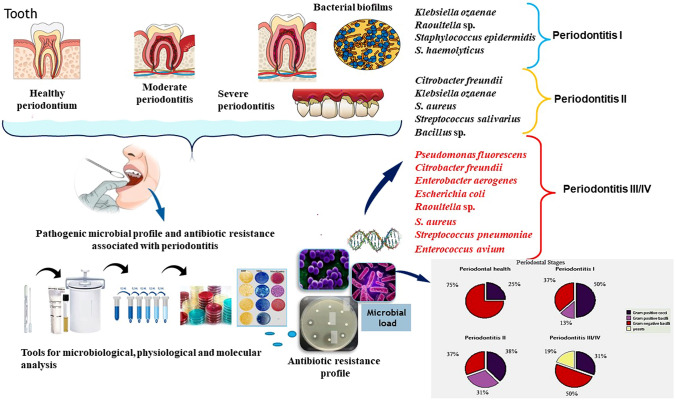
Abstract
Abstract
Phenotyping based on conventional microbiological, physiological, and molecular analysis by using ARDRA technique was developed with the aim to assess the pathogenic microbial load associated with different stages of the periodontal disease. In addition, in the face of the global issue of antimicrobial resistance, the isolated bacterial strains were evaluated for their antibiotic susceptibility profile. The pathogenic bacterial community was predominantly of Gram-negative strains (66.66%). The most common species were Citrobacter freundii, Bacillus sp., Raoutella sp., Klebsiella ozaenae and Pseudomonas sp. However, except for the healthy control group, Staphylococcus spp. was isolated from all stages of periodontitis. Multidrug resistance to beta-lactam antibiotics was observed for Streptococcus pneumoniae, Raoutella sp. and Enterococcus avium. Here, we verify a statistically significant relationship between periodontitis stages and the diversity of the bacterial community. Patients with periodontitis showed a more diverse and numerous bacterial community compared to healthy patients. In this sense, we reinforce that biofilms that harbour multidrug-resistant bacteria are a major concern in relation to restoring patient health. Thus, prophylactic measures for maintaining oral health are still the best option for reduce the risk of disease.
Graphic Abstract
Keywords: Bacterial community, β-lactam resistance, ARDRA, Periodontal disease
Introduction
A diverse and numerous microbial community that including species that interact with each other and with the host is present in the human oral cavity [1]. Biofilms can result from this interaction and harbour microorganisms anaerobic, facultative aerobes and microaerophiles from different genera [2] such as Streptococcus, Veillonella, Granulicatella, Neisseria, Haemophilus, Corynebacterium, Rothia, Actinomyces, Prevotella, Capnocytophaga, Porphyromonas, Fusobacterium and Lactobacillus species [3]. Changes that affect the balance between the commensal microbiota and the host can lead to dysbiosis and consequently to pathologies, such as periodontitis [4]. Periodontitis is an inflammatory clinical condition of multifactorial nature that affect the periodontium, that is, the tissue that surround and support the teeth, such as the gums, alveolar bone, and periodontal ligament [5, 6]. It is caused mainly by pathogenic bacteria [7]. According to the current classification for the periodontal conditions [8] two major groups are defined: periodontal and gingival health and periodontitis and its three forms: necrotizing periodontitis, periodontitis as a manifestation of systemic disease, and only periodontitis, previously characterized by chronic and aggressive forms. Periodontitis is also subdivided based on: (a) severity and complexity of administration: stage I-initial; stage II-moderate; stage III-severe, with potential for additional tooth loss, and stage IV-severe with potential for loss of teeth; (b) extension and distribution; (b) degrees—according to evidence or risk of rapid progression/early response to treatment: (a) grade A—low rate of progression; (b) grade B—moderate rate of progression; (c) grade C—rapid rate of progression [8].
Unlike classic bacterial infections, in most cases, the diversity of the microbiota increases as periodontal disease progresses and making it difficult the diagnosis of the causative agent [6]. In addition, periodontitis is constantly associated with the progression of chronic systemic pathologies, such as diabetes mellitus and cardiovascular diseases, and can be associated with the progression of atherosclerosis, i.e. a risk of respiratory infections [9]. Dental biofilm is a factor of resistance to the activity of antimicrobial agents, it is therefore a great concern, mainly because the presence of multidrug-resistant microorganism is an additional obstacle to the treatment of periodontal disease.
Our hypothesis is that if we consider that the progression of periodontitis is related to the pathogenic microbial load, then the more advanced the periodontitis stage, the more diverse and numerous the pathogenic microbiota will be. Thus, the aim of this study was to evaluate the microbial profile associated with different stages of periodontitis as well as to analyse the susceptibility profile to antimicrobials of pathogenic agents.
Material and Methods
This study was approved by the human subject ethics board of National Health Council via Brazil Platform and was conducted in accordance with the Helsinki Declaration of 1975 and revised in 2013 according to World Medical Association [10]. The individuals of this study were aged between 13 and 67 years old, of both sexes, regardless of race and social class who agreed to participate of this research including the disclosure of the data. All patients were undergoing dental evaluation at a municipal health center which underwent screening and previous diagnosis of periodontitis according to medical records. No reference was made to the identity of each patient or any information of a strictly personal nature. Thus, all individuals signed an informed consent form. After the patients' health screening, all were effectively diagnosed by a health professional (dentist) who is also co-author of the present study.
Population of Study
The population sample was composed by 18 patients: n = 15 individuals with periodontal disease (periodontitis stages I or II or III and IV) and n = 3 individuals healthy which constituted the control group. As an inclusion/exclusion criterion, only participants with a diagnosis of periodontitis were included besides no woman was pregnant or breastfeeding.
The sample was composed by individuals of both sexes who had not undergone periodontal therapy in the last 6 months. Except for swab collections, in the oral cavity of each patient there was no periodontal intervention by the study researchers.
The swabs samples were collected from oral cavity of each individual and transported in Stuart's medium until laboratory analysis.
Isolation of Microorganisms and Biochemical Characterization
The samples were enriched in Brain–Heart Infusion broth (BHI, Acumedia®, USA) incubated in anaerobic jars at 35 °C ± 1 °C over night. Then, aliquots of each sample were inoculated in petri dishes containing different culture media and supplements. All agars and culture broths were from Acumedia® (USA), except for BG and VRB (Oxoid UK). BHI Agar and Columbia Blood Agar Base (BA), enriched with 5% lamb's blood (Newprov, Brazil) both indicated for isolation and cultivation of a wide variety of microorganisms; Eosin Methylene Blue Agar (EMB), Brilliant Green Agar (BG), Violet Red Bile agar (VRBL) and MacConkey Agar (MC) used for the isolation and differentiation of Gram-negative bacilli; Baird Parker Agar (BP), plus 1% tellurite and 5% Egg Yolk and Agar Salt Mannitol (SM), indicated for isolation, detection and enumeration of Staphylococcus aureus; Bile Aesculin Agar (BAe) and Agar Mitis Salivarius (MS) selective isolation and differentiation of mixed cultures from Enterococcus sp. and Streptococcus sp.. All cultures were incubated under anaerobic conditions at 35 °C/24 h.
The morphological characteristics of the colonies growth in each culture medium were analyzed after Gram staining by optical microscopy (Nikon, Japan, obj.16x/100x).
Each bacteria isolate was subjected to biochemical analysis such as catalase test by addition of 3% hydrogen peroxide, oxidase test using reactive tapes for oxidase and hemolysis reaction (α, β or λ) on blood agar. In addition, the ability of microorganisms to assimilate sugars was evaluated to glucose, sucrose, and lactose. Each test was performed by using phenol red broth plus 1% carbohydrate.
Molecular Identification of Microorganisms
The genomic DNA of each bacterial isolates was extracted with GenElute™ Bacterial Genomic DNA Kit (Micromed, Brazil) as described by the manufacturer. The extracted DNAs were analyzed by electrophoresis on 1% agarose gel, followed by the amplification of the 16S rDNA region by the polymerase chain reaction (PCR) technique. The molecular profile was performed by using ARDRA (Amplified 16S Ribosomal DNA Restriction Analysis), a molecular technique based on the restriction fragment length polymorphism of the 16S ribosomal genes amplified by a polymerase chain reaction (PCR) [11, 12]. The amplified genetic material was cleaved with the restriction endonucleases AluI, Rsa I or Hha I. The restriction endonucleases were selected by Vector NTI Version 4.0 bioinformatics program with sequences of 16S ribosomal gene of species from GenBank. To perform the cleavage reaction, for each 200 ng of DNA we added 1.5 µL of reaction buffer (10x) and 0.5 µL of restriction enzyme. Then, sterile ultrapure water (MilliQ, Merck, Millipore, German) was added until reaching a final volume of 15 µL. After the reaction, the generated DNA fragments were separated by electrophoresis in 1.5% agarose gel for 4 h at 50v, exposed to UV light [12] and the picture taken with a gel documentation system.
The molecular profiles were analyzed by using the Vector NTI Version 4.0 The sequences used for comparative analysis were obtained from GenBank via BLAST Program of the National Center for Biotechnology Information-NCBI [13]. The samples that were not identified by ARDRA technique had their genetic material amplified, as mentioned above, purified and proceeded for sequencing the region 16S rDNA by conventional methods [12] and the nucleotide sequences were compared as mentioned before considering the statistical significance.
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing of the bacterial isolates was performed by Kirby Bauer method according to the Clinical Laboratory Standards Institute [14]. From each of the bacterial genera obtained during species identification, a representative strain was chosen at random and followed for the evaluation of the susceptibility profile to antimicrobials. Disks impregnated with antimicrobials (Laborclin, Brazil) were inserted on the agar: Tetracycline 30 µg; Chloramphenicol 30 µg; Imipenem 10 µg; Amoxicillin 10 µg; Azithromycin 15 µg; Cephalothin 30 µg; Clindamycin 2 µg and Cefotaxime 30 µg. The agar plates were incubated at 35 °C/24 h.
Statistical Analysis
When necessary, statistical analysis based on average ± SD, an ANOVA and chi-square tests were performed to assess whether there was difference at the 0.05 significance level.
Results and Discussion
In this study, we look for a possible relationship between periodontal stage and the microbiota pathogenic. According to the stage of periodontal disease we found that 16.6% of patients had periodontitis stage I, 3.3%, periodontitis stage II and 33.3% periodontitis stages III/IV. The average age of patients with periodontal disease was 43.8 years old and 36 years old for the control group.
According Zaura et al. [15] the two types of surface of oral cavity present conditions for microbial colonization, i.e., the mucosa and the teeth which provide a diverse and abundant community. In this sense, an imbalance between the microbiota and its host results in disease.
Taken together, recent findings support the hypothesis that the complex microbiota associated with common periodontitis is the result of a slow, continuous process, taking place in a habitat with favorable ecological conditions, whereas the microbiota in highly active lesions, or in subjects with severe medical conditions, evolves under ecologic pressure and thus has a low diversity.
Morphological, Biochemical, and Molecular Characterization
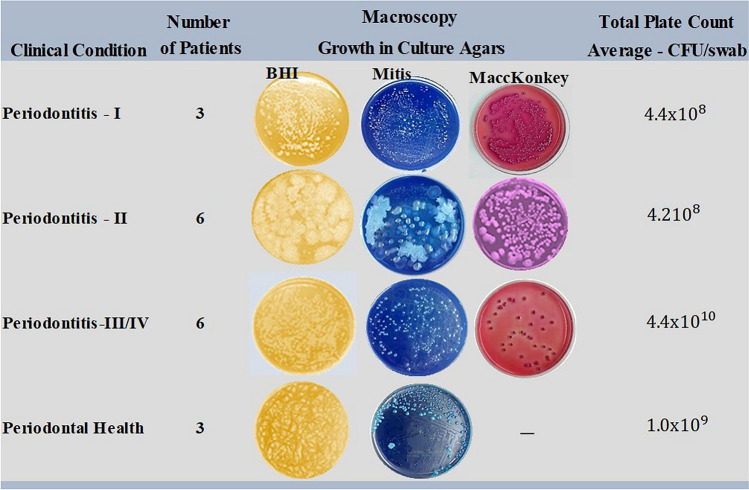
The Fig. 1 shows the macroscopic characteristics of the bacterial isolates in three types of agar plates and the respective clinical condition of patients.
Fig. 1.
Macroscopic characteristics and load of bacterial isolates in agars plates related to periodontal clinical condition of patients
There was microbiological diversity even for individuals of the control group (with periodontal health) whose average microbial load of Gram positive and Gram-negative bacterial isolates was respectively 1.82 × 109 and 1.0 × 102 CFU/patient. Thus, the microbial load in healthy patients was high, but the average number of Gram-negative pathogens for that group was relatively low. For patients with periodontitis, the microbial load of Gram-positive pathogens was 1.7 × 108 CFU/patient and for Gram-negative the microbial load was 2.9 × 107 CFU/patient. It is also observed that individuals with periodontitis stages III/IV had a higher microbial load compared to the other stages.
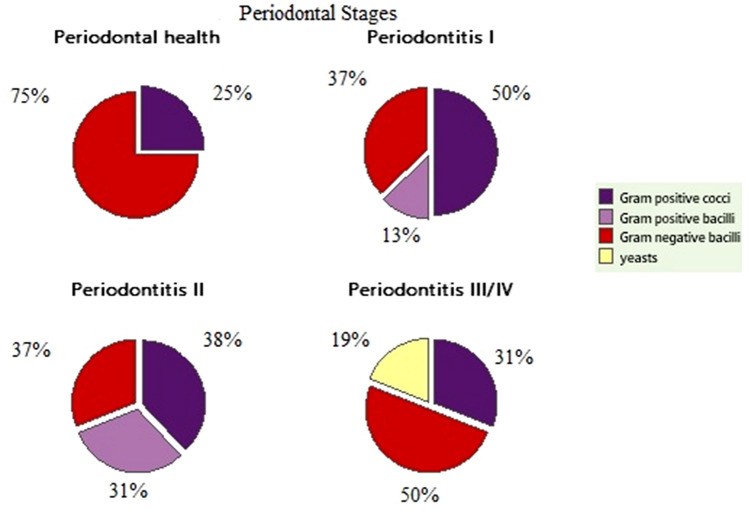
The Fig. 2 indicates the percentage relationship between the groups of microorganisms and the clinical condition of the patients used in this study. Regarding the percentage of Gram-positive cocci, consisting of 50% for individuals with periodontitis stage I, 38% for periodontitis stage II and 31% for periodontitis stages III and IV. The differences between the percentage profiles in relation to the periodontitis stages were significant (p < 0.05).
Fig. 2.
Comparative percentage between groups of microorganisms and clinical periodontal condition of patients
It is estimated that as the microbial biofilm progresses there is a transition from the aerobic environment to an anaerobic environment. So, aerobic gram-positive cocci are gradually replaced by facultative anaerobic or anaerobic Gram-negative bacilli, with greater pathogenic potential [15]. In this way, microbial diversity becomes greater for periodontal disease patients than in healthy individuals, i.e., increasing as the disease progresses with complex changes in the patients' microbiota [6, 15, 16]. In the present study, the high number of Gram-negative bacteria in the control group i.e. 75% of the total isolates indicates that they also occur in the oral cavity of healthy individuals. In this way, they can compete with other or even behave like opportunists when immunity is low.
All isolates were considered for initial characterization by biochemical analysis. This step was necessary since it allowed the initial differentiation of groups based on the assimilation profile of amino acids and carbohydrates (Table 1). About 78.04% were positive catalase, while 19.5% were positive oxidase. In addition, 95.12% of isolates were able to use glucose as a carbon source, 58.53% sucrose and 29.26% fermented lactose. About 51.2% of the microorganisms showed γ-hemolysis reaction in blood agar, 41.4% β-hemolysis and 7.4% α-hemolysis reaction. Twenty-five (n = 25) isolates were identified at species level by molecular analysis. For the others, there was no significant percentage of similarity, preferring here to consider them as undetermined, and that way they were stored for further study.
Table 1.
Biochemical and morphological profile of microorganisms isolates from oral cavity of patients with periodontitis
| Isolates | Tests | Sugar assimilation/gas (+/−) | |||||
|---|---|---|---|---|---|---|---|
| No. | Morphology/Gram (+/−) | Catalase | Oxidase | Hemolysis | Gly | Suc | Lac |
| 1 | bacilli G− | − | − | γ | +/+ | +/+ | +/+ |
| 2 | bacilli G− | − | − | γ | +/− | −/− | −/− |
| 3 | bacilli G− | − | − | γ | +/+ | +/+ | +/+ |
| 4 | bacilli G− | − | + | β | +/+ | +/+ | +/+ |
| 5 | bacilli G− | + | + | γ | +/− | +/+ | −/− |
| 6 | bacilli G− | + | + | β | v/− | −/− | −/− |
| 7 | bacilli G− | + | + | β | +/+ | −/− | −/− |
| 8 | bacilli G− | + | − | α | +/− | −/− | −/− |
| 9 | bacilli G− | + | − | β | +/+ | +/+ | −/− |
| 10 | bacilli G− | + | − | γ | +/+ | +/+ | +/+ |
| 11 | bacilli G− | + | − | γ | +/− | +/− | −/− |
| 12 | bacilli G− | + | − | γ | +/− | −/− | −/− |
| 13 | bacilli G− | + | − | γ | +/+ | +/+ | −/− |
| 14 | bacilli G− | + | − | γ | +/+ | +/+ | +/+ |
| 15 | bacilli G− | + | − | γ | +/+ | −/− | −/− |
| 16 | bacilli G− | + | − | γ | +/− | +/− | −/− |
| 17 | bacilli G− | + | − | γ | +/+ | +/+ | +/+ |
| 18 | bacilli G− | + | − | γ | +/+ | +/+ | +/+ |
| 19 | bacilli G− | + | − | β | −/− | −/− | −/− |
| 20 | bacilli G+ | + | − | β | +/− | −/− | −/− |
| 21 | bacilli G+ | + | − | β | +/− | −/− | −/− |
| 22 | bacilli G+ | + | − | β | +/− | −/− | −/− |
| 23 | cocci G+ | − | + | α | +/− | −/− | +/− |
| 24 | cocci G+ | − | − | β | +/− | +/− | +/− |
| 25 | cocci G+ | − | − | γ | +/− | +/− | −/− |
| 26 | cocci G+ | − | − | γ | +/− | +/− | −/− |
| 27 | cocci G+ | + | − | β | +/− | −/− | −/− |
| 28 | cocci G+ | + | − | γ | +/− | −/− | −/− |
| 29 | cocci G+ | + | − | β | +/− | +/− | −/− |
| 30 | cocci G+ | + | − | β | +/− | +/− | −/− |
| 31 | cocci G+ | + | − | β | +/− | +/− | −/− |
| 32 | cocci G+ | + | − | β | +/− | +/− | −/− |
| 33 | cocci G+ | + | − | γ | +/− | +/− | −/− |
| 34 | cocci G+ | + | − | β | +/− | +/+ | −/− |
| 35 | cocci G+ | + | − | β | +/− | +/− | +/− |
| 36 | cocci G+ | + | − | β | +/− | +/− | +/− |
| 37 | cocci G+ | + | − | γ | +/− | +/+ | +/+ |
| 38 | yeast | − | + | α | +/+ | −/− | −/− |
| 39 | yeast | + | − | γ | +/+ | −/− | −/− |
| 40 | yeast | + | + | γ | +/+ | −/− | −/− |
| 41 | yeast | + | + | γ | +/+ | −/− | −/− |
The number of bacterial genera was higher in the advanced stages of periodontal disease, i.e., three genera were identified in patients with periodontitis stage I, five genera in patients with periodontitis stage II and eight genera identified in patients with periodontitis stages III and IV. Staphylococcus sp. was the most frequent genera, having been isolated from patients with periodontitis stages II, III and IV. High prevalence rates of Staphylococcus spp. have been reported as an oral colonizer in saliva samples of hospitalized patients [17]. Likewise, Colombo et al. [18] observed that the proportion of S. aureus was significantly higher in patients with periodontal disease than in healthy individuals.
Other species as Citrobacter freundii, Bacillus sp., Raoutella sp., Klebsiella ozaenae, Streptococcus sp. and Pseudomonas sp. were also isolated. Raoutella sp., Escherichia coli and Citrobacter freundii are also related to periodontal inflammation and tissue destruction and they are potential risk factors for the development of several systemic diseases [19]. Raoutella sp., Klebsiella pneumoniae, P. aeruginosa and Staphylococcus sp., may be involved in breathing problems, like pneumonia [9, 18, 20].
P. aeruginosa was also isolated from control group (periodontal health). However, according Colombo et al. [18] there was no difference in the amount of P. aeruginosa when comparing individuals with periodontitis and healthy individuals. Regarding the species Lactococcus lactis, it was isolated only from control group.
Molecular Profile
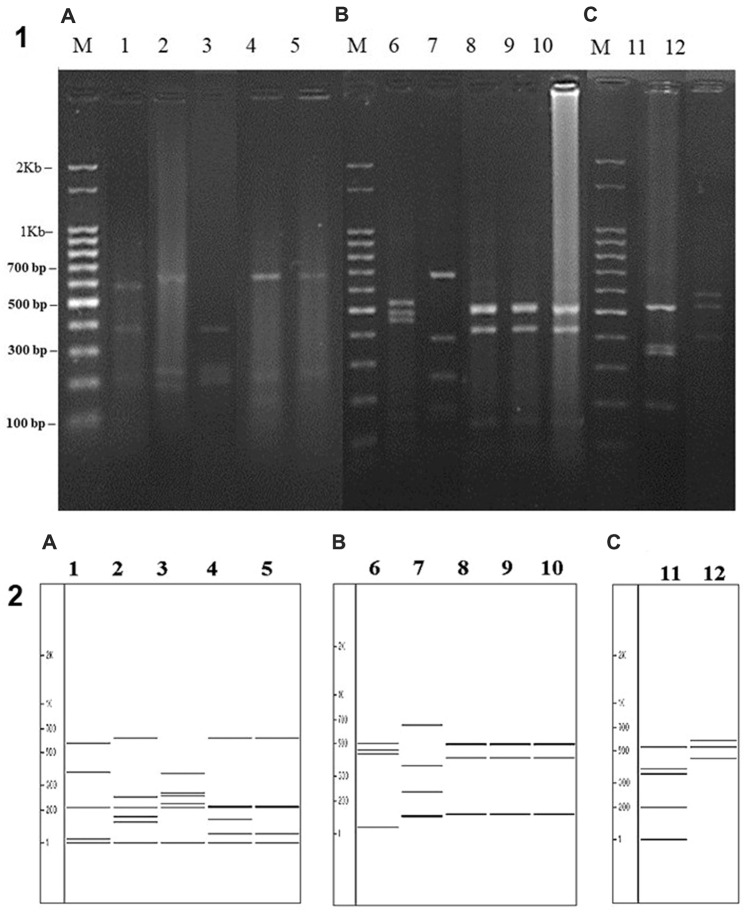
The Fig. 3 shows agarose gel with DNA fragments of PCR products and an in silico gel obtained with the aid of the Vector NTI 4.0 Program. The molecular profile by ARDRA of twelve bacterial isolates was determined from the comparison of sequences of the GenBank via BLAST Program [13]. Five of the isolates had their genetic material cleaved with restriction enzyme Alu I (A), five with restriction enzyme Rsa I (B) and two with restriction enzyme HhaI (C). Thus, the molecular profiles determined were respectively: A- Klebsiella ozaenae (NCBI access number: CP027612.1); Enterococcus avium (NCBI access number: AF061008.1); Lactococcus lactis (NCBI access number: CP028160.1); Staphylococcus haemolyticus (NCBI access number: CP025396.1); S. epidermidis (NCBI access number: CP000029.1); B- Escherichia coli (NCBI access number: NR_024570.1); Pseudomonas aeruginosa (NCBI access number: CP031677.1); S. aureus (NCBI access number: MRSA107, three isolates); (C) Klebsiella azaenae (NCBI access number: CP027612.1); Streptococcus salivarius (NCBI access number: CP020451.2).
Fig. 3.
A stained ethidium bromide agarose gel showing fragments of DNA from PCR products (16S rDNA) by ARDRA technique of bacterial isolates of oral cavity of patients with periodontitis. Lane M contains a 2 Kb plus DNA ladder (Ludwig Biotech) and strategic fragments are identified. Legend: (1) 1.5% agarose gel with PCR products; (2) molecular profile of in silico gel obtained with the aid of the Vector NTI 4.0 program: Lanes 1–12 the bacterial isolates had their genetic material cleaved with the restriction enzymes: Alu I (A), Rsa I (B) and (C) HhaI (C). Lanes: 1 Klebsiella ozaenae (NCBI access number: CP027612.1); 2 Enterococcus avium (NCBI access number: AF061008.1); 3 Lactococcus lactis (NCBI access number: CP028160.1); 4 Staphylococcus haemolyticus (NCBI access number: CP025396.1); 5 S. epidermidis (NCBI access number: CP000029.1); 6 Escherichia coli (NCBI access number: NR_024570.1); 7 Pseudomonas aeruginosa (NCBI access number: CP031677.1); 8–10 S. aureus (NCBI access number: MRSA107, three isolates); 11 Klebsiella azaenae (NCBI access number: CP027612.1); 12 Streptococcus salivarius (NCBI access number: CP020451.2)
Antimicrobial Susceptibility Profile
Seven isolates (41.2%) presented resistance for two or more antimicrobials of clinical use (Table 2). Amoxicillin was responsible for the highest resistance profile, i.e. 80% of the strains (n = 12) were resistant to that antibiotic. However, the chloramphenicol was effective in inhibiting in vitro all microorganisms. Streptococcus sp., Raoultella sp. (one isolate) and Enterococcus avium isolated from patients with periodontitis stages III and IV were resistant respectively to n = 4, n = 5 e n = 6 antibiotics. They also presented multidrug resistance profile. In addition, resistance to β-lactam antibiotics was observed for n = 13 strains, besides multidrug-resistance in two of them: Raoutella sp. and Enterococcus avium. This last one was resistant to four β-lactam antibiotics.
Table 2.
Antimicrobial susceptibility profile of bacterial strains isolated from patients with periodontitis
| Bacterial strains | Clinical condition | Antimicrobials | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Clindamycin | Azithromycin | Amoxicillin | Cephalothin | Cefotaxime | Tetracyclin | Chloramphenicol | Imipenem | ||
| Klebsiella ozaenae | Periodontitis I | – | S | R | S | S | S | S | I |
| Raoultella sp. | Periodontitis I | – | S | R | S | S | S | S | S |
| Staphylococcus haemolyticus | Periodontitis I | S | S | R | S | S | S | S | S |
| Citrobacter freundii | Periodontitis II | – | S | S | S | S | S | S | S |
| Klebsiella ozaenae | Periodontitis II | – | S | R | S | S | S | S | I |
| Staphylococcus aureus | Periodontitis II | S | R | R | S | S | S | S | S |
| Streptococcus salivarius | Periodontitis II | S | S | S | S | S | S | S | S |
| Bacillus sp. | Periodontitis II | – | – | – | – | – | – | – | – |
| Pseudomonas fluorescens | Periodontitis III /IV | – | – | – | – | I | R | S | R |
| Citrobacter freundii | Periodontitis III /IV | – | S | I | S | S | S | S | S |
| Enterobacter aerogenes | Periodontitis III /IV | – | S | R | S | S | S | S | I |
| Escherichia coli | Periodontitis III /IV | – | S | R | R | S | S | S | I |
| Raoultella sp. | Periodontitis III /IV | – | S | R | R | R | R | S | R |
| Staphylococcus aureus | Periodontitis III /IV | S | R | R | S | S | S | S | S |
| Streptococcus pneumoniae | Periodontitis III /IV | R | R | R | R | S | S | I | S |
| Enterococcus avium | Periodontitis III /IV | R | R | R | R | R | S | I | R |
| Pseudomonas aeruginosa | Periodontal health | – | – | – | – | S | I | I | S |
| Citrobacter freundii | Periodontal health | – | S | R | S | S | S | S | S |
| Lactococcus lactis | Periodontal health | – | – | – | – | – | – | – | – |
R Resistance; S Susceptible; – not determined
Regarding the use of antibiotics, the main concern is the loss of effectiveness mainly due to the horizontal transfer of genes [21] and, consequently, the development of bacterial resistance [20]. Dental biofilm can inhibit the action of several drugs and constitutes a resistance factor to antimicrobial agents [1]. Thus, the presence of microorganisms resistant to the drugs in common use is another obstacle to the treatment of periodontal disease.
At the present study one strain of E. avian showed resistance to six of the eight antimicrobials evaluated, and of these, four were β-lactams. E. avian is not a common human pathogen, however a few cases of bacteremia have been reported [22]. Although the patients involved in this study are workers from a region of intense livestock activity, whose main activity is the poultry production for food sector, it is not possible to affirm that there is a relationship on origin of contamination by E. avian, but in this regard future investigations will be carried out.
For Enterococcus sp., the resistance to β-lactam antibiotics is related to increased production of PBP5 (Penicillin-binding protein 5) and the spread of strains with mutant pbp5 genes through horizontal gene transfer [23, 24]. Studies claim that enterococcal plasmids during horizontal gene transfer can undergo genetic recombination processes, contributing to increased diversity and the evolution of antibiotic resistance [24, 25]. Molecular mechanisms such as gene that code for efflux pumps, antibiotic inactivating enzymes and other factors that assist in the low permeability of the membrane contribute directly to bacterial resistance [26, 27]. Pseudomonas aeruginosa, Enterococcus faecalis and species of Enterobacteriaceae (including Klebsiella and E. coli) were multidrug resistance, so they constitute a major threat to human health. Due to this problem, the mentioned microorganisms are included in the list of priority pathogens by the World Health Organization for research and development of new antibiotics [22].
The genus Staphylococcus was present in the samples referring to periodontitis stages I, II, and III/IV, but only showed resistance to azithromycin and/or amoxicillin. Unlike this, Kim and Lee [28] evaluated that none of the isolates of S. aureus (n = 18) from the oral cavity of patients with periodontitis presented resistance to chloramphenicol, clindamycin, or imipenem, but 7,31% were resistant to tetracycline.
Although several microorganisms are susceptible to several antimicrobials, none of the drugs administered alone can combat all pathogens related to periodontal diseases [1]. It is also believed that the biofilm can block the action of several antimicrobials, hindering the permeability of drugs [27]. As a result, there is a concern for example, a patient with gingivitis can revert to a state of health, however, those who have periodontitis will remain with the disease, even after therapy [8, 29].
New research on colonization and microbiological interactions existing in the oral environment are fundamental for a greater perception of the influence that such pathogens have on periodontal health [19]. Thus, although dental biofilm can harbor a small percentage of pathogenic bacteria, it can still cause damage to the periodontal structure [2]. In periodontal disease there is an interaction between the subgingival biofilm composition and the host response, more than being caused by a single pathogen [30]. Furthermore, pathogens and multidrug resistance are factors of great concern. In addition, despite the importance of a microbiological approach for understanding the dynamics of the microbiota associated with periodontal disease, there is no specific effective antibacterial therapeutic support for the treatment of periodontal disease [6].
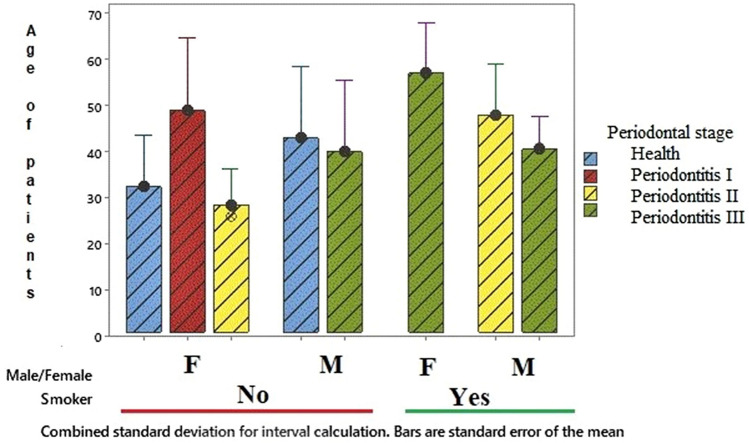
In addition to the results obtained in the present study, a preliminary evaluation was also carried out to verify an association between smoking, age, and sex with the microbial load of patients with periodontitis. Thus, of the patients analyzed, 46.66% were smokers and 53.34% were non-smokers. It is established that smoking is considered a risk factor for periodontitis in terms of onset, extension, or worsening of the disease [16, 31]. In our study for the group of patients here analyzed and based on the microbial load there was no statistically significant correlation (p > 0.05) in relation to smoking, age, or even the patient's gender between each of these factors and the periodontitis stage. In addition, there was also no statistically significant difference between the periodontal stage for male or female patients. However, stage I periodontitis was observed only for non-smoking women (Fig. 4).
Fig. 4.
Group of patients with periodontal disease or healthy according to age, sex, and smoking habit. Legend: smokers: 43.66%; non-smokers 53.34%. There was no statistically significant correlation (p > 0.05) between periodontal stage, male/female (M/F) and smoking
Conclusions
Based on present study, we consider that there was an association between microbial load, species diversity and the stage of periodontal disease. Periodontitis cases, specially in the early stages (I, II) need to be safely evaluated and followed up to contribute to the prevention of other diseases arising.
The main bacterial strains isolated from patients with periodontitis stages III and IV were Citrobacter freundii, Bacillus sp., Raoutella sp., Klebsiella azaenae, Streptococcus pneumoniae. and Pseudomonas spp. Multidrug resistance was observed in strains of Streptococcus pneumoniae, Raoutella sp. and Enterococcus avium. Thus, biofilms formed by a pathogenic and multidrug-resistant microbiota can constitute an additional concern regarding the restoration of periodontitis patient health since antibiotic therapy can result in the infection not being controlled. The knowledge of the determinant causes/conditions and stage of periodontal disease as well as prophylactic measures for maintaining oral health are still the best option to reduce the risk of disease. In addition, the evaluation of effective antimicrobials by avoiding indiscriminate administration is also suggestive of good conduct practices.
Acknowledgements
We thanks to the National Council for Technological and Scientific Development (CNPq, Brazil) by partial financial support. The authors would like also to thank all the technical professionals of the microbiology and molecular biology laboratories of the Biotechnology Center in which this study was carried out.
Authors Contribution
All authors contributed to this study. Thus, RA conducted the microbiological and molecular experiments. JMLNG defined experimental design, analysis, and text of the manuscript. MSc. QLS collected samples as well as made the medical diagnostic the patients involved in the study. CMB and CAA contributed to the molecular identification of the microorganisms. CL reviewed the manuscript text and the theoretical study. The final manuscript was approved by all authors.
Funding
This study was supported by a grant from CNPq, Brazil.
Compliance with Ethical Standards
Conflict of interest
All authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Marsh PD, Zaura E. Dental biofilm: ecological interactions in health and disease. J Clin Periodontol. 2017;44:12–22. doi: 10.1111/jcpe.12679. [DOI] [PubMed] [Google Scholar]
- 2.Popova C, Dosseva-Panova V, Panov V. Microbiology of periodontal diseases A Review. Biotechnol Biotechnol Equip. 2013;27:3754–3759. doi: 10.5504/BBEQ.2013.0027. [DOI] [Google Scholar]
- 3.Larsen T, Fiehn NE. Dental biofilm infections–an update. Apmis. 2017;125:376–384. doi: 10.1111/apm.12688. [DOI] [PubMed] [Google Scholar]
- 4.Lasserre JF, Brecs MC, Toma S. Oral microbes, biofilms and their role in periodontal and peri-implant diseases. Materials. 2018;11:1802. doi: 10.3390/ma11101802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kinane DF, Stathopoulou PG, Papapanou PN. Periodontal diseases. Nat Rev Dis Primers. 2017;3:17038. doi: 10.1038/nrdp.2017.38. [DOI] [PubMed] [Google Scholar]
- 6.Mombelli A. Microbial colonization of the periodontal pocket and its significance for periodontal therapy. Periodontol 2000. 2017;76:85–96. doi: 10.1111/prd.12147. [DOI] [PubMed] [Google Scholar]
- 7.Kagiya T. MicroRNAs: potential biomarkers and therapeutic targets for alveolar bone loss in periodontal disease. Int J Mol Sci. 2016;17:1317. doi: 10.3390/ijms17081317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caton J, Armitage G, Berglundh T, Chapple ILC, Kenneth SJ, Kornman S, et al. A new classification scheme for periodontal and peri-implant diseases and conditions—Introduction and key changes from the 1999 classification. J Periodontol. 2018;89:S1–S8. doi: 10.1002/JPER.18-0157. [DOI] [PubMed] [Google Scholar]
- 9.Hong CHL, Aung MM, Kanagasabai K, Lim CA, Liang S, Tan KS. The association between oral health status and respiratory pathogen colonization with pneumonia risk in institutionalized adults. Int J Dent Hyg. 2018;16:96–102. doi: 10.1111/idh.12321. [DOI] [PubMed] [Google Scholar]
- 10.World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310:2191–2194. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
- 11.Mendoza-Espinoza A, Koga Y, Zavaleta AI. Amplified 16S ribosomal DNA restriction analysis for identification of Avibacterium paragallinarum. Avian Dis. 2008;52:54–58. doi: 10.1637/8036-062507-Reg. [DOI] [PubMed] [Google Scholar]
- 12.Sambrook J, Russel DW. Molecular cloning: a laboratory manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 13.NCBI (2019) Nucleotide. https://www.ncbi.nlm.nih.gov/nuccore/. Accessed 20 Nov 2019
- 14.CLSI . Laboratory standards for antimicrobial susceptibility testing. 29. Wayne, PA: Clinical Laboratory Standards Institute (CLSI); 2019. [Google Scholar]
- 15.Zaura E, Keijser BJ, Huse SM, Crielaard W. Defining the healthy" core microbiome" of oral microbial communities. BMC Microbiol. 2009;9:259. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Camelo-Castillo AJ, Mira A, Pico A, Nibali L, Henderson B, Donos N, et al. Subgingival microbiota in health compared to periodontitis and the influence of smoking. Front Microbiol. 2015;6:119. doi: 10.3389/fmicb.2015.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuanazzi D, Souto R, Mattos MBA, Zuanazzi MR, Tura BR, Sansone C, Colombo APV. Prevalence of potential bacterial respiratory pathogens in the oral cavity of hospitalised individuals. Arch Oral Biol. 2010;55:21–28. doi: 10.1016/j.archoralbio.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Colombo AV, Barbosa GM, Higashi D, Micheli GD, Rodrigues PH, Simionato MRL. Quantitative detection of Staphylococcus aureus, Enterococcus faecalis and Pseudomonas aeruginosa in human oral epithelial cells from subjects with periodontitis and periodontal health. J Med Microbiol. 2013;62:1592–1600. doi: 10.1099/jmm.0.055830-0. [DOI] [PubMed] [Google Scholar]
- 19.Colombo APV, Magalhães CB, Hartenbach FARR, Souto RM, Silva-Boghossian CM. Periodontal-disease-associated biofilm: a reservoir for pathogens of medical importance. Microb Pathog. 2016;94:27–34. doi: 10.1016/j.micpath.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 20.Sękowska A. Raoultella spp. clinical significance, infections, and susceptibility to antibiotics. Folia Microbiol. 2017;62:221–227. doi: 10.1007/s12223-016-0490-7. [DOI] [PubMed] [Google Scholar]
- 21.Lerminiaux NA, Cameron ADS. Horizontal transfer of antibiotic resistance genes in clinical environments. Can J Microbiol. 2019;65:34–44. doi: 10.1139/cjm-2018-0275. [DOI] [PubMed] [Google Scholar]
- 22.World Health Organization (2017) WHO publishes list of bacteria for which new antibiotics are urgently needed. https://www.who.int/news-room/detail/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed.
- 23.Layton BA, Walters SP, Lam LH, Boehm AB. Enterococcus species distribution among human and animal hosts using multiplex PCR. J Appl Microbiol. 2010;109:539–547. doi: 10.1111/j.1365-2672.2010.04675.x. [DOI] [PubMed] [Google Scholar]
- 24.Gagetti P, Bonofiglio L, Gabarrot GG, Kaufman S, Mollerach M, Vigliarolo L, et al. Resistance to β-lactams in enterococci. Rev Argent Microbiol. 2019;51:179–183. doi: 10.1016/j.ram.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 25.Sante L, Morroni G, Brenciani A, Vignaroli C, Antonelli A, D’Andrea MM, et al. pHTβ-promoted mobilization of non-conjugative resistance plasmids from Enterococcus faecium to Enterococcus faecalis. J Antimicrob Chemother. 2017;72:2447–2453. doi: 10.1093/jac/dkx197. [DOI] [PubMed] [Google Scholar]
- 26.Sánchez MB. Antibiotic resistance in the opportunistic pathogen Stenotrophomonas maltophilia. Front Microbiol. 2015;6:658. doi: 10.3389/fmicb.2015.00658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyle J, Chapple I. Molecular aspects of the pathogenesis of periodontitis. Periodontol 2000. 2015;69:7–17. doi: 10.1111/prd.12104. [DOI] [PubMed] [Google Scholar]
- 28.Kim GY, Lee CH. Antimicrobial susceptibility and pathogenic genes of Staphylococcus aureus isolated from the oral cavity of patients with periodontitis. J Periodontal Implant Sci. 2015;45:223–228. doi: 10.5051/jpis.2015.45.6.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chapple ILC, Mealey BL, Van Dyke TE, Bartold PM, Dommisch H, Eickholz P, et al. Periodontal health and gingival diseases and conditions on an intact and a reduced periodontium: consensus report of workgroup 1 of the 2017 world workshop on the classification of periodontal and peri-implant diseases and conditions. J Periodontol. 2018;89:S74–S84. doi: 10.1002/JPER.17-0719. [DOI] [PubMed] [Google Scholar]
- 30.Mombelli A. Microbial colonization of the periodontal pocket and its significance for periodontal therapy. Periodontol. 2018;76:85–96. doi: 10.1111/prd.12147. [DOI] [PubMed] [Google Scholar]
- 31.Borojevic T. Smoking and periodontal disease. Mater Sociomed. 2012;24:274–276. doi: 10.5455/msm.2012.24.274-276. [DOI] [PMC free article] [PubMed] [Google Scholar]