Graphical abstract
Keywords: Freshwater fish, Carnobacterium, Protein hydrolysis, sterile raw shovelnose catfish extract
Highlights
-
•
Carnobacterium and Vagococcus genera were identified from Paraná River freshwater fishes.
-
•
Low acidifying and antilisterial Carnobacterium strains were selected.
-
•
Proteolysis of raw fish extract by C. maltaromaticum strains was confirmed.
-
•
C. maltaromaticum strains may be used as functional cultures to develop novel LPFP.
Abstract
Lactic acid bacteria (LAB) isolated from freshwater fish (hatcheries and captures) from Paraná river (Argentina) were analyzed by using culture-dependent approaches. The species belonging to Carnobacterium (C.) divergens, C. inhibens, C. maltaromaticum, C. viridans and Vagococcus (V.) salmoninarum were identify as predominant by RAPD-PCR and 16 s rRNA gene sequencing. C. maltaromaticum (H-17, S-30, B-42 and S-44) grew in raw fish extract and slightly reduced the medium pH (5.81–5.91). These strains exhibited moderate fish sarcoplasmic protein degradation (≤ 73 %) releasing small peptides and free amino acids, being alanine, glycine, asparagine and arginine concentrations increased in a higher extent (17.84, 1.47, 1.26 and 0.47 mg/100 mL, respectively) by S-44 strain at 96 h incubation. Interestingly C. maltaromaticum H-17 was able to inhibit Listeria monocytogenes. Results suggest that these strains would contribute to the development of new safe and healthy fishery products with improved nutritional and sensory characteristics.
1. Introduction
In the last years, deep changes in consumer demands for animal proteins occurred. Because of the high quality of fish proteins, the association between its consumption and health was perceived as a healthy alternative to other meats [1]. Therefore, recent efforts are concerned with the development of new products that should retain nutritional and safety properties. Different strategies have been applied to increase the shelf life of lightly preserved fish products (LPFP). Among technologies, salt addition, smoked, refrigeration, vacuum and modified atmospheres during packaging were applied. These procedures modify the environment of fish flesh favoring the development of lactic acid bacteria (LAB) over Gram negative bacteria which are mostly aerobic and osmotic sensitive [2]. Different LAB genera have been isolated from fish, seafood and LPFP, being Carnobacterium, Lactobacillus, Leuconostoc and Lactococcus the predominant [3,4]. The fact that LAB were found in fish stored under these technologies increased the interest of their use as biopreservative and/or functional cultures in fish and seafood [5]. Most of the studies have been obtained on the inhibition of Listeria spp. by different species of LAB, which is due to either bacteriocin production or competition mechanisms [[6], [7], [8], [9]]. Psychrotrophic LAB strains isolated from fish and seafood are very promising as functional cultures in refrigerated products with scarce impact on sensory properties of final products (Duffes et al. 1999; Saraoui, Leroi, Björkroth, and Pilet, 2016). Particularly, Carnobacterium species have been reported to grow in fish as low carbohydrates matrix [10,2,11]), thus exhibiting low acidification rates which would prevent fishery products from sensory defects.
Although LAB in fish flesh has long been disregarded, the current consumer demands for convenient foods lead the food industry to consider LAB as promising agents for fishy products diversification. In addition to their antimicrobial ability, LAB may diversify through their proteolytic activity to obtain novel fish derived products such as LPFP. However, the role of LAB in fish products is complex, depending on fish species, treatment and storage conditions, and interaction between present bacteria. Even when LAB have no particular negative effect, they may be responsible for strong sensory degradation, leading to rejection of the products [2]. Increasing numbers of studies are aiming to exploit LAB ability to control safety and quality for fish applications. However, fish technological developments are still in its infancy compared to dairy and meat products.
By using the proteolytic system of LAB, new products with functional properties and specific formulations based on fish proteins may be developed through a moderate protein degradation with the release of short peptides and/or bioactive peptides as well as essential amino acids. Fish flesh proteolysis by LAB has been studied in Asian traditional fermented products supplemented or not with salt and carbohydrates which are mainly composed of freshwater fish species [12,13]. Under these conditions, the pH decreased and the sarcoplasmic and myofibrillar proteins were degraded with the concomitant increase in α-amino nitrogen, soluble peptides and free amino acids, this process affecting the product sensorial quality due to protein aggregates formation of by covalent bounds between proteins [12,14]. Nevertheless, many studies have concluded that initial breakdown of muscle protein is mainly attributed to endogenous proteinases, followed by the action of LAB peptidases which further degrade the protein fragments to small peptides and free amino acids [12,13,15,16]. The presence of exopeptidases in LAB may also contribute to the generation of free amino acids from the N-amino terminal of fish muscle proteins and peptides [12]. Hydrolytic activity on fish soluble proteins was reported for Lactobacillus plantarum, Leuconostoc mesenteroides and Tetragenococcus halophilus with an important role in the generation of small peptides, amino acids and volatile compounds [[17], [18], [19]]. The aim of this study was to identify the predominant LAB isolated from freshwater fishes and their subsequent ability to growth and degrade fish protein in sterile raw fish extract.
2. Material and methods
2.1. Isolation of lactic acid bacteria
Samples of different freshwater fish specimens (Table 1) from the Parana river obtained from local markets at Posadas city (Misiones, Argentina) were analyzed. The skin, gills and flesh from eviscerated frozen (about - 18 °C) wild and farmed fish species were used for the isolation of LAB. Fish species included Pseudoplatystoma corruscans (shovelnose catfish or surubí), Megaleporinus obtusidens (headstander or boga), Piaractus mesopotamicus (small-scaled pacu or pacú), Hoplias mbigua (trahira or tararira) and Pachyurus bonaerensis (La Plata croaker or corvina de río). Fish samples were processed both, within 24 h (fresh samples) and after stored in airtight sealed bags at 6 ± 1 °C during 10 days (spoiled samples). Samples were homogenized in peptone water (Britania, Argentina), and aliquots of 10-fold serial dilutions were spread plated onto Man Rogosa Sharpe agar (MRS agar, Biokar, France) and tryptone soy agar (TSA, Biokar) supplemented with 0.5 % yeast extract (Britania), (TSA-YE). All plates were incubated in duplicate in anaerobic jars with AnaeroPack-Anaero (Mitsubishi Gas Chemical, Japan) at 6 ± 1 °C for 8 days. In addition, plates of MRS agar were incubated in duplicate under aerobic conditions at 29 ± 1 °C for 48−72 h. An average of 15 colonies were randomly picked from plates containing ≤ 300 colonies. Gram-positive and catalase negative bacteria (presumptive LAB) were purified by successive streaking on MRS agar plates, and stored in sterile skimmed milk (Svelty, Nestlé, Argentina) supplemented with glycerol (20 %) at −20 °C for further experimentations.
Table 1.
Freshwater fish LAB isolates biotypes and sequence information by using RAPD.
| Isolated Nº | Fish sample |
Identified LAB biotypes | Accession Nº | |
|---|---|---|---|---|
| Species | Condition | |||
| S-21 | Wild shovelnose catfish | spoiled | C. divergens | MT452566 |
| S-22 | Wild shovelnose catfish | spoiled | C. divergens | MT452567 |
| S-26 | Wild shovelnose catfish | spoiled | C. divergens | MT452568 |
| S-34 | Wild shovelnose catfish | spoiled | C. divergens | MT452569 |
| S-29 | Wild shovelnose catfish | spoiled | C. maltaromaticum | MT452890 |
| S-30 | Wild shovelnose catfish | spoiled | C. maltaromaticum | MT452891 |
| S-32 | Wild shovelnose catfish | spoiled | C. maltaromaticum | MT452892 |
| S-44 | Wild shovelnose catfish | spoiled | C. maltaromaticum | MT452893 |
| H-17 | Wild trahira | spoiled | C. maltaromaticum | MT451944 |
| H-18 | Wild trahira | spoiled | C. maltaromaticum | MT452514 |
| B-42 | Wild headstander | spoiled | C. maltaromaticum | MT452897 |
| B-40 | Wild headstander | spoiled | V. salmoninarum | MT452898 |
| B-41 | Wild headstander | spoiled | V. salmoninarum | MT452899 |
| B-35 | Farmed headstander | fresh | C. viridans | MT452895 |
| B-37 | Farmed headstander | fresh | C. inhibens | MT452896 |
| P-16 | Wild La Plata croaker | spoiled | C. inhibens | MT452894 |
C: Carnobacterium; V: Vagococcus.
2.2. Molecular identification by culture-dependent methods
Potential LAB isolates were subjected to genotypic characterization by RAPD-PCR analysis and identification by sequence analysis of 16S rRNA genes. Strain differentiation was performed by RAPD-PCR in a 25 μl reaction mix using single primers P16 (5′- TCG CCA GCC A - 3′) [20] and M13 (5′- GAG GGT GGC GGT TCT - 3′) [21]. Resulting amplicons were separated by electrophoresis on 1.5 % (w/v) agarose gel and visualized by UV transillumination after staining with GelRed™ Nucleic Acid Gel Stain (Biotium, Hayward, CA). The 100 pb Plus DNA ladder (Genbiotech, Buenos Aires, Argentina) was used as a molecular size marker. RAPD-PCR profiles were analyzed with the Scientific Image Processing ImageJ 1.47v software (National Institutes of Health, Bethesda, MD). LAB isolates were subjected to RAPD-PCR analysis at least twice. Molecular identification of LAB isolates with different RAPD-PCR patterns was carried out by partial 16S rRNA gene sequencing by using universal oligonucleotide primers (27 F 5′ AGA GTT TGA TCC TGG CT 3′ and 1492R 5′ TAC GGT TAC CTT GTT ACG ACT T 3′). Genomic DNA was obtained by colony PCR as reported by [22] with modifications. Briefly, 1 mL aliquots of LAB overnight cultures were washed twice with sterile physiological solution (0.85 % w/v NaCl) and suspended in the same solution. A 0.5 μL portion of cell suspensions were treated three times for 30 s in a microwave at 800 W and used for PCR reactions that were performed in a final volume of 50 μL containing identical concentration of the reaction mixture as detailed above. Thirty-five thermal cycles with the following steps were used: denaturation at 94 °C for 60 s, annealing at 48 °C for 30 s, and elongation at 72 °C for 90 s. rRNA gene sequencing was performed at the DNA-sequencing center (Macrogen, Korea) and identification queries were fulfilled by a BLAST search in GenBank (http://www.ncbi.nlm.nih.gov/GenBank/).
2.3. LAB growth and acidification
Lactic acid bacteria growth and acidification were evaluated in vitro by using a sterile raw fish extract prepared as follows: fresh shovelnose catfish dorsal muscle was cut into thin pieces under aseptic conditions, suspended in distilled water (1:10 w/v) in sterile bags (Whirl Pak, Nasco, USA) and homogenized (Stomacher 400, Seward, Worthing, UK) during 5 min. The obtained slurry was subjected to solids separation by filtering through a three-layer-cloth and kept as raw fish extract (RFE); the liquid extract was then successively filtered through 10−12 μm, 0.45 μm and 0.22 μm filters (Steritop® Filter Unit, Millipore, USA) using a vacuum filtration system (Sterifil®, Millipore, USA). The absence of bacterial growth in raw sarcoplasmic protein extract (RFE) was confirmed by plating on Plate Count Agar (PCA) and pH was determined. Overnight LAB cultures in MRS broth (Biokar; 29 ± 1 °C) were centrifuged (1–14, Sigma, Germany; 5000 g, 5 min), harvested cells washed twice in sterile saline solution (0.85 % NaCl) and suspended in the same solution. Cell suspensions (0.5 mL) were used to inoculate 50 mL of RFE. Samples were incubated at 29 ± 1 °C for 96 h. Non-inoculated samples were used as control. The growth of LAB strains was evaluated by optical density in a L-Vis-400 spectrophotometer (Labnics Equipaments, USA) at 600 nm. pH measurements of RFE after 96 h were performed by an Adwa pH-meter (Adwa AD1030, Hungary) equipped with a refillable pH electrode with glass body and BNC connector (Adwa AD1131B).
2.4. Antimicrobial activity
Antibacterial activity of LAB isolates was determined against Listeria monocytogenes LM (clinical isolate from Parque de la Salud Dr. Ramón Madariaga, Misiones, Argentina) cultured in trypticase soy broth supplemented with 0.5 % yeast extract (TSB-YE, Biokar, France) at 30 °C for 24 h, as sensitive strain by the spot-on-the-lawn test, according to literature [23], with modification. Briefly, 2 μL of an LAB overnight culture grown in MRS broth was spotted on TSA-YE agar plate and incubated at 29 ± 1 °C for 24 h. After incubation period, the plates were overlaid with 10 mL of soft TSA agar (0.8 % w/v bacteriological agar, Oxoid) seeded with L. monocytogenes (ca. 106 CFU/mL) followed by incubation at 29 ± 1 °C for 24 h. The presence of a clear inhibition zone around the spots was considered as a positive antagonistic effect. The diameters of the inhibition zones were measured and results expressed in millimeters.
2.5. Proteolytic activity of LAB isolates
All of proteolytic activity analysis were carried out in the cell free supernatants of RFE cultures centrifuged at 10000 xg, 10 min, 4 °C (IEC multiRF,Thermo Electron, MA, EE.UU.) at the beginning (0 h) and after 96 h of incubation (29 ± 1 °C).
2.5.1. Determination of soluble proteins concentration on RFE
The concentration of soluble proteins was determined by Bradford method [24] using bovine serum albumin (Sigma) as standard. The concentration was determined spectrophotometrically (595 nm) using a microplate reader (VersaMax ELISA Microplate Reader, Molecular Devices, USA) and commercial Bradford solution (Bio-Rad Laboratories, Hercules, CA). Protein concentration was expressed as mg of soluble proteins per 100 mL at the beginning (0 h) and 96 h of incubation (29 ± 1 °C).
2.5.2. Free amino acid analysis
Free amino acids were measured according to the o-phthaldialdehyde (OPA) test [25], measuring the increase in OD at 340 nm relative to the control using a VERSAmax™ Tunable Microplate reader (Sunnyvale, CA, USA). The OPA solution contained 2.5 mL 20 % (w/v) SDS, 25 mL 100 mM sodium tetraborate, 40 mg OPA (previously dissolved in 1 mL methanol), 100 μL 2-mercaptoethanol and distilled water up to a 50 mL final volume. Samples were incubated with 0.75 M trichloroacetic acid (1:3) at 4 °C for 30 min and centrifuged (5000g, 10 min). A 10 μL supernatant aliquot of this mixture was added to 0.2 mL OPA reagent and incubated at room temperature for 5 min until reading. Results were calculated using a standard curve of l-leucine (BDH Chemicals Ltd Poole, England) and free amino acids (FAA) expressed as mEq Leu /100 mL of RFE at the beginning (0 h) and at 96 h of incubation (29 ± 1 °C). All the amino acids except for proline, cysteine and methionine were determined by this method [26].
2.5.3. Gel electrophoresis SDS-PAGE
Fish protein degradation was evaluated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) [27] and performed as follows: cultured supernatants samples (8 μl) were suspended in 5 μl Laemmli buffer and heated at 100 °C for 5 min. The corresponding blanks (non-fermented, heated fish extract), and the molecular weight marker (10.0–250 kDa; Presition Plus, Biorad) were loaded, separately. SDS-PAGE were carried out on 12 % polyacrylamide gels on vertical slab electrophoresis cells (BIO RAD Mini PROTEAN® ll System, Hercules, CA, USA) for 3 h at 70 V. Coomassie brilliant blue R250 (0.1 % w/v) was used for bands visualization. The molecular weight of the fractions was calculated by comparison with bands of molecular weight marker mobility in the gel using the QuantiScan software (BIOSOFT 1.5, USA).
2.5.4. Protein hydrolysis by RP-HPLC
Degradation of fish proteins was monitored by RP-HPLC using a TotalChrom v6.2.0.0.1 with LC Instrument Control (Perkin Elmer series 200, USA) with an UV detector fitted with a C18 column (4.6 × 250 mm, 100 A, 5 μm, Phenomenex, USA). LAB culture samples were filtered (0.45-μm filters; Ministart high flow, Sartorius) and solvent A, water/trifluoroacetic acid (100/0.05, v/v) and solvent B acetonitrile/water/trifluoroacetic acid (60/40/0.05, v/v) with a flow rate of 1 mL/min were used. Before injection, the column was equilibrated with 1 % solvent B for 2.5 min. The filtered (0.45-μm filters; Ministart high flow, Sartorius) samples (20 μL) were eluted as follows: 0–20 min, linear gradient from 1 to 80 % solvent B; 20–33 min, linear gradient from 80 to 100 % solvent B; and 33–38 min linear gradient from 100 to 1 % solvent B. Eluted peaks in the chromatograms were detected at 214 nm. The RFE protein hydrolysis was evaluated by using the QuantiScan software (BIOSOFT 1.5, USA). The peaks height displayed by the software from the gel bands intensity were compared and expressed as percentage of degradation.
2.5.5. Free amino acids by RP-HPLC
Free amino acids were determined in the supernatants of filtered (0.45-μm filters; Ministart high flow, Sartorius) fish extracts. These supernatants were transformed into o-phthaldialdehyde (OPA) derivatives and the concentration of amino acids was determined by RP-HPLC. The OPA reagent contained 200 mg of OPA, 9 mL methanol, 1 mL sodium borate buffer (0.4 mmol/L, pH10), and 160 μL β-mercaptoethanol. This reagent was prepared 24 h before use and kept at 8 °C. The amino acids used as standards (Sigma) were treated with OPA reagent in the same way as the samples. The chromatographic separation was carried out using a Shimadzu liquid chromatograph equipped with a C18 column (Gemini 5μ, C18 110A, 150 × 4.6 mm) and a Shimadzu fluorescence detector (excitation, 340 nm; emission, 460 nm). The binary solvent system consisted of solvent A, sodium phosphate buffer (40 mmol/L), pH 6.0, and solvent B, acetonitrile/methanol/water (45:45:10, v/v/v). The elution gradient was carried out at 30 °C with a flow rate of 1 mL/min. The injection volume of the derivatized amino acids was 10 μL. The amino acids proline, cysteine and methionine were not detected by this method [26].
2.6. Statistical analysis
All assays were carried out in triplicate, and results were expressed as mean values with standard deviations. Statistical analyses were performed using Minitab 14 software (PA, USA). Comparisons were accomplished by ANOVA general linear model followed by Tukey’s post-hoc test and p < 0.05 was considered significant.
3. Results and discussion
3.1. Isolation and identification of lactic acid bacteria
Fish species used in this study (shovelnose catfish, headstander, small-scaled pacu, trahira and La Plata croaker) were analyzed fresh and frozen (wild/farmed fish within 24 h after fishing) and spoiled (after 10 days at 6 ± 1 °C). The bacterial counts on MRS and TSB-YE displayed values of 4–5 log CFU/g and 8–9 CFU/g for fresh and spoiled fish samples (data not shown). As expected higher total numbers were found for spoiled fish. Cultivable bacteria population in freshwater fresh fishes (gills, skin and filleted flesh) were similar to that reported for Atlantic fresh anchovies, salmon and mackerel [28,29]. However, only about 10 % of the cultivable microbiota here analyzed was represented by LAB since 33 isolates out of 373 picked and suspected colonies were Gram positive and catalase negative (data not shown). The presence of LAB in fish species is well documented and constitute a part of indigenous microbiota from the gills, skin and gut content [30,11,31]. Bacteria populations on the surface of fish approximate that of the surrounding water and was reported to be low, while gill tissue has been found to harbor high bacterial populations [32]. In contrast to marine environment (high salt and pressures presence), river water is more abundant in dead and living biota, which is a rich source of nutrients necessary for the growth of heterotrophic microorganisms [33].
Presumptive LAB isolates were subjected to RAPD-PCR analysis by using the primers P16 and M13. Strains showing identical RAPD band patterns were considered as one RAPD-biotype; at least one representative from each biotype was identified by partial 16S rRNA gene sequencing. In Table 1, biotype information for RAPD-PCR obtained with the used primers of fish isolates were associated with Carnobacterium (C). divergens (4), C. inhibens (2), C. maltaromaticum (7), C. viridans (1) and Vagococcus (V). salmoninarum (2). Fish samples were analyzed within 24 h (fresh samples) or after 10 days under chilling conditions to allow the development of natural microbiota (spoiled samples); this procedure allowed the isolation of most of the LAB (ca. 80 %). The dominance of species from Carnobacterium genus among fish samples is in agreement with that widely reported for freshwater and marine different fish species, both wild and farmed [[34], [35], [36]]. Indeed, Carnobacterium have been reported to be the dominant LAB genus in Atlantic salmonids and river trouts accounting for up to 15 % of the viable culturable populations [35]. Here, four species from Carnobacterium genus were present in freshwater fish samples; C. maltaromaticum isolated from shovelnose catfish (surubí), trahira (tararira) and headstander (boga) is in coincidence with that reported from freshwater and marine fishes [30,37,38]. In addition, C. divergens isolated from shovelnose catfish was also reported from rainbow trout and Artic charr [34,39,40], whereas C. inhibens species identified from La Plata croaker (corvina de río) and farmed headstander is in coincidence with that reported for Atlantic salmon [41]. C. viridans, isolated from farmed headstander and represented by only one strain has been mainly associated with spoilage (H2O2 production) in the form of green discoloration in cured meat products [42]. Besides Carnobacterium, V. salmoninarum was isolated from freshwater fishes, particularly from spoiled headstander; this LAB species was reported as a bacterium of pathological significance in salmonid farms; the co-occurrence of these two genera were found to be phylogenetically related [31]. In addition, Carnobacterium and Vagococcus genera were recently reported as part of the core bacterial affiliated with the Firmicutes phylum for fresh- and salt-water farmed salmon [43].
3.2. Growth of LAB in raw fish extract (RFE) and anti-listerial activity
A medium contained shovelnose catfish muscle without added carbohydrates (pH 6.45) was used to grow LAB isolates. Results showed that all LAB were able to grow and pH at 96 h was differently decreased (Table 2). According to their growth (OD600) they may be divided into three groups, those LAB strains with higher growth (OD600 = 0.32−0.41) represented by C. divergens S-22, C. maltaromaticum H-17/H-18 and V. salmoninarum B-40/B-41; those with intermediate growth (OD600 = 0.19−0.29) as C. divergens S-29 and C. maltaromaticum S-30/B-42/S-44 and the rest of the strains exhibiting poor growth (OD600 ≤ 0.16). Reduction of pH at 96 h because the fermentation of carbohydrates present in RFE was variable and uncorrelated with growth; Carnobacterium strains S-22/H-18 and V. salmoninanrum showed the highest fermentative activity with a final pH between 5.13 and 5.22. As most fish contain very low concentration of carbohydrates (< 0.5 %) in the muscle tissue and small amounts of lactic acid is produced post-mortem, fish flesh has a pH > 6.0 (Gram and Huss, 1996). In addition to glucose, the LAB growth and pH changes in RFE may be assigned to ribose fermentation originated from ATP degradation and to arginine-deiminase pathway that is expressed by most Carnobacterium [42,44]. Despite the small pH reduction by strains of this genus here found, as arginine was absent in RFE medium (Table 3), it would not have been used. Nevertheless, a low fermentative activity of LAB strains would be appropriated for their use as functional culture in the development of LPFP products with increased safety and nutritional value avoiding the occurrence of sensory alterations by excessive acidification. Indeed, C. maltaromaticum H-17, S-30, B-42 and S-44 strains showed adequate growth ability (0.22−0.32) combined with low acidification activity (5.81–5.91) in RFE, so that they could be interesting candidates for their use in LPFP development.
Table 2.
Growth and pH in RFE (96 h, 29 ± 1 °C) and anti-listerial activity of freshwater fish LAB strains.
| Isolates | Growth (OD600nm) | pH | Antilisterial activity (mm) |
|---|---|---|---|
| S-21 | 0.05 ± 0.03 | 5.88 ± 0.12 | 10 ± 0.71 |
| S-22 | 0.36 ± 0.05 | 5.22 ± 0.09 | 11 ± 1.41 |
| S-26 | 0.11 ± 0.04 | 5.94 ± 0.06 | – |
| S-34 | 0.07 ± 0.01 | 5.87 ± 0.08 | – |
| S-29 | 0.19 ± 0.00 | 5.83 ± 0.13 | 10 ± 1.06 |
| S-30 | 0.22 ± 0.03 | 5.81 ± 0.22 | 08 ± 1.41 |
| S-32 | 0.16 ± 0.03 | 6.20 ± 0.17 | 16 ± 2.12 |
| S-44 | 0.25 ± 0.01 | 5.84 ± 0.06 | – |
| H-17 | 0.32 ± 0.02 | 5.88 ± 0.17 | 18 ± 0.00 |
| H-18 | 0.41 ± 0.01 | 5.17 ± 0.11 | 17 ± 0.71 |
| B-42 | 0.29 ± 0.04 | 5.91 ± 0.14 | – |
| B-40 | 0.32 ± 0.08 | 5.27 ± 0.13 | – |
| B-41 | 0.38 ± 0.03 | 5.13 ± 0.23 | – |
| B-35 | 0.04 ± 0.00 | 6.10 ± 0.18 | – |
| B-37 | 0.05 ± 0.03 | 6.25 ± 0.03 | – |
| P-16 | 0.02 ± 0.00 | 6.32 ± 0.01 | – |
In bold is highlighted selected strain for protein hydrolysis analysis. Data are shown as mean values ± standard deviation.
Table 3.
Amino acid profiles obtained by RP-HPLC at 96 h of incubation (29 ± 1 °C) in raw fish extracts inoculated with C. maltaromaticum strains.
 |
Grey columns show the net change relative to respective control at initial time (0 h).
When anti-listerial activity of freshwater fish LAB was addressed, many strains (C. maltaromaticum H-17/H-18/S-32/S-29/S30 and C. divergens S-21/S-22), were able to inhibit L. monocytogenes in different extension (Table 2); among strains C. maltaromaticum H-17 exhibited the high inhibition activity. In coincidence, it was reported that these two Carnobacterium species have been extensively studied as anti-listerial protective cultures in fish and meat products [7,42].
3.3. Proteolytic activity of C. maltaromaticum strains in raw shovelnose catfish extract (RFE)
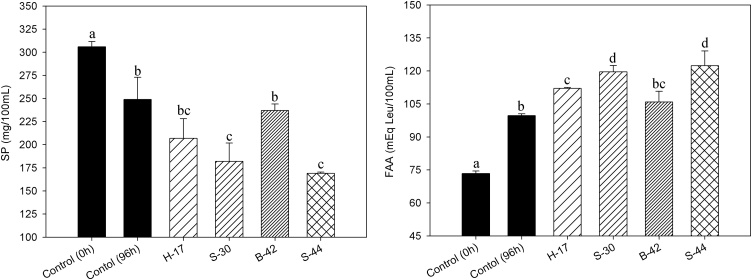
C. maltaromticum H-17, S-30, B-42 and S-44 were selected for their god growth (OD600 ≥ 0.22), low acidification and inhibitory properties (H-17 and S-30) to analyze their proteolytic activity on RFE. When changes of soluble proteins (SP) in control and inoculated extracts after 96 h of growth were evaluated, a decrease (ΔSP = 57–137 mg/100 mL) in their concentration represented by lower values of SP were observed in both control and C. maltaromaticum strains (Fig. 1). On the other hand, free amino acids (FAA, OPA analysis) in fish extracts also indicated that control and the four assayed strains were able to release peptides and amino acids (ΔFAA = 26.3–49.0 mEq Leu/100 mL) from sarcoplasmic fish proteins at 96 h. Indeed, soluble proteins degradation would be carried out by both, endogenous and LAB proteolytic enzymes since significantly (p ≤ 0.05) higher activities were observed in the inoculated samples (Fig. 1). However, the drop pH (5.8–5.9) in cultures samples, about 0.6 points respect to control samples, could have produced some isoelectric precipitation of soluble proteins and caused a decreasing on SP values measured by Bradford method [45]. On the other hand, this little drop pH in culture samples could also improves the activity of acid muscle proteinases which optimum pH is lower than 6.0 [16]. Muscle tissue postmortem evolution was characterized by successive biochemical reactions and autolytic modifications resulting in changes of the muscular structure. Numerous studies have concluded that initial breakdown of muscle protein is mainly attributed to endogenous proteinases such as cathepsins, followed by the action of microbial peptidases which further degrade the protein fragments to small peptides and free amino acids [12,46]. It is known that most LABs does not exhibit cell-wall proteinase activity, primary muscle proteolysis being dependent on endogenous enzymes, LABs increasing the concentration of peptides and amino acids during growth predominantly by the activity of strain-specific intracellular peptidases. However, a recent comparative genomic analysis of C. maltaromaticum strains from different ecological niches revealed the conserved presence of oligopeptides transporter systems (OppABCDF and DtpT) and intracellular peptidases [47]. These data suggest that this LAB species is indeed able to exploit amino acids from the proteins present in the environments as a contribution to fish protein degradation.
Fig. 1.
Soluble proteins (SP, by Bradford) and free amino acids (FAA, by OPA) after incubation (96 h, 29 ± 1 °C) of raw fish extracts inoculated with C. maltaromaticum strains. The results sharing the superscript letter are not significantly different (P < 0.05).
3.4. Protein analysis by SDS-PAGE
Hydrolysis of RFE by C. maltaromaticum strains was evaluated and compared by gel electrophoresis (Fig. 2). Since fish proteins were extracted with water, it would contain only sarcoplasmic water-soluble proteins. Control (uninoculated) extract samples resulted in 12 major protein bands similar to that described for sea bass sarcoplasmic fraction [48]. In this study, electrophoretic separation of hydrolytic products after 96 h in control samples showed degradation (14 %) of the dense band between 41 and 39 kD, that was referred as creatine kinase and aldolase, resulting in a visible dissociation into two bands. In addition, a decrease in intensity (52 %) of a faint band between 34 and 27 kD was also observed in the control at 96 h. These results are in accordance with that previously reported for other uninoculated fish sarcoplasmic extracts [16,48]. Particularly, since pH of RFE was about 6.45, approaching to the natural pH of fish muscle, activity of endogenous proteinases were probably inhibited, as most protein bands in the control remained unchanged [19]. On the other hand, when fish extracts were inoculated with Carnobacterium strains, a degradation of the 41−39 kDa fractions to lower molecular weight (MW) bands was produced after 96 h of incubation, this degradation being greater (49–73 %) than that occurred in control sample. Because of this activity, a new polypeptide band of about 35 kDa was detected in all inoculated fish extracts and was not present in the control sample. In addition, the 17 kDa band, indicated as nucleoside diphosphate kinase by Ladrat et al. [48], increased its intensity (20–60 %) in all cultures samples, mainly in the H-17, after 96 h. The presence of ca. 35 kDa polypeptide only in the inoculated samples suggests that a carnobacterial proteinase could have exerted some activity on sarcoplasmic fish proteins. The presence of a similar MW band in silver carp sausages inoculated with Pediococcus pentosaceus was previously reported [49]. It has been already reported that Lactobacillus and Pediococcus are able to degrade mainly the sarcoplasmic proteins from fish and meat muscle with a strain-dependent activity, with a marked decrease in intensity or even the disappearance of 39−41 kDa bands [12,16,19,50]. However, the role of LAB proteases are not always clearly demonstrable in complex media such as fresh muscle with the coexistence of endogenous enzymes. Fadda et al. [51] showed the degradation of sarcoplasmic and myofibrillar proteins by L. plantarum in a sterile bufferated system (pH = 6.5–7.0), where endogenous acid proteases were partially inhibited. Although in our work the drop of pH was not pronounced (5.81–5.91) in the inoculated samples, it was probably enough to improve the activity of endogenous cathepsins.
Fig. 2.
Fish sarcoplasmic protein profiles obtained by SDS-PAGE after incubation (96 h; 29 ± 1 °C) with C. maltaromaticum strains. M: molecular marker. Molecular weight of bands as referred by Ladrat et al. [48] are indicated with dotted arrows.
3.5. Peptide analysis
Chromatograms (RP-HPLC) resulting from the proteolytic activity of Carnobacterium strains on sarcoplasmic proteins are shown in Fig. 3. The control profiles presented some minor changes after 96 h with respect to the initial polypeptide profile (0 h); only a slight increase (28–40 %) of peaks 2, 5 and 6 while a decrease in peak 6 occurred after 96 h. These changes indicated a poor proteolytic activity on uninoculated fish extract in coincidence with the results observed for SP, FAA (Table 3) and SDS-PAGE (Fig. 2) at 96 h of incubation. When RFE was inoculated with C. maltaromaticum strains, some more changes were observed such as the new peaks 3 and 4 present in all the inoculated samples. In addition, peak 1 only appeared after the inoculation of C. maltaromaticum B-42 and S-44, while polypeptides corresponding to peaks 6 and 7 (which were present in the control sample) were almost completely degraded after 96 h in all inoculated samples. Changes of RP-HPLC polypeptide profiles were comparable to those obtained during the fermentation of fish sausage by Lactobacillus plantarum by Wang et al. (2017), but lower than that reported during meat degradation by lactobacilli [51,52]. Peptides and polypeptides released by C. maltaromaticum strains had hydrophilic nature and contributed to fish protein hydrolysis. However, the fact that Carnobacterium strains were more hydrolytic on polypeptides released by endogenous proteases than those from muscle proteins would indicate a moderate effect on muscle integrity, thus on the sensorial quality of fish. On the other hand, low MW and small size peptides, which could be released by Carnobacterium, could be related to desirable sensory characteristics and could exhibit biological activities [53,54].
Fig. 3.
Peptide profiles by RP-HPLC after incubation (96 h, 29 ± 1 °C) in raw shovelnose catfish extracts inoculated with C. maltaromaticum strains.
3.6. Amino acid analysis
In this work only eight amino acids were evaluated in raw shovelnose catfish extracts since the other ones were not detected in the chromatogram profiles (Fig. S1). Possibly the non-observed amino acids were under the detection limit except for Pro, Met, and Cys, which are not detected by the methodology used [26]. The release of the detected amino acids resulting from the activity of Carnobacterium strains on fish extract is shown in Table 3. Amino acids in control (uninoculated) samples showed small changes after 96 h; slight reduction for Asp and Ser and increases for the remained amino acids with the exception of the two-fold increase for Ala concentration, were observed. On the contrary, when the fish extract was inoculated with C. maltaromaticum strains, a net increase in the amino acids Ala, Gly, Arg and Glu occurred, while Asp, Ser and His were consumed. Nevertheless, the amount of total amino acids released by bacteria was higher than that consumed. C. maltaromaticum S-44 showed the highest release of free amino acids (21.38 mg/100 mL) mainly due to the accumulation of Ala, Gly and Asn in the extract. It is worth noting that Ala was the predominant amino acid representing more than 80 % of total free amino acids concentration in all inoculated extracts. Similarly, net increase of Ala was also reported for sarcoplasmic pork and fish proteins extracts inoculated with L. plantarum ([51]; Wang et al., 2017). On the other hand, in this study, an increase in the amino acid Arg was observed only in the inoculated samples in agreement to that reported by Nie et al. [12] for fermented fish sausage. This fact could be assigned to LAB capacity to form Arg by transamination of other amino acids [55]. Although Arg and Gly are not essential amino acids, several health and sensorial benefits have been attributed to their consumption. The supply of Gly with the diet especially contributes to the synthesis of glutathione and collagen, increasing the activity of multiple liver enzymes, which may prevent or decrease several metabolic disorders in individuals with cardiovascular and inflammatory diseases, cancers, diabetes, and obesity. Also, Gly has the property to enhance the quality of sleep and neurological function [56]. In addition, Arg has been related to improve reproductive, pulmonary, renal, cardiovascular, liver and immune functions. Moreover a positive effect was observed on healing and increasing insulin sensitivity, which could have health promoting impact on diabetes and metabolic syndrome treatments [57].
4. Conclusion
Carnobacterium and Vagococcus salmoninarum were determined as predominant among the identified LAB from freshwater fish in Argentinean northeast region. These LAB strains showed variable growth and acidification capacity in raw fish extract free of exogenous carbohydrates, while five C. maltaromaticum and two C. divergens strains exhibited antilisterial activity. When the LAB strains selected by their growth and low acidifying properties were assayed for their hydrolytic activity on fish proteins, a moderate protein degradation as well as peptide and amino acids release was shown. Although high molecular weight protein hydrolysis could not be exclusively attributed to the activity of carnobacterial proteolytic system, a positive effect on fish proteins degradation may be assigned; a small peptides and amino acids enrichment occurred. These promising results suggest that C. maltaromaticum H-17 (major antibacterial compound producer) and S-44 (higher amino acid releaser) may be selected to improve safety, nutritional and sensory profile for novel formulation of lightly preserved fish products.
CRediT authorship contribution statement
Andrea Micaela Dallagnol: Conceptualization, Methodology, Investigation, Writing - original draft, Writing - review & editing. Micaela Pescuma: Methodology, Investigation, Writing - original draft, Writing - review & editing. Natalia Gamarra Espínola: Investigation. Mariela Vera: Investigation. Graciela Margarita Vignolo: Conceptualization, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
All the Authors declare no conflict of interest.
Acknowledgements
This study was supported by PICT 2015 No. 1746 from Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT), and by Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Argentina.
We warmly thank Ana Thea and Luis A. Brumovsky from the Universidad Nacional de Misiones (UNaM) for their kind assistance during the use of the RP-HPLC system. We also gratefully thank the Laboratorio de Genetica Evolutiva (LGE, UNaM) for their assistance and allow us to make use of all available equipment and reagents.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.btre.2021.e00589.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Conte F., Passantino A., Longo S., Voslářová E. Consumers’ attitude towards fish meat. Ital. J. Food Saf. 2014;3:178–181. doi: 10.4081/ijfs.2014.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leroi F. Occurrence and role of lactic acid bacteria in seafood products. Food Microbiol. 2010;27:698–709. doi: 10.1016/j.fm.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 3.González-Rodríguez M.N., Sanz J.J., Santos J.A., Otero A., García-López M.L. Numbers and types of microorganisms in vacuum-packed cold-smoked freshwater fish at the retail level. Int. J. Food Microbiol. 2002;77:161–168. doi: 10.1016/s0168-1605(02)00048-x. [DOI] [PubMed] [Google Scholar]
- 4.Silbande A., Cornet J., Cardinal M., Chevalier F., Rochefort K., Smith-Ravin J., Adenet S., Leroi F. Characterization of the spoilage potential of pure and mixed cultures of bacterial species isolated from tropical yellowfin tuna (Thunnus albacares) J. Appl. Microbiol. 2018;124:559–571. doi: 10.1111/jam.13663. [DOI] [PubMed] [Google Scholar]
- 5.Pilet M.-F., Leroi F. Protective Cultures, Antimicrobial Metabolites and Bacteriophages for Food and Beverage Biopreservation. Elsevier; 2011. Applications of protective cultures, bacteriocins and bacteriophages in fresh seafood and seafood products; pp. 324–347. [DOI] [Google Scholar]
- 6.Alves V.F., De Martinis E.C.P., Destro M.T., Vogel B.F., Gram L. Antilisterial activity of a Carnobacterium piscicola isolated from Brazilian smoked fish (surubim [Pseudoplatystoma sp.]) and its activity against a persistent strain of Listeria monocytogenes isolated from surubim. J. Food Prot. 2005;68:2068–2077. doi: 10.4315/0362-028X-68.10.2068. [DOI] [PubMed] [Google Scholar]
- 7.Matamoros S., Pilet M.F., Gigout F., Prévost H., Leroi F. Selection and evaluation of seafood-borne psychrotrophic lactic acid bacteria as inhibitors of pathogenic and spoilage bacteria. Food Microbiol. 2009;26:638–644. doi: 10.1016/j.fm.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 8.Nilsson L., Hansen T.B., Garrido P., Buchrieser C., Glaser P., Knøchel S., Gram L., Gravesen A. Growth inhibition of Listeria monocytogenes by a nonbacteriocinogenic Carnobacterium piscicola. J. Appl. Microbiol. 2005;98:172–183. doi: 10.1111/j.1365-2672.2004.02438.x. [DOI] [PubMed] [Google Scholar]
- 9.Vescovo M., Scolari G., Zacconi C. Inhibition of Listeria innocua growth by antimicrobial-producing lactic acid cultures in vacuum-packed cold-smoked salmon. Food Microbiol. 2006;23:689–693. doi: 10.1016/j.fm.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 10.dos Reis F.B., de Souza V.M., Thomaz M.R.S., Fernandes L.P., de Oliveira W.P., De Martinis E.C.P. Use of Carnobacterium maltaromaticum cultures and hydroalcoholic extract of Lippia sidoides Cham. against Listeria monocytogenes in fish model systems. Int. J. Food Microbiol. 2011;146:228–234. doi: 10.1016/j.ijfoodmicro.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 11.Gómez-Sala B., Herranz C., Díaz-Freitas B., Hernández P.E., Sala A., Cintas L.M. Strategies to increase the hygienic and economic value of fresh fish: biopreservation using lactic acid bacteria of marine origin. Int. J. Food Microbiol. 2016;223:41–49. doi: 10.1016/j.ijfoodmicro.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 12.Nie X., Lin S., Zhang Q. Proteolytic characterisation in grass carp sausage inoculated with Lactobacillus plantarum and Pediococcus pentosaceus. Food Chem. 2014;145:840–844. doi: 10.1016/j.foodchem.2013.08.096. [DOI] [PubMed] [Google Scholar]
- 13.Zeng X., Xia W., Jiang Q., Guan L. Biochemical and sensory characteristics of whole carp inoculated with autochthonous starter cultures. J. Aquat. Food Prod. Technol. 2015;24:52–67. doi: 10.1080/10498850.2012.754535. [DOI] [Google Scholar]
- 14.Xu Y., Xia W., Yang F., Moon J., Nie X. Effect of fermentation temperature on the microbial and physicochemical properties of silver carp sausages inoculated with Pediococcus pentosaceus. Food Chem. 2010;118:512–518. doi: 10.1016/j.foodchem.2009.05.008. [DOI] [Google Scholar]
- 15.Wang W., Xia W., Gao P., Xu Y., Jiang Q. Proteolysis during fermentation of Suanyu as a traditional fermented fish product of China. Int. J. Food Prop. 2017;20:S166–S176. doi: 10.1080/10942912.2017.1293089. [DOI] [Google Scholar]
- 16.Yang F., Xia W.S., Zhang X.W., Xu Y.S., Jiang Q.X. A comparison of endogenous and microbial proteolytic activities during fast fermentation of silver carp inoculated with Lactobacillus plantarum. Food Chem. 2016;207:86–92. doi: 10.1016/j.foodchem.2016.03.049. [DOI] [PubMed] [Google Scholar]
- 17.Gelman A., Drabkin V., Glatman L. Evaluation of lactic acid bacteria, isolated from lightly preserved fish products, as starter cultures for new fish-based food products. Innov. Food Sci. Emerg. Technol. 2000;1:219–226. doi: 10.1016/S1466-8564(00)00023-0. [DOI] [Google Scholar]
- 18.Udomsil N., Rodtong S., Tanasupawat S., Yongsawatdigul J. Proteinase-producing halophilic lactic acid bacteria isolated from fish sauce fermentation and their ability to produce volatile compounds. Int. J. Food Microbiol. 2010;141:186–194. doi: 10.1016/j.ijfoodmicro.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 19.Wang W., Xia W., Gao P., Xu Y. Sarcoplasmic protein hydrolysis activity of Lactobacillus plantarum 120 isolated from suanyu: a traditional Chinese low Salt fermented fish. J. Food Process. Preserv. 2017;41 doi: 10.1111/jfpp.12821. [DOI] [Google Scholar]
- 20.Plumed-Ferrer C., Barberio A., Franklin-Guild R., Werner B., McDonough P., Bennett J., Gioia G., Rota N., Welcome F., Nydam D.V., Moroni P. Antimicrobial susceptibilities and random amplified polymorphic DNA-PCR fingerprint characterization of Lactococcus lactis ssp. lactis and Lactococcus garvieae isolated from bovine intramammary infections. J. Dairy Sci. 2015;98:6216–6225. doi: 10.3168/jds.2015-9579. [DOI] [PubMed] [Google Scholar]
- 21.Vera-Pingitore E., Jimenez M.E., Dallagnol A., Belfiore C., Fontana C., Fontana P., von Wright A., Vignolo G., Plumed-Ferrer C. Screening and characterization of potential probiotic and starter bacteria for plant fermentations. LWT - Food Sci. Technol. 2016;71:288–294. doi: 10.1016/j.lwt.2016.03.046. [DOI] [Google Scholar]
- 22.Smit B.A., van Hylckama Vlieg J.E.T., Engels W.J.M., Meijer L., Wouters J.T.M., Smit G. Identification, cloning, and characterization of a Lactococcus lactis branched-chain-keto acid decarboxylase involved in flavor formation. Appl. Environ. Microbiol. 2005;71:303–311. doi: 10.1128/AEM.71.1.303-311.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tulini F.L., Winkelströter L.K., De Martinis E.C.P. Identification and evaluation of the probiotic potential of Lactobacillus paraplantarum FT259, a bacteriocinogenic strain isolated from Brazilian semi-hard artisanal cheese. Anaerobe. 2013;22:57–63. doi: 10.1016/j.anaerobe.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 24.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 25.Church F.C., Swaisgood H.E., Porter D.H., Catignani G.L. Spectrophotometric assay using o-phthaldialdehyde for determination of proteolysis in milk and isolated milk proteins. J. Dairy Sci. 1983;66:1219–1227. doi: 10.3168/jds.S0022-0302(83)81926-2. [DOI] [Google Scholar]
- 26.Pescuma M., Hébert M.E., MozzI F., Font de Valdez G. Functional fermented whey-based beverage using lactic acid bacteria. Int. J. Food Microbiol. 2010;141:73–81. doi: 10.1016/j.ijfoodmicro.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Belfiore C., Björkroth J., Vihavainen E., Raya R., Vignolo G. Characterization of Leuconostoc strains isolated from fresh anchovy (Engraulis anchoita) J. Gen. Appl. Microbiol. 2010;56:175–180. doi: 10.2323/jgam.56.175. [DOI] [PubMed] [Google Scholar]
- 29.Svanevik C.S., Lunestad B.T. Characterisation of the microbiota of Atlantic mackerel (Scomber scombrus) Int. J. Food Microbiol. 2011;151:164–170. doi: 10.1016/j.ijfoodmicro.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 30.Balcázar J.L., de Blas I., Ruiz-Zarzuela I., Vendrell D., Gironés O., Muzquiz J.L. Sequencing of variable regions of the 16S rRNA gene for identification of lactic acid bacteria isolated from the intestinal microbiota of healthy salmonids. Comp. Immunol. Microbiol. Infect. Dis. 2007;30:111–118. doi: 10.1016/j.cimid.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 31.Michel C., Pelletier C., Boussaha M., Douet D.-G., Lautraite A., Tailliez P. Diversity of lactic acid bacteria associated with fish and the fish farm environment, established by amplified rRNA gene restriction analysis. Appl. Environ. Microbiol. 2007;73:2947–2955. doi: 10.1128/AEM.01852-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Austin B. The bacterial microflora of fish. Rrevised. Sci. World J. 2006;6:931–945. doi: 10.1100/tsw.2006.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sica M.G., Olivera N.L., Brugnoni L.I., Marucci P.L., López-Cazorla A.C., Cubitto M.A. Isolation, identification and antimicrobial activity of lactic acid bacteria from the Bahía Blanca Estuary. Rev. Biol. Mar. Oceanogr. 2010;45:389–397. doi: 10.4067/s0718-19572010000300003. [DOI] [Google Scholar]
- 34.González C.J., Encinas J.P., García-López M.L., Otero A. Characterization and identification of lactic acid bacteria from freshwater fishes. Food Microbiol. 2000;17:383–391. doi: 10.1006/fmic.1999.0330. [DOI] [Google Scholar]
- 35.Merrifield D.L., Balcázar J.L., Daniels C., Zhou Z., Carnevali O., Sun Y.-Z., Hoseinifar S.H., Ringø E. Aquaculture Nutrition. John Wiley & Sons, Ltd; Chichester, UK: 2014. Indigenous lactic acid bacteria in fish and crustaceans; pp. 128–168. ch6. [DOI] [Google Scholar]
- 36.Ringø E., Hoseinifar S.H., Ghosh K., Doan H.Van, Beck B.R., Song S.K. Lactic acid bacteria in finfish—an update. Front. Microbiol. 2018;9:1–37. doi: 10.3389/fmicb.2018.01818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mansfield G.S., Desai A.R., Nilson S.A., Van Kessel A.G., Drew M.D., Hill J.E. Characterization of rainbow trout (Oncorhynchus mykiss) intestinal microbiota and inflammatory marker gene expression in a recirculating aquaculture system. Aquaculture. 2010;307:95–104. doi: 10.1016/j.aquaculture.2010.07.014. [DOI] [Google Scholar]
- 38.Skrodenyte-Arbačiauskiene V., Sruoga A., Butkauskas D., Skrupskelis K. Phylogenetic analysis of intestinal bacteria of freshwater salmon Salmo salar and sea trout Salmo trutta trutta and diet. Fish. Sci. 2008;74:1307–1314. doi: 10.1111/j.1444-2906.2008.01656.x. [DOI] [Google Scholar]
- 39.Bruni L., Pastorelli R., Viti C., Gasco L., Parisi G. Characterisation of the intestinal microbial communities of rainbow trout (Oncorhynchus mykiss) fed with Hermetia illucens (black soldier fly) partially defatted larva meal as partial dietary protein source. Aquaculture. 2018;487:56–63. doi: 10.1016/j.aquaculture.2018.01.006. [DOI] [Google Scholar]
- 40.Ringø E., Wesmajervi M.S., Bendiksen H.R., Berg A., Olsen R.E., Johnsen T., Mikkelsen H., Seppola M., Strøm E., Holzapfel W. Identification and characterization of carnobacteria isolated from fish intestine. Syst. Appl. Microbiol. 2001;24:183–191. doi: 10.1078/0723-2020-00020. [DOI] [PubMed] [Google Scholar]
- 41.Joborn A., Dorsch M., Olsson J.C., Westerdahl A., Kjelleberg S. Carnobacterium inhibens sp. nov., isolated from the intestine of Atlantic salmon (Salmo salar) Int. J. Syst. Bacteriol. 1999;49:1891–1898. doi: 10.1099/00207713-49-4-1891. [DOI] [PubMed] [Google Scholar]
- 42.Leisner J.J., Laursen B.G., Prévost H., Drider D., Dalgaard P. Carnobacterium: positive and negative effects in the environment and in foods. FEMS Microbiol. Rev. 2007;31:592–613. doi: 10.1111/j.1574-6976.2007.00080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rudi K., Angell I.L., Pope P.B., Vik J.O., Sandve S.R., Snipen L.-G. Stable core gut microbiota across the freshwater-to-saltwater transition for farmed atlantic salmon. Appl. Environ. Microbiol. 2017;84:1–9. doi: 10.1128/AEM.01974-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Venugopal V., Shahidi F. Structure and composition of fish muscle. Food Rev. Int. 1996;12:175–197. doi: 10.1080/87559129609541074. [DOI] [Google Scholar]
- 45.Tahergorabi R., Matak K.E., Jaczynski J. Fish protein isolate: development of functional foods with nutraceutical ingredients. J. Funct. Foods. 2015;18:746–756. doi: 10.1016/j.jff.2014.05.006. [DOI] [Google Scholar]
- 46.Fadda S., López C., Vignolo G. Role of lactic acid bacteria during meat conditioning and fermentation: peptides generated as sensorial and hygienic biomarkers. Meat Sci. 2010;86:66–79. doi: 10.1016/j.meatsci.2010.04.023. [DOI] [PubMed] [Google Scholar]
- 47.Iskandar C.F., Borges F., Taminiau B., Daube G., Zagorec M., Remenant B., Leisner J.J., Hansen M.A., Sørensen S.J., Mangavel C., Cailliez-Grimal C., Revol-Junelles A.-M. Comparative genomic analysis reveals ecological differentiation in the genus Carnobacterium. Front. Microbiol. 2017;8:1–14. doi: 10.3389/fmicb.2017.00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ladrat C., Verrez-Bagnis V., Noël J., Fleurence J. In vitro proteolysis of myofibrillar and sarcoplasmic proteins of white muscle of sea bass (Dicentrarchus labrax L.): effects of cathepsins B, D and L. Food Chem. 2003;81:517–525. doi: 10.1016/S0308-8146(02)00481-8. [DOI] [Google Scholar]
- 49.Xu Y., Xia W., Yang F., Nie X. Physical and chemical changes of silver carp sausages during fermentation with Pediococcus pentosaceus. Food Chem. 2010;122:633–637. doi: 10.1016/j.foodchem.2010.03.023. [DOI] [Google Scholar]
- 50.Aro Aro J.M., Nyam-Osor P., Tsuji K., Shimada K., Fukushima M., Sekikawa M. The effect of starter cultures on proteolytic changes and amino acid content in fermented sausages. Food Chem. 2010;119:279–285. doi: 10.1016/j.foodchem.2009.06.025. [DOI] [Google Scholar]
- 51.Fadda S., Sanz Y., Vignolo G., Aristoy M., Oliver G., Toldrá F. Characterization of muscle sarcoplasmic and myofibrillar protein hydrolysis caused by Lactobacillus plantarum. Appl. Environ. Microbiol. 1999;65:3540–3546. doi: 10.1128/aem.65.8.3540-3546.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanz Y., Fadda S., Vignolo G., Aristoy M.C., Oliver G., Toldrá F. Hydrolytic action of Lactobacillus casei CRL 705 on pork muscle sarcoplasmic and myofibrillar proteins. J. Agric. Food Chem. 1999;47:3441–3448. doi: 10.1021/jf981255n. [DOI] [PubMed] [Google Scholar]
- 53.Sánchez A., Vázquez A. Bioactive peptides: a review. Food Qual. Saf. 2017;1:29–46. doi: 10.1093/fqsafe/fyx006. [DOI] [Google Scholar]
- 54.Temussi P. Advances in Food and Nutrition Research. 2007. The sweet taste receptor: a single receptor with multiple sites and modes of interaction; pp. 199–239. [DOI] [PubMed] [Google Scholar]
- 55.Tavaria F.K., Dahl S., Carballo F.J., Malcata F.X. Amino acid catabolism and generation of volatiles by lactic acid bacteria. J. Dairy Sci. 2002;85:2462–2470. doi: 10.3168/jds.S0022-0302(02)74328-2. [DOI] [PubMed] [Google Scholar]
- 56.Razak M.A., Begum P.S., Viswanath B., Rajagopal S. Multifarious beneficial effect of nonessential amino acid, glycine: a review. Oxid. Med. Cell. Longev. 2017;2017:1–8. doi: 10.1155/2017/1716701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu G., Bazer F.W., Davis T.A., Kim S.W., Li P., Marc Rhoads J., Carey Satterfield M., Smith S.B., Spencer T.E., Yin Y. Arginine metabolism and nutrition in growth, health and disease. Amino Acids. 2009;37:153–168. doi: 10.1007/s00726-008-0210-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.