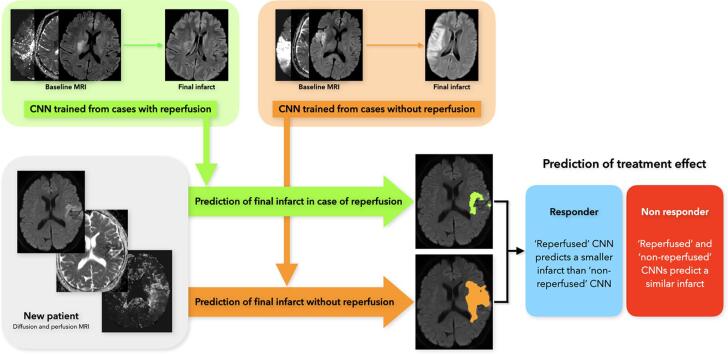
Graphical abstract
Keywords: Stroke, Prediction, Convolutional neural network, Magnetic resonance imaging, Reperfusion status
Highlights
-
•
Deep learning predictive models of the final stroke infarct may have better performances when trained on reperfusion status specific subsets.
-
•
The proposed predictive models outperformed the standards threshold-based perfusion-diffusion mismatch models.
-
•
Comparing the predicted infarct in case of a successful vs failed reperfusion may help in estimating the treatment effect and guiding therapeutic decisions in selected patients.
Abstract
Background
Predictive maps of the final infarct may help therapeutic decisions in acute ischemic stroke patients. Our objectives were to assess whether integrating the reperfusion status into deep learning models would improve their performance, and to compare them to current clinical prediction methods.
Methods
We trained and tested convolutional neural networks (CNNs) to predict the final infarct in acute ischemic stroke patients treated by thrombectomy in our center. When training the CNNs, non-reperfused patients from a non-thrombectomized cohort were added to the training set to increase the size of this group. Baseline diffusion and perfusion-weighted magnetic resonance imaging (MRI) were used as inputs, and the lesion segmented on day-6 MRI served as the ground truth for the final infarct. The cohort was dichotomized into two subsets, reperfused and non-reperfused patients, from which reperfusion status specific CNNs were developed and compared to one another, and to the clinically-used perfusion-diffusion mismatch model. Evaluation metrics included the Dice similarity coefficient (DSC), precision, recall, volumetric similarity, Hausdorff distance and area-under-the-curve (AUC).
Results
We analyzed 109 patients, including 35 without reperfusion. The highest DSC were achieved in both reperfused and non-reperfused patients (DSC = 0.44 ± 0.25 and 0.47 ± 0.17, respectively) when using the corresponding reperfusion status-specific CNN. CNN-based models achieved higher DSC and AUC values compared to those of perfusion-diffusion mismatch models (reperfused patients: AUC = 0.87 ± 0.13 vs 0.79 ± 0.17, P < 0.001; non-reperfused patients: AUC = 0.81 ± 0.13 vs 0.73 ± 0.14, P < 0.01, in CNN vs perfusion-diffusion mismatch models, respectively).
Conclusion
The performance of deep learning models improved when the reperfusion status was incorporated in their training. CNN-based models outperformed the clinically-used perfusion-diffusion mismatch model. Comparing the predicted infarct in case of successful vs failed reperfusion may help in estimating the treatment effect and guiding therapeutic decisions in selected patients.
1. Introduction
Early reperfusion, by means of intravenous thrombolysis or thrombectomy, is the main therapeutic goal in acute ischemic stroke (Powers et al., 2019). Acute treatment decisions have increasingly incorporated advanced neuroimaging to estimate patients’ prognosis and likelihood of benefiting from revascularization procedures (Nogueira et al., 2018, Albers et al., 2018). Currently, both computed-tomography (CT) and Magnetic Resonance Imaging (MRI) entail threshold-based methods to delineate the still salvageable brain (i.e. ischemic penumbra) from the already lost tissue (infarct core). Specifically in MRI, criteria for the infarct core is based on Apparent Diffusion Coefficient (ADC) extracted from Diffusion-Weighted Imaging (DWI), and criteria for the ischemic penumbra is based on Time to maximum of the residue function (Tmax) extracted from perfusion-weighted imaging. Precisely, infarct core is defined as ADC voxel values < 600620x10−6 mm2/s, and ischemic penumbra is defined as Tmax voxel values 6 s (Kidwell et al., 2013, Olivot et al., 2009). Patients with a large penumbra and limited ischemic core (so-called ‘target mismatch’ profile) have a high probability of benefiting from reperfusion, even in late time windows (Nogueira et al., 2018, Albers et al., 2018). However, these fixed-threshold methods may fail to capture the significant interindividual heterogeneity observed in stroke progression (Rekik et al., 2012). While the clinical and imaging characteristics of some patients may clearly indicate urgent reperfusion therapies, the benefit/risk balance in others can appear more uncertain. Thus, personalized probability maps of the final infarct would be of high clinical value to guide acute revascularization decisions and possibly help evaluate novel neuroprotective strategies.
Convolutional neural networks (CNNs), a subtype of machine learning, are flexible, data-driven methods capable of automatic non-linear feature extraction, with promising results in stroke lesion segmentation (Qiu et al., 2020). A well-acknowledged limitation of CNNs is the large quantity of data required for their training and validation. Only a limited number of studies, with heterogeneous treatment paradigms and evaluations metrics, have evaluated CNNs for the prediction of the final stroke lesion from baseline MRI (Winzeck et al., 2018, Pinto et al., 2018, Nielsen et al., 2018, Yu et al., 2020) or CT (Robben et al., 2020). Sample size and performance were modest (50 to 200 patients, Dice similarity coefficient 0.50 or lower), illustrating both the inherent difficulty of prediction tasks and scarcity of high-quality data, compared to simpler image segmentation tasks.
In the present work, we evaluated the impact of integrating the reperfusion status on the performance of CNNs for predicting the final infarct in patients with proximal intracranial occlusions treated by thrombectomy. Reperfusion is the single most important clinical metadata known to influence the progression of ischemic lesions from the baseline imaging (used as inputs to CNN) to the final infarct (Tsai and Albers, 2015). Previous studies have investigated direct integration of the reperfusion status during the learning process of CNN-based methods (Pinto et al., 2018, Robben et al., 2020). Another dichotomized the training set according to the reperfusion status with random forest-based methods (McKinley et al., 2017), but has not been evaluated with CNNs. We hypothesized that training CNNs from reperfusion status-specific subcohorts could improve their performance. Our objectives were: (1) to assess the impact of the reperfusion status on CNN-based predictive models; (2) to compare the predictive value of these CNNs against the threshold-based perfusion-diffusion mismatch models. An ancillary objective was to assess the relative predictive importance of the MRI inputs with an ablation study.
2. Material and methods
2.1. Data
We describe the HIBISCUS-STROKE and I-KNOW cohorts, from which the final stroke lesion was assessed. This section details the MRI protocol, patient inclusion criteria and image post-processing steps (upsampling, registration, normalization).
2.1.1. Patients and imaging protocol
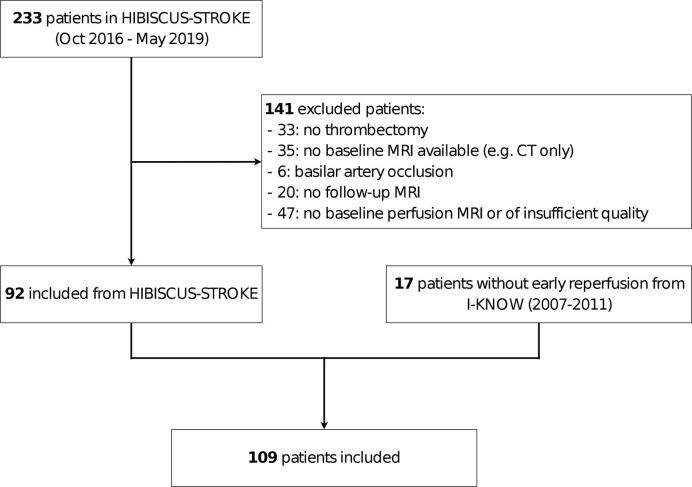
Patients were included from the HIBISCUS-STROKE and I-KNOW cohorts. HIBISCUS-STROKE is an ongoing monocentric observational cohort enrolling patients with a large intracranial artery occlusion treated by thrombectomy, following a baseline diffusion-perfusion MRI. I-KNOW (2007–2011) was a prospective multicenter observational study of stroke patients with both admission and several follow-up MRI. A subset of these patients underwent an acute follow-up perfusion MRI (3 h from the baseline MRI) to assess early reperfusion (Cho et al., 2015). In total, 109 patients were analyzed as shown in Fig. 1. Early reperfusion was observed in 74 patients, while 35 had no reperfusion (17 from I-KNOW and 18 from HIBISCUS-STROKE). Baseline patients’ characteristics are summarized in A.2. The inclusion and exclusion criteria for both cohorts are detailed in A.1. All patients from both cohorts gave their informed consent and the imaging protocol was approved by the regional ethics committee.
Fig. 1.
Patient inclusion flowchart.
In both cohorts, all patients underwent the following MRI protocol on admission: diffusion-weighted-imaging (DWI), T2-weighted fluid-attenuated-inversion-recovery (FLAIR), T2-gradient echo, MR-angiography and dynamic susceptibility-contrast perfusion imaging (DSC-PWI). A follow-up FLAIR was performed several days after admission (specifically, 6 and 30 days in HIBISCUS-STROKE and I-KNOW, respectively). MRI acquisition parameters are described in A.3.
2.1.2. Image post-processing
Parametric maps were extracted from the DSC-PWI by circular singular value decomposition of the tissue concentration curves (Olea Sphere, Olea Medical, La Ciotat, France): cerebral blood flow (CBF), cerebral blood volume (CBV), mean transit time (MTT), time to maximum (Tmax) and time to peak (TTP). Lesions on the baseline DWI and final FLAIR were segmented by an expert (THC) blinded to the clinical data with a semi-automated method (3D Slicer, https://www.slicer.org/). Specifically, a region-of-interest-controlled thresholding was used with manual corrections when required (for the DWI lesion, an ADC upper threshold of 620x10−6 mm2/s was used).
Images from HIBISCUS-STROKE were coregistered within subjects to the baseline DWI MRI using non-linear registration with Ants (Avants et al., 2011). Images from I-KNOW were coregistered within subjects to the PWI-DSC MRI (matrix 128x128) using affine registration with Statistical Parametric Mapping 8. Once co-registration was performed, HIBISCUS-STROKE patients had all MRI slices of size 192x192 compared to 128x128 for I-KNOW patients. As I-KNOW patients were largely in the minority (17 patients out of the 109 total patients), we up-sampled the images of I-KNOW patients to 192x192. The skull from all patients was removed using FSL (Smith et al., 2001). Finally, images were normalized between 0 and 1 to ensure inter-patient standardization.
2.2. Early reperfusion and training sets
We describe reperfusion criteria and we define the training sets.
2.2.1. Assessment of early reperfusion
In HIBISCUS-STROKE, early reperfusion was assessed at the end of the endovascular procedure with the modified Thrombolysis in Cerebral Infarction (mTICI) score (grade 0: no reperfusion; grade 1: anterograde reperfusion past the initial occlusion, but limited distal branch filling with little or slow distal reperfusion; grade 2a: anterograde reperfusion of less than half of the occluded target artery previously ischemic territory; grade 2b: anterograde reperfusion of more than half of the previously occluded target artery ischemic territory; grade 2c: near complete reperfusion, i.e. 90% but less than mTICI 3; grade 3: complete anterograde reperfusion) (Zaidat et al., 2013). Angiographic reperfusion was defined by mTICI scores of 2b-3, while patients without reperfusion had mTICI scores of 0-2a.
In I-KNOW, no patient was treated by endovascular procedures. Early reperfusion was assessed 3 h after the first MRI (H3) and was defined as voxels with Tmax 6 s at admission (H0) and Tmax < 6 s at H3. Acute reperfusion was defined by a reperfusion ratio (volume of reperfused voxels at H3/perfusion lesion volume at H0) of 50%.
2.2.2. Training sets
Three distinct training sets and corresponding models were built to assess the impact of reperfusion on the accuracy of final infarct prediction: a ‘general’ model, trained on the entire cohort irrespective of the reperfusion status (all training set); a ‘reperfused’ model, trained only with reperfused patients (reperfused training set); a ‘non-reperfused’ model, trained only with non-reperfused patients (non reperfused training set). Given the high rate of angiographic success in patients treated by thrombectomy (mTICI score of 2b-3 in 70% of patients) (Goyal et al., 2016), we expected a limited proportion of non-reperfused patients from HIBISCUS-STROKE. We thus included patients without early reperfusion from I-KNOW (identified by the H3 perfusion MRI follow-up) in order to improve this imbalance. I-KNOW patients were only included in the training set of the general and the non-reperfused models, but were not included in any testing set.
2.3. Proposed CNN architecture
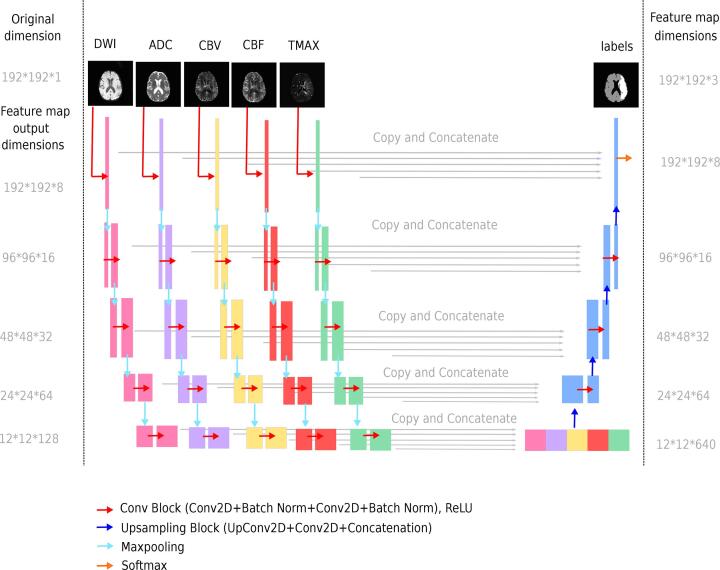
We used a U-Net architecture, a multi-scale network that has already shown its potential for infarct prediction tasks (Winzeck et al., 2018, Yu et al., 2020). Perfusion and diffusion MRI were used as inputs, as both modalities are complementary to evaluate the risk of infarction (Barber et al., 1998). More precisely, a total of five inputs were used: DWI and ADC for diffusion MRI, as well as Tmax, CBF and CBV for perfusion MRI. Previous studies in other medical applications have evaluated methods for combining the input data into CNNs, showing the merit of late fusion strategies (Nie et al., 2016, Aygün et al., 2018, Dolz et al., 2018, Dolz et al., 2018). Late fusion incorporates each input independently into distinct convolutional branches, subsequently merging features at a higher level. This strategy was chosen for its potential to better integrate each MRI input and the impact of reperfusion status. The comparison of the early and late fusion strategies is presented in C.
The five inputs (DWI, ADC, Tmax, CBV, CBF) were fed into our late fusion network of 5 distinct convolution branches. The proposed architecture is depicted in Fig. 2, and its encoding layers are detailed in Table 1. Each input consisted of whole 2D images (192x192). No patches were used in order to secure a large spatial context for lesion prediction. The network produced probability maps with 3 classes: lesion, healthy tissue, background. The lesion probability map was thresholded at 0.5 to define the final infarct. Training and configuration of the network are detailed in B.
Fig. 2.
Overview of the proposed deep learning architecture. Top left: The network takes five MRI images (2D slices from DWI, ADC, CBV, CBF, Tmax volumes) as input. Below: Each input image is processed independently on 5 separate branches. Pink, purple, yellow, red and green feature maps result from 2D-convolutions and maxpooling. The output of the 5 branches are then concatenated, and upsampled through 2D-deconvolution layers. The network produces an output map with 3 classes (lesion, healthy tissue and background). Top Right: The predicted lesion has to be compared to the true lesion from the final FLAIR.
Table 1.
Encoding layers of the proposed late fusion U-net. The encoder is composed of 5 convolution blocks (Conv Block), maxpooling operations (2D MaxPooling) and dropout. The Conv Block is made of: 2D convolution (3*3)+ batch normalization + 2D convolution (3*3)+ batch normalization.
| Layer (type) | Output shape |
|---|---|
| Conv Block 1 | 192*192*8 |
| 2D MaxPooling | 96*96*8 |
| Conv Block 2 | 96*96*16 |
| 2D MaxPooling | 48*48*16 |
| Conv Block 3 | 48*48*32 |
| 2D MaxPooling | 24*24*32 |
| Conv Block 4 | 24*24*64 |
| Dropout + 2D Maxpooling | 12*12*64 |
| Conv Block 5 + Dropout | 12*12*128 |
| Concatenation | 12*12*640 |
2.4. Evaluation
2.4.1. Ground truth
The final lesion is given by the FLAIR MRI, which was performed several days after admission (specifically, 6 and 30 days in HIBISCUS-STROKE and I-KNOW, respectively). The brain mask and the final lesion on the FLAIR MRI were segmented by experts using semi-automatic intensity-based thresholding. The ground truth for each patient was therefore a 3D mask with 3 classes: one class for background, one class for healthy tissues and one class for the lesion.
2.4.2. Metrics
Standard metrics for assessing image segmentation/prediction tasks were used: the Dice similarity coefficient (DSC), precision, recall, volumetric similarity (VS), and Hausdorff distance (HD) (Taha and Hanbury, 2015). The DSC measures the relative overlap of the prediction with the ground truth ( and FP are respectively the true positive, false negative and false positive voxels):
| (1) |
Precision (also know as positive predictive value) measures the percentage of voxels identified as lesion that have been classified correctly, while recall (also know as sensitivity) measures the percentage of actual lesion voxels that have been classified correctly:
| (2) |
| (3) |
The VS gives a relative ratio between the prediction and the ground truth volumes, without considering any overlap of the two volumes:
| (4) |
The HD is a measure of the distance of the largest error between the prediction (A) and ground truth (B):
| (5) |
The area-under-the-curve (AUC) is widely used in medical evaluation. Based on the ROC curve (Hajian-Tilaki, 2013), it provides an aggregated performance measure of an image modality or parametric map across all possible threshold values. However, the overwhelming number of non-infarcted voxels relative to infarcted ones can drive high AUC values while the extent and location of the infarct is poorly predicted (Jonsdottir et al., 2009). Several studies thus favored the DSC, which is more specific for lesion prediction (Winder et al., 2019, Yu et al., 2020). We presented AUC values in order to facilitate comparisons with some previous studies, notably when comparing CNN-based models and the clinical perfusion-diffusion mismatch model (Nielsen et al., 2018, Yu et al., 2020).
2.4.3. Perfusion-diffusion mismatch model
Our CNN-based predictive models were compared with the current reference method used in clinical practice. According to the perfusion-diffusion mismatch model, the projected final infarct can be defined as follows: (1) in reperfused patients, the final infarct is represented by the baseline diffusion lesion; (2) in non-reperfused patients, the final infarct is defined as the union of the acute diffusion lesion and the ischemic penumbra (voxels with a Tmax 6 s and normal DWI)(Olivot et al., 2009). The AUC of the perfusion-diffusion mismatch model to predict the final infarct was assessed in patients with and without reperfusion. Non-infarcted voxels were those not included in the diffusion lesion in reperfused patients, and those not included in the diffusion penumbra in non-reperfused patients. Infarcted voxels were the complementary voxels. The AUC was computed as in Jonsdottir et al., 2009.
2.4.4. Statistical analyses
A two-sided Wilcoxon signed-rank test was performed in order to compare the performances of: (1) reperfused vs general, non-reperfused vs general and reperfused vs non-reperfused models; (2) models with all MRI inputs vs models with ablation of one or more MRI inputs; (3) reperfused model vs diffusion lesion model; (4) non-reperfused model vs diffusion penumbra lesion model. Statistical analyses were performed using R version 3.5.1.
3. Results
3.1. Performance of the general, reperfused and non-reperfused CNNs
The performances and comparisons of the general, reperfused and non-reperfused models tested in reperfused and non-reperfused patients are presented in Table 2.
Table 2.
Performance metrics of the general, reperfused and non-reperfused models among (a) reperfused and (c) non-reperfused patients (average values ± standard deviation). Bold values correspond to the best value of the respective evaluation metric (column-wise). P-values from two-sided wilcoxon signed-rank tests comparing the general, reperfused and non-reperfused models among (b) reperfused and (d) non-reperfused patients. Bold values correspond to significant differences, with (*) indicating P < 0.05, (**) indicating P < 0.01 and (***) indicating P < 0.001. Note that tests were not corrected for multiple comparisons, and correspond to independent two-by-two comparisons.
|
(a) Performance metrics among reperfused patients | |||||
|---|---|---|---|---|---|
| Model | DSC | VS | Precision | Recall | HD |
| General | 0.43 ± 0.24 | 0.69 ± 0.27 | 0.55 ± 0.28 | 0.43 ± 0.25 | 33.23 ± 15.6 |
| Reperfused | 0.44 ± 0.25 | 0.70 ± 0.27 | 0.50 ± 0.27 | 0.50 ± 0.26 | 38.58 ± 18.1 |
| Non-reperfused | 0.35 ± 0.21 | 0.57 ± 0.28 | 0.60 ± 0.25 | 0.31 ± 0.24 | 40.05 ± 15.6 |
|
(b) Model comparisons among reperfused patients | |||||
|---|---|---|---|---|---|
| Two-sided Test | DSC P-value | VS P-value | Precision P-value | Recall P-value | HD P-value |
| General vs Reperfused | 0.43 | 0.53 | 3.7e-6 (***) | 1.4e-6 (***) | 0.048 (*) |
| General vs Non-Reperfused | 1.4e-8 (***) | 4.3e-6 (***) | 0.0069 (**) | 1.0e-10 (***) | 0.0041 (**) |
| Reperfused vs Non-Reperfused | 2.3e-8 (***) | 1.6e-5 (***) | 2.9e-7 (***) | 2.7e-11 (***) | 0.65 |
|
(c) Model performance among non-reperfused patients | |||||
|---|---|---|---|---|---|
| Model | DSC | VS | Precision | Recall | HD |
| General | 0.44 ± 0.21 | 0.66 ± 0.26 | 0.39 ± 0.25 | 0.63 ± 0.21 | 30.61 ± 16.1 |
| Reperfused | 0.44 ± 0.22 | 0.63 ± 0.25 | 0.36 ± 0.23 | 0.69 ± 0.22 | 44.53 ± 16.7 |
| Non-reperfused | 0.47 ± 0.17 | 0.74 ± 0.13 | 0.49 ± 0.22 | 0. 52 ± 0.21 | 37.70 ± 17.7 |
|
(d) Model comparisons among non-reperfused patients | |||||
|---|---|---|---|---|---|
| Two-sided Test | DSC P-value | VS P-value | Precision P-value | Recall P-value | HD P-value |
| General vs Reperfused | 0.93 | 0.55 | 0.13 | 0.021 (*) | 0.0023 (**) |
| General vs Non-Reperfused | 0.17 | 0.21 | 0.0016 (**) | 0.00084 (***) | 0.11 |
| Reperfused vs Non-Reperfused | 0.13 | 0.034 (*) | 0.00067 (***) | 5.3e-5 (***) | 0.12 |
Among reperfused patients, the non-reperfused model was inferior to either the reperfused or general models for all metrics except for precision (Table 2, Table 2-b). The model seems to predict many false negative voxels (low recall), many outlier voxels (high hausdorff distance), and a different volume than expected (low VS). Conversely, no clear-cut performance difference was found between the reperfused and general models.
Among non-reperfused patients, the non-reperfused model had better or similar performance than the reperfused model for all metrics except for recall (Table 2, Table 2-d). The model seems to predict the lesion well in terms of volume and localisation (high VS and high DSC), with few false positive voxels (high precision) but some false negative voxels (medium recall). No clear overall difference was observed between the non-reperfused and general models, or between the reperfused and general models.
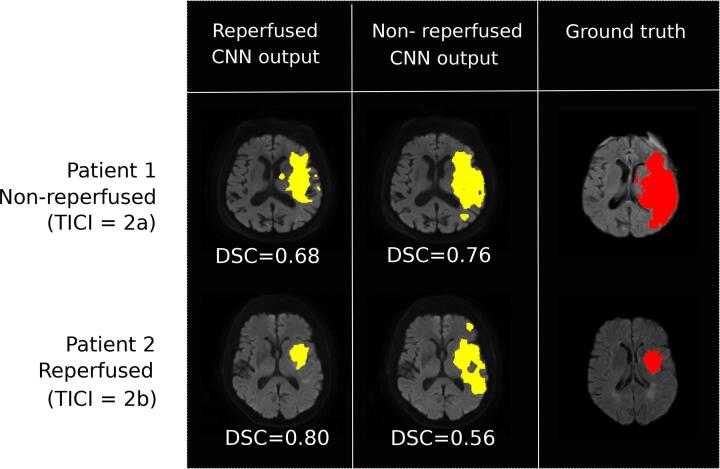
The predicted infarct volumes were significantly larger with the non-reperfused compared to the reperfused model (39.7 mL (61.3–20) vs 17.5 mL (28–5.1), for the non-reperfused and reperfused models, respectively; median with interquartile range). Accordingly, significant differences of VS between these two models were observed (Tables 2-b and -d). Fig. 3 illustrates and compares the output of the two CNNs (reperfused and non-reperfused) for two patients with distinct reperfusion status.
Fig. 3.
CNN-based predictions of the final infarct using the reperfused and non-reperfused models, applied in: patient 1 (no reperfusion, TICI = 2a); patient 2 (reperfused, TICI = 2b).
3.2. Comparison of CNN-based models and the perfusion-diffusion mismatch model
In both reperfused and non-reperfused patients, the DSC, VS and recall of CNN-based models were superior to those of the perfusion-diffusion mismatch models (Table 3). Final lesion predicted by CNNs are therefore more spatially and volumetrically coherent (high DSC and VS), and have fewer false negative voxels than the mismatch model. At the patient level, higher DSC values were achieved with CNN-based models in 68% and 89% of the reperfused and non-reperfused patients, respectively. Conversely, the precision of mismatch models was higher than that of CNN, suggesting more false positive voxels with the latter methods.
Table 3.
Comparison of CNN-based and perfusion-diffusion mismatch models. Among reperfused patients (upper rows), the CNN-based reperfused model was compared to the threshold-based diffusion lesion. Among non-reperfused patients (lower rows), the CNN-based non-reperfused model was compared to the threshold-based diffusion penumbra lesion. Bold values correspond to the best value of the respective evaluation metric (column-wise). A two-sided wilcoxon signed-rank test was performed between the proposed models and the clinical models, with (.) indicating P < 0.10, (*) indicating P < 0.05, (**) indicating P < 0.01 and (***) indicating P < 0.001.
|
Reperfused patients | |||||
|---|---|---|---|---|---|
| Model | DSC | VS | Precision | Recall | HD |
| CNN | 0.44 ± 0.21 (*) | 0.66 ± 0.26 (***) | 0.39 ± 0.25 | 0.63 ± 0.21 (***) | 30.61 ± 16.1 |
| Perfusion-diffusion mismatch | 0.41 ± 0.23 | 0.56 ± 0.27 | 0.71 ± 0.31 (***) | 0.33 ± 0.20 | 19.34 ± 10.3 (***) |
|
Non-reperfused patients | |||||
|---|---|---|---|---|---|
| Model | DSC | VS | Precision | Recall | HD |
| CNN | 0.47 ± 0.17 (***) | 0.74 ± 0.13 (***) | 0.49 ± 0.22 | 0.52 ± 0.21 (***) | 37.70 ± 17.7 (***) |
| Perfusion-diffusion mismatch | 0.26 ± 0.17 | 0.31 ± 0.21 | 0.84 ± 0.16 (***) | 0.17 ± 0.13 | 69.15 ± 7.7 |
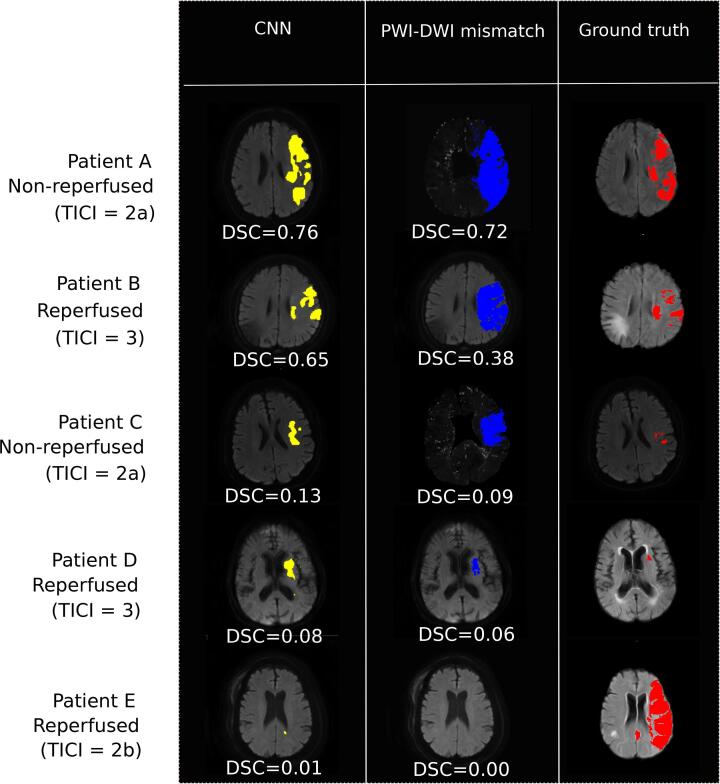
CNN-based models achieved higher AUC values compared to those of perfusion-diffusion mismatch models (reperfused patients: 0.87 ± 0.13 vs 0.79 ± 0.17, P < 0.001; non-reperfused patients: 0.81 ± 0.13 vs 0.73 ± 0.14, P < 0.01, in CNN vs perfusion-diffusion mismatch models, respectively). Cases illustrating successful or suboptimal outputs from CNN and mismatch models are presented in Fig. 4.
Fig. 4.
Output predictions from CNN models compared with the PWI-DWI mismatch model. Five tested patients are shown: two successful cases when CNN models outperform PWI-DWI mismatch in reperfused and non-reperfused patients (patient A with TICI = 2a and patient B with TICI = 3) and three difficult patients to predict, for both CNN and PWI-DWI mismatch models (patient C with TICI = 2a, patient D with TICI = 3 and patient E with TICI = 2b). For each prediction model, patient-wide DSC is specified.
The comparison of CNNs and perfusion-diffusion mismatch model was included as the latter remains the reference method in clinical practice. The mismatch model only provides a crude threshold-based segmentation of baseline images, and may not match the feature extraction potential of CNNs. Also, the mismatch model is only based on ADC and Tmax in order to predict the final lesion outcome, whereas our model is based on more inputs (DWI, ADC, Tmax, CBV, CBF).
3.3. Value of the MRI inputs for predicting the final infarct
An ablation study was performed with the reperfused and non-reperfused models (tested only in reperfused and non-reperfused patients, respectively) in order to evaluate the relative importance of the different MRI inputs for predicting the final infarct. In both reperfused and non-reperfused patients, the full CNN models (i.e. including DWI, ADC, Tmax, CBF and CBV) had similar performances compared to models without CBF and CBV, suggesting these latter inputs had limited predictive value (lines 1 and 2 from Table 4, Table 4-b). Conversely, adding the diffusion data (DWI and ADC) to Tmax maps significantly increased the DSC of these CNNs. This performance increase was more pronounced among reperfused patients compared to those without reperfusion.
Table 4.
Evaluation metrics of the reperfused and non-reperfused models after successive ablation of the MRI inputs, tested among (a) reperfused and (b) non-reperfused patients, respectively (average values ± standard deviation). Bold values correspond to the best value of the respective evaluation metric (column-wise). A two-sided wilcoxon signed-rank test was performed between the full models with all 5 MRI inputs and the ablated ones, with (.) indicating P < 0.10, (*) indicating P < 0.05, (**) indicating P < 0.01 and (***) indicating P < 0.001.
|
(a) Reperfused model: ablation study among reperfused patients | |||||
|---|---|---|---|---|---|
| Input MRI | DSC | VS | Precision | Recall | HD |
| DWI + ADC + Tmax+CBF + CBV | 0.44 ± 0.21 | 0.66 ± 0.26 | 0.39 ± 0.25 | 0.63 ± 0.21 | 30.61 ± 16.1 |
| DWI + ADC + Tmax | 0.44 ± 0.25 | 0.70 ± 0.26 | 0.54 ± 0.28 (***) | 0.46 ± 0.27 (**) | 35.13 ± 15.6 (.) |
| DWI | 0.42 ± 0.24 (*) | 0.70 ± 0.26 | 0.51 ± 0.28 | 0.44 ± 0.27 (***) | 31.28 ± 16.1 (**) |
| ADC | 0.40 ± 0.24 (***) | 0.67 ± 0.28 (.) | 0.47 ± 0.27 (.) | 0.43 ± 0.27 (***) | 34.35 ± 20.4 (*) |
| Tmax | 0.32 ± 0.20 (***) | 0.63 ± 0.30 (*) | 0.44 ± 0.25 (*) | 0.35 ± 0.25 (***) | 29.99 ± 13.7 (**) |
|
(b) Non-reperfused model: ablation study among non-reperfused patients | |||||
|---|---|---|---|---|---|
| Input MRI | DSC | VS | Precision | Recall | HD |
| DWI + ADC + Tmax+CBF + CBV | 0.47 ± 0.17 | 0.74 ± 0.13 | 0.49 ± 0.22 | 0.52 ± 0.21 | 37.70 ± 17.7 |
| DWI + ADC + Tmax | 0.47 ± 0.18 | 0.74 ± 0.16 | 0.52 ± 0.22 | 0.50 ± 0.22 | 35.77 ± 20.2 |
| DWI | 0.45 ± 0.17 | 0.71 ± 0.17 | 0.50 ± 0.22 | 0.50 ± 0.25 | 33.20 ± 17.2 |
| ADC | 0.42 ± 0.15 (.) | 0.73 ± 0.23 | 0.47 ± 0.18 | 0.46 ± 0.21 (.) | 28.35 ± 12.9 (**) |
| Tmax | 0.40 ± 0.19 (*) | 0.65 ± 0.21 | 0.50 ± 0.29 | 0.46 ± 0.24 (**) | 26.86 ± 13.3 (.) |
4. Discussion
4.1. Impact of the reperfusion status on CNN performance
Our study showed that the performance of CNN-based models improved when trained from reperfusion status-specific subgroups. The predicted lesion had better overlap (i.e. higher DSC) with the final infarct in both reperfused and non-reperfused patients, when using the corresponding reperfusion status-specific CNN.
Baseline imaging features do have significant predictive value, and CNNs trained without data on reperfusion can successfully predict the final lesion in some patients (Yu et al., 2020). This may in part reflect the mostly homogenous profile of patients currently treated by thrombectomy (i.e. limited cerebral damage at baseline and successful reperfusion). Indeed, the training set for our general CNN consisted of 70% of reperfused patients, and this case-mix likely accounts for the lack of significant difference between the general and reperfused models.
Still, the pathophysiological rationale for integrating the reperfusion status in predictive models is strong. Timely reperfusion is closely associated with increased penumbra salvage and reduced final infarct size (Cho et al., 2015). We propose that a new patient’s eligibility to treatment could be assessed by using both CNNs (the one trained from reperfused and the other from non-reperfused patients). The clinician would thus have a dual set of predictive maps allowing a comparison of the projected infarct with and without reperfusion, and an estimation of the treatment effect. A mismatch between these two models (i.e. a smaller infarct in case of a successful thrombectomy that achieved reperfusion, than in the no-reperfusion model) would indicate that this patient is likely to benefit from therapy (responder). Conversely, a similar output from the reperfused and non-reperfused models would suggest a limited effect of therapy (non-responder). In our selected dataset, the final predicted infarct was substantially larger with the non-reperfused CNN in 53 (50%) patients when considering the following criteria: DSC between the two CNNs < 0.5 and non-reperfused CNN lesion volume 20% larger than the output of the reperfused CNN. Conversely, the absence of a clear difference between the two models would suggest limited benefit from reperfusion therapies. Reliable predictions of the final infarct may also help in evaluating novel neuroprotection strategies, by comparing the projected vs observed infarct size in patients with ischemia–reperfusion (Hougaard et al., 2013). This approach may facilitate the screening of a larger number of putative neuroprotectants at lesser cost than full-sized controlled trials.
Our results indicate that CNN can successfully take into account reperfusion by conditioning the training dataset according to this clinical status, in order to achieve more robust predictions. The full validation of this approach will require a multicentric collaboration in order to collect high quality longitudinal data, including cases without reperfusion.
4.2. Comparison to current clinical prediction methods
Our CNN models achieved higher AUC and DSC than the perfusion-diffusion mismatch models currently used in clinical practice (patient A and B in Fig. 4 are illustrative cases). Our results were in the same range as those of recently reported CNNs: the best model of the ISLES challenge achieved a DSC of 0.38 (Winzeck et al., 2018, Nielsen et al., 2018 reported a mean AUC of 0.88, while Yu et al., 2020 reported a mean DSC and AUC of 0.53 and 0.89, respectively. However, a strict comparison is not possible as the cited studies were all performed on different datasets, and in the light of different time-windows of prediction.
We also confirmed that predicting the final infarct remains a challenging task. Mean DSC were modest (0.44 and 0.47 for the reperfused and non-reperfused model, respectively), corresponding to an assortment of highly accurate predictions (DSC0.7) and failure of both CNNs and perfusion-diffusion mismatch models in other cases (e.g. patient C, D and E in Fig. 4). Partial and sometimes extensive reversal of the diffusion lesion can be observed (patients C and D in Fig. 4), especially in the event of early reperfusion (Yoo et al., 2019). This phenomenon may particularly affect patients with small baseline DWI lesion, in whom even limited discrepancies between the predicted and observed infarct may result in very low DSC values. Still, no significant correlation was found between the DSC and baseline DWI lesion volume (r = 0.038, p = 0.72). Also, baseline imaging cannot account for subsequent events that may alter the progression of ischemic lesions (e.g. patient E in Fig. 4: a possible case of reocclusion after a successful reperfusion). These patients illustrate the heterogeneity and complexity of stroke lesion progression. Reinforcement learning could help improve the performance of CNNs by training more specifically on these underrepresented patients (Arulkumaran et al., 2017).
4.3. Predictive value of the MRI inputs
The ablation study showed that CBF and CBV had limited impact on the performance of our CNN. This result is in line with the common qualitative observation that the perfusion lesion is less conspicuous on CBF or CBV maps compared to Tmax maps. A previous voxel and threshold-based study had also observed that these parameters were poor predictors of the final infarct (Christensen et al., 2009).
Thus, ADC, DWI and Tmax could constitute the main inputs for the network predicting the final infarct. Similarly, Livne et al. have shown that both perfusion parameters and DWI made significant predictive contributions, albeit with a different method (extreme gradient tree boosting) and among patients who were not treated by thrombectomy and thus had a significantly lower rate of reperfusion (Livne et al., 2018). Our study was conducted among thrombectomy-treated patients with a reperfusion rate of 80%, in whom the baseline DWI lesion is known to have a strong correlation with the final infarct. Our results further suggest that Tmax maps may have a greater predictive value among non-reperfused patients, which would be consistent with previously available data. Wheeler et al., 2013 had shown a strong correlation between the baseline diffusion lesion and final infarct volume in reperfused patients, and a high correlation between the Tmax 6 s lesion and final infarct volume for non-reperfused patients.
These observations support our chosen deep learning architecture. The late fusion configuration allows for better integration of the distinct information contained in perfusion and diffusion imaging. Training reperfusion status-specific models entail assigning distinct weights to each MRI input. The performance of CNNs built with an early fusion configuration are presented in C. Early fusion had overall worse performance than late fusion. Fewer performance differences were also observed between the general, reperfused and non-reperfused models, suggesting that early fusion may overlook the reperfusion status.
4.4. Limitations
Our study presents several limitations. Patients were included from two cohorts with different treatment protocols: HIBISCUS-STROKE involved patients treated by thrombectomy, whereas I-KNOW was a multicentric observational study of patients managed conservatively or with intravenous thrombolysis without any endovascular procedure. However, I-KNOW only contributed patients with proximal occlusions without reperfusion, who likely have a very similar course to failed thrombectomy cases. Methods for assessing early reperfusion differed between these two cohorts. Nevertheless, as proposed in a previous study, MRI and angiographic data can be pooled when evaluating reperfusion (Marks et al., 2014). Several precautions were observed to limit potential biases: (i) TICI score assessment strictly followed standard recommandations (Zaidat et al., 2013) and was thus not a surrogate for recanalization; (ii) both TICI score and DSC-PWI assess tissue perfusion; similar criteria for both methods were used to identify reperfusion (TICI 2b and DSC-PWI reperfusion ratio 50%); (iii) in I-KNOW, the follow-up DSC-PWI used to assess reperfusion was performed with a median delay of 170 min from the baseline MRI, and was thus in a similar ultra-early time frame as HIBISCUS patients undergoing endovascular treatment. Furthermore, no significant difference was found between the non-reperfused patients of the two cohorts for the following baseline variables: gender, age, baseline NIHSS score, time from symptoms onset to MRI, baseline DWI lesion size. The HIBISCUS cohort had a majority of M1 occlusions (15/18; 3 patients had a M2 occlusion), while most I-KNOW patients had M2 occlusions (12/17; 5 had a M1 occlusion. This significant difference in occlusion level (p = 0.002, Fisher’s exact test) is likely related to the distinct inclusion criteria of these two cohorts (HIBISCUS specifically included patients with proximal intracranial occlusions). Other clinical parameters such as age and time from symptoms onset to imaging and reperfusion are recognized prognostic factors. Their integration in predictive CNNs may enhance model performance and warrants further investigation. Finally, the interval between stroke onset and the follow-up MRI was 6 days. Other studies used different or similar delays: 3 to 7 days (Yu et al., 2020), 1-month (Nielsen et al., 2018) or 90 days (Winzeck et al., 2018). A previous study has shown that the 24-h DWI lesion volume was well correlated with day 90 FLAIR lesion volume (Campbell et al., 2012). Infarct volume at either time points predicted functional outcome. Studies using different intervals may be compared provided a successful coregistration of baseline and final images was achieved.
5. Conclusion
The performance of deep learning models improved when the reperfusion status was incorporated in their training. CNN-based models outperformed the clinically-used perfusion-diffusion mismatch model. Comparing the predicted infarct in case of a successful vs failed reperfusion may help in estimating the treatment effect and guiding therapeutic decisions in selected patients.
Author Contributions
Noëlie Debs: investigation, methodology, writing - original draft. Tae-Hee Cho: conceptualization, data acquisition and annotation, validation, critical revision of the manuscript. David Rousseau: conceptualization, validation, critical revision of the manuscript. Yves Berthezène: data acquisition; critical revision of the manuscript. Marielle Buisson: project administration. Omer Eker: data acquisition, critical revision of the manuscript. Laura Mechtouff: data acquisition, critical revision of the manuscript. Norbert Nighoghossian: project administration, data acquisition, critical revision of the manuscript. Michel Ovize: project administration, critical revision of the manuscript. Carole Frindel: conceptualization, validation, critical revision of the manuscript, supervision.
Acknowledgment and funding sources
This work was supported by the RHU MARVELOUS (ANR-16-RHUS-0009) of Université Claude Bernard Lyon-1 (UCBL), and was also performed within the framework of the RHU BOOSTER (ANR-18-RHUS-0001), within the program ”Investissements d’Avenir“ operated by the French National Research Agency (ANR).
Contributor Information
Noëlie Debs, Email: noelie.debs@creatis.insa-lyon.fr.
Tae-Hee Cho, Email: tae-hee.cho@chu-lyon.fr.
David Rousseau, Email: david.rousseau@univ-angers.fr.
Yves Berthezène, Email: yves.berthezene@chu-lyon.fr.
Marielle Buisson, Email: marielle.buisson01@chu-lyon.fr.
Omer Eker, Email: omer.eker@chu-lyon.fr.
Laura Mechtouff, Email: laura.mechtouff@chu-lyon.fr.
Norbert Nighoghossian, Email: norbert.nighoghossian@chu-lyon.fr.
Michel Ovize, Email: michel.ovize@chu-lyon.fr.
Carole Frindel, Email: carole.frindel@creatis.insa-lyon.fr.
Appendix A. Data
A.1. Inclusion criteria of HIBISCUS-STROKE and I-KNOW
Inclusion criteria for HIBISCUS-STROKE were: (1) patients with an anterior circulation stroke related to a proximal intracranial occlusion (internal carotid artery, M1 or M2 occlusion), directly admitted to our comprehensive stroke unit (‘mothership’ paradigm); (2) diffusion and perfusion MRI as baseline imaging; (3) patients treated by thrombectomy with or without intravenous thrombolysis.
Inclusion and exclusion criteria for I-KNOW were: (1) NIHSS 4; (2) diffusion and perfusion MRI consistent with an acute anterior circulation ischemic stroke; and (3) admission MRI completed within 6 h for patients treated with intravenous thrombolysis, or within 12 h for those managed without thrombolysis. Patients with lacunar or posterior circulation stroke, unknown time of onset or intracerebral hemorrhage were excluded. No patient received intra-arterial therapy. For the present study, additional inclusion criteria were applied, as follows: (1) both admission and acute follow-up diffusion and perfusion MRI obtained 3 h after initial imaging (H3) available and assessable; (2) visible occlusion on the baseline MRA; and (3) H3 perfusion without significant reperfusion. (see Table A.5).
Table A.5.
Baseline characteristics (median with interquartile range, unless otherwise indicated). NIHSS: National Institutes of Health Stroke Scale; DWI: diffusion-weighted imaging; ICA: internal carotid artery.
| Clinical variables | |
|---|---|
| Women, n (percentage) | 45 (41.3) |
| Age | 70 (57–79) |
| NIHSS score | 15 (10–19) |
| Time from symptoms onset to MRI | 105 (78–154) |
| Intravenous tPA, n (percentage) | 59 (54.1) |
| Site of occlusion, n (percentage): | |
| intracranial ICA + M1 | 27 (24.8) |
| M1 | 54 (49.5) |
| intracranial ICA + M2 | 23 (21.1) |
| M2 | 5 (4.6) |
| cervical ICA, n (percentage) | 19 (17.4) |
| DWI lesion size, mL | 24.9 (7.4–50.9) |
A.2. Patients’ baseline characteristics
.
A.3. MRI protocol
All patients underwent DWI (IKNOW: repetition time 6000 ms, field of view 24 cm, matrix 128128 (IKNOW) or 192192 (HIBISCUS-STROKE), slice thickness 5 mm), Fluid-attenuated-inversion-recovery (repetition time 8690 ms, echo time 109 ms, inversion time 2500 ms, field of view 21 cm, matrix 224256, section thickness 5 mm), T2-weighted gradient echo (repetition time 800 ms, echo time 28 ms, flip angle 20, field of view 230 mm, matrix 512512, section thickness of 5 mm), MRA and DSC-PWI (echo time 40 ms, repetition time 1500 ms, field of view 24 cm, matrix 128128, slice thickness 5 mm; gadolinium contrast at 0.1 mmol/kg), both for the admission and follow-up MRI.
Appendix B. Network training and parameters
Only slices including the final infarct were used to train the U-net and no data augmentation was employed. We used a multi-class Dice function as a loss function (Milletari et al., 2016), for which the lesion class was assigned a weight 8 times higher than those of healthy and background classes. We used the Adam optimizer ( and ) and a batch size of 12. To prevent overfitting, we applied dropout (set to 0.5), used a L2 regularizer reg at each convolution layer () and the number of epochs (set to 500) was regulated by early stopping (i.e. the training was stopped once the best validation multi-class dice did not increase more than 0.005 on 100 epochs). The evaluation of each model was performed using a 5-fold cross-validation. Note that patients from I-KNOW dataset were added in the training set of the general and the non-reperfused models for data-augmentation purposes, but were not used in the testing set. Specifically, the number of training patients was, depending on the fold: between 89 and 91 patients for the general model, between 59 and 60 patients for the reperfused model, and between 30 and 31 patients for the non-reperfused model. The number of test patients varied between 17 and 19 (reperfused and non-reperfused patients combined).
The number of parameters is proportional to the number of U-Net path: thus, the number of trainable parameters is 1997851 for a U-Net architecture with 5 MRI sequence inputs, 1242603 for 3 MRI inputs, and 487355 when using only one input. The higher the number of paths, the less the information is compressed and the more the architecture offers the possibility of learning different information on each input data. Thus, we chose not to balance the number of parameters between each architecture. However, to ensure a fair comparison, each network’s hyperparameters were independently fine-tuned on a fixed search space. The best parameters were found to be the same in all tested architectures. We used Keras 2.1.3 library with Python 3.6.3 interface. The training phase took approximately 1 h on a work station with an NVIDIA GeForce GTX 1080 GPU with 128 GB memory.
Appendix C. Impact of the multiple MRI fusion configuration
We compared our proposed late fusion deep learning architecture to an early fusion one, where all patient input images are combined at the beginning of the CNN. This fusion strategy reduces both the computational complexity and training parameters (Chen et al., 2019). Each patient being represented by DWI, ADC, Tmax, CBV, CBF, the early fusion architecture stacks channel-wise these 5 MRI inputs and does not process them independently. Results are shown in Table C.6.
Table C.6.
Evaluation metrics after training models on different training set (all, reperfused, and non-reperfused) with different fusion strategies (early and late) and evaluating them on reperfused testing patients (a) and non-reperfused testing patients (b) (average values ± standard deviation). Bold values correspond to the best value of the respective evaluation metric (column-wise). A two-sided wilcoxon signed-rank test was performed between global model and the two other models (reperfused and non-reperfused) for a given fusion strategy, with (.) indicating P < 0.10, (*) indicating P < 0.05, (**) indicating P < 0.01 and (***) indicating P < 0.001.
|
(a) Evaluation on reperfused testing patients | ||||||
|---|---|---|---|---|---|---|
| Fusion | Training | DSC | VS | Precision | Recall | HD |
| early | all | 0.39 ± 0.25 | 0.59 ± 0.30 | 0.56 ± 0.31 | 0.40 ± 0.26 | 29.51 ± 16.26 |
| early | reperfused | 0.41 ± 0.25 | 0.64 ± 0.30 | 0.46 ± 0.29 (***) | 0.49 ± 0.30 (***) | 31.24 ± 15.61 |
| early | non-reperfused | 0.36 ± 0.22 (*) | 0.63 ± 0.27 | 0.54 ± 0.26 | 0.33 ± 0.24 (***) | 26.64 ± 11.16 |
| late | all | 0.43 ± 0.24 | 0.69 ± 0.27 | 0.55 ± 0.28 | 0.43 ± 0.25 | 33.23 ± 15.64 |
| late | reperfused | 0.44 ± 0.25 | 0.70 ± 0.27 | 0.50 ± 0.27 | 0.50 ± 0.26 (***) | 38.58 ± 18.15 |
| late | non-reperfused | 0.35 ± 0.21 (***) | 0.57 ± 0.28 (***) | 0.60 ± 0.25 (***) | 0.31 ± 0.24 (***) | 40.05 ± 15.66 (**) |
|
(b) Evaluation on non-reperfused testing patients | ||||||
|---|---|---|---|---|---|---|
| Fusion | Training | DSC | VS | Precision | Recall | HD |
| early | all | 0.42 ± 0.24 | 0.62 ± 0.27 | 0.42 ± 0.28 | 0.55 ± 0.29 | 30.98 ± 18.23 |
| early | reperfused | 0.41 ± 0.26 | 0.51 ± 0.31 | 0.36 ± 0.29 | 0.69 ± 0.24 | 30.94 ± 16.30 |
| early | non-reperfused | 0.42 ± 0.18 | 0.66 ± 0.17 | 0.42 ± 0.24 | 0.55 ± 0.22 | 28.48 ± 13.63 |
| late | all | 0.44 ± 0.21 | 0.66 ± 0.26 | 0.39 ± 0.25 | 0.63 ± 0.21 | 30.61 ± 16.15 |
| late | reperfused | 0.44 ± 0.22 | 0.63 ± 0.25 | 0.36 ± 0.23 | 0.69 ± 0.22 (*) | 44.53 ± 16.79 (**) |
| late | non-reperfused | 0.47 ± 0.17 | 0.74 ± 0.13 | 0.49 ± 0.22 (**) | 0. 52 ± 0.21 (***) | 37.70 ± 17.74 |
It appears that best metric values are obtained when performing a late fusion strategy rather than an early fusion: average values of DSC, VS, precision and recall are higher whatever the training set (all, reperfused, non reperfused). However, lowest values for HD metric are obtained when performing early fusion. Early fusion seems to offer a better spatial delineation of the final lesion: fewer outliers seem to be predicted, which drastically decreases HD values.
With early fusion configuration, differences observed between the global model and the reperfused and non-reperfused submodels are smaller and not significant. This type of architecture seems less adapted to take into account the status of reperfusion.
References
- Albers G. Thrombectomy for stroke at 6 to 16 hours with selection by perfusion imaging. N. Engl. J. Med. 2018;378:708–718. doi: 10.1056/NEJMoa1713973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arulkumaran K., Deisenroth M.P., Brundage M., Bharath A.A. Deep reinforcement learning: a brief survey. IEEE Signal Process. Mag. 2017;34:26–38. [Google Scholar]
- Avants B.B., Tustison N.J., Song G., Cook P.A., Klein A., Gee J.C. A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage. 2011;54:2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aygün, M., Şahin, Y.H., Ünal, G., 2018. Multi modal convolutional neural networks for brain tumor segmentation. arXiv preprint arXiv:1809.06191.
- Barber P., Darby D., Desmond P., Yang Q., Gerraty R., Jolley D., Donnan G., Tress B., Davis S.M. Prediction of stroke outcome with echoplanar perfusion- and diffusion-weighted MRI. Neurology. 1998;51:418–426. doi: 10.1212/wnl.51.2.418. [DOI] [PubMed] [Google Scholar]
- Campbell, B.C.V., Tu, H.T.H., Christensen, S., Desmond, P.M., Levi, C.R., Bladin, C.F., Hjort, N., Ashkanian, M., SAlling, C., Donnan, G.A., Davis, S.M., Ostergaard, L., Parsons, M.W., 2012. Assessing response to stroke thrombolysis: validation of 24-hour multimodal magnetic resonance imaging. Arch. Neurol. 69, 46-50. [DOI] [PubMed]
- Chen Y., Wang K., Liao X., Qian Y., Wang Q., Yuan Z., Heng P.A. Channel-UNet: a spatial channel-wise convolutional neural network for liver and tumors segmentation. Front. Genet. 2019;10 doi: 10.3389/fgene.2019.01110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho T.H., Nighoghossian N., Mikkelsen I.K., Derex L., Hermier M., Pedraza S., Fiehler J., Østergaard L., Berthezène Y., Baron J.C. Reperfusion within 6 hours outperforms recanalization in predicting penumbra salvage, lesion growth, final infarct, and clinical outcome. Stroke. 2015;46:1582–1589. doi: 10.1161/STROKEAHA.114.007964. [DOI] [PubMed] [Google Scholar]
- Christensen, S., Mouridsen, K., Wu, O., Hjort, N., Karstoft, H., Thomalla, G., Röther, J., Fiehler, J., Kucinski, T., Østergaard, L., 2009. Comparison of 10 perfusion MRI parameters in 97 sub-6-hour stroke patients using voxel-based receiver operating characteristics analysis. Stroke 40, 2055–2061. [DOI] [PubMed]
- Dolz J., Ayed I.B., Desrosiers C. Dense multi-path U-Net for ischemic stroke lesion segmentation in multiple image modalities. International MICCAI Brainlesion Workshop, Springer. 2018:271–282. [Google Scholar]
- Dolz J., Desrosiers C., Ayed I.B. IVD-Net: Intervertebral disc localization and segmentation in MRI with a multi-modal UNet. International Workshop and Challenge on Computational Methods and Clinical Applications for Spine Imaging, Springer. 2018:130–143. [Google Scholar]
- Goyal M., Menon B.K., van Zwam W.H., Dippel D.W., Mitchell P.J., Demchuk A.M., Dávalos A., Majoie C.B., van der Lugt A., De Miquel M.A. Endovascular thrombectomy after large-vessel ischaemic stroke: a meta-analysis of individual patient data from five randomised trials. Lancet. 2016;387:1723–1731. doi: 10.1016/S0140-6736(16)00163-X. [DOI] [PubMed] [Google Scholar]
- Hajian-Tilaki K. Receiver operating characteristic (ROC) curve analysis for medical diagnostic test evaluation. Caspian J. Internal Med. 2013;4:627. [PMC free article] [PubMed] [Google Scholar]
- Hougaard, K.D., Hjort, N., Zeidler, D., SA,rensen, L., NA,rgaard, A., Hansen, T.M., von Weitzel-Mudersbach, P., Simonsen, C.Z., Damgaard, D., Gottrup, H., Svendsen, K., Rasmussen, P.V., Ribe, L.R., Mikkelsen, I.K., Nagenthiraja, K., Cho, T.H., Redington, A.N., BA,tker, H.E., Ostergaard, L., Mouridsen, K., Andersen, G., 2013. Remote ischemic perconditioning as an adjunct therapy to thrombolysis in patients with acute ischemic stroke: a randomized trial. Stroke 45, 159-167. [DOI] [PubMed]
- Jonsdottir K.Y., Østergaard L., Mouridsen K. Predicting tissue outcome from acute stroke magnetic resonance imaging: improving model performance by optimal sampling of training data. Stroke. 2009;40:3006–3011. doi: 10.1161/STROKEAHA.109.552216. [DOI] [PubMed] [Google Scholar]
- Kidwell C.S., Wintermark M., De Silva D.A., Schaewe T.J., Jahan R., Starkman S., Jovin T., Hom J., Jumaa M., Schreier J. Multiparametric MRI and CT models of infarct core and favorable penumbral imaging patterns in acute ischemic stroke. Stroke. 2013;44:73–79. doi: 10.1161/STROKEAHA.112.670034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livne M., Boldsen J.K., Mikkelsen I.K., Fiebach J.B., Sobesky J., Mouridsen K. Boosted tree model reforms multimodal magnetic resonance imaging infarct prediction in acute stroke. Stroke. 2018;49:912–918. doi: 10.1161/STROKEAHA.117.019440. [DOI] [PubMed] [Google Scholar]
- Marks M.P., Lansberg M.G., Mlynash M., Kemp S., McTaggart R., Zaharchuk G., Bammer R., Albers G.W. Correlation of AOL recanalization, TIMI reperfusion and TICI reperfusion with infarct growth and clinical outcome. J. Neurointerventional Surgery. 2014;6:724–728. doi: 10.1136/neurintsurg-2013-010973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley R., Häni L., Gralla J., El-Koussy M., Bauer S., Arnold M., Fischer U., Jung S., Mattmann K., Reyes M. Fully automated stroke tissue estimation using random forest classifiers (FASTER) J. Cerebral Blood Flow Metab. 2017;37:2728–2741. doi: 10.1177/0271678X16674221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milletari F., Navab N., Ahmadi S.A. in: 2016 Fourth International Conference on 3D Vision (3DV), IEEE. 2016. V-Net: fully convolutional neural networks for volumetric medical image segmentation; pp. 565–571. [Google Scholar]
- Nie D., Wang L., Gao Y., Shen D. 2016 IEEE 13th international symposium on biomedical imaging (ISBI), IEEE. 2016. Fully convolutional networks for multi-modality isointense infant brain image segmentation; pp. 1342–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen A., Hansen M.B., Tietze A., Mouridsen K. Prediction of tissue outcome and assessment of treatment effect in acute ischemic stroke using deep learning. Stroke. 2018;49:1394–1401. doi: 10.1161/STROKEAHA.117.019740. [DOI] [PubMed] [Google Scholar]
- Nogueira R.G. Thrombectomy 6 to 24 hours after stroke with a mismatch between deficit and infarct. N. Engl. J. Med. 2018;378:11–21. doi: 10.1056/NEJMoa1706442. [DOI] [PubMed] [Google Scholar]
- Olivot J.M., Mlynash M., Thijs V.N., Kemp S., Lansberg M.G., Wechsler L., Bammer R., Marks M.P., Albers G.W. Optimal Tmax threshold for predicting penumbral tissue in acute stroke. Stroke. 2009;40:469–475. doi: 10.1161/STROKEAHA.108.526954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto A., Mckinley R., Alves V., Wiest R., Silva C.A., Reyes M. Stroke lesion outcome prediction based on MRI imaging combined with clinical information. Front. Neurol. 2018;9:1060. doi: 10.3389/fneur.2018.01060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers W.J. Guidelines for the early management of patients with acute ischemic stroke: 2019 update to the 2018 guidelines for the early management of acute ischemic stroke: a guideline for healthcare professionals from the american heart association/american stroke association. Stroke. 2019;50:e344–e418. doi: 10.1161/STR.0000000000000211. [DOI] [PubMed] [Google Scholar]
- Qiu, W., Kuang, H., Teleg, E., Ospel, J.M., Sohn, S.I., Almekhlafi, M., Goyal, M., Hill, M.D., Demchuk, A.M., Menon, B.K., 2020. Machine learning for detecting early infarction in acute stroke with non-contrast-enhanced CT. Radiology, 191193. [DOI] [PubMed]
- Rekik I., Allassonnière S., Carpenter T.K., Wardlaw J.M. Medical image analysis methods in MR/CT-imaged acute-subacute ischemic stroke lesion: segmentation, prediction and insights into dynamic evolution simulation models. A critical appraisal. NeuroImage: Clinical. 2012;1:164–178. doi: 10.1016/j.nicl.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robben D., Boers A.M., Marquering H.A., Langezaal L.L., Roos Y.B., van Oostenbrugge R.J., van Zwam W.H., Dippel D.W., Majoie C.B., van der Lugt A. Prediction of final infarct volume from native CT perfusion and treatment parameters using deep learning. Med. Image Anal. 2020;59 doi: 10.1016/j.media.2019.101589. 101589. [DOI] [PubMed] [Google Scholar]
- Smith S., Bannister P.R., Beckmann C., Brady M., Clare S., Flitney D., Hansen P., Jenkinson M., Leibovici D., Ripley B. FSL: New tools for functional and structural brain image analysis. NeuroImage. 2001;13:249. [Google Scholar]
- Taha A.A., Hanbury A. Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. BMC Med. Imaging. 2015;15:29. doi: 10.1186/s12880-015-0068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, J.P., Albers, G.W., 2015. Reperfusion versus recanalization: the winner is. [DOI] [PubMed]
- Wheeler H.M., Mlynash M., Inoue M., Tipirneni A., Liggins J., Zaharchuk G., Straka M., Kemp S., Bammer R., Lansberg M.G. Early diffusion-weighted imaging and perfusion-weighted imaging lesion volumes forecast final infarct size in DEFUSE 2. Stroke. 2013;44:681–685. doi: 10.1161/STROKEAHA.111.000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winder A.J., Siemonsen S., Flottmann F., Thomalla G., Fiehler J., Forkert N.D. Technical considerations of multi-parametric tissue outcome prediction methods in acute ischemic stroke patients. Sci. Rep. 2019;9:1–12. doi: 10.1038/s41598-019-49460-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeck S., Hakim A., McKinley R., Pinto J.A., Alves V., Silva C., Pisov M., Krivov E., Belyaev M., Monteiro M. ISLES 2016 and 2017-benchmarking ischemic stroke lesion outcome prediction based on multispectral MRI. Front. Neurol. 2018;9:679. doi: 10.3389/fneur.2018.00679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo J., Choi J.W., Lee S.J., Hong J.M., Hong J.H., Kim C.H., Kim Y.W., Kang D.H., Kim Y.S., Hwang Y.H., Ovbiagele B., Demchuk A.M., Lee J.S., Sohn S.I. Ischemic diffusion lesion reversal after endovascular treatment. Stroke. 2019;50:1504–1509. doi: 10.1161/STROKEAHA.118.024263. [DOI] [PubMed] [Google Scholar]
- Yu Y., Xie Y., Thamm T., Gong E., Ouyang J., Huang C., Christensen S., Marks M.P., Lansberg M.G., Albers G.W. Use of deep learning to predict final ischemic stroke lesions from initial magnetic resonance imaging. JAMA Network Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.0772. e200772–e200772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidat O.O., Yoo A.J., Khatri P., Tomsick T.A., Von Kummer R., Saver J.L., Marks M.P., Prabhakaran S., Kallmes D.F., Fitzsimmons B.F.M. Recommendations on angiographic revascularization grading standards for acute ischemic stroke: a consensus statement. Stroke. 2013;44:2650–2663. doi: 10.1161/STROKEAHA.113.001972. [DOI] [PMC free article] [PubMed] [Google Scholar]