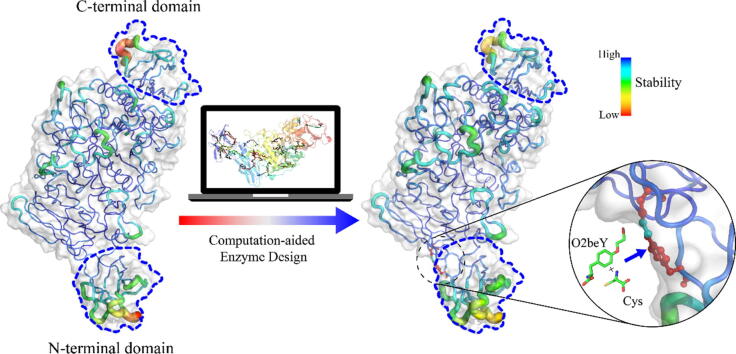
Graphical abstract
Keywords: Computational design, Thermostabilization, Noncanonical amino acids, Thioether staple, Multi-modular enzyme
Abstract
Enzyme thermostabilization is considered a critical and often obligatory step in biosynthesis, because thermostability is a significant property of enzymes that can be used to evaluate their feasibility for industrial applications. However, conventional strategies for thermostabilizing enzymes generally introduce non-covalent interactions and/or natural covalent bonds caused by natural amino acid substitutions, and the trade-off between the activity and stability of enzymes remains a challenge. Here, we developed a computationally guided strategy for constructing thioether staples by incorporating noncanonical amino acid (ncAA) into the more flexible N/C-terminal domains of the multi-modular pullulanase from Bacillus thermoleovorans (BtPul) to enhance its thermostability. First, potential thioether staples located in the N/C-terminal domains of BtPul were predicted using RosettaMatch. Next, eight variants involving stable thioether staples were precisely predicted using FoldX and Rosetta ddg_monomer. Six positive variants were obtained, of which T73(O2beY)-171C had a 157% longer half-life at 70 °C and an increase of 7.0 °C in Tm, when compared with the wild-type (WT). T73(O2beY)-171C/T126F/A72R exhibited an even more improved thermostability, with a 211% increase in half-life at 70 °C and a 44% enhancement in enzyme activity compared with the WT, which was attributed to further optimization of the local interaction network. This work introduces and validates an efficient strategy for enhancing the thermostability and activity of multi-modular enzymes.
1. Introduction
Enzyme thermostabilization remains a topic of interest in biocatalysis field, because thermostability is a significant property of enzymes that can be used to evaluate their feasibility for industrial applications [1]. Enzymes with considerable thermostability exhibit remarkable advantages in accelerating reactions, increasing evolutionary potential, and reducing microbial contamination and production cost [2], [3]. Presently, developing thermostable enzymes is a fundamental priority for green manufacturing processes, including pharmaceutical development, fine chemical production, and biofuel synthesis. However, these green biosynthesis and chemical manufacturing processes generally occur under harsh conditions, and the thermostability of natural enzymes is often insufficient [4]. Therefore, it is of great significance to the field of green chemistry to improve enzyme thermostability and reduce the increasingly prominent environmental pollution and resource depletion.
Enzyme thermostabilization against thermal denaturation has been an urgent and long-standing goal in enzyme design and engineering. Directed evolution [5] is a well-known strategy in protein engineering for enzyme thermostabilization [6] that is generally applicable with the aid of high-throughput screening [7]. Although it is often effective, directed evolution is labor-intensive and time-consuming. For instance, multiple rounds of mutation and screening are required to obtain desired variants with obvious improvements in thermostability [7]. During the third wave of biocatalysis, the process of protein thermostabilization via computational design has been promoted based on a large amount of protein structure analysis data and increased bioinformatics tool development [4], [8]. Compared with irrational directed evolution, rational computational design is more rapid and targeted, with a comparably small library generally containing <102 variants [9], [10]. Computational design of enzyme stabilization focuses on optimizing native interactions or reducing the configurational entropy of the unfolded state ensemble to stabilize proteins with the help of computational tools that utilize appropriate scoring functions, search algorithms, and amino acid rotamer libraries to predict potential stabilizing mutations [11], [12]. Among computational design strategies including consensus sequence-based site-directed mutagenesis [13], proline introduction [14], disulfide bond introduction [15], and surface-charge optimization [16], disulfide bond introduction is regarded as an effective approach for enhancing enzyme thermostability [17].
A disulfide bond stabilizes the three-dimensional protein structure mainly by decreasing the configurational entropy of the unfolded state ensemble, making a critical contribution to enzyme thermostability [18], [19], [20], [21]. A covalent disulfide bond possesses a dissociation energy of 250 kJ/mol, which is notably stronger than that of non-covalent bonds including hydrogen bonds and hydrophobic interactions [22]. However, disulfide bonds are reversible and homotypic, which may introduce undesired phenomena. First, disulfide bonds may be disconnected or reformed owing to changes in the external environment after their formation and may only remain relatively stable [23]. Second, disulfide scrambling sometimes occurs, thereby accelerating the formation of inclusion bodies and affecting the soluble expression of proteins [24]. Finally, proteins with disulfide bonds agglomerate and precipitate more easily, which is not conducive to enzyme storage [25].
Seeking chemically stable covalent bonds to replace reversible disulfide bonds for protein stabilization poses a novel challenge. Incorporating noncanonical amino acid (ncAA) into enzymes can improve enzymatic properties, such as thermostability [26]. Unnatural thioether linkages can be formed as a result of the cross-linking reaction between a cysteine and a noncanonical O-2-bromoethyl tyrosine (O2beY), introducing unique properties of redox-stability and irreversibility [27]. Moore et al. [27] have computationally predicted and constructed thioether linkages via RosettaMatch for enzyme stabilization of the single-domain myoglobin [28]. However, for large multi-modular proteins, RosettaMatch predicts a large number of variants from which to identify positive mutants, resulting in a heavy workload. The thermostabilization of large proteins has always been difficult owing to certain restrictions [29]. For instance, some researchers have sought to thermostabilize proteins by using various engineering strategies to target the N/C terminal region, including cyclization, truncation, and splicing [29], [30]. However, although the N/C terminal regions are generally the most flexible parts of the protein backbone, it is still difficult to improve the thermostability of a large multi-modular protein via the N/C terminal cyclization strategy [30], [31]. Modifying the N/C terminal region by using an unnatural amino acid staple to thermostabilize proteins remains an untested possibility.
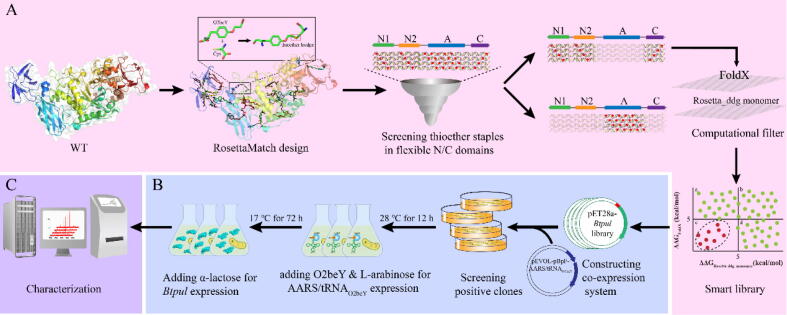
Herein, to develop a method for enzyme thermostabilization, pullulanase from Bacillus thermoleovorans (BtPul), an enzyme with a large molecular weight (160 kDa) and a multi-domain structure, was selected as a model system [32]. Distinct from other pullulanases, the neutral thermophilic BtPul specifically catalyzes the hydrolysis of the α-1, 6-glucosidic linkages of unmodified substrates and is widely used for industrial saccharification, polymeric material processing, organometallic chemistry, and drug formulation [33], [34], [35], [36]. The potential thioether stapling sites were first identified using RosettaMatch. To filter out unstable staples, the conformational stability of mutants with stapling sites at the flexible N/C-terminal domains was evaluated using different algorithm predictors, FoldX [37] and Rosetta ddg_monomer [38]. Then, the targeted variants were constructed via a co-expression system for further thermostability and activity characterization. In the co-expression system, pEVOL-pBpF-AARS/tRNAO2beY referred to the pEVOL-pBpF plasmid encoding the orthogonal aminoacyl-tRNA synthetase (AARS)/tRNA that was used specifically for O2beY incorporation (AARS/tRNAO2beY), and pET28a-Btpul referred to the pET28a plasmid bearing the BtPul gene (Btpul). The approach for enzyme thermostabilization via computational design is illustrated in Fig. 1.
Fig. 1.
Scheme of the computational design method for enzyme thermostabilization. (A) The smart library generated via computational tools. RosettaMatch was utilized to design potential stapling sites. N1, N2, and C were the N/C structural domains, which were notably more flexible than structural domain A. Computational filtering was performed via FoldX and Rosetta ddg_monomer. The red ball located in the c region displays the ΔΔG values that were in the allowable range when computed by both FoldX and Rosetta ddg_monomer, indicating that these variants in the c region were more likely to increase the BtPul thermostability than the variants in the other three regions (a, b, and d). (B) Construction and expression of variants in the smart library. Co-expression systems were expressed in self-inducing medium. (C) Characterization of enzymatic properties. Verification of the thioether linkage was conducted using quadrupole time-of-flight mass spectrometry (Q-TOF MS). Thermostability was assessed by determining the apparent melting temperature (Tm) and half-life (t1/2). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2. Materials and methods
2.1. Materials
E. coli JM109 and E. coli BL21 (DE3) were used as the hosts for recombinant gene cloning and expression, respectively. The recombinant plasmid pET28a-Btpul (GenBank Accession No. EMBL AJ315595) was constructed by Genewiz (Suzhou, China). The plasmid pEVOL-pBpF encoding AARS/tRNA was purchased from Addgene (Beijing, China). PrimeSTAR HS DNA polymerase, which was used for polymerase chain reaction (PCR), was purchased from TaKaRa Biotechnology Co., Ltd. (Beijing, China). Primers were all synthesized by Genewiz (Suzhou, China). The ClonExpress II One Step Cloning Kit for homologous recombination was purchased from the Vazyme Biotech Company, Ltd. (Nanjing, China). DNA sequencing was completed by Genewiz (Suzhou, China). The HisTrap HP affinity column and disposable PD-10 desalting column that were respectively used for protein purification and desalination were purchased from GE Healthcare Life Sciences (Beijing, China). O2beY, pullulan, and l-arabinose were purchased from Tokyo AMATEK (Suzhou, China), Chemical Industry (Tokyo, Japan), and Ourchem (Shanghai, China), respectively. Premixed Protein Marker (High) and 4× Protein SDS-PAGE Loading Buffer were purchased from TaKaRa Biotechnology Co., Ltd. (Beijing, China). All other materials were purchased from Sigma-Aldrich (St. Louis, MO, USA) or Sinopharm Chemical Reagent (Shanghai, China).
2.2. Homology modeling
We selected a wild-type (WT) enzyme (PDB ID: 3wdj) with 57.08% homology to BtPul as a template and used SWISS-MODEL to model BtPul, while models of mutants harboring thioether linkages were directly generated via RosettaMatch. Models were evaluated using MolProbity [39] and PROCHECK [40]. The model with the lowest MolProbity score and with more than 95% allowed area in the Ramachandran plots was identified as the qualified model and used for further study and analysis [41].
2.3. Computational design
RosettaMatch was utilized to theoretically compute all spatial positions where the O2beY/cysteine (O2beY/Cys) pair could be suitably stapled in BtPul. The output had multiple structural files with different compatible geometries. From the potential stapling sites, we only selected the ones located in the N/C-terminal of BtPul. FoldX and Rosetta ddg_monomer were utilized to compute the Gibbs free energy changes (ΔΔG) of each remaining stapling site. Negative values obtained via the two predictors indicated stable mutations, while positive values indicated unstable mutations. Stapling sites with a value of ΔΔG in the range of ≤5 kcal mol−1 were chosen, while the others were eliminated. As the last parameter, we set the length of the primary sequence between O2beY and Cys to ≥90 amino acids. All relative codes for computation were listed in Supplementary Materials.
2.4. Construction of variants
Variants were constructed using homologous recombination. The short gene fragments encoding Btpul or AARS/tRNA were amplified using overlap extension PCR (the primers are listed in Supplementary Table S1). Instead of incorporating p-benzoylphenylalanine, three mutations were introduced to AARS/tRNA (T158A, S159I, and L162A) to establish specificity for O2beY. The long fragment was amplified using pET28a or pEVOL-pBpF as a template, with reverse primers (Supplementary Table S1). After amplification, the obtained short fragment was recombined with the long fragment using the ClonExpress II One Step Cloning Kit. The obtained recombination plasmids, pET28a-Btpul and pEVOL-pBpF-AARS/tRNAO2beY, were respectively transformed into E. coli JM109 for confirmation by sequencing, and then the two recombination plasmids were co-transformed into E. coli BL21 (DE3) for expression. Positive cells were seeded onto LB agar plates containing kanamycin (50 μg·mL−−1) and chloramphenicol (34 mg·L−1).
2.5. Recombinant protein expression
Recombinant strains were incubated overnight at 37 °C and 200 rpm in 5 mL of LB culture medium containing kanamycin (50 μg·mL−1) and chloramphenicol (34 mg·L−1). Overnight cultures were transferred to 50 mL of self-inducing medium without α-lactose (5.0 g·L−1 yeast extract, 10 g·L−1 tryptone, 5.0 g·L−1 glycerol, 1.0 g·L−1glucose, 7.1 g·L−1 Na2HPO4, 6.8 g·L−1 KH2PO4, 2.67 g·L−1 NH4Cl, 0.71 g·L−1 Na2SO4, and 0.25 g·L−1 MgSO4; pH 7.5) supplemented with the same antibiotics. After 2.5–3 h of incubation (37 °C, 200 rpm), 4 mM l-arabinose and 1 mM O2beY were added to establish the expression of charged AARS/tRNAO2beY. The strains were then incubated at 28 °C and 200 rpm for 12 h. Finally, α-lactose at a final concentration of 10 g·L−1 was added to establish the expression of the BtPul variants, and the strains were incubated again at 17 °C and 200 rpm for 48–60 h.
2.6. Recombinant protein purification and characterization
Cells were harvested by centrifugation at 8228 g for 5 min followed by sonication via ultrasonic breaker (Scientz, China). The lysate was centrifuged at 4 °C and 18,514g for 30 min in a refrigerated centrifuge. After filtering through a 0.22 μm filter membrane, the supernatant of the lysate was loaded onto a HisTrap HP affinity column equilibrated with loading buffer (20 mM Tris-HCl, 150 mM NaCl, 20 mM imidazole; pH 7.5). Pure target protein was obtained by eluting with elution buffer (20 mM Tris-HCl, 150 mM NaCl, 1 M imidazole; pH 7.5) at 2–3 mL·min−1. Desalination was conducted with low-salt buffer (10 mM Tris-HCl, 0.1 M NaCl; pH 7.5) using a disposable PD-10 desalting column. Characterization was performed using 10% (w/v) sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) with the addition of 20% (v/v) β-mercaptoethanol, followed by the accurate determination of protein mass using Q-TOF MS (Waters, USA) with ultra-pure water as the solvent.
2.7. Apparent melting temperature and half-life assays
Differential scanning fluorimetry (DSF) [42] was employed to determine the apparent melting temperature (Tm), which characterizes the thermodynamic stability of proteins. For the DSF experiment, 5 μL of 100 × SSYPRO® Orange dye (Sigma; S5692) was added to a 20 μL enzyme sample, which was pre-dissolved in low-salt buffer and had an enzyme concentration of 50 μg·mL−1. Then, the mixture was heated from 25 °C to 95 °C at a rate of 1 °C·min−1 in the StepOnePlus™ real-time PCR (Life Technologies, USA).
Half-life (t1/2), one of the characteristic parameters of protein kinetic stability, was determined by incubating the enzyme solution with an enzyme concentration of 50 μg·mL−1 at 70 °C for different time intervals (0, 30, 60, 90, 120, 180, and 240 min) and measuring residual enzyme activity. Then, the first-order rate constant (kd) was obtained by determining the value of ln (residual enzyme activity)/incubation time (t). The half-life was finally calculated using the formula t1/2 = ln2/kd.
2.8. Activity and kinetics assays
Pullulanase activity was determined using the 3, 5-dinitrosalicylic acid (DNS) method described by Wang et al [43]. A 200 μL pullulanase solution diluted in advance with 0.2 M sodium phosphate buffer (pH 6.0) was mixed thoroughly with an equal volume of 2% (w/v) pullulan dissolved in the same buffer and then incubated at 70 ℃ for 20 min to induce the reaction. The negative control was established by adding 2% (w/v) pullulan after the reaction. Finally, the amount of reducing sugar was determined by measuring the absorbance at 540 nm. One unit of pullulanase activity was defined as the amount of pullulanase that released 1 μmol of reducing sugar (equivalent to glucose) per minute under the specified assay conditions.
To determine the kinetic characteristics of the enzyme, the concentration of the pullulan substrate was varied from 0.25 to 16 mg·mL−1 (0.25, 0.5, 1.0, 2.0, 4.0, 6.0, 8.0, 10.0, 12.0, 14.0, and 16.0 mg·mL−1). Pullulanase activity was determined as described above, but with 5 min of incubation rather than 20 min. Using GraphPad Prism 5, kinetic parameters were obtained via linear fitting of the Michaelis-Menten equation [44].
2.9. Secondary structure assays
Secondary structures of enzyme proteins were analyzed via circular dichroism (CD) spectrum. Purified enzymes dissolved in ultra-pure water was diluted to a protein concentration of 0.25 mg·mL−1. Then, 300 μL dilute solution was inhaled to a quartz colorimetric dish whose thickness is an optical path length of 1 mm. Structural elements of enzyme proteins could be obtained at the spectrum range of 190–250 nm with a scanning speed of 30 nm·min−1.
3. Results and discussion
3.1. Prediction and construction of protein stapling sites
The WT Ramachandran plot showed that up to 99.8% of residues were located in allowed regions [45], indicating that the three-dimensional WT model was reasonable enough to be the platform for subsequent computations. One hundred and ninety-six positions that could accommodate the formation of a thioether linkage via the O2beY/Cys pair were identified using RosettaMatch. The general rules for choosing the suitable sites are proposed as: (1) the stapling sites located in the flexible region outside active center are generally prior [31]; (2) the stapling sites predicted to have energetically favorable interactions with surrounding residues should be selected, for which the range of ΔΔG should be enlarged properly to have potential positive mutants; (3) the stapling sites of the O2beY/Cys pair far apart in the first-order sequence but close in space generally give the thioether cross-links with positive effect on enzyme thermostability [27].
To maximize the positive effect on enzyme thermostability, in this work, 86 stapling sites located in the N/C terminal domain were selected, due to the flexibility of the N/C terminal region [31]. The ΔΔG values of the selected stapling sites were then computed using FoldX and Rosetta ddg_monomer. The two predictors could only perform calculations if tyrosine replaced O2beY, potentially overshadowing the positive effect of the thioether cross-link. Therefore, we enlarged the range of ΔΔG to minimize the omission of stable mutants and then selected 53 stapling sites with a ΔΔG of ≤ 5 kcal mol−1 (Supplementary Table S2). Thioether cross-links have a more obvious positive effect on enzyme thermostability when the components of the O2beY/Cys pair are far apart in sequence but very close in space [27]. Therefore, 8 stapling sites with a primary sequence distance between O2beY and Cys of ≥ 90 amino acids were reserved (Supplementary Table S2), to further maximize thermostabilization. Meanwhile, we were interested in one particular stapling site (with residues at 711 and 699) located in loop 690–700 that was found to have a strongly positive effect on BtPul thermostability [45]. Hence, W711(O2beY)-A699C was also added to the final smart library, although the components were only 11 amino acids apart.
3.2. Expression and identification of variants
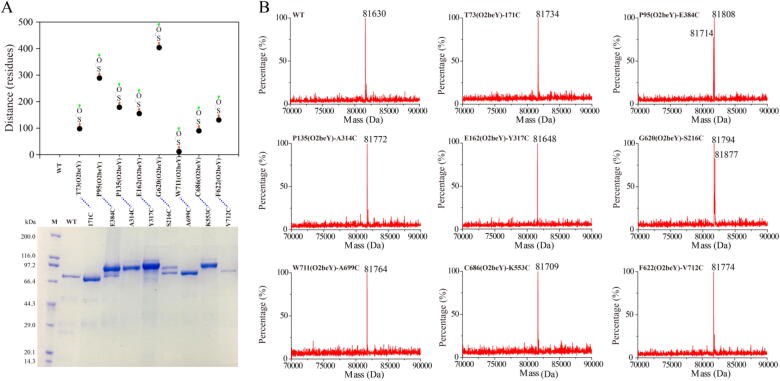
Soluble expression of all variants was successfully obtained via amber stop codon suppression in E. coli BL21 cells containing the AARS/tRNAO2beY system. The results of SDS-PAGE analysis (Fig. 2A) indicated that the variants exhibited different structural variations under the conditions of denaturation and reduction (SDS + mercaptoethanol). For instance, the presence or absence of the O2beY/Cys cross-link and the diverse positions of the stapling sites produced different behaviors in the analysis [27], [46]. There were lags in the electrophoretic mobility of five of the eight variants, namely P95(O2beY)-E384C, P135(O2beY)-A314C, E162(O2beY)-Y317C, G620(O2beY)-S216C, and C686(O2beY)-K553C, compared with that of the WT.
Fig. 2.
Identification of stapled BtPul variants. (A) SDS-PAGE of the wild-type (WT) and its variants. “Distance” refers to the number of amino acids between the components of the O2beY/Cys pair in the primary sequence. M represents the lane of the protein marker. (B) Q-TOF MS spectrum of the WT and its variants. Theoretical masses: T73(O2beY)-171C: 81821 Da for not cross-linked, 81739 Da for cross-linked; P95(O2beY)-E384C: 81799 Da for not cross-linked, 81717 Da for cross-linked; P135(O2beY)-A314C: 81857 Da for not cross-linked, 81775 Da for cross-linked; E162(O2beY)-Y317C: 81733 Da for not cross-linked, 81651 Da for cross-linked; G620(O2beY)-S216C: 81881 Da for not cross-linked, 81799 Da for cross-linked; W711(O2beY)-A699C: 81768 Da for not cross-linked, 81686 Da for cross-linked; C686(O2beY)-K553C: 81794 Da for not cross-linked, 81712 Da for cross-linked; F622(O2beY)-V712C: 81779 Da for not cross-linked, 81697 Da for cross-linked.
Simultaneously, among these five variants, the O2beY/Cys cross-linking reaction between P95(O2beY)-E384C and G620(O2beY)-S216C occurred only partially, which implied that the thioether linkage formation was a dynamic process and that the conformational changes may have affected the cross-linking reaction. Sometimes, the redox stability and irreversibility of thioether linkages can also keep the overall enzyme structure compact. However, this requires the presence of a denaturing and reductive agent, which increases electrophoretic mobility, as was the case with T73(O2beY)-171C. For F622(O2beY)-V712C and W711(O2beY)-A699C, the O2beY/Cys cross-linking reaction did not occur, even though the O2beY/Cys pair was close enough in spatial proximity, indicating that close spatial proximity is a necessary but insufficient condition for a cross-linking reaction to occur. The results of RosettaMatch were based on the compatibility of the sulfide product with the position of the selected residue in the natural structure. The ethyl group regarded as an independent ligand was restrained to enable placement in a compatible geometry between deprotonated Tyr and Cys, forming O-C and S-C bonds with Tyr and Cys, respectively. However, the cross-linking of the thioether also depends on whether those compatible geometries are accessible for the nucleophilic attack of the Cys side-chain thiol group on the O2beY alkyl bromide group.
The results of the SDS-PAGE analyses were also corroborated by the results of Q-TOF MS (Fig. 2B). All single signals near 81 kDa exhibited prospective changes in molecular weight as a result of mutations. For instance, the O2beY/Cys cross-linking reaction resulted in an extra 82 Da decrease in molecular weight, caused by the loss of HBr. For P95(O2beY)-E384C and G620(O2beY)-S216C, there were two signals visible in the mass spectrum that were attributed to the stapled and unstapled forms of the two variants, which is also consistent with the SDS-PAGE results.
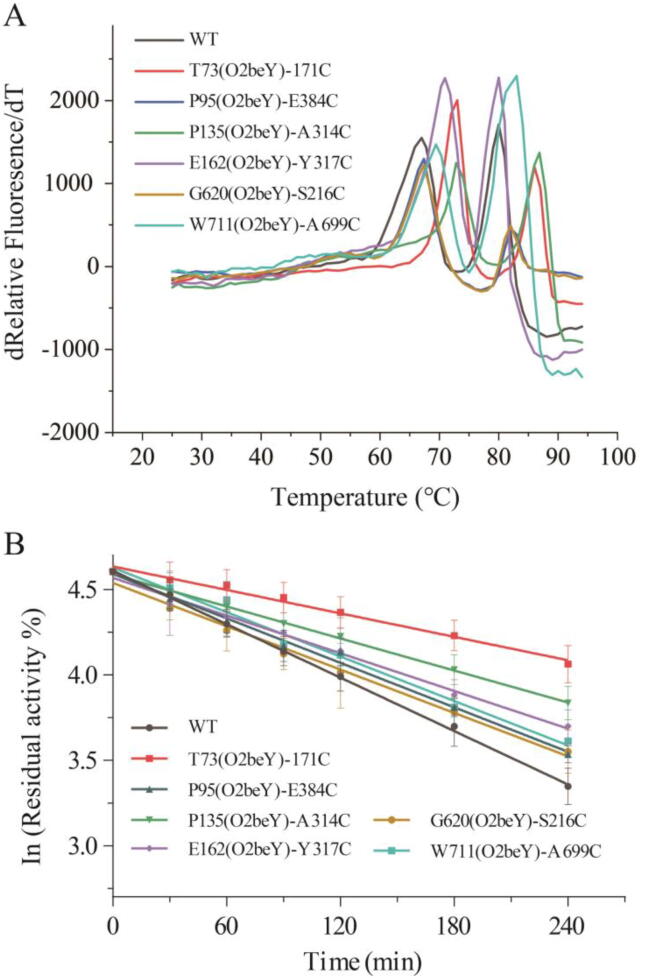
3.3. Thermostability of variants
We next investigated the thermostability characteristics of the eight variants. Protein thermostability is divided into thermodynamic stability and kinetic stability. Thermodynamic stability refers to a protein’s tendency towards irreversible unfolding, which involves changes in the overall structure of the protein [47]. The denaturation midpoint, Tm, is one of the characteristic parameters of thermodynamic stability and refers to the temperature at which 50% of a protein is in the unfolded state. In our study, six positive variants were identified by directly determining Tm via the DSF method. Compared with the WT, the six variants exhibited increases in Tm of 1.8–7.0 °C (Fig. 3A, Supplementary Table S3). This result indicated that the O2beY/Cys cross-link enhanced the enzyme’s ability to resist high temperatures, thus increasing the Tm. In contrast, the stapling variant C686(O2beY)-K553C did not exhibit an increase in Tm even though the cross-linking reaction occurred, implying that the choice of stapling site for the thioether linkage is particularly critical for the success of thermostability modifications. Further structural insight (Supplementary Fig. S1) indicated the distance between 686-O2beY and the centroid of the benzene ring on 608-Tyr was only 0.9 Å after the cross-linking reaction of thioether linkage between 686-O2beY and 553-Cys. This structural feature may seriously affect the positional conformation of Tyr608 and bring a large steric hindrance, thereby result in the instability of the local structure and even the whole structure of C686(O2beY)-K553C.
Fig. 3.
Comparison of the thermostability characteristics of the wild-type and positive variants. (A) Apparent melting temperature curve detected via the differential scanning fluorimetry method. (B) The first-order deactivation curve measured under standard conditions.
The Tm of W711(O2beY)-A699C also increased to a certain extent, although the cross-linking reaction did not occur as expected, which demonstrated that the 690–700 loop played a significant role in protein thermostabilization. The relevant improvement mechanism is shown in Supplementary Fig. S2. A larger and stronger hydrophobic network was generated, owing to the mutations at the residue sites 711 and 699 (analyzed using the Ring server). Hydrophobic interactions, which can stabilize the tertiary protein structure, play a critical part in stabilizing the entire macromolecular protein structure [48]. Furthermore, more hydrophobic interactions usually facilitate a decrease in the thermal unfolding rate at high temperatures, since hydrophobic interactions can keep proteins stable and robust at increased temperatures [49], [50].
Sometimes, protein unfolding is related to protein inactivation (in general, the former determines the latter). However, thermodynamic stability and kinetic stability are not exactly the same, and sometimes changes in the overall structure of a protein do not significantly affect the local structure of an active protein region [47]. To further investigate the effect of the thioether staple, it was necessary to evaluate the parameters characterizing kinetic stability, of which the half-life (t1/2) is considered to be more common than the others [51]. As shown in Fig. 3B and Supplementary Table S3, the six positive variants exhibited increases in t1/2, with t1/2 values of 330 min for T73(O2beY)-171C, 171 min for P95(O2beY)-E384C, 243 min for P135(O2beY)-A314C, 197 min for E162(O2beY)-Y317C, 175 min for G620(O2beY)-S216C, and 181 min for W711(O2beY)-A699C. Among them, the most stable variant, T73(O2beY)-171C, exhibited a half-life that was 157% longer than that of the WT. These results demonstrated that the ability of all six variants to sustain their activity improved, since the length of time and temperature at which a protein remains active while undergoing irreversible denaturation indicate the protein’s kinetic stability.
3.4. Catalytic activity of variants
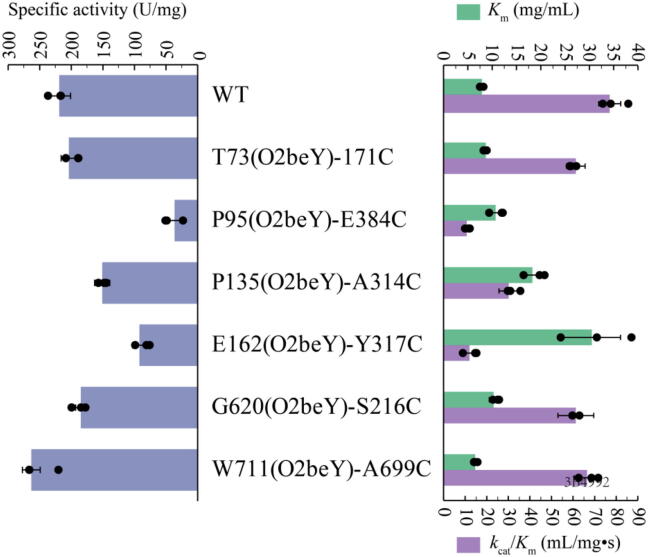
Thermostability improvements via protein engineering is often accompanied by the loss of catalytic activity, since the selective pressure to maintain function is absent [52]. Therefore, the effect of thioether staples on catalytic properties, including specific activity and kinetic parameters, was assessed. Whereas P95(O2beY)-E384C, P135(O2beY)-A314C, and E162(O2beY)-Y317C exhibited obvious reductions in specific activity compared with the WT (Fig. 4, Supplementary Table S3), both T73(O2beY)-171C and G620(O2beY)-S216C exhibited slightly decreased activity compared with the parent protein.
Fig. 4.
Comparison of specific activity (blue bars, left), Km (green bars, right), and kcat/Km (purple bars, right) of the wild-type and positive variants. All final values are shown as the mean ± standard deviation from three independent repeated experiments. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Further analysis of kinetic parameters indicated that the Km and kcat/Km of T73 (O2beY)-171C and G620 (O2beY)-S216C only exhibited slightly unsatisfactory fluctuations, while those of P95 (O2beY)-E384C, P135(O2beY)-A314C, and E162 (O2beY)-Y317C displayed obvious changes (Fig. 4, Supplementary Table S3). These results indicated that the thioether staple may sometimes have negative effects on the enzyme activity of BtPul, reducing the affinity of BtPul for the pullulan substrate and adversely affecting its catalytic efficiency. This may be explained by the opposing effects of flexibility and rigidity. Enzymes need sufficient conformational flexibility to exert catalytic activity towards a substrate [53], but a rigid structure confers considerable thermostability. For W711(O2beY)-A699C, the specific activity increased, accompanied by decreases in Km and kcat/Km. Engineering modifications in the 690–700 loop could be a useful strategy, as enzyme activity was almost unaffected by the mutation in this loop in our previous research [45].
3.5. Combinatorial mutations
Combinatorial mutations were induced in W711(O2beY)-A699C and variants in which the cross-linking reaction of the thioether linkage occurred completely. However, all combinatorial variants exhibited lower enzyme activities (less than 120 U mg−1) than the WT (Supplementary Fig. S3). Subsequently, structure-based engineering was applied to the best staple mutant, T73(O2beY)-171C. First, for residue site 126, T was substituted by F to form a potential π-π stacking with 73(O2beY), namely T73(O2beY)-171C/T126F. Meanwhile, given that both T73(O2beY) and T126F were mutated to hydrophobic residues, residues near sites 73 and 126 (at sites 72, 75, and 127) were substituted by a hydrophilic R in the T73(O2beY)-171C/T126F variant to make up for the potential activity loss arising from the hydrophobic surface and improve the thermostability further. The R residue bears a guanylyl group, which can improve the thermostability by forming an electrostatic network and long-range hydrogen bonds with surrounding negatively charged groups, and was shown to comprise a larger proportion of the surface of thermophilic enzymes than of the surface of mesophilic enzymes [54], [55]. Hence, another seven variants were constructed, namely T73(O2beY)-171C/T126F/A72R, T73(O2beY)-171C/T126F/W75R, T73(O2beY)-171C/T126F/A127R, T73(O2beY)-171C/T126F/A72R/W75R, T73(O2beY)-171C/T126F/A127R/A72R, T73(O2beY)-171C/T126F/A127R/W75R, and T73(O2beY)-171C/T126F/A127R/A72R/W75R.
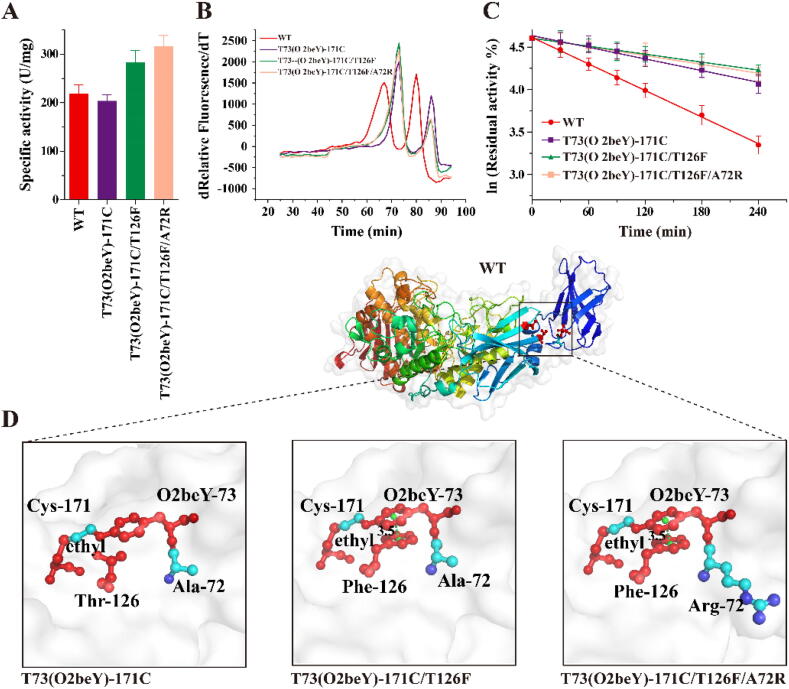
Among the eight combinatorial variants, T73(O2beY)-171C/T126F and T73(O2beY)-171C/T126F/A72R exhibited enzyme activity that increased by 29% and 44%, respectively, compared with that of the WT (Fig. 5A), which could be explained by their obviously reduced values of Km (Supplementary Table S3). Both had values of Tm similar to that of T73(O2beY)-171C (Fig. 5B), and their half-lives increased by 232% and 211%, respectively, compared with that of the WT (Fig. 5C). Structural visualization indicated that the introduction of 126F formed a π-π stacking, with a centroid distance of 3.5 Å between the two aromatic rings. π-π stacking, a kind of noncovalent aromatic interaction with a distance of less than 7 Å between two aromatic rings, is considered an important mechanism of protein structure stabilization [31]. Moreover, further introduction of 72R might increase the local hydrophilicity (Fig. 5D), thereby increasing enzyme activity. Considering that the incorporation of ncAA may affect the correct folding of the enzyme, we also determined the secondary structure of the WT and final combinatorial variant T73(O2beY)-171C/T126F/A72R. As shown in the CD spectrum (Supplementary Fig. S4), compared with the WT, there was no obvious change in the structural elements of T73(O2beY)-171C/T126F/A72R, indicating that the enzyme still has the right structure after incorporating the ncAA. However, the results of SDS-PAGE and Q-TOF MS analysis indicated that the thioether cross-link was impacted by the introduction of either F or R (Supplementary Fig. S5).
Fig. 5.
Comparison of enzymatic properties and structure visualization of positive combinatorial variants. (A) Specific activity. (B) Apparent melting temperature curve detected via the differential scanning fluorimetry method. (C) The first-order deactivation curve measured under standard conditions. (D) Structure visualization of T73(O2beY)-171C and a positive combinatorial variant. The green dashed line refers to the centroid distance between the two aromatic rings. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Conclusion
Following recent developments in biotechnology, computational technology, and other platform technologies, ncAA incorporation is becoming an increasingly attractive prospect in the biological field. Biocatalysts with new catalytic functions and structural properties could be obtained via ncAA incorporation. To enhance thermostability of multi-modular enzymes, in this work, we developed an effective computation-guided strategy for constructing thioether staples by incorporating ncAA into the N/C-terminal domains with high flexibility. In addition to RosettaMatch, different computational tools were integratedly employed for prediction of potential positive variants with increased reliability and reduced workload. Then the multi-modular pullulanase thermostabilization was performed by genetically constructing noncanonical O2beY and forming thioether linkages. The single variant T73(O2beY)-171C exhibited a 157% longer half-life and 7.0 °C higher Tm than the WT. Furthermore, the activity-stability trade-off of the enzyme was achieved by further introducing hydrophilic R to increase the local hydrophilicity at the surrounding hydrophobic surface of unnatural thioether linkage. Consequently, the combinatorial variant T73(O2beY)-171C/T126F/A72R exhibited a 211% increase in half-life at 70 °C and a 44% increase in enzyme catalytic activity, compared with WT. Therefore, this study provides a reference for modifying the thermostability of other enzymes, especially multi-modular enzymes. Computer-assisted rational design of unnatural staples will further expand thermostabilization strategy and greatly promote the development of protein modifications with ncAA for the design of thermostable proteins.
Author Contributions
The manuscript was prepared through contributions of all authors. J. Bi, Y. Nie, and Y. Xu conceived and designed the experiments. J. Bi, X. Jing, L. Wu, X. Zhou, and J. Gu performed the experiments. J. Bi and Y. Nie analyzed experimental data. J. Bi and Y. Nie wrote the main manuscript with input from co-authors. All authors have given approval to the final version of the manuscript.
Funding sources
This work was supported by the National Natural Science Foundation of China (31872891, 21676120), the Program of Introducing Talents of Discipline to Universities (111-2-06), the High-End Foreign Experts Recruitment Program (G20190010083), the Program for Advanced Talents within Six Industries of Jiangsu Province (2015-NY-007), the National Program for Support of Top-notch Young Professionals, the Fundamental Research Funds for the Central Universities (JUSRP51504), the Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, Top-notch Academic Programs Project of Jiangsu Higher Education Institutions, the Program for the Key Laboratory of Enzymes of Suqian (M201803), and the National First-Class Discipline Program of Light Industry Technology and Engineering (LITE2018-09).
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
CRediT authorship contribution statement
Jiahua Bi: Conceptualization, Methodology, Software, Formal analysis, Investigation, Writing - original draft, Visualization. Xiaoran Jing: Methodology, Investigation, Data curation. Lunjie Wu: Investigation, Software, Data curation. Xia Zhou: Investigation, Formal analysis. Jie Gu: Investigation, Formal analysis. Yao Nie: Conceptualization, Supervision, Funding acquisition, Resources, Project administration, Writing - review & editing. Yan Xu: Supervision, Funding acquisition, Resources, Project administration, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.12.043.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Bommarius A.S., Blum J.K., Abrahamson M.J. Status of protein engineering for biocatalysts: how to design an industrially useful biocatalyst. Curr Opin Chem Biol. 2011;15:194–200. doi: 10.1016/j.cbpa.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 2.Bloom J.D., Labthavikul S.T., Otey C.R., Arnold F.H. Protein stability promotes evolvability. Proc Natl Acad Sci U S A. 2006;103:5869–5874. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hua H., Luo H., Bai Y., Wang K., Niu C. A thermostable glucoamylase from Bispora sp. MEY-1 with stability over a broad pH range and significant starch hydrolysis capacity. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0113581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bornscheuer U.T., Huisman G.W., Kazlauskas R.J., Lutz S., Moore J.C. Engineering the third wave of biocatalysis. Nature. 2012;485:185–194. doi: 10.1038/nature11117. [DOI] [PubMed] [Google Scholar]
- 5.Arnold F.H. Innovation by evolution: bringing new chemistry to life (Nobel Lecture) Angew Chem Int Ed. 2019;58:14420–14426. doi: 10.1002/anie.201907729. [DOI] [PubMed] [Google Scholar]
- 6.Denard C.A., Ren H., Zhao H. Improving and repurposing biocatalysts via directed evolution. Curr Opin Chem Biol. 2015;25:55–64. doi: 10.1016/j.cbpa.2014.12.036. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y., Yu X., Zhao H. Biosystems design by directed evolution. AIChE J. 2020;66 [Google Scholar]
- 8.Musil M., Konegger H., Hon J., Bednar D., Damborsky J. Computational design of stable and soluble biocatalysts. ACS Catal. 2019;9:1033–1054. [Google Scholar]
- 9.Bednar D., Beerens K., Sebestova E., Bendl J., Khare S. FireProt: Energy- and evolution-based computational design of thermostable multiple-point mutants. PLoS Comput Biol. 2015;11 doi: 10.1371/journal.pcbi.1004556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guanlin L., Yuan C., Xingrong F., Feng S., Li X. Identification of a hot-spot to enhance Candida rugosa lipase thermostability by rational design methods. RSC Adv. 2018;8 doi: 10.1039/c7ra11679a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng Q., Capra N., Palacio C.M., Lanfranchi E., Otzen M. Robust ω-transaminases by computational stabilization of the subunit interface. ACS Catal. 2020;10:2915–2928. doi: 10.1021/acscatal.9b05223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou J., Song B., Simmerling C., Raleigh D. Experimental and computational analysis of protein stabilization by Gly-to-D-Ala substitution: a convolution of native state and unfolded state effects. J Am Chem Soc. 2016;138:15682–15689. doi: 10.1021/jacs.6b09511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duan X., Chen J., Wu J. Improving the thermostability and catalytic efficiency of Bacillus deramificans pullulanase by site-directed mutagenesis. Appl Environ Microbiol. 2013;79:4072–4077. doi: 10.1128/AEM.00457-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moroder L., Budisa N. Synthetic biology of protein folding. ChemPhysChem. 2010;11:1181–1187. doi: 10.1002/cphc.201000035. [DOI] [PubMed] [Google Scholar]
- 15.Mahadevi A.S., Sastry G.N. Cation-π interaction: its role and relevance in chemistry, biology, and material science. Chem Rev. 2013;113:2100–2138. doi: 10.1021/cr300222d. [DOI] [PubMed] [Google Scholar]
- 16.Gribenko A.V., Patel M.M., Liu J., McCallum S.A., Wang C. Rational stabilization of enzymes by computational redesign of surface charge-charge interactions. Proc Natl Acad Sci U S A. 2009;106:2601–2606. doi: 10.1073/pnas.0808220106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gihaz S., Bash Y., Rush I., Shahar A., Pazy Y. Bridges to stability: engineering disulfide bonds towards enhanced lipase biodiesel synthesis. ChemCatChem. 2020;12:181–192. [Google Scholar]
- 18.Moroder L., Musiol H.J., Renner C. Engineered disulfide bonds for protein design. In: Luis M., Johannes B., editors. Oxidative folding of peptides and proteins. The Royal Society of Chemistry; 2009. pp. 236–252. [Google Scholar]
- 19.He J., Tang F., Chen D.W., Yu B., Luo Y. Design, expression and functional characterization of a thermostable xylanase from Trichoderma reesei. PLoS ONE. 2019;14 doi: 10.1371/journal.pone.0210548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Q.A., Joo J.C., Yoo Y.J., Kim Y.H. Development of thermostable Candida antarctica lipase B through novel in silico design of disulfide bridge. Biotechnol Bioeng. 2012;109:867–876. doi: 10.1002/bit.24371. [DOI] [PubMed] [Google Scholar]
- 21.Liu L., Deng Z., Yang H., Li J., Shin H.D. In silico rational design and systems engineering of disulfide bridges in the catalytic domain of an alkaline α-amylase from Alkalimonas amylolytica to improve thermostability. Appl Environ Microbiol. 2014;80:798–807. doi: 10.1128/AEM.03045-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morgan B, Herrmann J (2017) Disulfide bonds. In eLS, John Wiley & Sons, Ltd (Ed.). pp. 1-10.
- 23.Tyagi V., Fasan R. Myoglobin-catalyzed olefination of aldehydes. Angew Chem Int Ed. 2016;55:2512–2516. doi: 10.1002/anie.201508817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nozach H., Fruchart-Gaillard C., Fenaille F., Beau F., Ramos O.H. High throughput screening identifies disulfide isomerase DsbC as a very efficient partner for recombinant expression of small disulfide-rich proteins in E. coli. Microb Cell Fact. 2013;12:37. doi: 10.1186/1475-2859-12-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanghe M., Danneels B., Last M., Beerens K., Stals I. Disulfide bridges as essential elements for the thermostability of lytic polysaccharide monooxygenase LPMO10C from Streptomyces coelicolor. Protein Eng Des Sel. 2017;30:401–408. doi: 10.1093/protein/gzx014. [DOI] [PubMed] [Google Scholar]
- 26.Li J.C., Liu T., Wang Y., Mehta A.P., Schultz P.G. Enhancing protein stability with genetically encoded noncanonical amino acids. J Am Chem Soc. 2018;140:15997–16000. doi: 10.1021/jacs.8b07157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore E.J., Zorine D., Hansen W.A., Khare S.D., Fasan R. Enzyme stabilization via computationally guided protein stapling. Proc Natl Acad Sci U S A. 2017;114:12472–12477. doi: 10.1073/pnas.1708907114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zanghellini A., Jiang L., Wollacott A.M., Cheng G., Meiler J. New algorithms and an in silico benchmark for computational enzyme design. Protein Sci. 2006;15:2785–2794. doi: 10.1110/ps.062353106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sammond D.W., Kastelowitz N., Donohoe B.S., Alahuhta M., Lunin V.V. An iterative computational design approach to increase the thermal endurance of a mesophilic enzyme. Biotechnol Biofuels. 2018;11:189. doi: 10.1186/s13068-018-1178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schoene C., Fierer J.O., Bennett S.P., Howarth M. SpyTag/SpyCatcher cyclization confers resilience to boiling on a mesophilic enzyme. Angew Chem Int Ed. 2014;53:6101–6104. doi: 10.1002/anie.201402519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Z., Cen Y.K., Zou S.P., Xue Y.P., Zheng Y.G. Recent advances in the improvement of enzyme thermostability by structure modification. Crit Rev Biotechnol. 2020;40:83–98. doi: 10.1080/07388551.2019.1682963. [DOI] [PubMed] [Google Scholar]
- 32.Zouari Ayadi D., Ben Ali M., Jemli S., Ben Mabrouk S., Mezghani M. Heterologous expression, secretion and characterization of the Geobacillus thermoleovorans US105 type I pullulanase. Appl Microbiol Biotechnol. 2008;78:473–481. doi: 10.1007/s00253-007-1318-9. [DOI] [PubMed] [Google Scholar]
- 33.Akassou M., Groleau D. Advances and challenges in the production of extracellular thermoduric pullulanases by wild-type and recombinant microorganisms: a review. Crit Rev Biotechnol. 2019;39:337–350. doi: 10.1080/07388551.2019.1566202. [DOI] [PubMed] [Google Scholar]
- 34.Wang X., Nie Y., Xu Y. Industrially produced pullulanases with thermostability: discovery, engineering, and heterologous expression. Bioresource Technol. 2019;278:360–371. doi: 10.1016/j.biortech.2019.01.098. [DOI] [PubMed] [Google Scholar]
- 35.Li L., Dong F., Lin L., He D., Wei W. N-terminal domain truncation and domain insertion-based engineering of a novel thermostable type I pullulanase from Geobacillus thermocatenulatus. J Agric Food Chem. 2018;66:10788–10798. doi: 10.1021/acs.jafc.8b03331. [DOI] [PubMed] [Google Scholar]
- 36.Zhong C., You C., Wei P., Zhang Y.-H.P. Thermal cycling cascade biocatalysis of myo-inositol synthesis from sucrose. ACS Catal. 2017;7:5992–5999. [Google Scholar]
- 37.Guerois R., Nielsen J.E., Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 38.Kellogg E.H., Leaver-Fay A., Baker D. Role of conformational sampling in computing mutation-induced changes in protein structure and stability. Proteins. 2011;79:830–838. doi: 10.1002/prot.22921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen V.B., Arendall W.B., Headd J.J., Keedy D.A., Immormino R.M. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laskowski R., Macarthur M.W., Moss D.S., Thornton J. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 41.Li G., Fang X., Su F., Chen Y., Xu L. Enhancing the thermostability of Rhizomucor miehei lipase with a limited screening library by rational design point mutations and disulfide bonds. Appl Environ Microbiol. 2017;84 doi: 10.1128/AEM.02129-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ericsson U.B., Hallberg B.M., Detitta G.T., Dekker N., Nordlund P. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal Biochem. 2006;357:289–298. doi: 10.1016/j.ab.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 43.Wang X., Nie Y., Xu Y. Improvement of the activity and stability of starch-debranching pullulanase from Bacillus naganoensis via tailoring of the active sites lining the catalytic pocket. J Agric Food Chem. 2018;66:13236–13242. doi: 10.1021/acs.jafc.8b06002. [DOI] [PubMed] [Google Scholar]
- 44.Swift M. GraphPad prism, data analysis, and scientific graphing. J Chem Inf Comput Sci. 1997;37:411–412. [Google Scholar]
- 45.Bi J., Chen S., Zhao X., Nie Y., Xu Y. Computation-aided engineering of starch-debranching pullulanase from Bacillus thermoleovorans for enhanced thermostability. Appl Microbiol Biotechnol. 2020;104:7551–7562. doi: 10.1007/s00253-020-10764-z. [DOI] [PubMed] [Google Scholar]
- 46.Chen X.H., Xiang Z., Hu Y.S., Lacey V.K., Cang H. Genetically encoding an electrophilic amino acid for protein stapling and covalent binding to native receptors. ACS Chem Biol. 2014;9:1956–1961. doi: 10.1021/cb500453a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alves M., Vieira N.S.M., Rebelo L.P.N., Araújo J.M.M., Pereiro A.B. Fluorinated ionic liquids for protein drug delivery systems: Investigating their impact on the structure and function of lysozyme. Int J Pharm. 2017;526:309–320. doi: 10.1016/j.ijpharm.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 48.Gromiha M.M., Pathak M.C., Saraboji K., Ortlund E.A., Gaucher E.A. Hydrophobic environment is a key factor for the stability of thermophilic proteins. Proteins. 2013;81:715–721. doi: 10.1002/prot.24232. [DOI] [PubMed] [Google Scholar]
- 49.Chakravorty D., Khan M.F., Patra S. Multifactorial level of extremostability of proteins: can they be exploited for protein engineering? Extremophiles. 2017;21:419–444. doi: 10.1007/s00792-016-0908-9. [DOI] [PubMed] [Google Scholar]
- 50.Rathi P.C., Jaeger K.E., Gohlke H. Structural rigidity and protein thermostability in variants of lipase A from Bacillus subtilis. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0130289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Polizzi K.M., Bommarius A.S., Broering J.M., Chaparro-Riggers J.F. Stability of biocatalysts. Curr Opin Chem Biol. 2007;11:220–225. doi: 10.1016/j.cbpa.2007.01.685. [DOI] [PubMed] [Google Scholar]
- 52.Zhong C.Q., Song S., Fang N., Liang X., Zhu H. Improvement of low-temperature caseinolytic activity of a thermophilic subtilase by directed evolution and site-directed mutagenesis. Biotechnol Bioeng. 2009;104:862–870. doi: 10.1002/bit.22473. [DOI] [PubMed] [Google Scholar]
- 53.Yu H., Dalby P.A. Exploiting correlated molecular-dynamics networks to counteract enzyme activity-stability trade-off. Proc Natl Acad Sci U S A. 2018;115:e12192–e12200. doi: 10.1073/pnas.1812204115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kang K.S., Lee Y.J., Park J.H., Yokozawa T. The effects of glycine and L-arginine on heat stability of ginsenoside Rb1. Biol Pharm Bull. 2007;30:1975–1978. doi: 10.1248/bpb.30.1975. [DOI] [PubMed] [Google Scholar]
- 55.Li L., Liao H., Yang Y., Gong J., Liu J. Improving the thermostability by introduction of arginines on the surface of α-L-rhamnosidase (r-Rha1) from Aspergillus niger. Int J Biol Macromol. 2018;112:14–21. doi: 10.1016/j.ijbiomac.2018.01.078. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.