Abstract
Together with their undeniable role in the ecology of arid and semiarid ecosystems, Agave species are emerging as a model to dissect the relationships between crassulacean acid metabolism and high efficiency of light and water use, and as an energy crop for bioethanol production. Transcriptome resources from economically valuable Agaves species, such as Agave tequilana and A. salmiana, as well as hybrids for fibers, are now available, and multiple gene expression landscape analyses have been reported. Key components in molecular mechanisms underlying drought tolerance could be uncovered by analyzing gene expression patterns of roots. This study describes an efficient protocol for high-quality total RNA isolation from phenolic compounds-rich Agave roots. Our methodology involves suitable root handling and collecting in the field and using saving-time commercial kits available. RNA isolated from roots free of lignified out-layers and clean cortex showed high values of quality and integrity according to electrophoresis and microfluidics-based platform. Synthesis of long full-length cDNAs and PCR amplification tested the suitability for downstream applications of extracted RNA. The protocol was applied successfully to A. tequilana roots but can be used for other Agave species that also develop lignified epidermis/exodermis in roots.
Keywords: Agave, Full-length cDNA, Lignin, RNA extraction, Roots
Introduction
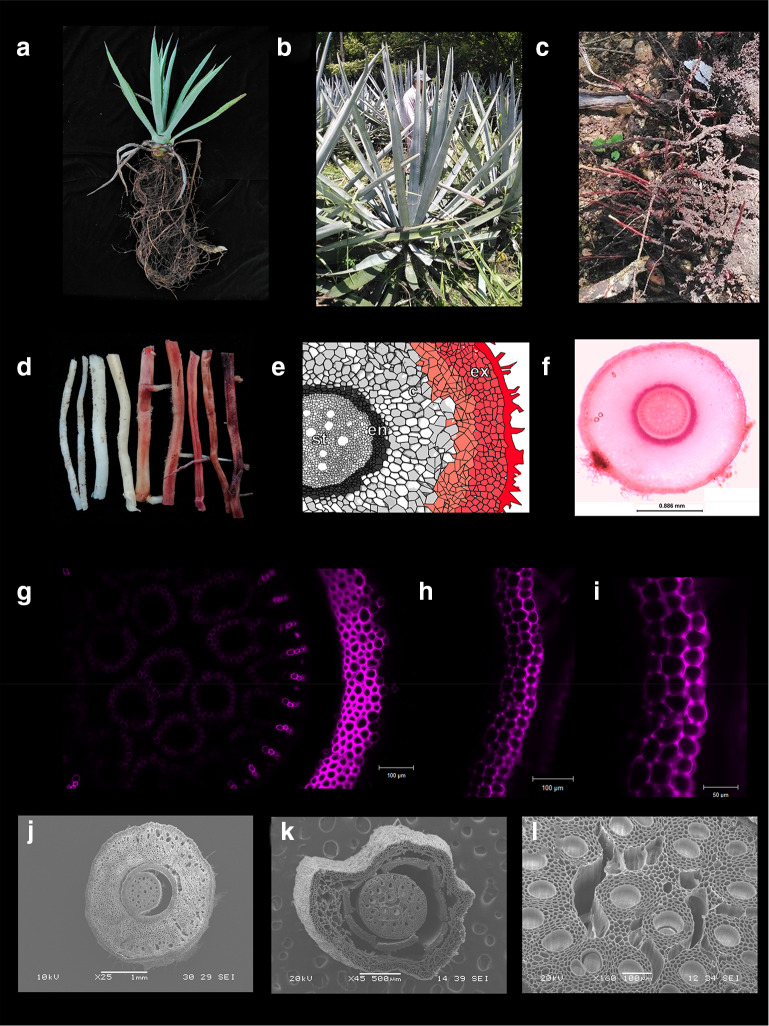
Agave species (Agavaceae) are xeric monocots considered keystones species in arid and semiarid regions and endemic to America. The genus Agave groups about 166 species, most of them are native to Mexico where they grow in different habits, from sea level to an elevation of 3400 m (Good-Avila et al. 2006). Traditionally, Agave plants have been used for beverages, food, fiber, medicines, shelter, and as ornamentals (Niechayev et al. 2019). Because of their remarkable tolerance to long drought periods, high light efficiency, minimal water needs, high biomass productivity, and high contents of soluble sugar and lignocellulose, Agave species are considered an emerging biofuel crop (Davis et al. 2011; Escamilla-Treviño 2012; Gross et al. 2013; Corbin et al. 2015). Additionally, analysis of the crassulacean acid metabolism (CAM) in Agave plants is opening up a new avenue of research to create more efficient crops based on CAM traits (Abraham et al. 2016). This growing interest, which began in the last decade, is contrasted with the relatively scarce transcriptomic resources available for some Agave species (Table 1), and with the fact that there are no sequenced reference genomes thus far. Due to a well-developed tequila industry and a potential for bioethanol production, transcriptomics data have mostly been reported for A. tequilana leaves, and recently also for the Agave hybrid No. 11648 [(A. amaniensis × A. angustifolia) × A. amaniensis], used for fiber production (Table 1). Transcriptomics landscapes for other organs like roots have been less explored; however, Agave tolerance to drought could be directly controlled by physiology and morpho-anatomical traits of roots, modulated by gene expression. The Agave root system is fairly shallow (Nobel 1976) (Fig. 1a–c). This root system is originated from a short-lived primary root. At maturity, the system is composed by multiple shallow roots that grow vertically and give rise to usually horizontal lateral roots, which are extended in a radial pattern (Dubrovsky 1997; Cruz-Ramírez et al. 2009). Connected to this network, it is usual to find rhizomes, which are not roots but underground stems. In A. deserti, alternated periods of wet conditions and drought induce suberization of the epidermis, hypodermis, and endodermis of young lateral roots (Huang and Nobel 1992), and lipid accumulation and lignification of the epidermis/exodermis in older roots (North and Nobel 1991, 1995). Cardiac and saponin glycosides have also been reported in A. americana roots (Kadam et al. 2012). Although RNA isolation is considered a routine protocol, for Agave roots, this step could be a bit tricky. Our laboratory experience with total RNA isolation from mature lateral roots of A. tequilana shows that lignin accumulates on out-layers and cortex (Fig. 1a–l) and can co-precipitate and yield low-quality RNA if they are not removed. Here, we report a detailed procedure for collecting and handling Agave root samples from soil and a protocol for total RNA extraction at micro-scale. This protocol is based on the standard guanidine thiocyanate-based method widely used for total RNA isolation in Agave species (Table 1), and yields total RNA with suitable quality for downstream applications such as gene expression analysis, cloning or sequencing.
Table 1.
Protocols used for total RNA isolation in several Agave species
| Agave specie/tissue/analyses | Protocol used | References |
|---|---|---|
| A. tequilana Weber var. azul/pines and plants/gene cloning | Aurum Total RNA Mini kit (Bio-Rad, 7326820) | Ávila-Fernández et al. (2007) |
| A. tequilana/leaf, pine, and floral tissues/transcriptome sequencing |
TRIzol (Thermo Fisher Scientific, 15596026) Concert (Thermo Fisher Scientific) |
Martínez et al. (2010) |
| A. tequilana and A. deserti/roots, pine and leaf/transcriptome sequencing | TRIzol (Thermo Fisher Scientific, 15596026) | Gross et al. (2013) |
| Agave hybrid No. 11648/leaf/RT-PCR analysis | E.Z.N.A.® Plant RNA (Omega, R6827-01) | Gao et al. (2014) |
| A. tequilana, A. victoriae-reginae, and A. striata/roots, pine, leaf and floral tissues/transcriptome sequencing and RT-qPCR analysis | TRIzol (Thermo Fisher Scientific, 15596026) | Avila-de Dios et al. (2015) |
| A. tequilana/bulbs/RT-qPCR analysis | TRIzol (Thermo Fisher Scientific, 15596026) | Abraham et al. (2015) |
| Agave americana/meristem, rhizome, root, and pine/transcriptome sequencing | Spectrum™ Plant Total RNA (Sigma-aldrich, STRN10) | Abraham et al. (2016) |
| Agave salmiana/callus, leaves, and root/transcriptome sequencing and RT-qPCR analysis | TRIzol (Thermo Fisher Scientific, 15596026) | Cervantes-Pérez et al. (2018) |
| Agave hybrid No. 11648, A. americana, A. deserti, and A. tequilana/leaf/transcriptome sequencing | RNAprep Pure Plant Kit (Tiangen, DP432) | Huang et al. (2019a; b) |
| Agave sisalana/leaf/transcriptome sequencing | Phenol-based RNA isolation protocol | Batcho et al. (2019) |
| Agave sisalana/leaf/transcriptome sequencing | TRIzol (Thermo Fisher Scientific, 15596026) | Sarwar et al. (2019) |
| A. tequilana/shoot apical meristems and leaves/transcriptome sequencing and RT-qPCR analysis | TRIzol (Thermo Fisher Scientific, 15596026) | Avila-de Dios et al. (2019) |
| A. tequilana/leaf, roots, and pine/gene cloning | Trizol (Sigma-aldrich, T9424) | Sierra et al. (2019) |
| Agave hybrid No. 11648/pine and leaf/RT-qPCR analysis | RNAprep Pure Plant Kit (Tiangen, DP432) | Deng et al. (2019) |
| Agave angustifolia/leaf/RT-qPCR analysis | Trizol (Sigma-aldrich, T9424) | Sara et al. (2020) |
Fig. 1.
Root system of A. tequilana. Root system of 2-year-old plant (a) and 6-year-old plants (b, c). In mature roots (red roots, d), high accumulation of lignin in cortex (e–g) and epidermis/exodermis (e, h, i) was revealed by staining with basic Fuchsin, and imagined under light microscopy (f) and confocal microscopy (g–i). This gradual lignin deposition from young (j) to mature roots (k) alters viability out-layers cells. A slight lignin deposition (g) is also observed in vascular tissues (g, l). Diagram of a cross section of agave root was re-drawn of North and Nobel 1995
Materials and methods
Plant material and lignin deposition analysis
Samples of lateral roots of four 6-year-old plants of A. tequilana Weber var. Azul were collected at field. Mature root samples for lignin deposition analysis or morphology by electronic scanning microscopy (SEM) was collected and kept in water or fixed in 4% paraformaldehyde solution, respectively. Lignin deposition was revealed by basic fuchsin staining (Sigma-Aldrich, 215597) as reported (Kapp et al. 2015) and imaged under a stereomicroscope (Leica M165 C, Leica, Germany) coupled with a Leica DFC290 camera, and confocal microscopy (Zeiss AXIO Imager.Z2, Carl Zeiss, Germany). For SEM analysis, root samples were gradually dehydrated with ethanol and frozen using liquid nitrogen. Micrographs were obtained using a JEOL JSM-6360LV Scanning Electron Microscope (Japan), equipped with a cold plate and an integrated digital image acquisition system. For RNA extraction, root samples collected in the field were immediately frozen in liquid nitrogen and stored at − 80 °C until processed. All common solutions and standard lab ware were prepared as recommended for routine RNA isolation protocols, and reagent setup as suggested by manufacture’s instructions.
Recommendations for collecting and handling Agave root samples from soil
Agave plants are very susceptible to fungal pathogens if soil is not well drained; be careful to sample only healthy plants.
Clean harvested roots using dry paper towels and then cut them quickly into 5-cm long fragments using pruning shears.
When collecting mature roots, eliminate lignified epidermis/exodermis using tweezers as possible, and remove the red layer deposited on cortex using a paper towel soaked with absolute ethanol (clean roots). This red layer is highly soluble in absolute ethanol (Figs. 1, 2).
Place clean root fragments into a Falcon 50 ml conical tube and freeze using liquid nitrogen. Store at -80ºC until use.
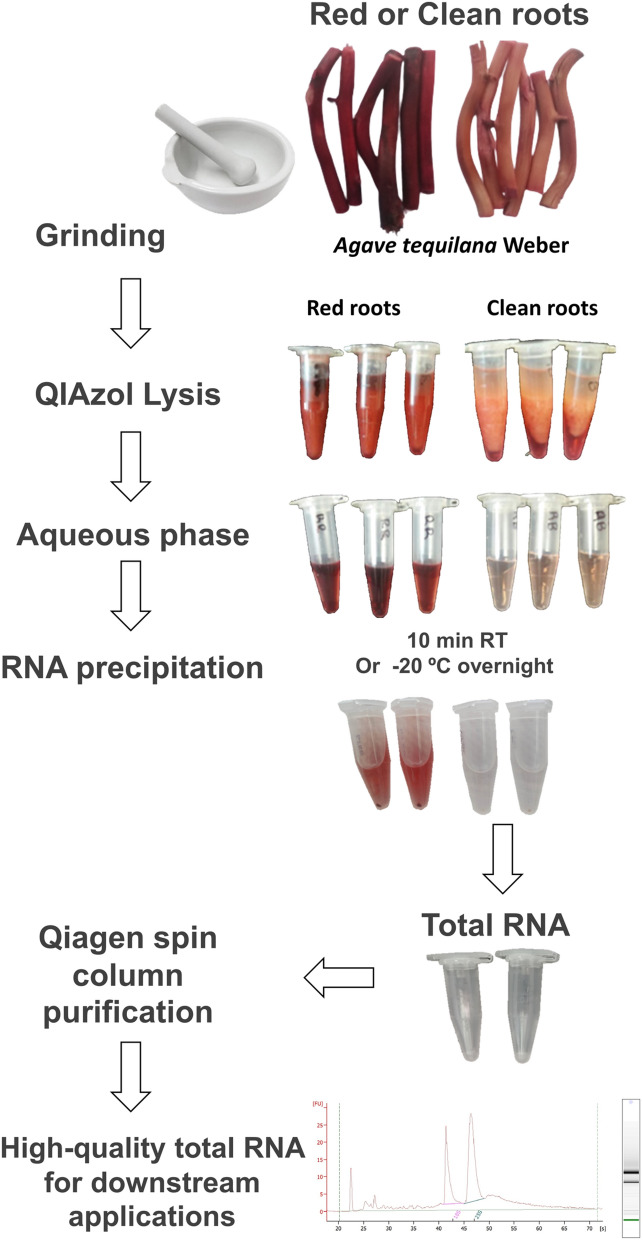
Fig. 2.
Workflow diagram of the principal steps for total RNA isolation. Contamination by phenolic compounds through the protocol is illustrated for control roots (red roots)
Total RNA extraction protocol (a workflow diagram is shown in Fig. 2)
This protocol enables up to 8–10 mini-extractions to be performed simultaneously. We set up the protocol using QIazol lysis reagent (Qiagen, 79306), but other kits such as TRIzol reagent (Thermo Fisher Scientific) also work:
Total RNA isolation
Pour liquid nitrogen into a sterile porcelain mortar and pestle to pre-cool them. Repeat this step as necessary. Mortar and pestle will be ready for use when a small volume of liquid nitrogen remains at the bottom of the mortar.
Transfer the root sample collected to the pre-cooled mortar and grind to get a homogenous and fine powder. Tip: do not grind large volume of samples. Small samples will yield better results.
Transfer the powder to 1.5 ml RNAse-free microcentrifuge tubes. If possible, weigh 100 mg aliquot per tube. Tips: the powder can be transferred using a sterile small spatula or 1 ml RNA-free tips; in both cases, spatula or tips have to be previously pre-cooled. If using tips, use one tip per tube to reduce contamination risks. Add powder to fill ¾ volume of the conical space of the tube. This volume will be enough to get approximately 100 mg.
Add 1 ml of QIazol lysis reagent per 100 mg of powder. Cap the microcentrifuge tube and immediately vortex until getting a defrosted homogenate. Keep at room temperature for 5 min. Stop: if necessary, keep the tubes at − 80 for maximum 1 week and then go to step 5.
Add 200 μl chloroform. Cap the tube and vortex briefly. Place the tube on the benchtop at room temperature for 2–3 min.
Centrifuge at 12,000 × g for 15 min at 4 °C.
Transfer the upper aqueous phase into a fresh tube. Be careful to avoid the interphase to avoid contamination.
Add 500 μl isopropanol per 1 ml QIAzol reagent used in step 1. Mix thoroughly by inverting the tube 4–6 times. Do not mix by pipetting or vortexing.
Let the tube sit on the benchtop at room temperature for 10 min for RNA precipitation. Optional: RNA precipitation can occur at – 20 ºC overnight.
Centrifuge at 12,000 × g for 10 min at 4 °C.
Decant the supernatant and wash the pellet adding at least 1 ml of 75% ethanol, and vortex briefly (not more than 5 seg).
Centrifuge at 7500 × g for 5 min at 4 °C.
Decant the supernatant, spin the tube and remove the ethanol completely by pipetting. If necessary, repeat the spin and pipetting. Caution: be careful not to touch the pellet.
Dry the pellet by air for 15–20 min.
Re-suspend the pellet with DEPC-water.
Quantification and analysis of quality of total RNA
Concentration and purity parameters (A260/280 and A260/230) of isolated RNA can be assessed using NanoDrop microvolume spectrometer and before standard electrophoresis analysis.
Total RNA purification by silica-based spin column
To remove DNA and other contaminants that could be co-precipitated during RNA isolation, a purification procedure is a must for downstream applications. Here, we purified Agave total RNA samples using the RNeasy® MinElute® Cleanup Kit (Qiagen, 74204) that is based on RNA-adsorption in silica spin columns. Below, the standard protocol is described and some suggestions are included to improve the performance of RNA purification and quality.
According to the manufacturer’s suggestion, up to 45 μg RNA can be purified per column. Tip: mix the previously isolated RNA stocks and up to 100 μl with DEPC-water. Use preferentially DEPC-water from double-autoclaved aliquots.
Add 100 μl cold 70% ethanol and mix well by pipetting 4–6 times.
Transfer immediately the mix to the RNAeasy mini spin column (pink column) placed in a 2 ml collection tube (supplied by the manufacturer).
Centrifuge for 15 s at 8000 × g. Discard the flow-through.
Add 700 μl buffer RW1 to the spin column. Close the lid and centrifuge for 15 at 8000 × g. Discard the flow-through.
Add 500 μl buffer RPE to the spin column. Close the lid and centrifuge for 15 at 8000 × g. Discard the flow-through.
Add 500 μl buffer RPE to the spin column. Close the lid and centrifuge for 1 min at 8000 × g. Discard the flow-through. Place the spin column in a fresh 2 ml collection tube. CAUTION: some mini spin columns can have an internal circle shoulder just above the silica filter. Remove any ethanol or wash buffer trapped in this shoulder by pipetting.
Dry the spin column by centrifuging for 2 min at full speed. Keep the tube open during centrifuging. Optional: cut the cap if needed.
Discard the collection tube and place the spin column in new fresh 1.5 ml tube. Add 30–50 μl RNAse-free water directly to the spin column membrane. Let rest for 1 min on the benchtop. Tip: the manufacturer supplies RNAse-free water or you could use previously prepared DEPC-water.
To elute the RNA, centrifuge for 1 min at full speed.
Hereafter, keep the RNA samples in ice. Quantify RNA concentration using a microvolume spectrometer and check RNA integrity by electrophoresis. Store purified total RNA samples at – 20 ºC until use.
Evaluation of amenability for downstream applications
After spin column purification, 1 μg total RNA from clean roots was used to synthesize cDNA using reverse transcriptase and oligo (dT) (10 μM), following the manufacturer’s protocol (SuperScript III, Invitrogen, 18080093). Specific primers for CELLULOSE SYNTHASE7-like gene (CESA7) forward: 5′-ATGTCCTTGATGGAGTCTGGTCTG-3′ and reverse: 5′-TTAGCACTCCACTCCACACTGTTTAAG-3′) and for SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN1-like gene (SND1) (forward: 5′-ATGAGCATCACTGTGAATGGCCA-3′ and reverse: 5′-TTATATTGAGGCGTGGGGCACG-3′) were used to amplify full-length cDNAs using a proofreading DNA polymerase (Platinum SuperFi, Invitrogene, 12351010). The PCR program for CESA7-like gene was used as follows: 98 ºC for 3 min; 35 cycles of 98 ºC for 30 s, 62 ºC for 30 s, and 72 ºC for 1.5 min, and final extension at 72 ºC for 5 min. The PCR program for SND1-like gene was used as follows: 98 ºC for 3 min; 35 cycles of 98 ºC for 30 s, 62 ºC for 30 s, and 72 ºC for 30 s, and final extension at 72 ºC for 5 min. PCR products (5 μl) were analyzed in agarose gels and visualized as described above.
Statistical analyses and images processing
All data were analyzed as completely randomized factorial design. One-way independent ANOVA analyses were carried out test using SAS software (SAS Institute, USA) and Tukey’s honestly significant difference (HSD) tests were applied to find out which treatment’s means (compared with each other) are different (P ≤ 0.05). All images were prepared using Remove.bg (Kaleido Al GmbH, Austria), Microsoft PowerPoint (Microsoft, USA) and GIMP 2.10.12 free software (The GIMP Development Team, USA).
Results and discussion
High-quality RNA is a key prerequisite for successful gene cloning and expression analysis during molecular studies of plant development or responses to environmental clues. Although reliable protocols and a range of commercial kits are available, isolation of high-quality RNA is still challenging when plants are enriched with polysaccharides, polyphenols and others secondary metabolites. Most of these contaminants can degrade or co-precipitate with RNA due to physical interactions or similar chemical properties (Alemzadeh et al. 2005; Guan et al. 2019). This has led to optimize protocols previously reported according to plant species or even tissue-specific (Rubio-Piña and Zapata-Pérez 2011; Guan et al. 2019). In the case of Agave species, roots are organs with secondary metabolites, most likely polyphenols, and even lignified (Fig. 1). Our protocol for total RNA extraction of such roots simplifies the procedure of eliminating double precipitation by high LiCl concentrations or using polyvinylpyrrolidone (PVP) or polyethylene glycol (PEG), by doing just a suitable handling of Agave roots previous RNA isolation (Fig. 2). Removing lignified epidermis/exodermis and metabolites accumulated in the root cortex (clean roots) substantially reduced phenolic compounds that remained in the supernatant and precipitated with RNA isolated, as is shown by extractions of clean or control (red) roots processed with the protocol proposed here (Fig. 2). A comparison of RNA precipitation during 10 min at root temperature (RT) or at − 20 ºC overnight showed that RNA precipitation from polyphenol-rich samples provides better RNA yield when is carried out for short time (10 min) (Fig. 3a, b). Moreover, quality of isolated RNA can be increased if polyphenolic compounds are previously eliminated of the processed root samples (Fig. 3a, b). This statement is supported by our RNA quality data from red roots (control) or clean roots, which were determined by spectrophotometry (Fig. 3b, left), and confirmed by high-resolution automated electrophoresis (Agilent 2100 Bioanalyzer system) (Fig. 3c). Although purification by silica-based column can reduce the level of polyphenols/carbohydrates, in our case, RNAs isolated from lignified root (red) samples kept low levels of purity and quality even after purification as confirmed by RNA integrity number (RIN) values obtained by Bioanalyzer analysis (Fig. 3c). Taken together, these data indicate that removing lignified epidermis and metabolites accumulated in epidermis/exodermis and cortex is an essential step to get high-quality RNA from Agave roots, as reported in other tropical crops such as coffee (Huded et al. 2018). In our protocol, electrophoretic patterns of RNAs, visualized by standard electrophoresis and microfluidics-based platform, suggest no RNA degradation but likely an irreversible binding of polyphenol-derived quinones to RNA molecules as previously reported (Graham et al. 1993; Huded et al. 2018).
Fig. 3.
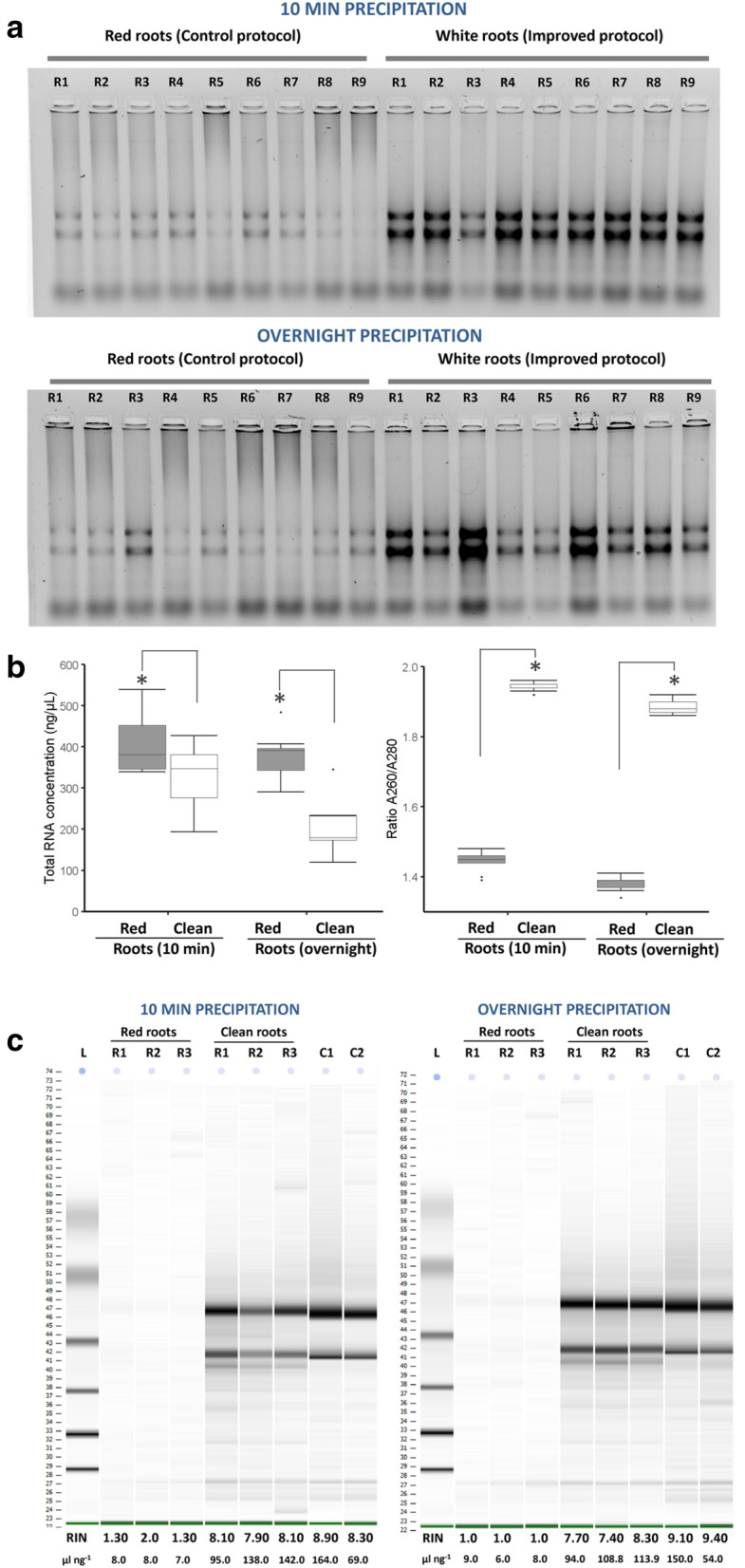
Removing lignin and phenolic compounds in Agave roots is an essential step to get high-quality RNA. a Electrophoretic resolution patterns from ethidium bromide-stained geles and b yield parameters (quantity and quality) of total RNA samples isolated from control roots (red roots) and clean roots. Two RNA precipitation conditions (10 min at room temperature and − 20 ºC overnight) were also contrasted. Box and whisker plots represent mean value ± SD. Asterisks indicate significant statistical differences between treatments determined by Tukey’s HSD test (P ≤ 0.05). c Agilent bioanalyzer gel-like images of total RNA purified by spin column
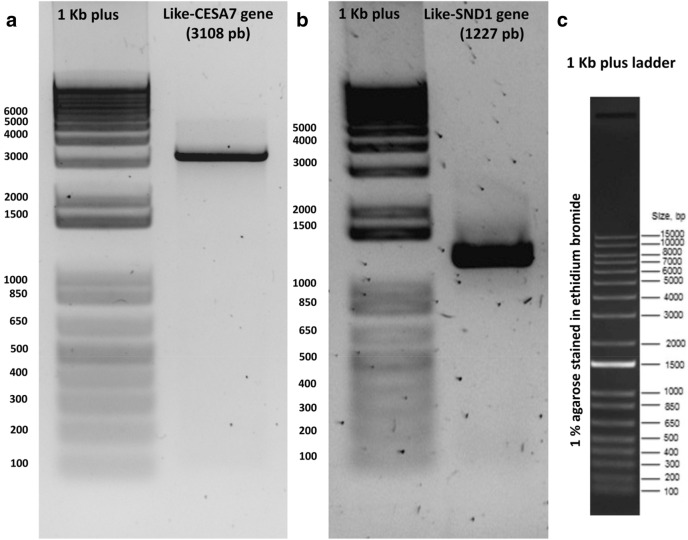
In Arabidopsis thaliana, CELLULOSE SYNTHASE7 or AtCESA7 (At5g17420) is involved in cellulose biosynthesis, while SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN1 or AtSDN1 (At1g32770) is a master modulator of lignin formation (reviewed in Handakumbura and Hazen 2012; McCahill and Hazen 2019). Here, we evaluated the suitability of RNA isolated from clean agave roots by amplifying the full-length cDNAs of CESA7-like gene and SDN1-like genes, found after mining Agave transcriptomes previously reported (Gross et al. 2013; Avila de Dios et al. 2019). Electrophoresis analysis showed a specific amplified PCR product of 3108 pb for CESA7, and a 1227 pb-length product for the like-SND1 gene, as expected for amplification from full-length cDNAs (Fig. 4). No extra re-amplification PCR rounds were needed for the subsequent cDNA cloning.
Fig. 4.
PCR amplification of full-length cDNAs of two agave genes related to cell wall using cDNA from total RNA of clean roots. Unique and abundant PCR amplicons were observed for the (a) CESA7-like gene and (b) SDN1-like gene, indicating no DNA contamination in isolated RNA samples. c 1 kb plus (Thermo Fisher Scientific, 10787018) is shown to help to determine the PCR product length
Conclusion
Our improved protocol for handling and RNA extraction allowed the isolation of high-quality total RNA from lignified and high-polyphenol accumulating roots from A. tequilana. This effective and simple protocol yielded suitable RNA for synthesis of long cDNAs. Removing outer dead epidermis of roots did not compromise the root transcriptomic landscape isolated, as we were able to amplify full-length cDNAs related to secondary cell wall formation. The protocol developed was used successfully in A. tequilana roots but can be used for other Agave species that also develop lignified epidermis/exodermis in roots.
Acknowledgements
This work was funded by CONACyT (Grant 1049) and Colpos. Luis F. Maceda-Lopez is grateful for a M.Sc. fellowship from CONACyT. Eleazar Garcia-Hernández and Dalia C. Moran Velazquez were supported by an undergraduate scholarship from CONACyT (Grant 1049). Authors especially grateful to Victor H. Fernandez-Carrillo, manager of Santa Genoveva farm, for supporting during Agave samples collecting.
Author contributions
LFML, JLVA, DCMV, and FAC designed and performed the RNA extraction, quantification, and quality RNA analysis. JLVA, EGH, LRL, DHD, and MACV performed RT-PCR analysis. Lignin deposition and SEM analysis were carried out by LFML, SBAC, LRL, and FAC. Data mining was performed by LFML, EAD, JS, LT, ILR, EGC, and FAC. LFML, LT, EGC, ILR, JS, and FAC discussed results. LFML and FAC conceived and designed the research and wrote the paper. All authors read and approved the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Ethical approval
Each of the authors confirms that this manuscript is original, has not been previously published, and is not currently under consideration by any other journal. Additionally, all of the authors have approved the contents of this paper and have agreed to the 3 Biotech’s submission policies.
Contributor Information
Luis F. Maceda-López, Email: maceda.luis@outlook.com
José L. Villalpando-Aguilar, Email: nachfornez@gmail.com
Eleazar García-Hernández, Email: garcia.eleazar@colpos.mx.
Emmanuel Ávila de Dios, Email: aviladedios@gmail.com.
Silvia B. Andrade-Canto, Email: sbac@cicy.mx
Dalia C. Morán-Velázquez, Email: moran.dalia@colpos.mx
Lorena Rodríguez-López, Email: lodoyarena96@gmail.com.
Demetrio Hernández-Díaz, Email: demet2422@gmail.com.
Manuel A. Chablé-Vega, Email: chable.manuel@colpos.mx
Laura Trejo, Email: laura.trejo@st.ib.unam.mx.
Elsa Góngora-Castillo, Email: elsa.gongora@cicy.mx.
Itzel López-Rosas, Email: itzel.rosas@colpos.mx.
June Simpson, Email: june.simpson@cinvestav.mx.
Fulgencio Alatorre-Cobos, Email: fulgencio@colpos.mx.
References
- Abraham PE, Yin H, Borland AM, et al. Transcript, protein and metabolite temporal dynamics in the CAM plant Agave. Nat Plants. 2016;2:16178. doi: 10.1038/nplants.2016.178. [DOI] [PubMed] [Google Scholar]
- Abraham-Juárez MJ, Hernández-Cárdenas R, Santoyo-Villa JN, et al. Functionally different PIN proteins control auxin flux during bulbil development in Agave tequilana. J Exp Bot. 2015;66:3893–3905. doi: 10.1093/jxb/erv191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alemzadeh A, Fujie M, Usami S, Yamada T. Isolation of high-quality RNA from high-phenolic tissues of eelgrass (Zostera marina L.) by keeping temperature low. Plant Mol Biol Rep. 2005;23:421. doi: 10.1007/BF02788892. [DOI] [Google Scholar]
- Avila-de Dios E, Gomez-Vargas AD, Damian-Santos ML, Simpson J. New insights into plant glycoside hydrolase family 32 in Agave species. Front Plant Sci. 2015;6:594. doi: 10.3389/fpls.2015.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila-de Dios E, Delaye L, Simpson J. Transcriptome analysis of bolting in A. tequilana reveals roles for florigen, MADS, fructans and gibberellins. BMC Genomics. 2019;20:473. doi: 10.1186/s12864-019-5808-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ávila-Fernández Á, Olvera-Carranza C, Rudiño-Piñera E, et al. Molecular characterization of sucrose: sucrose 1-fructosyltransferase (1-SST) from Agave tequilana Weber var. azul. Plant Sci. 2007;173:478–486. doi: 10.1016/j.plantsci.2007.07.009. [DOI] [Google Scholar]
- Batcho-Agossa A, Sarwar-Muhammad B, Tariq L, et al. Identification and characterisation of heat shock protein gene (HSP70) family and its expression in Agave sisalana under heat stress. J Hortic Sci Biotechnol. 2019;1–13:470–482. doi: 10.1080/14620316.2019.1685412. [DOI] [Google Scholar]
- Cervantes-Pérez SA, Espinal-Centeno A, Oropeza-Aburto A, et al. Transcriptional profiling of the CAM plant Agave salmiana reveals conservation of a genetic program for regeneration. Dev Biol. 2018;442:28–39. doi: 10.1016/j.ydbio.2018.04.018. [DOI] [PubMed] [Google Scholar]
- Corbin-Kendall R, Byrt-Caitlin S, Bauer S, et al. Prospecting for energy-rich renewable raw materials: agave leaf case study. PLoS ONE. 2015;10:e0135382. doi: 10.1371/journal.pone.0135382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Ramírez A, Calderón-Vázquez C, Herrera-Estrella L (2009) Effect of nutrient availability on root system development. Annual Plant Reviews Volume 37: Root Development 288–324. 10.1002/9781444310023.ch11.
- Davis-Sarah C, Dohleman-Frank G, Long-Stephen P. The global potential for Agave as a biofuel feedstock. Gcb Bioenergy. 2011 doi: 10.1111/j.1757-1707.2010.01077.x. [DOI] [Google Scholar]
- Deng G, Huang X, Xie L, et al. Identification and expression of SAUR Genes in the CAM Plant Agave. Genes. 2019;10:555. doi: 10.3390/genes10070555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky JG. Determinate primary-root growth in seedlings of Sonoran desert Cactaceae; its organization, cellular basis, and ecological significance. Planta. 1997;203:85–92. doi: 10.1007/s00050168. [DOI] [Google Scholar]
- Escamilla-Treviño LL. Potential of plants from the genus Agave as bioenergy crops. BioEnergy Res. 2012;5:1–9. doi: 10.1007/s12155-011-9159-x. [DOI] [Google Scholar]
- Gao J, Yang F, Zhang S, et al. Expression of a hevein-like gene in transgenic Agave hybrid No. 11648 enhances tolerance against zebra stripe disease. Plant Cell Tissue Organ Cult. 2014;119:579–585. doi: 10.1007/s11240-014-0557-6. [DOI] [Google Scholar]
- Good-Avila SV, Souza V, Gaut BS, Eguiarte LE. Timing and rate of speciation in Agave (Agavaceae) Proc Natl Acad Sci USA. 2006;103:9124–9129. doi: 10.1073/pnas.0603312103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham GC. A method for extraction of total RNA from Pinus radiata and other conifers. Plant Mol Biol Rep. 1993;11:32–37. doi: 10.1007/BF02670557. [DOI] [Google Scholar]
- Gross SM, Martin JA, Simpson J, et al. De novo transcriptome assembly of drought tolerant CAM plants, Agave deserti and Agave tequilana. BMC Genomics. 2013;14:563. doi: 10.1186/1471-2164-14-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan L, Ma X, Zhou X, et al. An optimized method to obtain high-quality RNA from cassava storage root. 3 Biotech. 2019;9:118. doi: 10.1007/s13205-019-1608-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handakumbura PP, Hazen SP. Transcriptional regulation of grass secondary cell wall biosynthesis: playing catch-up with Arabidopsis thaliana. Front Plant Sci. 2012;3:74. doi: 10.3389/fpls.2012.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Castellano S, Garruña-Hernández R, Us-Camas R, et al. Agave angustifolia albino plantlets lose stomatal physiology function by changing the development of the stomatal complex due to a molecular disruption. Mol Genet Genomics. 2020 doi: 10.1007/s00438-019-01643-y. [DOI] [PubMed] [Google Scholar]
- Huang B, Nobel PS. Hydraulic conductivity and anatomy for lateral roots of Agave deserti during root growth and drought-induced abscission. J Exp Bot. 1992;43:1441–1449. doi: 10.1093/jxb/43.11.1441. [DOI] [Google Scholar]
- Huang X, Xiao M, Xi J, et al. De novo transcriptome assembly of Agave H11648 by Illumina sequencing and identification of cellulose synthase genes in Agave species. Genes. 2019;10:103. doi: 10.3390/genes10020103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Xie L, Gbokie T, et al. Transcriptome dataset of leaf tissue in Agave H11648. Brown Univ Dig Addict Theory Appl. 2019;4:62. doi: 10.3390/data4020062. [DOI] [Google Scholar]
- Huded AKC, Jingade P, Mishra MK. A rapid and efficient SDS-based RNA isolation protocol from different tissues of coffee. 3 Biotech. 2018;8:183. doi: 10.1007/s13205-018-1209-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam PV, Yadav KN, Deoda RS, et al. Pharmacognostic and phytochemical studies on roots of Agave Americana (Agavaceae) Intern J Pharmacogn Phytochem Res. 2012;4:92–96. [Google Scholar]
- Kapp N, Barnes WJ, Richard TL, Anderson CT. Imaging with the fluorogenic dye Basic Fuchsin reveals subcellular patterning and ecotype variation of lignification in Brachypodium distachyon. J Exp Bot. 2015;66:4295–4304. doi: 10.1093/jxb/erv158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Hernández A, Mena-Espino M, Herrera-Estrella AH, et al. Construcción de bibliotecas de ADNc y análisis de expresión génica por RT-PCR en agaves. Revista latinoamericana de química. 2010;38:21–44. [Google Scholar]
- McCahill IW, Hazen SP. Regulation of cell wall thickening by a medley of mechanisms. Trends Plant Sci. 2019 doi: 10.1016/j.tplants.2019.05.012. [DOI] [PubMed] [Google Scholar]
- Niechayev NA, Jones AM, Rosenthal DM, Davis SC. A model of environmental limitations on production of Agave americana L. grown as a biofuel crop in semi-arid regions. J Exp Bot. 2019;70:6549–6559. doi: 10.1093/jxb/ery383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobel PS. Water relations and photosynthesis of a desert CAM plant, Agave deserti. Plant Physiol. 1976;58:576–582. doi: 10.1104/pp.58.4.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North GB, Nobel PS. Changes in hydraulic conductivity and anatomy caused by drying and rewetting roots of Agave deserti (Agavaceae) Am J Bot. 1991;78:906–915. doi: 10.1002/j.1537-2197.1991.tb14494.x. [DOI] [Google Scholar]
- North GB, Nobel PS. Hydraulic conductivity of concentric root tissues of Agave deserti Engelm. under wet and drying conditions. New Phytol. 1995;130:47–57. doi: 10.1111/j.1469-8137.1995.tb01813.x. [DOI] [Google Scholar]
- Rubio-Piña JA, Zapata-Pérez O (2011) Isolation of total RNA from tissues rich in polyphenols and polysaccharides of mangrove plants. Electron J Biotechnol
- Sara HC, René GH, Rosa UC, et al. Agave angustifolia albino plantlets lose stomatal physiology function by changing the development of the stomatal complex due to a molecular disruption. Mol Genet Genomics. 2020;295:787–805. doi: 10.1007/s00438-019-01643-y. [DOI] [PubMed] [Google Scholar]
- Sarwar MB, Ahmad Z, Rashid B, et al. De novo assembly of Agave sisalana transcriptome in response to drought stress provides insight into the tolerance mechanisms. Sci Rep. 2019;9:396. doi: 10.1038/s41598-018-35891-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra-Gómez Y, Rodríguez-Hernández A, Cano-Sánchez P, et al. A biophysical and structural study of two chitinases from Agave tequilana and their potential role as defense proteins. FEBS J. 2019;286:4778–4796. doi: 10.1111/febs.14993. [DOI] [PubMed] [Google Scholar]