Abstract
In sectors like healthcare and hospitality, it has been realized that fabrics play a pivotal role in transfer of nosocomial infections. However, there is a major gap in drawing correlation between different fibre types and their interaction with microorganisms. Such information is important to formulate guidelines for textile materials for use in these sectors. In the current study, the adherence of four important bacteria, Staphylococcus aureus, Acinetobacter calcoaceticus, Escherichia coli, and Pseudomonas aeruginosa was studied on six different fibre types namely polyester, wool, polypropylene, viscose, silk and cotton. Among these fibres, viscose showed maximum adherence while silk fibres showed the least attachment of bacterial strains. Bacterial adhesion was correlated with the surface characteristics (surface charge, hydrophobicity etc.) of bacteria, and nanoroughness of fibres. Adhesion of these bacteria was tested on five hydrocarbons of different hydrophobicities. E. coli, the weakest biofilm producer, and with the highest surface energy and lowest hydrophobicity amongst the bacteria compared in the study, had the lowest load on all fibres. Scanning electron microscopy revealed non-uniform binding of gram-negative and gram-positive bacteria. Nanoroughness of fibres favored bacterial adhesion. The study showed correlation between surface properties and adherence of bacteria on fibres, with the results being of direct significance to medical and hospitality sectors.
Electronic supplementary material
The online version of this article (10.1007/s12088-020-00903-5) contains supplementary material, which is available to authorized users.
Keywords: Textile, Infection, Bacterial adhesion, Surface morphology, Contact angle
Introduction
Textile surfaces exhibit a potent role in microbial adhesion and transfer [1–3]. Specifically, uniform of health care practitioners has been reported to mediate the transfer of nosocomial infection in health care sectors with reports on an increasing trend in cases of antibiotic resistant bacteria in the hospital environment [4]. The microbial load on healthcare textiles can serve as a route for the transfer of pathogenic microorganisms as soft surfaces serve as a suitable habitat for the growth and multiplication of organisms [3, 5]. The same is true for the hospitality sector. Hence it is imperative to study how textile properties affect microbial load. This will help in strategizing measures to combat the spread, and develop guidelines for application in healthcare and hospitality sectors.
The interaction of microorganisms with textile depends on various factors like type of microorganism, surface characteristics of microbe and various environmental factors (physical and chemical). The zeta potential on the surface of bacteria depends on the nature of bacterial species, growth medium, surface architecture, and age of the bacteria [6]. Bacterial adherence and biofilm development on the surfaces of materials may lead to various health complications [7]. Microbial adhesion on textile is also influenced by material surface characteristics like chemical composition, roughness, the surface charge of textiles etc. In the case of soft surfaces like textiles, zeta potential is a parameter describing the nature of functional group and its dissociation. The surface charge on fibre depends on molecular as well as supramolecular structure. Composition and concentration of adsorbate also plays a role in determining the surface charge [8]. Bacterial species carry negative charge on its surface under physiological conditions [9]. Also textile surfaces have negative charge, thus bacteria mostly experience the double electric layer repulsion when approaching these textile surfaces [6]. Interaction between these double electric layers plays a pivotal role in bacterial adhesion on textile surfaces with adherence depending on the ionic stability and pH of the solution.
The present study aimed to study the interaction of six selected fibres, viz. viscose, polypropylene, polyester, cotton, silk and wool, with four important bacteria normally found as a part of human skin flora, viz. Staphylococcus aureus, Acinetobacter calcoaceticus, Escherichia coli and Pseudomonas aeruginosa. A comprehensive analysis of the surface properties of bacteria and fibres was performed to draw a correlation between the two.
Materials and Methods
Materials
Cultures of S. aureus, A. calcoaceticus, E. coli and P. aeruginosa were assessed from the culture collection of Department of Biochemical Engineering and Biotechnology, Indian Institute of Technology Delhi. Luria Bertani (LB) broth and LB agar was used to grow bacterial cultures. Cotton, polypropylene, polyester, viscose, silk and wool fibres were procured locally.
Nanoroughness of Fibres
Nanoroughness of fibres was determined by Atomic Force Microscopy (AFM, Asylum Research MFP3D-SA, Oxford Instruments, Manchester, UK). Ten fibres of each type (length of 3 cm) mounted on a glass slide was analysed by AFM probe generating a surface topography map, which reflected surface roughness.
Microbial Adhesion to Hydrocarbon (MATH) Test
Bacterial cultures (grown overnight) were used for MATH test with the hydrocarbons n-octane, p-xylene, n-hexane, toluene, and n-hexadecane as per standard protocol [10].
Contact Angle Measurement of Bacteria
For measurement of contact angle of bacteria a cellulose membrane filter was placed over the bacterial colonies such that the colonies shifted to the filter. A tiny drop of water was placed carefully on a colony in such a way that the latter did not wash away from the agar [11]. Then the contact angle of water with bacterial colony was measured using Michell Tilting Pad JYSP-360 v Contact Angle Goniometer (Beijing United Test Co., Ltd, Beijing). Six independent measurements were taken for each sample.
Surface Charge on Cells
Bacterial cell pellets (centrifuged from culture broth) were mixed with phosphate-buffered saline (PBS) and analysed using Dynamic Light Scattering (Malvern Panalytical, UK).
Biofilm Formation
Quantification of biofilm of S. aureus, A. calcoaceticus, E. coli and P. aeruginosa were performed in a microtiter plate assay based on measurement of optical density (OD) at 570 nm [12]. The experiment was performed in triplicate. The average OD values were calculated for negative control and the bacterial strains [13]. Cut off value (ODc) was determined (Eq. 1) and final OD of tested strains were calculated (Eq. 2).
| 1 |
| 2 |
If the obtained value was negative, it was considered as zero, while a positive value represented biofilm formation.
For interpretation of result, strains were divided into various categories:
OD ≤ ODc = No biofilm producer ……………. (i)
4 × ODc < OD = Strong biofilm producer .…………….(ii).
2 × ODc < OD ≤ 4 × ODc = Moderate biofilm producer .……………(iii).
ODc < OD ≤ 2 × ODc = Weak biofilm producer .…………….(iv).
.
Bacterial Load on Fibres
Fibres (10 mg of each type) were sterilized by autoclaving. Pure culture of bacteria was inoculated in 100 ml of LB broth and incubated at 37 °C with rotation at a speed of 150 rpm for 3.5 h. 10 mg of each sterile fibre (cotton, viscose, polypropylene, polyester, silk and wool) was incubated individually at 37 °C with 2.5 ml of each bacteria, viz. S. aureus, A. calcoaceticus, E. coli, P. aeruginosa [~(2.5 ± 0.22)*107 CFU/ml] in 50 ml centrifuge tubes. Controls included culture broths without fibres. Sampling was performed at four time points (45, 90, 135 and 180 min) by agitating in a shaker at 50 rpm. Subsequently broth was drained from treated fibres, 5 ml of PBS was added to each treatment centrifuge tube, and vortexed to dislodge bacteria from fibres. The supernatant was plated on LB agar to obtain CFU/mg of fibres. All experiments were performed in triplicates (for both control and treatment samples).
Scanning Electron Microscopy (SEM)
Visualization of bacterial adhesion on different fibres was performed by SEM. 10 mg of sterilized fibres were incubated with 10 ml of log phase culture in centrifuge tubes and incubated at 37 °C (50 rpm) overnight. Samples were further treated with 5 ml of Karnovsky fixative (1 g paraformaldehyde crystals, 12.5 ml double distilled water, 1.25 ml 50% glutaraldehyde, 11.25 ml PBS) and incubated for 60 min at room temperature, followed by washing of samples in an increasing concentration of ethanol (20%, 40%, 70% and then 100% ethanol). Air-drying of samples was done for 1 h at room temperature. Dried samples were attached to aluminum coupons by using double-sided sticky carbon tape. The samples were coated with a thin layer (~ 200 Å) of gold so that the samples would become conductive.
Statistical Analysis
One-way ANOVA was used to analyse the data by using SPSS Statistical System (SPSS 16.0 for Windows). Duncan’s multiple range test (DMRT) at p < 0.05 was used to compare mean values [14].
Results
Upon observing the 3D topography of fibres, viscose fibre exhibited the highest nanoroughness (258.2 nm), followed by polyester (203 nm), cotton (158.2 nm), wool (102.9 nm), polypropylene (86.8 nm), while silk fibre had least nanoroughness (55.3 nm) (Fig. 1).
Fig. 1.
3D surface topography of different fibres: cotton (a), polyester (b), polypropylene (c), silk (d), viscose (e), and wool (f)
Based on MATH test and contact angle measurements, P. aeruginosa showed the highest hydrophobicity followed by A. calcoaceticus, S. aureus, and E. coli (Table 1). The trend was the opposite in terms of zeta potential with E. coli exhibiting the highest zeta potential (− 27.7 mV), followed by S. aureus (− 18.7 mV), A. calcoaceticus (− 9.7 mV) and P. aeruginosa (− 6.6 mV). Similar trend was observed for cell surface charge with E. coli displaying the highest and P. aeruginosa the least charge on cell surface.
Table 1.
Properties of different bacteria
| Property | P. aeruginosa | A. calcoaceticus | S. aureus | E. coli |
|---|---|---|---|---|
| Hydrocarbon (MATH test) | % removal of cells from PBS | |||
| n-octane | 14.2 | 12 | 6.3 | 6.2 |
| p-xylene | 12.9 | 13 | 8.7 | 6 |
| n-hexane | 6.3 | 6.1 | 3.6 | 3 |
| Toluene | 6.2 | 6.4 | 3.2 | 2.5 |
| n-hexadecane | 18 | 17.9 | 13.9 | 7.1 |
| Contact Angle | 38.1 ± 1.5 | 32.0 ± 1.5 | 26.9 ± 1.8 | 23.5 ± 0.7 |
|
Surface Energy (mJ/m2) (Geometric mean approach) |
60.11 | 64.91 | 66.72 | 67.64 |
| Zeta potential (mV) | − 6.60 | − 9.7 | − 18.7 | − 27.7 |
|
Biofilm Producer (O.D) |
Strong (1.896 ± 0.19) |
Moderate (0.552 ± 0.007) |
Strong (1.672 ± 0.15) |
Weak (0.239 ± 0.003) |
Values are mean of 3 replicates
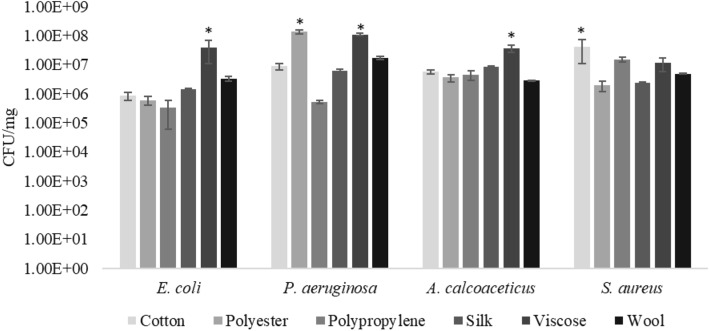
The load of four bacterial strains on six fibres was expressed in CFU/mg of fibres. Results clearly indicated that bacterial load on fibres was dependent on both the type of bacteria as well as fibre (Fig. 2). E. coli showed lowest adhesion on cotton, polypropylene, polyester, silk and wool with higher adhesion on viscose fibres (107 CFU/mg) (Suppl Fig. 1a). P. aeruginosa exhibited low adhesion on cotton, polypropylene, silk and wool, with intermediate adhesion on viscose, and highest adhesion on polyester. A. calcoaceticus had higher adhesion on viscose, while S. aureus was found to have highest load on cotton (Suppl Fig. 1b). On comparison between E. coli (gram-negative) bacteria and S. aureus (gram-positive), adhesion of gram-positive bacteria was mostly observed to be higher than gram-negative (Supplementary Fig. 1).
Fig. 2.
Bacterial counts on different fibres; mean count of bacterial cells in supernatant after dislodging of fibres (cotton, polyester, polypropylene, silk, viscose, wool) in PBS, incubated with different bacterial species (E. coli, P. aeruginosa, A. calcoaceticus, S. aureus). Error bars denote standard deviation (n = 03); *represent significant difference between the different fibres for each bacterial species based on one-way ANOVA
Amongst the four bacteria under study, count of P. aeruginosa was highest (2.7 × 108 CFU/mg) followed by S. aureus (7.8 × 107 CFU/mg), A. calcoaceticus (6.2 × 107 CFU/mg) and E. coli (4.5 × 107 CFU/mg). E. coli was found to have the lowest load on all fibres except viscose. In the context of bacterial abundance on different fibres, it was found that viscose fibre attracted higher number of bacteria with a count in the order of 107–108 CFU/mg in comparison to polyester, cotton, wool, polypropylene and silk which attracted in the order of 106–107 CFU/mg. Biofilm assay of S. aureus, A. calcoaceticus, P. aeruginosa and E. coli showed that S. aureus and P. aeruginosa were strong biofilm producers, A. calcoaceticus was moderate biofilm producer, while E. coli was observed to be a weak biofilm producer. SEM analysis was carried out to assess the mode of adherence of bacteria on fibres (Supplementary Fig. 2). Clumping of S. aureus, a cocci shaped bacteria, was observed on almost all the fibres, while E. coli, a rod-shaped bacteria, was observed to be irregularly arranged. Bacterial adhesion on fibres was not uniform.
Discussion
Bacteria-textile surface interaction is a complex process, which is affected by physicochemical characteristics of the textile surface, as well as surface properties of bacteria [15–17]. During the interaction between bacteria and the surface of textile, proteins and other biomolecules present in the surrounding medium can affect the interface between bacteria and textile by absorbing onto the surface of the textile [18].
It has been reported that measurement of the hydrophobicity of different cell surfaces is useful to get an insight into its adhesion on various surfaces. Super-hydrophilic substrate having negative zeta potential showed limited binding of bacteria due to the less hydrophobic interaction between the surface of bacteria and material. This strategy can be used to design the surface such as to prevent bacterial adhesion, subsequently solving the problem related to antibiotic administration [19]. In the present study it was observed that E. coli had the lowest hydrophobicity and highest zeta potential, while P. aeruginosa showed the highest hydrophobicity and lowest zeta potential value.
Surface nanoroughness has been reported to be an influential factor in terms of adhesion [20]. In the present study viscose fibre showed highest nanoroughness while silk had the least value. This correlated well with the trend of bacterial binding on fibres, in which maximum adhesion was observed on viscose fibres, while the least was observed on silk fibres. Silk has been earlier reported to discourage microbial adhesion [21], and is considered to be the most resistant among all types of natural fibres [1].
Takashima et al. [22] compared the binding ratio of S. aureus and P. aeruginosa on cotton, polyester, acrylic, nylon and wool fibres. It was found that S. aureus bound to polyester and acrylic fibres at a high ratio (96.2% and 87.6%), on to wool at an intermediate ratio (63.2%), and least binding ratio on cotton and nylon fibres (2.0% and 0.9%) was observed. However, P. aeruginosa bound to polyester and acrylic fibres at a high ratio (99.9% and 95.4%), to wool at an intermediate ratio of 84.7%, while least binding ratio was observed on nylon and cotton fibres (14.9% and 8.1%). The results of our study concur with the results obtained by Takashima et al. [22]. In both the studies, adhesion of P. aeruginosa was higher than S. aureus on textile fibres. In our study, the surface properties of bacteria as well as nanoroughness of fibres has been explored to study the different factors involved in bacterial adhesion on textile fibres. In the current study, introducing a dislodging step, provided a more sensitive approach for determination of CFU count on textile fibres, thereby enhancing the traditional method of CFU count. The present method thus, seems to be more accurate, reliable and reproducible for assessing the adherence of bacterial species on different fibres. On evaluating the CFU counts, the results showed the count to be statistically different (p < 0.05) with fibre type, which reflects that the surface properties of fibres like nanoroughness affect adhesion of microorganisms.
The direct contact between bacterial cells and surface of the fibres is generally prevented due to repulsive forces, however, significant contact can occur because of the cell surface appendages like fimbriae, pilli, exopolysaccharide fibrils and flagella [23]. P. aeruginosa [24] and S. aureus [25], A. calcoaceticus [26] and E. coli [27] have been reported to form biofilm on substrates. Biofilm formation depends on the proteins present on the surface of bacteria secreted by the secretome, which is a protein secretion system in the bacterial cells. These protein secretion systems are responsible for making changes in the cell envelope of gram-negative and gram-positive bacteria [28]. Due to the difference in the cell walls of gram positive and gram negative bacteria biofilm formation may differ [29].
SEM analysis showed irregular arrangement of bacterial cells on different fibres. Surface topography like crevices and grooves favor bacterial adhesion as they provide more surface area to adhere. In the case of viscose fibres, bacterial adhesion was observed on grooves and crevices. Silk fibre has a smooth and cylindrical surface. Bacterial adhesion was found to be more in the crevices/undulations of fibres. Interaction of fibre surface with single as well as multiple layers of cells was also observed. The type of fibre may also influence the clumping of cells. In our study, it was observed that bacteria did not cover the surface of fibres uniformly. Similar results of bacterial adhesion were obtained at fabric level through SEM analysis [1], which gives only a qualitative assessment of adhesion on different fibres [30]. The morphological examination of bacterial adhesion by SEM inferred that the factors other than hydrophilicity, surface roughness and absorbance of water could also significantly affect the extent of bacterial adhesion to the fibres [31]. There is also a need to check the effect of incubation time on adhesion. In the present study, shaking during incubation of fibres with cultures, and the relatively short incubation time would not have favoured the formation of biofilm, which has been reported to take a longer duration [32]. However, the initial phase of initiation of biofilm formation on the fibres (as is also evident from SEM images, Suppl Fig. 2) cannot be ruled out. For an accurate estimation of formation of mature biofilm on the fibres such an experiment should be carried out for a longer incubation period [33].
Despite the fact that this was an in vitro study, and findings might differ in real life samples, a positive correlation could be drawn between the nanoroughness of fibres and their bacterial load. Also, ability of bacteria to form biofilm and their hydrophobicity correlated positively with the load of bacteria of fibres. Further studies on the assessment of the survival and growth of bacteria on fibres will provide information about the associated risks in healthcare settings. Also, quantification of genes and transcripts involved in biofilm formation will reflect on the contribution of biofilm formation in adherence to fibres. Strategies to discourage biofilm formation by treatment of hospital textiles with anti-biofilm substances [34] should be worked upon using the most feasible and economical technique.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by FIRP Scheme (MI01681G) and Grand Challenge Scheme (MI1798G) of IIT Delhi. SV acknowledges fellowship received from DST-INSPIRE towards her doctoral work.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bajpai V, Dey A, Ghosh S, Bajpai S, Jha MK. Quantification of bacterial adherence on different textile fabrics. Int Biodeterior Biodegrad. 2011;65:1169–1174. doi: 10.1016/j.ibiod.2011.04.012. [DOI] [Google Scholar]
- 2.Banu A, Anand M, Nagi N. White coats as a vehicle for bacterial dissemination. J Clinic Diagnos Res. 2012;6:1381–1384. doi: 10.7860/jcdr/2012/4286.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta P, Bairagi N, Priyadarshini R, Singh A, Chauhan D, Gupta D. Bacterial contamination of nurses’ white coats made from polyester and blend fabric. J Hos Infect. 2016;94:92–94. doi: 10.1016/j.jhin.2016.05.016. [DOI] [PubMed] [Google Scholar]
- 4.Sailo CV, Pandey P, Mukherjee S, Zami Z, Lalremruata R, Nemi L, Senthil Kumar N. Pathogenic microbes contaminating mobile phones in hospital environment in Northeast India: incidence and antibiotic resistance. Trop Med Health. 2019;47:59. doi: 10.1186/s41182-019-0190-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subbalaxmi MVS, Lakshmi V, Lavanya V. Antibiotic resistance—experience in a tertiary care hospital in South India. J Assoc Phys India. 2010;58:Suppl.18–S22. [PubMed] [Google Scholar]
- 6.Oh JK, Yegin Y, Yang F, Zhang M, Li J, Huang S, Verkhoturov SV, Schweikert EA, Perez-Lewis K, Scholar EA, Taylor TM, Castillo A, Cisneros-Zevallos A, Min Y, Akbulut M. The influence of surface chemistry on the kinetics and thermodynamics of bacterial adhesion. Sci Rep. 2018;8:17247. doi: 10.1038/s41598-018-35343-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lebeaux D, Ghigo JM, Beloin C. Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol Mol Biol Rev. 2014;78:510–543. doi: 10.1128/mmbr.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grancarić AM, Ristić N, Tarbuk A, Ristić I. Electrokinetic phenomena of cationised cotton and its dyeability with reactive dyes. Fibres Text East Eur. 2013;21:106–110. [Google Scholar]
- 9.Wu J, Liu H, Wang P, Zhang D, Sun Y, Li E. Oxygen reduction reaction affected by sulfate-reducing bacteria: different roles of bacterial cells and metabolites. Ind J Microbiol. 2017;57:344–350. doi: 10.1007/s12088-014-0488-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goel A, Halami PM, Tamang J. Genome analysis of Lactobacillus plantarum isolated from some Indian fermented foods for bacteriocin production and probiotic marker genes. Front Microbiol. 2020;11:40. doi: 10.3389/fmicb.2020.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dixit D, Soppina V, Ghoroi C. A non-electric and affordable surface engineered particle (SEP) based point-of-use (POU) water disinfection system. Sci Rep. 2019;9:18245. doi: 10.1038/s41598-019-54602-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stepanović S, Vuković D, Hola V, Bonaventura GD, Djukić S, Ćirković I, Ruzicka F. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by Staphylococci. Apmis. 2007;115:891–899. doi: 10.1111/j.1600-0463.2007.apm_630.x. [DOI] [PubMed] [Google Scholar]
- 13.Rodrigues LB, dos Santos LR, Tagliari VZ, Rizzo NN, Trenhago G, de Oilveira AP, Goetz F, do Nascimento VP. Quantification of biofilm production on polystyrene by Listeria, Escherichia coli and Staphylococcus aureus isolated from a poultry slaughterhouse. Braz J Microbiol. 2010;41:1082–1085. doi: 10.1590/S1517-83822010000400029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Little TM, Hills FC. Agricultural Experimentation. Hoboken: Wiley; 1978. [Google Scholar]
- 15.Kelleher SM, Habimana O, Lawler J, O’reilly B, Daniels S, Casey E, Cowley A. Cicada wing surface topography: An investigation into the bactericidal properties of nanostructural features. ACS Appl Mater Interfaces. 2016;8:14966–14974. doi: 10.1021/acsami.5b08309. [DOI] [PubMed] [Google Scholar]
- 16.Abrigo M, Kingshott P, McArthur SL. Electrospun polystyrene fiber diameter influencing bacterial attachment, proliferation, and growth. ACS Appl Mater Interfaces. 2015;7:7644–7652. doi: 10.1021/acsami.5b00453. [DOI] [PubMed] [Google Scholar]
- 17.Oh JK, Yegin Y, Yang F, Zhang M, Li J, Huang S, Taylor TM. The influence of surface chemistry on the kinetics and thermodynamics of bacterial adhesion. Sci Rep. 2018;8:1–13. doi: 10.1038/s41598-018-35343-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ploux L, Ponche A, Anselme K. Bacteria/material interfaces: role of the material and cell wall properties (2010) J Adhes Sci Technol 24:2165 – 201.10.1163/016942410X511079
- 19.Yuan Y, Hays MP, Hardwidge PR, Kim J. Surface characteristics influencing bacterial adhesion to polymeric substrates. RSC Adv. 2017;7:14254–14261. doi: 10.1039/C7RA01571B. [DOI] [Google Scholar]
- 20.Bagherifard S, Hickey DJ, de Luca AC, Malheiro VN, Markaki AE, Guagliano M, Webster TJ. The influence of nanostructured features on bacterial adhesion and bone cell functions on severely shot peened 316L stainless steel. Biomaterials. 2015;73:185–197. doi: 10.1016/j.biomaterials.2015.09.019. [DOI] [PubMed] [Google Scholar]
- 21.Krasowska A, Sigler K. How microorganisms use hydrophobicity and what does this mean for human needs? Front Cell Infect Microbiol. 2014;4:1–7. doi: 10.3389/fcimb.2014.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takashima M, Shirai F, Sageshima M, Ikeda N, Okamoto Y, Dohi Y. Distinctive bacteria-binding property of cloth material. Am J Infect Cont. 2004;32:27–30. doi: 10.1016/j.ajic.2003.05.003. [DOI] [PubMed] [Google Scholar]
- 23.Berne C, Ducret A, Hardy GG, Brun YV (2015) Adhesins involved in attachment to abiotic surfaces by Gram-negative bacteria. Microbial Biofilms, 163–199. 10.1128/9781555817466.ch9 [DOI] [PMC free article] [PubMed]
- 24.Gupta K, Singh SP, Manhar AK, Saikia D, Namsa ND, Konwar BK, Mandal M. Inhibition of Staphylococcus aureus and Pseudomonas aeruginosa biofilm and virulence by active fraction of Syzygium cumini (L.) skeels leaf extract: in-vitro and in silico studies. Ind J Microbiol. 2019;59:13–21. doi: 10.1007/s12088-018-0770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gui Z, Wang H, Ding T, Zhu W, Zhuang X, Chu W. Azithromycin reduces the production of α-hemolysin and biofilm formation in Staphylococcus aureus. Ind J Microbiol. 2014;54:114–117. doi: 10.1007/s12088-013-0438-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martí S, Rodríguez-Baño J, Catel-Ferreira M, Jouenne T, Vila J, Seifert H, Dé E. Biofilm formation at the solid-liquid and air-liquid interfaces by Acinetobacter species. BMC Res Notes. 2011;4:1–5. doi: 10.1186/1756-0500-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma G, Sharma S, Sharma P, Chandola D, Dang S, Gupta S, Gabrani R. Escherichia coli biofilm: development and therapeutic strategies. J Appl Microbiol. 2016;121:309–319. doi: 10.1111/jam.13078. [DOI] [PubMed] [Google Scholar]
- 28.Chagnot C, Zorgani MA, Astruc T, Desvaux M. Proteinaceous determinants of surface colonization in bacteria: bacterial adhesion and biofilm formation from a protein secretion perspective. Front Microbiol. 2013;4:1–26. doi: 10.3389/fmicb.2013.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ploux L, Ponche A, Anselme K. Bacteria/material interfaces: role of the material and cell wall properties. J Adhesi Sci Technol. 2010;24:2165–2201. doi: 10.1163/016942410X511079. [DOI] [Google Scholar]
- 30.Schmidt-Emrich S, Stiefel P, Rupper P, Katzenmeier H, Amberg C, Maniura-Weber K, Ren Q. Rapid assay to assess bacterial adhesion on textiles. Materials. 2016;9:1–13. doi: 10.3390/ma9040249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azam MT, Khan AS, Muzzafar D, Faryal R, Siddiqi SA, Ahmad R, Rehman IU. Structural, surface, in vitro bacterial adhesion and biofilm formation analysis of three dental restorative composites. Materials. 2015;8:3221–3237. doi: 10.3390/ma8063221. [DOI] [Google Scholar]
- 32.Kurmoo Y, Hook AL, Harvey D, Dubern J-F, Williams P, Morgan SP, Korposh S, Alexander MR. Real time monitoring of biofilm formation on coated medical devices for the reduction and interception of bacterial infections. Biomater Sci. 2020;8:1464–1477. doi: 10.1039/C9BM00875F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pericolini E, Colombari B, Ferretti G, Iseppi R, Ardizzoni A, Girardis M, Sala A, Peppoloni S, Blasi E. Real-time monitoring of Pseudomonas aeruginosa biofilm formation on endotracheal tubes in vitro. BMC Microbiol. 2018;18:84. doi: 10.1186/s12866-018-1224-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saxena P, Joshi Y, Rawat K, Bisht R. Biofilms: Architecture, resistance, quorum sensing and control mechanisms. Ind J Microbiol. 2019;59:3–12. doi: 10.1007/s12088-018-0757-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.