Abstract
Inherited retinal diseases (IRDs), defined by dysfunction or progressive loss of photoreceptors, are disorders characterized by elevated heterogeneity, both at the clinical and genetic levels. Our main goal was to address the genetic landscape of IRD in the largest cohort of Spanish patients reported to date. A retrospective hospital-based cross-sectional study was carried out on 6089 IRD affected individuals (from 4403 unrelated families), referred for genetic testing from all the Spanish autonomous communities. Clinical, demographic and familiar data were collected from each patient, including family pedigree, age of appearance of visual symptoms, presence of any systemic findings and geographical origin. Genetic studies were performed to the 3951 families with available DNA using different molecular techniques. Overall, 53.2% (2100/3951) of the studied families were genetically characterized, and 1549 different likely causative variants in 142 genes were identified. The most common phenotype encountered is retinitis pigmentosa (RP) (55.6% of families, 2447/4403). The most recurrently mutated genes were PRPH2, ABCA4 and RS1 in autosomal dominant (AD), autosomal recessive (AR) and X-linked (XL) NON-RP cases, respectively; RHO, USH2A and RPGR in AD, AR and XL for non-syndromic RP; and USH2A and MYO7A in syndromic IRD. Pathogenic variants c.3386G > T (p.Arg1129Leu) in ABCA4 and c.2276G > T (p.Cys759Phe) in USH2A were the most frequent variants identified. Our study provides the general landscape for IRD in Spain, reporting the largest cohort ever presented. Our results have important implications for genetic diagnosis, counselling and new therapeutic strategies to both the Spanish population and other related populations.
Subject terms: Molecular medicine, Genetics research, Clinical genetics, Hereditary eye disease
Introduction
Inherited retinal diseases (IRDs) are one of the most heterogeneous clinical and genetical disorders known among all human medical conditions, characterized by the progressive loss of photoreceptor cells, resulting in severe visual impairment1. IRDs are classified as rare diseases, and their estimated prevalence is about 1 in 10002–40001. IRDs can be classified according to different clinical or genetic criteria, based upon the primary retinal cell affected (rods, cones, retinal pigment epithelium (RPE), bipolar cells or ganglion cells), the ophthalmological findings and/or the affected gene found after the genetic testing.
All modes of inheritance can be observed (autosomal dominant (AD), autosomal recessive (AR), X-linked (XL), including rare non-Mendelian forms such as mitochondrial or digenic inheritance patterns). Age of onset of first symptoms (from early childhood to adulthood), rate of progression, association with extra-ocular symptoms (non-syndromic versus syndromic forms) or causative gene can also help to subclassify the different phenotypes3. The most prevailing form of IRDs is Retinitis Pigmentosa (RP [MIM: 268000]), which is estimated to affect approximately 1.5 million people worldwide4. RP begins with the degeneration of rod photoreceptors, resulting in night blindness and characteristic pigmentary changes in the peripheral retina. This is also considered a rod-cone dystrophy due to the subsequent cone photoreceptor death in later stages. Other forms of IRD, including cone-dominated diseases, are characterized by photophobia, reduced visual acuity and impaired colour vision (i.e. cone dystrophy (CD), achromatopsia, blue cone monochromatism), as well as generalized retinal degeneration involving simultaneously both cones and rods such as in rod-cone or cone-rod dystrophies (CRD). The most severe form of non-syndromic IRDs is Leber congenital amaurosis (LCA) characterized by congenital or early childhood blindness. Other IRD forms are characterized by central vision loss affecting primarily the macula and are therefore acknowledged as Macular Dystrophies (MD), such as Stargardt disease (STGD1 [MIM: 248200]) and Best Vitelliform Macular Dystrophy (VMD2 [MIM: 153700])1.
Conversely, syndromic IRDs are subclassified according to the type of syndrome. The most prevalent is Usher syndrome (sensorineural hearing loss and RP) which can be further subclassified to Usher type I (USH1 [MIM: 276900]), type II (USH2 [MIM: 276901]) and type III (USH3 [MIM: 276902]), and other syndromes such as Bardet-Biedl (BBS [MIM: 209900]) or Alström (ALMS [MIM: 203800])3.
Since the identification of rhodopsin (RHO [MIM: 180380]) in 1990 as the first gene involved in the development of AD-RP5, 270 genes have been additionally described as causative of IRDs (RetNet, Retinal Information Network; https://sph.uth.edu/retnet/; accessed on March 2020). Some of these genes have been reported in only few families worldwide; therefore, their individual contribution to IRD prevalence is relatively small. The most prevalent IRD-causing genes across all populations are ABCA4 (MIM: 601691), RHO, USH2A (MIM: 608400) and RPGR (MIM: 312610), which account for high percentages of some of the IRD subtypes, i.e. 70–71% in STGD1/AR-MD/AR-CRD6, 19–25% of AD-RP, 10% of AR-RP, and 70% of XL-RP, respectively1. In addition, although the majority of the causative variants are private, some are more frequent, especially in Spanish families, such as USH2A (GenBank: NM_206933.3) c.2299delG (p.Glu767SerfsTer21) and c.2276G > T (p.Cys759Phe)7 or ABCA4 (GenBank: NM_000350.3) c.3386G > T (p.Arg1129Leu) and c.5882G > A (p.Gly1961Glu)6.
The aim of this study is to present a comprehensive overview of the largest cohort of IRD patients ever reported worldwide and related to IRD in the Spanish population. The presented data includes the presumed inheritance pattern for the different phenotypic subtypes, mutational spectrum, prevalence of genes carrying likely pathogenic variants and the recurrence of disease related variants.
Results
Prevalence of IRD in Spain
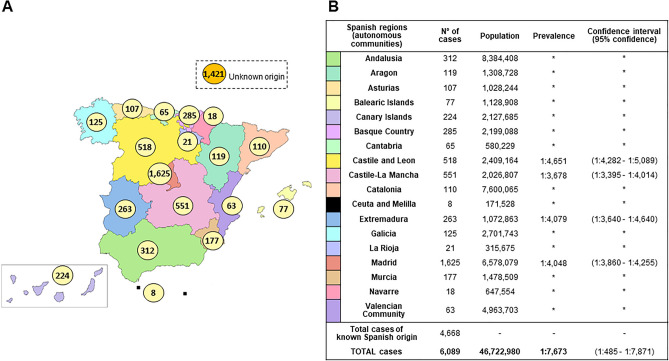
The number of cases diagnosed as having IRD in our hospital (until August 2019) was 6089, and the last Spanish population registry accounted for 46,722,980 habitants giving us a minimal prevalence of 1:7673 (confidence interval (CI):1:7485–1:7871). Regional distribution of cases and prevalence can be seen in Fig. 1A,B. Considering a worldwide IRD prevalence of 1:10002–40001, our cohort would represent 20–53% of the total patients with IRD in Spain as shown in Supplementary Table S1.
Figure 1.
IRD affected cases distribution and estimated prevalence in Spain. (A) Distribution of RP/NON-RP affected cases across Spain. Total cases: 6089 (known Spanish origin: 4668; unknown origin: 1421). Spanish map modified using image editor from https://www.veomapas.com/mapa-mudo-de-las-comunidades-autonomas-de-espana-m103.html. (B) IRD estimated prevalence in Spanish regions. Data was obtained from the number of cases we had in our Hospital Service and the recorded population in the different regions. *Inconclusive data.
For non-syndromic IRD, our cohort was grouped in 2 categories: 1703 affected individuals (1335 families), in cone-dominated phenotypes—hereafter “NON-RP”—and 3561 affected cases from 2447 unrelated families within primarily rod affection—hereafter “RP”. This resulted in a minimal prevalence of 1:27,436 (CI:1:26,192–1:28,804) and 1:13,121 (CI:12,704–13,566) for NON-RP and RP, respectively. Additionally, syndromic IRD forms represented the smallest fraction accounting for 13.6% of the patients with 825 affected individuals (621 families), resulting in a minimal prevalence of 1:56,634 (CI:1:53,016–1:60,781).
In our cohort, we have a higher proportion of cases from Madrid area (26.7%; 1625/6089).
Initial classification of IRD families by clinical type and suspected mode of inheritance prior to genetic testing
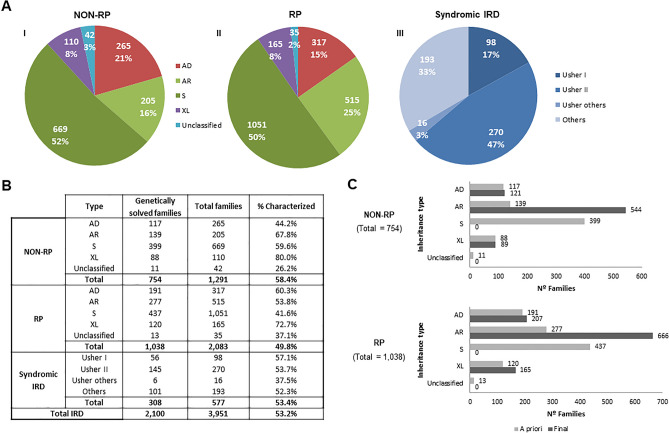
Non-syndromic NON-RP and RP cases were categorized by the mode of inheritance (Fig. 2A-I and A-II). Syndromic IRD were categorized by the specific type of suspected syndrome (Fig. 2A-III), instead of inheritance type, given that most of them were sporadic (53.6%) or had recessive inheritance (40.2%). The remaining 6.2% corresponded to dominant (0.5%), X-linked (0.7%), mitochondrial inherited disease (0.2%) or non-classificable cases (4.8%).
Figure 2.
“A priori” and final classification of IRD affected cases. (A) “A priori” classification of IRD families with data obtained from the clinical and family history of the patients, before performing molecular tests: NON-RP (I), RP (II), and syndromic IRD (III), and subclassification according to the inheritance type in case of RP and NON-RP, and type of syndrome in case of syndromic IRD. Data obtained with the clinical and familiar history of the patients, before performing molecular tests. AD Autosomal Dominant, AR autosomal recessive, S sporadic, XL X-linked. (B) Proportion of genetically solved NON-RP, RP, syndromic IRD and total IRD. The diagnostic ratio in the different group of NON-RP and RP by the type of “a priori” inheritance and in the different group of syndromic IRD by type of syndrome is indicated. (C) Genetically solved families. Comparison of inheritance classification before (light gray) and after (dark gray) the molecular study was performed.
According to this “a priori” diagnosis based on the clinical and familial history of the patients, the main inheritance pattern in non-syndromic NON-RP and RP was recessive or sporadic, representing the 68% and 75% of cases, respectively. Autosomal dominant and X-linked forms accounted for 21% and 8% for NON-RP and 15% and 8% for RP, respectively (Fig. 2A-I). Families with no familiar data were annotated as unclassified. Non-syndromic RP represents the most common phenotype, representing 55.6% of families in our cohort.
In the present cohort, 47% of the syndromic IRD index cases (270/577) suffered from USH2, followed by 17% USH1 (98/577), as well as other very rare syndromes like some atypical forms of Usher syndrome (3%; 16/577) and ciliopathies such as BBS or ALMS (16%; 90/577). A miscellanea of non-ciliopathic syndromes or unclassified symptoms were presented in 103 index cases.
Molecular studies
Diagnostic yield
Genetic testing was performed in a total of 3951 index cases with available DNA6–11 (89.7% of the total cohort), including 1291 NON-RP, 2083 RP, and 577 syndromic IRD patients as shown in Supplementary Fig. S1. The genetic analysis procedures evolved over time since novel genetic approaches were implemented in our laboratory. A definite genetic diagnosis was established for 53.2% (2100/3951) of cases. For NON-RP families, 754 out of 1291 (58.4%) obtained a genetic diagnosis; in RP the molecular cause was identified in 1038 out of 2083 (49.8%). Similarly, 53.4% (308/577) of syndromic IRD families were genetically solved (Fig. 2B).
Final inheritance pattern and reclassification of characterized families
The identification of the causative gene allowed us to reclassify the inheritance mode in 8.2% (146/1792) of the NON-RP and RP families, thus establishing the final inheritance type (Supplementary Table S2). A comparison between the “a priori” suspected inheritance based on the pedigree and the final inheritance suggested by the molecular diagnosis was performed in characterized NON-RP and RP families (Fig. 2C). As expected, most sporadic NON-RP (n = 378) and RP cases (n = 379) were confirmed as having AR inheritance after the genetic testing. The rest of the S cases were reclassified to AD (n = 43) and XL (n = 36) (Supplementary Table S2). Twenty-four cases (11 NON-RP and 13 RP) with an initial unknown mode of inheritance were classified as: AD (n = 5), AR (n = 16), and XL (n = 3) after the molecular testing.
Gene landscape
In total, 1549 different pathogenic and likely pathogenic variants were identified in 142 different genes. These included SNVs (Single Nucleotide Variants) and CNVs (Copy Number Variants). As showed in Figs. 3, 4, 5, there was a wide spectrum of genes implicated in IRD, 121 of them represented in 1% or less of the cohort. The 5 most frequent mutated genes were ABCA4, USH2A, RS1 (MIM: 300839), CRB1 (MIM: 604210) and RHO.
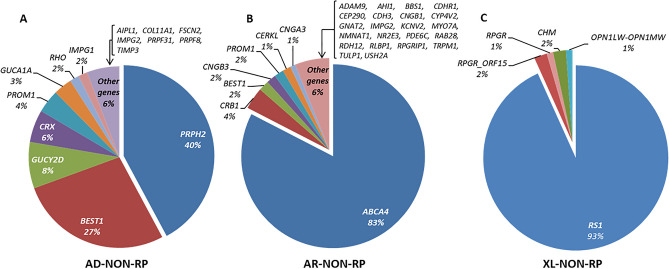
Figure 3.
Classification of mutated genes in the genetically solved NON-RP affected cases. Below each gene is given the percentage that each gene was mutated in the cohort. Total characterized families: AD-NON-RP: 121; AR-NON-RP: 544; XL-NON-RP: 89.
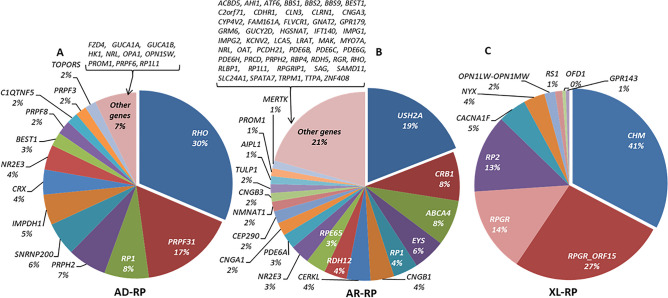
Figure 4.
Classification of mutated genes in the genetically solved RP affected cases. Below each gene is given the percentage that each gene was mutated in the cohort. Total characterized families: AD-RP: 207; AR-RP: 666; XL-RP: 165.
Figure 5.
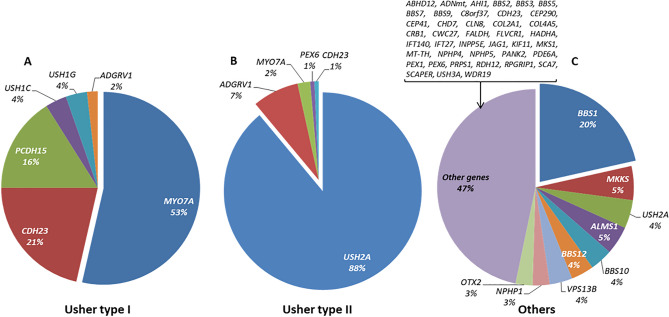
Classification of mutated genes in the genetically solved syndromic IRD affected cases. Below each gene is given the percentage that each gene was mutated in the cohort. Total characterized families: Usher I: 56; Usher II: 145; Others (including atypical Usher): 107.
In NON-RP families, PRPH2 (MIM: 179605), ABCA4 and RS1 were the most commonly mutated genes, explaining 42.2%, 82.7% and 93.3% of the AD, AR and XL forms, respectively as shown in Supplementary Table S3.
For non-syndromic RP, 207 AD-RP families were genetically characterized with heterozygous variants identified in one of 23 genes found. The most frequent mutated genes were RHO (65/207; 31.4%) and PRPF31 (MIM: 606419) (34/207; 16.4%). In AR-RP families, the most common gene mutated was USH2A (127/666; 19.1%). However, the number of other disease genes detected was very high (N = 70) in the 666 families characterized. For XL-RP families, both the number of cases and variety of genes were low (N = 9) as shown in Supplementary Table S4. For these patients, 41.2% (68/165) of the index cases (all males) carried a hemizygous pathogenic variant in RPGR (44 RPGR_ORF15 and 24 in the rest of RPGR regions), and 32.7% (54/165) in CHM (MIM: 300390).
A total of 53 cases out of the total 2100 (2.5%) were clinically reconsidered and reclassified after genetic testing: 12 NON-RP were reclassified as RP (9/754; 1.2%; characterized with RHO, FSCN2, PRPF8, AHI1, CNGB1, TRPM1 and CHM) or as syndromic IRD (3/754; 0.4%; COL11A1, CDH3 and MYO7A) and 37 RP as NON-RP (32/1038; 3.1%; C1QTNF5, GUCA1A, CNGB3, CNGA3, ACBD5, GNAT2, PDE6C, ATF6, PDE6H, RS1 and OPN1LW-OPN1MW), syndromic IRD (1/1038; 0.1%; MYO7A) and other visual diseases, such as exudative familiar vitreoretinopathy, optic atrophy, albinism and gyrate atrophy (4/1038; 0.4%; FZD4, OPA1, GPR143 and OAT). Moreover, 4 syndromic cases were reclassified to RP group (4/308; 1.3%; RDH12, PDE6A and RPGRIP1).
Among syndromic IRD families, the causative gene was identified in 56 families with a diagnosis of USH1, 145 of USH2, 6 of atypical Usher, and 101 of other syndromes including Bardet-Biedl or Alström syndrome, respectively. In USH1, biallelic variants in MYO7A (MIM: 276903) were identified in 30 out of 56 patients (53.5%). USH2A defects were the main cause of USH2 in 90% (129/145) of the patients. The group “others” was a clinically heterogeneous group of non-Usher cases with a total of 48 involved genes, with BBS1 (MIM: 209901) being the most frequent one (N = 23), as shown in Supplementary Table S5.
In addition to clinical and genetic heterogeneity, the group of “others” included in syndromic IRD also presented unusual modes of inheritance, such as triallelism. In our cohort, 4 possible triallelic cases have been identified, all of them diagnosed with BBS, 3 patients carrying biallelic BBS1 variants together with one allele in MKKS (MIM: 604896)9,12 and one additional case carrying biallelic MKKS variants and one allele in BBS5 (MIM: 603650). The phenotypic modifier effect of the triallelism could only be stablished in two of the 4 families. One has two affected siblings with different phenotypic severity that correlates with the presence of the third allele; and in the other both affected showed triallelism and have the same clinical manifestation. Although, it could not be stablished in the rest of the families, as they were sporadic cases.
Most frequent variants
Our findings reflect the high allelic heterogeneity in IRD. We identified 458 different disease-causing variants in 45 genes in cases “a priori” classified as NON-RP, as well as 836 in 94 genes in the “a priori” RP cases and 295 in 55 genes in the “a priori” syndromic IRD. The most common pathogenic variant detected in our NON-RP cohort was the previously known missense change ABCA4 c.3386G > T (p.Arg1129Leu) (180 mutated alleles of 3,618; 5% of the total pathogenic alleles), presented in 21.5% (162/754) of the characterized families, in homozygous or compound heterozygous state in 18 and 144 families, respectively (Supplementary Table S6). Among the RP families, the most prevalent pathogenic variant was the missense change USH2A c.2276G > T (p.Cys759Phe), identified in 106 alleles in 8.4% of the solved families (87/1038); in 19 cases in homozygous and in 68 cases in compound heterozygous state. In addition, there were 15 other variants present in more than 10 genes (Supplementary Table S6). Some of the mutated genes overlap in NON-RP (Fig. 3), RP (Fig. 4) and syndromic IRD (Fig. 5).
Disease-causing variant distribution in Spain
Analysis of causing variants by the different Spanish regions resulted in a wide variety of disease-causing variants. Table 1 shows variants detected in more than 5% of characterized families, by Spanish regions. All these variants are depicted in the Supplementary Table S6 of NON-RP and RP most frequent causing variants, except for the nonsense variant PRCD (OMIM: 610598) (GenBank: NM_001077620.3) c.64C > T (p.Arg22Ter), that was found in homozygosity in 3 families, representing 7.2% (6/83) of the identified alleles from Murcia.
Table 1.
Variants detected in > 5% of NON-RP, RP and syndromic IRD by Spanish regions.
| Spanish region | Gene | Nucleotide change | Amino acid change | Nº of alleles | Nº of total alleles of this gene found in the region | Frequency in the region | |
|---|---|---|---|---|---|---|---|
| NON-RP | Basque Country | ABCA4 | c.3386G > T | p.Arg1129Leu | 18 | 93 | 19.40% |
| ABCA4 | c.5882G > A | p.Gly1961Glu | 5 | 5.40% | |||
| Castile-La Mancha | ABCA4 | c.3386G > T | p.Arg1129Leu | 8 | 60 | 13.30% | |
| Madrid | ABCA4 | c.3386G > T | p.Arg1129Leu | 47 | 373 | 12.60% | |
| Murcia | ABCA4 | c.3386G > T | p.Arg1129Leu | 8 | 71 | 11.30% | |
| ABCA4 | c.5882G > A | p.Gly1961Glu | 7 | 9.90% | |||
| Non-syndromic RP and syndromic IRD | Murcia | USH2A | c.2276G > T | p.Cys759Phe | 11 | 83 | 13.30% |
| PRCD | c.64C > T | p.Arg22Ter | 6 | 7.20% | |||
| Canary Islands | NR2E3 | c.932G > A | p.Arg311Gln | 8 | 83 | 9.60% | |
| Castile-La Mancha | USH2A | c.2276G > T | p.Cys759Phe | 18 | 202 | 8.90% | |
| Madrid | USH2A | c.2276G > T | p.Cys759Phe | 50 | 714 | 7.00% | |
| Extremadura | USH2A | c.2276G > T | p.Cys759Phe | 5 | 87 | 5.70% |
Regions with variants detected below this percentage are not represented in the table, and neither are regions with less than 50 mutated alleles in total or variants that were presented less than 5 times. In bold the variant which not appeared as one of the most frequent variants in Supplementary Table S6.
Discussion
This is the first and largest comprehensive study addressing the prevalence and epidemiology of IRD in the Spanish population. The cohort here described, comprising 6089 cases from 4403 unrelated families, is not based on a national registry of IRD patients, but it is the outcome of a very wide recruiting effort of a single center over the last 28 years. An increasing number of centers are currently performing clinical and/or genetic diagnosis of IRD in Spain, therefore our cohort did not reflect all of IRD patients in our country. Hence, to date, no accurate data about the IRD prevalence in the Spanish population is available.
In terms of representation of patients from the different Spanish regions, our cohort reflects a biased recruitment, being enriched with patients from Madrid and the surrounding regions (i.e. Castile and Leon, Castile-La Mancha, and Extremadura) probably due to the fact that our hospital has been their referral center during most of the time of the study. Other areas like Andalusia, Catalonia, Navarre or the Valencian Community had different referral centers and genetic testing is performed locally. In spite of these limitations, the large sample size of our cohort and the exhaustive molecular analysis performed over the years, together with an overall low genetic heterogeneity in the Spanish population13, have allowed a straightforward extrapolation of prevalent genes and/or variants in IRD. Considering a worldwide prevalence of 1:10002–40001 and an estimated Spanish population of 46.7 million, our cohort would represent 20–53% of the total patients with IRD in Spain. Despite numerous studies about the characteristics of the different IRDs in Spain, such as NON-RP and RP have been partially published, still no global overview of NON-RP and RP diseases using a representative cohort has been addressed yet before in our country.
Several studies on IRD have been performed globally (Supplementary Table S7) and, in the two last years, some including big cohorts14–16 or meta-analysis17 have been published, reporting more than 125 genes explaining 55–62% of the families using several molecular techniques to achieve that14–16 as it could be also seen in this study.
Other studies focused on stablishing the prevalence of IRD in certain regions has been performed in Western countries and in cohorts of non-syndromic RP, including Western Australia (1:6000)18 or Maine (1:4756)19, as well as in cohorts with general IRD and an estimated prevalence of 1:3454 in Denmark20 or 1:3856 in Norway21. However, this prevalence has been reported in areas and populations with low rates of consanguinity and could be higher when consanguinity rate increases2, which is not the common scenario in Spain nowadays.
Our study identified AR inheritance as the most common mode of inheritance for non-syndromic IRD, explaining up to 70–75% NON-RP and RP subcohorts (Fig. 2A-I and A-II). By contrast, only 7% of our NON-RP and RP families are explained by X-linked genes. These results were consistent with previous studies published18–22. Besides, some cases could be explained by different molecular mechanisms as the pseudodominance, incomplete penetrance or the presence of two variants in an AR gene in AD a priori families8, so extended segregation analysis within these families are needed. Additional non-Mendelian transmission patterns were only found in exceptionally rare cases with syndromic IRD, including 3 families carrying variants affecting the mitochondrial DNA and 4 cases with apparent triallelism in BBS-associated genes. Within syndromic IRD group, most of the cases were explained by AR biallelic monogenic inheritance. Similar to previous published studies from other countries3,23, Usher syndrome was the most prevalent form of syndromic IRD in our cohort, and more specifically, USH2, representing almost half of the total syndromic IRD families.
The overall diagnostic rate of 53.2% obtained here is similar to other studies previously reported (50–70%)24–26. Molecular studies allowed the identification of the genes responsible for the disease and the reclassification of the inheritance type. In our work, 8.2% of the patients were reclassified after the detection of the disease-causing variants in genes with a specific inheritance pattern. All of those were or have been previously validated. Moreover, in all the characterized sporadic cases a more accurate genetic classification and counselling could be done8. Additionally, a 2.5% were clinically reclassified after the genetic testing, due to a poor clinical data acquisition at the origin center. So, identification of the genetic cause of the disease represents a hallmark for the patients, firstly, regarding genetic counselling and the risk of affectation for other relatives; and secondly, given the possibility of future recruitments for clinical trials targeting specific genes and variants.
A total of 142 different genes were identified as the cause of IRD in our study, but it is important to notice that each subgroup of the cohort (AD, AR and XL NON-RP and RP) has an enrichment of characterized cases in specific genes.
For instance, PRPH2 was mutated in more than a third of AD-NON-RP families, followed by BEST1 (MIM: 607854). As expected, ABCA4 was the most prevalent gene in AR-NON-RP families. Recent studies in Norway21 and Korea22 also identified this gene as one of the most prevalent mutated genes. A study published by Birtel et al.27 in patients with MD and cone/cone-rod dystrophy showed a similar distribution of mutated genes, with ABCA4, PRPH2 and BEST1 responsible for 74% of their solved cases. For the XL-NON-RP subcohort, RS1 was the most frequently mutated gene.
Non-syndromic RP presented a wider spectrum of causative genes, with RHO, USH2A and RPGR (RPGR_ORF15 and the rest of RPGR regions) being the most prevalent ones in AD-RP, AR-RP and XL-RP subcohorts, respectively. Our findings are in line with those published in other studies8,28. For instance, Hartong et al.3, showed as well MYO7A, USH2A and BBS1 to be the most frequently mutated genes in USH1, USH2 and BBS, respectively. Other studies in different populations highlighted different genes as the most representative in their IRD cohorts. For example, Eisenberger et al.24 described RP1 (MIM: 603937) (11.3%) and EYS (MIM: 612424) (9.4%) as the most frequent genes in German patients with AR-RP, and Kim et al.22 detected that EYS (22%) and PDE6B (MIM: 180072) (17%) are most frequently involved in AR-RP in Korean patients. EYS was also the most prevalent causative gene in the Japanese population studied by Maeda et al.29, implicated in 21 out of 33 AR-RP patients (63.6%), whereas in our population EYS was mutated in 5.5% of the families with “a priori” AR-RP diagnosis, being the fourth most frequent gene, after USH2A, CRB1 and ABCA4. However, the order of the causative genes in AR-RP changes after reviewing the clinical data of ABCA4 related IRD patients, since they were mostly reclassified as NON-RP, downgrading EYS as the third most common gene in AR-RP in our population. This result supports an eastward gradient in the frequency of EYS variants in patients throughout the world and within Europe, being more frequent in Germany than in Spain.
The most frequent causing variants detected in our study appeared, as expected, in ABCA4 and USH2A, the most prevalent mutated genes in the Spanish population6–8,30. ABCA4 c.3386G > T (p.Arg1129Leu) is a variant almost exclusively found in Spanish NON-RP patients6,30, being probably a Spanish founder mutation31,32. However, USH2A c.2276G > T (p.Cys759Phe) is not exclusive from the Spanish population and has been reported in other populations33.
According to the geographical distribution of the variants within the country, no differences between regions were observed. In NON-RP, the two most common ABCA4 variants were also the most represented in regions with variant frequencies above 5%. Meanwhile, in RP we found a higher representation of the most common USH2A variant, which appeared above 5% of the total alleles in four regions. Finally, two variants appeared to be more frequent in some regions, i.e. PRCD c.64C > T (p.Arg22Ter) in Murcia and NR2E3 (MIM: 604485) (GenBank: NM_014249.4) c.932G > A (p.Arg311Gln) in the Canary Islands, where a founder effect could be happening.
Our results delineate the genetic background of the Spanish IRD patients, indicating a wide range of causative genes involved in the disease. Some of the causing variants identified are also frequent in Europe. Some examples include the ABCA4 c.5882G > A (p.Gly1961Glu), reported with high prevalence in the Italian, German and Spanish populations30,34,35; the c.2276G > T (p.Cys759Phe) as one of the most frequent variants in USH2A, especially in European countries36, USH2A c.2299delG (p.Glu767SerfsTer21), which is possibly an ancestral European pathogenic variant37; BBS1 (GenBank: NM_024649.5) c.1169T > G (p.Met390Arg), identified previously by Mykytyn et al.38 in 22 North American BBS probands with North European ancestry; and CRB1 (GenBank: NM_201253.3) c.2843G > A (p.Cys948Tyr), identified repeatedly in different European countries39,40. Finally, RHO (GenBank: NM_000539.3) c.1040C > T (p.Pro347Leu), described in the Italian and French populations41,42 and also in non-European cohorts5,43,44.
Variants found in individuals from East Mediterranean and Middle-Eastern countries also appeared in our Spanish cohort: PRCD c.64C > T (p.Arg22Ter) found by Sharon et al.14 in homozygosis in 15 Israeli Muslim Arab families, and by Beheshtian et al.45 in a Persian family; and FAM161A (MIM: 613596) (GenBank: NM_001201543.2) c.1355_1356delCA (p.Thr452SerfsTer3), which was identified in Jewish families mainly originating from North African countries46. As mentioned above, the variant in PRCD was found with a higher frequency in the region of Murcia, and this could be due to the settlement of Muslim populations during several centuries during the Middle Ages13. FAM161A does not have a significant specific geographical distribution in Spain.
Remarkably, we identified three pathogenic variants with high frequency in Spain: ABCA4 c.3386G > T (p.Arg1129Leu), previously mentioned; CERKL (MIM: 608381) (GenBank: NM_201548.5) c.847C > T (p.Leu283Phe), first described by Tuson et al.47, and RP1 c.1625C > G (p.Ser542Ter) previously described originally as a Spanish founder pathogenic variant. These three variants had been scarcely reported outside the Spanish population. In the case of RP1 c.1625C > G (p.Ser542Ter) variant48, because of its presence in 11 out of 244 unrelated families, we can extrapolate that it may very well account for approximately 4.5% of all AR-RP cases in the Spanish population. Other groups also identified this variant in Swiss patients26.
In conclusion, this study shows the general landscape of the genetic underpinnings of IRD in Spain and will help design clinical and preventive healthcare approaches to this disorder in our country.
Materials and methods
Cohort description
A retrospective analysis was performed including all IRD patients from our Spanish registry at the Fundación Jiménez Díaz University Hospital (FJD, Madrid, Spain) from 1991 until August 2019. This patient registry includes: all patients referred to the Genetic Service at the FJD for genetic diagnostic testing and/or counselling due to a previous clinical suspicion of IRD, and patients without genetic analysis in our unit but identified in the shared electronic clinical history of our same-company hospitals using ICD (International Classification of Diseases) terms. The complete cohort contains 6089 IRD affected cases (including index cases and affected relatives) belonging to 4403 unrelated families as shown in Supplementary Fig. S1.
This study was approved by the Ethics Committee of the FJD under approval number 134/2016_FJD and fulfilled all the tenets of the Declaration of Helsinki and its further reviews. A written informed consent form was obtained from all the patients or their legal guardians.
IRD classification and diagnostic criteria
During this study, different clinical, demographic and familiar data were collected, including (i) family pedigree; (ii) age at onset of visual acuity loss, extent of visual field loss, night blindness and/or other early symptoms of retinal dystrophy; (iii) presence of any systemic findings suggestive of syndromic forms of IRD; (iv) geographical origin of the patients.
Clinical diagnosis was based on ophthalmic examination, including measurement of best-corrected visual acuity, visual field testing, fundus examination and, if possible, full-field electroretinography, fundus autofluorescence and spectral domain optical coherence tomography scan. NON-RP and RP include non-syndromic IRD, and their clinical classification was done according to previously described criteria6,8. NON-RP group include most patients with CD, CRD and achromatopsia, although some of them were included in the RP group due to incomplete phenotyping at the moment of the diagnosis. Non-syndromic LCA cases were also included in RP.
For NON-RP and RP families, an “a priori” inheritance pattern (AD/AR/XL/sporadic (S)) was established according to previously described criteria1. The subgroup of XL-RP also included choroideremia cases.
For cases not extensively described in the first referral, a generic classification was made as NON-RP or RP.
Criteria for syndromic IRD diagnosis were previously described7,9.
Information about the geographic origin of all the IRD cases from Spain was available in 4668. They are distributed throughout the 17 different Spanish communities (Fig. 1A).
Inheritance reclassification of IRD cases
After molecular diagnosis, inherited patterns were reviewed and compared with “a priori” data of each family. Statistical analysis between these data sets to assess the global association in the NON-RP group was made using the Fisher’s exact test with a p equal to 0.497. Whereas for the RP group, Chi-square test was used with a p below 0.001. Comparisons for each type of inheritance have also been made with the Fisher’s exact test in the NON-RP Unclassified subgroup and with the Chi-square test in the rest. Fisher’s exact test was used in those cases in which more than 20% of expected values were below than 5, or at least one of the expected frequencies was below 1. Regarding the significance levels chosen, we have a global comparison for which the significance level is the usual threshold of 0.05 and p-value is not corrected, and several post-hoc comparisons for which Bonferroni’s multiple comparisons adjustment is applied, multiplying the p-values by the number of comparisons.
Prevalence of IRD
In this study, we performed a retrospective analysis of the largest cohort of patients with IRD from Spain, whom were recruited during a period of 28 years by a single center, the FJD. The FJD is a center of reference for molecular diagnosis of IRD from all over the country, especially in some specific autonomous communities, like Castile and Leon, Castile-La Mancha, Extremadura or Madrid. On the other hand, as we take into account other Spanish regions, there are other centers of reference and the prevalence data obtained is inconclusive.
Prevalence was calculated for each clinical type of the disease by dividing the total number of diagnosed IRD cases by the total population in Spain, published by the Spanish Statistical Office (INE; http://www.ine.es) in January 2019.
Genomic screening
Genomic DNA samples were obtained from the FJD Biobank from a total of 3951 families (89.7%), including 1291 NON-RP, 2083 non-syndromic RP and 577 syndromic IRD families. Molecular studies were performed using different molecular techniques as shown in Supplementary Table S8. According to the technology available and the knowledge on the genetic determinants of IRD at the time of the diagnosis, a maximum of 291 different genes involved in IRD were processed for the molecular characterization (Supplementary S1 Appendix). In these studies, index cases were initially screened, analysed following the American College of Medical Genetics and Genomics (ACMG; https://www.acmg.net/docs/standards_guidelines_for_the_interpretation_of_sequence_variants.pdf) variants classification guidelines. If potentially disease-causing variants were found, segregation analysis was performed when DNA samples from relatives were available.
In the general description of mutated genes and frequent pathogenic variants, only fully molecularly characterized index cases were considered. Patients with a heterozygote allele in a recessive gene were counted as uncharacterized.
The frequency of recurrent IRD causing variants was established considering not only the total Spanish population, but also the different geographical regions of Spain (Fig. 1A), in order to assess the possibility of identifying any endemic or founder effects. Pathogenic variants with a prevalence above 5% in a particular region were recorded, and only those with a higher prevalence were considered for further analysis.
Supplementary Information
Acknowledgements
The authors would like to thank Ignacio Mahillo for the statistical support, and all patients who participated in the study.
Author contributions
Conceptualization, I.P.R., G.G., I.F.I., M.P.V., B.A., F.B.K., E.C., B.J.R., I.M.M., R.R.A., B.G.S., P.M., A.A.F., M.C. and C.A.; Data curation, I.P.R., G.G., I.F.I., M.P.V., L.O., L.P.A., C.L. and C.A.; Formal analysis, I.P.R., G.G., I.F.I., M.P.V., B.A., F.B.K., A.B.A., R.C.M., E.C., R.F.S., J.G.M., I.G.V., A.G.P., L.H.B., F.I.B., B.J.R., E.L., M.A.L.M., R.L.R., I.L.S., I.M.M., A.M.R., L.O., L.P.A., R.R.A., M.R.A., E.R.P., C.S.J., S.T.S., M.J.T.T., C.V.M., C.V., O.Z., D.A.G., A.A., R.P.C., I.S.N., R.A., S.B., F.P.M.C., R.W.J.C., E.D.B., H.H., S.K., C.R., D.S., G.A., M.B., M.C., J.M.M., D.V., B.G.S., P.M., A.A.F., M.C. and C.A.; Funding acquisition, C.A.; Investigation, I.P.R., G.G., I.F.I., M.P.V., B.A., F.B.K., A.B.A., R.C.M., E.C., R.F.S., J.G.M., I.G.V., A.G.P., L.H.B., F.I.B., B.J.R., E.L., M.A.L.M., R.L.R., I.L.S., I.M.M., A.M.R., L.O., L.P.A., R.R.A., M.R.A., E.R.P., C.S.J., S.T.S., M.J.T.T., C.V.M., C.V., O.Z., D.A.G., J.A.L., A.A., D.C., P.F.S.J., L.G.M., M.G.H., C.L., M.I.L.M., R.P.C., L.R.J.D.S., C.R., R.S.A., I.S.N., S.D.T., E.V., R.A., S.B., F.P.M.C., R.W.J.C., E.D.B., H.H., S.K., C.R., D.S., G.A., M.B., M.C., J.M.M., D.V., B.G.S., P.M., A.A.F., M.C. and C.A.; Methodology, I.P.R., G.G., I.F.I., M.P.V., D.A.G., J.A.L., A.A., D.C., P.F.S.J., L.G.M., M.G.H., C.L., M.I.L.M., R.P.C., L.R.J.D.S., C.R., R.S.A., I.S.N., S.D.T., E.V., E.A., M.J.G., G.G.G., I.H., T.J., R.A., S.B., F.P.M.C., R.W.J.C., E.D.B., H.H., S.K., C.R., D.S., G.A., M.B., M.C., J.M.M., D.V., B.G.S., P.M., A.A.F., M.C. and C.A.; Project administration, C.A.; Resources, F.B.K., E.C., B.J.R., I.L.S., E.R.P., S.T.S., E.A., M.C.A.C., M.J.B.M., S.B., C.B.L., S.B., J.C.M., C.C., C.C.P, M.F.B., A.F.R., E.G.G., M.J.G., M.G.B., L.M.G.C., G.G.G., B.G., B.G.F., N.G., E.G.N., I.H., I.H.A., C.I., S.I.A., T.J., I.L.R., M.A.L.A., V.L.G., F.L.G., L.M., P.M.P., M.M.I., R.O.I., F.P.M., G.P.N., F.J.R.F., R.R.L., M.R.P., L.R.P., B.R.S., J.R., N.R., R.S.V., A.S., I.V.P., E.V.D., R.A., S.B., F.P.M.C., R.W.J.C., E.D.B., H.H., S.K., C.R., D.S., G.A., M.B., M.C., J.M.M., D.V., B.G.S., P.M. and C.A.; Software, I.F.I., B.A., R.A., S.A., F.P.M.C., R.W.J.C., E.D.B., H.H., S.K., C.R., D.S. and P.M.; Supervision, M.C. and C.A.; Validation, I.P.R., G.G., I.F.I., M.P.V., B.A., F.B.K., A.B.A., R.C.M., E.C., R.F.S., J.G.M., I.G.V., A.G.P., L.H.B., F.I.B., B.J.R., E.L., M.A.L.M., R.L.R., I.L.S., I.M.M., A.M.R., L.O., L.P.A., R.R.A., M.R.A., E.R.P., C.S.J., S.T.S., M.J.T.T., C.V.M., C.V., O.Z., D.A.G., J.A.L., A.A., D.C., P.F.S.J., L.G.M., M.G.H., C.L., M.I.L.M., R.P.C., L.R.J.D.S., C.R., R.S.A., I.S.N., S.D.T., E.V., E.A., M.C.A.C., M.J.B.M., S.B., C.B.L., S.B., J.C.M., C.C., C.C.P., M.F.B., A.F.R., E.G.G., M.J.G., M.G.B., L.M.G.C., G.G.G., B.G., B.G.F., N.G., E.G.N., I.H., I.H.A., C.I., S.I.A., T.J., I.L.R., M.A.L.A., V.L.G., F.L.G., L.M., P.M.P., M.M.I., R.O.I., F.P.M., G.P.N., F.J.R.F., R.R.L., M.R.P., L.R.P., B.R.S., J.R., N.R., R.S.V., A.S., I.V.P., E.V.D., G.A., M.B., M.C., J.M.M., D.V., B.G.S., P.M., A.A.F., M.C. and C.A.; Visualization, I.P.R., G.G., I.F.I., M.C. and C.A.; Writing—original draft, I.P.R., G.G., I.F.I., M.P.V., M.G.H., M.C. and C.A.; Writing—review & editing, all authors.
Funding
This work was supported by the Instituto de Salud Carlos III (ISCIII) of the Spanish Ministry of Health (FIS; PI16/00425 and PI19/00321), Centro de Investigación Biomédica en Red Enfermedades Raras (CIBERER, 06/07/0036), IIS-FJD BioBank (PT13/0010/0012), Comunidad de Madrid (CAM, RAREGenomics Project, B2017/BMD-3721), European Regional Development Fund (FEDER), the Organización Nacional de Ciegos Españoles (ONCE), Fundación Ramón Areces, Fundación Conchita Rábago and the University Chair UAM-IIS-FJD of Genomic Medicine. Irene Perea-Romero is supported by a PhD fellowship from the predoctoral Program from ISCIII (FI17/00192). Ionut F. Iancu is supported by a grant from the Comunidad de Madrid (CAM, PEJ-2017-AI/BMD7256). Marta del Pozo-Valero is supported by a PhD grant from the Fundación Conchita Rábago. Berta Almoguera is supported by a Juan Rodes program from ISCIII (JR17/00020). Pablo Minguez is supported by a Miguel Servet program from ISCIII (CP16/00116). Marta Corton is supported by a Miguel Servet program from ISCIII (CPII17/00006). The funders played no role in study design, data collection, data analysis, manuscript preparation and/or publication decisions.
Data availability
Part of the NGS data are available in public, open access repositories such as the European Genome-Phenome Archive (EGA; https://www.ebi.ac.uk/ega/home; EGAD00001005746 and EGAD00001005498), RD-Connect (https://rd-connect.eu/) and the Collaborative Spanish Variant Server (CSVS; http://csvs.babelomics.org/) as aggregated data. The rest of the data are available upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Irene Perea-Romero, Gema Gordo, Ionut F. Iancu, Marta Corton and Carmen Ayuso.
A list of authors and their affiliations appears at the end of the paper.
Change history
5/10/2021
A Correction to this paper has been published: 10.1038/s41598-021-89275-4
Contributor Information
Marta Corton, Email: mcorton@quironsalud.es.
Carmen Ayuso, Email: cayuso@fjd.es.
The ESRETNET Study Group:
Ana Bustamante-Aragones, Rocio Cardero-Merlo, Ruth Fernandez-Sanchez, Jesus Gallego-Merlo, Ines Garcia-Vara, Ascension Gimenez-Pardo, Laura Horcajada-Burgos, Fernando Infantes-Barbero, Esther Lantero, Miguel A. Lopez-Martinez, Andrea Martinez-Ramas, Lorena Ondo, Marta Rodriguez de Alba, Carolina Sanchez-Jimeno, Camilo Velez-Monsalve, Cristina Villaverde, Olga Zurita, Domingo Aguilera-Garcia, Jana Aguirre-Lamban, Ana Arteche, Diego Cantalapiedra, Patricia Fernandez-San Jose, Liliana Galbis-Martinez, Maria Garcia-Hoyos, Carlos Lombardia, Maria I. Lopez-Molina, Raquel Perez-Carro, Luciana R. J. Da Silva, Carmen Ramos, Rocio Sanchez-Alcudia, Iker Sanchez-Navarro, Sorina D. Tatu, Elena Vallespin, Elena Aller, Sara Bernal, Maria J. Gamundi, Gema Garcia-Garcia, Inmaculada Hernan, Teresa Jaijo, Guillermo Antiñolo, Montserrat Baiget, Miguel Carballo, Jose M. Millan, and Diana Valverde
The ERDC Study Group:
Rando Allikmets, Sandro Banfi, Frans P. M. Cremers, Rob W. J. Collin, Elfride De Baere, Hakon Hakonarson, Susanne Kohl, Carlo Rivolta, and Dror Sharon
The Associated Clinical Study Group:
Maria C. Alonso-Cerezo, Maria J. Ballesta-Martinez, Sergi Beltran, Carmen Benito Lopez, Jaume Català-Mora, Claudio Catalli, Carmen Cotarelo-Perez, Miguel Fernandez-Burriel, Ana Fontalba-Romero, Enrique Galán-Gómez, Maria Garcia-Barcina, Loida M. Garcia-Cruz, Blanca Gener, Belen Gil-Fournier, Nancy Govea, Encarna Guillen-Navarro, Ines Hernando Acero, Cristina Irigoyen, Silvia Izquierdo-Álvarez, Isabel Llano-Rivas, Maria A. López-Ariztegui, Vanesa Lopez-Gonzalez, Fermina Lopez-Grondona, Loreto Martorell, Pilar Mendez-Perez, Maria Moreno-Igoa, Raluca Oancea-Ionescu, Francesc Palau-Martinez, Guiomar Perez de Nanclares, Feliciano J. Ramos-Fuentes, Raquel Rodriguez-Lopez, Montserrat Rodriguez-Pedreira, Lydia Rodriguez-Peña, Berta Rodriguez-Sanchez, Jordi Rosell, Noemi Rosello, Raquel Saez-Villaverde, Alfredo Santana, Irene Valenzuela-Palafoll, and Eva Villota-Deleu
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-81093-y.
References
- 1.Ayuso C, Millan JM. Retinitis pigmentosa and allied conditions today: A paradigm of translational research. Genome Med. 2010;2:34. doi: 10.1186/gm155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanany M, Rivolta C, Sharon D. Worldwide carrier frequency and genetic prevalence of autosomal recessive inherited retinal diseases. Proc. Natl. Acad. Sci. U. S. A. 2020;117:2710–2716. doi: 10.1073/pnas.1913179117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368:1795–1809. doi: 10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- 4.Vaidya P, Vaidya A. Retinitis pigmentosa: Disease encumbrance in the eurozone. Int. J. Ophthalmol. Clin. Res. 2015;2:030. doi: 10.23937/2378-346x/1410030. [DOI] [Google Scholar]
- 5.Dryja TP, et al. Mutations within the rhodopsin gene in patients with autosomal dominant retinitis pigmentosa. N. Engl. J. Med. 1990;323:1302–1307. doi: 10.1056/NEJM199011083231903. [DOI] [PubMed] [Google Scholar]
- 6.Riveiro-Alvarez R, et al. Outcome of ABCA4 disease-associated alleles in autosomal recessive retinal dystrophies: Retrospective analysis in 420 Spanish families. Ophthalmology. 2013;120:2332–2337. doi: 10.1016/j.ophtha.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanco-Kelly F, et al. Clinical aspects of Usher syndrome and the USH2A gene in a cohort of 433 patients. JAMA Ophthalmol. 2015;133:157–164. doi: 10.1001/jamaophthalmol.2014.4498. [DOI] [PubMed] [Google Scholar]
- 8.Martin-Merida I, et al. Genomic landscape of sporadic retinitis pigmentosa: Findings from 877 Spanish Cases. Ophthalmology. 2019;126:1181–1188. doi: 10.1016/j.ophtha.2019.03.018. [DOI] [PubMed] [Google Scholar]
- 9.Sanchez-Navarro I, et al. Combining targeted panel-based resequencing and copy-number variation analysis for the diagnosis of inherited syndromic retinopathies and associated ciliopathies. Sci. Rep. 2018;8:5285. doi: 10.1038/s41598-018-23520-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin-Merida I, et al. Toward the mutational landscape of autosomal dominant retinitis pigmentosa: A comprehensive analysis of 258 Spanish families. Investig. Ophthalmol. Vis. Sci. 2018;59:2345–2354. doi: 10.1167/iovs.18-23854. [DOI] [PubMed] [Google Scholar]
- 11.Avila-Fernandez A, et al. Mutation analysis of 272 Spanish families affected by autosomal recessive retinitis pigmentosa using a genotyping microarray. Mol. Vis. 2010;16:2550–2558. [PMC free article] [PubMed] [Google Scholar]
- 12.Castro-Sanchez S, et al. Exploring genotype-phenotype relationships in Bardet–Biedl syndrome families. J. Med. Genet. 2015;52:503–513. doi: 10.1136/jmedgenet-2015-103099. [DOI] [PubMed] [Google Scholar]
- 13.Gayan J, et al. Genetic structure of the Spanish population. BMC Genom. 2010;11:326. doi: 10.1186/1471-2164-11-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharon D, et al. A nationwide genetic analysis of inherited retinal diseases in Israel as assessed by the Israeli inherited retinal disease consortium (IIRDC) Hum. Mutat. 2019 doi: 10.1002/humu.23903. [DOI] [PubMed] [Google Scholar]
- 15.Pontikos N, et al. Genetic basis of inherited retinal disease in a molecularly characterized cohort of more than 3000 families from the United Kingdom. Ophthalmology. 2020;127:1384–1394. doi: 10.1016/j.ophtha.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goetz KE, et al. Genetic testing for inherited eye conditions in over 6000 individuals through the eyeGENE network. Am. J. Med. Genet. C Semin. Med. Genet. 2020;184:828–837. doi: 10.1002/ajmg.c.31843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaffal L, et al. The genetics of rod-cone dystrophy in Arab countries: A systematic review. Eur. J. Hum. Genet. 2020 doi: 10.1038/s41431-020-00754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chelva E, et al. A retinitis pigmentosa register for western Australia. Aust. N. Z. J. Ophthalmol. 1992;20:311–317. doi: 10.1111/j.1442-9071.1992.tb00743.x. [DOI] [PubMed] [Google Scholar]
- 19.Bunker CH, Berson EL, Bromley WC, Hayes RP, Roderick TH. Prevalence of retinitis pigmentosa in Maine. Am. J. Ophthalmol. 1984;97:357–365. doi: 10.1016/0002-9394(84)90636-6. [DOI] [PubMed] [Google Scholar]
- 20.Bertelsen M, Jensen H, Bregnhoj JF, Rosenberg T. Prevalence of generalized retinal dystrophy in Denmark. Ophthalmic Epidemiol. 2014;21:217–223. doi: 10.3109/09286586.2014.929710. [DOI] [PubMed] [Google Scholar]
- 21.Holtan JP, Selmer KK, Heimdal KR, Bragadottir R. Inherited retinal disease in Norway—A characterization of current clinical and genetic knowledge. Acta Ophthalmol. 2019 doi: 10.1111/aos.14218. [DOI] [PubMed] [Google Scholar]
- 22.Kim MS, et al. Genetic mutation profiles in Korean patients with inherited retinal diseases. J. Korean Med. Sci. 2019;34:e161. doi: 10.3346/jkms.2019.34.e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Motta FL, Martin RP, Filippelli-Silva R, Salles MV, Sallum JMF. Relative frequency of inherited retinal dystrophies in Brazil. Sci. Rep. 2018;8:15939. doi: 10.1038/s41598-018-34380-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eisenberger T, et al. Increasing the yield in targeted next-generation sequencing by implicating CNV analysis, non-coding exons and the overall variant load: The example of retinal dystrophies. PLoS ONE. 2013;8:e78496. doi: 10.1371/journal.pone.0078496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang XF, et al. Genotype–phenotype correlation and mutation spectrum in a large cohort of patients with inherited retinal dystrophy revealed by next-generation sequencing. Genet. Med. 2015;17:271–278. doi: 10.1038/gim.2014.138. [DOI] [PubMed] [Google Scholar]
- 26.Tiwari A, et al. Next generation sequencing based identification of disease-associated mutations in Swiss patients with retinal dystrophies. Sci. Rep. 2016;6:28755. doi: 10.1038/srep28755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Birtel J, et al. Clinical and genetic characteristics of 251 consecutive patients with macular and cone/cone-rod dystrophy. Sci. Rep. 2018;8:4824. doi: 10.1038/s41598-018-22096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bravo-Gil N, et al. Unravelling the genetic basis of simplex Retinitis Pigmentosa cases. Sci. Rep. 2017;7:41937. doi: 10.1038/srep41937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maeda A, et al. Development of a molecular diagnostic test for Retinitis Pigmentosa in the Japanese population. Jpn. J. Ophthalmol. 2018;62:451–457. doi: 10.1007/s10384-018-0601-x. [DOI] [PubMed] [Google Scholar]
- 30.Riveiro-Alvarez R, et al. Frequency of ABCA4 mutations in 278 Spanish controls: An insight into the prevalence of autosomal recessive Stargardt disease. Br. J. Ophthalmol. 2009;93:1359–1364. doi: 10.1136/bjo.2008.148155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Del Pozo-Valero M, et al. Genotype–phenotype correlations in a Spanish cohort of 506 families with biallelic ABCA4 pathogenic variants. Am. J. Ophthalmol. 2020;219:195–204. doi: 10.1016/j.ajo.2020.06.027. [DOI] [PubMed] [Google Scholar]
- 32.Cremers FPM, Lee W, Collin RWJ, Allikmets R. Clinical spectrum, genetic complexity and therapeutic approaches for retinal disease caused by ABCA4 mutations. Prog. Retin. Eye Res. 2020;79:100861. doi: 10.1016/j.preteyeres.2020.100861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seyedahmadi BJ, Rivolta C, Keene JA, Berson EL, Dryja TP. Comprehensive screening of the USH2A gene in Usher syndrome type II and non-syndromic recessive retinitis pigmentosa. Exp. Eye Res. 2004;79:167–173. doi: 10.1016/j.exer.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 34.Rivera A, et al. A comprehensive survey of sequence variation in the ABCA4 (ABCR) gene in Stargardt disease and age-related macular degeneration. Am. J. Hum. Genet. 2000;67:800–813. doi: 10.1086/303090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Passerini I, et al. Novel mutations in of the ABCR gene in Italian patients with Stargardt disease. Eye (Lond) 2010;24:158–164. doi: 10.1038/eye.2009.35. [DOI] [PubMed] [Google Scholar]
- 36.Dreyer B, et al. Identification of novel USH2A mutations: Implications for the structure of USH2A protein. Eur. J. Hum. Genet0. 2000;8:500–506. doi: 10.1038/sj.ejhg.5200491. [DOI] [PubMed] [Google Scholar]
- 37.Aller E, et al. The USH2A c.2299delG mutation: dating its common origin in a Southern European population. Eur. J. Hum. Genet. 2010;18:788–793. doi: 10.1038/ejhg.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mykytyn K, et al. Identification of the gene (BBS1) most commonly involved in Bardet–Biedl syndrome, a complex human obesity syndrome. Nat. Genet. 2002;31:435–438. doi: 10.1038/ng935. [DOI] [PubMed] [Google Scholar]
- 39.Kousal B, et al. Phenotypic features of CRB1-associated early-onset severe retinal dystrophy and the different molecular approaches to identifying the disease-causing variants. Graefes Arch. Clin. Exp. Ophthalmol. 2016;254:1833–1839. doi: 10.1007/s00417-016-3358-2. [DOI] [PubMed] [Google Scholar]
- 40.Bujakowska K, et al. CRB1 mutations in inherited retinal dystrophies. Hum. Mutat. 2012;33:306–315. doi: 10.1002/humu.21653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ziviello C, et al. Molecular genetics of autosomal dominant retinitis pigmentosa (ADRP): A comprehensive study of 43 Italian families. J. Med. Genet. 2005;42:e47. doi: 10.1136/jmg.2005.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Audo I, et al. Spectrum of rhodopsin mutations in French autosomal dominant rod-cone dystrophy patients. Investig. Ophthalmol. Vis. Sci. 2010;51:3687–3700. doi: 10.1167/iovs.09-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roshandel D, et al. Rhodopsin gene mutation analysis in Iranian patients with autosomal dominant retinitis pigmentosa. Int. Ophthalmol. 2019 doi: 10.1007/s10792-019-01099-4. [DOI] [PubMed] [Google Scholar]
- 44.Yang G, et al. Spectrum of rhodopsin gene mutations in Chinese patients with retinitis pigmentosa. Mol. Vis. 2014;20:1132–1136. [PMC free article] [PubMed] [Google Scholar]
- 45.Beheshtian M, et al. Impact of whole exome sequencing among Iranian patients with autosomal recessive retinitis pigmentosa. Arch. Iran. Med. 2015;18:776–785. [PubMed] [Google Scholar]
- 46.Bandah-Rozenfeld D, et al. Homozygosity mapping reveals null mutations in FAM161A as a cause of autosomal-recessive retinitis pigmentosa. Am. J. Hum. Genet. 2010;87:382–391. doi: 10.1016/j.ajhg.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tuson M, Marfany G, Gonzalez-Duarte R. Mutation of CERKL, a novel human ceramide kinase gene, causes autosomal recessive retinitis pigmentosa (RP26) Am. J. Hum. Genet. 2004;74:128–138. doi: 10.1086/381055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Avila-Fernandez A, et al. Identification of an RP1 prevalent founder mutation and related phenotype in Spanish patients with early-onset autosomal recessive retinitis. Ophthalmology. 2012;119:2616–2621. doi: 10.1016/j.ophtha.2012.06.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Part of the NGS data are available in public, open access repositories such as the European Genome-Phenome Archive (EGA; https://www.ebi.ac.uk/ega/home; EGAD00001005746 and EGAD00001005498), RD-Connect (https://rd-connect.eu/) and the Collaborative Spanish Variant Server (CSVS; http://csvs.babelomics.org/) as aggregated data. The rest of the data are available upon reasonable request.