Abstract
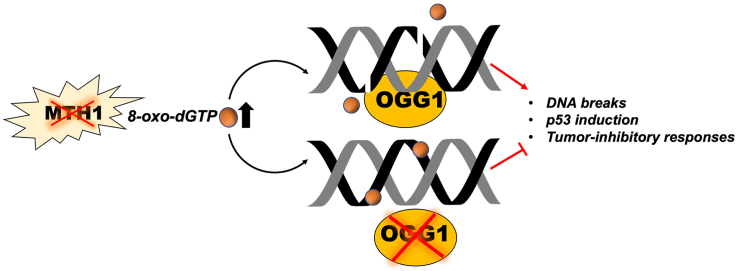
Cancer cells develop protective adaptations against oxidative DNA damage, providing a strong rationale for targeting DNA repair proteins. There has been a high degree of recent interest in inhibiting the mammalian Nudix pyrophosphatase MutT Homolog 1 (MTH1). MTH1 degrades 8-oxo-dGTP, thus limiting its incorporation into genomic DNA. MTH1 inhibition has variously been shown to induce genomic 8-oxo-dG elevation, genotoxic strand breaks in p53-functional cells, and tumor-inhibitory outcomes. Genomically incorporated 8-oxo-dG is excised by the base excision repair enzyme, 8-oxo-dG glycosylase 1 (OGG1). Thus, OGG1 inhibitors have been developed with the idea that their combination with MTH1 inhibitors will have anti-tumor effects by increasing genomic oxidative DNA damage. However, contradictory to this idea, we found that human lung adenocarcinoma with low OGG1 and MTH1 were robustly represented in patient datasets. Furthermore, OGG1 co-depletion mitigated the extent of DNA strand breaks and cellular senescence in MTH1-depleted p53-wildtype lung adenocarcinoma cells. Similarly, shMTH1-transduced cells were less sensitive to the OGG1 inhibitor, SU0268, than shGFP-transduced counterparts. Although the dual OGG1/MTH1 inhibitor, SU0383, induced greater cytotoxicity than equivalent combined or single doses of its parent scaffold MTH1 and OGG1 inhibitors, IACS-4759 and SU0268, this effect was only observed at the highest concentration assessed. Collectively, using both genetic depletion as well as small molecule inhibitors, our findings suggest that OGG1/MTH1 co-inhibition is unlikely to yield significant tumor-suppressive benefit. Instead such co-inhibition may exert tumor-protective effects by preventing base excision repair-induced DNA nicks and p53 induction, thus potentially conferring a survival advantage to the treated tumors.
Keywords: OGG1 inhibition, MTH1 inhibition, oxidative DNA damage, lung cancer, p53, senescence
Graphical abstract
Highlights
-
•
Low MTH1/low OGG1 tumors are robustly represented in patient lung adenocarcinoma datasets but low MTH1/high OGG1 are not.
-
•
Co-depletion of OGG1 in lung adenocarcinoma cells mitigates shMTH1-induced DNA strand breaks and p53-induced senescence.
-
•
p53-null tumor cells have lower OGG1 vs. wt p53 counterparts and are more resistant to MTH1 loss-induced anti-tumor effects.
-
•
Pharmacologic co-inhibition of OGG1 and MTH1 does not enhance cytotoxicity over the respective single inhibitors.
1. Introduction
Due to the hallmark genomic instability in cancer, targeting DNA repair pathways in tumors is a clinically promising paradigm (e.g. ATR inhibitors, PARP inhibitors) [1]. In the past five years, there has been great interest in targeting the mammalian 8-oxo-dGTPase, MutT Homolog 1 (MTH1) [2,3]. MTH1 depletion has been shown to promote DNA breaks in the context of oncogenically-induced oxidative stress [[4], [5], [6]] and uncapping of short telomeres in cancer cell lines which manifests as double strand breaks [7]. The first-in-class MTH1 inhibitors, TH588 and TH287, have been shown to also produce DNA double strand break (53BP1) foci [8]. However in subsequent studies, the cytotoxicity of these inhibitors as well as their best-in-class derivative, TH1579 (karonudib) [9], has been attributed to their enhanced genomic 8-oxo-dG incorporation [[9], [10], [11], [12]], as significant enhancement of DNA breaks relative to control conditions are observed only if recombinant OGG1 is added to the treated cells during the comet assay. However, a body of in vitro and in vivo work has shown that genomic 8-oxo-dG, in and of itself, is not a genotoxic lesion but instead a mildly mutagenic one [13]. In contrast to the TH inhibitors, other independently developed MTH1 inhibitors have been unable to produce robust anti-tumor effects, despite comparable or better target engagement [[14], [15], [16], [17], [18]] and similar inhibition of MTH1 8-oxo-dGTPase activity and genomic 8-oxo-dG incorporation [19]. We and others have surmised that the cytotoxicity of TH588 and TH287 is not due to their inhibition of MTH1 8-oxo-dGTPase activity but rather due to their direct production of reactive oxygen species (ROS) or other off-target activity [[19], [20], [21]]. As karonudib is derived from the same scaffold [9], this may be true for karonudib as well.
ROS can exacerbate MTH1 functional loss by increasing genomic 8-oxo-dG through situ oxidation of dG as well as by generating oxidized nucleotides [[22], [23], [24]], which can then be incorporated by DNA polymerases with varying degrees of fidelity [[25], [26], [27]]. Genomic 8-oxo-dG is repaired by the base excision repair (BER) enzyme, human 8-oxoguanine glycosylase (OGG1). We reasoned that co-inhibition of OGG1 along with MTH1 would recapitulate the enhanced genomic levels presumed to underlie the cytotoxicity of the TH inhibitors, if this is indeed their main mode of action. OGG1-knockout animals are viable and pathologically unremarkable [28], suggesting normal cells tolerate its deficiency, and several OGG1 small molecule inhibitors have been developed that show excellent target engagement [29,30]. However, to the best of our knowledge, to date there are no studies that have investigated the specific molecular effects of OGG1 co-inhibition on the mechanisms and anti-tumor outcomes of MTH1 inhibition. Here, we investigate how OGG1 co-inhibition in cancer cell lines alters the well-characterized effects of MTH1 inhibition [4]. To this end, we utilized MTH1/OGG1 co-depletion via shRNA, co-treatment with OGG1 and MTH1 small molecule inhibitors as well as treatment with a novel OGG1/MTH1 dual inhibitor developed by members of our team [31]. Our results support that, rather than increasing cytotoxicity, OGG1 co-inhibition protects against the anti-tumor effects of targeting MTH1 by limiting the DNA strand breaks, p53 induction, and cellular senescence evoked by MTH1 inhibition.
Our studies, therefore, indicate that OGG1 is critical for inducing the tumor-suppressive outcomes that arise from suppressing functional MTH1. Complementarily, we postulate that preventing repair of genomically incorporated 8-oxo-dG (leading to steady-state increases in 8-oxo-dG incorporation) by OGG1 inhibition antagonizes the tumor-suppressive effects of MTH1 inhibition. Further, our results suggest that the low or mutated OGG1 observed in some human cancers [[32], [33], [34], [35], [36]] may serve as a tumor-protective adaptation against irreparable DNA breaks that can occur as byproducts of BER.
2. Results
2.1. Lung adenocarcinomas do not exhibit elevated OGG1 in the context of low MTH1 expression
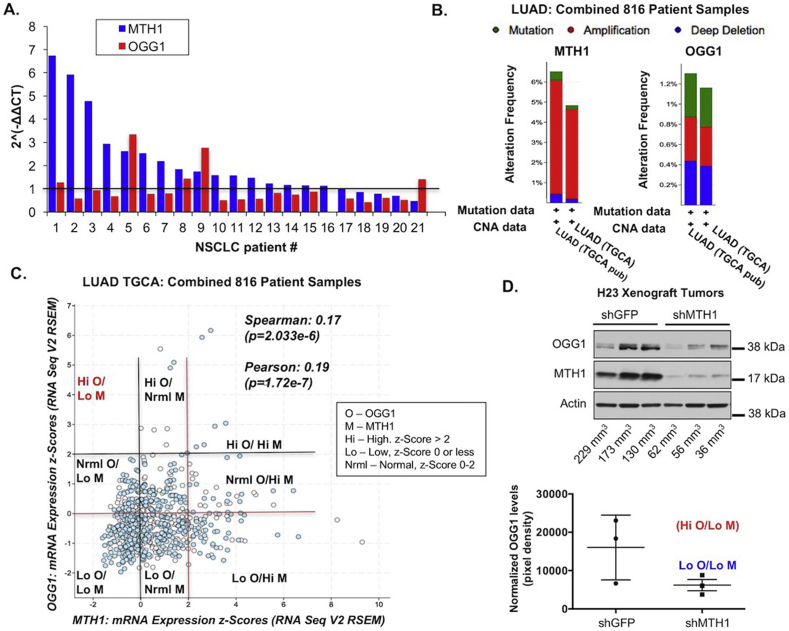
We have previously shown that MTH1 expression and 8-oxo-dGTPase activity are elevated in human lung adenocarcinomas relative to counterpart normal tissues [4,19,37,38]. To assess whether OGG1/MTH1 co-inhibition is likely to enhance anti-tumor outcomes, we evaluated whether OGG1 levels were elevated globally in human lung adenocarcinoma specimens or in tumor subsets without enhanced MTH1 expression. Using paired non-small cell lung cancer mRNA specimens, we found that unlike MTH1, OGG1 levels were not pervasively elevated in the tumor relative to its normal tissue counterpart (Fig. 1A).
Fig. 1.
Lung adenocarcinomas show a selective disadvantage against high OGG1 levels in the context of low MTH1 (NUDT1) status.
A. MTH1 and OGG1 mRNA expression levels in paired normal/tumor non-small cell lung cancer (NSCLC) patient-derived tissues. Each number represents a specimen pair from a distinct patient. Normalized tumor values (each sample value divided by the value from its normal counterpart, set to 1 as per the horizontal line) are shown for OGG1 and MTH1. B. Frequency (percentage) of OGG1 vs. MTH1 mutations or copy number alterations (CNA). Gene amplifications (red), mutations (green), and deletions (blue) are shown for lung adenocarcinoma specimens from 816 patients. Graphs are derived from The Cancer Genome Atlas (TCGA) lung adenocarcinoma (LUAD) datasets at cbioportal.org. C. Correlations between MTH1 and OGG1 levels in the above LUAD dataset. Spearman and Pearson correlation coefficients are shown (generated automatically at cbioportal.org). Expression levels are indicated by z-scores, which represent the numbers of standard deviations above or below the mean raw gene expression. The black gridlines demarcate low MTH1 (Lo M, z-scores 0 or lower) vs. robust OGG1 (Hi O, z-scores 2 or higher) levels and the red gridlines demarcate low OGG1 (Lo O, z-scores 0 or lower) vs. robust MTH1 (Hi M, z-scores 2 or higher) levels. Normal (Nrml) values are designated for z-scores greater than 0 and less than 2. Quadrants made by the intersecting gridlines are labeled according to the legend on the right of the graph. Note the significant representation of tumors with low OGG1 and low MTH1 (Lo O/Lo M) and the lack of Hi O/Lo M tumors. D. Immunoblot showing OGG1 and MTH1 levels in H23 xenograft tumors. Flash-frozen tumors were lysed and total protein lysate probed against the indicated antibodies, with actin as the loading control. Tumor sizes (in mm3) corresponding to the samples are indicated below the immunoblot lanes. Quantitated OGG1 levels (normalized to their actin signals) from the shGFP and shMTH1 tumors are shown below the immunoblot (error bars indicated ±SEM). Note that the variation in tumor OGG1 levels observed in the shGFP samples are collapsed to the low OGG1/low MTH1 (Lo O/Lo M) category in the shMTH1 samples. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Similarly, the percentage of lung adenocarcinomas in The Cancer Genome Atlas (TCGA) dataset (cbioportal.org) with OGG1 amplifications were far lower than those with MTH1 amplifications (Fig. 1B; ~0.4% vs. ~6% respectively, in red). Indeed, changes in OGG1 were nearly equally distributed between amplifications, mutations, and deletions whereas the dominant change in MTH1 was amplification (Fig. 1B). When we determined the inter-relationship between MTH1 and OGG1 expression in lung adenocarcinoma, there was a small but significant positive correlation between the two, and tumors with both low MTH1 and low OGG1 were robustly represented in this 816 patient cohort (Fig. 1C). This suggested to us that lung adenocarcinomas with low co-expression of these genes (Lo O/Lo M) were not particularly underrepresented in the disease spectrum, as might be expected if low levels of both gene products selected adversely against tumor growth and/or progression. Indeed, as seen from the co-expression quadrants based on z-scores (as defined in Fig. 1C), the only tumors not represented in the patient samples were those with high OGG1 and low MTH1 expression (Hi O/Lo M).
We also evaluated OGG1 expression in protein lysates derived from xenograft tumors formed by the H23 lung adenocarcinoma line in shGFP- and shMTH1-transduced counterparts [4]. As previously reported by us [4], the tumor take by the H23 shMTH1 is very low and approximately 30% of injection sites form tumors. Whereas there was a variation in OGG1 expression in the control shGFP tumors, we found that the only tumors formed by MTH1-depleted cells had all selected for overall low OGG1 expression (categorized as Lo O/Lo M, Fig. 1D) compared to shGFP tumors. Interestingly, the largest tumor in the control shGFP group had the lowest OGG1 expression (Fig. 1D).
Collectively, these findings in human patient and xenograft tumors presaged that low OGG1 would not necessarily produce tumor-inhibitory effects either alone or in conjunction with low MTH1. Rather they suggested that robust OGG1 function may in fact be detrimental to tumors with low or inhibited MTH1.
2.2. Co-depletion of OGG1 mitigates shMTH1-induced DNA strand breaks, cellular senescence and increased p53 levels
To model MTH1/OGG1 co-inhibition and explore its molecular sequelae in vitro, we utilized A549 lung adenocarcinoma cells. We have extensively characterized the in vitro and in vivo effects of MTH1 depletion in this cell line in a prior study [4]. We previously reported that MTH1 depletion via shRNA produces a profound and rapid senescent phenotype accompanied by DNA strand breaks and p53 induction in these cells [4]. We have also previously characterized MTH1 depletion in two p53 nonfunctional lung adenocarcinoma cell lines, H23 and H3584. However, for these proof-of-principle studies, we chose A549, as wildtype p53 is required for easily quantifiable and on-target outcomes associated with MTH1 depletion, namely DNA strand breaks and cell senescence.
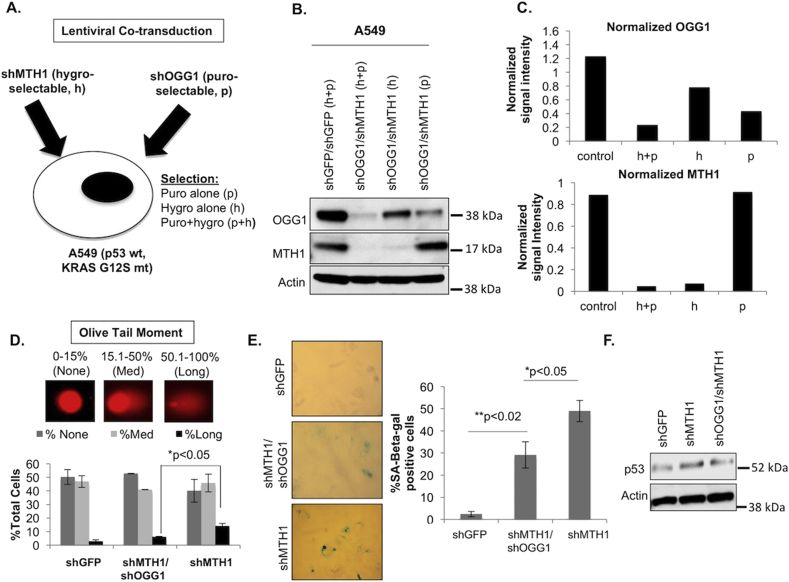
Because of the rapid proliferation arrest induced by MTH1 depletion [[4], [5], [6]], instead of conducting sequential lentiviral transductions with shMTH1 and shOGG1, we opted to assess effects of MTH1/OGG1 co-depletion on clonal selection associated with the relative levels of these two proteins. To this end, we co-transduced A549 cells with viral supernatants containing a hygromycin (h)-selectable pLKO.shMTH1 construct and a puromycin (p)-selectable pLKO.shOGG1 construct (Fig. 2A). We also co-transduced A549 cells with control supernatants containing hygromycin- and puromycin-selectable pLKO.shGFP constructs. We then selected the counterpart cultures in either puromycin (p), hygromycin (h) or both (h + p).
Fig. 2.
Co-depletion of OGG1 in A549 lung adenocarcinoma cells protects against MTH1 depletion-induced DNA breaks and p53-induced senescence.
A. Simultaneous lentiviral co-transduction and selection schemes employed in this experiment. The pLKO.shMTH1 construct is under hygromycin (h) selection and the pLKO.shOGG1 construct is under puromycin (p) selection. Control samples consist of co-transduced puromycin- and hygromycin-selectable pLKO.shGFP constructs. B. Immunoblot showing protein levels of OGG1 and MTH1 under the various individual or co-selection schemes denoted in (A) in the shRNA co-transduced cells. Actin is shown as the loading control. C. Quantitation of actin-normalized OGG1 and MTH1 protein expression levels from (B) under the different selection schemes. Normalized quantifications are based on pixel densities of immunoblot bands obtained via ImageJ. D. Alkaline comet assay. Representative olive tail moments (10× magnification) are indicated for the types of comets that were scored (no/medium/long tails) in the indicated cultures. Error bars (±SD) and p-values from an unpaired Student t-test are shown from two independent experiments. E. Senescence-associated beta-galactosidase (SA-beta-gal) activity assay. Cell senescence was assessed in the indicated cultures. Positive staining was quantified in approximately 50 cells per condition. Data represent two experimental replicates. Error bars (±SD) and p-values from an unpaired Student's t-test significance are shown. F. Immunoblot showing p53 and actin (loading control) protein levels in the indicated samples.
When we immunoblotted lysates from these three counterparts as well as the control, we noticed that selecting for MTH1 inhibition alone (h) led to levels of OGG1 that were nevertheless lower than the baseline levels in the shGFP control cultures (Fig. 2B and C). On the other hand, selecting for OGG1 inhibition alone (p) led to MTH1 levels in this culture that were almost identical to the shGFP culture. The lowest OGG1 levels were in the shOGG/shMTH1 co-selected cultures (h + p) (Fig. 2B and C). These observations suggested to us that there was an advantage to having low baseline OGG1 levels when MTH1 was depleted, which in turn promoted fitness-based selection of the co-depleted populations, even when antibiotic selection favored only MTH1 knockdown.
Using the alkaline single-cell gel electrophoresis (comet) assay, we next assessed the aggregate levels of abasic sites (due to incomplete BER) and DNA single and double strand breaks in control, shMTH1 and co-selected shOGG1/shMTH1 cultures. Our results indicated that co-depletion of OGG1/MTH1 reduced the extent of irreparable fragmented DNA (as exemplified by the percentage of cells with ‘long-tailed comets’) (Fig. 2D). We did not anticipate a complete rescue of this phenotype as there are other enzymes also capable of causing DNA breaks at the sites of 8-oxo-dG [39], and because we could not achieve 100% knockdown of OGG1 levels. Nevertheless, the extent of reduction we observed was significant. We did not observe morphological signs of death in any of the cultures (consistent with our previous studies involving MTH1 knockdown [4]). However, consistent with the lower levels of irreparable DNA strand breaks, we found that the OGG1/MTH1 co-depleted cells also showed a significantly lower extent of senescence-associated beta-galactosidase (SA-beta-gal) activity as well as a less prominent increase in p53 levels compared to the shMTH1 counterparts (Fig. 2E and F). Taken together, these results indicated that low OGG1 levels protect against MTH1 depletion-induced irreparable DNA breaks and concomitant p53-associated senescence that we previously reported [[4], [5], [6]].
2.3. p53-deficient cell lines that are resistant to shMTH1-induced DNA strand breaks and proliferative arrest exhibit lower OGG1 levels relative to their p53-competent isogenic counterparts
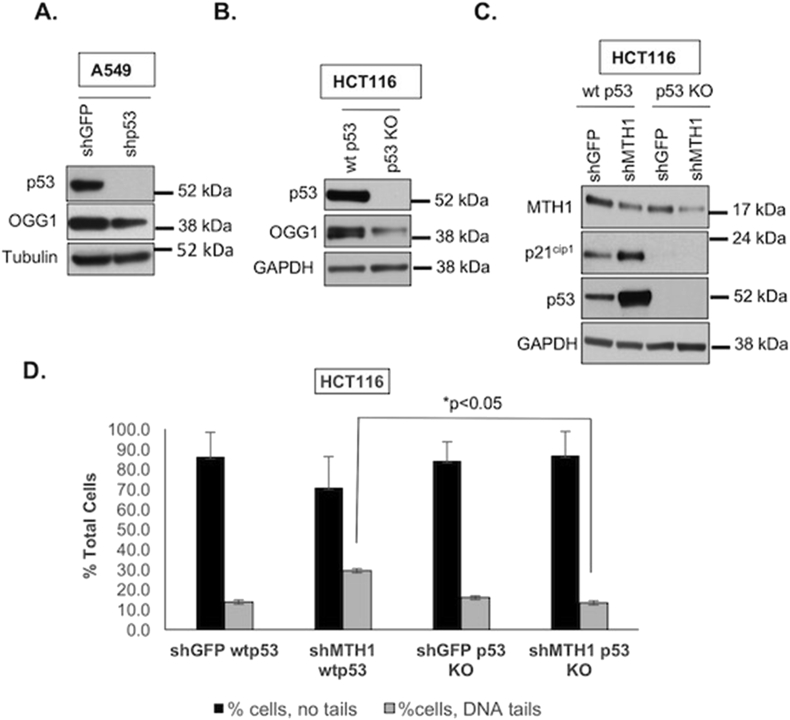
We have previously shown that the ability of MTH1 inhibition to induce DNA strand breaks depends on wildtype (wt) p53, and that cells with non-functional p53 do not respond to MTH1 depletion with DNA breaks unless p53 function is restored [4]. Similarly, wt p53 cells lose the ability to sustain irreparable DNA strand breaks under MTH1 depletion if p53 is knocked down via shRNA [4]. Interestingly, prior studies have shown that p53 controls both OGG1 expression and function, and that loss of p53 prevents initiation of BER by OGG1 [40]. Thus, in p53-nonfunctional cancers, low or functionally aberrant OGG1 may represent a tumor-protective advantage for preventing genotoxic stress arising from BER-mediated DNA breaks at genomic base oxidation sites. To explore this idea, we assessed OGG1 expression in p53-depleted (shp53) A549 cells that we had previously shown lose the ability to induce DNA breaks or cell senescence upon MTH1 depletion [4]. We found that upon stable p53 depletion via lentiviral shRNA, these A549 cells exhibited lower OGG1 expression compared to their isogenic shGFP counterpart cells (Fig. 3A).
Fig. 3.
Mitigation of DNA strand breaks and proliferative arrest observed upon MTH1 depletion in p53-null cells correlates with low OGG1 expression.
A. Loss of p53 in A549 cells corresponds to decreased OGG1 levels. Immunoblotted total protein lysates were probed with the indicated antibodies. Tubulin is shown as the loading control. B. Loss of p53 in HCT116 colorectal cells corresponds to decreased OGG1 levels. Isogenic HCT116 cells either wildtype (wt) or null (KO) for p53 were immunoblotted against the indicated antibodies with GAPDH as the loading control. C. HCT116 cells lacking p53 do not express the cell cycle arrest marker, p21cip1 upon MTH1 depletion. Immunoblotted total protein lysates were probed with the indicated antibodies. GADPH is shown as the loading control. D. Alkaline comet assay. Percentage of cells with no tails (as in Fig. 2E) vs. those with DNA tails (indicating DNA breaks) are graphed for the indicated samples. Note we did not observe the variations in DNA olive tail moments seen for A549 (Fig. 2E) to warrant separation of HCT116 cells into ‘medium’ and ‘long’ tails. Error bars (± SD) and p-values from an unpaired Student t test are shown from two independent experiments.
We next assessed OGG1 levels in an established isogenic HCT116 wt p53 and p53-knockout (KO) pair [41] and found that the p53 KO counterparts have much lower OGG1 expression (Fig. 3B). This finding is consistent with prior reports of low OGG1 expression and function in this cell line [40]. We then transduced these matched HCT116 p53 wt/KO lines with shMTH1 or control shGFP constructs, and found that the G1/S cell cycle arrest marker, p21cip1, was only induced in the wt p53 counterparts upon MTH1 depletion (Fig. 3C). Indeed, loss of p53 (and potentially lower OGG1) increased the tolerance for MTH1 inhibition (as in Fig. 2) as seen by the greater degree of MTH1 knockdown in the p53 KO vs. wt counterparts (Fig. 3C). Because we used the same viral supernatant aliquots and transduction process for both isogenic lines, the lower MTH1 knockdown efficiency in the p53 wt cells was not due to suboptimal viral production or transduction.
Furthermore, similar to the shOGG1 co-transduced counterparts in Fig. 2, the MTH1-depleted HCT116 p53 KO cells also exhibited lower DNA breaks vs. their p53 wt isogenic counterparts, as detected by the alkaline comet assay (Fig. 3D). Altogether, these findings in the HCT116 cells support our results with the A549 cells (Fig. 2) that low OGG1 is associated with protection against MTH1 inhibition-induced DNA breaks and proliferation arrest. They further suggest the intriguing possibility that loss of functional p53 (an extremely common tumor alteration) protects against genotoxic tumor-suppressive stress by mitigating BER-induced DNA breaks in the face of elevated oncogenic ROS levels.
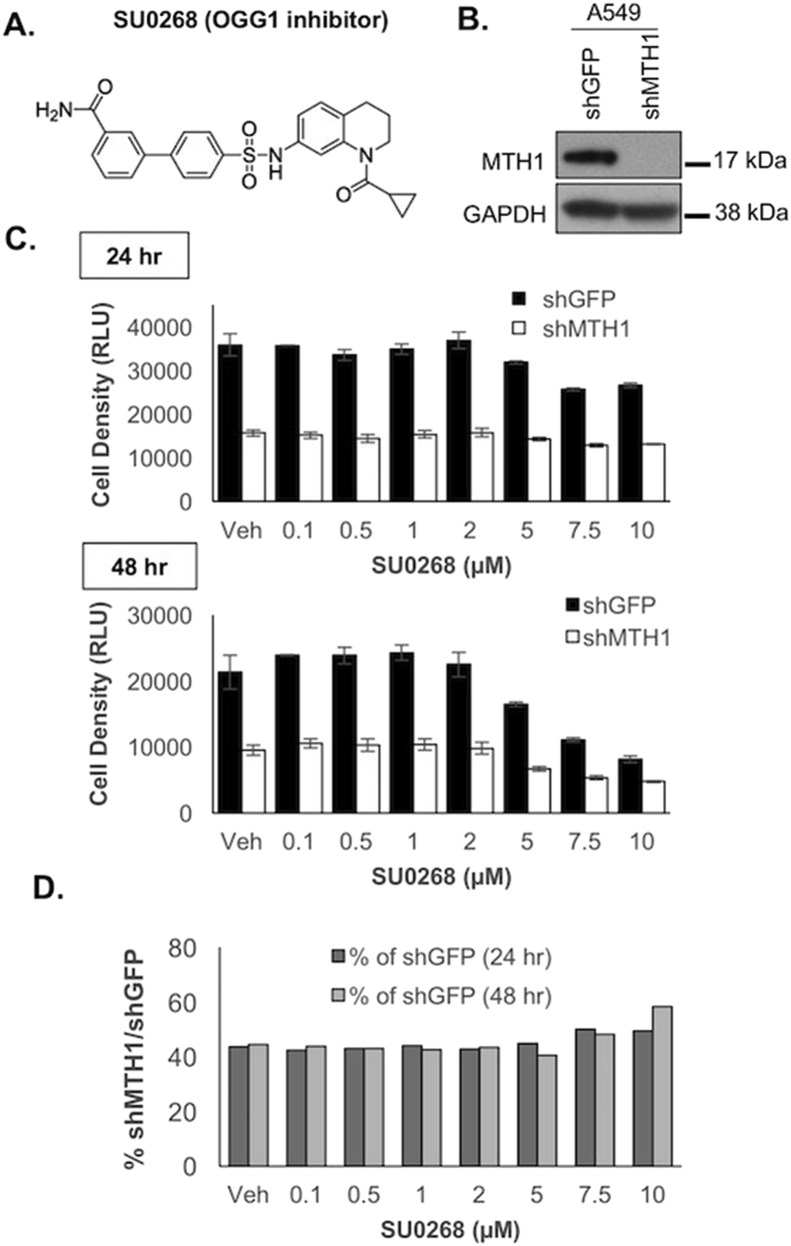
2.4. MTH1-depleted cells are less sensitive to the OGG1-specific inhibitor, SU0268, than their control shGFP counterparts
To further verify our findings with MTH1 and OGG1 shRNA co-transduction, we acutely treated shMTH1-transduced A549 cells with the specific and potent OGG1 inhibitor, SU026829 (Fig. 4A and B) within 2 days of transduction, right before onset of the senescence phenotype. Cell numbers were assessed via the ATP-based CellTiter-Glo assay, which outputs a linear relationship between cell density and luminescence between 0 and 50,000 cells (encompassing our experimental range of 2000 cells). As anticipated, by the end of the experimental period, the A549 shMTH1 cells were about half the density of the control shGFP cells (Fig. 4C). However, the shGFP cells were more susceptible to OGG1 inhibition via increasing SU0268 doses than the shMTH1 cells, especially at the 48 time-point (Fig. 4C). In fact, when the relative cell density-associated luminescent signals for treated shGFP and shMTH1 cells were examined, the shMTH1 cells exhibited increasing relative viability compared to the shGFP at the higher SU0268 concentrations and at the later treatment timepoint (Fig. 4D). As BER activity is not affected by cell proliferation [42], the greater resistance to SU0268 cannot be ascribed to a lesser effect of inhibiting OGG1 on the slower-dividing shMTH1 cells. These findings recapitulated those in Fig. 2, namely that OGG1 inhibition provided a viability advantage in MTH1-depleted vs. control cells, leading to their selection and enrichment over time.
Fig. 4.
The OGG1-specific small molecule inhibitor, SU0268, is less effective in shMTH1-transduced cells vs. control shGFP-transduced cells.
A. Published chemical structure of SU0268. B. Immunoblot showing MTH1 and GAPDH (loading control) protein expression levels in A549 shGFP and shMTH1 cultures, to establish levels of MTH1 protein knockdown. C. Change in cell density under shMTH1, SU0268 or the combined treatment conditions. A549 shMTH1 or shGFP cells were dosed with the indicated concentrations of SU0268 in triplicate. Cell density across the treatment conditions was assessed at 24 and 48 h
post-treatment via the luminescence signal from CellTiter-Glo®. Error bars represent ± SD. D. Relative change in shMTH1 vs. shGFP cell density under treatment conditions. Cell densities for shMTH1 normalized to the corresponding shGFP values in (C) are shown at 24 and 48 h. The increase in normalized shMTH1 values over time and increasing SU0268 doses indicate an enhanced survival advantage of shMTH1 cells with OGG1 inhibition vs. shGFP cells with OGG1 inhibition.
2.5. Treatment of multiple cancer cell lines with combined OGG1/MTH1 small molecule inhibition shows no consistent cytotoxic advantage over treatment with the corresponding single agent inhibitors
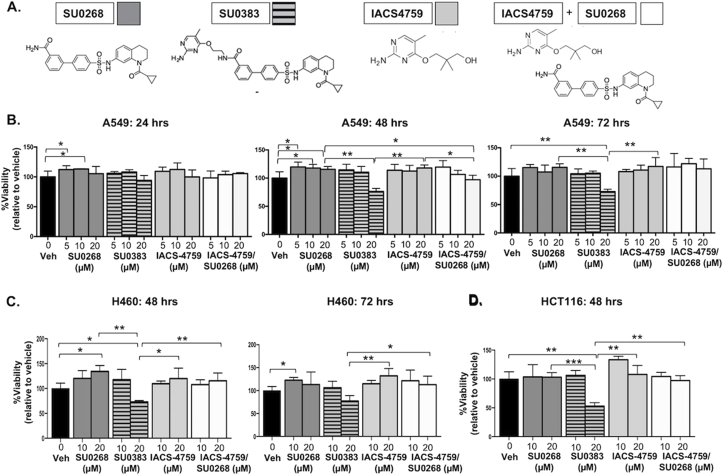
To further investigate the molecular effects of MTH1/OGG1 co-inhibition, we treated A549 cells with a novel small molecule OGG1/MTH1 co-inhibitor SU038331 as well as with SU0268, IACS-4759 (a second-generation MTH1 inhibitor with minimal off-target effects [15,19]) or with a combination of SU0268/IACS-4759, the two molecules which make up the SU0383 scaffold (Fig. 5A). We evaluated dose response over three timepoints (24, 48, and 72 h). This array of treatments indicated that over 24 h there were no significant decreases in viability relative to the DMSO control from any treatment (Fig. 5B). Furthermore, no treatment decreased cell viability below 50% under any assessed condition (Fig. 5B). As we have previously published, IACS-4759 alone also did not produce any significant anti-tumor cytotoxicity, presumably due to its inability to target non-MTH1 8-oxo-dGTPase activity [19]. In general, across the majority of treatment conditions, we did not observe any results to suggest an enhanced anti-tumor advantage to co-inhibiting MTH1/OGG1 over the single inhibitor treatments (Fig. 5B). We observed a noticeable decline in cell viability only in the 20 μM (but not 10 μM) SU0383-treated cells compared to the other treatment groups. This decline in viability, however, was not recapitulated to a similar extent in the SU0268/IACS-4759 combinatorially-treated cells, and the cytotoxic effect remained well under 50% at all treatment durations (Fig. 5B).
Fig. 5.
Small molecule-based co-inhibition of OGG1 and MTH1 does not exhibit consistent enhanced cytotoxicity in tumor cells vs. individual OGG1 or MTH1 inhibitor treatment.
A. Published chemical structures of SU0268 (OGG1 inhibitor), SU0383 (MTH1/OGG1 dual inhibitor), IACS-4759 (MTH1 inhibitor). Each shaded square represents the corresponding treatment condition data graphed for the viability assay in (B). B. Normalized cell viability. A549 cells were dosed, in triplicate, with SU0268, SU0383, IACS-4759 or SU0268/IACS-4759 at the indicated concentrations. Vehicle represents DMSO. Dose response was evaluated at 24, 48, and 72 h following treatment via the vehicle-normalized luminescence signal in a Cell Titer-Glo® assay. Error bars (±SD) and p-values are indicated within the graphs (*p < 0.05, **p < 0.01). Results shown are representative of two independent experiments. C. Normalized cell viability. H460 cells were dosed, in triplicate, with SU0268, SU0383, IACS-4759 or SU0268/IACS-4759 at the indicated concentrations. Vehicle represents DMSO. Dose response was evaluated at 48 and 72 h following treatment via the vehicle-normalized luminescence signal in a Cell Titer-Glo® assay. Error bars (±SD) and p-values are indicated within the graphs (*p < 0.05, **p < 0.01). D. Normalized cell viability. HCT116 cells were dosed, in triplicate, with SU0268, SU0383, IACS-4759 or SU0268/IACS-4759 at the indicated concentrations. Vehicle represents DMSO. Dose response was evaluated at 48 h following treatment via the vehicle-normalized luminescence signal in a Cell Titer-Glo® assay. Error bars (±SD) and p-values are indicated within the graphs (*p < 0.05, **p < 0.01, ***p < 0.001).
Similar results as with A549 were obtained with the p53-competent H460 lung adenocarcinoma and HCT116 colorectal cancer cell lines (Fig. 5C and D). In the H460 cells (Fig. 5C), as in the A549 cells (Fig. 5B), SU0268 treatment alone produced a small but statistically significant increase in viability relative to DMSO, supporting our other observations herein that lung adenocarcinoma cells ostensibly select for low OGG1 function. As with A549, both H460 and HCT116 cells also only exhibited noticeably decreased viability with the 20 μM SU0383 treatment relative to the other treatment groups (Fig. 5C and D). Again, this reduction in cell viability was not recapitulated by combined treatment with the individual MTH1/OGG1 inhibitors.
SU0383 has a slightly lower efflux ratio (5.1) vs. SU0268 (6.2)29, 31 suggesting it may be better retained intracellularly and thus exhibit somewhat more potent cytotoxicity, as its effects on cell viability are noticeably seen only at the 48- and 72-h timepoints (Fig. 5B and C). However, this difference in efflux ratio is modest, and as the cytotoxic effects do not change materially with longer treatment durations, another explanation is that the pronounced cytotoxicity at 20 μM SU0383 treatment is due to off-target effects. This is particularly likely as the reported IC50s of SU0383 for MTH1 (0.034 μM) and OGG1 (0.49 μM) as well as the prior reported cytotoxic dose (10 μM) [31] are below the 20 μM at which we see cytotoxicity in our experiments (Fig. 5C and D). In our treatment series, we did not find that the SU0268/IACS-4759 co-treated cells were more viable than the correspondingly-treated IACS-4759- or SU0268-treated cells (Fig. 5B–D). We posit that the incomplete penetrance of on-target effects relative to stable protein depletion or, alternatively, potential off-target cytotoxic effects from the small molecules competed with any survival advantage from dual MTH1/OGG1 inhibition. However, the fact that there was no consistently deleterious effect from combining MTH1 and OGG1 inhibition, in any of the three cancer cell lines evaluated, supports our overall premise that this co-inhibition is unlikely to be pervasively beneficial in a therapeutic setting.
3. Discussion
In this study, we showed that co-inhibition of MTH1 and OGG1, either by genetic or pharmacologic means, did not induce significant anti-tumor effects in cancer cells. Instead, under stable genetic knockdown in a cell population, OGG1 co-depletion conferred a protective advantage by mitigating the DNA damage and cell senescence induced by MTH1 depletion. Interestingly, in the TCGA LUAD patient dataset we analyzed, the only quadrant with no tumors was the one with low MTH1 and high OGG1 (Hi O/Lo M), encompassing tumors with MTH1 z-scores <0 and OGG1 z-scores 2 or higher (Fig. 1C), further supporting the idea that OGG1 action becomes deleterious to tumors when MTH1 is absent. Similarly, in an shMTH1 lung adenocarcinoma cell line-derived xenograft tumor model [4], we found palpable tumor formation only occurred in cohorts which had selected for low OGG1 expression upon MTH1 depletion (Fig. 1D). Furthermore, we did not see enhanced cytotoxicity through co-treatment with potent MTH1 and OGG1 small molecule inhibitors or treatment with a dual OGG1/MTH1 inhibitor relative to control or single agent treatments. Our findings suggest that BER processing of incorporated 8-oxo-dG induces MTH1 inhibition-mediated DNA breaks and tumor-inhibitory responses. Our findings further indicate that there may not be therapeutic benefit to combining OGG1 and MTH1 inhibitors in a clinical setting, without further consideration of tumor OGG1 status or that of other 8-oxo-dG repairing/preventive mechanisms.
Although here we describe proof-of-principle results on how compromising OGG1 function alters the effects of inhibiting 8-oxo-dGTP detoxification in tumor cells, the broader implications of our findings are supported by established observations. Several prior studies have shown that OGG1 is either mutated or otherwise functionally impaired in tumors without apparent tumor-suppressive consequences [[32], [33], [34], [35], [36]], similar to what we found through assessment of OGG1 levels in patient specimens and mining of patient datasets. Curiously, the late-onset spontaneous tumors (including lung tumors) [43] observed in MTH1-null animals are suppressed in the OGG1/MTH1 double-knockout animals [44]. At first glance, this finding may support the potential for anti-tumor effects from MTH1/OGG1 co-repression. However, the MTH1-null tumors occur with relatively low frequency and only in mice at 18 months or older, and could thus represent tumor-promoting phenomena from the aging tissue environment. We have shown MTH1 loss accelerates senescence [5,6], and that MTH1 overexpression overcomes oncogene-induced senescence [6]. MTH1-overexpressing mice are reported to show improved functional aging [45]. Senescent cells produce a distinct secretome known as the senescence-associated secretory phenotype (SASP) that has tumor-promoting capabilities [46]. In this regard, it is possible the late-onset tumors in MTH1-null animals are a result of paracrine factors produced by senescent cells/tissue induced by this MTH1 loss. As we see in our study, OGG1 depletion is able to partially rescue the MTH1 loss-related senescent phenotype. Similarly, the tissue environment of the OGG1/MTH1 knockout animals may also be protected from aging-associated alterations, thus mitigating late-onset tumor development.
Our results support that OGG1 function is necessary for MTH1 depletion-induced DNA strand breaks. Unrepaired genomic 8-oxo-dG is not considered a genotoxic lesion and does not cause catastrophic stalling of DNA polymerases [13]. Thus, the sequelae from harboring low OGG1 (or similar 8-oxo-dG-processing repair enzymes) are most likely to be mutagenic events, which are more tolerable (or even beneficial) for tumor survival than excessive repair-induced breaks in the DNA backbone. BER enzymes, including OGG1, induce DNA nicks and abasic sites upon 8-oxo-dG repair initiation. Resealing of these repair-mediated breaks by the dRP lyase function of DNA polymerase beta is the rate limiting step in completing BER. Accordingly, initiation of BER has been implicated in production of irreparable DNA breaks, particularly under conditions of oxidative stress [[47], [48], [49], [50]]. These observations, in conjunction our findings here, collectively support that OGG1-mediated repair initiation at sites of incorporated 8-oxo-dGTP translate MTH1 depletion into irreparable DNA breaks and tumor-inhibitory responses. Hence, we observed partial rescue of MTH1-induced DNA breaks when OGG1 was co-depleted (Fig. 2). This explanation is logically more consistent than the idea that extant and unprocessed genomic 8-oxo-dG is the source of MTH1 inhibition-induced cytotoxicity, as has been previously suggested [[9], [10], [11], [12]]. Our findings point to robust OGG1, and more generally robust BER activity, as an important determinant for tumor suppression arising from MTH1 inhibition. Further consistent with our findings that OGG1 inhibition protects against the cytotoxic effects of oxidative insult, two different OGG1 inhibitors have been shown to reduce inflammation-induced adverse cellular effects [30,51].
A critical role for p53 in BER has previously been comprehensively described, with p53 depletion reducing repair activity [52]. Introducing p53 into HCT116 p53 KO cells has been reported to increase OGG1 levels, suggesting direct control of OGG1 expression by p53 [40]. Furthermore 8-oxo-dG levels are increased in the p53 KO cells relative to p53 wt counterparts [53], suggesting p53 also controls OGG1 BER function. Thus, our findings that the p53-null A5494 and HCT116 cells (Fig. 3C and D) are less susceptible to MTH1 depletion-induced DNA breaks and proliferative arrest, and also exhibit lower OGG1 expression vs. their p53 wt counterparts (Fig. 3A and B) additionally support our assertion that co-inhibition of OGG1 function protects against MTH1 inhibition-induced tumor-inhibitory effects. Given the commonality of p53 mutations and deletions in cancer, there may be potential benefit to restoring OGG1 function in such tumors particularly in conjunction with oxidative stressors, in order to evoke genotoxic tumor-suppressive responses.
Our study is the first to describe the molecular effects of co-targeting MTH1 and OGG1 in tumor cells. Overall, our findings strongly suggest that MTH1 inhibition requires robust OGG1 (and/or other 8-oxo-dG-repairing BER function) to compromise tumor cell viability by translating increased genomic incorporation of 8-oxo-dG into DNA strand breaks. Further studies of OGG1 expression and function in tumors treated with on-target MTH1 inhibitors or therapeutics intended to restore p53 function will be required to shed definitive light on the clinical potential of targeting OGG1 in tumors.
4. Materials and methods
4.1. MTH1 and OGG1 gene expression analysis in human tumors
All human subjects research was carried out following written consent obtained from the patients and according to the protocols approved by the University of Miami Institutional Review Board (IRB# 20100200). Retrospective frozen tissue samples consisting of deidentified matched normal and tumor pairs were obtained from untreated patients diagnosed with stage 1, 2, or 3 NSCLC at the Sylvester Comprehensive Cancer Center (Miami, FL). Samples were processed and analyzed for mRNA levels as previously described [4]. The mRNA expression levels of MTH1 and OGG1 were examined in lung adenocarcinoma TCGA datasets using publicly available datasets from cBioportal (http://www.cbioportal.org/).
4.2. Cell lines and culture
A549 and H460 cells were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). The HCT116 p53 wildtype (wt) and p53 knockout (KO) isogenic human colon cancer cells were obtained from Dr. Bert Vogelstein's laboratory (Johns Hopkins University, Baltimore, MD). A549 cells were authenticated by short tandem repeat (STR) profiling and mycoplasma testing via PCR (GeneticaCorp) in August 2018 and cultures used in this study were within 2 passages of the STR-validated line. The H460 and HCT116 lines were similarly validated by STR-profiling and for lack of mycoplasma in December 2020. All cells were maintained in RPMI-1640 complete base media (Thermo Scientific, Grand Island, NY) supplemented with 10% fetal bovine serum (HyClone, Cytiva, Marlborough, MA) and 100 units per ml of penicillin streptomycin (Thermo Scientific, Grand Island, NY). Cells were cultured at 37 °C in a humidified incubator at 21% oxygen, 5% CO2 (HeraCell Tri-Gas, Thermo Fisher Scientific, Inc).
4.3. DNA constructs and viral transduction
The pLKO.shGFP, pLKO.hygro shMTH1 and pLKO.shp53 constructs have been described previously [[4], [5], [6]]. The pLKO.puro shOGG1 lentiviral constructs were purchased from Sigma. The following are the targeting sequences used in this study:
shGFP: 5′ GCAAGCTGACCCTGAAGTTCA 3’.
shMTH1: 5′ GAAATTCCACGGGTACTTCAA 3’.
shOGG1: 5′ CGGATCAAGTATGGACACTGA 3’.
shp53: 5′ GACTCCAGTGGTAATCTACTT 3’.
Plasmids were sequenced prior to use (Source Biosciences). Lentiviral and retroviral supernatant production was carried out in HEK 293T cells (ATCC), and infection of target cells was performed as described previously [54]. Prior to carrying out experiments, transduced cells were selected in media containing either 2.5 μg/mL puromycin (Sigma, P7255) or 100 μg/mL hygromycin (Sigma, H3274) or both, for a minimum duration corresponding to the time taken for untransduced cells to die in antibiotic-supplemented media. Protein knockdown or overexpression in the transduced cells was verified via Western blotting.
4.4. Comet assay
The alkaline-modified comet assay was carried out according to the comet assay kit (Trevigen, cat no: 15140-122, Gaithersburg, MD) instructions for alkaline unwinding and electrophoresis conditions. Gel electrophoresis was carried out at 21 V for 30 min at 4 °C. Positive and negative control cells (Trevigen) were run along with each sample to ensure that lack of ‘tails’ in a sample or long ‘tails’ were not due to technical issues. A minimum of 100 individual cells per sample were scored in duplicate from two independent experiments, with the DNA tail lengths being visually categorized as either long, medium or none in double-blind scoring. The p-values were established using an unpaired Student t-test, with Welch correction applied if variances were found to be unequal.
4.5. Western blotting
Western blots were performed using standard procedures. Cell culture pellets were lysed in a sodium fluoride (NaF) buffer, as previously described [54]. Protein lysates from xenograft tumor chunks were made using RIPA buffer (Thermo Scientific, 89900), supplemented with a protease inhibitor cocktail (Roche, 11697498001). Protein concentrations were measured using the Bradford Protein Assay Dye Reagent (Biorad, 5000006). Approximately 10–30 μg of total protein was run on a 4–12% Bis-Tris pre-cast NuPage gel (Thermo Scientific, NP0321BOX) on the Novex gel system and subsequently transferred onto a section of PVDF membrane (Immobilon, EMD Millipore, IPVH000010) at 30 V at 4 °C. Blots were probed with antibodies against OGG1 (Abcam, ab124741, rabbit polyclonal), MTH1 (Novus Biologicals, NB100-109, rabbit polyclonal), GAPDH (Abcam, ab9485, rabbit polyclonal), actin (Millipore Sigma, A2066, rabbit polyclonal), tubulin (Millipore Sigma, T6557, mouse monoclonal), p53 (Santa Cruz Biotech, DO-1 sc-126, mouse monoclonal), p21cip1 (Santa Cruz Biotech, sc-817, mouse monoclonal). Immunoblot membranes were stripped between separate antibody incubations as needed, and loss of the original signal was checked on film prior to probing with a subsequent antibody. Molecular weight standards (Full Range molecular weight markers in kiloDalton (kDa), GE, RPN800E) were indicated to the right of the immunoblot bands. Western blotting images represent data consistent with a minimum of two independent runs. Densitometry of images was carried out via the ImageJ Analyze Gels (NIH) module and normalized to the loading signal for each band.
4.6. Senescence-associated beta-galactosidase (SA-beta-gal) assay
SA-beta-gal staining was carried out as previously described [4]. All chemicals were purchased from Sigma-Aldrich. Briefly, cells were washed in PBS, fixed in 0.2% glutaraldehyde for 5 min at room temperature, washed once in PBS and incubated overnight in freshly-made staining solution (1 mg/mL 5-bromo-4-chloro-3-indolyl-β-galactoside, 150 mM NaCl, 2 mM MgCl2, 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 40 mM NaPi, pH 6.0). The next day, the cells were incubated for a further hour at 37 °C to intensify staining and then washed and stored in PBS at 4 °C till image acquisition. To quantify positive staining, at least 50 cells were counted for each sample over multiple fields of view, excluding fields at the very edge. Results represent data from a minimum of two independent experiments.
4.7. MTH1 and OGG1 inhibitors
IACS-4759 was a kind gift from Dr. Alessia Petrocchi, MD Anderson.
SU0268 [29] and SU0383 [31] were synthesized in Dr. Eric Kool's lab. Inhibitor working stock solutions were prepared in DMSO (Sigma, D2650).
4.8. Drug treatment and cell viability assays
A549 shGFP and shMTH1 (2000 cells per well) were plated in triplicate in 96-well plates in RPMI-1640 culture medium. At 24 h after plating, cells were changed to their corresponding culture medium at a concentration of 0.1 μM, 0.5 μM, 1 μM, 2.5 μM, 5 μM, 7.5 μM, 10 μM of SU0268 or SU0383 or DMSO (Sigma, D2650) as the vehicle control. After 24 or 48 h post-drug treatment, luminescence was measured using the Cell Titer-Glo® Kit (Promega, G7571) on a SpectraMax iD3 Microplate Reader (Molecular Devices, LLC).
For the single and combinatorial drug treatments (Fig. 5), A549 cells were plated (750 cells per well) in 96-well plates and, 24 h after plating, culture media was replaced in the wells with either SU0268, SU0383, IACS-4759, or SU0268/IACS-4759 at 5 μM, 10 μM or 20 μM concentrations. Vehicle wells were treated with DMSO (Sigma, D2650). Luminescence was measured after 24, 48, and 72 h. H460 and HCT116 (at 750 cells per well) were similarly treated with either SU0268, SU0383, IACS-4759, or SU0268/IACS-4759 added at 10 μM or 20 μM concentrations. Luminescence was measured after 48 and 72 h for H460 and after 48 h for HCT116. Data were normalized to luminescence values from vehicle-treated controls (DMSO) within each group, and plotted as % viability.
4.9. Statistical analysis
Standard deviation from the mean (SD) or standard error of the mean (SEM) were assessed as indicated in the data shown. The significance of observed differences was evaluated by an unpaired two-tailed Student's t-test and p values < 0.05 were deemed to be significant. Statistical analyses were carried out using GraphPad Prism.
Declaration of competing interest
All authors confirm there is no conflict of interest to declare.
Acknowledgements
We thank current members of the Rai lab, Abel Sousa and Clara Troccoli, as well as former University of Miami/Rai lab undergraduate research volunteers, Christina Jayaraj and Brooke Zarouri, for technical assistance. This work was supported by grants from the National Institutes of Health/National Cancer Institute (United States of America) R01CA175086 (to PR) and R01CA217809 (to ETK). This work was also supported by Sylvester Comprehensive Cancer Center research funding to PR.
References
- 1.Brown J.S., O'Carrigan B., Jackson S.P., Yap T.A. Targeting DNA repair in cancer: beyond PARP inhibitors. Canc. Discov. 2017;7:20–37. doi: 10.1158/2159-8290.CD-16-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maki H., Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- 3.Mo J.Y., Maki H., Sekiguchi M. Hydrolytic elimination of a mutagenic nucleotide, 8-oxo-dGTP, by human 18-kilodalton protein: sanitization of nucleotide pool. Proc. Natl. Acad. Sci. U. S. A. 1992;89:11021–11025. doi: 10.1073/pnas.89.22.11021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel A., Burton D.G., Halvorsen K., Balkan W., Reiner T., Perez-Stable C. MutT Homolog 1 (MTH1) maintains multiple KRAS-driven pro-malignant pathways. Oncogene. 2014 doi: 10.1038/onc.2014.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rai P., Onder T.T., Young J.J., McFaline J.L., Pang B., Dedon P.C. Continuous elimination of oxidized nucleotides is necessary to prevent rapid onset of cellular senescence. Proc. Natl. Acad. Sci. U. S. A. 2009;106:169–174. doi: 10.1073/pnas.0809834106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rai P., Young J.J., Burton D.G., Giribaldi M.G., Onder T.T., Weinberg R.A. Enhanced elimination of oxidized guanine nucleotides inhibits oncogenic RAS-induced DNA damage and premature senescence. Oncogene. 2011;30:1489–1496. doi: 10.1038/onc.2010.520. [DOI] [PubMed] [Google Scholar]
- 7.Fouquerel E., Lormand J., Bose A., Lee H.T., Kim G.S., Li J. Oxidative guanine base damage regulates human telomerase activity. Nat. Struct. Mol. Biol. 2016;23:1092–1100. doi: 10.1038/nsmb.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gad H., Koolmeister T., Jemth A.S., Eshtad S., Jacques S.A., Strom C.E. MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool. Nature. 2014;508:215–221. doi: 10.1038/nature13181. [DOI] [PubMed] [Google Scholar]
- 9.Warpman Berglund U., Sanjiv K., Gad H., Kalderen C., Koolmeister T., Pham T. Validation and development of MTH1 inhibitors for treatment of cancer. Ann. Oncol. : official journal of the European Society for Medical Oncology. 2016;27:2275–2283. doi: 10.1093/annonc/mdw429. [DOI] [PubMed] [Google Scholar]
- 10.Einarsdottir B.O., Karlsson J., Soderberg E.M.V., Lindberg M.F., Funck-Brentano E., Jespersen H. A patient-derived xenograft pre-clinical trial reveals treatment responses and a resistance mechanism to karonudib in metastatic melanoma. Cell Death Dis. 2018;9:810. doi: 10.1038/s41419-018-0865-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hua X., Sanjiv K., Gad H., Pham T., Gokturk C., Rasti A. Karonudib is a promising anticancer therapy in hepatocellular carcinoma. Ther Adv Med Oncol. 2019;11 doi: 10.1177/1758835919866960. 1758835919866960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moukengue B., Brown H.K., Charrier C., Battaglia S., Baud'huin M., Quillard T. TH1579, MTH1 inhibitor, delays tumour growth and inhibits metastases development in osteosarcoma model. EBioMedicine. 2020;53:102704. doi: 10.1016/j.ebiom.2020.102704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delaney S., Jarem D.A., Volle C.B., Yennie C.J. Chemical and biological consequences of oxidatively damaged guanine in DNA. Free Radic. Res. 2012;46:420–441. doi: 10.3109/10715762.2011.653968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Samaranayake G.J., Huynh M., Rai P. MTH1 as a chemotherapeutic target: the elephant in the room. Cancers. 2017;9 doi: 10.3390/cancers9050047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petrocchi A., Leo E., Reyna N.J., Hamilton M.M., Shi X., Parker C.A. Identification of potent and selective MTH1 inhibitors. Bioorg. Med. Chem. Lett. 2016;26:1503–1507. doi: 10.1016/j.bmcl.2016.02.026. [DOI] [PubMed] [Google Scholar]
- 16.Kettle J.G., Alwan H., Bista M., Breed J., Davies N.L., Eckersley K. Potent and selective inhibitors of MTH1 probe its role in cancer cell survival. J. Med. Chem. 2016;59:2346–2361. doi: 10.1021/acs.jmedchem.5b01760. [DOI] [PubMed] [Google Scholar]
- 17.Ellermann M., Eheim A., Rahm F., Viklund J., Guenther J., Andersson M. Novel class of potent and cellularly active inhibitors devalidates MTH1 as broad-spectrum cancer target. ACS Chem. Biol. 2017;12:1986–1992. doi: 10.1021/acschembio.7b00370. [DOI] [PubMed] [Google Scholar]
- 18.Kawamura T., Kawatani M., Muroi M., Kondoh Y., Futamura Y., Aono H. Proteomic profiling of small-molecule inhibitors reveals dispensability of MTH1 for cancer cell survival. Sci. Rep. 2016;6:26521. doi: 10.1038/srep26521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samaranayake G.J., Troccoli C.I., Zhang L., Huynh M., Jayaraj C.J., Ji D. The existence of MTH1-independent 8-oxo-dGTPase activity in cancer cells as a compensatory mechanism against on-target effects of MTH1 inhibitors. Mol. Canc. Therapeut. 2020;19:432–446. doi: 10.1158/1535-7163.MCT-19-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Waals L.M., Laoukili J., Jongen J.M.J., Raats D.A., Borel Rinkes I.H.M., Kranenburg O. Differential anti-tumour effects of MTH1 inhibitors in patient-derived 3D colorectal cancer cultures. Sci. Rep. 2019;9:819. doi: 10.1038/s41598-018-37316-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J.Y., Jin L., Yan X.G., Sherwin S., Farrelly M., Zhang Y.Y. Reactive oxygen species dictate the apoptotic response of melanoma cells to TH588. J. Invest. Dermatol. 2016;136:2277–2286. doi: 10.1016/j.jid.2016.06.625. [DOI] [PubMed] [Google Scholar]
- 22.Rai P. Oxidation in the nucleotide pool, the DNA damage response and cellular senescence: defective bricks build a defective house. Mutat. Res. 2010;703:71–81. doi: 10.1016/j.mrgentox.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 23.Russo M.T., Blasi M.F., Chiera F., Fortini P., Degan P., Macpherson P. The oxidized deoxynucleoside triphosphate pool is a significant contributor to genetic instability in mismatch repair-deficient cells. Mol. Cell Biol. 2004;24:465–474. doi: 10.1128/MCB.24.1.465-474.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sangsuwan T., Haghdoost S. The nucleotide pool, a target for low-dose gamma-ray-induced oxidative stress. Radiat. Res. 2008;170:776–783. doi: 10.1667/RR1399.1. [DOI] [PubMed] [Google Scholar]
- 25.Shibutani S., Takeshita M., Grollman A.P. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxo-dG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 26.Pavlov Y.I., Minnick D.T., Izuta S., Kunkel T.A. DNA replication fidelity with 8-oxodeoxyguanosine triphosphate. Biochemistry. 1994;33:4695–4701. doi: 10.1021/bi00181a029. [DOI] [PubMed] [Google Scholar]
- 27.Shimizu M., Gruz P., Kamiya H., Kim S.R., Pisani F.M., Masutani C. Erroneous incorporation of oxidized DNA precursors by Y-family DNA polymerases. EMBO Rep. 2003;4:269–273. doi: 10.1038/sj.embor.embor765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klungland A., Rosewell I., Hollenbach S., Larsen E., Daly G., Epe B. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc. Natl. Acad. Sci. U. S. A. 1999;96:13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tahara Y.K., Auld D., Ji D., Beharry A.A., Kietrys A.M., Wilson D.L. Potent and selective inhibitors of 8-oxoguanine DNA glycosylase. J. Am. Chem. Soc. 2018;140:2105–2114. doi: 10.1021/jacs.7b09316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Visnes T., Cazares-Korner A., Hao W., Wallner O., Masuyer G., Loseva O. Small-molecule inhibitor of OGG1 suppresses proinflammatory gene expression and inflammation. Science. 2018;362:834–839. doi: 10.1126/science.aar8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tahara Y.K., Kietrys A.M., Hebenbrock M., Lee Y., Wilson D.L., Kool E.T. Dual inhibitors of 8-oxoguanine surveillance by OGG1 and NUDT1. ACS Chem. Biol. 2019;14:2606–2615. doi: 10.1021/acschembio.9b00490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paz-Elizur T., Ben-Yosef R., Elinger D., Vexler A., Krupsky M., Berrebi A. Reduced repair of the oxidative 8-oxoguanine DNA damage and risk of head and neck cancer. Canc. Res. 2006;66:11683–11689. doi: 10.1158/0008-5472.CAN-06-2294. [DOI] [PubMed] [Google Scholar]
- 33.Speina E., Arczewska K.D., Gackowski D., Zielinska M., Siomek A., Kowalewski J. Contribution of hMTH1 to the maintenance of 8-oxoguanine levels in lung DNA of non-small-cell lung cancer patients. J. Natl. Cancer Inst. 2005;97:384–395. doi: 10.1093/jnci/dji058. [DOI] [PubMed] [Google Scholar]
- 34.Paz-Elizur T., Sevilya Z., Leitner-Dagan Y., Elinger D., Roisman L.C., Livneh Z. DNA repair of oxidative DNA damage in human carcinogenesis: potential application for cancer risk assessment and prevention. Canc. Lett. 2008;266:60–72. doi: 10.1016/j.canlet.2008.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Audebert M., Chevillard S., Levalois C., Gyapay G., Vieillefond A., Klijanienko J. Alterations of the DNA repair gene OGG1 in human clear cell carcinomas of the kidney. Canc. Res. 2000;60:4740–4744. [PubMed] [Google Scholar]
- 36.Chevillard S., Radicella J.P., Levalois C., Lebeau J., Poupon M.F., Oudard S. Mutations in OGG1, a gene involved in the repair of oxidative DNA damage, are found in human lung and kidney tumours. Oncogene. 1998;16:3083–3086. doi: 10.1038/sj.onc.1202096. [DOI] [PubMed] [Google Scholar]
- 37.Giribaldi M.G., Munoz A., Halvorsen K., Patel A., Rai P. MTH1 expression is required for effective transformation by oncogenic HRAS. Oncotarget. 2015;6:11519–11529. doi: 10.18632/oncotarget.3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McPherson L.A., Troccoli C.I., Ji D., Bowles A.E., Gardiner M.L., Mohsen M.G. Increased MTH1-specific 8-oxo-dGTPase activity is a hallmark of cancer in colon, lung and pancreatic tissue. DNA Repair. 2019;83:102644. doi: 10.1016/j.dnarep.2019.102644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.David S.S., O'Shea V.L., Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chatterjee A., Mambo E., Osada M., Upadhyay S., Sidransky D. The effect of p53-RNAi and p53 knockout on human 8-oxoguanine DNA glycosylase (hOgg1) activity. Faseb. J. 2006;20:112–114. doi: 10.1096/fj.04-3423fje. [DOI] [PubMed] [Google Scholar]
- 41.Sur S., Pagliarini R., Bunz F., Rago C., Diaz L.A., Jr., Kinzler K.W. A panel of isogenic human cancer cells suggests a therapeutic approach for cancers with inactivated p53. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3964–3969. doi: 10.1073/pnas.0813333106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iyama T., Wilson D.M., 3rd DNA repair mechanisms in dividing and non-dividing cells. DNA Repair. 2013;12:620–636. doi: 10.1016/j.dnarep.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsuzuki T., Egashira A., Igarashi H., Iwakuma T., Nakatsuru Y., Tominaga Y. Spontaneous tumorigenesis in mice defective in the MTH1 gene encoding 8-oxo-dGTPase. Proc. Natl. Acad. Sci. U. S. A. 2001;98:11456–11461. doi: 10.1073/pnas.191086798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakumi K., Tominaga Y., Furuichi M., Xu P., Tsuzuki T., Sekiguchi M. Ogg1 knockout-associated lung tumorigenesis and its suppression by Mth1 gene disruption. Canc. Res. 2003;63:902–905. [PubMed] [Google Scholar]
- 45.De Luca G., Ventura I., Sanghez V., Russo M.T., Ajmone-Cat M.A., Cacci E. Prolonged lifespan with enhanced exploratory behavior in mice overexpressing the oxidized nucleoside triphosphatase hMTH1. Aging Cell. 2013;12:695–705. doi: 10.1111/acel.12094. [DOI] [PubMed] [Google Scholar]
- 46.Coppe J.P., Patil C.K., Rodier F., Sun Y., Munoz D.P., Goldstein J. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ensminger M., Iloff L., Ebel C., Nikolova T., Kaina B., Lbrich M. DNA breaks and chromosomal aberrations arise when replication meets base excision repair. J. Cell Biol. 2014;206:29–43. doi: 10.1083/jcb.201312078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang R., Li C., Qiao P., Xue Y., Zheng X., Chen H. OGG1-initiated base excision repair exacerbates oxidative stress-induced parthanatos. Cell Death Dis. 2018;9:628. doi: 10.1038/s41419-018-0680-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kidane D., Murphy D.L., Sweasy J.B. Accumulation of abasic sites induces genomic instability in normal human gastric epithelial cells during Helicobacter pylori infection. Oncogenesis. 2014;3:e128. doi: 10.1038/oncsis.2014.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chou W.C., Hu L.Y., Hsiung C.N., Shen C.Y. Initiation of the ATM-Chk2 DNA damage response through the base excision repair pathway. Carcinogenesis. 2015;36:832–840. doi: 10.1093/carcin/bgv079. [DOI] [PubMed] [Google Scholar]
- 51.Qin S., Lin P., Wu Q., Pu Q., Zhou C., Wang B. Small-molecule inhibitor of 8-oxoguanine DNA glycosylase 1 regulates inflammatory responses during Pseudomonas aeruginosa infection. J. Immunol. 2020;205:2231–2242. doi: 10.4049/jimmunol.1901533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou J., Ahn J., Wilson S.H., Prives C. A role for p53 in base excision repair. EMBO J. 2001;20:914–923. doi: 10.1093/emboj/20.4.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Achanta G., Huang P. Role of p53 in sensing oxidative DNA damage in response to reactive oxygen species-generating agents. Canc. Res. 2004;64:6233–6239. doi: 10.1158/0008-5472.CAN-04-0494. [DOI] [PubMed] [Google Scholar]
- 54.Samaranayake G.J., Troccoli C.I., Huynh M., Lyles R.D.Z., Kage K., Win A. Thioredoxin-1 protects against androgen receptor-induced redox vulnerability in castration-resistant prostate cancer. Nat. Commun. 2017;8:1204. doi: 10.1038/s41467-017-01269-x. [DOI] [PMC free article] [PubMed] [Google Scholar]