Abstract
Predicting the therapeutic response to ocular hypotensive drugs is crucial for the clinical treatment and management of glaucoma. Our aim was to identify a possible genetic contribution to the response to current pharmacological treatments of choice in a white Mediterranean population with primary open-angle glaucoma (POAG) or ocular hypertension (OH). We conducted a prospective, controlled, randomized, partial crossover study that included 151 patients of both genders, aged 18 years and older, diagnosed with and requiring pharmacological treatment for POAG or OH in one or both eyes. We sought to identify copy number variants (CNVs) associated with differences in pharmacological response, using a DNA pooling strategy of carefully phenotyped treatment responders and non-responders, treated for a minimum of 6 weeks with a beta-blocker (timolol maleate) and/or prostaglandin analog (latanoprost). Diurnal intraocular pressure reduction and comparative genome wide CNVs were analyzed. Our finding that copy number alleles of an intronic portion of the MLIP gene is a predictor of pharmacological response to beta blockers and prostaglandin analogs could be used as a biomarker to guide first-tier POAG and OH treatment. Our finding improves understanding of the genetic factors modulating pharmacological response in POAG and OH, and represents an important contribution to the establishment of a personalized approach to the treatment of glaucoma.
Subject terms: Genetic markers, Genetics, Biomarkers, Diseases, Health care, Medical research, Molecular medicine
Introduction
Glaucoma, a leading cause of irreversible blindness worldwide, is characterized by an optic nerve neuropathy that can develop at any age, with Mendelian inheritance typical for early-onset disease, and complex inheritance typical of adult-onset disease (before and after age 40, respectively)1. Primary open-angle glaucoma (POAG), the most prevalent form of the disease, is related to increased intraocular pressure (IOP). Its treatment poses challenges, and interestingly, clinical and therapeutic response can also be influenced by environmental and genetic factors2,3. Pharmacological treatment is the main therapeutic pillar for glaucoma, with topical beta-blockers and prostaglandin analogs as the first-line hypotensive drugs used to treat ocular hypertension (OH)4. Expected hypotensive capacity, according to several studies, is 20–25% for beta-blockers, and 25–35% for prostaglandins4–6. However, the response profile to pharmacological treatment differs in patients. The above-mentioned drugs (in monotherapy or in combination) have also been found to cause intolerance as a result of both topical and systemic undesirable adverse effects4.
Pharmacogenomics aims to study the way individual genome variations influence drug response. Genetic variations can affect drug bioavailability, metabolism, sensitivity, toxicity, and dosage. Detailed knowledge of drug pharmacogenetics enables patient management to be optimized by dosage customized according to an individual’s genetic profile7,8. The main medical value of this approach is that it is possible to predict, for each individual, whether a drug at a specific dose will be effective or whether it should be avoided due to a high risk of toxicity or lack of response8,9. To date, the most common and most extensively studied inter-individual genetic variations that shape pharmacological response are single nucleotide variations together with small insertions/deletions (indels). Other studies have focused on genome structural variations, including inversions, translocations, and copy number variants (CNVs), which have also been found to be a rich source of genetic variability10–12. The consequence of structural variation is usually that part of or a whole gene sequence is rearranged (inverted, duplicated, deleted, etc.), commonly resulting in interference with gene functioning. Functionally relevant CNVs have been previously described in genes related to drug absorption, distribution, metabolism, and excretion, including cytochrome P450 (CYP) and gluthatione S-trasferase (GST), with significantly different frequencies across human populations8,12,13. Also of interest are genome-wide association studies (GWAS), as these have successfully identified loci contributing to complex diseases and traits14–16, including pharmacogenomic response. However, apart from other limitations, the high cost and the need for very large cohorts have hampered the generalization of such approaches.
Recently, various studies have tried to elucidate a genetic basis for drug response to glaucoma treatment. In Chinese patients with glaucoma, for example, a differential response to latanoprost has been described for single-nucleotide polymorphisms (SNPs) in the prostaglandin F2-alpha receptor (PTGFR) and solute carrier organic anion transporter family member 2A1 (SLCO2A1) genes17, with the study concluding that 2 SNPs-rs3766355 in PTGFR and rs4241366 in SLCO2A1—correlated with a good drug response over both the short- and long-term17. More recently, a genotyping study of a cohort of Mediterranean patients with POAG identified 5 SNPs related to response to latanoprost, with PTGFR SNPs associated with good (rs6686438, rs10786455) and negative (rs3753380, rs6672484, rs11578155) responses. Interestingly, rs3753380 has previously been reported to be related to a poor response to latanoprost in healthy Japanese subjects18,19. The matrix metalloproteinase 1 (MMP-1) gene has also been related to refractoriness to latanoprost20.
Given the lack of studies exploring the contribution of CNVs to individual pharmacological treatment responses, our aim was to examine the effect of this type of genetic variation in the individual response to latanoprost (prostaglandin) or timolol maleate (beta-blocker) in a white Mediterranean population treated for POAG or OH. We observed that a CNV located in an intronic portion of the muscle-enriched A-type lamin-interacting protein (encoded by the MLIP gene) predicts a better response to prostaglandins and, concomitantly, a poorer response to beta-blockers. Analysis of this genetic variant could contribute to better clinical management of glaucoma patients.
Material and methods
This study followed the ethical precepts of the Declaration of Helsinki (Fortaleza, Brazil, Oct 2013) and was approved by a local ethics committee (CEIC del Centro de Oftalmología Barraquer Barcelona; Ref. COB033). Human samples were obtained, processed, and analyzed in accordance with current EU regulations on the collection and preservation of human tissues and samples (2004/23/CE and 2006/17/CE), and in accordance with the protocol and legal requirements on the use of biological samples for biomedical research in Spain (Law 14/2007 and Royal Decree 1716/2011). The patients received clinical information and gave their informed consent to participate in the study. The use, protection, communication, and transfer of the personal data of participating patients complied with local regulations (Organic Law 5/2018).
Patients
Included in the study were patients aged 18 and older of both genders, without prior diagnoses and presenting with POAG or OH in one or both eyes requiring pharmacological treatment. IOP was measured using a Goldmann applanation tonometer (Carl Zeiss, Inc., Jena, Germany) mounted on a slit-lamp biomicroscope. IOP measurements were always made at the same time (9 am ± 1 h) by the same researcher (MIC) using the same tonometer, calibrated monthly according to protocol. As diagnostic criteria we considered the following: for POAG, IOP > 21 mmHg associated with optic disc or retinal nerve fiber layer abnormalities, and/or reproducible visual field abnormalities ([4]); and for OH, IOP > 21 mmHg without optic disc or retinal nerve fiber layer abnormalities.
Exclusions from the study were as follows: patients with corneal alterations that could hamper tonometer measurements; patients with pachymetry > 600 µm or < 500 µm; patients with pseudoexfoliative, pigmentary, or secondary glaucoma; patients who had undergone ocular surgery, with the exception of uncomplicated cataract surgery or laser procedures concerning the anterior segment if performed more than 6 months previously; patients with active ocular diseases; and patients currently treated or treated in the previous 6 months with topical, dermatological, intramuscular, or systemic steroids (Table S1, supplementary data).
Experimental design, cohort, and evaluation
A single-center prospective controlled randomized interventional partial crossover study was conducted (Fig. 1). An initial (baseline) tonometry measurement was made before starting treatment, and a second tonometry measurement to evaluate treatment response was made after 6 weeks (42 ± 3 days). According to consensus criteria, the efficacy of baseline IOP reduction attributable to prostaglandin derivatives and to beta-blockers is 25–35% and 20–25%, respectively4–6.
Figure 1.
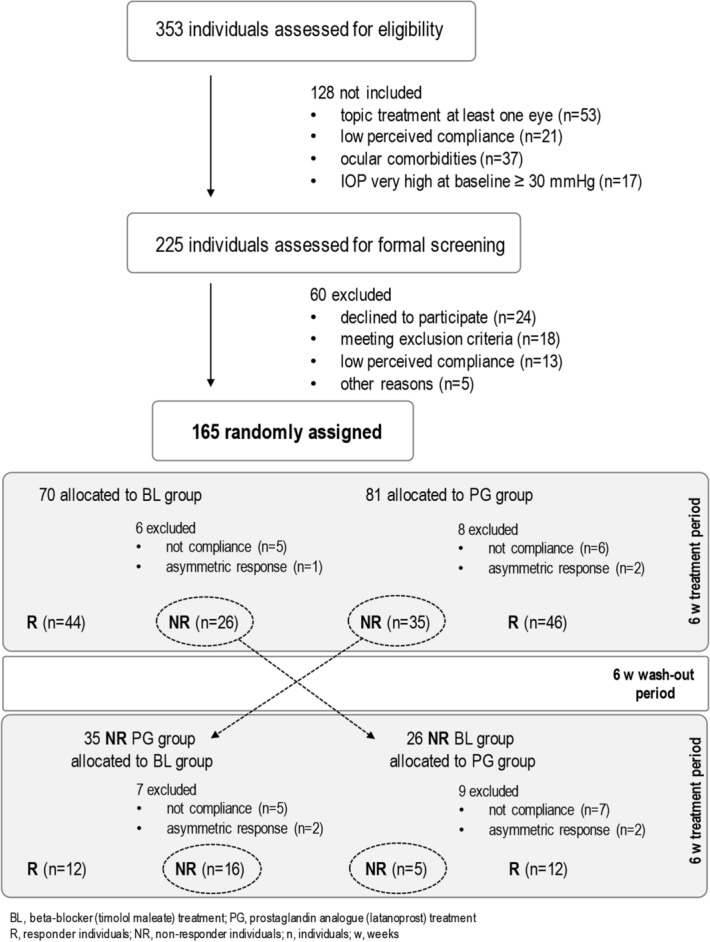
Study flowchart, design and cohort.
Patients were randomized to 2 groups receiving hypotensive treatment as follows. PG group patients received the topical prostaglandin derivative latanoprost (Xalatan, 50 μg/mL) at night, and response and non-response were defined as IOP ≥ 30% and IOP < 25% reduction over baseline, respectively. BL group patients received the topical beta-blocker timolol maleate (Timoftol, 5 mg/mL) every 12 h, and response and non-response were defined as IOP ≥ 25% and IOP < 20% reductions over baseline, respectively4–6. After the second tonometry measurement, non-responders were assigned to a wash-out period of 6 weeks and their treatment was crossed over (to timolol maleate for latanoprost non-responders, and to latanoprost for timolol maleate non-responders) for a further 6 weeks, after which they were reassigned to the corresponding group according to response (Fig. 1). Patients with elevated IOP in both eyes, who presented an unsatisfactory therapeutic response in one of them4, were not considered in the final analysis.
DNA samples and genetic analysis
The GWAS analysis of CNVs was conducted in 2 sets of pooled samples meeting the inclusion criteria: for PG group responders and non-responders, and for BL group responders and non-responders. DNA samples were collected at a screening visit before the intervention. Genomic DNA was isolated from 2 mL of saliva using the Oragene collector (ORAGENE-DNA, OG-500; DNA Genotek Inc, Ottawa, Canada), following manufacturer’s recommendations. DNA concentration and purity were measured, using a Denovix DS-11 device (Wilmington, DE, USA), after reading optical density at 260/280 and 260/230 ratios along a continuous wavelength. Equimolar amounts (100 ng each) of the DNA samples were mixed together to form each of the pools. We used 21 and 14 different DNA samples to prepare the PG responder and non-responder pools, respectively. To prepare the BL responder and non-responder pools, 20 and 13 different DNA samples were used, respectively. High-resolution array-based comparative genomic hybridization (aCGH) (Agilent 1 M, AMADID 021,529) was used to hybridize the Cy3-labelled DNA responder pool against the Cy5-labelled non-responder DNA pool. CNV calling was done using the ADM-2 algorithm, with parameters as follows: threshold 6, and 3 consecutive probes with absolute log2ratio values > 0.25. This ensured an approximate average resolution of 9–12 kb for CNV detection (file Table S2, supplementary data).
This microarray-based discovery assay identified 7 regions as having log2ratio values above or below the threshold, reflecting CNVs for responders and non-responders, respectively. A follow-up validation study using a custom multiplex ligation-dependent probe amplification (MLPA) assay was used to genotype the number of copies for each of the 7 loci in a larger cohort of patients, following standard procedures21,22. The MLPA mix included 4 control probes targeting copy-number neutral and non-variable regions, plus 7 probes targeting the identified variable regions. Control DNA samples were used to normalize values for each probe. In the validation assay with the custom MLPA mix, we analyzed 189 samples from a total of 151 individuals. CNV was estimated from the multimodal distribution of normalized probe heights, which recapitulated the different combinations of copy number alleles and genotypes.
Statistical analysis
Quantitative data was described in terms of means and standard deviation while qualitative data was described in terms of frequency distribution. The Kolmogorov–Smirnov test was used to determine distribution normality for the measured variables. A 2-tailed Student t-test was run and p < 0.05 was considered statistically significant. Those analyses were performed using the Statistical Package for Social Sciences v18.0 (SPSS Inc., Chicago, IL, USA).
Copy-number genotype distributions between responders and non-responders were compared using Fisher’s exact test, in 3 × 2 contingency tables constructed according to the frequency of the different genotypes (AA, AB, and BB). The null hypothesis, rejected if p < 0.05, was that allele frequency distributions were no different between the 2 treatment groups. Association analyses between copy numbers for different loci and responses to the 2 treatments were performed, using the SNPassoc R package23, by assessing interactions between CNV alleles and treatment. The Bonferroni correction was used to identify which CNVs were significantly associated with response to treatment. A generalized linear model was used to compute the probability of interaction between MLIP CNV status and treatment.
Results
A total of 151 patients were studied, 81 and 70 of whom received prostaglandin treatment with latanoprost (PG group) and beta-blocker treatment with timolol maleate (BL group), respectively. All patients were treated for at least 6 weeks. In the PG group, 46 (56.7%) and 35 (43.3%) patients were responders and non-responders, respectively, while the corresponding figures for the BL group were 44 (62.8%) and 26 (37.2%), respectively. Baseline IOP values for the BL and PG groups were 24.3 ± 2.9 mmHg and 24.4 ± 2.2 mmHg, respectively, reduced after treatment to 19.0 ± 3.1 mmHg and 18.9 ± 3.3 mmHg, respectively. Age, sex, and best corrected visual acuity were not response-related for either drug. Table 1 summarizes the demographic characteristics of the study cohort and features of the treatment response.
Table 1.
Demographic and clinical characteristics of study cohort.
| Total (N) | R (n) | NR (n) | p | |
|---|---|---|---|---|
| BL | 72 | 44 | 28 | |
| Age, years (M ± SD) | 50.0 ± 15.3 | 58.2 ± 14.3 | 52.5 ± 16.5 | 0.126 |
| Female (%) | 39 (54.1%) | 26 (59.1%) | 13 (46.4%) | 0.293 |
| Male (%) | 33 (45.9%) | 18 (40.9%) | 15 (53.6%) | |
| IOP1, mmHg (M ± SD) | 24.3 ± 2.9 | 24.8 ± 3.2 | 23.4 ± 2.1 | 0.063 |
| IOP2, mmHg (M ± SD) | 19.0 ± 3.1 | 17.2 ± 2.3 | 21.6 ± 2.4 | 0.0001 |
| BCVA (M ± SD) | 0.88 ± 0.25 | 0.92 ± 0.24 | 0.83 ± 0.26 | 0.161 |
| PG | 80 | 45 | 35 | |
| Age, years (M ± SD) | 57.8 ± 13.4 | 57.6 ± 13.6 | 57.9 ± 13.4 | 0.914 |
| Female (%) | 43 (54%) | 23 (51%) | 20 (57%) | 0.447 |
| Male (%) | 37 (46%) | 22 (49%) | 15 (43%) | |
| IOP1, mmHg (M ± SD) | 24.4 ± 2.2 | 24.6 ± 3.7 | 24.3 ± 3.1 | 0.701 |
| IOP2, mmHg (M ± SD) | 18.9 ± 3.3 | 16.8 ± 2.5 | 21.3 ± 3.1 | 0.0001 |
| BCVA (M ± SD) | 0.90 ± 0.29 | 0.99 ± 0.21 | 0.80 ± 0.34 | 0.006 |
BL, beta-blocker (timolol maleate) treatment group; PG, prostaglandin analogue (latanoprost) treatment group; R, responder individuals; NR, non-responder individuals; (N)(n), number of individuals; IOP1, intraocular pressure before treatment; IOP2, after treatment; BCVA, best corrected visual acuity (decimal); M ± SD, mean ± standard deviation; p, p-value for responders vs non-responders.
Patients not responding to the initial treatment after 6 weeks underwent a wash-out period of 6 weeks, after which treatment was crossed over to the alternative drug (see Fig. 1); these patients were re-evaluated after 6 weeks (42 ± 3 days) according to the established protocol, and their response to the alternative treatment was recorded. This strategy increased the size of the cohort to 189 samples and validated part of the pharmacological response of our cohort: 35 PG non-responders were re-treated with timolol maleate, of whom 12 were reassigned to the BL responder group, 16 were classified as non-responders, and 7 were excluded (not compliance or asymmetric response) from final analysis. In BL group, 26 non-responders were re-treated with latanoprost, of whom 12 were reassigned to the PG responder group, 5 were classified as non-responders, and 9 were excluded (not compliance or asymmetric response). Table 2 summarizes genotype frequencies for MLIP copy numbers together with the percentage average response in IOP reduction in our final cohort (i.e., timolol maleate and latanoprost responders and non-responders).
Table 2.
Percentage average IOP reduction for responder and non-responder genotype frequencies for MLIP.
| MLIP | BL group | PG group | NR group |
|---|---|---|---|
| Total n (%) | 56 (29.5%) | 58 (31.7%) | 21 (9.0%) |
| A/A | 2 (23.3%) | 9 (33.8%) | 1 (0.0%) |
| A/B | 21 (30.5%) | 29 (30.2%) | 13 (10.2%) |
| B/B | 33 (29.3%) | 20 (32.8%) | 7 (7.9%) |
MLIP, muscular LMNA interacting protein; A, 1 copy allele; B, no copies allele; BL, beta-blocker (timolol maleate) treatment group; PG, prostaglandin analogue (latanoprost) treatment group; NR, patients not responsive for any treatment in monotherapy; n, number of individuals; (%), percentage average IOP reduction.
An initial analysis of global CNVs using the high-resolution microarray identified 7 genomic regions (3.8–46 kb in size) – 13q21, glutathione S-transferase theta-1 (GSTT1), late cornified envelope 1D (LCE1D), phosphatase and actin regulator 1 (PHACTR1), MLIP, 1p31, and 2q22 – that showed CNVs between the responder and non-responder pooled groups (Table 3 and file Table S2, supplementary data). The custom MLPA validation assay showed that 2 of the 7 candidate loci exhibited significant CNVs between responders and non-responders, namely, 1p31 and MLIP. As for the other 5 loci, we could not verify significant differences, possibly due to sample selection bias originating in the relatively small number of individuals pooled for the discovery-phase microarray assay.
Table 3.
Genomic loci and differences in copy numbers for responder and non-responder pooled groups (genomic assembly GRCh37).
| Locus | Chromosome | Cytoband | Start | End | Size (Kb) | p |
|---|---|---|---|---|---|---|
| 13q21 | 13 | 13q21.1 | 57759370 | 57788921 | 29 | 2001E−23 |
| GSTT1 | 22 | 22q11.23 | 24349305 | 24395353 | 46 | 8545E−18 |
| LCE1D | 1 | 1q21.3 | 152762076 | 152769870 | 7.7 | 4322E−07 |
| PHACTR1 | 6 | 6p24.1 | 13156200 | 13159684 | 3.8 | 1543E−07 |
| MLIP | 6 | 6p12.1 | 53929240 | 53934834 | 5.5 | 4883E−10 |
| 1p31 | 1 | 1p31.1 | 72766555 | 72801950 | 35 | 3764E−31 |
| 2q22 | 2 | 2q22.3 | 146865725 | 146876364 | 10 | 263E−12 |
GSTT1, glutathione S-transferase theta 1; LCE1D, late cornified envelope 1D; PHACTR1, phosphatase and actin regulator 1; MLIP, muscular LMNA interacting protein.
For beta-blockers we found more CNVs for non-responders than for responders (Fisher’s exact test; p = 0.0039). On further investigation, we found that, under an additive model, each copy of the 1p31 variant allele conferred a negative effect on beta-blocker response in terms of IOP reduction capacity (odds ratio, OR = 0.37; p = 0.0009). A similar result was observed for CNVs for the MLIP locus, with significant differences in genotype distributions between responders and non-responders (Fisher’s exact test; p = 0.02448). Similarly, the more copies of the MLIP variant, the weaker the capacity of beta-blockers to reduce IOP (OR = 0.40; p = 0.0057) (Fig. 2A).
Figure 2.
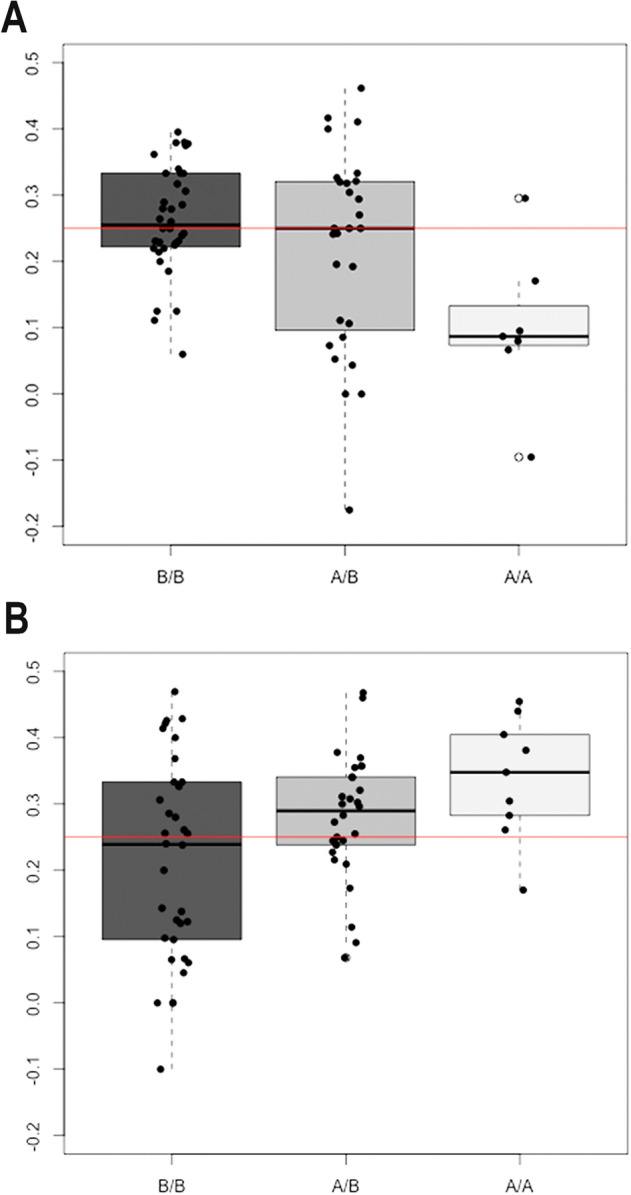
Correlation between copy number of the MLIP locus and intraocular pressure (IOP) reduction response to timolol maleate (A) and to latanoprost (B). Boxplots in each panel shows the percentage distribution of IOP reductions for individuals (black dots) grouped according to their MLIP genotype (B: no copy; A: 1 copy). The horizontal red line marks a 25% IOP reduction that differentiates responders from non-responders. Panel A. IOP reduction in response to a beta-blocker (timolol maleate). More copy numbers (AA genotype) tend to be associated with a smaller percentage IOP reduction, while the median response is higher among individuals with 1 or no MLIP variant copy. Panel B. Individual responses to prostaglandins (latanoprost) and their genotypes. Individuals with fewer copy numbers show a lower median response to latanoprostf in terms of relative IOP reduction.
Interestingly, for the PG group the distribution of MLIP copy number alleles was significantly different for responders and non-responders (Fisher’s exact test; p = 0.03814), with additional CNVs for MLIP producing a significantly greater IOP reduction in PG group responders (OR = 2.34, p = 0.01057) (Fig. 2B).
When overall predicted responses were calculated for both the PG and BL groups (Fig. 3A) and related to CNVs for MLIP (Fig. 3B), we found response probabilities of almost 90% to latanoprost (PG group) for AA genotypes (2 copies) and of almost 70% to timolol maleate (BL group) for BB genotypes (no copies) (Fig. 3B). However, 21 patients overall (14%) were non-responders to either drug in a monotherapy regimen: 13 AB genotypes (1 copy), 7 BB genotypes (0 copies), and 1 AA genotype (2 copies).
Figure 3.
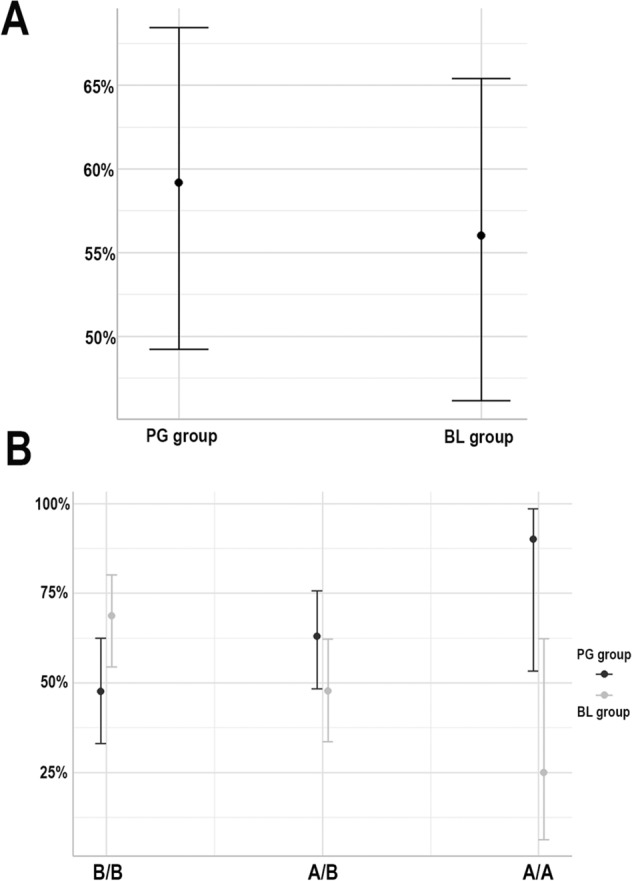
Response probabilities for timolol maleate and latanoprost. (A) Overall response probability for both therapeutic groups. (B) Response probability related to copy number variant (CNV) genotyping BB (no copy), AB (1 allelic copy), and AA (2 allelic copies) for the MLIP locus and the therapeutic groups.
Discussion
The current concept of personalized medicine means that a patient’s response profile can be anticipated prior to drug treatment, thereby avoiding undesirable systemic and/or local adverse effects, whether mild or severe24–26. Genomic analyses have identified key genes contributing to a glaucoma pathophysiology, with glaucoma-related genes defining key biological pathways and processes as potential targets for novel gene-based therapies1,2. Personalized glaucoma management relies on a description of genetic variants that impinge on treatment efficacy, minimize pharmacological adverse effects, and ameliorate disease prognosis and progression. Although clinical pharmacogenetic tests are as yet not widely used in ophthalmology, simple and rapid approaches could be applied to therapeutic decision-making.
Our study followed a multi-step approach to identifying and validating the role played by previously unexplored genetic variations in the pharmacogenomic response to two first-line pharmacological treatments for POAG and OH (prostaglandins and beta-blockers). More specifically, we explored the contribution of CNVs, using a strategy based on pooled genomes that aimed to minimize individual CNVs and enrich the common variants present in our group of patients27–29. DNA samples from responders and non-responders to prostaglandin and beta-blockers (according to clinical criteria) were independently pooled and analyzed using high-resolution aCGH which identified the 7 loci that exhibited copy number differences between responders and non-responders. A custom MLPA assay was used to genotype individual CNV status in a larger cohort of patients.
Our findings illustrate the efficacy of our approach to identifying possible biomarkers of a genetic predisposition to particular pharmacological responses associated with POAG and OH. Our results indicate that initial screening based on studying equimolar DNA pools of carefully phenotyped patients, followed by subsequent individual validation of a larger cohort, is effective.
The validation study showed that pharmacological response was largely influenced by the copy numbers in a 5.6-kb region that overlaps with a known polymorphic CNV30, located within intron 1 of the RefSeq MLIP gene (accession number NM_138569). Patients higher copy number of MLIP variant showed a good response to latanoprost (OR = 2.3), and a poor response to timolol maleate. Conversely, patients with no copies of MLIP variant showed a poor response to latanoprost, and a good response to timolol maleate. This interesting finding led us to hypothesize that the two drug types might act through different pathways.
MLIP is encoded in the short arm of chromosome 6, and its product interacts with muscular laminin-A (encoded by the LMNA gene), a structural component of the nuclear lamina known to be related to familial dilated cardiomyopathy31–33. MLIP has been proposed as a key regulator of cardiomyopathy and is reported to have a potential as a therapeutic target to attenuate heart failure progression32,34. In humans, MLIP is strongly expressed in the heart, smooth and skeletal muscle, and brain, weakly expressed in the liver, and not expressed in other human tissues examined to date32–35. We were unable to detect mRNA expression for MLIP in peripheral blood (data not shown); had we done so, this would have added further value to its potential as a biomarker of therapeutic response.
Interestingly, the region of variability coincides with a recently annotated long non-coding RNA (lncRNA) called MLIP antisense RNA 1 (MLIP-AS1). While lncRNAs were initially thought to be transcriptional noise, accumulated and recent evidence points to regulated expression in particular contexts, during both embryonic stem-cell differentiation and adult tissue differentiation36. It has also been suggested that lncRNAs are involved in several regulatory processes, including chromatin modification, transcriptional regulation, and post-transcriptional regulation37. It therefore cannot be ruled out that absence of this regulation could lead to misregulation of MLIP and interfere in drug response at the ocular level. It has also been reported that alternative splicing patterns of MLIP mRNA may define versatile and stable regions of MLIP likely to modulate its interactions and functions in different tissues35; altogether, therefore, further complementary research would be justified to identify its role in eye-related tissues.
As mentioned, previous reports suggest that MLIP might play a crucial role in heart disease. While an MLIP knock-out mouse model did not show an impact on cardiac function or structure, it did lead to myocardial-specific metabolic abnormalities and cardiac-specific protein kinase B (PKB, or Akt) pathway overactivation, inhibited, in contrast, by cardiac-specific MLIP overexpression32; this is evidence in favor of a direct impact of MLIP on that key signaling pathway. Interestingly, the phosphoinositide-3 kinase (PI3K)/Akt pathway promotes survival and neuroprotection in neurons by inhibiting pro-apoptotic B-cell lymphoma 2 (Bcl-2). Several neuroprotective drugs acting through different mechanisms, including prostaglandin analogs, have been demonstrated to reduce retinal ganglion cell loss and structural damage (caused by increased IOP) through the PI3K/Akt pathway38. It is already known that prostaglandin analogs facilitate aqueous humor drainage via uveoscleral flow, as, acting on metalloproteinases and degrading the extracellular matrix, increased uveoscleral outflow in the ciliary muscle reduces IOP39. We therefore cannot rule out MLIP and Akt signaling pathway participation in IOP response, nor in neuroprotective mechanisms for the optic nerve or different retinal cell populations. Future studies are needed to clarify ciliary muscle and trabecular meshwork protein activation by the PI3K/Akt and Akt/mammalian target of rapamycin (mTOR) signaling pathways in patients with different CNV genotypes for MLIP.
To date, no known genes or regulatory elements are affected by the CNV region located in the 1p31 band. The closest gene is human neuronal growth regulator 1 (NEGR1), located several kb upstream of the CNVs identified as linked to the response to prostaglandins. NEGR1 has been associated with a large number of phenotypes in numerous GWAS analyses of complex phenotypes, including body mass index/weight gain, the hemostatic phenotype, immunity, and developmental delay40,41. Given the existing evidence, and since a possible physiological effect of these CNVs on NEGR1 (or other gene) expression has not been conclusively demonstrated, drawing conclusions for a possible relationship with the studied phenotype might be risky.
In conclusion, our findings contribute to knowledge on the genetic factors affecting prostaglandin and beta-blocker response in glaucoma treatment, providing new insights into the genetic architecture and pathways involved in POAG and OH. Nonetheless, since the clinical utility of this finding remains uncertain, high-quality clinical trials are necessary to accumulate sufficient evidence before this knowledge can be transitioned to the clinical setting.
Supplementary Information
Acknowledgements
We would like to thank Mr. Andrew Hudson for his critical review of the text, and Dr. Jose Pulido (Professor Emeritus of the Departments of Ophthalmology and Molecular Medicine at Mayo Clinic/Vickie and Jack Farber Vision Research Center at Wills Eye Hospital) for suggestions and commentary. We would like to thank the Centro de Oftalmología Barraquer (Barcelona, Spain) for it support in this research. Part of this work was granted by the Spanish Ministry of Science and Innovation (Torres Quevedo project—PTQ-11-04953), also co-financed by the European Social Fund.
Author contributions
M.I.C.: conceptualization, data curation, investigation, methodology, formal analysis, validation, writing—original draft and review manuscript. O.V.: conceptualization, data curation, methodology and review manuscript. B.K.: data curation, investigation, methodology, formal analysis and review manuscript. H.M. and I.B.: investigation and review manuscript. J.R.G.: statistical support and review manuscript. L.A.: conceptualization, data curation, methodology, formal analysis, and review manuscript. R.P.C.-M.: conceptualization, methodology, supervision, visualization, project administration, writing original draft, review and editing manuscript.
Funding
The methodology described in this work is protected by patents 18195545.1 (European Patent Office, 19 Sept 2018) and PCT/ES2019/070606 (13 Sept 2019).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: María I. Canut and Olaya Villa.
Change history
4/9/2021
A Correction to this paper has been published: 10.1038/s41598-021-87653-6
Contributor Information
Lluís Armengol, Email: luis.armengol@qgenomics.com.
Ricardo P. Casaroli-Marano, Email: rcasaroli@ub.edu
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-020-80954-2.
References
- 1.Wiggs JL, Pasquale LR. Genetics of glaucoma. Hum. Mol. Genet. 2017;26:R21–R27. doi: 10.1093/hmg/ddx184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shiga Y, Akiyama M, Nishiguchi KM, Sato K, Shimozawa N, et al. Genome-wide association study identifies seven novel susceptibility loci for primary open-angle glaucoma. Hum. Mol. Genet. 2018;27:1486–1496. doi: 10.1093/hmg/ddy053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gharahkhani P, Burdon KP, Cooke Bailey JN, Hewitt AW, Law MH, et al. Analysis combining correlated glaucoma traits identifies five new risk loci for open-angle glaucoma. Sci. Rep. 2018;8:3124. doi: 10.1038/s41598-018-20435-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.European Glaucoma Society Terminology and Guidelines for Glaucoma 4th Edition. Br. J. Ophthalmol. 2017;101:73–195. doi: 10.1136/bjophthalmol-2016-EGSguideline.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camras CB, Hedman K. Rate of response to latanoprost or timolol in patients with ocular hypertension or glaucoma. J. Glaucoma. 2003;12:466–469. doi: 10.1097/00061198-200312000-00004. [DOI] [PubMed] [Google Scholar]
- 6.Magacho L, Reis R, Shetty RK, Santos LC, Ávila MP. Efficacy of latanoprost or fixed-combination latanoprost-timolol in patients switched from a combination of timolol and a nonprostaglandin medication. Ophthalmology. 2006;113:442–445. doi: 10.1016/j.ophtha.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 7.Klein ME, Parvez MM, Shin JG. Clinical implementation of pharmacogenomics for personalized precision medicine: barriers and solutions. J. Pharm. Sci. 2017;106:2368–2379. doi: 10.1016/j.xphs.2017.04.051. [DOI] [PubMed] [Google Scholar]
- 8.Whirl-Carrillo M, McDonagh EM, Hebert JM, Gong L, Sangkuhl K, et al. Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 2012;92:414–417. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.from pharmacogenetic biomarkers to therapeutic and dosage optimization Ong, F.S., Kuo, J.Z., Wu, W.-C., Cheng, C.-Y., Blackwell, W-LB., et al. Personalized medicine in ophthalmology. J. Pers. Med. 2013;3:40–69. doi: 10.3390/jpm3010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Girirajan S, Campbell CD, Eichler EE. Human copy number variation and complex genetic disease. Annu. Rev. Genet. 2011;45:203–226. doi: 10.1146/annurev-genet-102209-163544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He Y, Hoskins JM, McLeod HL. Copy number variants in pharmacogenetic genes. Trends Mol. Med. 2011;17:244–251. doi: 10.1016/j.molmed.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaedigk A, Sangkuhl K, Whirl-Carrillo M, Klein T, Leeder JS. Prediction of CYP2D6 phenotype from genotype across world populations. Genet Med. 2017;19:69–76. doi: 10.1038/gim.2016.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirschhorn JN. Genome wide Association Studies-Illuminating biologic pathways. N. Engl. J. Med. 2009;360:1699–1701. doi: 10.1056/NEJMp0808934. [DOI] [PubMed] [Google Scholar]
- 16.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 17.Zhang P, Jiang B, Xie L, Huang W. PTGFR and SLCO2A1 gene polymorphisms determine intraocular pressure response to latanoprost in Han Chinese patients with glaucoma. Curr. Eye Res. 2016;41:561–1565. doi: 10.3109/02713683.2016.1143013. [DOI] [PubMed] [Google Scholar]
- 18.Sakurai M, Higashide T, Takahashi M, Sugiyama K. Association between genetic polymorphisms of the prostaglandin F2alpha receptor gene and response to latanoprost. Ophthalmology. 2007;114:1039–1045. doi: 10.1016/j.ophtha.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 19.Sakurai M, Higashide T, Ohkubo S, Takeda H, Sugiyama K. Association between genetic polymorphisms of the prostaglandin F2alpha receptor gene, and response to latanoprost in patients with glaucoma and ocular hypertension. Br. J. Ophthalmol. 2014;98:469–473. doi: 10.1136/bjophthalmol-2013-304267. [DOI] [PubMed] [Google Scholar]
- 20.Ussa F, Fernandez I, Brion M, Carracedo A, Blazquez F, et al. Association between SNPs of metalloproteinases and prostaglandin F2alpha receptor genes and latanoprost response in open-angle glaucoma. Ophthalmology. 2015;122:1040–1048. doi: 10.1016/j.ophtha.2014.12.038. [DOI] [PubMed] [Google Scholar]
- 21.Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, et al. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcinkowska-Swojak M, Uszczynska B, Figlerowicz M, Kozlowski P. An MLPA-based strategy for discrete CNV genotyping: CNV-miRNAs as an example. Hum. Mutat. 2013;34:763–773. doi: 10.1002/humu.22288. [DOI] [PubMed] [Google Scholar]
- 23.González JR, Armengol L, Solé X, Guinó E, Mercader JM, et al. SNPassoc: An R package to perform whole genome association studies. Bioinformatics. 2007;23:654–655. doi: 10.1093/bioinformatics/btm025. [DOI] [PubMed] [Google Scholar]
- 24.Korte, J. M., Kaila, T., & Saari, K. M. Systemic bioavailability and cardiopulmonary effects of 0.5% timolol eyedrops. Graefe’s Arch. Clin. Exp. Ophthalmol.240, 430–435 (2002). [DOI] [PubMed]
- 25.Cramer JA. Effect of partial compliance on cardiovascular medication effectiveness. Heart. 2002;88:203–206. doi: 10.1136/heart.88.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwartz GF, Quigley HA. Adherence and persistence with glaucoma therapy. Surv. Ophthalmol. 2008;53(Suppl 1):S57–S68. doi: 10.1016/j.survophthal.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 27.Earp, M. A., Rahmani, M., Chew, K., & Brooks-Wilson, A. Estimates of array and pool-construction variance for planning efficient DNA-pooling genome wide association studies. BMC Med. Genomics.4, 81 (2019). [DOI] [PMC free article] [PubMed]
- 28.Castro-Giner F, Bustamante M, Ramon González J, Kogevinas M, Jarvis D, et al. A pooling-based genome-wide analysis identifies new potential candidate genes for atopy in the European Community Respiratory Health Survey (ECRHS) BMC Med. Genet. 2009;10:128. doi: 10.1186/1471-2350-10-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forstbauer LM, Brockschmidt FF, Moskvina V, Herold C, Redler S, et al. Genome-wide pooling approach identifies SPATA5 as a new susceptibility locus for alopecia areata. Eur. J. Hum. Genet. 2012;20:326–332. doi: 10.1038/ejhg.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacDonald JR, Ziman R, Yuen RK, Feuk L, Scherer SW. The database of genomic variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42:D986–D992. doi: 10.1093/nar/gkt958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Esslinger U, Garnier S, Korniat A, Proust C, Kararigas G, et al. Exome-wide association study reveals novel susceptibility genes to sporadic dilated cardiomyopathy. PLoS ONE. 2017;12:e0172995. doi: 10.1371/journal.pone.0172995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cattin M, Wang J, Weldrick J, Roeske C, Mak E, et al. Deletion of MLIP (muscle-enriched A-type lamin-interacting protein) leads to cardiac hyperactivation of Akt/mammalian target of rapamycin (mTOR) and impaired cardiac adaptation. J. Biol. Chem. 2015;44:26699–26714. doi: 10.1074/jbc.M115.678433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahmady E, Deeke SA, Rabaa S, Kouri L, Kenney L, et al. Identification of a novel muscle A-type lamin-interacting protein (MLIP) J. Biol. Chem. 2011;286:19702–19713. doi: 10.1074/jbc.M110.165548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang ZP, Kataoka M, Chen J, Wu G, Ding J, et al. Cardiomyocyte-enriched protein CIP protects against pathophysiological stresses and regulates cardiac homeostasis. J. Clin. Invest. 2015;125:4122–4134. doi: 10.1172/JCI82423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cattin ME, Deeke SA, Dick SA, Verret-Borsos ZJA, Tennakoon G, et al. Expression of murine muscle-enriched A-type lamin-interacting protein (MLIP) is regulated by tissue-specific alternative transcription start sites. J. Biol. Chem. 2018;293:19761–19770. doi: 10.1074/jbc.RA118.003758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol. Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 37.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 38.Gauthier AC, Liu J. Epigenetics and signaling pathways in glaucoma. Biomed. Res. Int. 2017;2017:5712341. doi: 10.1155/2017/5712341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Denis P, Lafuma A, Khoshnood B, Mimaud V, Berdeaux G. A meta-analysis of topical prostaglandin analogues intra-ocular pressure lowering in glaucoma therapy. Curr. Med. Res. Opin. 2007;23:601–608. doi: 10.1185/030079907X178720. [DOI] [PubMed] [Google Scholar]
- 40.Speakman JR. Functional analysis of seven genes linked to body mass index and adiposity by genome-wide association studies: a review. Hum. Hered. 2013;75:57–79. doi: 10.1159/000353585. [DOI] [PubMed] [Google Scholar]
- 41.Singh K, Jayaram M, Kaare M, Leidmaa E, Jagomäe T, et al. Neural cell adhesion molecule Negr1 deficiency in mouse results in structural brain endophenotypes and behavioral deviations related to psychiatric disorders. Sci. Rep. 2019;9:5457. doi: 10.1038/s41598-019-41991-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


