in the dorsal neocortex and that these representations express a similar degree of reliance of hippocampal feedback. In the framework of the “index theory” (Teyler and DiScenna, 1986; Teyler and Rudy, 2007; McNaughton, 2010), our results may suggest that, without the hippocampus, the cortex fails to express a unique code for each experience, and this failure likely explains the failure of the cortex to bind together attributes of an experience into unique episodic memories. This possibility remains to be investigated by placing animals in familiar and novel environments before and after hippocampal lesion.
Footnotes
This work was supported by the Canadian Institutes of Health Research (Grant PJT 156040 to B.L.M.; Grant PJT 390930 to M.H.M.)], by the US Defense Advanced Research Projects Agency (Grant HR0011-18-2-0021 to B.L.M.), and by the Natural Sciences and Engineering Research Council of Canada (Grant 1631465 to B.L.M. and Grant #40352 to M.H.M.). This research was enabled in part by support provided by WestGrid (www.westgrid.ca) and Compute Canada (www.computecanada.ca). We thank Isabelle Gauthier, Di Shao, animal care staff, and all other members of the Animal Welfare Committee at the University of Lethbridge for making animal experiments possible. We also thank Rui Pais and Andrew McNaughton for valuable comments. Further, we also thank Amanda Mauthe-Kaddoura for providing logistics support, Valerie Lapointe and Karim Ali for technical support, and Dun Mao for his valuable help during the experiments.
The authors declare no competing financial interests.
References
- Ammassari-Teule M, Passino E (1997) The dorsal hippocampus is selectively involved in the processing of spatial information even in mice with a genetic hippocampal dysfunction. Psychobiology 25:118–125. [Google Scholar]
- Arns M, Sauvage M, Steckler T (1999) Excitotoxic hippocampal lesions disrupt allocentric spatial learning in mice: effects of strain and task demands. Behav Brain Res 106:151–164. 10.1016/S0166-4328(99)00103-5 [DOI] [PubMed] [Google Scholar]
- Bonin V, Histed MH, Yurgenson S, Clay Reid R (2011) Local diversity and fine-scale organization of receptive fields in mouse visual cortex. J Neurosci 31:18506–18521. 10.1523/JNEUROSCI.2974-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnevie T, Dunn B, Fyhn M, Hafting T, Derdikman D, Kubie JL, Roudi Y, Moser EI, Moser M-B (2013) Grid cells require excitatory drive from the hippocampus. Nat Neurosci 16:309–317. 10.1038/nn.3311 [DOI] [PubMed] [Google Scholar]
- Brun VH, Leutgeb S, Wu H-Q, Schwarcz R, Witter MP, Moser EI, Moser M-B (2008) Impaired spatial representation in CA1 after lesion of direct input from entorhinal cortex. Neuron 57:290–302. 10.1016/j.neuron.2007.11.034 [DOI] [PubMed] [Google Scholar]
- Burgess N, Jeffery KJ, O'Keefe J (1999) The hippocampal and parietal foundations of spatial cognition. Oxford: Oxford UP. [Google Scholar]
- Calton JL, Stackman RW, Goodridge JP, Archey WB, Dudchenko PA, Taube JS (2003) Hippocampal place cell instability after lesions of the head direction cell network. J Neurosci 23:9719–9731. 10.1523/JNEUROSCI.23-30-09719.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HRan, Esteves IM, Neumann AR, Sun J, Mohajerani MH, McNaughton BL (2020) Coordinated activities of retrosplenial ensembles during resting-state encode spatial landmarks. Philos Trans R Soc Lond B Biol Sci 375:20190228. 10.1098/rstb.2019.0228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RE, Broadbent NJ, Squire LR (2005) Hippocampus and remote spatial memory in rats. Hippocampus 15:260–272. 10.1002/hipo.20056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon RMJ, Rawlins JNP (2005) Hippocampal lesions, species-typical behaviours and anxiety in mice. Behav Brain Res 156:241–249. 10.1016/j.bbr.2004.05.027 [DOI] [PubMed] [Google Scholar]
- Deacon RMJ, Croucher A, Rawlins JNP (2002) Hippocampal cytotoxic lesion effects on species-typical behaviours in mice. Behav Brain Res 132:203–213. 10.1016/S0166-4328(01)00401-6 [DOI] [PubMed] [Google Scholar]
- Faraji J, Lehmann H, Metz GA, Sutherland RJ (2008) Rats with hippocampal lesion show impaired learning and memory in the ziggurat task: a new task to evaluate spatial behavior. Behav Brain Res 189:17–31. 10.1016/j.bbr.2007.12.002 [DOI] [PubMed] [Google Scholar]
- Fiser A, Mahringer D, Oyibo H, Petersen A, Leinweber M, Keller G (2016) Experience-dependent spatial expectations in mouse visual cortex. Nat Neurosci 19:1658–1664. 10.1038/nn.4385 [DOI] [PubMed] [Google Scholar]
- Gener T, Perez-Mendez L, Sanchez-Vives MV (2013) Tactile modulation of hippocampal place fields. Hippocampus 23:1453–1462. 10.1002/hipo.22198 [DOI] [PubMed] [Google Scholar]
- Grieves RM, Jeffery KJ (2017) The representation of space in the brain. Behav Processes 135:113–131. 10.1016/j.beproc.2016.12.012 [DOI] [PubMed] [Google Scholar]
- Harvey CD, Coen P, Tank DW (2012) Choice-specific sequences in parietal cortex during a virtual-navigation decision task. Nature 484:62–68. 10.1038/nature10918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill AJ. (1978) First occurrence of hippocampal spatial firing in a new environment. Exp Neurol 62:282–297. 10.1016/0014-4886(78)90058-4 [DOI] [PubMed] [Google Scholar]
- Jung MW, Wiener SI, McNaughton BL (1994) Comparison of spatial firing characteristics of units in dorsal and ventral hippocampus of the rat. J Neurosci 14:7347–7356. 10.1523/JNEUROSCI.14-12-07347.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkcaldie MTK. (2012) Neocortex. In: The mouse nervous system (Watson C, Paxinos G, Puelles L, eds), pp 52–111. San Diego: Academic. [Google Scholar]
- Knierim J. (2006) Neural representations of location outside the hippocampus. Learn Mem 13:405–415. 10.1101/lm.224606 [DOI] [PubMed] [Google Scholar]
- Kolb B, Sutherland RJ, Whishaw IQ (1983) A comparison of the contributions of the frontal and parietal association cortex to spatial localization in rats. Behav Neurosci 97:13–27. 10.1037/0735-7044.97.1.13 [DOI] [PubMed] [Google Scholar]
- Leutgeb S, Leutgeb JK, Barnes CA, Moser EI, McNaughton BL, Moser M-B (2005) Independent codes for spatial and episodic memory in hippocampal neuronal ensembles. Science 309:619–623. 10.1126/science.1114037 [DOI] [PubMed] [Google Scholar]
- Mao D, Kandler S, McNaughton BL, Bonin V (2017) Sparse orthogonal population representation of spatial context in the retrosplenial cortex. Nat Commun 8:243. 10.1038/s41467-017-00180-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao D, Neumann AR, Sun J, Bonin V, Mohajerani MH, McNaughton BL (2018) Hippocampus-dependent emergence of spatial sequence coding in retrosplenial cortex. Proc Natl Acad Sci U S A 115:8015–8018. 10.1073/pnas.1803224115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNaughton BL. (2010) Cortical hierarchies, sleep, and the extraction of knowledge from memory. Artif Intell 174:205–214. 10.1016/j.artint.2009.11.013 [DOI] [Google Scholar]
- McNaughton BL, Leonard B, Chen L (1989) Cortical-hippocampal interactions and cognitive mapping: a hypothesis based on reintegration of the parietal and inferotemporal pathways for visual processing. Psychobiology 17:230–246. 10.1007/BF03337774 [DOI] [Google Scholar]
- McNaughton BL, Mizumori SJY, Barnes CA, Leonard BJ, Marquis M, Green EJ (1994) Cortical representation of motion during unrestrained spatial navigation in the rat. Cereb Cortex 4:27–39. 10.1093/cercor/4.1.27 [DOI] [PubMed] [Google Scholar]
- McNaughton BL, Battaglia FP, Jensen O, Moser EI, Moser M-B (2006) Path integration and the neural basis of the “cognitive map”. Nat Rev Neurosci 7:663–678. 10.1038/nrn1932 [DOI] [PubMed] [Google Scholar]
- Mehta MR, Barnes CA, McNaughton BL (1997) Experience-dependent, asymmetric expansion of hippocampal place fields. Proc Natl Acad Sci U S A 94:8918–8921. 10.1073/pnas.94.16.8918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller VM, Best PJ (1980) Spatial correlates of hippocampal unit activity are altered by lesions of the fornix and entorhinal cortex. Brain Res 194:311–323. 10.1016/0006-8993(80)91214-7 [DOI] [PubMed] [Google Scholar]
- Minderer M, Brown KD, Harvey CD (2019) The spatial structure of neural encoding in mouse posterior cortex during navigation. Neuron 102:232–248.e11. 10.1016/j.neuron.2019.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moita MAP, Rosis S, Zhou Y, LeDoux JE, Blair HT (2003) Hippocampal place cells acquire location-specific responses to the conditioned stimulus during auditory fear conditioning. Neuron 37:485–497. 10.1016/S0896-6273(03)00033-3 [DOI] [PubMed] [Google Scholar]
- Moser M-B, Rowland DC, Moser EI (2015) Place cells, grid cells, and memory. Cold Spring Harb Perspect Biol 7:a021808. 10.1101/cshperspect.a021808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir GM, Bilkey DK (2001) Instability in the place field location of hippocampal place cells after lesions centered on the perirhinal cortex. J Neurosci 21:4016–4025. 10.1523/JNEUROSCI.21-11-04016.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller R, Kubie J (1987) The effects of changes in the environment on the spatial firing of hippocampal complex-spike cells. J Neurosci 7:1951–1968. 10.1523/JNEUROSCI.07-07-01951.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumby DG, Gaskin S, Glenn MJ, Schramek TE, Lehmann H (2002) Hippocampal damage and exploratory preferences in rats: memory for objects, places, and contexts. Learn Mem 9:49–57. 10.1101/lm.41302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadel L. (1968) Dorsal and ventral hippocampal lesions and behavior. Physiology and Behavior 3:891–900. 10.1016/0031-9384(68)90174-1 [DOI] [Google Scholar]
- Nerad L, Liu P, Bilkey DK (2009) Bilateral NMDA lesions centered on the postrhinal cortex have minimal effects on hippocampal place cell firing. Hippocampus 19:221–227. 10.1002/hipo.20517 [DOI] [PubMed] [Google Scholar]
- Nitz DA. (2006) Tracking route progression in the posterior parietal cortex. Neuron 49:747–756. 10.1016/j.neuron.2006.01.037 [DOI] [PubMed] [Google Scholar]
- Nitz DA. (2009) Parietal cortex, navigation, and the construction of arbitrary reference frames for spatial information. Neurobiol Learn Mem 91:179–185. 10.1016/j.nlm.2008.08.007 [DOI] [PubMed] [Google Scholar]
- Nitz DA. (2012) Spaces within spaces: rat parietal cortex neurons register position across three reference frames. Nat Neurosci 15:1365–1367. 10.1038/nn.3213 [DOI] [PubMed] [Google Scholar]
- O'Keefe J, Conway DH (1978) Hippocampal place units in the freely moving rat: why they fire where they fire. Exp Brain Res 31:573–590. 10.1007/BF00239813 [DOI] [PubMed] [Google Scholar]
- O'Keefe J, Dostrovsky J (1971) The hippocampus as a spatial map. Preliminary evidence from unit activity in the freely-moving rat. Brain Res 34:171–175. 10.1016/0006-8993(71)90358-1 [DOI] [PubMed] [Google Scholar]
- O'Keefe J, Speakman A (1987) Single unit activity in the rat hippocampus during a spatial memory task. Exp Brain Res 68:1–27. 10.1007/BF00255230 [DOI] [PubMed] [Google Scholar]
- Ocampo AC, Squire LR, Clark RE (2018) The beneficial effect of prior experience on the acquisition of spatial memory in rats with CA1, but not large hippocampal lesions: a possible role for schema formation. Learn Mem 25:115–121. 10.1101/lm.046482.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachitariu M, Stringer C, Dipoppa M, Schröder S, Rossi L, Dalgleish H, Carandini M, Harris KD (2017) Suite2p: beyond 10,000 neurons with standard two-photon microscopy. BioRxiv. Advance online publication. Retrieved November 18, 2020. doi: 10.1101/061507. [DOI] [Google Scholar]
- Pakan JMP, Currie SP, Fischer L, Rochefort NL (2018) The impact of visual cues, reward, and motor feedback on the representation of behaviorally relevant spatial locations in primary visual cortex. Cell Rep 24:2521–2528. 10.1016/j.celrep.2018.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parron C, Save E (2004) Evidence for entorhinal and parietal cortices involvement in path integration in the rat. Exp Brain Res 159:349–359. 10.1007/s00221-004-1960-8 [DOI] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ (2003) The mouse brain in stereotaxic coordinates, Ed 2 San Diego: Academic. [Google Scholar]
- Pnevmatikakis EA, Soudry D, Gao Y, Machado TA, Merel J, Pfau D, Reardon T, Mu Y, Lacefield C, Yang W, Ahrens M, Bruno R, Jessell TM, Peterka DS, Yuste R, Paninski L (2016) Simultaneous denoising, deconvolution, and demixing of calcium imaging data. Neuron 89:285–299. 10.1016/j.neuron.2015.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirk GJ, Muller RU, Kubie JL (1990) The firing of hippocampal place cells in the dark depends on the rat's recent experience. J Neurosci 10:2008–2017. 10.1523/JNEUROSCI.10-06-02008.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem AB, Diamanti EM, Fournier J, Harris KD, Carandini M (2018) Coherent encoding of subjective spatial position in visual cortex and hippocampus. Nature 562:124–127. 10.1038/s41586-018-0516-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Save E, Nerad L, Poucet B (2000) Contribution of multiple sensory information to place field stability in hippocampal place cells. Hippocampus 10:64–76. [DOI] [PubMed] [Google Scholar]
- Schlesiger MI, Cannova CC, Boublil BL, Hales JB, Mankin EA, Brandon MP, Leutgeb JK, Leibold C, Leutgeb S (2015) The medial entorhinal cortex is necessary for temporal organization of hippocampal neuronal activity. Nat Neurosci 18:1123–1132. 10.1038/nn.4056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheintuch L, Geva N, Baumer H, Rechavi Y, Rubin A, Ziv Y (2020) Multiple maps of the same spatial context can stably coexist in the mouse hippocampus. Curr Biol 30:1467–1476.e6. 10.1016/j.cub.2020.02.018 [DOI] [PubMed] [Google Scholar]
- Skaggs WE, McNaughton BL, Gothard KM (1992) An information-theoretic approach to deciphering the hippocampal code. In: NIPS'92: Proceedings of the 5th International Conference on Neural Information Processing Systems (Hanson SJ, Cowan JD, Giles CL, eds), pp 1030–1037. San Francisco: Morgan Kaufmann. [Google Scholar]
- Skelin I, Kilianski S, McNaughton BL (2019) Hippocampal coupling with cortical and subcortical structures in the context of memory consolidation. Neurobiol Learn Mem 160:21–31. 10.1016/j.nlm.2018.04.004 [DOI] [PubMed] [Google Scholar]
- Strange BA, Witter MP, Lein ES, Moser EI (2014) Functional organization of the hippocampal longitudinal axis. Nat Rev Neurosci 15:655–669. 10.1038/nrn3785 [DOI] [PubMed] [Google Scholar]
- Teyler TJ, DiScenna P (1986) The hippocampal memory indexing theory. Behav Neurosci 100:147–154. 10.1037/0735-7044.100.2.147 [DOI] [PubMed] [Google Scholar]
- Teyler TJ, Rudy JW (2007) The hippocampal indexing theory and episodic memory: updating the index. Hippocampus 17:1158–1169. 10.1002/hipo.20350 [DOI] [PubMed] [Google Scholar]
- Whitlock JR, Pfuhl G, Dagslott N, Moser M-B, Moser EI (2012) Functional split between parietal and entorhinal cortices in the rat. Neuron 73:789–802. 10.1016/j.neuron.2011.12.028 [DOI] [PubMed] [Google Scholar]
- Wilber AA, Clark BJ, Forster TC, Tatsuno M, McNaughton BL (2014) Interaction of egocentric and world-centered reference frames in the rat posterior parietal cortex. J Neurosci 34:5431–5446. 10.1523/JNEUROSCI.0511-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MA, McNaughton BL (1993) Dynamics of the hippocampal ensemble code for space. Science 261:1055–1058. 10.1126/science.8351520 [DOI] [PubMed] [Google Scholar]
- Zhang KI, Ginzburg BL, McNaughton TJ Sejnowski (1998) Interpreting neuronal population activity by reconstruction: unified framework with application to hippocampal place cells. J Neurophysiol 79:1017–1044. 10.1152/jn.1998.79.2.1017 [DOI] [PubMed] [Google Scholar]