Abstract
Background
Cancer cells could show the characteristics of cancer stem cells (CSCs) through epithelial-mesenchymal transition (EMT). EZH2 was associated with EMT. Ultrasound-targeted microbubble destruction (UTMD) could enhance gene transfection efficiency. Here, we explored the effect of UTMD-mediated shEZH2 on liver CSCs.
Methods
EZH2 expression in liver cancer and the overall survival of liver cancer patients were analyzed by bioinformatics. Liver CSCs (CD133+HuH7) were sorted by flow cytometry. After transfection of shEZH2 through UTMD (UTMD-shEZH2) or liposome (LIP-shEZH2), the viability, proliferation, sphere formation, migration, and invasion of CD133+HuH7 cells were detected by MTT, colony formation, tumor-sphere formation, wound healing, and transwell assays, respectively. A mice subcutaneous-xenotransplant tumor model was established by injecting CD133+HuH7 or CD133−HuH7 cells into the limbs of mice. Tumor weight and volume were documented. The expressions of EZH2, EMT-related factors, and STAT3/PI3K/AKT pathway-related factors in CD133+HuH7 cells or tumor tissues were detected by RT-qPCR, Western blot, or immunohistochemical.
Results
EZH2 was high-expressed in liver cancer, and the patients with high expression of EZH2 had a poor survival. CD133+ HuH7 cells had higher EZH2 expression, higher viability, and stronger sphere-forming and tumor-forming abilities than CD133− HuH7 cells. ShEZH2 inhibited the viability, proliferation, sphere formation, migration, and invasion of CD133+ HuH7 cells, decreased the weight and volume of the xenotransplant tumor, inhibited the expressions of EZH2, Vimentin, N-Cadherin, Twist-1, p-STAT3, p-PI3K, and p-AKT, and increased E-Cadherin expression. UTMD-shEZH2 caused a stronger effect on CD133+ HuH7 cells than LIP-shEZH2.
Conclusion
UTMD-mediated shEZH2 inhibited the stemness and EMT of liver CSCs in vitro and in vivo through regulating the STAT3/PI3K/AKT pathway.
Keywords: liver cancer, cancer stem cells, ultrasound-targeted microbubble destruction, epithelial-mesenchymal transition, xenotransplant
Introduction
Liver cancer is one of the most common human malignant tumors, and its incidence and mortality rates rank top among all malignant tumors.1,2 At present, the main approaches to liver cancer treatment are surgery, liver transplantation, radiotherapy, chemotherapy and biotherapy.3,4 Despite advances in the treatment of liver cancer, the long-term survival of liver cancer patients after treatment is still significantly poor due to the high malignancy, rapid progression, and easy recurrence and metastasis of the disease.5,6
Cancer stem cells (CSCs) are stem cell-like cells with the biological potential of self-renewal, replication, non-directional differentiation, high tumorigenicity, resistance to radiotherapy and chemotherapy, and so on.7,8 With these characteristics that maintain and promote tumor growth, CSCs were considered to be a cause of cancer occurrence, metastasis, recurrence and treatment resistance.5,9,10 An increasing number of studies have proved that there is a close relationship between epithelial-mesenchymal transition (EMT) and CSCs.11 EMT refers to the phenomenon that epithelial cells transform into mesenchymal cells in a specific physiological or pathological state.12 After EMT, tumor cells lose the characteristics of epithelial cells, and show the nature of interstitial cells, with enhanced abilities of invasion and metastasis.11,13 Therefore, during the progression of EMT, cancer cells can show the characteristics of CSCs.11,13–15 In addition, more and more researches have reported that EZH2 has a regulatory effect on the progression of EMT; for example, Zhang et al proved that EZH2 was a enhancer of EMT in endometriosis;16 Gan et al reported that EZH2 could induce the EMT and pluripotent phenotype of gastric cancer cells.17 In addition, EZH2 has also been prove to own the ability to regulate EMT in pancreatic cancer.18 However, whether EZH2 had an effect on the progression of EMT in liver cancer requires more investigation.
As a novel transfection approach, ultrasound-targeted microbubble destruction (UTMD) has been gradually widely used in various research.19,20 UTMD refers to the technology that under the action of ultrasound, microbubbles break up and release a large number of microbubbles which form reversible micropores on the cell membrane and increase cell membrane permeability, so that exogenous target genes can enter the cell.21 Moreover, it was reported that UTMD has the advantages of low immunogenicity, low toxicity, reusability and easy operation.21,22
On the basis of these backgrounds, we transfected UTMD-mediated shEZH2 into liver CSCs to explore whether EZH2 had an effect on the progression of EMT in liver CSCs.
Materials and Methods
Ethics Statement
Animal experiments in this research were approved by the Committee of Experimental Animals of The First Affiliated Hospital of Zhengzhou University Hospital (Z20190603G). All experiments involving animals were performed in The First Affiliated Hospital of Zhengzhou University.
Cell Culture
Human liver epithelial cell THLE-3 (CRL-11233) and human liver cancer cells Hep3B (HB-8064) and SK-HEP-1 (HTB-52) were bought from ATCC (Rockville, MD, USA). Human liver cancer cells HuH7 (CL-0120) and LM3 (CL-0278) were bought from Procell (Wuhan, China, https://www.procell.com.cn/). Human liver cancer cell MHCC97H was brought from Jennio-bio (Guangzhou, China, http://www.jennio-bio.com/products_detail/andproductId=217.html). All cells were cultured in complete medium which consisted of RPMI 1640 medium (C11875500BT, Gibco, MA, USA) containing 10% fetal bovine serum (FBS; 16140071, Gibco) in a 37°C, humidified atmosphere with 5% CO2.
Flow Cytometry
Flow cytometry was used to sort liver cancer stem cells from HuH7 cells. In brief, the HuH7 cells were collected and resuspended in PBS (C10010500BT, Gibco). Then, the cells were incubated with the human-specific CD133 antibody (ab19898, Abcam, CA, USA) for 30 min in the dark. The isotype mouse IgG (ab190475, Abcam) was used as a control antibody to incubate the cells under the same condition. Afterwards, the labeled HuH7 cells were sorted using the flow cytometer (FACS-LSR II, Becton-Dickinson, NJ, USA). Finally, the freshly sorted and isolated CD133+HuH7 cells were collected and cultured in RPMI 1640 medium without FBS in a 37°C, humidified atmosphere with 5% CO2.
Liposome Transfection
ShRNAs for EZH2 (shEZH2; stB0002755A-1-5) and negative control (shNC; siN0000001-1-5) were synthesized by RIBOBIO (Guangzhou, China). The interference sequence of shEZH2 was synthesized as 5ʹ-CCGGCCCAACATAGATGGACCAAATCTCGAGATTTGGTCCATCTATGTTGGGTTTTTG-3ʹ; the interference sequence of shNC was synthesized as 5ʹ-AATTCTCCGAACGTGTCACGT-3ʹ. Before transfection, 2.0 × 105 CD133+HuH7 cells in 2 mL of complete medium were seeded into a 6-well plate. After the cells reached about 80% confluence, 100 μL of OPTI MEM (31985070, Gibco) was used to dissolve 2 μg of shEZH2 and shNC; meantime, another 100 μL of OPTI MEM was used to dilute 2 μL of Lipofectamine 2000 (11668019, Invitrogen, MA, USA). Then the two OPTI MEM solutions were mixed and incubated for 10 min at room temperature. After the previous complete medium was discarded, the CD133+HuH7 cells in each well were added with the mixed OPTI MEM, and subsequently added with 1.8 mL of RPMI 1640 medium for an additional 48-h incubation. The transfection ways of shEZH2 and shNC into CD133+HuH7 cells through Lipofectamine 2000 were named LIP-shEZH2 and LIP-shNC.
UTMD Transfection
Before transfection, 2.0 × 105 CD133+HuH7 cells in 2 mL of complete medium were seeded into a 6-well plate. Normal saline (R21479, OKA, Beijing, China, http://www.bjoka-vip.com/) was used to dilute SonoVue (J20130045, Bracco Imaging B.V., Milan, Italy, https://imaging.bracco.com/) to a concentration of 2×108 bubbles/mL. After the confluence of the cells reached about 80%, 200 μL of SonoVue was used to dissolve 2 μg of shEZH2 and shNC. After the complete medium was discarded, the 200 μL SonoVue containing shRNAs was mixed with 1.8 mL of RPMI 1640 medium and subsequently added into each well. Then the cells were subjected to ultrasound irradiation at 1 MHz and 1W/cm2 for 30 s using a therapeutic AH6 ultrasound machine (Bandelin, Berlin, Germany). After the cells were cultured in a 37°C, humidified atmosphere with 5% CO2 for 3 h, the original complete medium was refreshed for an additional 48-h culture. The transfection ways of shEZH2 and shNC into CD133+HuH7 cells through UTMD were named UTMD-shEZH2 and UTMD-shNC.
MTT Assays
CD133+HuH7 cells transfected with shRNA, CD133+HuH7 cells, or CD133−HuH7 cells were seeded into a 96-well plate, and each well contained 1.0 × 104 cells in 100 μL of complete medium. After the cells were grown for 24, 48, or 72 h, the medium was discarded and 100 μL of 0.5 mg/mL MTT (B7777, APExBIO, Houston, USA) was added to incubate the cells in each well for 4 h. Then 100 μL of DMSO (ST038, Beyotime, Shanghai, China) was added to each well after the MTT solution was removed. Finally, the absorbance of each well was measured at 570 nm with a microplate reader (Imark, Bio-Rad, CA, USA).
Tumor-Sphere Formation Assay
CD133+HuH7 cells transfected with shRNA, CD133+HuH7 cells, or CD133−HuH7 cells were seeded into a 6-well plate, each well containing 1000 cells in 2 mL of complete medium. After the cells were cultured for two weeks, tumor spheres were formed. Finally, the tumor spheres were observed and documented under an optical microscope (DM4M, Leica, Solms, Germany) at a magnification of 200 ×. According to the diameters of the tumor spheres that were calculated using Image J 1.8.0 software, the tumor sphere size was categorised into three types: 50–100 μm, 100–150 μm, and over 150 μm.
Colony Formation Assay
CD133+HuH7 cells transfected with shRNA were seeded into a 6-well plate, each well containing 1000 cells in 2 mL of complete medium. After two weeks of culture, the medium was discarded and 4% formaldehyde (P804536, Macklin, Shanghai, China) was added into each well to fix the formed cell colonies for 10 min. After removing the formaldehyde, 0.3% crystal violet (C110704, Aladdin, Shanghai, China) was used to stain the colonies for 15 min. Subsequently, the stained colonies were washed three times with PBS to remove the redundant crystal violet. Finally, the colony number in each well was counted and analyzed using Image J 1.8.0 software.
Wound Healing Assays
CD133+HuH7 cells transfected with shRNA were seeded into a 6-well plate, each well containing 3.5 × 105 cells in 2 mL of complete medium. After the cells of each well reached 95% confluence, a vertical wound was created and 2 mL of RPMI 1640 medium without FBS was added into each well. Images of the vertical wounds in each well were collected at 0 and 24 h using an optical microscope (DM4M, Leica, Solms, Germany) at a magnification of 100 ×. Image J software 1.8.0 was used to analyze the images in this assay.
Transwell Assays
CD133+HuH7 cells (1.5 × 105) transfected with shRNA were resuspended in 200 μL of RPMI 1640 medium without FBS. The cells were then seeded into transwell chambers (3422, Corning Life Sciences, NY, USA) which were pre-coated with 200 mg/mL Matrigel (354203, Corning Life Sciences). And the transwell chambers embedded into a 24-well plate, with each well containing 700 μL of complete medium. After the cells were incubated for24 h, the inner layer of the chambers was wiped off, and the cells in the outer layer of the chamber were fixed by 4% paraformaldehyde and stained with crystal violet at room temperature for 15 min. Lastly, the number of cells that invaded to the lower membrane was calculated from four fields of view under an optical microscope (DM4M, Leica, Solms, Germany) at a magnification of 250×23 Image J 1.8.0 software was used to count the cells that invaded the outer layer of the chamber from the inner layer.
Animals and Subcutaneously Xenograft
In this study, 42 six-week-old male BALB/c nude mice (weight: 20–22 g) were obtained from SLAC Laboratory Animal Technology (Shanghai, China). All experimental animals were fed in the same animal feeding unit and maintained on a 12 hour dark/light cycle in an SPF-controlled environment. The animal experiment consisted of two parts.
For the first part, 12 nude mice were selected and randomly divided into two groups (n=6): 103group and 104 group. Firstly, CD133+HuH7 cells and CD133−HuH7 cells in PBS were mixed with an equal volume of Matrigel (354248, Corning Life Sciences) to adjust the cell concentrations to 1 × 103/200 μL and 1 × 104/200 μL respectively. Then, 200 μL of the mixed solution containing 1 × 103 CD133+HuH7 cells was subcutaneously injected into the left hind limb of the mice in the 103 group; while 200 μL of the mixture containing 1 × 103 CD133−HuH7 cells was subcutaneously injected into the right hind limb of the mice in103 group. Meantime, 200 μL of the mixed solution containing 1 × 104 CD133+HuH7 cells was subcutaneously injected into the left hind limb of the mice in group 104; while 200 μL of the mixture containing 1 × 104 CD133−HuH7 cells was subcutaneously injected into the right hind limb of the mice in group 104. After being normally housed for 42 days, all mice were anesthetized with 2% sodium pentobarbital (50 mg/kg) (B005, Jiancheng, Nanjing) and sacrificed by cervical dislocation. Finally, the subcutaneous tumorigenesis of CD133+HuH7 cells and CD133−HuH7 cells was documented. During the normally feeding, the subcutaneous tumorigenesis in the mice was observed and documented every 7 day: the major diameter (L) and minor diameter (W) of the tumor were measured and recorded with a vernier caliper (ASO-CC-1052-22, Asone, kanto, Japan). Tumor volume was calculated by the formula: tumor volume = major diameter (L) × (minor diameter)2 (W2)/2 mm3, and the tumor volume changes of each group of mice were finally presented in the form of a curve.
For the second part, the remaining 30 nude mice were randomly divided into five groups (n=6): Control group, LIP-shNC group, LIP-shEZH2 group, UTMD-shNC group, and UTMD-shEZH2 group. Firstly, CD133+HuH7 cells, CD133+HuH7 cells transfected with LIP-shNC, CD133+HuH7 cells transfected with LIP-shEZH2, CD133+HuH7 cells transfected with UTMD-shNC, and CD133+HuH7 cells transfected with UTMD-shEZH2 in PBS were mixed with an equal volume of Matrigel to adjust the cell concentration to 2 × 105/200 μL. Then, 200 μL of the mixed solution containing 2 × 105 CD133+HuH7 cells were subcutaneously injected into the right hind limb of the mice in the Control, LIP-shNC, LIP-shEZH2, UTMD-shNC, and UTMD-shEZH2 groups separately, and afterwards, the mice were normally housed. The subcutaneous tumorigenesis in the mice was observed and documented every other day: the tumor weight was measured with an AE1202 electronic balance (SOPTOP, Shanghai, China, http://www.hengping.com/productshow_47.html), and the L and W of the tumor were measured and recorded. Tumor volume was calculated by the formula: tumor volume = major diameter (L) × (minor diameter)2 (W2)/2 mm3, and the tumor volume changes of each group of mice were finally presented in the form of a curve. Finally, all mice were anesthetized with 2% sodium pentobarbital (50 mg/kg) and sacrificed by cervical dislocation after housing for four weeks, and tumor tissues were then harvested, weighed, and photographed.
Immunohistochemical
Paraffin (S25190, Yuanye, Shanghai, China) was used to embed the tumor tissues previously harvested. The tissues were cut into 4 μm thick slices under a microtome (RM2235, Leica, Solms, Germany) and the slices were further fixed on a glass slide (P105-2001, MeVid, Jiangsu, China, http://www.nt-mevid.com/ProDetail.aspx?ProId=54). After deparaffinization, the tissue slides were incubated with antigen repair solution (p0081, Beyotime) for 10 min at room temperature, followed by incubation with endogenous peroxidase blocker (BF06060, Biodragon, Beijing, China) for another 10 min at room temperature. After incubation with 5% FBS for 1 h at room temperature, the tissue slides were incubated with E-cadherin (1:100, ab1416, Abcam) or Vimentin (1:500, ab92547, Abcam) antibody overnight at 4°C. The next day, all tissue slides were incubated with a corresponding secondary antibody (G-21234, 1:500, Thermo Scientific) for 30 min and treated with the DBA reagent (SFQ004, 4A Biotech, Beijing, China) for 30 min. Then, the tissue slides were treated with hematoxylin (B25380, Yuanye) for 10 min. Finally, the detected indexes were observed and documented using an optical microscope (DM4M, Leica, Solms, Germany) at magnifications of 200 × and 400 ×. Furthermore, the quantification of immunohistochemical assay was evaluated through the ratio of the number of positive cells to the number of total cells in five randomly selected fields from each slide.
RNA Extraction and RT-qPCR
Total mRNAs in liver cancer cells and the tumor tissues of nude mice were extracted using TRIzol (15596, Invitrogen, MA, USA). In brief, 200 µL and 500 µL of TRIzol was used to lyse 3.5 × 106 liver cancer cells and 20 mg of tumor tissues for 15 min at room temperature, respectively. After the cells were centrifuged for 20 min (14,000 × g), about 200 µL of isopropanol (H822173, Macklin, Shanghai) was added to the supernatant (about 200 µL), mixed, and centrifuged for 5 min (10,000 × g). Then RNase-free H2O (RF001, Real-Times, Beijing, China) was used to dilute the mRNA sediment. EasyScript First-Strand cDNA Synthesis SuperMix (AE301-02, TransGen Biotech) was further used to reverse-transcribe the mRNA into cDNA in the light of the instructions. Then cDNA amplification was performed using PerfectStart Green qPCR SuperMix (AQ601-01, TransGen Biotech) and the gene primers in the QuantStudio6 system (Applied Biosystems, CA, USA), and the reaction condition was set as: at 94°Cfor 30 s, at 94°Cfor 30 s for 40 cycles, and at 60°Cfor 30 s for 40 cycles. Finally, RNA was quantified using the 2−ΔΔCT method. The primer sequences are showed in Table 1.
Table 1.
RT-qPCR Primers
| Target Gene | Forward Primers, 5ʹ-3’ | Reverse Primers, 5ʹ-3’ |
|---|---|---|
| EZH2 | GGACCACAGTGTTACCAGCAT | GTGGGGTCTTTATCCGCTCAG |
| E-Cadherin | CGAGAGCTACACGTTCACGG | GGGTGTCGAGGGAAAAATAGG |
|
N-Cadherin Vimentin Twist-1 |
TCAGGCGTCTGTAGAGGCTT GACGCCATCAACACCGAGTT GTCCGCAGTCTTACGAGGAG |
ATGCACATCCTTCGATAAGACTG CTTTGTCGTTGGTTAGCTGGT GCTTGAGGGTCTGAATCTTGCT |
| GAPDH | AGGTCGGTGTGAACGGATTTG | GGGGTCGTTGATGGCAACA |
Western Blot Assays
Total protein in liver cancer cells and the tumor tissues of nude mice were extracted using NP-40 (P0013F, Beyotime). In brief, 250 µL of NP-40 was used to incubate 3.5 × 106 liver cancer cells and 20 mg of tumor tissues for 15 min at 4°C. After the cells were centrifuged for 30 min (14,000×g), the total protein (supernatant) was collected, and its concentration was subsequently measured using a BCA kit (P0009, Beyotime). Afterwards, 40 µg of the total protein and 2 µL of marker (PR1910, Solarbio, Beijing, China) were added into each lane on SDS-PAGE gels (P0052A, Beyotime). The protein was then separated from the gels by electrophoresis at 35 V for 10 min and at 100 V for 90 min, and transferred to PVDF membranes (ISEQ00010, Millipore, MA, USA) which had been incubated with methanol (M813895, Macklin). After being blocked with 5% non-fat milk for 2 h, the membranes were incubated with the following primary antibodies at 4°C overnight: EZH2 (1:2000, 85kD, ab186006, Abcam), E-Cadherin (1:3000, 110kD, ab1416, Abcam), Vimentin (1:1000, 54kD, ab92547, Abcam), N-Cadherin (1:2500, 130kD, ab18203, Abcam), Twist-1 (1:1000, 22kD, ab49254, Abcam), p-STAT3 (1:2500, 88kD, ab76315, Abcam), STAT3 (1:5000, 88kD, ab119352, Abcam), p-PI3K (1:1500, 85kD, #4228, Cell Signaling Technology, MA, USA), PI3K (1:1500, 85kD, #4292, Cell Signaling Technology), p-AKT (1:1000, 56kD, ab38449, Abcam), AKT (1:2500, 56kD, ab179463, Abcam), β-actin (1:5000, 42kD, ab179467, Abcam), and GAPDH (1:5000, 36kD, ab181602, Abcam). The next day, the corresponding secondary antibody (1:5000, ab205718, Abcam) was used to incubate the membranes for 2 h at normal atmosphere temperature. Finally, after the surface of each membrane was covered with a developer solution (P0019, Beyotime), Image Lab 3.0 Software (Bio-Ras, CA, USA) was used to detect the signal of the membranes and perform densitometric analysis of the protein.
Statistical Analysis
All data involved in this study were analyzed by Student’s t-test and one-way ANOVA using SPSS 20.0 software. LSD and Dunnet’s were still used as post-hoc tests using SPSS 20.0 software. Statistical data were expressed as mean ± standard deviation. P < 0.05 was considered as statistically significant.
Results
EZH2 Was High-Expressed in Liver Cancer and Associated with Poor Survival
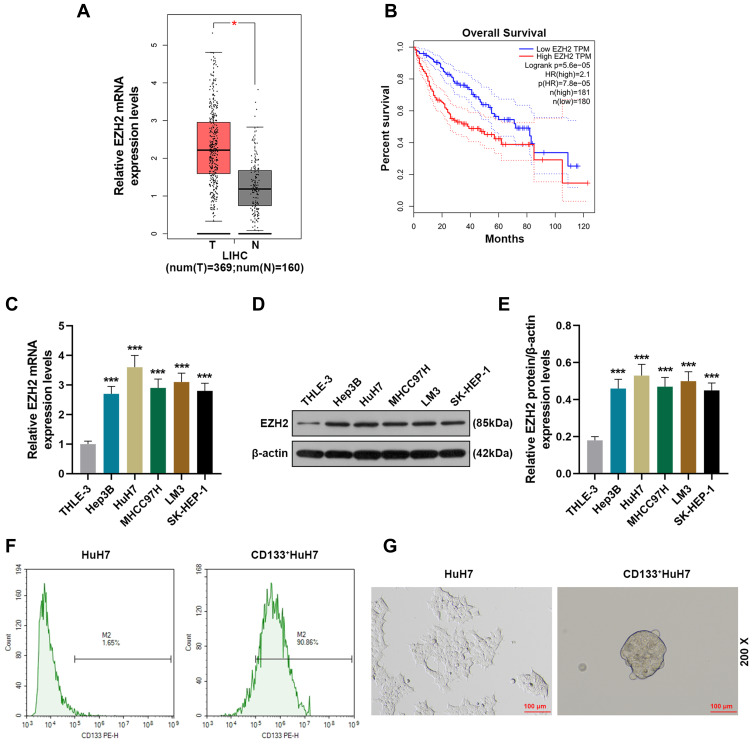
We first analyzed 369 tissue samples of liver hepatocellular carcinoma (LIHC) and 160 normal tissue samples retrieved from the TCGA database through GEPIA (http://gepia.cancer-pku.cn/index.html) to evaluate the level of EZH2 in liver cancer, and the analysis results showed that EZH2 was highly expressed in LIHC tissues as compared with normal tissues (Figure 1A). In addition, we further analyzed the overall survival of LIHC patients. As shown in Figure 1B, the overall survival of the patients with high expression of EZH2 was poorer than those with low expression of EZH2. Mechanically, we detected the expression of EZH2 in a series of liver cancer cell lines (Figure 1C-E). Similarly, it turned out that the transcription and translation levels of EZH2 in liver cancer cells were significantly up-regulated compared with normal liver epithelial cells (THLE-3). Considering that EZH2 had the highest expression in HuH7 cells among the liver cancer cell lines, HuH7 cells were chosen for the next experiments.
Figure 1.
EZH2 was high-expressed in liver cancer and associated with poor survival and CD133+ HuH7 cells had a strong sphere-forming ability. (A) The expression of EZH2 in liver cancer tissues retrieved from the TCGA database was analyzed using GEPIA (http://gepia.cancer-pku.cn/index.html) (*P< 0.001, vs T). (B) The overall survival of liver cancer patients retrieved from the TCGA database was analyzed. (C) The expressions of EZH2 in normal liver epithelial cells and liver cancer cells were detected by RT-qPCR; GAPDH was used as an internal control (***P< 0.001, vs THLE3).(D–E) The expressions of EZH2 in normal liver epithelial cells and liver cancer cells were detected by Western blot; β-actin was used as an internal control (***P< 0.05, vs THLE3). (F) CD133+HuH7 cells in HuH7 cells were sorted out by flow cytometry. (G) The sphere-forming abilities of HuH7 and CD133+HuH7 cells were detected by tumor sphere formation assay (200× Magnification).
Abbreviations: LIHC, liver hepatocellular carcinoma; T, tumor tissues.
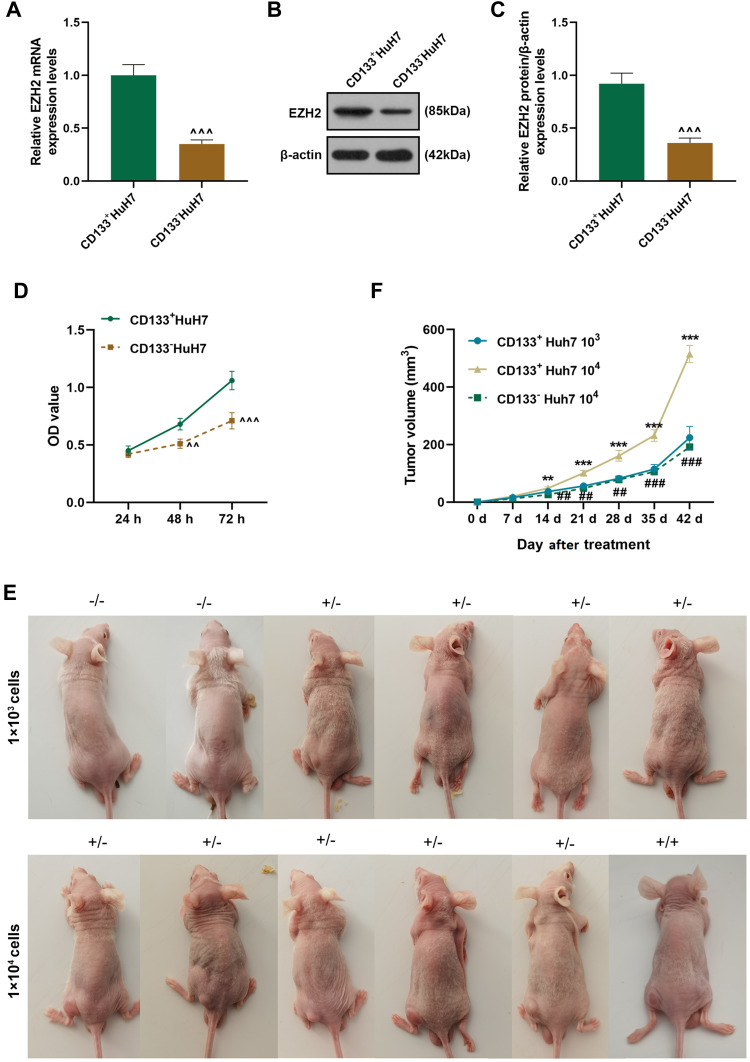
CD133+ HuH7 Cells with High Expression of EZH2 Had High Viability and Strong Sphere-Forming and Tumor-Forming Abilities
Considering that CD133 is a marker of liver cancer stem cells, flow cytometry was used to sortCD133+HuH7 cells (Figure 1F). Tumor sphere formation assay (Figure 1G) was performed to conduct a long-term culture of CD133+HuH7 cells with cancer stem cell-like properties. After 14 days of culture, the cells grew in aggregate clusters, and their size and number were both increased. These cells were collected and cultured for use in the following experiments. Then the expression levels of EZH2 in CD133+HuH7 cells and CD133−HuH7 cells were further detected. As shown in Figure 2A-C, the transcription and translation levels of EZH2 in CD133−HuH7 cells were significantly lower than those in CD133+HuH7 cells. In addition, the viability of CD133−HuH7 cells was also lower than that of CD133+HuH7 cells after culture for 48 and 72 h (Figure 2D). To examine the tumor-forming ability of CD133+HuH7 cells and CD133−HuH7 cells, the subcutaneously xenograft tumor model was established (Figure 2E-F). It was found that tumors grew in 4 mice which injected with 1 × 103 CD133+HuH7 cells, no any tumors grew in mice which injected with 1 × 103 CD133−HuH7 cells, tumors grew in all mice which injected with 1 × 104 CD133+HuH7 cells, only one tumor grew in mice which injected with 1 × 104 CD133−HuH7 cells (Figure 2E). Besides, the tumor volume in mice injected with 104 CD133+HuH7 cells was bigger than the tumor volume in mice injected with 103 CD133+HuH7 cells, and the tumor volume in mice injected with CD133+HuH7 cells was bigger than the tumor volume in mice injected with CD133−HuH7 cells (Figure 2F). Therefore, CD133+HuH7 cells were used for further experiments in this study.
Figure 2.
CD133+ HuH7 cells with high expression of EZH2 had high viability and a strong tumor-forming ability. (A) The expressions of EZH2 in CD133+HuH7 and CD133−HuH7 cells were detected by RT-qPCR; GAPDH was used as an internal control. (B–C) The expressions of EZH2 in CD133+HuH7 and CD133−HuH7 cells were detected by Western blot; β-actin was used as an internal control. (D) The viabilities of CD133+HuH7 and CD133−HuH7 cells were detected by MTT assay. (E) The xenograft tumor nude mice model was established by subcutaneous injection of CD133+HuH7 and CD133−HuH7 cells.(F) The tumor volume of the xenograft tumor nude mice was calculated every 7 days. (^^P< 0.01, ^^^P< 0.001, vs CD133+HuH7, **P< 0.01, ***P< 0.001, vs CD133+HuH7 103, ##P< 0.01, ###P< 0.001, vs CD133+HuH7 104).
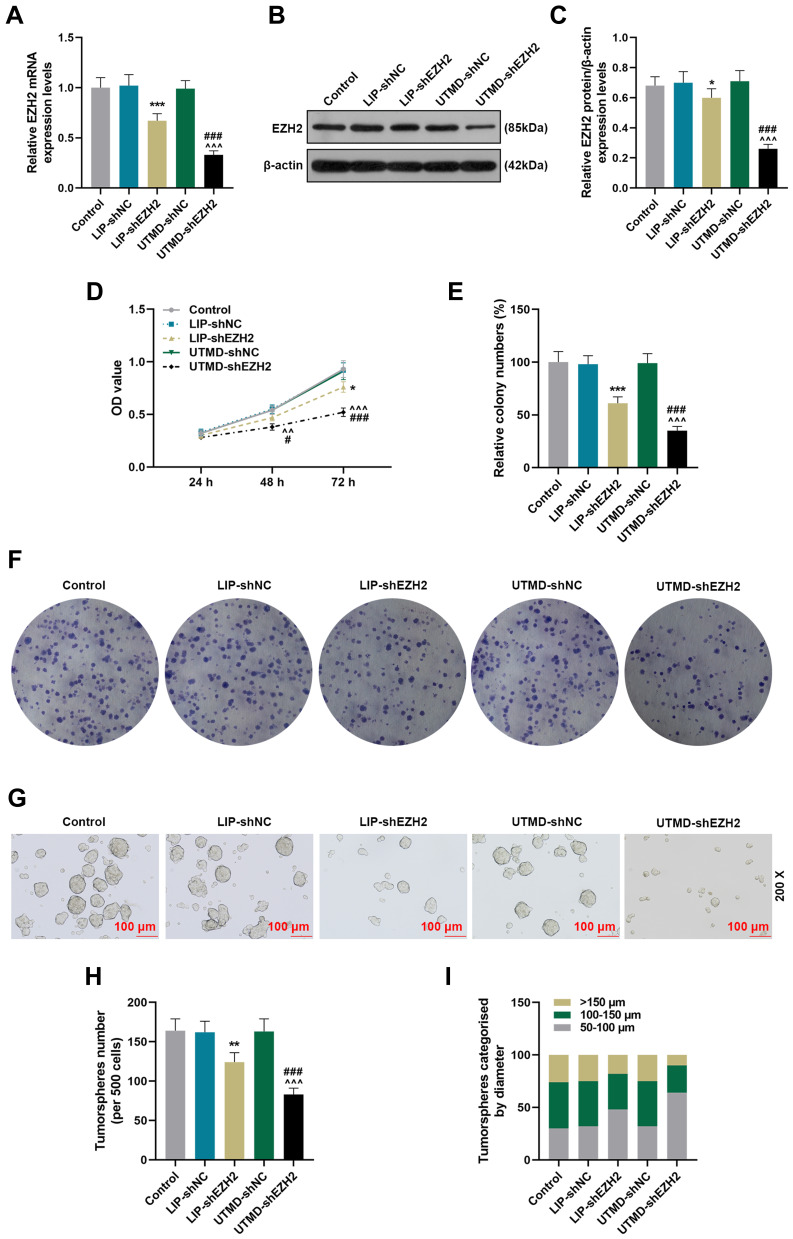
The Inhibitory Effect of UTMD-shEZH2 on the Viability, Proliferation, and Tumor Sphere Formation of CD133+ HuH7 Cells Was Stronger Than That of LIP-shEZH2
To compare the transfection efficiency of UTMD and LIP and detect the effect of EZH2 on CD133+ HuH7 cells, we transfected shEZH2 into CD133+ HuH7 cells through UTMD and LIP. As shown in Figure 3A-C, the gene and protein expressions of EZH2 were both decreased by LIP-shEZH2 and UTMD-shEZH2 as compared with LIP-shNC and UTMD-shNC respectively; and the effect of UTMD-shEZH2 on EZH2 expression was stronger than that of LIP-shEZH2, which revealed that the transfection efficiency of UTMD was better than LIP. Then we detected the changes in the viability and proliferation of CD133+ HuH7 cells after transfection with the two methods. As shown in Figure 3D-F, the viability and relative colony number of CD133+ HuH7 cells were decreased by LIP-shEZH2 and UTMD-shEZH2 as compared with LIP-shNC and UTMD-shNC respectively; and the effect of UTMD-shEZH2 was stronger than that of LIP-shEZH2. The results of tumorsphere formation assay (Figure 3G-I) demonstrated that the number and size of the tumorspheres of CD133+ HuH7 cells were also decreased by LIP-shEZH2 and UTMD-shEZH2 as compared with LIP-shNC and UTMD-shNC respectively; and the effect of UTMD-shEZH2 was stronger than that of LIP-shEZH2.
Figure 3.
The inhibitory effect of UTMD-shEZH2 on the viability, proliferation, and tumorsphere formation of CD133+ HuH7 cells was stronger than that of LIP-shEZH2. (A) The transfection efficiencies of UTMD-shEZH2 and LIP-shEZH2 were evaluated by RT-qPCR; GAPDH was used as an internal control. (B–C) The transfection efficiencies of UTMD-shEZH2 and LIP-shEZH2 were evaluated by Western blot; β-actin was used as an internal control. (D) The viability of CD133+ HuH7 cells was detected by MTT assay. (E–F) The proliferation of CD133+ HuH7 cells was detected by colony formation assay. (G–I) The tumorsphere formation ability of CD133+ HuH7 cells was detected by tumorsphere formation assay (200× Magnification). (*P< 0.05, **P< 0.01, ***P< 0.001, vs LIP-shNC; ^^P< 0.01, ^^^P< 0.001, vs UTMD-shNC; #P< 0.05, ###P< 0.001, vs LIP-shEZH2).
Abbreviations: UTMD, ultrasound-targeted microbubble destruction; LIP, liposome; NC, negative control.
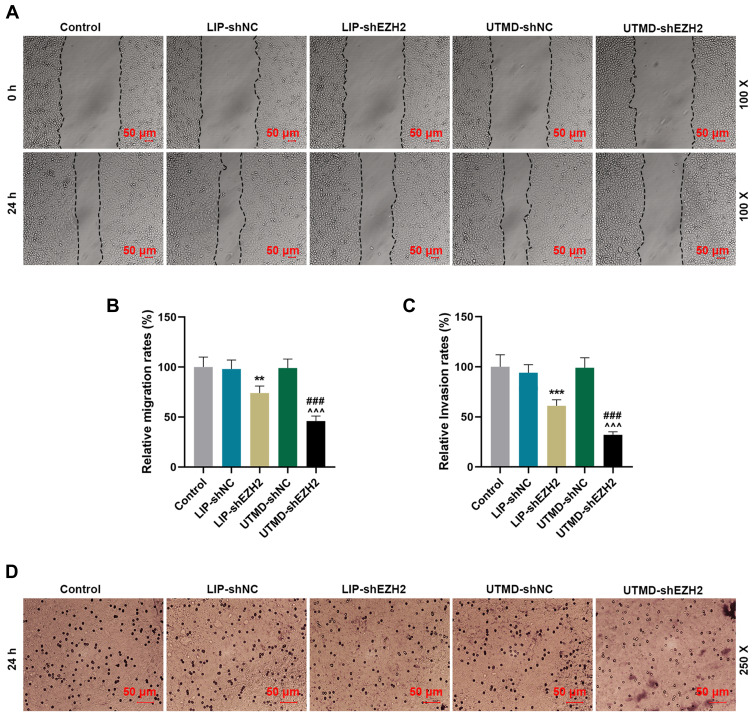
The Inhibitory Effect of UTMD-shEZH2 on the Migration and Invasion of CD133+ HuH7 Cells Was Stronger Than That of LIP-shEZH2
We further detected the changes in the ability of CD133+ HuH7 cells to migrate and invade after transfection. As shown in Figure 4A-B, the relative migration rate of CD133+ HuH7 cells was decreased by LIP-shEZH2 and UTMD-shEZH2 as compared with LIP-shNC and UTMD-shNC respectively; and the effect of UTMD-shEZH2 on the migration of CD133+ HuH7 cells was stronger than that of LIP-shEZH2. As shown Figure 4C-D, the relative invasion rate of CD133+ HuH7 cells was also decreased by LIP-shEZH2 and UTMD-shEZH2 as compared with LIP-shNC and UTMD-shNC respectively; and the effect of UTMD-shEZH2 on the invasion of CD133+ HuH7 cells was stronger than that of LIP-shEZH2.
Figure 4.
The inhibitory effect of UTMD-shEZH2 on the migration and invasion of CD133+ HuH7 cells was stronger than that of LIP-shEZH2. (A–B) The migration of CD133+ HuH7 cells was detected by wound healing assay (100× Magnification). (C–D) The invasion of CD133+ HuH7 cells was detected by transwell assay (250× Magnification). (**P< 0.01, ***P< 0.001, vs LIP-shNC; ^^^P< 0.001, vs UTMD-shNC; ###P< 0.001, vs LIP-shEZH2).
Abbreviations: UTMD, ultrasound-targeted microbubble destruction; LIP, liposome; NC, negative control.
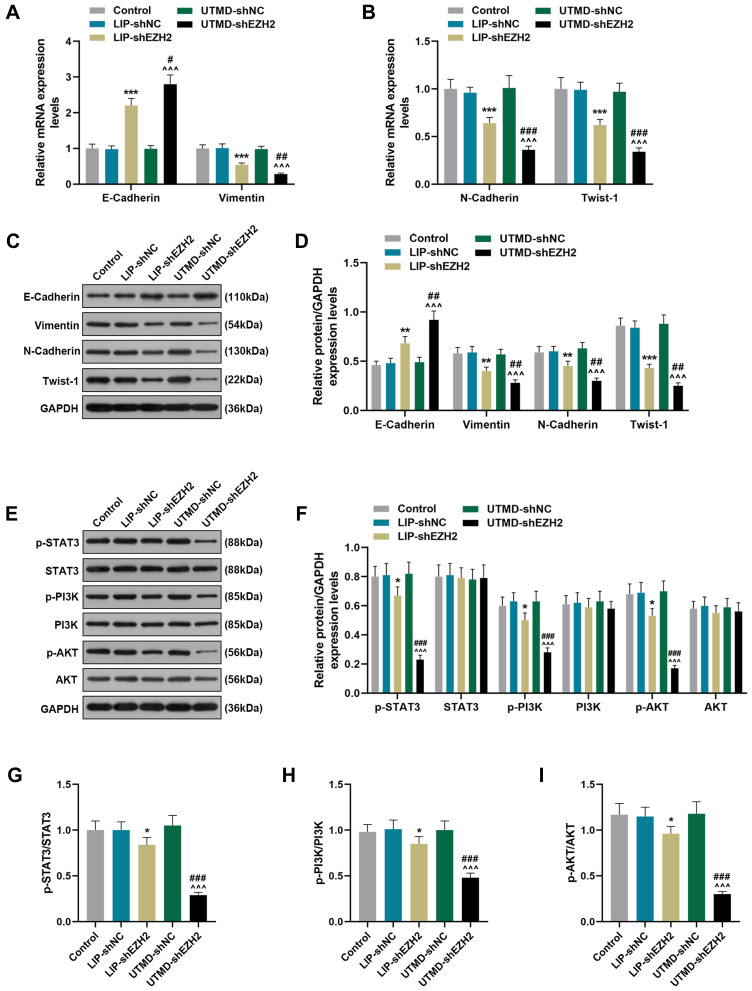
The Regulatory Effect of UTMD-shEZH2 on the Expressions of EMT-Related Factors and the Activation of the STAT3/PI3K/AKT Pathway in CD133+ HuH7 Cells Was Stronger Than That of LIP-shEZH2
To further verify the effect of EZH2 on CD133+ HuH7 cells and the potential mechanisms, we detected the expressions of key factors. As shown in Figure 5A-D, LIP-shEZH2 and UTMD-shEZH2 increased the expression of E-Cadherin and decreased the expressions of Vimentin, N-Cadherin, and Twist-1 at both the transcription and translation levels as compared with LIP-shNC and UTMD-shNC respectively; and the effect of UTMD-shEZH2 on the expressions of these factors was stronger than that of LIP-shEZH2. As shown Figure 5E-F, LIP-shEZH2 and UTMD-shEZH2 both down-regulated the expressions of p-STAT3, p-PI3K, and p-AKT as compared with LIP-shNC and UTMD-shNC respectively; and the effect of UTMD-shEZH2 on the expressions of these factors was stronger than that of LIP-shEZH2. In addition, the ratios of p-STAT3/STAT3, p-PI3K/PI3K, and p-AKT/AKT presented the same tendencies as p-STAT3, p-PI3K, and p-AKT did among all groups (Figure 5G-I), which indicated that shEZH2 could inhibit the activation of the STAT3/PI3K/AKT pathway in CD133+ HuH7 cells, and the effect of UTMD-shEZH2 was stronger than that of LIP-shEZH2.
Figure 5.
The regulatory effect of UTMD-shEZH2 on the expressions of EMT-related factors expressions and the activation of the STAT3/PI3K/AKT pathway in CD133+ HuH7 cells was stronger than that of LIP-shEZH2. (A–B) The expressions of E-Cadherin, Vimentin, N-Cadherin, and Twist-1 in CD133+ HuH7 cells were detected by RT-qPCR; GAPDH was used as an internal control. (C–D) The expression of E-Cadherin, Vimentin, N-Cadherin, and Twist-1 in CD133+ HuH7 cells were detected by Western blot; GAPDH was used as an internal control. (E–I) The expression of p-STAT3, STAT3, p-PI3K, PI3K, p-AKT, and AKT in CD133+ HuH7 cells were detected by Western blot; GAPDH was used as an internal control. (*P< 0.05, **P< 0.01, ***P< 0.001, vs LIP-shNC; ^^^P< 0.001, vs UTMD-shNC; #P< 0.05, ##P< 0.01, ###P< 0.001, vs LIP-shEZH2).
Abbreviations: UTMD, ultrasound-targeted microbubble destruction; EMT, epithelial-mesenchymal transition; LIP, liposome; NC, negative control.
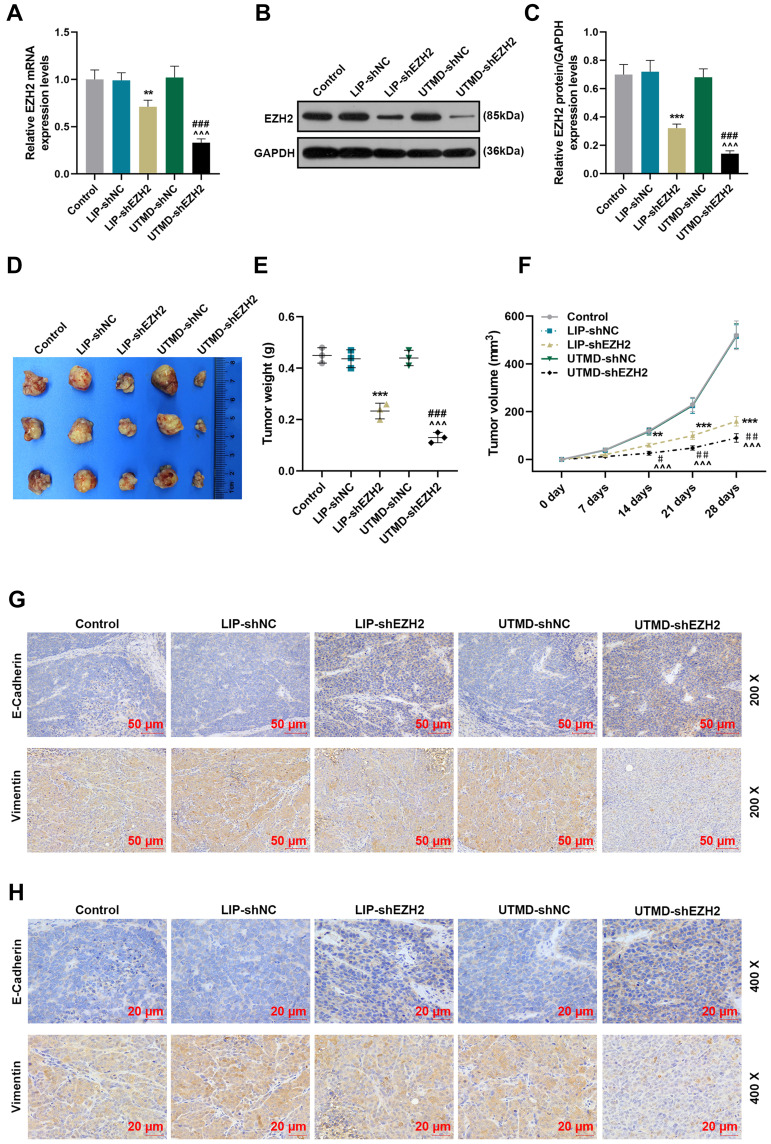
The Regulatory Effect of UTMD-shEZH2 on the Expressions of EZH2, E-Cadherin, and Vimentin and the Tumorigenicity of CD133+ HuH7 Cells in vivo Was Stronger Than That of LIP-shEZH2
To verify the above findings in vivo, we further established the xenograft tumor model by subcutaneously injecting CD133+ HuH7 cells into the nude mice. After the tumors were harvested, the expression of EZH2 was first detected. As exhibited in Figure 6A-C, the transcription and translation levels of EZH2 were inhibited by LIP-shEZH2 and UTMD-shEZH2 as compared with LIP-shNC and UTMD-shNC respectively; and the effect of UTMD-shEZH2 was stronger than that of LIP-shEZH2. In addition, the tumor morphology (Figure 6D), weight (Figure 6E), and volume (Figure 6F) were also documented. LIP-shEZH2 and UTMD-shEZH2 also reduced the tumor weight and volume as compared with LIP-shNC and UTMD-shNC respectively; and the effects of UTMD-shEZH2 on tumor weight and volume were stronger than those of LIP-shEZH2 (Figure 6E-F). Then, the expressions of E-Cadherin and Vimentin in tumor tissues were detected, and the results revealed that LIP-shEZH2 and UTMD-shEZH2 up-regulated the expression of E-Cadherin and down-regulated the expression of Vimentin as compared with LIP-shNC and UTMD-shNC respectively; and the effects of UTMD-shEZH2 on the expressions of E-Cadherin and Vimentin were stronger than those of LIP-shEZH2 (Figure 6G-H).
Figure 6.
The regulatory effect of UTMD-shEZH2 on the expressions of EZH2, E-Cadherin, and Vimentin and the tumorigenicity of CD133+ HuH7 cells in vivo was stronger than that of LIP-shEZH2. (A) The expression of EZH2 in the tumor tissues of nude mice was detected by RT-qPCR; GAPDH was used as an internal control. (B–C) The expression of EZH2 in the tumor tissues of nude mice was detected by Western blot; GAPDH was used as an internal control. (D) The tumor morphology of nude mice was photographed with a camera. (E) The tumor weights of the nude mice were measured and recorded with an electronic balance. (F) The tumor volume of the nude mice was measured and recorded using a vernier caliper. (G–H) The expressions of E-Cadherin and Vimentin in the tumor tissues of nude mice were detected by immunohistochemical (200× and 400× Magnification). (**P< 0.01, ***P< 0.001, vs LIP-shNC; ^^^P< 0.001, vs UTMD-shNC; #P< 0.05, ##P< 0.01, ###P< 0.001, vs LIP-shEZH2).
Abbreviations: UTMD, ultrasound-targeted microbubble destruction; LIP, liposome; NC, negative control.
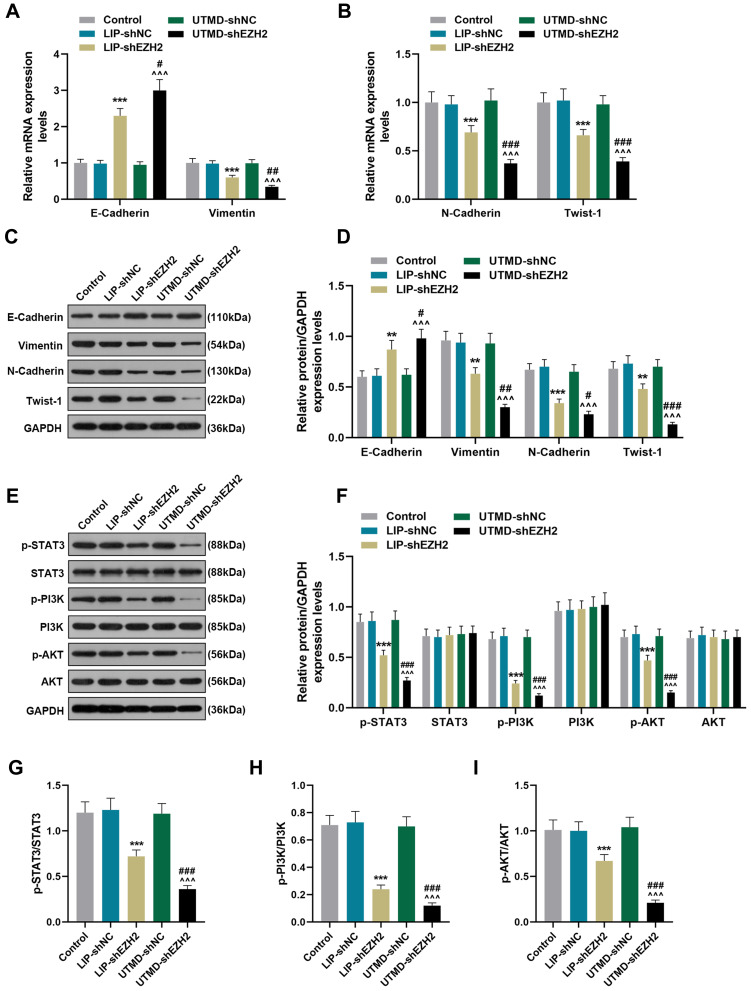
The Regulatory Effect of UTMD-shEZH2 on the Expressions of EMT-Related Factors and the Activation of the STAT3/PI3K/AKT Pathway of CD133+ HuH7 Cells in vivo Was Stronger Than That of LIP-shEZH2
As shown in Figure 7A-D, LIP-shEZH2 and UTMD-shEZH2 increased the gene and protein expressions of E-Cadherin and decreased the expressions of Vimentin, N-Cadherin, and Twist-1 in tumor tissues as compared with LIP-shNC and UTMD-shNC respectively; and the effect of UTMD-shEZH2 on the expressions of these factors was stronger than that of LIP-shEZH2. As shown Figure 7E-F, LIP-shEZH2 and UTMD-shEZH2 down-regulated the expressions of p-STAT3, p-PI3K, and p-AKT in tumor tissues as compared with LIP-shNC and UTMD-shNC respectively; and the effect of UTMD-shEZH2 on the expressions of these factors was stronger than that of LIP-shEZH2. In addition, the ratios of p-STAT3/STAT3, p-PI3K/PI3K, and p-AKT/AKT had the same tendency as p-STAT3, p-PI3K, and p-AKT did among all groups (Figure 7G-I), which further verified that shEZH2 inhibited the activation of the STAT3/PI3K/AKT pathway, and the effect of UTMD-shEZH2 was stronger than that of LIP-shEZH2.
Figure 7.
The regulatory effect of UTMD-shEZH2 on the expressions of EMT-related factors and the activation of the STAT3/PI3K/AKT pathway of CD133+ HuH7 cells in vivo was stronger than that of LIP-shEZH2. (A–B) The expressions of E-Cadherin, Vimentin, N-Cadherin, and Twist-1 in tumor tissues were detected by RT-qPCR; GAPDH was used as an internal control. (C–D) The expressions of E-Cadherin, Vimentin, N-Cadherin, and Twist-1 in tumor tissues were detected by Western blot; GAPDH was used as an internal control. (E–I) The expressions of p-STAT3, STAT3, p-PI3K, PI3K, p-AKT, and AKT in tumor tissues were detected by Western blot; GAPDH was used as an internal control. (**P< 0.01, ***P< 0.001, vs LIP-shNC; ^^^P< 0.001, vs UTMD-shNC; #P< 0.05, ##P< 0.01, ###P< 0.001, vs LIP-shEZH2).
Abbreviations: UTMD, ultrasound-targeted microbubble destruction; EMT, epithelial-mesenchymal transition; LIP, liposome; NC, negative control.
Discussion
CSCs, with the biological characteristics of self-renewal, replication, and high tumorigenicity, are considered to be related to the occurrence, development, recurrence, metastasis and drug resistance of malignant tumors.24,25 Some characteristic surface antigens have been found present on CSCs, such as CD13, CD44, and CD133.26,27 It was reported that CD133 was widely expressed in leukemia, liver cancer, lung cancer, prostate cancer and other CSCs, and could be used to identify and isolate a variety of CSCs including liver CSCs.28 CD133 functions as an important marker for CSCs, and has been proved to be closely related to the occurrence, development and prognosis of cancers.29 Therefore, we first sorted CD133+ HuH7 cells in this study. Previous research reported that CD133+ liver CSCs isolated from liver cancer cell lines have a stronger ability to form tumor and proliferate than CD133− liver cancer cells.29,30 Consistent with this finding, the CD133+ HuH7 cells sorted in this study also displayed a better ability to form spheres and proliferate than CD133− HuH7 cells in vitro and in vivo, which indicated that the liver CSCs in HuH7 cells (CD133+ HuH7) were successfully sorted and cultured.
Research has proved that cancer cells could show the characteristics of CSCs after EMT, a crucial process in embryo development and cancer invasion and metastasis; in addition, EZH2 was widely reported to have a regulatory effect on the progression of EMT in different cancers.16,17 EZH2 is a catalytic subunit of polycomb complex 2, and plays a key role in chromatin structure modification.31 It was also reported that EZH2 was highly expressed in many diseases including liver cancer.32 In this study, we further proved that EZH2 had a high expression level in liver cancer, which arouse our curiosity about whether the strong tumorigenicity of CD133+ HuH7 cells could be mediated by the regulatory effect of EZH2 on EMT.
We then transfected shEZH2 into CD133+ HuH7 cells to further detect the effect of EZH2 on the stemness (strong ability to proliferate, form tumor sphere, and migrate) of CD133+ HuH7 cells. There are many types of transfection and drug delivery methods, such as liposome-mediated transfection and drug delivery, calcium-phosphate-mediated transfection and drug delivery, and nanostructure-mediated transfection and drug delivery,33,34 and liposome-mediated transfection and drug delivery is the most widely used method in the laboratory.35 In recent decades, UTMD-mediated transfection has been discovered as a novel approach for transfection and drug delivery. However, although efforts have been made to explore the value of UTMD in transfection given its advantages of low immunogenicity, low toxicity, reusability, and easy operation,21,22 more research on UTMD-mediated transfection is still needed to ensure its effect. In this study, therefore, we employed both liposome transfection and UTMD to transfect shEZH2 into CD133+ HuH7 cells, and found that UTMD had a higher transfection efficiency than liposome transfection, which was consistent with previous research.36 In addition, we also found that shEZH2 decreased the strong ability of CD133+ HuH7 cells to proliferate and form tumor spheres in vitro, and reduced the weight and volume of the xenotransplant tumor in vivo, which confirmed that EZH2 did play a key role in regulating the tumorigenicity and stemness of CD133+ HuH7 cells. We then detected the migration and invasion of CD133+ HuH7 cells, and the results further revealed that shEZH2 decreased the ability of CD133+ HuH7 cells to migrate and invade; what’s more, the effect of UTMD-mediated shEZH2 was stronger than that of liposome-mediated shEZH2. During EMT, cancer cells lost the characteristics of epithelial cells and obtained the nature of mesothelial cells, which enhanced the invasion and metastasis abilities of cancer cells.13 Therefore, cancer cells would show the characteristics of CSCs after EMT; besides, these CSCs would lose the markers for epithelial cells (such as E-Cadherin) and further express the markers for mesothelial cells (such as N-Cadherin, Vimentin, and Twist-1).12 In this study, we found that shEZH2 decreased the expressions of N-Cadherin, Vimentin, and Twist-1 while increasing that of E-Cadherin in both CD133+ HuH7 cells and xenotransplant tumor tissues. These results confirmed that EZH2 regulated the progression of EMT both in vitro and in vivo during which the characteristics of CSCs in CD133+ HuH7 cells were potentiated, which further verified the regulatory effect of EZH2 on the tumorigenicity and stemness of CD133+ HuH7 cells. Furthermore, the effect of UTMD-mediated shEZH2 on the progression of EMT in CD133+ HuH7 cells was stronger than that of liposome-mediated shEZH2.
The mechanism of CD133 + cells in maintaining the characteristics of stem cells as well as the regulatory mechanisms of EMT have been substantially discussed.14,37 Many genes and related signal pathways are found involved in the gene expression profile of CD133 + subpopulation of various tumors, including the STAT3/PI3K/AKT pathway.14,38 In addition, the STAT3/PI3K/AKT pathway was also closely related to EZH2-mediated EMT progression.31,39 To understand the mechanism revealed in this study more clearly, or at least partly, we detected the activation of the STAT3/PI3K/AKT pathway in both CD133+ HuH7 cells and xenotransplant tumor tissues, and the results exhibited that shEZH2 inhibited the activation of the STAT3/PI3K/AKT pathway both in vitro and in vivo. In addition, the effect of UTMD-mediated shEZH2 on the STAT3/PI3K/AKT pathway was stronger than that of liposome -mediated shEZH2.
However, there are some other limitations in our current study that should be noted. Only one cell line was used in the in vitro experiments, if the current discoveries were proved in two or more cells lines, the results would be more convincing. Also, we solely centered on the role of EZH2on liver CSCs in vitro, but the effects in vivo needed to be addressed in detail. Future researches, therefore, are urgently required.
To sum up, through the investigation of the effect of EZH2 on CD133+ HuH7 cells based on the two types of transfection method, we not only proved the effect of EZH2 on liver CSCs, but also confirmed that the efficiency of UTMD-mediated transfection is better than LIP-mediated transfection; and these findings indicate that both UTMD-mediated transfection and drug delivery might be developed into promising methods for the treatment of diseases.
Conclusions
Based on the findings in our present research, we could conclude that UTMD-mediated shEZH2 inhibited the stemness and EMT of CD133+ HuH7 cells.
Funding Statement
No funding was received.
Ethics Approval and Informed Consent
All animal experiments were performed in accordance with the guidelines of the China Council on Animal Care and Use. This research was approved by the Committee of Experimental Animals of The First Affiliated Hospital of Zhengzhou University Hospital (Z20190603G). All experiments involving animals were performed in The First Affiliated Hospital of Zhengzhou University.
Disclosure
The authors declare that they have no conflicts of interest.
References
- 1.Song HJ, Jiang X, Henry L, Nguyen MH, Park H. Proton pump inhibitors and risk of liver cancer and mortality in patients with chronic liver disease: a systematic review and meta-analysis. Eur J Clin Pharmacol. 2020;14(10):020–02854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin Z-L, Ding J, Sun G-P, et al. Application of Paclitaxel-loaded EGFR peptide-conjugated magnetic polymeric liposomes for liver cancer therapy. Curr Med Sci. 2020;40(1):145–154. doi: 10.1007/s11596-020-2158-4 [DOI] [PubMed] [Google Scholar]
- 3.Ruman U, Fakurazi S, Masarudin MJ, Hussein MZ. Nanocarrier-based therapeutics and theranostics drug delivery systems for next generation of liver cancer nanodrug modalities. Int J Nanomedicine. 2020;15:1437–1456. doi: 10.2147/IJN.S236927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lo KM, Wu VW, Li Y, Jun XH. Factors affecting target motion in stereotactic body radiotherapy of liver cancer using CyberKnife. J Med Imaging Radiat Oncol. 2020;15(10):1754–9485. [DOI] [PubMed] [Google Scholar]
- 5.Liu Q, Sun J, Luo Q, Ju Y, Song G. Salinomycin suppresses tumorigenicity of liver cancer stem cells and Wnt/beta-catenin signaling. Curr Stem Cell Res Ther. 2020;23:103860. [DOI] [PubMed] [Google Scholar]
- 6.Wang LT, Wang SN, Chiou SS, et al. TIP60-dependent acetylation of the SPZ1-TWIST complex promotes epithelial-mesenchymal transition and metastasis in liver cancer. Oncogene. 2019;38(4):518–532. doi: 10.1038/s41388-018-0457-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li N, Zhu Y. Targeting liver cancer stem cells for the treatment of hepatocellular carcinoma. Therap Adv Gastroenterol. 2019;12:1756284818821560. doi: 10.1177/1756284818821560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishiguro K, Yan IK, Lewis-Tuffin L, Patel T. Targeting liver cancer stem cells using engineered biological nanoparticles for the treatment of hepatocellular cancer. Hepatol Commun. 2020;4(2):298–313. doi: 10.1002/hep4.1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inamura K, Komizu Y, Yamakuchi M, Ishida S, Matsumoto Y, Matsushita T. Inhibitory effect of hybrid liposomes on the growth of liver cancer stem cells. Biochem Biophys Res Commun. 2019;509(1):268–274. doi: 10.1016/j.bbrc.2018.12.118 [DOI] [PubMed] [Google Scholar]
- 10.Zhu Y, Tang H, Zhang L, et al. Suppression of miR-21-3p enhances TRAIL-mediated apoptosis in liver cancer stem cells by suppressing the PI3K/Akt/Bad cascade via regulating PTEN. Cancer Manag Res. 2019;11:955–968. doi: 10.2147/CMAR.S183328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H, Unternaehrer JJ. Epithelial-mesenchymal transition and cancer stem cells: at the crossroads of differentiation and dedifferentiation. Dev Dyn. 2019;248(1):10–20. doi: 10.1002/dvdy.24678 [DOI] [PubMed] [Google Scholar]
- 12.Tao C, Huang K, Shi J, Hu Q, Li K, Zhu X. Genomics and prognosis analysis of epithelial-mesenchymal transition in glioma. Front Oncol. 2020;10:183. doi: 10.3389/fonc.2020.00183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishiwata T. Cancer stem cells and epithelial-mesenchymal transition: novel therapeutic targets for cancer. Pathol Int. 2016;66(11):601–608. doi: 10.1111/pin.12447 [DOI] [PubMed] [Google Scholar]
- 14.Jin W. Role of JAK/STAT3 signaling in the regulation of metastasis, the transition of cancer stem cells, and chemoresistance of cancer by epithelial-mesenchymal transition. Cells. 2020;9:1. doi: 10.3390/cells9010217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang SW, Zhang ZG, Hao YX, et al. HIF-1alpha induces the epithelial-mesenchymal transition in gastric cancer stem cells through the Snail pathway. Oncotarget. 2017;8(6):9535–9545. doi: 10.18632/oncotarget.14484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Q, Dong P, Liu X, Sakuragi N, Guo SW. Enhancer of Zeste homolog 2 (EZH2) induces epithelial-mesenchymal transition in endometriosis. Sci Rep. 2017;7(1):017–06920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gan L, Xu M, Hua R, et al. The polycomb group protein EZH2 induces epithelial-mesenchymal transition and pluripotent phenotype of gastric cancer cells by binding to PTEN promoter. J Hematol Oncol. 2018;11(1):017–0547. doi: 10.1186/s13045-017-0547-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma J, Zhang J, Weng YC, Wang JC. EZH2-mediated microRNA-139-5p regulates epithelial-mesenchymal transition and lymph node metastasis of pancreatic cancer. Mol Cells. 2018;41(9):868–880. doi: 10.14348/molcells.2018.0109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu J, Wang Y, Li Z, Wang Q, Zhou X, Wu W. Ultrasound-Targeted Microbubble Destruction (UTMD) combined with liposome increases the effectiveness of suppressing proliferation, migration, invasion, and Epithelial- Mesenchymal Transition (EMT) via Targeting Metadherin (MTDH) by ShRNA. Med Sci Monit. 2019;25:2640–2648. doi: 10.12659/MSM.912955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang H, Sun Y, Wei J, et al. The effects of ultrasound-targeted microbubble destruction (UTMD) carrying IL-8 monoclonal antibody on the inflammatory responses and stability of atherosclerotic plaques. Biomed Pharmacother. 2019;118(109161):30. doi: 10.1016/j.biopha.2019.109161 [DOI] [PubMed] [Google Scholar]
- 21.Huang S, Ren Y, Wang X, et al. Application of ultrasound-targeted microbubble destruction-mediated exogenous gene transfer in treating various renal diseases. Hum Gene Ther. 2019;30(2):127–138. doi: 10.1089/hum.2018.070 [DOI] [PubMed] [Google Scholar]
- 22.Zhang D, Yang L, Tian H, et al. [Enhancement of gene transfection efficiency and therapeutic effect of ultrasound-targeted microbubble destruction in vivo with cationic microbubble]. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2018;32(2):228–236. doi: 10.7507/1002-1892.201706058.Chinese. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu L, Wu X, Chen M, et al. The effects and mechanism of YK-4-279 in combination with docetaxel on prostate cancer. Int J Med Sci. 2017;14(4):356–366. doi: 10.7150/ijms.18382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arnold CR, Mangesius J, Skvortsova -I-I, Ganswindt U. The role of cancer stem cells in radiation resistance. Front Oncol. 2020;10:10. doi: 10.3389/fonc.2020.00164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scriba LD, Bornstein SR, Santambrogio A, et al. Cancer stem cells in pheochromocytoma and paraganglioma. Front Endocrinol. 2020;11:79. doi: 10.3389/fendo.2020.00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rozeik MS, Hammam OA, Ali AI, et al. Evaluation of CD44 and CD133 as markers of liver cancer stem cells in Egyptian patients with HCV-induced chronic liver diseases versus hepatocellular carcinoma. Electron Physician. 2017;9(7):4708–4717. doi: 10.19082/4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang H, Jin Y, Jin S, Tan Z, Peng Z, Kuang Y. Arsenite inhibits the function of CD133(+) CD13(+) liver cancer stem cells by reducing PML and Oct4 protein expression. Tumour Biol. 2016;37(10):14103–14115. doi: 10.1007/s13277-016-5195-7 [DOI] [PubMed] [Google Scholar]
- 28.Liu YM, Li XF, Liu H, Wu XL. Ultrasound-targeted microbubble destruction-mediated downregulation of CD133 inhibits epithelial-mesenchymal transition, stemness and migratory ability of liver cancer stem cells. Oncol Rep. 2015;34(6):2977–2986. doi: 10.3892/or.2015.4270 [DOI] [PubMed] [Google Scholar]
- 29.Ma S. Biology and clinical implications of CD133(+) liver cancer stem cells. Exp Cell Res. 2013;319(2):126–132. doi: 10.1016/j.yexcr.2012.09.007 [DOI] [PubMed] [Google Scholar]
- 30.Huang H, Hu M, Li P, Lu C, Li M. Mir-152 inhibits cell proliferation and colony formation of CD133(+) liver cancer stem cells by targeting KIT. Tumour Biol. 2015;36(2):921–928. doi: 10.1007/s13277-014-2719-x [DOI] [PubMed] [Google Scholar]
- 31.Lu DG, Tang QL, Wei JH, He FY, Lu L, Tang YJ. Targeting EZH2 by microRNA-449a inhibits osteosarcoma cell proliferation, invasion and migration via regulation of PI3K/AKT signaling pathway and epithelial-mesenchymal transition. Eur Rev Med Pharmacol Sci. 2020;24(4):1656–1665. doi: 10.26355/eurrev_202002_20339 [DOI] [PubMed] [Google Scholar]
- 32.Wang S, Cai L, Zhang F, Shang X, Xiao R, Zhou H. Inhibition of EZH2 attenuates sorafenib resistance by targeting NOTCH1 activation-dependent liver cancer stem cells via NOTCH1-related MicroRNAs in hepatocellular carcinoma. Transl Oncol. 2020;13(3):100741. doi: 10.1016/j.tranon.2020.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bertrand N, Wu J, Xu X, Kamaly N, Farokhzad OC. Cancer nanotechnology: the impact of passive and active targeting in the era of modern cancer biology. Adv Drug Deliv Rev. 2014;66:2–25. doi: 10.1016/j.addr.2013.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saw PE, Xu X, Zhang M, Cao S, Farokhzad OC, Wu J. Nanostructure engineering by simple tuning of lipid combinations. Angew Chem Int Ed Engl. 2020;59(15):6249–6252. doi: 10.1002/anie.201916574 [DOI] [PubMed] [Google Scholar]
- 35.Kumar P, Nagarajan A, Uchil PD. Selective agents for stable transfection. Cold Spring Harb Protoc. 2018;2018:9. doi: 10.1101/pdb.top096230 [DOI] [PubMed] [Google Scholar]
- 36.Wu Y, Sun T, Tang J, Liu Y, Li F. Ultrasound-targeted microbubble destruction enhances the antitumor efficacy of doxorubicin in a mouse hepatocellular carcinoma model. Ultrasound Med Biol. 2020;46(3):679–689. doi: 10.1016/j.ultrasmedbio.2019.09.017 [DOI] [PubMed] [Google Scholar]
- 37.Chen Q, Lin W, Yin Z, et al. Melittin inhibits hypoxia-induced vasculogenic mimicry formation and epithelial-mesenchymal transition through suppression of HIF-1alpha/Akt pathway in liver cancer. Evid Based Complement Alternat Med. 2019;1:9602935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding S-M, Lu A-L, Lu J-F, et al. Macrovascular endothelial cells enhance the motility of liver cancer cells by up-regulation of MMP-3, activation of Integrin/FAK signaling pathway and induction of non-classical epithelial-mesenchymal transition. J Cancer. 2020;11(8):2044–2059. doi: 10.7150/jca.38209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen MJ, Deng J, Chen C, Hu W, Yuan YC, Xia ZK. LncRNA H19 promotes epithelial mesenchymal transition and metastasis of esophageal cancer via STAT3/EZH2 axis. Int J Biochem Cell Biol. 2019;113:27–36. doi: 10.1016/j.biocel.2019.05.011 [DOI] [PubMed] [Google Scholar]